Abstract
CD8+ T cells directed against beta cell autoantigens are considered relevant for the pathogenesis of type 1 diabetes. Using single cell T cell receptor sequencing of CD8+ T cells specific for the IGRP265-273 epitope, we examined whether there was expansion of clonotypes and sharing of T cell receptor chains in autoreactive CD8+ T cell repertoires. HLA-A*0201 positive type 1 diabetes patients (n = 19) and controls (n = 18) were analysed. TCR α- and β-chain sequences of 418 patient-derived IGRP265-273-multimer+ CD8+ T cells representing 48 clonotypes were obtained. Expanded populations of IGRP265-273-specific CD8+ T cells with dominant clonotypes that had TCR α-chains shared across patients were observed. The SGGSNYKLTF motif corresponding to TRAJ53 was contained in 384 (91.9%) cells, and in 20 (41.7%) patient-derived clonotypes. TRAJ53 together with TRAV29/DV5 was found in 15 (31.3%) clonotypes. Using next generation TCR α-chain sequencing, we found enrichment of one of these TCR α-chains in the memory CD8+ T cells of patients as compared to healthy controls. CD8+ T cell clones bearing the enriched motifs mediated antigen-specific target cell lysis. We provide the first evidence for restriction of T cell receptor motifs in the alpha chain of human CD8+ T cells with specificity to a beta cell antigen.
Autoreactive CD8+ T cells are in all likelihood key mediators of the pancreatic beta cell destruction leading to type 1 diabetes1,2,3,4,5,6. T cell receptor (TCR)-mediated recognition of (auto-) antigenic peptides presented on MHC class I molecules is a prerequisite for CD8+ T cell mediated target cell destruction. Several islet autoantigen epitopes presented on MHC class I molecules5,7,8,9,10,11,12,13 and assays to measure and quantify CD8+ T cell responses against these epitopes have been described14,15,16,17. However, information on the TCR repertoire of autoantigen specific CD8+ T cells in type 1 diabetes is so far limited to the TCR sequencing of propagated CD8+ T cell clones18, TCR sequence information of single TCR chains of isolated bulk autoantigen specific CD8+ T cells19, or CDR3 spectrotype data on bulk autoantigen specific CD8+ T cells20. These studies do not provide clonotype information and have not been able to show restricted TCR usage by the autoreactive CD8+ T cells.
TCRs are heterodimers consisting of TCR α- and β-chains and TCR diversity results from combinatorial rearrangements of variable (V), joining (J), and, for TCR β, also the diversity (D) gene segments. V-(D)-J sequences of both chains constitute the hypervariable complementary determining region 3 (CDR3) which provides the major contact point with the antigenic peptide and, therefore, determines antigen specificity of the T cell. The unique combination defines a clonotype. Although TCR clonotypes can be promiscuous in their binding to MHC-peptide complexes21, TCRs that recognize epitopes of viral and tumour antigens often have preferred CDR3 motifs or gene usage22,23,24,25,26, indicating that some structural restriction of the MHC-peptide binding region of the TCR plays an important role in the selection and expansion of clones.
In this study, we interrogated the TCR repertoire of CD8+ T cells directed against an epitope of an islet autoantigen using single cell TCR sequencing in order to determine whether there is TCR selection in islet autoantigen-specific CD8+ T cells. We chose the islet-specific glucose-6-phosphatase catalytic subunit related protein (IGRP) antigen as a model islet autoantigen, since an HLA A*0201 restricted peptide, IGRP265-273, has been identified and IGRP265-273 directed CD8+ T cells have been detected in the pancreatic islets of organ donors with type 1 diabetes27. Additionally, the occurrence and quantification of CD8+ T cells directed against the islet autoantigen IGRP has been demonstrated to have prognostic value on autoimmune diabetes development in NOD mice4,28. Our findings suggest that, as described for virus-specific CD8+ T cells, there is selection and expansion of a restricted TCR repertoire in islet-antigen specific CD8+ T cells.
Results
T cell receptor sequencing reveals dominant clonotypes and common alpha chains for IGRP-specific CD8+ T cells
We initially tested our TCR sequencing approach using CD8+ T cells that stained positive with MHC class I multimers loaded with a bona fide Influenza peptide epitope (Flu MP58-66; Supplementary Fig. S1a). From the analysed cells, we identified new as well as previously described22,23,29,30 Flu MP58-66-specific T cell receptor chains (see Supplementary Table S1). We noted inter-individual sharing of TCR α-chains among the analysed Flu-specific cells (Supplementary Fig. S1b) and, in accordance with previous reports22,23,29,31, we observed preferential usage of TRBV19 (72.2%), TRBJ2-7 (31.1%) and TRAJ42 (24.4%) genes in the analysed Flu MP58-66-specific CD8+ T clonotypes (Supplementary Fig. S1c). These findings endorse the approach taken to obtain antigen-specific TCR information.
We proceeded to analyse CD8+ T cells with specificity against the type 1 diabetes autoantigen epitope, IGRP265-273. Consistent with a previous report19, we observed no difference in the frequency of CD8+ T cells that stained positive with multimers loaded with the HLA-A*0201 restricted peptide epitope IGRP265-273 between healthy controls (median frequency, 0.02% of CD8+ T cells) and children with recent onset of type 1 diabetes (0.01%; P = 0.11) or long-standing type 1 diabetes (0.01%; P = 0.06; Fig. 1a; Supplementary Table S2). However, three patients showed a prominent and distinct population of IGRP265-273-specific CD8+ T cells (T1D-1, T1D-2, T1D-3; Fig. 1b). The IGRP265-273-specific CD8+ T cells in at least two of these patients had an antigen-experienced memory phenotype, which was in contrast to a predominantly naive phenotype of IGRP265-273-specific CD8+ T cells in other patients tested and in healthy controls (Supplementary Fig. S2).
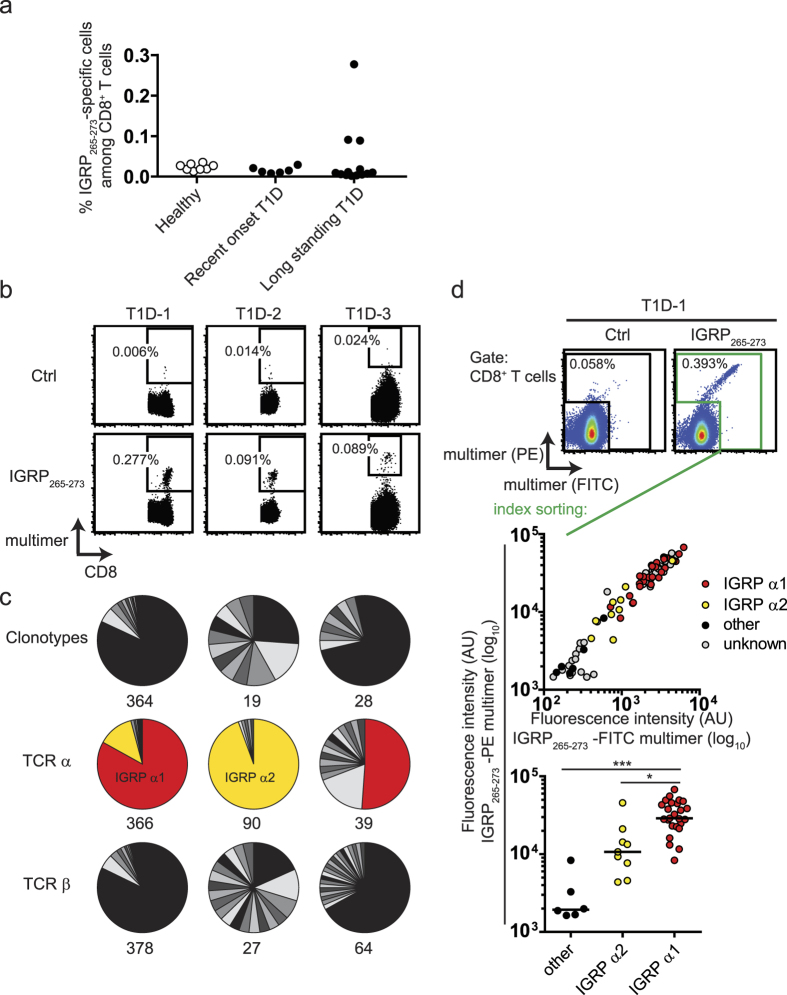
Figure 1. Identification and TCR repertoire analysis of IGRP265-273-specific CD8+ T cells.
(a) Frequencies of multimer positive IGRP265-273-specific cells (y axis) among CD8+ T cells in HLA A*0201 healthy children (n = 8) age matched to children with recent onset of type 1 diabetes (n = 6), and adult patients with long standing type 1 diabetes (n = 13) were determined via flow cytometry. (b) Representative multimer staining FACS plots of PBMC samples of donors with type 1 diabetes and highest frequencies of IGRP265-273-specific CD8+ T cells in (a). PBMC samples were stained with HLA-A2 multimers loaded with control peptide (HLA-A2140-149; top row) or IGRP265-273 (bottom row). Plots show cells in the CD8 gate; 105 (T1D-1, T1D-2) or 5 × 104 (T1D-3) CD8+ T cell events are shown. (c) TCR repertoire analysis upon TCR α- and β-chain sequencing of IGRP265-273-specific CD8+ T cells isolated as single cells from T1D-1 (left), T1D-2 (middle) and T1D-3 (right). Each piece of pie charts represents cells with the same TCR α-chain and TCR β-chain combination (clonotypes; upper row), TCR α-chain (middle row) or TCR β-chain (bottom row). Numbers of analyzed cells are indicated. Shades of gray represent private clonotypes or TCR chains, colors indicate sharing among individuals. Note: numbers and pie charts of donor 1 comprise information of all three analyzed time points shown in Supplementary Fig. S3. (d) Combined multimer analysis and TCR sequencing via single cell index sorting. PBMC from T1D-1 were stained with PE and FITC HLA-A2 multimers loaded with either control peptide (HLA-A2140-149; top left) or IGRP265-273 (top right). Using index sorting, multimer binding cells were isolated as single cells for TCR sequencing. TCR α-chain sequencing information was subsequently attributed to the individual sorted cells and visualized using index sorting fluorescence intensity information (middle). Cells for which no TCR α-chain information was obtained are depicted in gray. The bottom plot compares PE-multimer fluorescence intensities (y axis) of cells expressing the dominant TCR α-chains IGRP α1 or IGRP α2, and other TCR α-chains. Lines indicate median values and significant differences between groups according to one-way ANOVA using Dunn’s multiple comparison test are marked (*P = 0.01–0.05; ***P < 0.001).
Paired TCR α- and β-chain sequences of 411 IGRP265-273-specific CD8+ T cells were obtained from patients T1D-1, T1D-2, and T1D-3. Dominant TCR α- and β-chain combinations or clonotypes, defined as a unique combination of TCR α- and β-chains, were observed in each of the three patients (Fig. 1c and Supplementary Table S1). Moreover, the dominant IGRP265-273-specific CD8+ T cell clonotypes persisted in samples taken more than 10 months apart from patient T1D-1 (Supplementary Fig. S3). Of note, the major TCR α-chains were shared among the three patients, paired with a number of different TCR β-chains (Fig. 1c and Supplementary Table S1). Interestingly, as revealed by combined index sorting of IGRP265-273 multimer-positive CD8+ T cells and TCR sequencing for T1D-1, those cells that expressed the dominant TCR α-chain detected in T1D-1 and T1D-3 (IGRP α1; depicted as red piece of pie charts in Fig. 1c) had the highest median fluorescence intensity in the IGRP265-273-specific multimer staining (Fig. 1d) as compared to the IGRP α2-containing cells (depicted as yellow piece of pie charts in Fig. 1c; P < 0.05) and cells with other TCR α-chains (P < 0.001).
Taken together, by analysing the TCR repertoires of IGRP265-273 directed CD8+ T cells from patients with type 1 diabetes, we identified dominant IGRP265-273–specific clonotypes that were persistent in sequential samples and common usage of IGRP265-273–specific CD8+ T cell-derived TCR α-chains between individuals.
The TRAJ53 SGGSNYKLTF motif is frequent in HLA-A*0201 IGRP-specific clonotypes
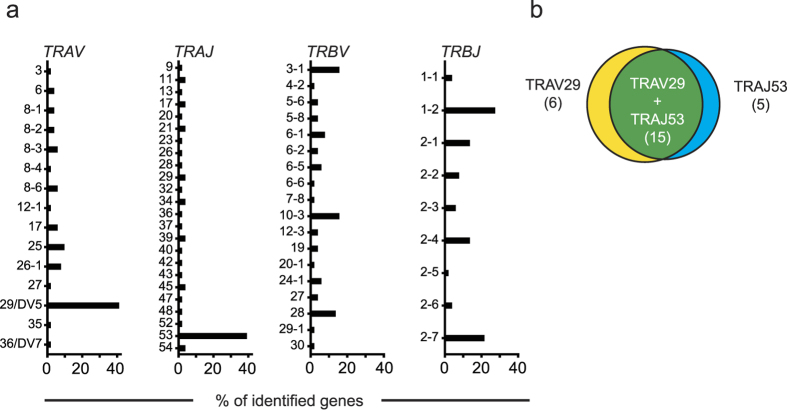
Examining the paired TCR α- and β-chains of single cell-sorted IGRP265-273-specific CD8+ T cells, we identified the CDR3 motif SGGSNYKLTF in the TCR α-chain of the dominant clonotypes of patients T1D-1 (299 cells), T1D-2 (5 cells), and T1D-3 (20 cells), and in a further 60 of 94 patient derived IGRP265-273-specific CD8+ T cells for which we obtained TCR α- and β-chain information. In total, 384 (91.9%) of 418 analysed patient-derived IGRP-specific CD8+ T cells for which we obtained TCR α- and β-chain information contained this CDR3 motif (6 patients had cells with full clonotype information; see Supplementary Table S1). The motif corresponds to the TRAJ53 gene, which together with TRAV29/DV5, are the predominantly used TRAJ and TRAV genes within all of the 48 identified patient-derived IGRP265-273-specific clonotypes (41.7% and 43.8%, respectively; Fig. 2a). Both TRAJ53 and TRAV29/DV5 genes were seen in 15 (31.3%) clonotypes (Fig. 2b), represented in 61 (14.6%) IGRP265-273-specific CD8+ T cells. Three clonotypes were identified in IGRP265-273-specific CD8+ T cells from healthy individuals. None of these had the SGGSNYKLTF motif in their TCR α-chain CDR3.
Figure 2. TRAV29/DV5 and TRAJ53 are frequent in IGRP265-273-specific CD8+ T cells.
(a) TCR gene usage identified in IGRP265-273-specific CD8+ T cells. Frequencies (x-axis) of TRAV, TRAJ, TRBV and TRBJ genes used in unique clonotypes (n = 51, of which 48 are from patient-derived cells) are shown. (b) TRAV29/DV5 and TRAJ53 usage coincide. Venn-diagram of IGRP-specific clonotypes expressing TRAV29/DV5 only (yellow), TRAJ53 only (blue) or both (green).
A part of the SGGSNYKLTF motif (GSNA/YKLT) was previously found in T lymphocytes isolated from pancreatic islets of NOD mice32,33,34, as well as in 2 of 53 CD4+ T cell clones isolated from islets of a HLA-A*0201 positive human patient with type 1 diabetes35. We, therefore, searched for the SGGSNYKLTF motif in our antigen-specific T cell database. Of the 51 IGRP265-273-specific CD8+ T cell clonotypes identified in both patients and controls, 19 (37.3%) had the SGGSNYKLTF motif. In comparison, 2 out of 90 (2.2%; P < 0.0001) of the TCR α-chains from Flu MP58-66-specific CD8+ T cell clonotypes, 0 out of 18 (0%; P = 0.0015) of the TCR α-chains from CMVpp65-specific CD8+ T cell clonotypes24, and 29 out of 1492 (1.9%, P < 0.0001) TCR α-chains from GAD65- or Tetanus toxoid-specific CD4+ T cell clonotypes analysed in previous studies36 had the SGGSNYKLTF motif arguing for an enrichment of this motif in IGRP-specific cells and against a general bias for this motif in antigen-specific T cells.
Dominant IGRP265-273-specific CD8+ T cell TCR α-chain and SGGSNYKLTF motif frequencies in CD8+ T cell repertoires
TCRs raised against specific antigens are proposed as potential disease biomarkers37. We were, therefore, interested in determining whether any of the TCR α-chains identified in IGRP265-273 directed cells were associated with type 1 diabetes. We first applied next generation sequencing of the TCR α-chain repertoire to determine whether the approach could detect the dominant IGRP265-273-specific chains IGRP α1 and IGRP α2 in bulk CD8+ T cells from the original patients T1D-1, T1D-2, and T1D-3 (Supplementary Fig. S4). Consistent with the single cell TCR sequencing results (Fig. 1), we retrieved message for IGRP α1 exclusively within the libraries of donors T1D-1 and T1D-3. Message for the dominant IGRP α2 was detected within both the libraries of T1D-1 and T1D-2.
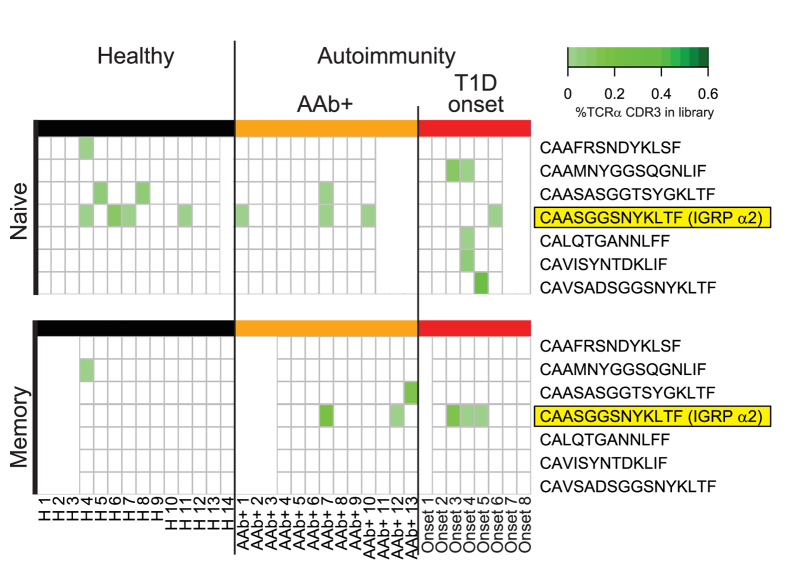
We, therefore, performed TCR α-chain next generation sequencing on naïve and memory CD8+ T cells from a new set of HLA A*0201 positive donors comprising control children (n = 14), multiple islet autoantibody (AAb+; n = 13) positive children, and recent onset patients with type 1 diabetes (n = 8). Seven of the 45 TCR α-chains identified in IGRP265-273 multimer-sorted single cells were detected in either the naïve (7 TCR α-chains) or memory (3 TCR α-chains) CD8+ T cells from these individuals (Fig. 3). Within memory CD8+ T cells, IGRP α2, the dominant TCR α-chain shared by patients T1D-1 and T1D-2, was detected in none of 11 control children as compared to 2 (20%) of 10 autoantibody positive children and 3 (43%) of 7 recent onset type 1 diabetes patients (P = 0.042 controls vs patients). Moreover, IGRP α2 read frequencies were higher in the memory CD8+ T cell repertoires of patients as compared to controls (P = 0.026). No other differences were observed.
Figure 3. Screening for IGRP265-273-specific TCR sequences in bulk sorted naïve and memory CD8+ T cells using TCR α-chain NGS.
Naïve (CCR7+CD45RA+) and memory (CCR7+CD45RA−, CCR7−CD45RA+/−) CD8+ T cells of healthy donors (n = 14) or donors with multiple islet autoantibodies (AAb+; n = 13) or recent onset type 1 diabetes (n = 8) were flow sorted from PBMC, their RNA extracted, individual cDNA libraries prepared and processed via next generation sequencing. TCR α-chain sequences were extracted and screened for those previously identified in IGRP265-273-specific single CD8+ T cells. TCR α-chain CDR3 region sequences retrieved from the donor libraries are listed on the right. Filled (green) boxes within the heatmaps indicate libraries in which the sequences were found, and shades of green visualize their abundance within the given libraries. White space holders in the heatmaps are shown if information was available only for the naïve or memory library.
We next examined whether the SGGSNYKLTF motif is per se associated with type 1 diabetes (Supplementary Fig. S5). TCR α-chains that contained the SGGSNYKLTF motif were detected in all donors and their abundance in naïve or memory CD8+ T cells was not significantly increased in AAb+ (median, naive 0.77% and memory 0.89%) or recent onset type 1 diabetes patients (median, 1.06% and 0.63%) as compared to control children (median, 1.07% and 0.73%), arguing against a general association of TCRs containing this motif with disease.
Dominant IGRP TCR α-chain expressing T cell clones specifically kill targets
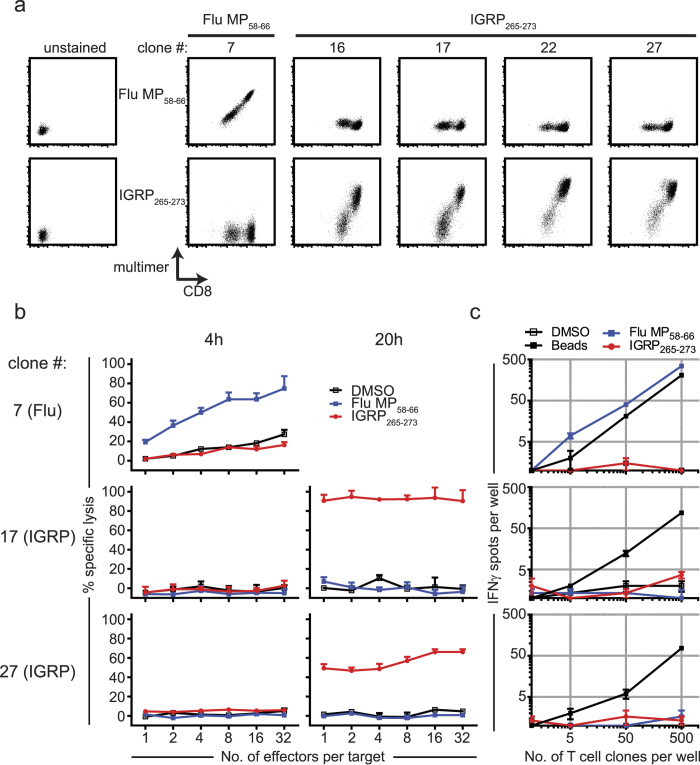
To further assess the peptide specificity of IGRP directed clonotypes containing the TRAJ53 SGGSNYKLTF motif, we expanded IGRP265-273 directed single cells and Flu MP58-66 directed cells from patient T1D-1 to obtain CD8+ T cell clones. Clones 16 and 17 expressed the dominant IGRP directed TCR of T1D-1 (comprising the TRAJ53-encoded IGRP α1) and clones 22 and 27 had identical TCRs comprising the TRAV29/DV5-TRAJ53-encoded IGRP α2 (see Supplementary Table S3 for TCR sequence information for CD8+ T cell clones). Binding to peptide loaded HLA-A*0201 multimers was confirmed for each of these IGRP265-273 directed clones and a Flu MP58-66 directed clone (clone 7; Fig. 4a). IGRP265-273 directed clones 17 and 27 were able to mediate peptide-specific target cell lysis, albeit at a later time point than the Flu MP58-66 directed clone 7 (Fig. 4b). Unlike the Flu MP58-66-directed clone 7, the IGRP265-273-specific clones did not yield IFNγ spot formation when stimulated with IGRP265-273 peptide, despite strong responses to a polyclonal stimulus (Fig. 4c). The single cell gene expression profiles of expanded Flu MP58-66- and IGRP265-273 directed clones also differed. While cells from all clones expressed many genes typical of activated CD8+ T cells such as GZMA and GZMB, we noted higher expression of GZMH (P = 7 × 10−8), and less TBX21 (P = 2.9 × 10−4), CD52 (P = 0.0012) and TNF (P = 0.0073) in the IGRP265-273 directed clones as compared to the Flu-specific clone (Supplementary Fig. S6).
Figure 4. Functional characterization of IGRP265-273-specific CD8+ T cell clones.
(a) IGRP265-273 (clones 16, 17, 22, 27) and Flu MP58-66 (clone 7) directed clones isolated from T1D-1 stained with anti-CD8 and HLA-A2 multimers loaded with Flu MP58-66 or IGRP265-273. Unstained samples served as controls (left column). Representative FACS plots of at least 3 independent experiments are shown. (b) IGRP-specific CD8+ T cell clones kill peptide loaded target cells. Flu MP58-66-specific clone 7 (top) and IGRP265-273-specific clones 17 (middle) and 27 (bottom) were titrated in a 51Cr release assay using K562/A*0201 cells as targets. Clones at increasing effector to target cell ratios (x axis) were stimulated with Flu MP58-66 peptide (blue), IGRP265-273 peptide (red) or solvent DMSO (black), and the percentages of specific lysis of target cells (y axis) were analysed after assay incubation for 4 h and 20 h. Representative data of three independent assays are shown. Data are presented as mean + SEM of triplicate wells. (c) IFNγ secretion by MP58-66 or IGRP265-273-specific CD8+ T cell clones. Titrated numbers of Flu MP58-66-specific (clone 7, top) and IGRP265-273-specific (clones 17 and 27, middle and bottom, respectively) clonal CD8+ T cells (x axis) were incubated in IFNγ ELISpot assays using K562/A*0201 cells for antigen presentation and stimulated with Flu MP58-66 peptide, IGRP265-273 peptide or control stimuli (solvent DMSO, anti-CD3/anti-CD28 coated beads). Data points in the analysis graphs show mean number of spots ± SEM of triplicate wells (y axis). Representative assays of three independent experiments are shown.
Discussion
We applied single cell TCR α- and β-chain sequencing to assess the TCR repertoire of CD8+ T cells directed against IGRP265-273 as a model target epitope of type 1 diabetes. We identified dominant IGRP-specific clonotypes in patients that were persistent in sequential samples, detected sharing of dominant TCR α-chains among patients, and identified a motif that is overrepresented in IGRP265-273-specific CD8+ T cells as compared to T cells with other antigen specificities.
The low frequency of islet-antigen-specific CD8+ T cells in the peripheral blood of type 1 diabetes patients has thus far hampered a detailed analysis of their TCR repertoire. The overall low frequency of IGRP265-273-specific cells in samples from both type 1 diabetes patients and controls observed in this study is in agreement with previous reports17,19 that looked at frequencies and phenotypes of CD8+ T cells specific for IGRP and additional autoantigen-specific epitopes, although one of these reports suggested increased frequencies of IGRP-specific CD8+ T cells in patients as compared to controls17. The identification of three patients with expanded populations of memory CD8+ T cells specific to IGRP265-273 allowed us to isolate and analyse a large number of IGRP265-273-specific cells. Although it is possible that our findings on TCR from these patients are not representative of all subjects, it is remarkable that we found sharing of TCR α-chains between the patients and that sharing was marked by a TRAJ53 encoded motif. These findings, as well as the impressive correlation between TCR α-chain and multimer staining intensity, the persistency of expanded clonotypes over time, and the ability of clones derived from multimer-positive CD8+ T cells to antigen-specifically lyse target cells speak for bona fide antigen-specific cells. Further supporting the methodological approach, we find similar TCR gene usage for Flu MP58-66-specific CD8+ T cells to that reported in previous reports22,23,29,30 and identical TCR chain CDR3 sequences and public motifs contained in them38.
Selection of antigen-specific T cells in our study was based upon multimer staining. As we demonstrated, different TCRs varied in their multimer binding intensities, suggesting high and low affinity TCRs. Studies in the NOD mouse have demonstrated that TCR avidity against IGRP is relevant to disease stage, with higher avidity clones marking a feature of disease progression39. Studies in the NOD mouse argue against IGRP as a primary target in the autoimmune disease process40 and describe a predominant presence of IGRP directed CD8+ T cells in later stages of disease28. It is, therefore, potentially interesting to find expansion of high affinity IGRP265-273-specific TCRs in the patient with long-standing type 1 diabetes. We did not, however, study sufficient numbers of patients to conclude that the expanded population with a memory phenotype is related to disease duration or accumulation over time.
The repertoire of IGRP-specific cells was highly enriched in TCR α-chains comprising the motif SGGSNYKLTF that is encoded by the TRAJ53 gene, especially when combined with the TRAV29/DV5 gene. Interestingly, this motif, and a highly similar motif encoded by Traj42 (SGGSNAKLTF), has been identified previously in autoreactive and pancreatic islet derived T cells in NOD mice32,34, including CD8+ T cells reactive to Igrp peptide33 and CD4+ T cells reactive to the Insulin peptide B:9-2341. Moreover, TCRs targeting the insulin B:9–23 peptide presented by the I-Ag7 MHC class II molecule frequently use the Vα gene segment Trav5D-4 rearranged to the Jα gene segments Traj53 and Traj4242,43. Thus, islet antigen reactive T cells in NOD mice and man appear to have a recurring preference for certain TCR α-chain genes. Such selection, if true, could be explained by structural similarities across multiple epitope/MHC targets.
Finally, in an attempt to identify molecular markers of type 1 diabetes, we identified one TCR α-chain that, in memory CD8+ T cells, was restricted to patients with type 1 diabetes or pre-type 1 diabetes. The association was not striking, but suggests that it may be possible to identify panels of clonotypes that mark type 1 diabetes-relevant CD8+ T cell activity. A limitation is that we did not perform next generation sequencing for the TCR β-chain. Nevertheless, our findings support the use of approaches similar to those used in this study to extend the knowledge of beta cell antigen-specific TCR repertoires. Moreover, it is possible that both the expansion of autoreactive T cells we observed for IGRP and an apparent preference for certain TCR α-chain genes in responses to different autoantigens reflects cross-reactivity to unknown non-self antigens44, which may be favoured by selection of T cells that bear TCRs with a moderate ability to cross-react with multiple peptides45.
Methods
Subjects
Samples were obtained from 75 individuals who had the HLA-A*0201 allele (Supplementary Table S2). These included 27 patients with type 1 diabetes (16 female, 11 male; median age at blood sampling: 22.9 years; range, 5.3–45.9 years), 14 of whom had a diabetes duration of less than one year (median disease duration: 0.04 years; range, 0.01–0.94 years) and 13 patients with a type 1 diabetes duration of more than one year (median disease duration: 14.33 years; range, 1.1–41.5 years); 13 non-diabetic autoantibody positive relatives of patients with type 1 diabetes (3 female, 10 male; median age: 11.6 years; range, 3.8–18.7 years) who had autoantibodies against at least two of four islet antigens (Insulin, GAD65, ZnT8, IA-2), and 35 healthy controls (20 female, 15 male; median age 14.3 years, range, 8.3–63.0 years). All methods were performed in accordance with relevant guidelines and regulations. Samples were collected with informed consent as part of studies that were approved by the ethical committees of the Technische Universität München (No. 2149/08) or Bavaria, Germany (Bayerische Landesaerztekammer, Nr.08043).
Peptides
Peptides used in this study were purchased from Mimotopes, Australia at >95% purity, as confirmed using HPLC and mass spectrometry.
Cell staining and flow cytometry
Cells were stained using the following monoclonal antibody-fluorochrome combinations: CD4-APC (clone SK3), CD8-APC-Cy7 (clone SK1), CD14-APC (clone M5E2), CD19-APC (clone HIB19), CD56-APC (clone B159) from BD Pharmingen; CD8a-eFluor450 (clone SK1), CD8a-PE (clone SK1), CD45RA-eFluor605NC (clone HI100) from eBioscience; CD16-APC (clone 3G8), CD197(CCR7)-Brilliant Violet 710 from BioLegend; CD335-APC (clone 9E2) from Miltenyi Biotech. 7-Aminoactinomycin D (7AAD, BD Pharmingen) or Sytox Blue (Invitrogen) were used to exclude dead cells. FITC or PE labeled HLA-A*0201 multimers loaded with either Flu MP58-66 (GILGFVFTL), IGRP265-273 (VLFGLGFAI) or a control peptide of the HLA-A2 protein (HLA-A2140-149; YAYDGKDYIA) were purchased from Immudex (Copenhagen, Denmark). Cells were acquired on a Becton Dickinson ARIA II or ARIA III flow sorter with FACS Diva software (for index sorting experiments, FACS Diva 8 software was used) and analysed using FlowJo software (FlowJo LLC Ashland, OR, USA). Unless stated otherwise, cells were stained in PBS supplemented with 1% pooled human AB serum for 30 min on ice followed by at least two washing steps using the same buffer and staining with dead cell marker before cell analysis. Multimer staining was carried out on thawed or fresh PBMC samples after overnight resting at 37 °C/5% CO2 in X-VIVO 15 supplemented with 5% pooled human AB serum. Cells were resuspended in PBS supplemented with 5% FBS and 50 nM dasatinib and incubated for 30 min at 37 °C. 2 × 106 cells were stained with multimers for 10 min at room temperature in the same buffer followed by an additional 20 min incubation at 4 °C in the presence of the respective antibodies. Cells were washed twice with PBS/5% FBS and incubated with 7AAD or Sytox Blue for 10 min immediately before flow cytometry. Gating strategies for the analysis and cell sorting of multimer stained samples are exemplarily shown in Supplementary Fig. S7. Gates for antigen-specific CD8+ T cell frequency analysis were based on staining using the control multimers or, when available, according to staining data of PBMC spiked with the respective antigen-specific CD8+ T cell clone.
TCR sequencing
RT-PCR amplification and sequencing of expressed TCR α- and β-chain genes from single CD8+ T cells, as well as subsequent cloning of PCR fragments, were performed as previously described46. Analysis of TCR α- and TCR β-chain sequences and junction peptide amino acid sequence extraction was conducted with reference to the International ImMunoGeneTics Information System database47. Obtained junction peptides were then analyzed using KNIME 2.11.248.
The TCR α-chain repertoire of 2 × 105 flow-sorted total, naïve (CD45RA+CCR7+) or memory (CD45RA+/−CCR7− and CD45RA−CCR7+) peripheral blood derived CD8+ T cells from patients and controls was analysed using next generation sequencing as previously described36. Libraries containing < 0.5 Mio reads were excluded. TCR CDR3 region sequence extraction and PCR error correction was performed with MiTCR49. All additional sequence analysis was conducted in KNIME 2.11.2.
Cloning of antigen specific CD8+ T cells
Flu MP58-66- and IGRP265-273-specific CD8+ T cell clones were propagated from multimer positive single cells according to a previously described protocol5. In brief, purified CD8+ T cells were stained with the respective multimers and viable multimer positive CD8+ T cells were flow-sorted and seeded at 1 cell/well in 96-well plates containing 105 irradiated allogeneic PBMCs per well in X-VIVO15 supplemented with 5% Cellkine (Zeptometrix Corp., NY, USA), IL-7 (10 ng/ml), IL-15 (0.1 ng/ml) and PHA-M (5 μg/ml). CD8+ T cell clones were then stimulated every 14 days at a 1:5 clone to feeder ratio (or smaller in the initial expansion phase, i.e. until a 1:5 ratio per well of a 96 well plate was achieved) with irradiated allogeneic PBMCs that alternately were used either in combination with PHA-M as described above or were preloaded with peptide (FluMP58-66 or IGRP265-273; 10 μg/ml).
Cytotoxicity assay
Cytotoxicity of propagated clones was analysed by radioactive 51Cr release assays. HLA-A*0201 expressing K562 cells (K562/A*020150; kindly provided by Prof. Thomas Wölfel; Johannes Gutenberg Universität, Mainz, Germany) were used as target cells. Briefly, 5 × 103 51Cr labelled target cells were seeded into V-shaped 96 well plates followed by effector cells (CD8+ T cell clones) in varying ratios. Assays were conducted in a final volume of 200 μl /well in X-VIVO15 medium supplemented with 5% AB serum, IL-7 (10 ng/ml) and IL-15 (0.1 ng/ml). Assays were conducted in triplicate and cultures were incubated for 20 h at 37 °C/5%CO2 in the presence of FluMP58-66 or IGRP265-273 peptides (10 μg/ml) or solvent (DMSO) control. After 4 or 20 hours incubation, 25 μl of supernatant from each well was transferred into 96 well sample plates containing 150 μl scintillation liquid per well. After 5 minutes shaking at room temperature 51Cr release was measured with a scintillation counter (1450 MicroBeta TriLux, Perkin Elmer). Maximum and spontaneous 51Cr release was determined from cells treated with lysis buffer or assay medium, respectively. Specific cytotoxicity was calculated using the formula: % specific release = (experimental–spontaneous release) × 100/(maximum–spontaneous release).
ELISpot assay
ELISpot assays on CD8+ T cell clones were performed as previously described16 using K562/A*0201 cells for antigen presentation. In brief, 96-well polyvinylidine fluoride plates (Merck Millipore, Darmstadt, Germany) were coated overnight with an IFNγ directed antibody (U-CyTech biosciences, Utrecht, The Netherlands). Plates were subsequently washed and blocked with AIM-V plus 10% AB serum. Assays were carried out with 5 × 104 K562/A*0201 cells/well in a 200 μl/well format using AIM-V medium supplemented with 0.5 ng/ml IL-7. Titrated numbers of viable CD8+ T cell clones (5–500 cells) were flow-sorted directly into the wells and incubated for 20–24 h in the presence of peptide (IGRP265-273 or Flu MP58-66; 10 μg/ml) or solvent (DMSO). Following removal of cells, IFNγ secretion was visualized as described previously51,52 using biotin-conjugated anti-IFNγ detector antibody (U-CyTech biosciences, Utrecht, The Netherlands), alkaline phosphatase-conjugated ExtrAvidin, and Sigmafast 5-bromo-4-chloro-3-indolyl phosphate/nitro blue tetrazolium (BCIP/NBT) tablets (Sigma-Aldrich Chemie GmbH, Taufkirchen, Germany). Spots were counted using a Bioreader 5000 (BioSys, Karben, Germany) and mean ± SEM values of at least triplicate wells calculated.
Statistical analysis
Frequencies of multimer directed cells between groups were compared using the Mann Whitney U test. Fluorescence intensity values of index-sorted cells with different TCR α-chains were compared using one-way ANOVA with Dunn’s multiple comparison test. TCR α CDR3 motif frequencies between antigen-specific T cell clonotypes were compared using Fisher’s exact test. The presence or absence of TCR α-chain sequences in next generation sequencing information was compared between groups using Fisher’s exact test. Read frequencies were compared using Mann-Whitney U test. Statistical analysis was performed using GraphPad prism (version 5.04) or R (version 3.2.5).
Additional Information
How to cite this article: Fuchs, Y. F. et al. CD8+ T cells specific for the islet autoantigen IGRP are restricted in their T cell receptor chain usage. Sci. Rep. 7, 44661; doi: 10.1038/srep44661 (2017).
Publisher's note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Material
Acknowledgments
We thank Cihan Erkut (EMBL Heidelberg) for support in data analysis. The work was supported by grants from the Kompetenznetz Diabetes Mellitus (Competence Network for Diabetes Mellitus) funded by the Federal Ministry of Education and Research (FKZ 01GI0805) and the Juvenile Diabetes Research Foundation (JDRF; JDRF-No 17-2012-16), and by grants from the German Federal Ministry of Education and Research (BMBF) to the German Center for Diabetes Research (DZD e.V.). Y.F.F. is supported by a postdoctoral fellowship of the JDRF (3-PDF-2016-188-A-N). E.B. is supported by the DFG Research Center and Cluster of Excellence–Center for Regenerative Therapies Dresden (FZ 111).
Footnotes
The authors declare no competing financial interests.
Author Contributions Y.F.F., A.E., and E.B. contributed to the conduct of the study and the acquisition, analysis and interpretation of data, and drafted, reviewed and approved the manuscript. S.D., C.S., D.K., C.W., A.L., A.G., J.K., A.D. and A-G.Z. contributed to the acquisition, analysis and interpretation of data, and reviewed and approved the manuscript. E.B. is the guarantor of this work and, as such, had full access to all of the data in the study and takes responsibility for the integrity of the data and the accuracy of the data analysis.
References
- Bottazzo G. F. et al. In situ characterization of autoimmune phenomena and expression of HLA molecules in the pancreas in diabetic insulitis. N Engl J Med 313, 353–360, doi: 10.1056/NEJM198508083130604 (1985). [DOI] [PubMed] [Google Scholar]
- Serreze D. V., Leiter E. H., Christianson G. J., Greiner D. & Roopenian D. C. Major histocompatibility complex class I-deficient NOD-B2mnull mice are diabetes and insulitis resistant. Diabetes 43, 505–509, doi: 10.2337/diabetes.43.3.505 (1994). [DOI] [PubMed] [Google Scholar]
- Wong F. S., Visintin I., Wen L., Flavell R. A. & Janeway C. A. Jr. CD8 T cell clones from young nonobese diabetic (NOD) islets can transfer rapid onset of diabetes in NOD mice in the absence of CD4 cells. The Journal of experimental medicine 183, 67–76 (1996). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trudeau J. D. et al. Prediction of spontaneous autoimmune diabetes in NOD mice by quantification of autoreactive T cells in peripheral blood. Journal of Clinical Investigation 111, 217–223, doi: 10.1172/jci200316409 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skowera A. et al. CTLs are targeted to kill beta cells in patients with type 1 diabetes through recognition of a glucose-regulated preproinsulin epitope. The Journal of clinical investigation 118, 3390–3402, doi: 10.1172/JCI35449 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Babon J. A. et al. Analysis of self-antigen specificity of islet-infiltrating T cells from human donors with type 1 diabetes. Nature medicine, doi: 10.1038/nm.4203 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di Lorenzo T. P., Peakman M. & Roep B. O. Translational mini-review series on type 1 diabetes: Systematic analysis of T cell epitopes in autoimmune diabetes. Clinical and experimental immunology 148, 1–16, doi: CEI3244 [pii]10.1111/j.1365-2249.2006.03244.x (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blancou P. et al. Immunization of HLA class I transgenic mice identifies autoantigenic epitopes eliciting dominant responses in type 1 diabetes patients. J Immunol 178, 7458–7466 (2007). [DOI] [PubMed] [Google Scholar]
- Enee E. et al. ZnT8 is a major CD8+ T cell-recognized autoantigen in pediatric type 1 diabetes. Diabetes 61, 1779–1784, doi: 10.2337/db12-0071 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pinkse G. G. et al. Autoreactive CD8 T cells associated with beta cell destruction in type 1 diabetes. Proceedings of the National Academy of Sciences of the United States of America 102, 18425–18430, doi: 10.1073/pnas.0508621102 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ouyang Q. et al. Recognition of HLA class I-restricted beta-cell epitopes in type 1 diabetes. Diabetes 55, 3068–3074, doi: 10.2337/db06-0065 (2006). [DOI] [PubMed] [Google Scholar]
- Toma A. et al. Recognition of a subregion of human proinsulin by class I-restricted T cells in type 1 diabetic patients. Proceedings of the National Academy of Sciences of the United States of America 102, 10581–10586, doi: 10.1073/pnas.0504230102 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jarchum I., Nichol L., Trucco M., Santamaria P. & DiLorenzo T. P. Identification of novel IGRP epitopes targeted in type 1 diabetes patients. Clin Immunol 127, 359–365, doi: 10.1016/j.clim.2008.01.015 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mallone R. et al. CD8+ T-cell responses identify beta-cell autoimmunity in human type 1 diabetes. Diabetes 56, 613–621, doi: 10.2337/db06-1419 (2007). [DOI] [PubMed] [Google Scholar]
- Mannering S. I. et al. Current approaches to measuring human islet-antigen specific T cell function in type 1 diabetes. Clinical and experimental immunology 162, 197–209, doi: 10.1111/j.1365-2249.2010.04237.x (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuchs Y. F. et al. Vagaries of the ELISpot assay: specific detection of antigen responsive cells requires purified CD8(+) T cells and MHC class I expressing antigen presenting cell lines. Clin Immunol 157, 216–225, doi: 10.1016/j.clim.2015.02.012 (2015). [DOI] [PubMed] [Google Scholar]
- Velthuis J. H. et al. Simultaneous detection of circulating autoreactive CD8+ T-cells specific for different islet cell-associated epitopes using combinatorial MHC multimers. Diabetes 59, 1721–1730, doi: 10.2337/db09-1486 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Unger W. W. et al. Islet-specific CTL cloned from a type 1 diabetes patient cause beta-cell destruction after engraftment into HLA-A2 transgenic NOD/scid/IL2RG null mice. PloS one 7, e49213, doi: 10.1371/journal.pone.0049213 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skowera A. et al. beta-cell-specific CD8 T cell phenotype in type 1 diabetes reflects chronic autoantigen exposure. Diabetes 64, 916–925, doi: 10.2337/db14-0332 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fozza C. et al. T-cell receptor repertoire analysis in monozygotic twins concordant and discordant for type 1 diabetes. Immunobiology 217, 920–925, doi: 10.1016/j.imbio.2012.01.002 (2012). [DOI] [PubMed] [Google Scholar]
- Wooldridge L. et al. A single autoimmune T cell receptor recognizes more than a million different peptides. The Journal of biological chemistry 287, 1168–1177, doi: 10.1074/jbc.M111.289488 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lehner P. J. et al. Human HLA-A0201-restricted cytotoxic T lymphocyte recognition of influenza A is dominated by T cells bearing the V beta 17 gene segment. The Journal of experimental medicine 181, 79–91 (1995). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valkenburg S. A. et al. Molecular basis for universal HLA-A*0201-restricted CD8+ T-cell immunity against influenza viruses. Proceedings of the National Academy of Sciences of the United States of America 113, 4440–4445, doi: 10.1073/pnas.1603106113 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Link C. S. et al. Abundant cytomegalovirus (CMV) reactive clonotypes in the CD8(+) T cell receptor alpha repertoire following allogeneic transplantation. Clinical and experimental immunology 184, 389–402, doi: 10.1111/cei.12770 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sensi M. et al. Cytotoxic T-lymphocyte clones from different patients display limited T-cell-receptor variable-region gene usage in HLA-A2-restricted recognition of the melanoma antigen Melan-A/MART-1. Proceedings of the National Academy of Sciences of the United States of America 92, 5674–5678 (1995). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Serana F. et al. Identification of a public CDR3 motif and a biased utilization of T-cell receptor V beta and J beta chains in HLA-A2/Melan-A-specific T-cell clonotypes of melanoma patients. Journal of translational medicine 7, 21, doi: 10.1186/1479-5876-7-21 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coppieters K. T. et al. Demonstration of islet-autoreactive CD8 T cells in insulitic lesions from recent onset and long-term type 1 diabetes patients. The Journal of experimental medicine 209, 51–60, doi: 10.1084/jem.20111187 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ko H. J. et al. Functional cytotoxic T lymphocytes against IGRP206-214 predict diabetes in the non-obese diabetic mouse. Immunology and cell biology 92, 640–644, doi: 10.1038/icb.2014.29 (2014). [DOI] [PubMed] [Google Scholar]
- Naumov Y. N. et al. Multiple glycines in TCR alpha-chains determine clonally diverse nature of human T cell memory to influenza A virus. J Immunol 181, 7407–7419 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gil A., Yassai M. B., Naumov Y. N. & Selin L. K. Narrowing of human influenza A virus-specific T cell receptor alpha and beta repertoires with increasing age. Journal of virology 89, 4102–4116, doi: 10.1128/JVI.03020-14 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neller M. A. et al. Naive CD8(+) T-cell precursors display structured TCR repertoires and composite antigen-driven selection dynamics. Immunology and cell biology 93, 625–633, doi: 10.1038/icb.2015.17 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santamaria P. et al. Beta-cell-cytotoxic CD8+ T cells from nonobese diabetic mice use highly homologous T cell receptor alpha-chain CDR3 sequences. J Immunol 154, 2494–2503 (1995). [PubMed] [Google Scholar]
- Lieberman S. M. et al. Identification of the beta cell antigen targeted by a prevalent population of pathogenic CD8+ T cells in autoimmune diabetes. Proceedings of the National Academy of Sciences of the United States of America 100, 8384–8388, doi: 10.1073/pnas.0932778100 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- DiLorenzo T. P. et al. Major histocompatibility complex class I-restricted T cells are required for all but the end stages of diabetes development in nonobese diabetic mice and use a prevalent T cell receptor alpha chain gene rearrangement. Proceedings of the National Academy of Sciences of the United States of America 95, 12538–12543 (1998). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pathiraja V. et al. Proinsulin-specific, HLA-DQ8, and HLA-DQ8-transdimer-restricted CD4+ T cells infiltrate islets in type 1 diabetes. Diabetes 64, 172–182, doi: 10.2337/db14-0858 (2015). [DOI] [PubMed] [Google Scholar]
- Eugster A. et al. High diversity in the TCR repertoire of GAD65 autoantigen-specific human CD4+ T cells. J Immunol 194, 2531–2538, doi: 10.4049/jimmunol.1403031 (2015). [DOI] [PubMed] [Google Scholar]
- Attaf M., Huseby E. & Sewell A. K. alphabeta T cell receptors as predictors of health and disease. Cellular & molecular immunology 12, 391–399, doi: 10.1038/cmi.2014.134 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stewart-Jones G. B., McMichael A. J., Bell J. I., Stuart D. I. & Jones E. Y. A structural basis for immunodominant human T cell receptor recognition. Nature immunology 4, 657–663, doi: 10.1038/ni942 (2003). [DOI] [PubMed] [Google Scholar]
- Amrani A. et al. Progression of autoimmune diabetes driven by avidity maturation of a T-cell population. Nature 406, 739–742, doi: 10.1038/35021081 (2000). [DOI] [PubMed] [Google Scholar]
- Oeser J. K. et al. Deletion of the G6pc2 gene encoding the islet-specific glucose-6-phosphatase catalytic subunit-related protein does not affect the progression or incidence of type 1 diabetes in NOD/ShiLtJ mice. Diabetes 60, 2922–2927, doi: 10.2337/db11-0220 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daniel D., Gill R. G., Schloot N. & Wegmann D. Epitope specificity, cytokine production profile and diabetogenic activity of insulin-specific T cell clones isolated from NOD mice. European journal of immunology 25, 1056–1062, doi: 10.1002/eji.1830250430 (1995). [DOI] [PubMed] [Google Scholar]
- Abiru N. et al. Dual overlapping peptides recognized by insulin peptide B:9-23 T cell receptor AV13S3 T cell clones of the NOD mouse. Journal of autoimmunity 14, 231–237, doi: 10.1006/jaut.2000.0369 (2000). [DOI] [PubMed] [Google Scholar]
- Simone E. et al. T cell receptor restriction of diabetogenic autoimmune NOD T cells. Proceedings of the National Academy of Sciences of the United States of America 94, 2518–2521 (1997). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cole D. K. et al. Hotspot autoimmune T cell receptor binding underlies pathogen and insulin peptide cross-reactivity. The Journal of clinical investigation 126, 3626, doi: 10.1172/JCI89919 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parello C. S. & Huseby E. S. Indoctrinating T cells to attack pathogens through homeschooling. Trends in immunology 36, 337–343, doi: 10.1016/j.it.2015.04.004 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eugster A. et al. Measuring T cell receptor and T cell gene expression diversity in antigen-responsive human CD4+ T cells. Journal of immunological methods 400–401, 13–22, doi: 10.1016/j.jim.2013.11.003 (2013). [DOI] [PubMed] [Google Scholar]
- Lefranc M. P. et al. IMGT, the international ImMunoGeneTics information system. Nucleic acids research 37, D1006–1012, doi: 10.1093/nar/gkn838 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berthold M. R. et al. KNIME–the Konstanz information miner: version 2.0 and beyond. SIGKDD Explor. Newsl. 1931-0145 11, 26–31 (2009). [Google Scholar]
- Mamedov I. Z. et al. Preparing unbiased T-cell receptor and antibody cDNA libraries for the deep next generation sequencing profiling. Frontiers in immunology 4, 456, doi: 10.3389/fimmu.2013.00456 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Britten C. M. et al. The use of HLA-A*0201-transfected K562 as standard antigen-presenting cells for CD8(+) T lymphocytes in IFN-gamma ELISPOT assays. Journal of immunological methods 259, 95–110 (2002). [DOI] [PubMed] [Google Scholar]
- Martinuzzi E. et al. The frequency and immunodominance of islet-specific CD8+ T-cell responses change after type 1 diabetes diagnosis and treatment. Diabetes 57, 1312–1320, doi: 10.2337/db07-1594 (2008). [DOI] [PubMed] [Google Scholar]
- Fuchs Y. F., Adler K. & Bonifacio E. Beta-cell autoimmunity. Methods Mol Biol 933, 265–274, doi: 10.1007/978-1-62703-068-7_17 (2012). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.