Abstract
An important analytical control in molecular amplification-based methods is an internal amplification control (IAC), which should be included in each reaction mixture. An IAC is a nontarget nucleic acid sequence which is coamplified simultaneously with the target sequence. With negative results for the target nucleic acid, the absence of an IAC signal indicates that amplification has failed. A general strategy for the construction of an IAC for inclusion in molecular beacon-based real-time nucleic acid sequence-based amplification (NASBA) assays is presented. Construction proceeds in two phases. In the first phase, a double-stranded DNA molecule that contains nontarget sequences flanked by target sequences complementary to the NASBA primers is produced. At the 5′ end of this DNA molecule is a T7 RNA polymerase binding sequence. In the second phase of construction, RNA transcripts are produced from the DNA by T7 RNA polymerase. This RNA is the IAC; it is amplified by the target NASBA primers and is detected by a molecular beacon probe complementary to the internal nontarget sequences. As a practical example, an IAC for use in an assay for the detection of Mycobacterium avium subsp. paratuberculosis is described, its incorporation and optimization within the assay are detailed, and its application to spiked and natural clinical samples is shown to illustrate the correct interpretation of the diagnostic results.
Nucleic acid sequence-based amplification (NASBA) (7, 11) is becoming widely used for the detection of microorganisms in various sample types, e.g., clinical, environmental, and food samples (6, 8). The major product of a NASBA reaction is single-stranded RNA, and there are various means whereby this product can be detected to produce a NASBA signal, such as ethidium bromide staining (22), enzyme-linked gel assay (22), and electrochemiluminescence detection (25). Alternatively, NASBA products can specifically be detected in real time by using molecular beacons (20) included in the reaction mixture (13). Molecular beacons are single-stranded nucleic acid sequence molecules that possess a stem-and-loop structure which is double labeled with a fluorescent dye and a universal quencher at the 5′ end and the 3′ end, respectively (20). The loop region is a sequence complementary to a target sequence in the nucleic acid to be detected, and the stem is formed by the annealing of complementary arm sequences on the ends of the probe. When this structure is closed, the probe does not produce fluorescence because the energy is transferred to the quencher and is released as heat (19). However, during the NASBA reaction, the molecular beacon hybridizes to the target RNA, separating the reporter dye and the quencher, yielding a real-time measurable fluorescence emission directly proportional to the concentration of the target sequence (13). The use of diverse molecular beacons labeled with different fluorophores allows multiplex detection in a single reaction (21). An additional advantage of this detection system is that no post-NASBA step is needed, which reduces the risks of cross contamination.
The ultimate goal of the development of new analytical techniques is their adoption and establishment as routine diagnostic tools. The routine use of such tools necessitates the use of reliable controls to verify the accuracy of the results obtained. A very important control for molecular amplification-based methods is the internal amplification control (IAC) (1, 9, 10). An IAC is a nontarget nucleic acid sequence that is present in every reaction mixture and that can be coamplified simultaneously with the target sequence. In a reaction without an IAC, a negative response (no signal) can mean that no target sequence was present in the reaction mixture. It could also mean, however, that the reaction was inhibited due to an equipment malfunction, operator error, use of the incorrect reaction mixture, poor enzyme activity, or the presence of inhibitory substances in the original sample matrix (2, 3, 16) which are coextracted with the target sequence. In a reaction with an IAC, a control signal should always be produced when no target sequence is present. When no control or target nucleic acid is detected, this means that the reaction has failed, and the sample must be reanalyzed.
We describe here the principles of development of an RNA IAC for real-time molecular beacon NASBA and provide a practical example of its incorporation and optimization for use in a NASBA assay.
MATERIALS AND METHODS
Nucleic acid samples.
Clavibacter michiganensis subsp. sepedonicus strain NCPPB 4053 DNA was used for the construction of an IAC for the NASBA assay. Long-term maintenance of the strain was at −80°C on beads (Protect Biotrading Wilnis). DNA was extracted from strains grown on yeast glucose mineral agar for 2 days at 21°C by use of the DNeasy kit (Qiagen, Hilden, Germany). Mycobacterium avium subsp. paratuberculosis strain ATCC 19698 was used as the NASBA target in this study. This strain was cultured in BBL Herrold's egg yolk agar slant with mycobactin J and amphotericin B, nalidixic acid, and vancomycin (Becton Dickinson and Co., Sparks, Md.) and was incubated at 37°C without shaking. Nucleic acids were obtained by using the isolation module of the NucliSens basic kit (bioMérieux bv, Boxtel, The Netherlands), according to the recommendations of the manufacturer. The same kit was used to extract nucleic acids from 42 bacterial strains different from M. avium subsp. paratuberculosis previously grown in appropriate media.
Oligonucleotide primers and probes.
The oligonucleotides used in this study are shown in Table 1. Primers Af2 and Ar and molecular beacon MB CMS1 (specific sequence) were previously designed and tested by van Beckhoven et al. (24) and modified as indicated in Table 1. M. avium subsp. paratuberculosis-specific primers MAP57F and MAP57R and molecular beacon MAP57MB were designed from the M. avium subsp. paratuberculosis clone uni7 genomic sequence (GenBank accession no. AF445426) (5, 15). The BLAST-n (version 2.2.6) tool (National Center for Biotechnology Information [www.ncbi.nlm.nih.gov]) was used to confirm that none of the selected oligonucleotides recognized any registered sequence other than that of the target. All oligonucleotides were purchased from MWG Biotech AG (Ebersberg, Germany).
TABLE 1.
Oligonucleotides used in this study
| Oligo- nucleotide | Type | Sequencea | Reference or source |
|---|---|---|---|
| MAP57F | Forward primer | 5′-CAA CGA CGA CCA AGA CGA-3′ | 15 |
| MAP57R | Reverse primer | 5′-aat tct aat acg act cac tat agg gag aag gAG CAA ACC GAT CAC GAC A-3′ | 15 |
| MAP57MB | Molecular beacon | 5′-FAM-CGA TCG CTG ATG AAA CCG AGC TCG TCG ATC G-DABCYL-3′ | 15 |
| Af2 | Forward primer | 5′-CGA TGC AAC GCG AAG AAC-3′ | 24 |
| Ar | Reverse primer | 5′-GGT TGG CCC CGG CAG TCT-3′ | 24 |
| MBI | Molecular beacon | 5′-HEX-CGC AGG AAC GTG CAG AGA TGT GCG CCC CTG CG-DABCYL-3′ | 24 |
| IAC F | Forward primer | 5′-aat tct aat acg act cac tat agg gag aag gCA ACG ACG ACC AAG ACG ACG ATG CAA CGC G-3′ | This work |
| IAC R | Reverse primer | 5′-AGC AAA CCG ATC ACG ACA GGT TGG CCC CGG CAG TCT-3′ | This work |
The stem sequences of the MB1 molecular beacons were changed to avoid a G at the 5′ end. Lowercase letters indicate the T7 promoter sequence. Underlining indicates the stem sequences of the molecular beacons.
PCR.
C. michiganensis subsp. sepedonicus DNA was used as a template in a PCR with primers Af2 and Ar. The reaction was performed in a 50-μl volume containing 1× AmpliTaq Gold buffer, 1.5 mM MgCl2, 200 μM deoxynucleoside triphosphates, 300 nM primers, 1 U of AmpliTaq Gold DNA polymerase (Applied Biosystems, Foster City, Calif.), and 2 μl of the target DNA solution. Thermocycling was performed with a GeneAmp PCR System 9700 (Applied Biosystems) by use of the following program: 10 min at 95°C; 40 cycles of 1 min at 95°C, 1 min at 55°C, and 1 min at 72°C; and a final extension step of 7 min at 72°C. The reaction products were electrophoresed on a 2% (wt/vol) agarose gel and visualized by staining with 10 ng of ethidium bromide ml−1, followed by UV transillumination. The amplicon size was 217 bp. This amplicon was purified and used in a second PCR with primers IACF and IACR. The reaction conditions of the second reaction were identical to those of the first one. The amplicon size was 274 bp.
Purification of PCR amplicons.
The amplicon band was excised from the agarose gel and purified with a QIAEX II gel extraction kit (Qiagen), according to the recommendations of the manufacturer.
Production of IAC RNA.
The purified amplicons from the second PCR were used as a template for T7 RNA polymerase. Briefly, 10 μl (1 μg) of DNA template was added to a reaction mixture containing 20 μl of transcription-optimized 5× buffer (Promega, Madison, Wis.), 10 μl of 100 nM dithiothreitol, 20 μl of a recombinant ribonucleotide triphosphate mix at 2.5 mM, 2 μl of T7 RNA polymerase (20 U/μl), and 38 μl of nuclease-free water. The reaction mixture was incubated at 37°C for 2 h and was stopped by heating at 75°C for 10 min, as indicated by Ausubel et al. (4).
Purification and quantification of IAC RNA.
After the T7 reaction, the template DNA was removed by treatment with DNase with an RQ1 RNase-free DNase kit (Promega). The reaction mixture, which contained 50 μl of nucleic acid extract, 10 μl of 10× reaction buffer, 5 μl of DNase (1 U/μl), and 35 μl of nuclease-free water, was incubated at 37°C for 30 min and stopped with 10 μl of RQ1 DNase Stop solution. For purification of the RNA, the RNeasy MiniElute cleanup kit (Qiagen) was used according to the recommendations of the manufacturer. The concentration and quality of the RNA were determined by spectrophotometry at 260 and 280 nm (17).
NASBA.
The NASBA reactions were carried out with the reagents and protocols included in the NucliSens basic kit (bioMérieux bv). Briefly, 2.5 μl of target template solution was added to 12.5 μl of a NASBA reaction premixture, which contained 80 mM Tris-HCl (pH 8.5), 24 mM MgCl2, 140 mM KCl, 30% (vol/vol) dimethyl sulfoxide, a 2 mM concentration of each deoxyribonucleotide triphosphate, a 4 mM concentration of each ribonucleotide triphosphate, 200 nM (each) primers MAP57F and MAP57R, and 2.5 μl of the IAC. Two different molecular beacons (200 nM [each] MAP57MB and MBI) were used to simultaneously detect the template and the IAC (Table 1). The target molecular beacon was labeled at the 5′ end with the fluorophore 6-carboxyfluorescein (FAM), and the IAC molecular beacon was labeled with hexosaminidase A. These fluorophores were selected, as their fluorescent emission peaks do not overlap. For both molecular beacons, 4-(4′-dimethylaminophenylaso) benzoic acid (DABCYL) was used as the quencher at the 3′ end. The reaction mixture was incubated at 65°C for 5 min in order to destabilize the secondary structure of RNA. After the reaction mixture was cooled to 41°C for 5 min to allow primer annealing, 5 μl of enzyme mixture was added. The enzyme mixture contained (per reaction mixture) 0.08 U of RNase H per μl, 32 U of T7 RNA polymerase per μl, and 6.4 U of avian myeloblastosis virus retrotranscriptase per μl. The reactions were run at 42°C for 90 min in an ABIPrism 7700 sequence detection system (Applied Biosystems), which allowed real-time monitoring of the signals. The intensity of the fluorescence signal (ΔR), obtained by subtracting the initial fluorescence intensity from its final value, and the time to positivity (TP; in minutes) were examined (12) by using a ΔR value of 20 as the fluorescence threshold. Unless otherwise stated, all reactions were performed at least in triplicate.
Optimization of IAC-containing real-time NASBA. (i) Verification of IAC detection by real-time NASBA.
Reactions were performed with molecular beacon MB1 and 100 fg of IAC, but without the M. avium subsp. paratuberculosis template or the specific molecular beacons for this template.
(ii) IAC and template coamplification and detection.
Reactions were performed with 1 ng of M. avium subsp. paratuberculosis template, 100 fg of the IAC, and the two specific molecular beacons.
(iii) Optimization of IAC molecular beacon concentration.
NASBA reactions were performed with 1 fg of IAC and 100 pg of M. avium subsp. paratuberculosis nucleic acid as well as decreasing molecular beacon MB1 concentrations (400, 200, and 100 nM).
(iv) Determination of IAC detection limit.
The reaction mixtures included decreasing amounts of IAC (100, 10, and 1 fg and 100, 10, and 1 ag per reaction mixture) but not M. avium subsp. paratuberculosis nucleic acids.
(v) Determination of IAC inhibition capacity.
The NASBA reaction mixtures contained increasing amounts of IAC (i.e., 10 and 100 ag and 1 and 10 fg) and approximately 500 M. avium subsp. paratuberculosis cells.
Artificial contamination of fecal samples with M. avium subsp. paratuberculosis and treatment prior to RTi-NASBA.
Human fecal samples were taken from a healthy volunteer. One gram of feces was mixed with 1 ml of phosphate-buffered saline (PBS; Dulbecco's PBS; Gibco, Paisley, Scotland) containing approximately 108 M. avium subsp. paratuberculosis strain ATCC 19698 cells. Subsequently, the nucleic acids were extracted by use of the NucliSens basic kit (bioMérieux bv). To simulate a situation in which nucleic acid purification was poorly performed, 50 μl of unextracted fecal suspension was added to the nucleic acid extract.
Fecal samples were taken from 25 cows testing positive and 25 cows testing negative for M. avium subsp. paratuberculosis (by standard methods, i.e., blood enzyme-linked immunosorbent assay [Herdcheck Mycobacterium paratuberculosis Antibody Test kit; IDEXX Laboratories, Inc., Westbrook, Maine] and fecal culture verification [18]) on cattle farms in the northern part of Spain. One gram of each sample was mixed with 1 ml of PBS, and the mixture was used for nucleic acid extraction. To test the limit of detection of M. avium subsp. paratuberculosis, 1 g of feces was taken from each of three cows testing negative for M. avium subsp. paratuberculosis by standard methods and mixed with 1 ml of PBS containing approximately 106, 105, 104, 103, 102, 10, and 1 M. avium subsp. paratuberculosis strain ATCC 19698 cells prior to nucleic acid extraction.
RESULTS AND DISCUSSION
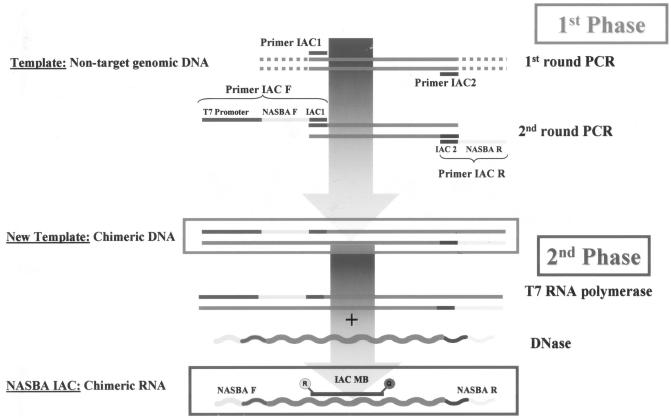
The principles of the construction of the NASBA IAC are summarized diagrammatically in Fig. 1. Briefly, construction proceeds in two phases. In the first phase, a double-stranded DNA fragment that contains nontarget sequences flanked by target sequences complementary to the NASBA primers is produced. At the 5′ end of this chimerical DNA is a T7 RNA polymerase binding sequence. In the second phase of IAC construction, RNA transcripts are produced from the chimerical DNA by T7 RNA polymerase. This RNA is the IAC; it is amplified by the target NASBA primers and is detected by a molecular beacon probe complementary to the internal nontarget sequences. The main features of the molecular beacon IAC for NASBA are the following: (i) it must be a single-stranded RNA, (ii) it must show a NASBA target forward primer sequence close to the 5′ end, (iii) the 3′ end must show the NASBA target reverse primer sequence (reversed and complementary), (iv) the internal sequence must not be homologous to the NASBA template, and (v) the internal sequence must show a molecular beacon hybridization site. It contains the sequences complementary to the target primers and a nontarget internal sequence which has a binding site for a molecular beacon labeled with a reporter different from that for the target molecular beacon probe.
FIG. 1.
General flowchart of construction of an IAC used for real-time NASBA with molecular beacon (MB) detection. Primer IAC1 and primer IAC2 are used to amplify the IAC internal sequence, which should display no homology to the template. NASBA F (forward) and NASBA R (reverse) are the specific primers for the intended target of the NASBA. The specific conditions of both PCRs in the first phase and the T7 RNA polymerase reaction and DNase treatment in the second phase are described in Materials and Methods. R, reporter fluorophore; Q, quencher.
After construction of the IAC, we recommend that the following steps subsequently be performed, in the following order: (i) verify that the IAC can be amplified and detected, (ii) establish that the IAC and template can be simultaneously amplified and detected, (iii) optimize the IAC molecular beacon concentration, (iv) determine the detection limit of the IAC in order to optimize the IAC concentration used in the NASBA reaction, and (v) verify that the system will operate with the sample matrix chosen.
As a practical example, an IAC was constructed for use in a molecular beacon-based real-time NASBA assay for M. avium subsp. paratuberculosis, according to the construction principles. Then, in accordance with the recommendations, it was verified that this IAC could be detected in the RTi-NASBA, with ΔR and TP values of 112.26 ± 21.42 and 46.02 ± 1.92 min, respectively. We then established that the IAC and template can be coamplified and detected simultaneously. ΔR and TP values were 201.05 ± 25.12 and 47.65 ± 2.96 min, respectively, for IAC and 210.26 ± 31.06 and 61.21 ± 2.55 min, respectively, for the M. avium subsp. paratuberculosis template.
The next step was to optimize the concentration of the IAC molecular beacon (MB1). We selected the molecular beacon concentration which gave the highest ΔR value and the shortest TP without affecting the signal of the target template. These criteria are similar to those recommended for duplex real-time PCR optimization, as indicated by the manufacturer of the real-time amplification system (User Bulletin 5, 1998; ABI Prism 7700 SDS; Applied Biosystems). The MB1 concentration that gave the best results (ΔR = 239.56 ± 32.40; TP = 53.25 ± 3.25 min) was 200 nM, and this concentration was selected for further use.
Determination of the optimal concentration of the IAC in the NASBA assay is a critical step. The IAC concentration should be kept as low as possible to avoid inhibition of the target-specific reaction. As seen in PCR assays (14), the use of an IAC concentration that is too high could negatively affect the limit of detection of the target. However, the IAC concentration should not be so low as to make it difficult to obtain an IAC amplification signal. We determined the detection limit of the IAC. The IAC could be detected to a concentration of 1 ag in the reaction mixture. However, below 10 ag, the ΔR values were very low (mean ΔR values, less than 40) and detectable amplification was delayed (TP values, greater than 80 min). We then observed that 1 fg of IAC was the largest amount of IAC that did not inhibit the M. avium subsp. paratuberculosis signal. Therefore, 1 fg of IAC was chosen for use in each reaction mixture. In the absence of the M. avium subsp. paratuberculosis target, this amount of IAC produced a TP value of 54.95 ± 4.26 min. As 67.72 min was the upper limit of the 99% probability confidence interval, we established 68 min as the threshold of the IAC TP, above which M. avium subsp. paratuberculosis-negative RTi-NASBA results were considered invalid and, thus, the samples needed to be retested. A series of NASBA experiments with nucleic acids from 42 nontarget bacterial strains further confirmed the IAC TP values (mean, 53.38 ± 3.66 min). The now fully optimized IAC-containing NASBA was capable of detecting 150 to 200 M. avium subsp. paratuberculosis cells per reaction mixture (data not shown).
Use of RTi-NASBA IAC with clinical samples.
Analyses of samples that possess a strong capacity to inhibit amplification increases the risk of false-negative results (3, 16). A complex matrix, feces, was selected to illustrate the use of the IAC to reveal inhibited reactions. Artificially added M. avium subsp. paratuberculosis cells could be detected in fecal samples by NASBA. Analysis of noncontaminated fecal samples showed positive IAC amplification. The inhibitors present in feces could adversely affect the NASBA detection of M. avium subsp. paratuberculosis, even when the bacterium was present at concentrations that were quite high (108 per g). In a simulation of a situation in which the purification of nucleic acids from feces had been poorly performed, both target and IAC signals were lost, revealing an inhibited reaction and the need to repeat the analysis. The IAC-containing NASBA-based method was capable of detecting approximately 103 M. avium subsp. paratuberculosis cells per g of artificially contaminated feces. In the absence of IAC in the NASBA, the same limit of detection was achieved. As a consequence, IAC can assist with the interpretation of RTi-NASBA results for a clinical sample without interfering with the results for the target.
The final optimized method was applied to 50 natural bovine fecal samples, 25 of which were obtained from a farm declared to be infected with M. avium subsp. paratuberculosis. The results were fully concordant with the ones obtained by the standard methods based on M. avium subsp. paratuberculosis culture in selective medium followed by serological confirmation. All 25 samples from the infected farm tested positive for M. avium subsp. paratuberculosis by standard methods and were positive for M. avium subsp. paratuberculosis by NASBA. In contrast, all 25 samples testing negative for M. avium subsp. paratuberculosis by standard methods were negative for M. avium subsp. paratuberculosis by NASBA. Thus, the RTi-NASBA-based method was capable of detecting realistic levels of M. avium subsp. paratuberculosis contamination. In addition, all IAC results were positive not only by M. avium subsp. paratuberculosis-negative NASBAs but also by M. avium subsp. paratuberculosis-positive NASBAs; i.e., the TP values were below the threshold (mean IAC TP values, 51.36 ± 4.48 and 48.50 ± 3.88 min, respectively). This was possibly due to low M. avium subsp. paratuberculosis loads in feces from infected animals. The lack of inhibition in any of the reactions suggests that the preamplification procedure used is effective with this type of sample.
The details presented above are intended to provide an illustrative example of the construction and optimization of an IAC for real-time NASBA. The principles are general and may be applied to real-time NASBA assays for other targets. Incidentally, the principles may, with little adaptation, also be used to construct IACs for other RNA detection methods, including reverse transcription-PCR.
In conclusion, we have presented the first report of a strategy for the construction of a real-time NASBA IAC and have described its use for assistance with the interpretation of NASBA results for artificially and naturally contaminated clinical samples. The method is simple and very easy to apply without the need for sophisticated molecular manipulation facilities. A molecular beacon NASBA is a highly useful development and should encourage investigators to use NASBA for routine diagnostics.
Acknowledgments
We thank E. Ferri for providing natural fecal samples and performing the standard culture-based M. avium subsp. paratuberculosis assays.
This work was supported by European Commission-funded project “SACROHN” N. QLK2-CT-2000-00928, a grant from the UK National Association for Colitis and Crohn's Disease, the “La Marató de TV3” Foundation, and the Spanish Ministerio de Ciencia y Tecnología (grants AGL2002-03496 and Inia CAL01-058-C2-2). D.R.-L. was supported by a University of Girona research fellowship.
REFERENCES
- 1.Abdulmawjood, A., S. Roth, and M. Bülte. 2002. Two methods for construction of internal amplification controls for the detection of Escherichia coli O157 by polymerase chain reaction. Mol. Cell. Probes 16:335-339. [DOI] [PubMed] [Google Scholar]
- 2.Al-Soud, W., and P. Radstrom. 2000. Effects of amplification facilitators on diagnostic PCR in the presence of blood, feces, and meat. J. Clin. Microbiol. 38:4463-4470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Al-Soud, W. A., and P. Radstrom. 2001. Purification and characterization of PCR-inhibitory components in blood cells. J. Clin. Microbiol. 39:485-493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ausubel, F. M., R. Brent, R. E. Kingston, D. D. Moore, J. G. Seidman, J. A. Smith, and K. Struhl. 1990. Enzymatic manipulation of DNA and RNA. In Current protocols in molecular biology, vol. 1. John Wiley & Sons, Inc., New York, N.Y.
- 5.Bannantine, J. P., E. Baechler, Q. Zhang, L. Li, and V. Kapur. 2002. Genome scale comparison of Mycobacterium avium subsp. paratuberculosis with Mycobacterium avium subsp. avium reveals potential diagnostic sequences. J. Clin. Microbiol. 40:1303-1310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chan, A. B., and J. D. Fox. 1999. NASBA and other transcription-based amplification methods for research and diagnostic microbiology. Rev. Med. Microbiol. 10:185-196. [Google Scholar]
- 7.Compton, J. 1991. Nucleic acid sequence-based amplification. Nature 350:91-92. [DOI] [PubMed] [Google Scholar]
- 8.Cook, N. 2003. The use of NASBA for the detection of microbial pathogens in food and environmental samples. J. Microbiol. Methods 53:165-174. [DOI] [PubMed] [Google Scholar]
- 9.Hoorfar, J., and N. Cook. 2003. Critical aspects of standardization of PCR, p. 51-64. In K. Sachse and J. Frey (ed.), Methods in molecular biology: PCR detection of microbial pathogens. Humana Press, Totowa, N.J. [DOI] [PubMed]
- 10.Hoorfar, J., N. Cook, B. Malorny, P. Rådström, D. De Medici, A. Abdulmawjood, and P. Fach. 2003. Diagnostic PCR: making internal amplification control mandatory. J. Clin. Microbiol. 41:5835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kievits, T., B. van Gemen, D. van Strijp, R. Schukkink, M. Dircks, H. Adriaanse, L. Malek, R. Sooknanan, and P. Lens. 1991. NASBA isothermal enzymatic in vitro nucleic acid amplification optimised for the diagnosis of HIV-1 infection. J. Virol. Methods 35:273-286. [DOI] [PubMed] [Google Scholar]
- 12.Lanciotti, R. S., and A. J. Kerst. 2001. Nucleic acid sequence-based amplification assays for rapid detection of West Nile and St. Louis encephalitis viruses. J. Clin. Microbiol. 39:4506-4513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Leone, G., H. van Schijndel, B. van Gemen, F. R. Kramer, and C. D. Schoen. 1998. Molecular beacon probes combined with amplification by NASBA enable homogeneous, real-time detection of RNA. Nucleic Acids Res. 26:2150-2155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Malorny, B., J. Hoorfar, C. Bunge, and R. Helmuth. 2003. Multicenter validation of the analytical accuracy of Salmonella PCR: towards an international standard. Appl. Environ. Microbiol. 69:290-296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Rodríguez, D., J. Lloyd, A. Herrewegh, J. Ikonomopoulos, M. D'Agostino, M. Pla, and N. Cook. 2004. A molecular beacon-based real-time NASBA assay for detection of Mycobacterium avium subsp. paratuberculosis in water and milk. FEMS Microbiol. Lett. 237:119-126. [DOI] [PubMed] [Google Scholar]
- 16.Rossen, L., P. Norskov, K. Holmstrom, and O. F. Rasmussen. 1992. Inhibition of PCR by components of food samples, microbial diagnostic assays and DNA-extraction solutions. Int. J. Food Microbiol. 17:37-45. [DOI] [PubMed] [Google Scholar]
- 17.Sambrook, J., and D. Russell. 2000. Molecular cloning: a laboratory manual, 3rd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 18.Sorensen, O., S. Rawluk, J. Wu, K. Manninen, and G. Ollis. 2003. Mycobacterium paratuberculosis in dairy herds in Alberta. Can. Vet. J. 44:221-226. [PMC free article] [PubMed] [Google Scholar]
- 19.Stryer, L. 1978. Fluorescence energy transfer as a spectroscopic ruler. Annu. Rev. Biochem. 47:819-846. [DOI] [PubMed] [Google Scholar]
- 20.Tyagi, S., and F. R. Kramer. 1996. Molecular beacons: probes that fluoresce upon hybridisation. Nat. Biotechnol. 14:303-308. [DOI] [PubMed] [Google Scholar]
- 21.Tyagi, S., D. P. Bratu, and F. R. Kramer. 1998. Multicolor molecular beacons for allele discrimination. Nat. Biotechnol. 16:49-53. [DOI] [PubMed] [Google Scholar]
- 22.Uyttendaele, M., R. Schukkink, B. van Gemen, and J. Debevere. 1994. Identification of Campylobacter jejuni, Campylobacter coli and Campylobacter lari by the nucleic acid amplification system NASBA. J. Appl. Bacteriol. 77:694-701. [DOI] [PubMed] [Google Scholar]
- 23.Uyttendaele, M., R. Schukkink, B. van Gemen, and J. Debevere. 1995. Development of NASBA, a nucleic acid amplification system, for identification of Listeria monocytogenes and comparison to ELISA and a modified FDA method. Int. J. Food Microbiol. 27:77-89. [DOI] [PubMed] [Google Scholar]
- 24.van Beckhoven, J. R., D. E. Stead, and J. M. van der Wolf. 2002. Detection of Clavibacter michiganensis subsp. sepedonicus by AmpliDet RNA, a new technology based on real time monitoring of NASBA amplicons with a molecular beacon. J. Appl. Microbiol. 93:840-849. [DOI] [PubMed] [Google Scholar]
- 25.van Gemen, B., R. van Beuningen, A. Nabbe, D. van Strijp, S. Jurriaans, P. Lens, and T. Kievits. 1994. A one-tube quantitative HIV-1 RNA NASBA nucleic acid amplification assay using electrochemiluminescent (ECL) labelled probes. J. Virol. Methods 49:157-167. [DOI] [PubMed] [Google Scholar]