Abstract
One-carbon (1C) metabolism, mediated by the folate cofactor, supports multiple physiological processes. These include biosynthesis (purines and thymidine), amino acid homeostasis (glycine, serine, and methionine), epigenetic maintenance, and redox defense. Both within eukaryotic cells and across organs, 1C metabolic reactions are compartmentalized. Here we review the fundamentals of mammalian 1C metabolism, including the pathways active in different compartments, cell types, and biological states. Emphasis is given to recent discoveries enabled by modern genetics, analytical chemistry, and isotope tracing. An emerging theme is the biological importance of mitochondrial 1C reactions, both for producing 1C units that are exported to the cytosol and for making additional products, including glycine and NADPH. Increased clarity regarding differential folate pathway usage in cancer, stem cells, development, and adult physiology is reviewed and highlights new opportunities for selective therapeutic intervention.
Introduction
Folate metabolism, which supports a broader set of transformations known as one-carbon (1C) metabolism, is a universal metabolic process that serves to activate and transfer 1C units for biosynthetic processes including purine and thymidine synthesis and homocysteine remethylation. Whereas most bacteria, yeast, and plants can synthesize folate, animals require dietary folate intake. In adults, insufficient dietary folate leads to anemia. In developing fetuses, it creates a disposition to birth defects known as neural tube defects, which involve failure of neural tube closure early in pregnancy. Outcomes range in severity from anencephaly, causing fetal loss, to spina bifida with partial leg paralysis (Copp et al., 2015).
Due to its essential role in nucleic acid synthesis, inhibition of folate metabolism blocks cellular proliferation, and inhibitors of bacterial folate synthesis and transformations (sulfamethoxazole and trimethoprim) or mammalian folate transformations (methotrexate and pemetrexed) are widely used antibiotics and chemo-therapeutics (Chattopadhyay et al., 2007). Folate metabolism also contributes to homocysteine remethylation, impacting epigenetics and possibly also cardiovascular health.
Folate Chemistry
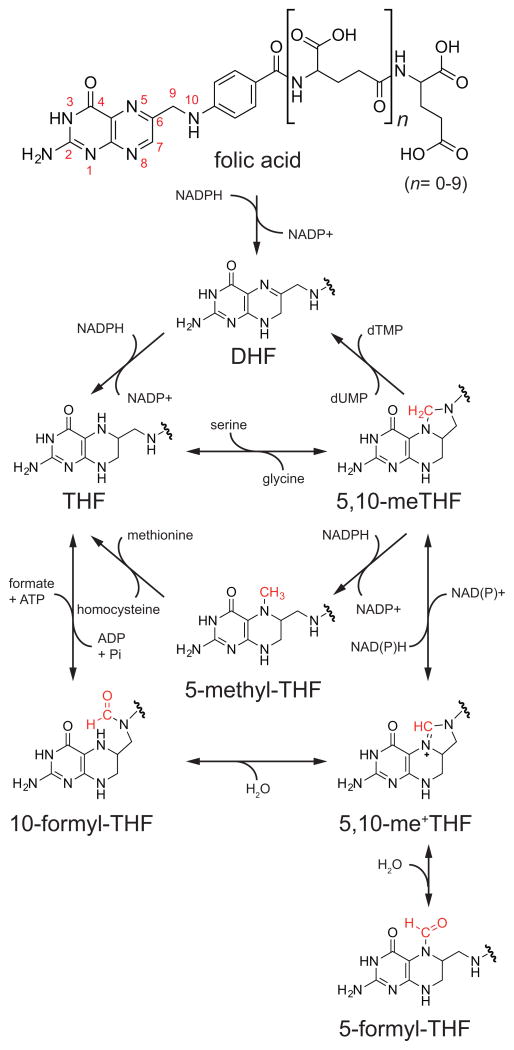
The term folate encompasses a complex set of molecules that share a common core structure involving three chemical moieties: a pteridine ring that can be reduced or oxidized, a para-aminobenzoic acid (PABA) linker that together with the pteridine ring binds 1C units, and a variable chain length polyglutamate tail that serves to localize the molecule within the cell (Figure 1). The biologically active form of folate is the reduced pteridine species, tetrahydrofolate (THF). Nearly all natural folate species in diet and in the body are present in the reduced form, typically 5-methyl-THF in humans (Wright et al., 2007). Folic acid (vitamin B9), a common synthetic food additive used to prevent neural tube defects, must be sequentially reduced to first dihydrofolate (DHF) and then THF before it can enter the folate cycle (Figure 1).
Figure 1. Chemical Transformations of Folates.
Folic acid is reduced to THF, which can then accept a 1C unit and undergo a series of oxidative/reductive transformations. DHF, dihydrofolate; THF, tetrahydrofolate; 5,10-meTHF, 5,10-methylene-THF; 5,10-me+THF, 5,10-methenyl-THF.
Folate molecules function as carriers for 1C units, allowing them to be manipulated and assembled in support of metabolic processes. To this end, 1C units are covalently bound to the 5-position nitrogen atom on the pterdine ring moiety and the 10-position nitrogen atom on the PABA moiety of THF (Figure 1). Through different covalent bonds to these nitrogen atoms, 1C units can be held in three different carbon oxidation states, each of which plays specific biosynthetic roles (Figure 1). New 1C units primarily enter the system as 5,10-methylene-THF, which can be made from the amino acids serine and glycine and the choline degradation products dimethylglycine and methylglycine (sarcosine). As 1C-loaded folates are not known to transfer across intracellular membranes, 5,10-methylene-THF must be generated in both the mitochondria and cytosol (Anderson et al., 2011). Once bound to THF, 1C units can be interconverted between different oxidation states, with 5,10-methylene-THF, 5-methyl-THF, and 10-formyl-THF each supporting distinct biosynthetic functions (Figure 1). Another formyl-THF species, 5-formyl-THF, does not play a direct biosynthetic role, but serves as a 1C reserve.
Outputs of Folate Metabolism
Products of 5,10-Methylene-THF: Thymidine and Serine
Thymidylate synthase (TYMS) converts deoxyuridine monophosphate (dUMP) to deoxythymidine monophosphate (dTMP) in a 5,10-methylene-THF-dependent reaction and is the target for the suicide inhibitor chemotherapeutic agent 5-fluorouracil (Figure 2).
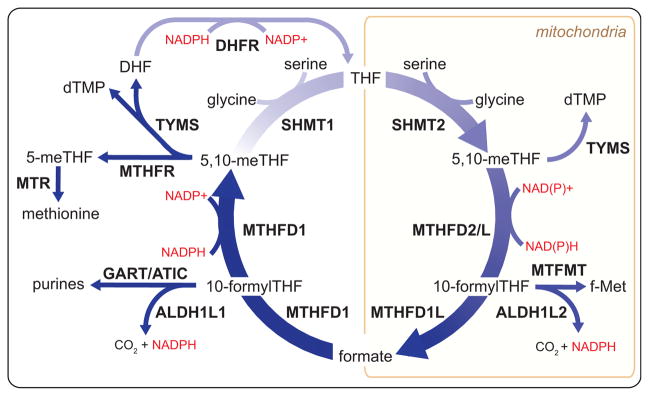
Figure 2. Products and Compartmentalization of Folate-Mediated 1C Metabolism.
Through an interlinked set of mitochondrial and cytosolic reactions, folate metabolism supports 1C anabolic reactions. All abbreviations are standard gene names. Certain descriptions utilize the common protein name for clarity. SHMT1/2, serine hydroxymethyl transferase, cytosolic(1)/mitochondrial (2); MTHFD1, methylenetetrahydrofolate dehydrogenase, cyclohydrolase, and formyltetrahydrofolate synthetase 1; MTHFD2/L, methylenetetrahydrofolate dehydrogenase 2/2-like; MTHFD1L, monofunctional tetrahydrofolate synthase, mitochondrial; MTFMT, mitochondrial methionyl-tRNA formyltransferase; TYMS, thymidylate synthetase; MTHFR, methylenetetrahydrofolate reductase; MTR, methionine synthase; DHFR, dihydrofolate reductase; GART, phosphoribosylglycinamide formyltransferase; ATIC, 5-aminoimidazole-4-carboxamide ribonucleotide formyltransferase/IMP cyclohydrolase; ALDH1L1/2, cytosolic (1)/mitochondrial (2) 10-formyltetrahydrofolate dehydrogenase.
Serine hydroxymethyltransferase (SHMT) uses 5,10-methylene-THF to convert glycine into serine. The reaction is reversible. Cells can use SHMT to make serine in one compartment and catabolize it in another, with the direction of flow depending upon the supply and demand of 1C units within each compartment (Ducker et al., 2016).
Product of 5-Methyl-THF: Methionine
The most reduced form of folate 1C unit, 5-methyl-THF, has a unique cellular fate, the remethylation of homocysteine to form methionine. 5-methyl-THF is produced by the cytosolic NADPH-dependent activity of methylene tetrahydrofolate reductase (MTHFR) andconsumedbythecobalamin (vitamin B12)-dependent enzyme methionine synthase (MTR) (Figure 2). Mammalian cells can also regenerate methionine from homocysteine in a folate-independent manner using betaine (trimethylglycine, a product of choline degradation) via the enzyme betaine-homocysteine methyltransferase (BHMT). Expression of BHMT is limited to liver and kidney (Pajares and Pérez-Sala, 2006), whereas widespread methionine synthase expression may enable homocysteine remethylation throughout the body.
Homocysteine remethylation is of particular physiological importance because methionine is the substrate for S-adenosylmethionine (SAM) synthetase. SAM, the reactive methyl carrier, is the second most common enzymatic cofactor after ATP and plays a major role in epigenetics; biosynthetic processes including phosphatidylcholine, creatine, and polyamine synthesis; and sulfur metabolism (Finkelstein, 1990; Mudd et al., 2007; Su et al., 2016). In fact, phosphatidylcholine synthesis from phosphoethanolamine is likely the largest 1C sink in adult mammals and consequently the largest source of S-adenosylhomocysteine (SAH) (Stead et al., 2006). Changes in methionine concentrations lead to changes in the ratio of SAM to SAH that impact many methylation reactions including histone methylation (Mentch et al., 2015). The SAM/SAH ratio is tightly regulated by metabolic mechanisms including inhibition of MTHFR by SAM (Kutzbach and Stokstad, 1971), inhibition of the SAM-consuming enzyme glycine-N-methyltransferase (GNMT) by 5-methyl-THF (Yeo and Wagner, 1992), and sequestration of 5-methyl-THF by cytosolic serine hydroxymethyltransferase (SHMT1) (Herbig et al., 2002; Stover and Schirch, 1991). In mice, SHMT1 loss increases, and SHMT1 overexpression decreases SAM levels (MacFarlane et al., 2008, 2011).
Products of 10-Formyl-THF: Purines, Formate, and CO2
The most oxidized form of folate carbon, 10-formyl-THF, is required for de novo synthesis of purines. In proliferating mammalian cells in culture, purine synthesis is the largest demand for folate 1C units. For every newly synthesized DNA or RNA base, an average of one 10-formyl-THF is required. (In comparison, 5,10-methylene-THF is needed only for DNA, at a rate of only one equivalent per four bases.) Purine synthesis is an 11-reaction process incorporating two 10-formyl-THF units into the purine backbone. The enzymes that perform this chemistry have been shown to co-localize in the single complex on the outside of mitochondria, termed the purinosome (An et al., 2008; French et al., 2016). Neither this complex nor its constituent enzymes have been identified within the mitochondrial matrix. However, there is a requirement for mitochondrial 10-formyl-THF. Like in bacteria, mitochondrial initiator methionine tRNAs are formylated. This formylation requires 10-formyl-THF and is required for translation of mitochondrially encoded proteins (Tucker et al., 2011).
To enable transfer of 1C units between compartments, 10-formyl-THF can be hydrolyzed to formate. Hydrolysis of 10-formyl-THF to free formate is coupled to ATP production and reversible. Finally, 10-formyl-THF 1C units can be fully oxidized to CO2 in an NADPH-generating reaction, enabling their terminal elimination (Krupenko et al., 2010).
Side Products of 1C Metabolism
The above sections describe the products made from the 1C unit itself. However, the reactions can also generate other important metabolic products. Glycine is a required precursor for many biosynthetic pathways including glutathione, purine, creatine, and heme synthesis. Cells can acquire glycine from the serum directly or by catabolism of choline, serine, or (in most mammals, but not humans) threonine. When exogenous glycine is unavailable, its synthesis from serine is essential to both purine and glutathione synthesis (Ducker et al., 2016). Serine can be synthesized from glucose, and the combination of the serine biosynthetic pathway and the SHMT reaction provides a route from carbohydrates to glycine.
Additionally, 1C transformations produce and consume redox equivalents. While many of these reactions are reversible, the oxidation of 10-formyl-THF to CO2 is not and generates an NADPH equivalent (Figure 2), with recent evidence suggesting that folate-mediated NADPH production may be particularly important in mitochondrial redox homeostasis (Fan et al., 2014; Piskounova et al., 2015).
Metabolic Cycles and Compartmentalization
Dihydrofolate-Tetrahydrofolate Cycle
Many folate enzymes alter the redox state of the 1C unit (Figure 2). Only a single one, however, alters THF itself: TYMS. Using 5,10-methylene-THF as the substrate, TYMS transfers a methyl unit to dUMP to make dTMP, abstracting from THF the two electrons required to reduce the 1C unit and producing DHF as the product. THF is regenerated from DHF by the NADPH-dependent activity of dihydrofolate reductase (DHFR), the canonical target of the antibacterial trimethoprim and the anticancer agent methotrexate. Each of these drugs leads to cellular toxicity in part by trapping folate as DHF and thereby depleting the cellular levels of THF required for other folate reactions.
Mitochondrial-Cytosolic 1C Cycle
A complete set of enzymes linking serine to formate exists in both the cytosol and mitochondria. This allows for not only parallel metabolic processes, but also for a complete oxidative/reductive cycle, which catabolizes serine in the mitochondria and synthesizes serine in the cytosol.
The subcellular compartmentalization of folate reactions has been thoroughly reviewed by Tibbetts and Appling (Tibbetts and Appling, 2010). Both compartments encode enzymes with 5,10-methylene-THF dehydrogenase activity, cyclohydrolase activity to interconvert the resulting 5,10-methenyl-THF (an intermediate with no biosynthetic role) with 10-formyl-THF, and formate-THF ligase activity that can release or reassimilate formate in an ATP-dependent manner. In the cytosol, each of these activities is localized to a single trifunctional protein, MTHFD1. In contrast, in mitochondria there are two enzymes with bifunctional dehydrogenase/cyclohydrolase activity (MTHFD2 and MTHFD2L) and a separate formate-THF ligase enzyme (MTHFD1L, which structurally resembles MTHFD1 but with loss of catalytic activity in both the dehydrogenase and cyclohydrolase domains).
The cycle that catabolizes serine in mitochondria and synthesizes serine in the cytosol is thermodynamically driven by the difference in electrochemical potential between mitochondrial NADH (and also NADPH) and cytosolic NADPH (Yang and MacKenzie, 1993). MTHFD1 uses NADPH and MTHFD2/L uses NADH, whereas MTHFD2L may use both substantially under physiological conditions, although cellular tracing studies are needed to confirm this (Shin et al., 2014). The high cytosolic NADPH/NADP+ ratio favors flux from formate toward serine in the cytosol.
Tracing Mitochondrial and Cytosolic Serine Catabolism
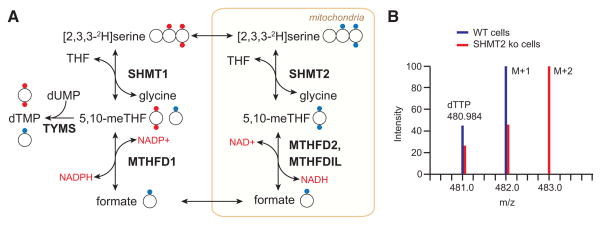
The introduction of deuterated serine ([2,3,3-2H]serine) as a tracer has enabled compartment-specific analysis of the flux of 1C units from serine, their main source, by mass spectrometry. When serine is metabolized within the mitochondria to formate, the oxidation of the 1C unit removes one of the deuterons; reductive incorporation of this 1C unit into thymidine or methionine in the cytosol results in their being singly labeled (M+1) (Figures 3A and 3B). Conversely, cytosolic metabolism of serine directly to 5,10-methylene-THF by SHMT1 involves no loss of deuterium and results accordingly in M+2 labeling of thymidine and methionine. This technique was developed by the Gregory and Stover groups, who first used it to trace the contribution of mitochondrial-derived serine to homocysteine remethylation in cell culture and in human volunteers (Gregory et al., 2000; Herbig et al., 2002). Both found that the majority of methionine 1C units originated from the mitochondria. When applied to MTHFD1L deletion mouse embryonic fibroblasts, this labeling technique demonstrates that in the absence of functional mitochondrial folate metabolism, cytosolic SHMT1 makes 5,10-methylene-THF from serine, and M+2 is now observed on methionine (Pike et al., 2010). Across a diverse panel of proliferating cell lines in culture, Ducker et al. found that most make 1C units originate solely in the mitochondria; however, several engaged in both cytosolic and mitochondrial serine catabolism (Ducker et al., 2016). Moreover, upon genetic ablation of the mitochondrial pathway, even cells that normally use only the mitochondrial pathway to catabolize serine will switch to making 1C units from serine in the cytosol.
Figure 3. [2,3,3-2H]Serine Is a Tracer for Mitochondrial Folate Pathway Activity.
(A) Compartment-specific metabolism of serine can be determined by following the incorporation of 2H from [2,3,3-2H]serine into terminal products of 1C metabolism such as thymidine. If serine is metabolized in the mitochondria, one proton attached to the 3-position carbon will be lost during oxidation to formate, as represented by the blue dots. Reductive incorporation of the formate into the cytosolic folate pool will result in singly deuterated thymidine monophosphate (TMP M+1). In contrast, a serine catabolized in the cytosol by SHMT1 will transfer both protons onto 5,10-methylene-THF and generate a doubly labeled TMP M+2, as represented by the red dots.
(B) Representative mass spectra of WT and SHMT2 deletion cells demonstrate the application of this tracing technique. Adapted from Ducker et al. (2016).
The deuterium from [2,3,3-2H]serine can be traced into NAD(P)H and from there into other metabolites (Fan et al., 2014). Lewis et al. demonstrated using a clever mutant isocitrate dehydrogenase (IDH) reporter strategy that folate metabolism selectively generates NADPH in mitochondria (Lewis et al., 2014). The authors transfected cells with IDH1 (cytosolic) or IDH2 (mitochondrial) constructs that carried active site mutations, leading to the production of the oncometabolite R(−)-2-hydroxyglutarate (2HG) from the NADPH catalyzed reduction of α-ketoglutarate (Dang et al., 2009). 2H-labeled 2HG was produced from [2,3,3-2H]serine in cells with the mutant mitochondrial IDH2, but not cytosolic IDH1. Thus, cytosolic folate metabolism proceeds in the reductive, NADPH-consuming direction.
This cofactor-mediated deuterium tracing can also be accomplished by direct monitoring of cellular metabolites without the introduction of a mutant enzyme. In cell lines with mutations that block mitochondrial folate metabolism, cytosolic folate-mediated NADPH production results in fatty acid labeling from 2H-serine. In cells with intact mitochondrial 1C metabolism, mitochondrial folate metabolism makes both NADH and NADPH, as evidenced by NADH-mediated labeling of malate and NADPH-mediated labeling of proline from 2H-serine (Ducker et al., 2016). Using clustered regularly interspaced short palindromic repeat (CRISPR) deletion mutants, this technique was able to isolate the source of most mitochondrial folate-derived NADPH to the activity of the formyl-THF oxidizing enzyme mitochondrial 10-formyltetrahydrofolate dehydrogenase (ALDH1L2).
Impact of NADH and NADPH Levels
Production of NADH by mitochondrial 5,10-methylene-THF dehydrogenase activity links 1C metabolism to the respiratory state of the cell. Mitochondrial NAD+/NADH ratios are maintained by oxidative phosphorylation, and early experiments in isolated mitochondria showed that formate production from serine was respiration dependent (García-Martínez and Appling, 1993). Conversely, in ρ0 cells, which are depleted for mitochondrial DNA, no alterations in folate metabolism were observed (Patel et al., 2003); however, these cells require exogenous pyruvate to grow, which functions in part to restore the NAD+/NADH ratio (Birsoy et al., 2015; Sullivan et al., 2015; Titov et al., 2016). The commonly used anti-diabetic agent metformin targets mitochondrial complex I and thus decreases the NAD+/NADH ratio. Metformin also inhibits cancer cell growth, in part through inhibition of biosynthetic metabolism (Griss et al., 2015). Metformin-induced growth inhibition can be partially rescued by supplementation with hypoxanthine and thymidine, products of 1C metabolism (Corominas-Faja et al., 2012). It remains unclear, however, if the impact of metformin on 1C metabolism is clinically significant at normal therapeutic doses.
Further evidence in support of a physiological link between mitochondrial NADH and 1C metabolism was provided by two recent studies of metabolic alterations induced by mitochondrial respiratory chain deficiency (Bao et al., 2016; Nikkanen et al., 2016). Using an inducible dominant-negative mitochondrial DNA polymerase, Bao et al. showed that mitochondrial DNA depletion triggers, via the transcription factor ATF4, expression of serine synthesis genes (PHGDH, PSAT1, and PSPH), thereby increasing serine to feed 1C metabolism. In related work, Nikkanen et al. investigated the outcome of mitochondrial DNA depletion in vivo. In a model of mitochondrial myopathy induced by mutation of the mitochondrial DNA helicase TWINKLE, reductions in tissue formate and 10-formyl-THF co-occurred with increases in expression of mitochondrial folate 1C enzymes (MTHFD2 and MTHFD1L). In the muscle and blood of progressive external ophthalmoplegia patients, increases in 1C-related metabolites, including serine and cystathionine, were observed. Together, these studies are consistent with a model whereby impaired electron chain transport activity lowers mitochondrial formate production, inducing an adaptive response to increase both 1C donors and mitochondrial 1C metabolic enzymes, but these changes are insufficient to regain full wild-type (WT) pathway functionality.
What role does cytosolic NADP+/NADPH play in driving reductive cytosolic folate metabolism? Unlike in the case of the NAD+ in the mitochondria, there is as of yet not an easy way to modulate steady-state cytosolic NADPH levels. However, the case of mitochondrial folate mutant cell lines is instructive. These cells now process 1C units oxidatively in the cytosol in order to produce 10-formyl-THF for purine biosynthesis. In engineered HEK293T cells deficient for MTHFD2, the NADP+/NADPH ratio does not significantly change between WT and mutant. Instead, there is a dramatic decrease in 10-formyl-THF. These results argue that changes in folate pools, as opposed to NADPH, control the thermodynamically favored direction of cytosolic 1C flux (Ducker et al., 2016).
Rationale for Compartmentalization
The differential electrochemical potential of NADH versus NADPH explains why folate metabolism, as constructed, involves mitochondrial formate shuttling to the cytosol. But it does not explain why the mitochondria need be included in the first place. If cytosolic MTHFD1 used NAD+ for the dehydrogenase reaction, it would favor the oxidative direction (West et al., 1996). Mitochondrial-specific 1C demand is small and could be met in theory by transport or a low-activity pathway. We believe that nature has chosen to localize most serine catabolism and 1C unit oxidation to mitochondria to uncouple 1C metabolism from glycolysis. Glucose fermentation to lactate is redox balanced, but this balance can be thrown off by any other NADH-generating cytosolic reaction. If 1C oxidation were to instead occur in the cytosol with concomitant NADH generation, cells could become depleted of cytosolic NAD+, blocking glycolysis. Thus, the mitochondrial localization of oxidative 1C metabolism enhances cellular robustness.
Polyglutamation Encodes Compartmentalization
The mitochondrial/cytosolic division of folate metabolism is encoded both genetically by the different isozymes and chemically by differences in the folates themselves. Folate is distributed by the circulation as THF-monoglutamate species (Wright et al., 2007). These monoglutamates are actively transported across the cell membrane using the anionic reduced folate transporter (RFC, SLC19A1) (Hou and Matherly, 2014). To retain folates within the cell, they are polyglutamated, which reduces their affinity for the transporter and enhances their affinity for folate enzymes (Kim et al., 1996). Folate polyglutamate synthetase (FPGS) activity is essential for cell proliferation (Freemantle et al., 1995). Compared to bacteria, the degree of polyglutamation is higher in eukaryotes (Leung et al., 2013; Lin et al., 1993; Lu et al., 2007). Changes in RFC and FPGS function are mechanisms of acquired resistance to antifolates (Zhao and Goldman, 2003). Mutations that impair RFC transport provide resistance to methotrexate in leukemic cells, whereas loss of FPGS expression leads to pemetrexed resistance, as polyglutamation is essential for its activity (Liani et al., 2003).
A mitochondrial folate transporter (SLC25A32) is responsible for translocation of THF into the mitochondria, and mutants deficient for this six-transmembrane helix protein are glycine auxotrophs, a common phenotype of mitochondrial folate pathway loss (Chasin et al., 1974; Titus and Moran, 2000). Folate polyglutamates are not thought to be substrates for this transporter, necessitating mitochondrial localization of FPGS activity, achieved by a mitochondria splice variant of the gene (Freemantle et al., 1995; Lowe et al., 1993). Mitochondrial folate polyglutamates are of larger average chain length, but this may only reflect the processive activity of FPGS on the trapped slow exchanging mitochondrial folate pool. The compartmental requirements for FPGS were recently elegantly confirmed by Lawrence and colleagues using inducible versions of either the mitochondrial or cytosolic isoforms of FPGS in an FPGS null cell line (Lawrence et al., 2014). When FPGS was expressed only in the cytosol, cells were glycine auxotrophs, whereas when expression was confined to the mitochondria, cells required thymidine and purines to circumvent their now defective cytosolic biosynthetic pathways.
Nuclear Folate Metabolism
Folate metabolism has long been identified in purified nuclei (Prem veer Reddy and Pardee, 1980). Isotope tracer studies in isolated nuclei show that dUMP is converted to dTMP and that formate can function as the 1C source (Anderson and Stover, 2009; Field et al., 2014). No nuclear isoforms of folate genes have been discovered. Rather, it has been proposed that cytosolic folate enzymes perform nuclear functions. SHMT1, SHMT2α (an alternatively spliced isoform of SHMT2), and MTHFD1 show partial nuclear localization as GFP-tagged overexpression constructs, and SHMT1 may associate with the nuclear lamin (Anderson et al., 2012b; Field et al., 2016), and its cell-cycle-specific sumoylation has been proposed to control the formation of a nuclear complex with TYMS (Anderson et al., 2012a). Such a complex, however, is not essential for maintaining nuclear thymidine pools, as HEK293 cells engineered via CRISPR to lack both SHMT1 and 2 (and thus lacking any SHMT-dependent nuclear complex) grow normally when supplemented with formate (Bao et al., 2016). More generally, the lack of nuclear-specific enzymes has hindered efforts to assign function to nuclear folate metabolism.
Sources of 1C Units
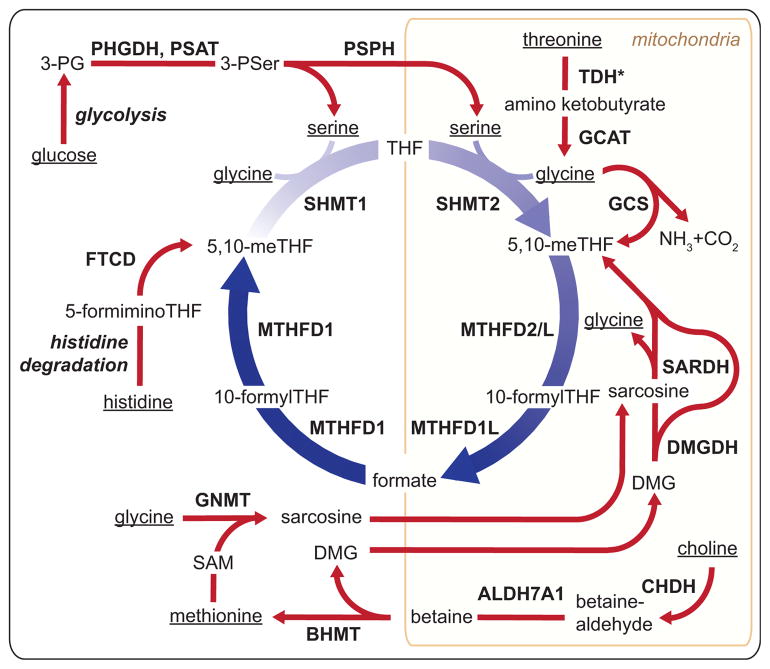
Eukaryotic cells mobilize multiple carbon sources to supply 1C metabolism. Serine, glycine, formate, histidine, and the choline-derived methylglycine species (sarcosine and dimethylglycine) can all directly contribute to the THF-bound 1C pool (Figure 4). Quantitatively, choline, serine, and glycine are the most important dietary sources of folate 1C units. Beyond supporting biosynthetic processes, 1C metabolism plays a pivotal role in catabolism of these nutrients.
Figure 4. Major Sources of 1C Units in the Mammalian Cell.
1C sources originate in the diet and are metabolized to generate 1C units. Dietary sources of 1C are underlined and include glucose, threonine, methionine, glycine, serine, histidine, and choline. TDH*, human TDH encodes a pseudogene without functional catalytic activity; 3PG, 3-phoshoglycerate; 3PSer, 3-phosphoserine; DMG, dimethylglycine; TDH, L-threonine dehydrogenase; GCAT, glycine C-acetyltransferase; GCS, glycine cleavage system; SARDH, sarcosine dehydrogenase; DMGDH, dimethylglycine dehydrogenase; CHDH, choline dehydrogenase; ALDH7A1, aldehyde dehydrogenase 7 family member A1; BHMT, betaine-homocysteine S-methyltransferase; GNMT, glycine N-methyltransferase; FTCD, formimidoyltransferase cyclodeaminase; PHGDH, phosphoglycerate dehydrogenase; PSAT, phosphoserine aminotransferase; PSPH, phosphoserine phosphatase.
Among the various 1C donors, only three are capable of directly feeding into the cytosolic folate-bound 1C pool: histidine, serine, and formate (Figure 4). Histidine degradation generates 5-formimino-THF, which is converted into 5,10-methylene-THF in the cytosol by formimidoyltransferase cyclodeaminase (Solans et al., 2000). Serine, which can be synthesized from glucose but under typical conditions in mammals mainly comes from dietary protein intake, can directly feed the cytosolic 1C pool via SHMT1. In rapidly proliferating cultured cells, however, the main direct source of cytosolic 1C units is formate released from their mitochondria.
Within the mitochondria, 1C units can be made from serine, glycine, sarcosine, or dimethylglycine and excreted into the cytosol as formate (Figure 4). SHMT2 converts serine into a 1C unit and glycine that can be further metabolized by the glycine cleavage system (GCS). Thus, SHMT2 and the GCS are poised to work in tandem. GCS is a mitochondrial-localized four-enzyme complex that decarboxylates glycine to form CO2 and deaminates the remaining carbon to form mitochondrial 5,10-methylene-THF (Kikuchi et al., 2008). This reaction is reversible in vitro (Kochi and Kikuchi, 1974). While not functional in humans, in all other mammals (including chimpanzees) glycine can also be produced in the mitochondria from threonine via threonine dehydrogenase (TDH) and glycine C-acetyltransferase (GCAT). While SHMT2 is the dominant 1C source, as determined by isotope tracing in most proliferating cells cultured in vitro, glycine catabolism is a major 1C source in stem cells, embryogenesis, and quiescent adult tissues.
Choline is an important dietary nutrient that supports both folate-independent methionine synthesis and, via the resulting dimethylglycine, folate-bound 1C pools. In the liver, choline can either be used for lipid head group synthesis or catabolized to dimethylglycine (Ueland, 2011). Choline catabolism begins with its two-step mitochondrial oxidation to trimethylglycine (betaine). In the cytosol, betaine remethylates homocysteine in a folate-independent reaction catalyzed by BHMT (Figure 4). After donating a methyl group to homocysteine, betaine becomes dimethylglycine, which can yield additional 1C units via folate-dependent mitochondrial reactions: dimethylglycine dehydrogenase (DMGDH) makes sarcosine and sarcosine dehydrogenase (SARDH) makes glycine.
In addition to being synthesized via the choline degradation pathway, sarcosine can be made in the cytosol by SAM-mediated methylation of glycine catalyzed by the enzyme GNMT (Luka et al., 2009). This provides a potential route from methionine to folate-bound 1C units. Sarcosine levels are elevated in aggressive and metastatic prostate cancer tumors (Sreekumar et al., 2009). This has raised great interest in sarcosine as a biomarker for prostate cancer; however, studies attempting to relate urine or blood sarcosine concentrations to prostate cancer have yielded inconsistent results (Kelly et al., 2016).
Circulating formate is another 1C source. In addition to being a product of folate metabolism, formate may also be produced from folate-independent processes including the kynurenine pathway of tryptophan degradation, the polyamine/methionine cycle, and the methanol detoxification pathway. Free formate concentrations in mammalian serum range from 20 to 50 μM and originate from both dietary sources and internal metabolic processes (Lamarre et al., 2013, 2014). The role for circulating formate in meeting normal 1C demand is unclear. Formate is typically considered a toxin and is responsible for metabolic acidosis resulting from methanol or formaldehyde poisoning. Flux from glycine to serine catalyzed by SHMT1 consumes 1C units and may serve the biological function of formate detoxification. Clearance of formate is known to be folate dependent, and its cross-species variance correlates with susceptibility to methanol poisoning (Sweeting et al., 2010). Thus, the folate system does not just support anabolism, but also has important catabolic and detoxification functions.
1C Metabolism in Development and Proliferative Adult Tissues
Decades of research have confirmed the essential requirement for dietary folate in both development and the adult. The best characterized function of folate metabolism is nucleic acid synthesis to support cellular proliferation, and folate metabolism is essential in proliferating tissues. What has only recently been uncovered, however, is the role specific folate-utilizing enzymes play in supporting growth and development, and which pathways are activated in fetal development, immune proliferation, and tissue homeostasis. Probably the most important insight over the past decade regards the role and importance of mitochondrial folate metabolism. Deletions in mitochondrial folate enzymes lead to neural tube defects and loss of viability. Many of these defects cannot be rescued metabolically, suggesting that mitochondrial folate metabolism is intrinsically required to supply 1C units in highly proliferative states, such as the embryo. This essentiality in development has hindered a rigorous examination of the requirement for mitochondrial folate metabolism in adult tissues, though expression patterns and limited tracing data suggest that it plays an important role in these contexts as well. Development of conditional and tissue-specific folate gene knockouts should eventually illuminate more precisely the roles of folate metabolism in adulthood.
Neural Tube Defects
Demand for 1C units is highest during fetal development, and folate deficiency leads to neural tube and congenital heart defects (Bailey and Berry, 2005; Beaudin and Stover, 2009). As the only enzyme capable of generating the cytosolic 10-formyl-THF required for purine synthesis, deletion of the trifunctional MTHFD1 enzyme is embryonic lethal (MacFarlane et al., 2009). The source of the embryonic 1C units appears to be substantially mitochondrial as SHMT1 deletion mice are viable and fertile (MacFarlane et al., 2008), although these animals do have vulnerabilities: in SHMT1−/− dams fed a standard diet no fetal abnormalities were observed; however, when folate and choline were removed from the diet, a fraction of fetuses developed neural tube defects (Beaudin et al., 2011). Maternal supplementation of the folate-poor diet with the thymidine precursor deoxyuridine completely rescued the neural tube defects, suggesting that in this instance, inadequate DNA synthesis was at least in part responsible for the neural tube defects (Martiniova et al., 2015). Surprisingly, however, thymidine was less effective as a rescue agent, and formate was not tested; thus, further investigation into the underlying mechanism is merited.
Mice defective in mitochondrial 1C metabolism are generally nonviable. MTHFD1L and GCS deletions are embryonic lethal due to neural tube defects (Momb et al., 2013; Narisawa et al., 2012). MTHFD1L−/− defects can be partially ameliorated through supplementation of dams with formate, but no viable offspring are produced (Momb et al., 2013). GCS mutants (deleted for the AMT gene) are nearly universally afflicted with neural tube defects. Maternal supplementation with methionine, but not thymidine, ameliorated some fetal deformities but was insufficient to recover viable pups. Interestingly, using a genetrap allele strategy targeting a different GCS component, GLDC, live mice deficient in GCS activity were created whose neural tube defects were formate rescuable (Pai et al., 2015). A genetrap allele is a selectable marker inserted into an early intron of a gene that contains both a splice acceptor site and stop codon followed by a polyadenylation sequence. Upon transcription, the unnatural splice site is incorporated into the nascent mRNA, resulting in early termination and functional deletion of the targeted gene, but unlike homozygous deletion techniques, some functional transcripts can be created due to missed splicing of the genetrap construct. Mice homozygous for the GLDC genetrap (GLDCGT1/GT1) were recovered at less than Mendelian ratios, and neural tube defects were common among fetuses. The live mice have brain defects including domed head, enlarged brain, and hydrocephalus leading to premature death. These abnormalities were largely rescuable by maternal formate supplementation, suggesting that the underlying neural tube defects are due to a lack of 1C units required to support neuronal proliferation (Pai et al., 2015). In humans, deficient GCS activity due to mutations in the AMT or GLDC genes causes nonketotic hyperglycinemia, which commonly presents as severe neonatal neurological dysfunction (Kure et al., 2006).
Although there has been no published attempt to make an SHMT2−/− mouse, serine appears to be a critical 1C source during development. Serine does not readily pass the blood-brain barrier, and accordingly, the main source of CNS serine is de novo synthesis from glucose. Knockout of the serine biosynthetic enzyme phosphoglycerate dehydrogenase (PHGDH) in mice causes severe neurological abnormalities including absence of the olfactory bulb, ganglionic eminence, and cerebellum (Yoshida et al., 2004). In humans, severe mutations in PHGDH or the downstream serine synthesis genes phosphoserine aminotransferase (PSAT1) and phosphoserine phosphatase (PSPH) result in the fatal Neu-Laxova syndrome, which is characterized by fetal malformation including microcephaly (Acuna-Hidalgo et al., 2014). Although definitive mouse genetic studies are lacking, methylglycine species appear to be less important as 1C sources, as a patient with an inactivating mutation in DMGDH has been identified and is apparently healthy (Binzak et al., 2001). Thus, mitochondrial generation of 1C units from both serine and glycine (but not methylated glycine species), supported by de novo serine synthesis from glucose, is required for normal mammalian development (Figure 5).
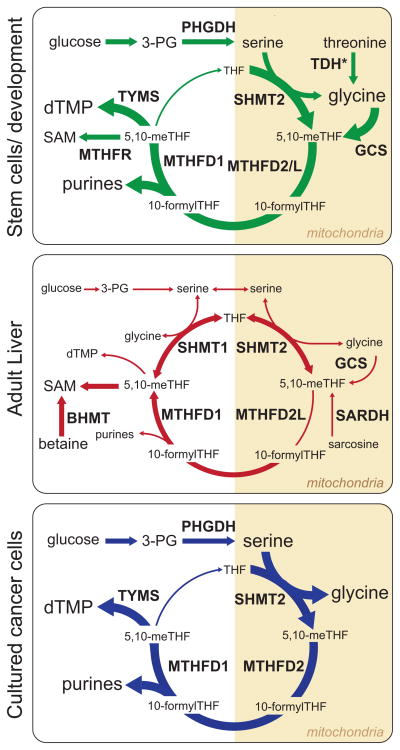
Figure 5. 1C Metabolism across Different Cell Types.
A cartoon representation of the major folate-dependent 1C metabolic pathways in three representative cell types. Variable font size and line widths represent the relative importance of each species or reaction in the indicated cell type. Reactions not drawn for an individual cell type are considered to be negligible in that context. TDH*, TDH activity is important in murine stem cells, but this enzyme is non-functional in humans.
MTHFD2, Hematopoiesis, and the Immune System
Hematopoiesis and immune response are among the most proliferative processes in the body and thus highly folate sensitive. In adults, dietary folate deficiency manifests as macrocytic anemia. Immune cells have long been noted to be sensitive to conventional antifolate therapies, limiting the effective doses that can be given for cancer therapy. Moreover, low-dose methotrexate is a mainstay in the treatment of certain autoimmune disorders including rheumatoid arthritis and lupus (Pincus et al., 2003). The downside of such therapy includes side effects in other proliferative tissues, including the gastrointestinal tract, and also in the liver. Thus, more selective inhibition of immune cell folate metabolism would be clinically useful.
Mouse genetics has uncovered a hematopoietic lineage-specific folate isozyme, MTHFD2. In contrast to mice lacking the closely related pathway genes MTHFD1L and AMT, those deleted for MTHFD2 do not show a neural tube defect but rather normal body development with a failure of the liver tissue to support hematopoiesis (Di Pietro et al., 2002). MTHFD2−/− embryos are smaller and paler than their WT littermates and lack normal liver redness. No rescue was observed when dams were supplemented with formate or a glycine-rich diet, and, in fact, supplemental glycine appeared to accelerate fetal death (Patel et al., 2003). One possible explanation for the lack of neural tube defect in this model is the presence of MTHFD2L, which is expressed throughout the embryo beginning at neural tube closure (Shin et al., 2014). This result suggests that MTHFD2 is of greatest functional importance in the hematopoietic lineage, including immune cells. The lineage specificity of MTHFD2 may be a function of the demand for 1C units for rapid proliferative growth; the reported Kcat/KM for MTHFD2 is 50-fold higher than for MTHFD2L (Shin et al., 2014).
Consistent with this, new work in T cell activation shows a requirement for rapid 1C metabolism. This is met by large changes in gene expression that serve both to mobilize existing stored 1C from 5-formyl-THF by 5-formyltetrahydrofolate cycloligase, and to produce new 1C units from serine via the SHMT2-MTHFD2 pathway (Ron-Harel et al., 2016). Interestingly, inhibition of T cell activation by SHMT2 short hairpin RNA (shRNA) was fully rescued by a combination of formate and the general antioxidant N-acetylcysteine, but not either alone, implicating the mitochondrial 1C pathway in T cells in both 1C unit generation and redox defense, as previously observed in HEK293T cells (Ducker et al., 2016). Given the widespread expression of MTHFD2L outside of the immune system, it seems likely that existing side effects of antifolate therapies could be mitigated by specific MTHFD2 inhibition. Accordingly, there is great promise in the development of MTHFD2-specific antifolates.
1C Metabolism in Stem Cells
Mouse embryonic stem cells (ESCs) divide at rates (4–5 hr doubling) far in excess of standard cultured cell lines, leading to unique metabolic stresses. Interestingly, murine ESCs selectively utilize glycine derived from TDH to generate 1C units for both nucleotide and methionine synthesis. Without threonine, mouse ESCs cannot synthesize DNA (Wang et al., 2009). The TDH reaction also generates acetyl-CoA, and threonine withdrawal alters histone modification by modulating both SAM and acetyl-CoA levels (Shyh-Chang et al., 2013). Threonine withdrawal can be rescued by a combination of pyruvate (as an acetyl-CoA source) and other 1C donors including glycine, dimethylglycine, and betaine.
The requirement of mouse ESCs for threonine raises two questions: Why the need for threonine as a 1C source in the mitochondria? And how is this demand met in humans given our lack of a functional TDH? Among all animals studied to date, humans appear to have uniquely lost the activity of the TDH gene (Edgar, 2002; Wang et al., 2009). In vitro studies have found that SHMT can break threonine into glycine via a retroaldol mechanism (Schirch and Szebenyi, 2005). Might such activity play a role in human ESCs? Or might murine, but not human, ESCs be defective in direct glycine uptake into mitochondria and/or mitochondrial serine catabolism, and therefore require threonine as a glycine source? Further research is needed to address these questions.
Folate Metabolism in Non-proliferative Adult Tissues
In development and in proliferative tissues, folate metabolism has an anabolic role in generating purines and thymidine, and in some cases also methionine, glycine, or serine. Among non-proliferative adult tissues, anabolic 1C reactions are largely localized to the liver and kidney, where they support the synthesis of the energy molecule creatine and the lipid head group choline, major organismal sinks for SAM-bound 1C units. In other non-proliferative tissues, the role of folate metabolism shifts to maintaining homeostasis of nucleotides and amino acids, and utilization of dietary choline, serine, and glycine as substrates for ATP and NADPH generation.
Folate metabolism is a major contributor to amino acid homeostasis (Brosnan et al., 2015). Isotopic tracer infusion studies in humans illustrate the high total body turnover in 1C-related amino acids. In one study, 13C-glycine was infused into healthy volunteers and GCS activity assayed by mass spectrometry. A large fraction of glycine was catabolized by the GCS and generated 1C units at over 20× the demand for homocysteine remethylation (Lamers et al., 2007). Similarly, in a different study to quantify the serine contribution to homocysteine remethylation, a small fraction of serine metabolic flux (~3%) could account for all of the homocysteine remethylation (Davis et al., 2004). These studies show that glycine and serine metabolism are dynamic but do not reveal the tissue specificity or function of the underlying reactions.
Amino Acid Homeostasis
1C metabolism directly controls the levels of three amino acids: methionine, serine, and glycine. Folate metabolism is the only direct route to catabolize glycine into the terminal waste products ammonia and CO2. 1C metabolism also indirectly controls cysteine levels via its effect on the transsulfuration pathway that transfers sulfur from homocysteine to serine to make cysteine.
Amino acid metabolism is constrained by that fact that any amino acids consumed in excess of immediate protein synthesis demand must be catabolized. The majority of amino acid catabolism is thought to occur in the liver and kidney where carbon can contribute to gluconeogenesis and nitrogen can be converted to urea for excretion (Brosnan, 2003). Quantitative measurements of the contributions of different organs to amino acid homeostasis are consistent with a major role for liver and kidney in 1C metabolism. These measurements are based on regional arterial-venous differences in serum amino acid concentrations, which reflect metabolic activity of the intervening organ. The liver is fed by both the arterial circulation and the portal vein, which runs from the small intestine and carries nutrients absorbed there. While sampling from the portal vein of humans is too invasive, the metabolic contribution of the liver can be determined in fasted individuals, where amino acid uptake from the intestines is minimal. During fasting, comparison of hepatic venous blood to arterial blood has revealed liver uptake of methionine and glycine that roughly matches their production from muscle (Felig, 1975). Thus, liver appears to be the major site of methionine and glycine utilization. In contrast, serine is consumed by many peripheral organs (Felig, 1975) and produced substantially from the kidney, which can make it from glucose, protein catabolism, and/or 1C-mediated conversion of glycine (Brosnan, 1987).
Liver Control of Systemic Methylation Potential
In parallel to consuming methionine, the liver also makes methionine. Folate-dependent homocysteine remethylation occurs throughout the body, but is likely most important in liver. Murine knockout of MTHFR, which produces the 5-methyl-THF required for homocysteine remethylation, results in homocystinuria (pathological elevation of homocysteine in the blood and urine), with early postnatal death and defects in brain development. Feeding of betaine to dams during pregnancy, which provides an alternative substrate for liver (but not brain) homocysteine remethylation, ameliorates the brain development defects and prevents death of the pups (Schwahn et al., 2004). MTHFR variants with severely reduced catalytic activity occur rarely in the human population and show similar phenotypes to the knockout mouse (Goyette et al., 1995). These observations suggest that brain development is dependent on liver 1C metabolism to maintain SAM homeostasis.
In contrast to the rarity of MTHFR alleles with severely reduced catalytic activity, alleles with moderately reduced catalytic activity are common in the human population and result in an elevation in circulating homocysteine levels without any developmental brain abnormalities. These low-activity alleles are associated with a modestly increased risk of cardiovascular disorders (Casas et al., 2005; Wald et al., 2002). Despite effectively lowering homocysteine levels, folate and cobalamin supplementation do not reduce the incidence of cardiovascular events in the general population, arguing against high homocysteine levels directly causing cardiovascular disease (Bazzano et al., 2006; Lonn et al., 2006; Zhou et al., 2011).
Increased homocysteine also occurs in liver disease, including non-alcoholic fatty liver disease (NAFLD) (Dai et al., 2016). In animals fed high-fat diets to induce NAFLD, liver 1C metabolism is dysregulated, as evidenced by intrahepatic increases in SAH and free homocysteine and decreases in methionine and the GNMT enzyme (Pacana et al., 2015). At the same time, deletions of 1C enzymes lead to development of liver disease. Mice lacking BHMT have elevated liver betaine and homocysteine and lower SAM, and develop hepatocellular carcinoma by 1 year of age (Teng et al., 2011).
Homocysteine concentrations are also regulated by the activity of the transsulfuration pathway, which is the only route to degrade homocysteine and thus irreversibly consume the methionine backbone. In this pathway, the homocysteine sulfur atom is transferred to serine to make cysteine. Transsulfuration occurs exclusively in the liver, brain, and pancreas; in the brain it is required to support glutathione synthesis (Vitvitsky et al., 2006). Cystathionine beta synthetase (CBS) is the first enzyme in this pathway and has an SAM regulatory site that activates the enzyme. Mutation of the site results in homocystinuria (Kluijtmans et al., 1996). Through the synthesis of reduced cysteine, this pathway contributes to glutathione synthesis and thus cellular redox homeostasis. By generating a reduced cysteine, it conserves an NADPH equivalent that would otherwise be required to reduce extracellular cystine taken in from the circulation (Eriksson et al., 2015).
Several mechanisms protect against excess buildup of methionine relative to homocysteine. These include SAM inhibition of MTHFR and disposal of excess SAM 1C units by GNMT. Mice deleted for GNMT develop progressive liver disease marked by steatosis and fibrosis, leading to hepatocellular carcinoma (Martínez-Chantar et al., 2008). Interestingly, while GNMT and BHMT deletions both cause progressive liver disease resulting in hepatocellular carcinoma, they have opposite metabolic effects: GNMT deletion causes high SAM and methionine and low homocysteine, consistent with human patients with GNMT deficiency (Liu et al., 2007; Luka et al., 2006). Apparently, either too high or too low liver SAM/SAH ratios can alter epigenetics and thereby promote cancer.
Mitochondria-to-Cytosol 1C Shuttling in Liver and Kidney
By combining expression data of 1C metabolic enzymes with flux constraints from arterial-venous amino acid measurements, it is possible to infer an approximate map of the active 1C pathways in the kidney and liver (Figure 5). BHMT is unique to the liver and kidney and together with methionine synthase supports the high liver demand for SAM, with liver estimated to carry out 80% of body total methylation (Stead et al., 2006). Enzymes catalyzing 1C unit generation from serine, glycine, and dimethylglycine, as well as the multifunctional cytosolic enzyme MTHFD1, show abundant expression in liver and kidney, with SHMT1 nearly exclusive to these two organs (Girgis et al., 1998) (Kure et al., 2001; Lang et al., 1994). Although mitochondria isolated from rat liver make formate, indicating a complete 1C pathway (Barlowe and Appling, 1988), rodent kidney and especially liver show very low expression of MTHFD2(L) and MTHFD1L (Bolusani et al., 2011; Shin et al., 2014). This raises the possibility that in these organs, mitochondrial 1C units are not completely oxidized and excreted as formate, but rather used to synthesize serine from glycine, with the serine exported to the cytosol and catabolized there via SHMT1. For supporting homocysteine remethylation, compared to 1C unit transfer via formate, transfer of 1C units from mitochondrial to cytosol via serine reduces the required number of enzymes and conserves cytosolic NADPH. Infusion of [2,3,3-2H]serine in human subjects produces serum methionine and liver-synthesized apoprotein B100 in both the M+1 and M+2 labeling states, indicating at least some cytosolic 1C production from serine (Gregory et al., 2000). Thus, liver and kidney may engage in an alternative 1C shuttle in which SHMT1 makes cytosolic 1C units.
Most Adult Tissues Express Both Mitochondrial and Cytosolic 1C Enzymes
The function of 1C metabolism in other adult non-proliferative tissues is unclear. Their modest demands for 1C units could potentially be met in a variety of ways, including circulating formate. Based on enzyme expression data, however, all adult tissues appear to have the ability to generate 1C units de novo from mitochondrial folate metabolism, transfer formate to the cytosol, synthesize thymidine and purines, and remethylate homocysteine. Excess folate-bound 1C units can be disposed of by the universally expressed enzyme cytosolic 10-formyltetrahydrofolate dehydrogenase (ALDH1L1), while the mitochondrial ALDH1L2 is largely confined to heart, pancreas, brain, and muscle (Krupenko et al., 2010). Beyond these housekeeping functions, little is known about relative fluxes and regulation. Given the roles of glycine and serine in neurotransmission, it seems likely the brain is an active site of folate metabolism. Expression data show high GCS transcripts in brain. SHMT1 is expressed selectively in the hippocampus, including the dentate gyrus and cornu ammonis, regions associated with cell proliferation (Abarinov et al., 2013). A polymorphism in MTHFD1L was recently associated with late-onset Alzheimer’s disease (Naj et al., 2010). Interestingly, elevated circulating homocysteine levels are associated with increased cognitive decline and progression to Alzheimer’s in multiple prospective trials, although both mechanistic understanding of this association and high-quality intervention studies to test the disease-modifying potential of lowering homocysteine are lacking (Smith and Refsum, 2016). In light of the confirmed fetal neurological defects associated with defective folate metabolism, and strong associations between 1C metabolism and cognition, detailed study using isotopic tracers in the brain is warranted.
Dietary Folate Intake and Adult Disease
While folate supplementation is recommended for pregnant women to prevent birth defects, the benefit of supplementation in other adults remains unclear. As noted above, elevated homocysteine levels are correlated with diseases of aging including cardiovascular disease and cognitive decline; however, clinical trials have not shown a benefit in supplemental vitamin B intake for these conditions. Across many studies using both experimental and epidemiological methods, gastrointestinal and in particular colorectal cancer incidence is inversely correlated with folate consumption (Giovannucci, 2002). Similar results have been reported in population-based studies examining breast and pancreatic cancer (Ericson et al., 2007; Larsson et al., 2006). However, in a clinical trial to test if supplemental folic acid would suppress the recurrence of colon polyps in patients recently diagnosed with at least one adenoma, no significant difference was observed between placebo and treatment groups, with a trend toward both more polyps and carcinoma in the folate group (Chae and Yun, 2007). These results suggest that adequate dietary folate over the long term may support genome integrity, but added folate may fuel the growth of existing lesions. The impact of the intake of the 1C donor vitamin choline is less studied, with a recent review not finding any well-proven associations between choline consumption and disease (Leermakers et al., 2015).
1C Metabolism Is Upregulated in Cancer
Targeting 1C metabolism led to the first selective chemotherapeutic agents, the drug aminopterin and its closely related analog, methotrexate, which is still used in treatment of leukemias today. 1C metabolism is also targeted by 5-fluorouracil, a suicide inhibitor of TYMS, and by the new antifolate pemetrexed, approved a decade ago for treatment of mesothelioma and now also a first-line treatment for lung cancer. Unlike methotrexate, which is a near perfect structural analog of folic acid, pemetrexed is more distinct, no longer containing the pterdine ring. Both methotrexate and pemetrexed interact with a wide diversity of folate enzymes. Methotrexate most potently blocks DHFR, while pemetrexed more potently inhibits TYMS and 5-aminoimidazole-4-carboxamide ribonucleotide (AICAR) transformylase (Chattopadhyay et al., 2007; Racanelli et al., 2009). Overexpression of DHFR and TYMS (as well loss of FPGS and RFC function, mentioned above) are mechanisms of acquired resistance to these agents. While pemetrexed is argued to have superior activity in solid tumors, neither fully fulfills the potential of 1C inhibition for anticancer therapy due to toxicities caused by inhibition of 1C metabolism in non-transformed cells, including those of the intestinal epithelium and bone marrow, resulting in gastric complications, anemia, and immune deficiency. While rescue therapy with 5-formyl-THF (folinic acid, marketed as leucovorin) or folate is commonly given in conjunction with these treatments, their therapeutic index remains modest.
Serine Synthesis and Mitochondrial Folate Genes Are Overexpressed in Cancer
New genomics and metabolomics approaches have highlighted distinctive aspects of 1C metabolism in cancer and rekindled interest in targeting this pathway with more selective antifolates (Amelio et al., 2014; Locasale, 2013). Multiple studies have identified increased expression of two 1C pathways in cancer: the de novo serine pathway and the mitochondrial 1C pathway. Expression of these pathways is controlled by common master transcription factors, as well as by genomic amplifications in the first enzyme of the serine pathway, PHGDH.
Consistent with the increased demand for DNA synthesis required for the proliferative cancer phenotype, multiple 1C metabolic enzymes are upregulated in cancer (Mehrmohamadi et al., 2014). Not surprisingly given their importance as drug targets, among all genes, TYMS and DHFR are some of the most overexpressed in cancer. Unexpectedly, mitochondrial folate enzymes are equally upregulated. A recent meta-analysis of tumor gene expression revealed MTHFD2 as the most consistently overexpressed metabolic gene in cancer (Nilsson et al., 2014). The gene for the mitochondrial enzyme directly upstream in the pathway, SHMT2, was identified as recurrently amplified and important for tumor growth (Lee et al., 2014). These two genes are co-overexpressed with TYMS in a large variety of human cancers, linking the DNA synthesis demands of tumors with the mitochondrial folate system. Intriguingly, a different meta-analysis found that across cell lines, expression of these and other mitochondrial folate enzymes was better correlated with sensitivity to methotrexate than expression of the cytosolic enzymes considered to be their primary targets (Vazquez et al., 2013). Total MTHFD2 expression correlates with invasiveness and poor prognosis in breast cancer (Lehtinen et al., 2013; Liu et al., 2014). In acute myeloid leukemia (AML), 1C genes such as MTHFD2 stand out as being among the most strongly repressed by differentiating agents such as all-trans retinoic acid (Pikman et al., 2016). Thus, cancer cells activate the immune/early embryogenesis-specific mitochondrial dehydrogenase, MTHFD2. Selective targeting of this enzyme may restrict antifolate associate toxicities to the immune system and improve therapeutic indices.
Concurrent with these increases in folate metabolic enzyme expression, cancers increase metabolism of 1C sources, namely serine and glycine (Figure 5). High net consumption of serine and glycine is nearly universal across the NCI-60 cancer panel (Jain et al., 2012). Isotope tracer analysis, however, shows that only serine, and not glycine, feeds into the 1C pool in cancer cells grown in culture (Jain et al., 2012; Labuschagne et al., 2014). Serine-derived 1C units are for nucleotide synthesis, as homocysteine remethylation is not observed in cultured cancer cell lines due to a combination of excess methionine and the lack of cobalamin in standard tissue culture media (Maddocks et al., 2016). p53−/− cells cultured without serine are deficient in growth, as are tumors in mice fed serine/glycine-free diets (Maddocks et al., 2013). In addition to serine uptake, de novo serine synthesis is commonly increased. In both breast cancer and melanoma, PHGDH is overexpressed at the protein level, due in part to genomic amplification of the locus (Locasale et al., 2011; Possemato et al., 2011). Patients with breast cancer tumors that overexpress de novo serine biosynthesis enzymes have worse prognoses, but this may reflect that PHGDH overexpression is highest in the hard-to-treat triple-negative breast cancer subtype (Antonov et al., 2014; Kim et al., 2014).
While genomic amplification has been observed with PHGDH, high expression of most folate pathway enzymes is due to aberrant transcriptional regulation. Recently, ATF4 has been identified as an upstream regulator of both de novo serine biosynthesis and mitochondrial folate enzyme transcription. In lung cancer, ATF4 is commonly activated by NRF2 due to mutations in the NRF2 suppressor KEAP1 (DeNicola et al., 2015; Kansanen et al., 2013). ATF4 can also be activated by mTOR, linking the PI3K signaling axis to overexpression of 1C genes (Ben-Sahra et al., 2016).
GCS activity appears to be important in some tumors. In lung cancer tumor-initiating cells, overexpression of GLDC is required for maintenance of the proliferative phenotype and is itself transforming (Zhang et al., 2012). This required the catalytic activity of GLDC and was associated with changes in 1C metabolites, most notably thymidine, but the authors were unable to prove whether glycine itself was supplying carbon or whether decreasing glycine levels promoted tumor growth through other mechanisms. Kim et al. found that glycine cleavage activity was essential for xenograft growth of the glioma cell line LN229 by preventing the buildup of toxic oxidative glycine byproducts (Kim et al., 2015).
Upregulation of mitochondrial 1C metabolism contributes to cancer independently of providing 1C units for biosynthesis, in part by supporting antioxidant defense. SHMT2 plays a protective role against hypoxic stress. In MYC amplified cancer cell lines grown under hypoxia, SHMT2 is induced and shRNA knockdown leads to decreases in NADPH and impaired survival (Ye et al., 2014). In human gliomas, SHMT2 expression is highest in ischemic tumor regions showing “pseudopalisading necrosis” (Kim et al., 2015). The authors proposed that part of the protective effect of SHMT overexpression was a downregulation of aerobic glycolysis, achieved by SHMT-mediated decreases in the PKM2 activators serine, succinyl-5-aminoimidazole-4-carboxamide-1-ribose-5′-phosphate (SAICAR), and fructose 1,6-bisphophate. In a herculean study of the determinants of metastasis in patient-derived xenografts from melanoma, Piskounova et al. identified a role for the mitochondrial enzyme that consumes the 1C unit of 10-formyl-THF to make NADPH (ALDH1L2) (Piskounova et al., 2015). Expression of this enzyme counteracts oxidative stress and thereby promotes tumor metastasis. In contrast to the cytosolic isoform ALDH1L1, expression of the mitochondrial ALDH1L2 is increased in many tumor types (Krupenko et al., 2010).
New Targets for Cancer Therapy
What then about selectively targeting particular 1C enzymes for the treatment of cancer? Cancer genetics suggest two obvious pathways not targeted by existing therapies, de novo serine synthesis and mitochondrial folate metabolism (Tedeschi et al., 2015). While few good inhibitors have been published, early studies employing genetics suggest that targeting these pathways will not be straightforward.
Nilsson et al. generated constitutive shRNA knockdown cells lines targeting PHGDH and assessed their ability to form tumors as xenografts (Nilsson et al., 2012). In KRAS mutant Colon 26 cell lines, knockdown of PHGDH did not impair xenograft implantation or growth. In the PHGDH amplified MDA-MB-468 breast cancer cells, shRNAs targeting PHGDH led to a significant growth defect that was not serine rescuable and impaired xenograft growth (Possemato et al., 2011). This is likely due to off-target effects of the shRNA, as CRISPR deletions have recently been made and are defective in growth only in the absence of serine. Researchers at Novartis reported that for two out of three separate PHGDH-expressing breast cancer cell lines, induced expression of shRNA constructs targeting PHGDH did not alter growth as xenograft, despite all three cell lines being potently inhibited by knockdown in culture (Chen et al., 2013). The development of small-molecule inhibitors of PHGDH should help further illuminate functional requirements for de novo serine biosynthesis in tumors and in the host (Mullarky et al., 2016; Pacold et al., 2016). It is likely that on-target inhibition of PHGDH will impact cancer growth only in serine-poor environments. The tumor microenvironment is low in serine (Kamphorst et al., 2015). Accordingly, specific PHGDH inhibitors that do not alter tumor cell growth in standard tissue culture media may nevertheless effectively slow tumor growth in vivo due in part to their systemic, rather than tumor-localized, activity.
Several groups have now reported results of experiments genetically targeting mitochondrial 1C genes. Nilsson et al. did not observe a change in growth rate when SHMT2 was knocked down in colon cancer xenografts (Nilsson et al., 2012). In contrast, Woo and colleagues reported robust inhibition of hepatocellular carcinoma xenograft growth using inducible shRNA against SHMT2 (Woo et al., 2016). In a murine model of AML, shRNA knockdown of MTHFD2 extended survival, but animals still succumbed to their disease (Pikman et al., 2016). Ducker et al. generated cancer cell lines with complete deletion for MTHFD2 using CRISPR. When grown as xenografts, only a modest decrease in final tumor volume was observed, with reversal of cytosolic 1C flux meeting 1C demand (Ducker et al., 2016). Simultaneous deletion of the cytosolic enzyme SHMT1 blocked xenograft formation, indicating a requirement for tumor-intrinsic 1C production. When evaluated within the context of relevant experiments from cultured cells, these in vivo studies suggest that although multiple 1C enzymes are highly overexpressed in cancer, no single enzyme’s catalytic activity is likely to be essential for tumorigenesis. Thus, rationally targeting this pathway may require inhibiting multiple enzymes. Alternatively, combined modulation of tumor and systemic metabolism may prove more effective than targeted interventions that hit only the tumor without also altering the availability of circulating nutrients.
Outlook
It has been 70 years since Sidney Farber’s seminal experiments with the antifolate aminopterin in pediatric leukemia proved the importance of folate metabolism to cancer cell growth, launching the modern science of chemotherapy. In parallel, the role of folate in 1C metabolic processes that contribute to fetal growth, bacterial infection, and immune activation were uncovered, all leading to significant medical advances. Nevertheless, our ability to selectively target 1C metabolism therapeutically remains crude. Two major obstacles have prevented the development of more specific 1C therapies. The first has been a lack of understanding of the specific ways in which 1C metabolism is altered in disease. New genetics data highlight unique and potentially actionable nodes of this metabolism in cancer, such as MTHFD2. Excitedly, new data on T cell activation may provide similar insight into the immune system, although overlap in tumor and T cell metabolism may limit therapeutic options for cancer. Second, and relatedly, there has been only a belated understanding of the complete scope of folate-supported reactions and products, and how non-1C “by-products” may contribute to pathogenesis. Whereas most existing 1C drugs have been focused on inhibiting the active transformation of folate 1C into DNA bases, such as inhibiting TYMS by 5-fluorouracil, new work highlights the critical roles 1C metabolism plays in supporting antioxidant defense through glutathione and NADPH. Importantly, these by-products have cell-type and context-dependent importance, potentially allowing for greater selectivity than general inhibition of nucleotide or DNA synthesis.
Metabolism in general and specifically folate metabolism have benefitted tremendously from the development of modern genetic and analytical tools. As these techniques have matured, our understanding of the complexity of these pathways has deepened and much is known beyond the “textbook” description. It is now time to leverage this knowledge for translational benefit, in the form of a new generation of effective folate inhibitors and modulators.
Acknowledgments
G.S.D. is supported by a postdoctoral fellowship from the American Cancer Society (PF-15-190-01-TBE). J.D.R. is supported by NIH grants R01CA163591 and P30DK019525 and Stand Up to Cancer. We thank Li Chen, Jonathan Ghergurovich, and all members of the J.D.R. lab for helpful discussions. J.D.R. is a founder and SAB member of Raze Therapeutics, which targets 1C metabolism.
References
- Abarinov EV, Beaudin AE, Field MS, Perry CA, Allen RH, Stabler SP, Stover PJ. Disruption of shmt1 impairs hippocampal neurogenesis and mnemonic function in mice. J Nutr. 2013;143:1028–1035. doi: 10.3945/jn.113.174417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Acuna-Hidalgo R, Schanze D, Kariminejad A, Nordgren A, Kariminejad MH, Conner P, Grigelioniene G, Nilsson D, Nordenskjöld M, Wedell A, et al. Neu-Laxova syndrome is a heterogeneous metabolic disorder caused by defects in enzymes of the L-serine biosynthesis pathway. Am J Hum Genet. 2014;95:285–293. doi: 10.1016/j.ajhg.2014.07.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amelio I, Cutruzzolá F, Antonov A, Agostini M, Melino G. Serine and glycine metabolism in cancer. Trends Biochem Sci. 2014;39:191–198. doi: 10.1016/j.tibs.2014.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- An S, Kumar R, Sheets ED, Benkovic SJ. Reversible compartmentalization of de novo purine biosynthetic complexes in living cells. Science. 2008;320:103–106. doi: 10.1126/science.1152241. [DOI] [PubMed] [Google Scholar]
- Anderson DD, Stover PJ. SHMT1 and SHMT2 are functionally redundant in nuclear de novo thymidylate biosynthesis. PLoS ONE. 2009;4:e5839. doi: 10.1371/journal.pone.0005839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson DD, Quintero CM, Stover PJ. Identification of a de novo thymidylate biosynthesis pathway in mammalian mitochondria. Proc Natl Acad Sci USA. 2011;108:15163–15168. doi: 10.1073/pnas.1103623108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson DD, Eom JY, Stover PJ. Competition between sumoylation and ubiquitination of serine hydroxymethyltransferase 1 determines its nuclear localization and its accumulation in the nucleus. J Biol Chem. 2012a;287:4790–4799. doi: 10.1074/jbc.M111.302174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson DD, Woeller CF, Chiang EP, Shane B, Stover PJ. Serine hydroxymethyltransferase anchors de novo thymidylate synthesis pathway to nuclear lamina for DNA synthesis. J Biol Chem. 2012b;287:7051–7062. doi: 10.1074/jbc.M111.333120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Antonov A, Agostini M, Morello M, Minieri M, Melino G, Amelio I. Bioinformatics analysis of the serine and glycine pathway in cancer cells. Oncotarget. 2014;5:11004–11013. doi: 10.18632/oncotarget.2668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bailey LB, Berry RJ. Folic acid supplementation and the occurrence of congenital heart defects, orofacial clefts, multiple births, and miscarriage. Am J Clin Nutr. 2005;81:1213S–1217S. doi: 10.1093/ajcn/81.5.1213. [DOI] [PubMed] [Google Scholar]
- Bao XR, Ong SE, Goldberger O, Peng J, Sharma R, Thompson DA, Vafai SB, Cox AG, Marutani E, Ichinose F, et al. Mitochondrial dysfunction remodels one-carbon metabolism in human cells. eLife. 2016;5:e10575. doi: 10.7554/eLife.10575. Published online June 16, 2016. http://dx.doi.org/10.7554/eLife.10575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barlowe CK, Appling DR. In vitro evidence for the involvement of mitochondrial folate metabolism in the supply of cytoplasmic one-carbon units. Biofactors. 1988;1:171–176. [PubMed] [Google Scholar]
- Bazzano LA, Reynolds K, Holder KN, He J. Effect of folic acid supplementation on risk of cardiovascular diseases: a meta-analysis of randomized controlled trials. JAMA. 2006;296:2720–2726. doi: 10.1001/jama.296.22.2720. [DOI] [PubMed] [Google Scholar]
- Beaudin AE, Stover PJ. Insights into metabolic mechanisms underlying folate-responsive neural tube defects: a minireview. Birth Defects Res A Clin Mol Teratol. 2009;85:274–284. doi: 10.1002/bdra.20553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beaudin AE, Abarinov EV, Noden DM, Perry CA, Chu S, Stabler SP, Allen RH, Stover PJ. Shmt1 and de novo thymidylate biosynthesis underlie folate-responsive neural tube defects in mice. Am J Clin Nutr. 2011;93:789–798. doi: 10.3945/ajcn.110.002766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ben-Sahra I, Hoxhaj G, Ricoult SJ, Asara JM, Manning BD. mTORC1 induces purine synthesis through control of the mitochondrial tetra-hydrofolate cycle. Science. 2016;351:728–733. doi: 10.1126/science.aad0489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Binzak BA, Wevers RA, Moolenaar SH, Lee YM, Hwu WL, Poggi-Bach J, Engelke UFH, Hoard HM, Vockley JG, Vockley J. Cloning of dimethylglycine dehydrogenase and a new human inborn error of metabolism, dimethylglycine dehydrogenase deficiency. Am J Hum Genet. 2001;68:839–847. doi: 10.1086/319520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Birsoy K, Wang T, Chen WW, Freinkman E, Abu-Remaileh M, Sabatini DM. An essential role of the mitochondrial electron transport chain in cell proliferation is to enable aspartate synthesis. Cell. 2015;162:540–551. doi: 10.1016/j.cell.2015.07.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolusani S, Young BA, Cole NA, Tibbetts AS, Momb J, Bryant JD, Solmonson A, Appling DR. Mammalian MTHFD2L encodes a mitochondrial methylenetetrahydrofolate dehydrogenase isozyme expressed in adult tissues. J Biol Chem. 2011;286:5166–5174. doi: 10.1074/jbc.M110.196840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brosnan JT. The 1986 Borden award lecture. The role of the kidney in amino acid metabolism and nutrition. Can J Physiol Pharmacol. 1987;65:2355–2362. doi: 10.1139/y87-373. [DOI] [PubMed] [Google Scholar]
- Brosnan JT. Interorgan amino acid transport and its regulation. J Nutr. 2003;133(6, Suppl 1):2068S–2072S. doi: 10.1093/jn/133.6.2068S. [DOI] [PubMed] [Google Scholar]
- Brosnan ME, MacMillan L, Stevens JR, Brosnan JT. Division of labour: how does folate metabolism partition between one-carbon metabolism and amino acid oxidation? Biochem J. 2015;472:135–146. doi: 10.1042/BJ20150837. [DOI] [PubMed] [Google Scholar]
- Casas JP, Bautista LE, Smeeth L, Sharma P, Hingorani AD. Homocysteine and stroke: evidence on a causal link from mendelian randomisation. Lancet. 2005;365:224–232. doi: 10.1016/S0140-6736(05)17742-3. [DOI] [PubMed] [Google Scholar]
- Chae YK, Yun JH. Folic acid and prevention of colorectal adenomas. JAMA. 2007;298:1397. doi: 10.1001/jama.298.12.1397-a. author reply 1397. [DOI] [PubMed] [Google Scholar]
- Chasin LA, Feldman A, Konstam M, Urlaub G. Reversion of a Chinese hamster cell auxotrophic mutant. Proc Natl Acad Sci USA. 1974;71:718–722. doi: 10.1073/pnas.71.3.718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chattopadhyay S, Moran RG, Goldman ID. Pemetrexed: biochemical and cellular pharmacology, mechanisms, and clinical applications. Mol Cancer Ther. 2007;6:404–417. doi: 10.1158/1535-7163.MCT-06-0343. [DOI] [PubMed] [Google Scholar]
- Chen J, Chung F, Yang G, Pu M, Gao H, Jiang W, Yin H, Capka V, Kasibhatla S, Laffitte B, et al. Phosphoglycerate dehydrogenase is dispensable for breast tumor maintenance and growth. Oncotarget. 2013;4:2502–2511. doi: 10.18632/oncotarget.1540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Copp AJ, Adzick NS, Chitty LS, Fletcher JM, Holmbeck GN, Shaw GM. Spina bifida. Nat Rev Dis Primers. 2015;1:15007. doi: 10.1038/nrdp.2015.7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corominas-Faja B, Quirantes-Piné R, Oliveras-Ferraros C, Vazquez-Martin A, Cufí S, Martin-Castillo B, Micol V, Joven J, Segura-Carretero A, Menendez JAJ. Metabolomic fingerprint reveals that metformin impairs one-carbon metabolism in a manner similar to the antifolate class of chemotherapy drugs. Aging (Albany, NY) 2012;4:480–498. doi: 10.18632/aging.100472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dai Y, Zhu J, Meng D, Yu C, Li Y. Association of homocysteine level with biopsy-proven non-alcoholic fatty liver disease: a meta-analysis. J Clin Biochem Nutr. 2016;58:76–83. doi: 10.3164/jcbn.15-54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dang L, White DW, Gross S, Bennett BD, Bittinger MA, Driggers EM, Fantin VR, Jang HG, Jin S, Keenan MC, et al. Cancer-associated IDH1 mutations produce 2-hydroxyglutarate. Nature. 2009;462:739–744. doi: 10.1038/nature08617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis SR, Stacpoole PW, Williamson J, Kick LS, Quinlivan EP, Coats BS, Shane B, Bailey LB, Gregory JF., 3rd Tracer-derived total and folate-dependent homocysteine remethylation and synthesis rates in humans indicate that serine is the main one-carbon donor. Am J Physiol Endocrinol Metab. 2004;286:E272–E279. doi: 10.1152/ajpendo.00351.2003. [DOI] [PubMed] [Google Scholar]
- DeNicola GM, Chen PH, Mullarky E, Sudderth JA, Hu Z, Wu D, Tang H, Xie Y, Asara JM, Huffman KE, et al. NRF2 regulates serine biosynthesis in non-small cell lung cancer. Nat Genet. 2015;47:1475–1481. doi: 10.1038/ng.3421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di Pietro E, Sirois J, Tremblay ML, MacKenzie RE. Mitochondrial NAD-dependent methylenetetrahydrofolate dehydrogenase-methenyltetrahydrofolate cyclohydrolase is essential for embryonic development. Mol Cell Biol. 2002;22:4158–4166. doi: 10.1128/MCB.22.12.4158-4166.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ducker GS, Chen L, Morscher RJ, Ghergurovich JM, Esposito M, Teng X, Kang Y, Rabinowitz JD. Reversal of cytosolic one-carbon flux compensates for loss of the mitochondrial folate pathway. Cell Metab. 2016;23:1140–1153. doi: 10.1016/j.cmet.2016.04.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edgar AJ. The human L-threonine 3-dehydrogenase gene is an expressed pseudogene. BMC Genet. 2002;3:18. doi: 10.1186/1471-2156-3-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ericson U, Sonestedt E, Gullberg B, Olsson H, Wirfält E. High folate intake is associated with lower breast cancer incidence in postmenopausal women in the Malmö Diet and Cancer cohort. Am J Clin Nutr. 2007;86:434–443. doi: 10.1093/ajcn/86.2.434. [DOI] [PubMed] [Google Scholar]
- Eriksson S, Prigge JR, Talago EA, Arnér ESJ, Schmidt EE. Dietary methionine can sustain cytosolic redox homeostasis in the mouse liver. Nat Commun. 2015;6:6479. doi: 10.1038/ncomms7479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan J, Ye J, Kamphorst JJ, Shlomi T, Thompson CB, Rabinowitz JD. Quantitative flux analysis reveals folate-dependent NADPH production. Nature. 2014;510:298–302. doi: 10.1038/nature13236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Felig P. Amino acid metabolism in man. Annu Rev Biochem. 1975;44:933–955. doi: 10.1146/annurev.bi.44.070175.004441. [DOI] [PubMed] [Google Scholar]
- Field MS, Kamynina E, Agunloye OC, Liebenthal RP, Lamarre SG, Brosnan ME, Brosnan JT, Stover PJ. Nuclear enrichment of folate cofactors and methylenetetrahydrofolate dehydrogenase 1 (MTHFD1) protect de novo thymidylate biosynthesis during folate deficiency. J Biol Chem. 2014;289:29642–29650. doi: 10.1074/jbc.M114.599589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Field MS, Kamynina E, Stover PJ. MTHFD1 regulates nuclear de novo thymidylate biosynthesis and genome stability. Biochimie. 2016;126:27–30. doi: 10.1016/j.biochi.2016.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finkelstein JD. Methionine metabolism in mammals. J Nutr Biochem. 1990;1:228–237. doi: 10.1016/0955-2863(90)90070-2. [DOI] [PubMed] [Google Scholar]
- Freemantle SJ, Taylor SM, Krystal G, Moran RG. Upstream organization of and multiple transcripts from the human folylpoly-gamma-glutamate synthetase gene. J Biol Chem. 1995;270:9579–9584. doi: 10.1074/jbc.270.16.9579. [DOI] [PubMed] [Google Scholar]
- French JB, Jones SA, Deng H, Pedley AM, Kim D, Chan CY, Hu H, Pugh RJ, Zhao H, Zhang Y, et al. Spatial colocalization and functional link of purinosomes with mitochondria. Science. 2016;351:733–737. doi: 10.1126/science.aac6054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- García-Martínez LF, Appling DR. Characterization of the folate-dependent mitochondrial oxidation of carbon 3 of serine. Biochemistry. 1993;32:4671–4676. doi: 10.1021/bi00068a027. [DOI] [PubMed] [Google Scholar]
- Giovannucci E. Epidemiologic studies of folate and colorectal neoplasia: a review. J Nutr. 2002;132(8, Suppl):2350S–2355S. doi: 10.1093/jn/132.8.2350S. [DOI] [PubMed] [Google Scholar]
- Girgis S, Nasrallah IM, Suh JR, Oppenheim E, Zanetti KA, Mastri MG, Stover PJ. Molecular cloning, characterization and alternative splicing of the human cytoplasmic serine hydroxymethyltransferase gene. Gene. 1998;210:315–324. doi: 10.1016/s0378-1119(98)00085-7. [DOI] [PubMed] [Google Scholar]
- Goyette P, Frosst P, Rosenblatt DS, Rozen R. Seven novel mutations in the methylenetetrahydrofolate reductase gene and genotype/phenotype correlations in severe methylenetetrahydrofolate reductase deficiency. Am J Hum Genet. 1995;56:1052–1059. [PMC free article] [PubMed] [Google Scholar]
- Gregory JF, 3rd, Cuskelly GJ, Shane B, Toth JP, Baumgartner TG, Stacpoole PW. Primed, constant infusion with [2H3]serine allows in vivo kinetic measurement of serine turnover, homocysteine remethylation, and transsulfuration processes in human one-carbon metabolism. Am J Clin Nutr. 2000;72:1535–1541. doi: 10.1093/ajcn/72.6.1535. [DOI] [PubMed] [Google Scholar]
- Griss T, Vincent EE, Egnatchik R, Chen J, Ma EH, Faubert B, Viollet B, DeBerardinis RJ, Jones RG. Metformin antagonizes cancer cell proliferation by suppressing mitochondrial-dependent biosynthesis. PLoS Biol. 2015;13:e1002309–e1002323. doi: 10.1371/journal.pbio.1002309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herbig K, Chiang EP, Lee LR, Hills J, Shane B, Stover PJ. Cytoplasmic serine hydroxymethyltransferase mediates competition between folate-dependent deoxyribonucleotide and S-adenosylmethionine biosyntheses. J Biol Chem. 2002;277:38381–38389. doi: 10.1074/jbc.M205000200. [DOI] [PubMed] [Google Scholar]
- Hou Z, Matherly LH. Biology of the Major Facilitative Folate Transporters SLC19A1 and SLC46A1. Elsevier Inc; 2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jain M, Nilsson R, Sharma S, Madhusudhan N, Kitami T, Souza AL, Kafri R, Kirschner MW, Clish CB, Mootha VK. Metabolite profiling identifies a key role for glycine in rapid cancer cell proliferation. Science. 2012;336:1040–1044. doi: 10.1126/science.1218595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamphorst JJ, Nofal M, Commisso C, Hackett SR, Lu W, Grabocka E, Vander Heiden MG, Miller G, Drebin JA, Bar-Sagi D, et al. Human pancreatic cancer tumors are nutrient poor and tumor cells actively scavenge extracellular protein. Cancer Res. 2015;75:544–553. doi: 10.1158/0008-5472.CAN-14-2211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kansanen E, Kuosmanen SM, Leinonen H, Levonen AL. The Keap1-Nrf2 pathway: Mechanisms of activation and dysregulation in cancer. Redox Biol. 2013;1:45–49. doi: 10.1016/j.redox.2012.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelly RS, Vander Heiden MG, Giovannucci E, Mucci LA. Metabolomic Biomarkers of prostate cancer: prediction, diagnosis, progression, prognosis, and recurrence. Cancer Epidemiol Biomarkers Prev. 2016;25:887–906. doi: 10.1158/1055-9965.EPI-15-1223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kikuchi G, Motokawa Y, Yoshida T, Hiraga K. Glycine cleavage system: reaction mechanism, physiological significance, and hyperglycinemia. Proc Jpn Acad, Ser B, Phys Biol Sci. 2008;84:246–263. doi: 10.2183/pjab/84.246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim DW, Huang T, Schirch D, Schirch V. Properties of tetra-hydropteroylpentaglutamate bound to 10-formyltetrahydrofolate dehydrogenase. Biochemistry. 1996;35:15772–15783. doi: 10.1021/bi9619684. [DOI] [PubMed] [Google Scholar]
- Kim SK, Jung WH, Koo JS. Differential expression of enzymes associated with serine/glycine metabolism in different breast cancer subtypes. PLoS ONE. 2014;9:e101004–e101014. doi: 10.1371/journal.pone.0101004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim D, Fiske BP, Birsoy K, Freinkman E, Kami K, Possemato RL, Chudnovsky Y, Pacold ME, Chen WW, Cantor JR, et al. SHMT2 drives glioma cell survival in ischaemia but imposes a dependence on glycine clearance. Nature. 2015;520:363–367. doi: 10.1038/nature14363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kluijtmans LA, Boers GH, Stevens EM, Renier WO, Kraus JP, Trijbels FJ, van den Heuvel LP, Blom HJ. Defective cystathionine beta-synthase regulation by S-adenosylmethionine in a partially pyridoxine responsive homocystinuria patient. J Clin Invest. 1996;98:285–289. doi: 10.1172/JCI118791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kochi H, Kikuchi G. Mechanism of the reversible glycine cleavage reaction in Arthrobacter globiformis. I Purification and function of protein components required for the reaction. J Biochem. 1974;75:1113–1127. doi: 10.1093/oxfordjournals.jbchem.a130483. [DOI] [PubMed] [Google Scholar]
- Krupenko NI, Dubard ME, Strickland KC, Moxley KM, Oleinik NV, Krupenko SA. ALDH1L2 is the mitochondrial homolog of 10-for-myltetrahydrofolate dehydrogenase. J Biol Chem. 2010;285:23056–23063. doi: 10.1074/jbc.M110.128843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kure S, Kojima K, Kudo T, Kanno K, Aoki Y, Suzuki Y, Shinka T, Sakata Y, Narisawa K, Matsubara Y. Chromosomal localization, structure, single-nucleotide polymorphisms, and expression of the human H-protein gene of the glycine cleavage system (GCSH), a candidate gene for nonketotic hyperglycinemia. J Hum Genet. 2001;46:378–384. doi: 10.1007/s100380170057. [DOI] [PubMed] [Google Scholar]
- Kure S, Kato K, Dinopoulos A, Gail C, DeGrauw TJ, Christodoulou J, Bzduch V, Kalmanchey R, Fekete G, Trojovsky A, et al. Comprehensive mutation analysis of GLDC, AMT, and GCSH in nonketotic hyperglycinemia. Hum Mutat. 2006;27:343–352. doi: 10.1002/humu.20293. [DOI] [PubMed] [Google Scholar]
- Kutzbach C, Stokstad EL. Mammalian methylenetetrahydrofolate reductase. Partial purification, properties, and inhibition by S-adenosylmethionine. Biochim Biophys Acta. 1971;250:459–477. doi: 10.1016/0005-2744(71)90247-6. [DOI] [PubMed] [Google Scholar]
- Labuschagne CF, van den Broek NJF, Mackay GM, Vousden KH, Maddocks ODK. Serine, but not glycine, supports one-carbon metabolism and proliferation of cancer cells. Cell Rep. 2014;7:1248–1258. doi: 10.1016/j.celrep.2014.04.045. [DOI] [PubMed] [Google Scholar]
- Lamarre SG, Morrow G, Macmillan L, Brosnan ME, Brosnan JT. Formate: an essential metabolite, a biomarker, or more? Clin Chem Lab Med. 2013;51:571–578. doi: 10.1515/cclm-2012-0552. [DOI] [PubMed] [Google Scholar]
- Lamarre SG, MacMillan L, Morrow GP, Randell E, Pongnopparat T, Brosnan ME, Brosnan JT. An isotope-dilution, GC-MS assay for formate and its application to human and animal metabolism. Amino Acids. 2014;46:1885–1891. doi: 10.1007/s00726-014-1738-7. [DOI] [PubMed] [Google Scholar]
- Lamers Y, Williamson J, Gilbert LR, Stacpoole PW, Gregory JF., 3rd Glycine turnover and decarboxylation rate quantified in healthy men and women using primed, constant infusions of [1,2-(13)C2]glycine and [(2)H3]leucine. J Nutr. 2007;137:2647–2652. doi: 10.1093/jn/137.12.2647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lang H, Minaian K, Freudenberg N, Hoffmann R, Brandsch R. Tissue specificity of rat mitochondrial dimethylglycine dehydrogenase expression. Biochem J. 1994;299:393–398. doi: 10.1042/bj2990393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larsson SC, Håkansson N, Giovannucci E, Wolk A. Folate intake and pancreatic cancer incidence: a prospective study of Swedish women and men. J Natl Cancer Inst. 2006;98:407–413. doi: 10.1093/jnci/djj094. [DOI] [PubMed] [Google Scholar]
- Lawrence SA, Titus SA, Ferguson J, Heineman AL, Taylor SM, Moran RG. Mammalian mitochondrial and cytosolic folylpolyglutamate synthetase maintain the subcellular compartmentalization of folates. J Biol Chem. 2014;289:29386–29396. doi: 10.1074/jbc.M114.593244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee GY, Haverty PM, Li L, Kljavin NM, Bourgon R, Lee J, Stern H, Modrusan Z, Seshagiri S, Zhang Z, et al. Comparative oncogenomics identifies PSMB4 and SHMT2 as potential cancer driver genes. Cancer Res. 2014;74:3114–3126. doi: 10.1158/0008-5472.CAN-13-2683. [DOI] [PubMed] [Google Scholar]
- Leermakers ETM, Moreira EM, Kiefte-de Jong JC, Darweesh SKL, Visser T, Voortman T, Bautista PK, Chowdhury R, Gorman D, Bramer WM, et al. Effects of choline on health across the life course: a systematic review. Nutr Rev. 2015;73:500–522. doi: 10.1093/nutrit/nuv010. [DOI] [PubMed] [Google Scholar]
- Lehtinen L, Ketola K, Mäkelä R, Mpindi JP, Viitala M, Kallioniemi O, Iljin K. High-throughput RNAi screening for novel modulators of vimentin expression identifies MTHFD2 as a regulator of breast cancer cell migration and invasion. Oncotarget. 2013;4:48–63. doi: 10.18632/oncotarget.756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leung KY, De Castro SCP, Cabreiro F, Gustavsson P, Copp AJ, Greene NDE. Folate metabolite profiling of different cell types and embryos suggests variation in folate one-carbon metabolism, including developmental changes in human embryonic brain. Mol Cell Biochem. 2013;378:229–236. doi: 10.1007/s11010-013-1613-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis CA, Parker SJ, Fiske BP, McCloskey D, Gui DY, Green CR, Vokes NI, Feist AM, Vander Heiden MG, Metallo CM. Tracing compartmentalized NADPH metabolism in the cytosol and mitochondria of mammalian cells. Mol Cell. 2014;55:253–263. doi: 10.1016/j.molcel.2014.05.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liani E, Rothem L, Bunni MA, Smith CA, Jansen G, Assaraf YG. Loss of folylpoly-gamma-glutamate synthetase activity is a dominant mechanism of resistance to polyglutamylation-dependent novel antifolates in multiple human leukemia sublines. Int J Cancer. 2003;103:587–599. doi: 10.1002/ijc.10829. [DOI] [PubMed] [Google Scholar]
- Lin BF, Huang RF, Shane B. Regulation of folate and one-carbon metabolism in mammalian cells. III Role of mitochondrial folylpoly-gamma-glutamate synthetase. J Biol Chem. 1993;268:21674–21679. [PubMed] [Google Scholar]
- Liu SP, Li YS, Chen YJ, Chiang EP, Li AFY, Lee YH, Tsai TF, Hsiao M, Huang SF, Chen YM. Glycine N-methyltransferase−/− mice develop chronic hepatitis and glycogen storage disease in the liver. Hepatology. 2007;46:1413–1425. doi: 10.1002/hep.21863. [DOI] [PubMed] [Google Scholar]
- Liu F, Liu Y, He C, Tao L, He X, Song H, Zhang G. Increased MTHFD2 expression is associated with poor prognosis in breast cancer. Tumour Biol. 2014;35:8685–8690. doi: 10.1007/s13277-014-2111-x. [DOI] [PubMed] [Google Scholar]
- Locasale JW. Serine, glycine and one-carbon units: cancer metabolism in full circle. Nat Rev Cancer. 2013;13:572–583. doi: 10.1038/nrc3557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Locasale JW, Grassian AR, Melman T, Lyssiotis CA, Mattaini KR, Bass AJ, Heffron G, Metallo CM, Muranen T, Sharfi H, et al. Phosphoglycerate dehydrogenase diverts glycolytic flux and contributes to oncogenesis. Nat Genet. 2011;43:869–874. doi: 10.1038/ng.890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lonn E, Yusuf S, Arnold MJ, Sheridan P, Pogue J, Micks M, McQueen MJ, Probstfield J, Fodor G, Held C, Genest J, Jr Heart Outcomes Prevention Evaluation (HOPE) 2 Investigators. Homocysteine lowering with folic acid and B vitamins in vascular disease. N Engl J Med. 2006;354:1567–1577. doi: 10.1056/NEJMoa060900. [DOI] [PubMed] [Google Scholar]
- Lowe KE, Osborne CB, Lin BF, Kim JS, Hsu JC, Shane B. Regulation of folate and one-carbon metabolism in mammalian cells. II Effect of folylpoly-gamma-glutamate synthetase substrate specificity and level on folate metabolism and folylpoly-gamma-glutamate specificity of metabolic cycles of one-carbon metabolism. J Biol Chem. 1993;268:21665–21673. [PubMed] [Google Scholar]
- Lu W, Kwon YK, Rabinowitz JD. Isotope ratio-based profiling of microbial folates. J Am Soc Mass Spectrom. 2007;18:898–909. doi: 10.1016/j.jasms.2007.01.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luka Z, Capdevila A, Mato JM, Wagner C. A glycine N-methyltransferase knockout mouse model for humans with deficiency of this enzyme. Transgenic Res. 2006;15:393–397. doi: 10.1007/s11248-006-0008-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luka Z, Mudd SH, Wagner C. Glycine N-methyltransferase and regulation of S-adenosylmethionine levels. J Biol Chem. 2009;284:22507–22511. doi: 10.1074/jbc.R109.019273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacFarlane AJ, Liu X, Perry CA, Flodby P, Allen RH, Stabler SP, Stover PJ. Cytoplasmic serine hydroxymethyltransferase regulates the metabolic partitioning of methylenetetrahydrofolate but is not essential in mice. J Biol Chem. 2008;283:25846–25853. doi: 10.1074/jbc.M802671200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacFarlane AJ, Perry CA, Girnary HH, Gao D, Allen RH, Stabler SP, Shane B, Stover PJ. Mthfd1 is an essential gene in mice and alters biomarkers of impaired one-carbon metabolism. J Biol Chem. 2009;284:1533–1539. doi: 10.1074/jbc.M808281200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacFarlane AJ, Anderson DD, Flodby P, Perry CA, Allen RH, Stabler SP, Stover PJ. Nuclear localization of de novo thymidylate biosynthesis pathway is required to prevent uracil accumulation in DNA. J Biol Chem. 2011;286:44015–44022. doi: 10.1074/jbc.M111.307629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maddocks ODK, Berkers CR, Mason SM, Zheng L, Blyth K, Gottlieb E, Vousden KH. Serine starvation induces stress and p53-dependent metabolic remodelling in cancer cells. Nature. 2013;493:542–546. doi: 10.1038/nature11743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maddocks ODK, Labuschagne CF, Adams PD, Vousden KH. Serine metabolism supports the methionine cycle and DNA/RNA methylation through de novo ATP synthesis in cancer cells. Mol Cell. 2016;61:210–221. doi: 10.1016/j.molcel.2015.12.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martínez-Chantar ML, Vázquez-Chantada M, Ariz U, Martínez N, Varela M, Luka Z, Capdevila A, Rodríguez J, Aransay AM, Matthiesen R, et al. Loss of the glycine N-methyltransferase gene leads to steatosis and hepatocellular carcinoma in mice. Hepatology. 2008;47:1191–1199. doi: 10.1002/hep.22159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martiniova L, Field MS, Finkelstein JL, Perry CA, Stover PJ. Maternal dietary uridine causes, and deoxyuridine prevents, neural tube closure defects in a mouse model of folate-responsive neural tube defects. Am J Clin Nutr. 2015;101:860–869. doi: 10.3945/ajcn.114.097279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mehrmohamadi M, Liu X, Shestov AA, Locasale JW. Characterization of the usage of the serine metabolic network in human cancer. Cell Rep. 2014;9:1507–1519. doi: 10.1016/j.celrep.2014.10.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mentch SJ, Mehrmohamadi M, Huang L, Liu X, Gupta D, Mattocks D, Gómez Padilla P, Ables G, Bamman MM, Thalacker-Mercer AE, et al. Histone methylation dynamics and gene regulation occur through the sensing of one-carbon metabolism. Cell Metab. 2015;22:861–873. doi: 10.1016/j.cmet.2015.08.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Momb J, Lewandowski JP, Bryant JD, Fitch R, Surman DR, Vokes SA, Appling DR. Deletion of Mthfd1l causes embryonic lethality and neural tube and craniofacial defects in mice. Proc Natl Acad Sci USA. 2013;110:549–554. doi: 10.1073/pnas.1211199110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mudd SH, Brosnan JT, Brosnan ME, Jacobs RL, Stabler SP, Allen RH, Vance DE, Wagner C. Methyl balance and transmethylation fluxes in humans. Am J Clin Nutr. 2007;85:19–25. doi: 10.1093/ajcn/85.1.19. [DOI] [PubMed] [Google Scholar]
- Mullarky E, Lucki NC, Beheshti Zavareh R, Anglin JL, Gomes AP, Nicolay BN, Wong JCY, Christen S, Takahashi H, Singh PK, et al. Identification of a small molecule inhibitor of 3-phosphoglycerate dehydrogenase to target serine biosynthesis in cancers. Proc Natl Acad Sci USA. 2016;113:1778–1783. doi: 10.1073/pnas.1521548113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naj AC, Beecham GW, Martin ER, Gallins PJ, Powell EH, Konidari I, Whitehead PL, Cai G, Haroutunian V, Scott WK, et al. Dementia revealed: novel chromosome 6 locus for late-onset Alzheimer disease provides genetic evidence for folate-pathway abnormalities. PLoS Genet. 2010;6:e1001130. doi: 10.1371/journal.pgen.1001130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Narisawa A, Komatsuzaki S, Kikuchi A, Niihori T, Aoki Y, Fujiwara K, Tanemura M, Hata A, Suzuki Y, Relton CL, et al. Mutations in genes encoding the glycine cleavage system predispose to neural tube defects in mice and humans. Hum Mol Genet. 2012;21:1496–1503. doi: 10.1093/hmg/ddr585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nikkanen J, Forsström S, Euro L, Paetau I, Kohnz RA, Wang L, Chilov D, Viinamäki J, Roivainen A, Marjamäki P, et al. Mitochondrial DNA replication defects disturb cellular dNTP pools and remodel one-carbon metabolism. Cell Metab. 2016;23:635–648. doi: 10.1016/j.cmet.2016.01.019. [DOI] [PubMed] [Google Scholar]
- Nilsson LM, Forshell TZ, Rimpi S, Kreutzer C, Pretsch W, Bornkamm GW, Nilsson JA. Mouse genetics suggests cell-context dependency for Myc-regulated metabolic enzymes during tumorigenesis. PLoS Genet. 2012;8:e1002573. doi: 10.1371/journal.pgen.1002573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nilsson R, Jain M, Madhusudhan N, Sheppard NG, Strittmatter L, Kampf C, Huang J, Asplund A, Mootha VK. Metabolic enzyme expression highlights a key role for MTHFD2 and the mitochondrial folate pathway in cancer. Nat Commun. 2014;5:3128. doi: 10.1038/ncomms4128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pacana T, Cazanave S, Verdianelli A, Patel V, Min HK, Mirshahi F, Quinlivan E, Sanyal AJ. Dysregulated hepatic methionine metabolism drives homocysteine elevation in diet-induced nonalcoholic fatty liver disease. PLoS ONE. 2015;10:e0136822. doi: 10.1371/journal.pone.0136822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pacold ME, Brimacombe KR, Chan SH, Rohde JM, Lewis CA, Swier LJYM, Possemato R, Chen WW, Sullivan LB, Fiske BP, et al. A PHGDH inhibitor reveals coordination of serine synthesis and one-carbon unit fate. Nat Chem Biol. 2016;12:452–458. doi: 10.1038/nchembio.2070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pai YJ, Leung KY, Savery D, Hutchin T, Prunty H, Heales S, Brosnan ME, Brosnan JT, Copp AJ, Greene NDE. Glycine decarboxylase deficiency causes neural tube defects and features of non-ketotic hyperglycinemia in mice. Nat Commun. 2015;6:6388. doi: 10.1038/ncomms7388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pajares MA, Pérez-Sala D. Betaine homocysteine S-methyltransferase: just a regulator of homocysteine metabolism? Cell Mol Life Sci. 2006;63:2792–2803. doi: 10.1007/s00018-006-6249-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patel H, Pietro ED, MacKenzie RE. Mammalian fibroblasts lacking mitochondrial NAD+-dependent methylenetetrahydrofolate dehydrogenase-cyclohydrolase are glycine auxotrophs. J Biol Chem. 2003;278:19436–19441. doi: 10.1074/jbc.M301718200. [DOI] [PubMed] [Google Scholar]
- Pike ST, Rajendra R, Artzt K, Appling DR. Mitochondrial C1-tetrahydrofolate synthase (MTHFD1L) supports the flow of mitochondrial one-carbon units into the methyl cycle in embryos. J Biol Chem. 2010;285:4612–4620. doi: 10.1074/jbc.M109.079855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pikman Y, Puissant A, Alexe G, Furman A, Chen LM, Frumm SM, Ross L, Fenouille N, Bassil CF, Lewis CA, et al. Targeting MTHFD2 in acute myeloid leukemia. J Exp Med. 2016;213:1285–1306. doi: 10.1084/jem.20151574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pincus T, Yazici Y, Sokka T, Aletaha D, Smolen JS. Methotrexate as the “anchor drug” for the treatment of early rheumatoid arthritis. Clin Exp Rheumatol. 2003;21(5, Suppl 31):S179–S185. [PubMed] [Google Scholar]
- Piskounova E, Agathocleous M, Murphy MM, Hu Z, Huddlestun SE, Zhao Z, Leitch AM, Johnson TM, DeBerardinis RJ, Morrison SJ. Oxidative stress inhibits distant metastasis by human melanoma cells. Nature. 2015;527:186–191. doi: 10.1038/nature15726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Possemato R, Marks KM, Shaul YD, Pacold ME, Kim D, Birsoy K, Sethumadhavan S, Woo HK, Jang HG, Jha AK, et al. Functional genomics reveal that the serine synthesis pathway is essential in breast cancer. Nature. 2011;476:346–350. doi: 10.1038/nature10350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prem veer Reddy G, Pardee AB. Multienzyme complex for metabolic channeling in mammalian DNA replication. Proc Natl Acad Sci USA. 1980;77:3312–3316. doi: 10.1073/pnas.77.6.3312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Racanelli AC, Rothbart SB, Heyer CL, Moran RG. Therapeutics by cytotoxic metabolite accumulation: pemetrexed causes ZMP accumulation, AMPK activation, and mammalian target of rapamycin inhibition. Cancer Res. 2009;69:5467–5474. doi: 10.1158/0008-5472.CAN-08-4979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ron-Harel N, Santos D, Ghergurovich JM, Sage PT, Reddy A, Lovitch SB, Dephoure N, Satterstrom FK, Sheffer M, Spinelli JB, et al. Mitochondrial biogenesis and proteome remodeling promote one-carbon metabolism for T cell activation. Cell Metab. 2016;24:104–117. doi: 10.1016/j.cmet.2016.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schirch V, Szebenyi DM. Serine hydroxymethyltransferase revisited. Curr Opin Chem Biol. 2005;9:482–487. doi: 10.1016/j.cbpa.2005.08.017. [DOI] [PubMed] [Google Scholar]
- Schwahn BC, Laryea MD, Chen Z, Melnyk S, Pogribny I, Garrow T, James SJ, Rozen R. Betaine rescue of an animal model with methylenetetrahydrofolate reductase deficiency. Biochem J. 2004;382:831–840. doi: 10.1042/BJ20030822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shin M, Bryant JD, Momb J, Appling DR. Mitochondrial MTHFD2L is a dual redox cofactor-specific methylenetetrahydrofolate dehydrogenase/methenyltetrahydrofolate cyclohydrolase expressed in both adult and embryonic tissues. J Biol Chem. 2014;289:15507–15517. doi: 10.1074/jbc.M114.555573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shyh-Chang N, Locasale JW, Lyssiotis CA, Zheng Y, Teo RY, Ratanasirintrawoot S, Zhang J, Onder T, Unternaehrer JJ, Zhu H, et al. Influence of threonine metabolism on S-adenosylmethionine and histone methylation. Science. 2013;339:222–226. doi: 10.1126/science.1226603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith AD, Refsum H. Homocysteine, B vitamins, and cognitive impairment. Annu Rev Nutr. 2016;36:211–239. doi: 10.1146/annurev-nutr-071715-050947. [DOI] [PubMed] [Google Scholar]
- Solans A, Estivill X, de la Luna S. Cloning and characterization of human FTCD on 21q22.3, a candidate gene for glutamate formiminotransferase deficiency. Cytogenet Cell Genet. 2000;88:43–49. doi: 10.1159/000015483. [DOI] [PubMed] [Google Scholar]
- Sreekumar A, Poisson LM, Rajendiran TM, Khan AP, Cao Q, Yu J, Laxman B, Mehra R, Lonigro RJ, Li Y, et al. Metabolomic profiles delineate potential role for sarcosine in prostate cancer progression. Nature. 2009;457:910–914. doi: 10.1038/nature07762. [DOI] [PMC free article] [PubMed] [Google Scholar] [Research Misconduct Found]
- Stead LM, Brosnan JT, Brosnan ME, Vance DE, Jacobs RL. Is it time to reevaluate methyl balance in humans? Am J Clin Nutr. 2006;83:5–10. doi: 10.1093/ajcn/83.1.5. [DOI] [PubMed] [Google Scholar]
- Stover P, Schirch V. 5-Formyltetrahydrofolate polyglutamates are slow tight binding inhibitors of serine hydroxymethyltransferase. J Biol Chem. 1991;266:1543–1550. [PubMed] [Google Scholar]
- Su X, Wellen KE, Rabinowitz JD. Metabolic control of methylation and acetylation. Curr Opin Chem Biol. 2016;30:52–60. doi: 10.1016/j.cbpa.2015.10.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sullivan LB, Gui DY, Hosios AM, Bush LN, Freinkman E, Vander Heiden MG. Supporting aspartate biosynthesis is an essential function of respiration in proliferating cells. Cell. 2015;162:552–563. doi: 10.1016/j.cell.2015.07.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sweeting JN, Siu M, McCallum GP, Miller L, Wells PG. Species differences in methanol and formic acid pharmacokinetics in mice, rabbits and primates. Toxicol Appl Pharmacol. 2010;247:28–35. doi: 10.1016/j.taap.2010.05.009. [DOI] [PubMed] [Google Scholar]
- Tedeschi PM, Vazquez A, Kerrigan JE, Bertino JR. Mitochondrial methylenetetrahydrofolate dehydrogenase (MTHFD2) overexpression is associated with tumor cell proliferation and is a novel target for drug development. Mol Cancer Res. 2015;13:1361–1366. doi: 10.1158/1541-7786.MCR-15-0117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teng YW, Mehedint MG, Garrow TA, Zeisel SH. Deletion of betaine-homocysteine S-methyltransferase in mice perturbs choline and 1-carbon metabolism, resulting in fatty liver and hepatocellular carcinomas. J Biol Chem. 2011;286:36258–36267. doi: 10.1074/jbc.M111.265348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tibbetts AS, Appling DR. Compartmentalization of Mammalian folate-mediated one-carbon metabolism. Annu Rev Nutr. 2010;30:57–81. doi: 10.1146/annurev.nutr.012809.104810. [DOI] [PubMed] [Google Scholar]
- Titov DV, Cracan V, Goodman RP, Peng J, Grabarek Z, Mootha VK. Complementation of mitochondrial electron transport chain by manipulation of the NAD+/NADH ratio. Science. 2016;352:231–235. doi: 10.1126/science.aad4017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Titus SA, Moran RG. Retrovirally mediated complementation of the glyB phenotype. Cloning of a human gene encoding the carrier for entry of folates into mitochondria. J Biol Chem. 2000;275:36811–36817. doi: 10.1074/jbc.M005163200. [DOI] [PubMed] [Google Scholar]
- Tucker EJ, Hershman SG, Köhrer C, Belcher-Timme CA, Patel J, Goldberger OA, Christodoulou J, Silberstein JM, McKenzie M, Ryan MT, et al. Mutations in MTFMT underlie a human disorder of formylation causing impaired mitochondrial translation. Cell Metab. 2011;14:428–434. doi: 10.1016/j.cmet.2011.07.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ueland PM. Choline and betaine in health and disease. J Inherit Metab Dis. 2011;34:3–15. doi: 10.1007/s10545-010-9088-4. [DOI] [PubMed] [Google Scholar]
- Vazquez A, Tedeschi PM, Bertino JR. Overexpression of the mitochondrial folate and glycine-serine pathway: a new determinant of methotrexate selectivity in tumors. Cancer Res. 2013;73:478–482. doi: 10.1158/0008-5472.CAN-12-3709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vitvitsky V, Thomas M, Ghorpade A, Gendelman HE, Banerjee R. A functional transsulfuration pathway in the brain links to glutathione homeostasis. J Biol Chem. 2006;281:35785–35793. doi: 10.1074/jbc.M602799200. [DOI] [PubMed] [Google Scholar]
- Wald DS, Law M, Morris JK. Homocysteine and cardiovascular disease: evidence on causality from a meta-analysis. BMJ. 2002;325:1202. doi: 10.1136/bmj.325.7374.1202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J, Alexander P, Wu L, Hammer R, Cleaver O, McKnight SL. Dependence of mouse embryonic stem cells on threonine catabolism. Science. 2009;325:435–439. doi: 10.1126/science.1173288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- West MG, Horne DW, Appling DR. Metabolic role of cytoplasmic isozymes of 5,10-methylenetetrahydrofolate dehydrogenase in Saccharomyces cerevisiae. Biochemistry. 1996;35:3122–3132. doi: 10.1021/bi952713d. [DOI] [PubMed] [Google Scholar]
- Woo CC, Chen WC, Teo XQ, Radda GK, Lee PTH. Down-regulating serine hydroxymethyltransferase 2 (SHMT2) suppresses tumorigenesis in human hepatocellular carcinoma. Oncotarget. 2016 doi: 10.18632/oncotarget.10415. Published online July 6, 2016 http://dx.doi.org/10.18632/oncotarget.10415. [DOI] [PMC free article] [PubMed]
- Wright AJA, Dainty JR, Finglas PM. Folic acid metabolism in human subjects revisited: potential implications for proposed mandatory folic acid fortification in the UK. Br J Nutr. 2007;98:667–675. doi: 10.1017/S0007114507777140. [DOI] [PubMed] [Google Scholar]
- Yang XM, MacKenzie RE. NAD-dependent methylenetetrahydrofolate dehydrogenase-methenyltetrahydrofolate cyclohydrolase is the mammalian homolog of the mitochondrial enzyme encoded by the yeast MIS1 gene. Biochemistry. 1993;32:11118–11123. doi: 10.1021/bi00092a022. [DOI] [PubMed] [Google Scholar]
- Ye J, Fan J, Venneti S, Wan YW, Pawel BR, Zhang J, Finley LWS, Lu C, Lindsten T, Cross JR, et al. Serine catabolism regulates mitochondrial redox control during hypoxia. Cancer Discov. 2014;4:1406–1417. doi: 10.1158/2159-8290.CD-14-0250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeo EJ, Wagner C. Purification and properties of pancreatic glycine N-methyltransferase. J Biol Chem. 1992;267:24669–24674. [PubMed] [Google Scholar]
- Yoshida K, Furuya S, Osuka S, Mitoma J, Shinoda Y, Watanabe M, Azuma N, Tanaka H, Hashikawa T, Itohara S, Hirabayashi Y. Targeted disruption of the mouse 3-phosphoglycerate dehydrogenase gene causes severe neurodevelopmental defects and results in embryonic lethality. J Biol Chem. 2004;279:3573–3577. doi: 10.1074/jbc.C300507200. [DOI] [PubMed] [Google Scholar]
- Zhang WC, Shyh-Chang N, Yang H, Rai A, Umashankar S, Ma S, Soh BS, Sun LL, Tai BC, Nga ME, et al. Glycine decarboxylase activity drives non-small cell lung cancer tumor-initiating cells and tumorigenesis. Cell. 2012;148:259–272. doi: 10.1016/j.cell.2011.11.050. [DOI] [PubMed] [Google Scholar]
- Zhao R, Goldman ID. Resistance to antifolates. Oncogene. 2003;22:7431–7457. doi: 10.1038/sj.onc.1206946. [DOI] [PubMed] [Google Scholar]
- Zhou YH, Tang JY, Wu MJ, Lu J, Wei X, Qin YY, Wang C, Xu JF, He J. Effect of folic acid supplementation on cardiovascular outcomes: a systematic review and meta-analysis. PLoS ONE. 2011;6:e25142. doi: 10.1371/journal.pone.0025142. [DOI] [PMC free article] [PubMed] [Google Scholar]