Abstract
Matrix assisted laser desorption/ionization (MALDI) imaging mass spectrometry (IMS) is an increasingly important technique for detection and spatial localization of phospholipids on tissue. Due to the high abundance and being easy-to-ionize of phosphatidylcholine (PC), therefore, selecting matrix to yield signals of other lipids has become the most crucial factor for a successful MALDI-IMS analysis of phospholipids. Herein, graphene oxide (GO) was proposed as a new matrix to selectively enhance the detection of other types of phospholipids that are frequently suppressed by the presence of PC in positive mode. Compared to the commonly used matrix DHB, GO matrix significantly improved signal-to-noise ratios of phospholipids as a result of its high desorption/ionization efficiency for nonpolar compounds. Also, GO afforded homogeneous crystallizations with analytes due to its monolayer structure and good dispersion, resulting in better reproducibility of shot-to-shot (CV < 13%) and spot-to-spot (CV < 14%) analysis. Finally, GO matrix was successfully applied to simultaneous imaging of PC, PE, PS and glycosphingolipid in the mouse brain, with a total of 65 phospholipids identified.
Lipids, especially phospholipids (PLs), are major constituents of cellular membranes and participate in many essential biological functions ranging from signal transduction, energy storage, cell adhesion and migration, metabolism, homeostasis, and apoptosis1,2. Defects in phospholipids metabolism are associated with numerous diseases including atherosclerosis, Alzheimer’s disease, cancer, and Niemann-Pick disease3. Thus, revealing their global distribution in vivo is crucial to disease diagnosis and treatment.
Matrix-assisted laser desorption/ionization imaging mass spectrometry (MALDI-IMS) is a well-established label-free technique that can simultaneously map a broad range of biomolecules in cells and tissues with high sensitivity, specificity and relative quantitative abilities4,5,6,7. In a typical MALDI-IMS workflow, a tissue is cryosectioned into slices, mounted onto a target and subsequently coated with a suitable matrix that extract analytes from the underlying tissue to form co-crystallization. Then specific regions of the tissue sections are scanned by MALDI MS in an array of discrete points and then images are reconstructed to form the mass spectrum charts from those points. So far, MALDI-IMS has proven to be a powerful technique for identification and spatial localization of phospholipids across a variety of biological tissues8, including mouse brain9, mouse liver10, rat heart11, and bovine lens12.
The successful MALDI-IMS analysis of phospholipids largely depends on the choice of matrix. Currently, the most frequently used matrix for phospholipid analysis is 2,5-dihydroxybenzoic acid (DHB)13. However, DHB tends to form heterogeneous crystals resulting in poor reproducibility and low resolution14. Moreover, phosphatidylchlines (PCs) and spingomyelins (SMs) frequently suppress the detection of other lipid classes in a positive mode15. To address this problem, a batch of matrices such as 9-Aminoacridine (9-AA)16, 4-phenyl-α-cyanocinnamic acid amide (Ph-CCA-NH2)17, N-(1-naphthyl)ethlenediamine dihydrochloride (NEDC)18, dithranol (DT)13, quercetin19, hydroxyflacones20, etc., have been demonstrated for profiling and imaging of phospholipids. Due to the high abundance and being easy-to-ionize of PC, it usually required two separate scan in positive and negative mode, separately, which was quite time-consuming and reagent-wasting. Therefore, searching for different matrices for simultaneous analysis of multiple phospholipids by MALDI-IMS remains to be a rather empirical task.
Recently, graphene (G) and graphene oxide (GO), as new members of carbon-based nanomaterials, have attracted considerable attention due to their unique properties, including simple monolayer structure, good thermal and chemical stability as well as excellent electronic properties21,22. In 2010, graphene was first introduced as a matrix for the facile analysis of amino acids, polyamines, anticancer drugs, nucleosides and steroids using MALDI-TOF MS23. Since then, G/GO have been employed as MALDI matrices for the facile analysis of small molecules24,25, proteins/peptides26,27, and polymers28. Compared with classical organic matrices, G/GO show several advantages, such as low background interference, enhanced ionization efficiency of analytes, rare hot-spot phenomenon, etc. In particular, G/GO are found to show a high desorption/ionization efficiency towards nonpolar compounds that are commonly difficult to ionize. This phenomenon would be attributed to the hydrophobic/hydrophobic interaction between nonpolar compounds and G/GO as well as the unique electronic property of G/GO for energy transfer28. Therefore, G/GO are promising matrices for lipid detection and imaging.
In this work, we present successful use of GO as a new matrix for MALDI-IMS analysis of phospholipids. The unique advantage of GO as matrix is that signal-to-noise (S/N) ratios of low-abundance phospholipid classes were selectively improved despite the presence of PC, which could provide a more comprehensive profile of phospholipids spatial distributions in mouse brain sections. In addition, compared to DHB, GO provides a homogeneous deposition on tissue, leading to higher signal intensities and better reproducibility. Finally, we present GO as novel MALDI-IMS matrix for imaging of various lipid classes in mouse brain tissue sections.
Results and Discussion
Evaluation of GO as matrix for MALDI analysis of phospholipids
The characterization of graphene oxide was kindly provided by the manufacturer. As shown in Figure S1–S4, graphene oxide synthesized through Hummer approach has a very small thickness (0.8–1.2 nm), indicating its one-atom-thick property. Typically, solvent and matrix concentration play critical roles in MALDI MS analysis29. Therefore, the preliminary experiment was conducted to optimize the solvent and matrix concentration. We investigated three solvents including H2O, C2H5OH/H2O (50:50 v/v) and ACN/H2O (50:50, v/v) containing 0.1% TFA, and a series of matrix concentrations ranging from 0.1 mg/mL to 10 mg/mL. It was found that GO dissolved in C2H5OH/H2O (50:50, v/v) at the concentration of 2 mg/mL could generate the best spectrum (data not shown).
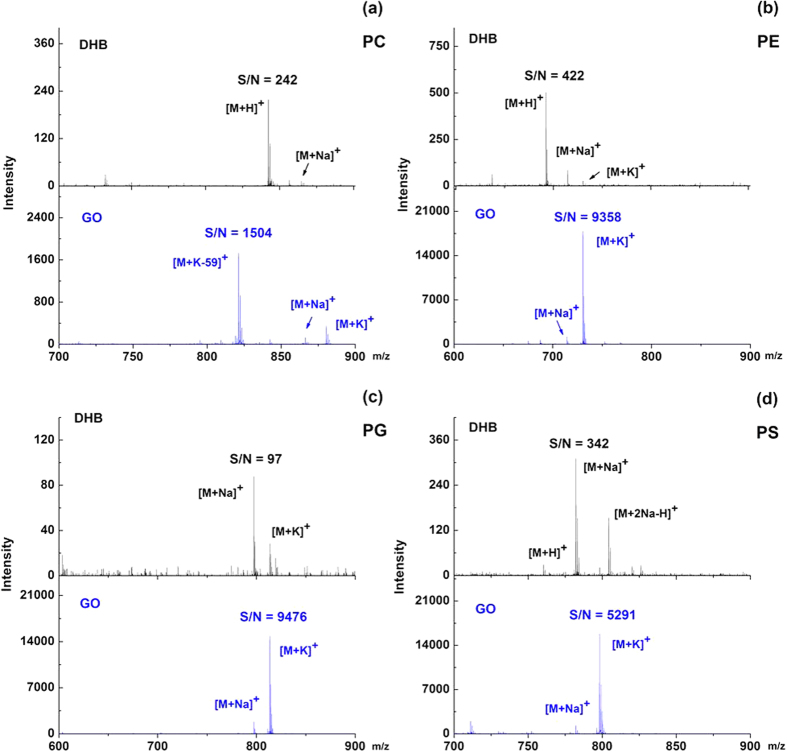
After optimization of the concentration and matrix buffer, GO was investigated as matrix to ionize different phospholipid classes. The main advantage of GO matrix is that it greatly enhanced the S/N ratios of phospholipids. Figure 1 displayed the MALDI mass spectra of four representative phospholipid species including PC (20:1/20:1), PE (16:0/16:0), PG (18:1/18:1) and PS (16:0/18:2) using DHB and GO as matrix, respectively. Compared to DHB matrix (Fig. 1, upper panel), GO matrix showed enhanced ionization (Fig. 1, lower panel) for all four phospholipid species. Specifically, the S/N ratios of PC, PE, PG and PS were enhanced by approximately 6, 22, 97 and 15 fold, respectively. It should be noted that when using DHB matrix, the signals of phospholipids were often observed in several forms of adducts like [M + H]+, [M + Na]+, [M + 2Na-H]+, and [M + K]+. However, GO preferred to generate predominant abundant [M + K]+ ion, which to some extent simplified the mass spectra and facilitated phospholipids identification. PC was generally detected in the form of [M + K-59]+, indicating that PC had lost its quaternary amine group to form a dehydrated diacylglycerol fragment ion. This phenomenon could probably be attributed to the strong interaction between the negative charges of GO and a permanent positive charge of PC’s head group. Subsequently, the limits of detection (LODs) for the most abundant peaks of PC, PE, PG and PS were determined with a dilution series of phospholipids. Compared with 1.8, 2.9, 12.3 and 2.5 pmol using DHB matrix, the LODs for PC, PE, PG and PS were 730, 93, 105 and 75 fmol using GO matrix, indicating the decrease of 2.5, 31, 117 and 33 fold, respectively.
Figure 1.
MALDI-TOF MS spectra of four individual phospholipid species (a) PC (20:1/20:1), (b) PE (16:0/16:0), (c) PG (18:1/18:1) and (d) PS (16:0/18:2) in positive mode using DHB (upper panel) and GO (lower panel) as matrix, respectively. The concentration of each phospholipid was 200 ng/μL.
The second advantage of GO matrix is that it could selectively improve the detection of other phospholipids in positive mode in the presence of PC. PC always suppresses the ionization of other phospholipids in positive mode due to its high abundance in eukaryotic membranes and its quaternary amine group22. A mixed solution containing four standard phospholipid species was used as a model sample to evaluate the ionization capability of GO towards other phospholipids. We blended PC, PE, PG and PS in molar ratios of 8:4:1:2 to mimic the composition of phospholipids in real biological tissues according to the previous literature30. As shown in Fig. 2, when using DHB matrix, the signal of PC predominated the mass spectra, and no peak of PG could be detected (Fig. 2a). By contrast, when using GO matrix, the S/N ratios of PC, PE and PS were increased by 10, 51 and 6 fold, while signals of PG at m/z 797.42 ([M + Na]+) and m/z 813.39 ([M + K]+) were detected with considerable S/N ratios (S/N = 50 and S/N = 172) (Fig. 2b, Table 1). This was probably because that PC’s quaternary amine group was cut off so that it could no longer suppress the ionization of other phospholipid classes, enabling the simultaneous detection of different phospholipid classes in biological samples.
Figure 2.
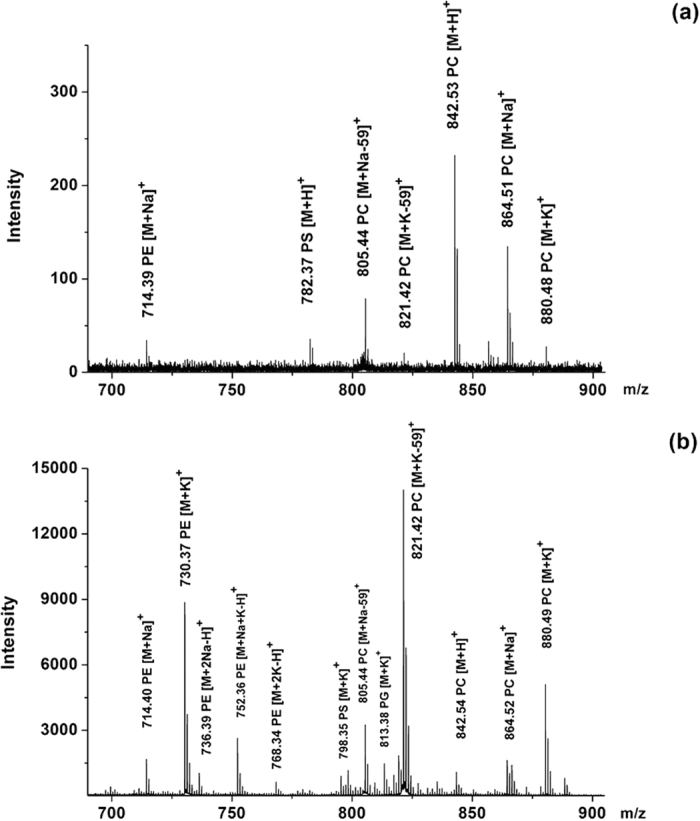
MALDI-TOF MS spectra of a mixture of four standard phospholipid species in positive mode using (a) DHB and (b) GO as matrix. Among the mixture, the molar ratio of PC (20:1/20:1), PE (16:0/16:0), PG (18:1/18:1) and PS (16:0/18:2) was 8:4:1:2.
Table 1. Comparison of DHB and GO matrix for MALDI-TOF MS analysis of a mixture of four standard phospholipid species in positive mode.
| Species | Signal | m/z | S/N ratio | Increase factor (GO/DHB) | |
|---|---|---|---|---|---|
| DHB | GO | ||||
| PC | [M + Na-59]+ | 805.44 | 48 | 367 | 10 |
| [M + K-59]+ | 821.42 | 8 | 1618 | ||
| [M + H]+ | 842.54 | 165 | 26 | ||
| [M + Na]+ | 864.52 | 102 | 190 | ||
| PE | [M + K]+ | 880.49 | 16 | 605 | 51 |
| [M + Na]+ | 714.39 | 20 | 185 | ||
| [M + K]+ | 730.38 | ND | 1027 | ||
| [M + Na + K-H]+ | 752.37 | ND | 287 | ||
| [M + 2K-H]+ | 768.34 | ND | 71 | ||
| PG | [M + Na]+ | 797.42 | ND | 50 | N/A |
| [M + K]+ | 813.39 | ND | 172 | ||
| PS | [M + Na]+ | 782.37 | 22 | 21 | 6 |
| [M + K]+ | 798.35 | ND | 125 | ||
The third advantage of GO matrix is that it could improve the signal reproducibility of phospholipids. As previously reported, DHB suffered from forming heterogeneous crystallization with analytes (Figure S5a), leading to wide variations of signal intensity (Figure S6 and S7). In contrast, as shown in Figure S5b, more uniform crystals were obtained by using GO matrix, indicating better reproducibility of signal intensity. For further confirmation, shot-to-shot experiment was conducted by acquiring 10 mass spectra in a single spot and spot-to-spot experiment was performed by acquiring 10 mass spectra in 10 spots. As illustrated in Figure S6 and S7, the results showed that GO produced much better shot-to-shot (CV < 13%) and spot-to-spot (CV < 14%) reproducibility for phospholipid analysis than DHB matrix.
Direct analysis of phospholipids in mouse brain tissue sections using GO
In order to further investigate the potential of GO for improved phospholipid imaging, mouse brain tissue sections were analysed directly. For comparison, DHB was used as a reference matrix. The LIPID MAPS DATABASE was used for a preliminary structure determination. As shown in Fig. 3, GO matrix showed significant advantages over DHB matrix in terms of sensitivity, selectivity and reproducibility.
Figure 3.
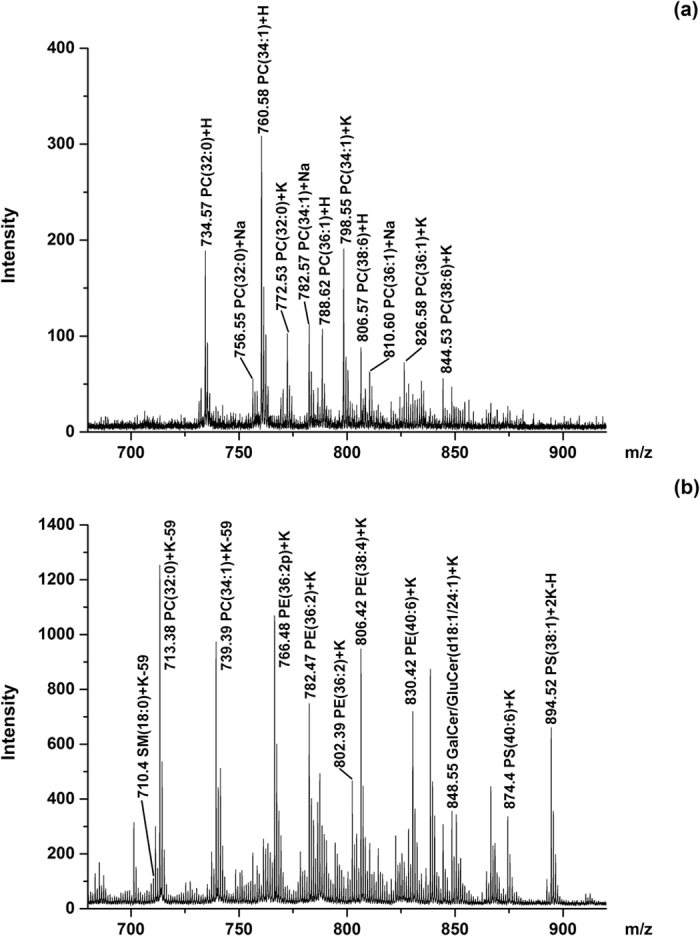
MALDI-TOF MS spectra of lipids in mouse brain tissue sections in positive mode using (a) DHB and (b) GO as matrix.
DHB matrix only generated peaks of PC and a few of SM (Fig. 3a), which was in agreement with previous reports31. In contrast, GO provided peaks of PC, PE, PS, SM, and glycosylsphingolipids (Fig. 3b). We detected a total of 65 lipid species using GO as matrix whereas only 13 lipid species were identified using DHB matrix. The detected phospholipids was listed in Table S1 (GO) and S2 (DHB). When using DHB matrix, protonated, sodiated and potassium adducts were detected. In contrast, when using GO matrix, the phospholipid species were primarily detected in the form of potassium adducts, which was in agreement with previous experiments.
Within all the identified PL groups using GO matrix, PC and PE were prominent peaks, since they were the two most abundant classes among phospholipid pools. Some important PC species such as PC(32:0), PC(34:1), and PC(36:1) generated peaks were detected with at least 3 times higher intensities when using GO matrix than using DHB matrix. Consistent with the literature previously reported,31 m/z values of the most prominent peaks corresponded to PC isoforms {m/z 713.4, [PC(32:0) + K-59]+; and m/z 739.4, [PC(34:1) + K-59]+} and PE isoforms {m/z 766.5, [PE(36:2p) + K]+; and m/z 806.4, [PE(38:4) + K]+} (Fig. 3b). All of the four peaks showed similar intensities, indicating that PE was not suppressed by PC in the extracts. The enhanced selectivity and sensitivity led to an increase in the total number of lipid species that could be detected and mapped.
Besides PC and PE, peaks corresponding to other PL classes such as SM species {m/z 731.5, [SM(d18:1/18:0) + K-59]+} and PS species {m/z 874.4, [PS(40:6) + K]+; and m/z 828.4, [PS(36:1) + K]+} were also observed, although with relatively weak intensities due to their relatively low abundance. Peaks corresponded to glycosphingolipid species also showed considerable intensities, including m/z 822.5, [GalCer/GluCer(d18:1/22:0) + K]+; m/z 848.6, [GalCer/GluCer(d18:1/24:1) + K]+; m/z 850.6, [GalCer/GluCer(d18:1/24:0) + K]+; and their derivatives32.
Characterization of phospholipids by tandem mass spectrometry
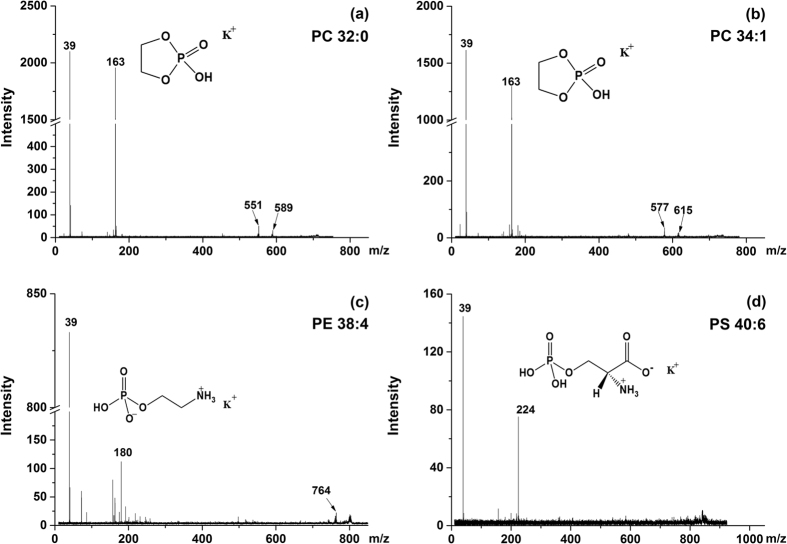
The above detected phospholipid identities were then further confirmed by in situ MALDI MS/MS as well as ESI MS/MS. Figure 4 showed the MS/MS spectra of for representative phospholipid species. For m/z 713.4, [PC(32:0) + K-59]+ and m/z 739.4, [PC(32:0) + K-59]+, m/z 163 was the diagnostic fragment of PC head group, with the quaternary amine group lost, while m/z 551 and m/z 577, along with their potassium adduct m/z 589 and m/z 615 were assigned to neutral loss of cyclophosphate, respectively (Fig. 4a and b). For m/z 806.4, [PE(38:4) + K]+, m/z 180 was identical diagnostic fragment of PE head group and m/z 764 was assigned to neutral loss of vinly amine (Fig. 4c). For m/z 874.4, [PS(40:6) + K]+, m/z 224 referred to the potassium adduct of PS head group (Fig. 4d).
Figure 4.
in situ MS/MS spectra of [M + K]+ ions of some typical phospholipids: (a) m/z 713.4, [PC(32:0) + K-59]+; (b) m/z 739.4, [PC(34:1) + K-59]+; (c) m/z 806.4, [PE(38:4) + K]+ and (d) m/z 874.4, [PS(40:6) + K]+.
To further confirm the presence of all lipids detected with GO matrix, crude extract of mouse brain was analysed by ESI-QqQ-MS/MS in a series of precursor ions (PI) and neutral loss (NL) scanning experiments (Figure S8). Structural confirmation of the lipids was also conducted by querying the literature previously reported33.
Imaging of phospholipids in mouse brain tissue section
Traditional phospholipid IMS analysis usually required two separate scans in both positive and negative mode since PC suppressed the signals of other phospholipid classes. Seeing that GO could selectively enhance the detection of other phospholipids in positive mode in the presence of PC, we attempted to map multiple phospholipid classes in a single positive scan. In this way, mouse brain tissue section were selected to illustrate the advantages of GO as matrix in phospholipid IMS analysis. GO solution [0.2 mg/mL in C2H5OH/H2O (50:50), v/v] was uniformly deposited onto the mouse brain tissue section by a TM-Sprayer. Herein, a more diluted matrix solution was used in case that the matrix solution could block the pipelines. The laser spot diameter was set to 50 μm. Therefore, a lateral resolution of 60 μm was achieved with no ablation overlap observed.
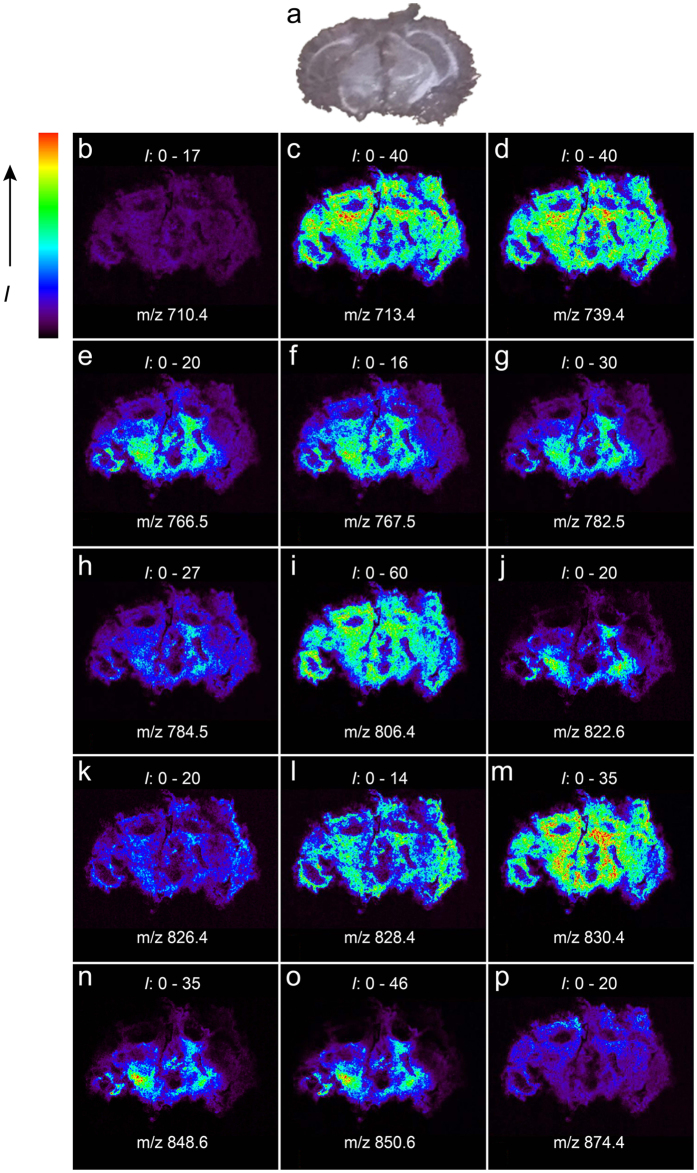
Figure 5(b–p) presented the IMS results from a mouse brain coronal section coated with GO matrix in the positive mode, which showed an excellent correlation with the corresponding optical image (Fig. 5a). Then we investigated some specific distribution patterns for PC, PE, SM, PS and glycosphingolipid species. For PC species, the ions at m/z 713.4, [PC(32:0) + K-59]+ and m/z 739.4, [PC(34:1) + K-59]+ appeared in the MALDI MS images of mouse brain with similar intensities in regions of cerebral cortex and thalamus, while the intensity of ion at m/z 767.5, [PC(36:1) + K-59]+ was greater in regions of thalamus than that in cerebral cortex. For PS species, the distribution of the ion at m/z 826.4, [PS(36:2) + K]+ showed a brighter depiction in regions of thalamus than in cerebral cortex, while the ion at m/z 874.4, [PS(40:6) + K]+ was mainly detected in the cerebral cortex. For glycospingolipid species, m/z 848.6, [GalCer/GluCer(d18:1/24:1) + K]+ were mainly detected in the cerebral peduncle. These results were basically consistent with the previous literatures reported34,35. Compared to traditional matrices, GO not only showed greatly improved sensitivity and selectivity, but also obtained simultaneous IMS analysis of multiple phospholipid classes in a single positive scan, thereby simplifying the workflow of IMS.
Figure 5.
Optical image (a) and MALDI-MS images (b–p) of a coronal mouse brain tissue section. Intensity (I) range is given for each ion image. (b) m/z 710.4, [SM(18:0) + K-59]+; (c) m/z 713.4 [PC(32:0) + K-59]+; (d) m/z 739.4 [PC(34:1) + K-59]+; (e) m/z 766.5 [PE(36:2p) + K]+; (f) m/z 767.5 [PC(36:1) + K-59]+; (g) m/z 782.5 [PE(36:2) + K]+; (h) m/z 784.5 [PE(36:1) + K]+; (i) m/z 806.4 [PE(38:4) + K]+; (j) m/z 822.6 [GalCer/GluCer(d18:1/22:0) + K]+; (k) m/z 826.4 [PS(36:2) + K]+; (l) m/z 828.4 [PS(36:1) + K]+; (m) m/z 830.4 [PE(40:6) + K]+; (n) m/z 848.6 [GalCer/GluCer(d18:1/24:1) + K]+; (o) m/z 850.6 [GalCer/GluCer(d18:1/24:0) + K]+; (p) m/z 874.4 [PS(40:6) + K]+.
Conclusions
In this study, GO has been successfully demonstrated to be a novel matrix to improve MALDI-IMS analysis of phospholipids. Compared with conventional DHB matrix, GO showed greater advantages on improved sensitivity, selectivity and reproducibility. In a single positive mode scan, a total of 65 lipid species could be successfully identified using GO in the mouse brain section, including PC, PE, PS, SM and glycosphingolipid species. Additionally, ion images of lipid classes above were also simultaneously mapped in the same scan, facilitating rapid screening of phospholipid biomarker candidates. Compared with previous study, which typically requires two separate scan in both positive and negative mode using other matrices such as binary matrix of CHCA and DHB, silver nanoparticles and 1, 5-diaminonaphthalene (DAN), using GO as matrix is time-saving, sample-saving and reagent-saving. Finally, when it comes to the cost of sample preparation, it requires approximately 32 μg GO (0.007 USD) for a given mouse brain section (about 0.5 cm2), which is less than 160 μg DHB (0.094 USD); 55 μg DAN (0.025 USD). In conclusion, it is believed that GO has great potential to be applied to simultaneous MALDI-IMS analysis of multiple and low abundant phospholipids in positive mode across various biological tissue sections.
Methods
Chemicals and Reagents
Phosphatidylcholine (PC) (20:1/20:1) and the sodium salt of phosphatidylserine (PS) (16:0/18:2) were purchased from Avanti Polar Lipid, Inc. (Alabaster, AL, USA). Phosphatidylethanolamine (PE) (16:0/16:0) and the sodium salt of phosphatidyglycerol (PG) (18:1/18:1) were purchased from Shanghai A.V.T. Pharmaceutical, Ltd. (Shanghai, China). Graphene oxide (GO) powder was obtained from Nanjing XFNANO Materials Tech Co., Ltd. 2,5-Dihydroxybenzoic acid (DHB) and trifluoroacetic acid (TFA) were obtained from Sigma-Aldrich (St. Louis, MO, USA). Methanol and acetonitrile (ACN) were HPLC grade. Ethanol and chloroform were analytical grade. Distilled water was purified by a Milli-Q system (Milford, MA, USA).
Sample preparation
Each phospholipid was dissolved in chloroform/methanol (50:50, v/v) at the concentration of 1.0 mg/mL as storage solution and diluted to a proper concentration as needed. All the standard solutions were kept at −20 °C for further use. DHB matrix was dissolved in ACN/H2O (50:50, v/v) containing 0.1% TFA at the concentration of 10 mg/mL. 2.0 mg GO was dispersed in a 1 mL solution of C2H5OH/H2O (50:50, v/v), and then sonicated for 5 min prior to use. 1 μL GO suspension and 1 μL sample solution were mixed, applied onto the MALDI target and then dried in the air for further analysis by MALDI-TOF MS.
Tissue dissection and matrix application
Adult male C57/BL6 mice were purchased from department of laboratory animal science (Fudan University) and were sacrificed by cervical dislocation. The brain was excised from the mouse immediately. Fresh frozen brain tissue was cut into 10 μm thickness sections using a Leica cryostat (Leica CM 3050 S, Leica Microsystems Inc, Wetzlar, Germany) at −20 °C. The serial slices were then thaw-mounted onto glass slides and dried in a desiccator. The tissue sections were coated with DHB matrix and GO using TM-SprayerTM (HTX Technologies). A Nikon scanner (Coolscan VED, Nikon, Tokyo, Japan) was used for optical scanning. All animal experiments were conducted in strict accordance with the National Institute of Health Guide for the Care and Use of Laboratory Animals and were approved by the Experimental Animal Ethics Committee of Shanghai Medical College, Fudan University.
MALDI MS analysis and data processing
All MALDI MS and MS/MS experiments were performed in positive ion reflection mode on a 5800 Proteomics Analyzer (Applied Biosystems, Framingham, MA, USA) equipped with a Nd:YAG laser (355 nm), a repetition rate of 400 Hz, and an acceleration voltage of 20 kV. Laser energy was set to 3500 and 3000 for DHB matrix and GO matrix, respectively. 1000 shots were accumulated for each spectrum. For tissue imaging, the laser step size was 60 μm. Assignment of detected lipids was based on the LIPID MAPS prediction tool (http://www.lipidmaps.org/tools/index.html), the literatures and MS/MS experiments. During the database search, the [M + H]+, [M + Na]+ and [M + K]+ ions were considered, with a mass tolerance of ±0.1 Da. Two-dimensional ion density maps were created using the image reconstruction software (BioMap, Novartis, Basel, Switzerland).
Lipid extraction and ESI QqQ-MS/MS Measurements
40 mg C57/BL6 mouse brain tissue aliquots were homogenized with a mortar and a pestle in liquid nitrogen. Next, 5 mL chloroform/methanol/formic acid (10:10:1, v/v/v) was added to each tube, and the extraction process was performed at −20 °C overnight. 2.2 mL buffer A (0.2 M H3PO4, 1 M KCl) was added into each tube and the tubes were then centrifuged (3000 rpm, 4 °C, 5 min) in a 5810 R centrifuge (Eppendorf, Germany). The lower organic phase was collected, lyophilized, re-dissolved with 1 mL isopropanol/hexane/100 mM ammonium acetate(aq) (50:48:2, v/v/v), and centrifuged (14,000 rpm, 4 °C, 40 min) in a Beckman Coulter Allegra 64 R centrifuge (Brea, CA). The supernatants were collected and stored at 4 °C until used.
For ESI QqQ-MS/MS measurements, all scans were performed on an API 4000 QTRAP tandem mass spectrometer (AB SCIEX). The details of the ESI QqQ-MS/MS method are described in the Supporting Information.
Additional Information
How to cite this article: Wang, Z. et al. Improved MALDI imaging MS analysis of phospholipids using graphene oxide as new matrix. Sci. Rep. 7, 44466; doi: 10.1038/srep44466 (2017).
Publisher's note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Material
Acknowledgments
The work was supported by the NST (Grants 2016YFA0501303), NSF (Grants 21335002 and 21675031), the Ph.D. Programs Foundation of Ministry of Education of China (20130071110034) and Shanghai Projects (Eastern Scholar, Shanghai Rising-star 15QA1400600, 15JC1400700 and B109).
Footnotes
The authors declare no competing financial interests.
Author Contributions Z.W. and Y.C. performed the experiments and wrote the manuscript. Y.W. and X.Z. helped to analyze data. Y.Z. and H.L. contributed to study design and overall supervision. All authors discussed the results and commented on the manuscript.
References
- Shevchenko A. & Simons K. Lipidomics: coming to grips with lipid diversity. Nat. Rev. Mol. Cell Biol. 11, 593–598 (2010). [DOI] [PubMed] [Google Scholar]
- Brown H. A. & Murphy R. C., Working towards an exegesis for lipids in biology. Nat. Chem. Biol. 5, 602–606 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maxfield F. R. & Tabas I. Role of cholesterol and lipid organization in disease. Nature 438, 612–621 (2005). [DOI] [PubMed] [Google Scholar]
- Berry K. A. et al. MALDI imaging of lipid biochemistry in tissues by mass spectrometry. Chem. Rev. 111, 6491–6512 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chughtai K. & Heeren R. M. Mass spectrometric imaging for biomedical tissue analysis. Chem. Rev. 110, 3237–3277 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen S. et al. Mass spectrometry imaging reveals the sub-organ distribution of carbon nanomaterials. Nat. Nanotechnol. 10, 176–182 (2015). [DOI] [PubMed] [Google Scholar]
- Zhang Y. et al. Combination of ESI and MALDI mass spectrometry for qualitative, semi-quantitative and in situ analysis of gangliosides in brain. Sci. Rep. 6, 25289, doi: 10.1038/srep25289 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gode D. & Volmer D. A. Lipid imaging by mass spectrometry - a review. Analyst 138, 1289–1315 (2013). [DOI] [PubMed] [Google Scholar]
- Hong J. H. et al. Global changes of phospholipids identified by MALDI imaging mass spectrometry in a mouse model of Alzheimer’s disease. J. Lipid Res. 57, 36–45 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shrivas K. et al. Ionic matrix for enhanced MALDI imaging mass spectrometry for identification of phospholipids in mouse liver and cerebellum tissue sections. Anal. Chem. 82, 8800–8806 (2010). [DOI] [PubMed] [Google Scholar]
- Jackson S. N. et al. Imaging of lipids in rat heart by MALDI-MS with silver nanoparticles. Anal. Bioanal. Chem. 406, 1377–1386 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le C. H., Han J. & Borchers C. H. Dithranol as a MALDI matrix for tissue imaging of lipids by Fourier transform ion cyclotron resonance mass spectrometry. Anal. Chem. 84, 8391–8398 (2012). [DOI] [PubMed] [Google Scholar]
- Wang H. Y., Jackson S. N., Post J. & Woods A. S. A Minimalist Approach to MALDI Imaging of Glycerophospholipids and Sphingolipids in Rat Brain Sections. Int. J. Mass Spectrom. 278, 143–149 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stauber J. et al. MALDI imaging of formalin-fixed paraffin-embedded tissues: application to model animals of Parkinson disease for biomarker hunting. J. Proteome Res. 7, 969–978 (2008). [DOI] [PubMed] [Google Scholar]
- Schiller J., Süss R., Petković M., Zschörnig O. & Arnold K. Negative-ion matrix-assisted laser desorption and ionization time-of-flight mass spectra of complex phospholipid mixtures in the presence of phosphatidylcholine: a cautionary note on peak assignment. Anal. Biochem. 309, 311–314 (2002). [DOI] [PubMed] [Google Scholar]
- Cerruti C. D., Benabdellah F., Laprevote O., Touboul D. & Brunelle A. MALDI imaging and structural analysis of rat brain lipid negative ions with 9-aminoacridine matrix. Anal. Chem. 84, 2164–2171 (2012). [DOI] [PubMed] [Google Scholar]
- Fülöp A. et al. 4-Phenyl-α-cyanocinnamic acid amide: screening for a negative ion matrix for MALDI-MS imaging of multiple lipid classes. Anal. Chem. 85, 9156–9163 (2013). [DOI] [PubMed] [Google Scholar]
- Wang J. et al. MALDI-TOF MS imaging of metabolites with a N-(1-naphthyl) ethylenediamine dihydro-chloride matrix and its application to colorectal cancer liver metastasis. Anal. Chem. 87, 422–430 (2015). [DOI] [PubMed] [Google Scholar]
- Wang X., Han J., Pan J. & Borchers C. H. Comprehensive imaging of porcine adrenal gland lipids by MALDI-FTMS using quercetin as a matrix. Anal. Chem. 86, 638–646 (2014). [DOI] [PubMed] [Google Scholar]
- Wang X. et al. Hydroxyflavones as a new family of matrices for MALDI tissue imaging. Anal. Chem. 85, 7566–7573 (2013). [DOI] [PubMed] [Google Scholar]
- Novoselov K. S. et al. Electric field effect in atomically thin carbon films. Science 306, 666–669 (2004). [DOI] [PubMed] [Google Scholar]
- Rao C. N., Sood A. K., Subrahmanyam K. S. & Govindaraj A. Graphene: the new two-dimensional nanomaterial. Angew. Chem. Int. Ed. 48, 7752–7777 (2009). [DOI] [PubMed] [Google Scholar]
- Dong X., Cheng J., Li J. & Wang Y. Graphene as a novel matrix for the analysis of small molecules by MALDI-TOF MS. Anal. Chem. 82, 6208–6214 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y. et al. High throughput identification of components from traditional Chinese medicine herbs by utilizing graphene or graphene oxide as MALDI-TOF-MS matrix. J. Mass Spectrom. 46, 804–815 (2011). [DOI] [PubMed] [Google Scholar]
- Zhang J. et al. Magnetic graphene composites as both an adsorbent for sample enrichment and a MALDI-TOF MS matrix for the detection of nitropolycyclic aromatic hydrocarbons in PM2.5. Analyst 140, 1711–1716 (2015). [DOI] [PubMed] [Google Scholar]
- Liu Q. et al. Preparation of graphene-encapsulated magnetic microspheres for protein/peptide enrichment and MALDI-TOF MS analysis. Chem. Commun. 48, 1874–1876 (2012). [DOI] [PubMed] [Google Scholar]
- Friesen W. L. et al. Two-dimensional graphene as a matrix for MALDI imaging mass spectrometry. J. Am. Soc. Mass Spectrom 26, 1963–1966 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu M., Lai Y., Chen G. & Cai Z. Laser desorption/ionization on the layer of graphene nanoparticles coupled with mass spectrometry for characterization of polymers. Chem. Commun. 47, 12807–12809 (2011). [DOI] [PubMed] [Google Scholar]
- Schwartz S. A., Reyzer M. L. & Caprioli R. M. Direct tissue analysis using matrix-assisted laser desorption/ionization mass spectrometry: practical aspects of sample preparation. J. Mass Spectrom. 38, 699–708 (2003). [DOI] [PubMed] [Google Scholar]
- van Meer G., Voelker D. R. & Feigenson G. W. Membrane lipids: where they are and how they behave. Nat. Rev. Mol. Cell Biol. 9, 112–124 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang J. H. & Caprioli R. M. Matrix precoated targets for direct lipid analysis and imaging of tissue. Anal. Chem. 85, 2907–2912 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan R. B. et al. Comparative lipidomic analysis of mouse and human brain with Alzheimer disease. J. Biol. Chem. 287, 2678–2688 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han X. & Gross R. W. Shotgun lipidomics: electrospray ionization mass spectrometric analysis and quantitation of cellular lipidomes directly from crude extracts of biological samples. Mass Spectrom Rev. 24, 367–412 (2005). [DOI] [PubMed] [Google Scholar]
- Hankin J. A. & Murphy R. C. Relationship between MALDI IMS intensity and measured quantity of selected phospholipids in rat brain sections. Anal. Chem. 82, 8476–8484 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Angel P. M., Spraggins J. M., Baldwin H. S. & Caprioli R. Enhanced sensitivity for high spatial resolution lipid analysis by negative ion mode matrix assisted laser desorption ionization imaging mass spectrometry. Anal. Chem. 84, 1557–1564 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.