Abstract
Chains of inorganic polyphosphate (poly-P) with hundreds of Pi residues linked by phosphoanhydride bonds, as in ATP, are found in every bacterial, fungal, plant, and animal cell, in which they perform various functions. In the spore-forming Bacillus cereus, we have identified three principal enzymes and genes involved in the metabolism of poly-P, namely, (i) poly-P kinase (PPK), which synthesizes poly-P reversibly from ATP, (ii) exopolyphosphatase (PPX), which hydrolyzes poly-P to Pi, and (iii) poly-P/AMP phosphotransferase (PAP), which uses poly-P as a donor to convert AMP to ADP, reversibly. In the null mutant of ppk, poly-P levels are reduced to <5% of the WT; in the ppx mutant, the PPK activity is elevated 10-fold, and the accumulation of poly-P is elevated ≈1,000-fold. All of the null mutants of ppk, ppx, and pap showed defects in motility and biofilm formation, but sporulation efficiency was impaired only in the ppx mutant. These enzymes and genes in B. cereus are nearly identical to those in the very closely related pathogen Bacillus anthracis, and, thus, they may provide attractive targets for the treatment of anthrax.
Keywords: polyphosphate kinase, exopolyphosphatase, polyphosphate/AMP phosphotransferase
Inorganic polyphosphate (poly-P) is a polymer of tens or hundreds of Pi residues linked by high-energy phosphoanhydride bonds. Poly-P is ubiquitous, having been found in all organisms (archaea, bacteria, fungi, plants, insects, and mammals) (1–3). Several poly-P-metabolizing enzymes have been purified and characterized, and the corresponding genes have been cloned and deleted (3–8). The enzyme that is primarily responsible for reversible poly-P synthesis in Escherichia coli is poly-P kinase 1 (PPK1), which catalyzes the polymerization of the γ phosphate of ATP into a poly-P chain (5). Poly-P can also be hydrolyzed to Pi either by exopolyphosphatase (PPX) (6) or by endopolyphosphatase (PPN) (7, 8). In E. coli, the genes for PPK1 and PPX are under the control of a common promoter. The inability to accumulate poly-P upon deletion of the ppkppx operon has produced several striking phenotypes in E. coli, including the loss of long-term survival in stationary phase; increased sensitivity to oxidative, osmotic, and thermal stress; and defects in adaptive growth in minimal medium after a shift from a rich medium (9–12). The ppk1 knockout mutant of Pseudomonas aeruginosa, PAO1, shows a dramatic deficiency in motility, an inability to form biofilms, and a loss of virulence (13). Also, poly-P stimulates the kinase activity of mTOR, which is the mammalian target of rapamycin that is involved in the proliferation of mammary cells (14).
Bacillus cereus, the spore-forming Gram-positive Bacillus found commonly in soil and many other sources, is an opportunistic pathogen that causes food poisoning manifested by diarrhoeal or emetic syndromes. It is related closely to the animal and human pathogen Bacillus anthracis (a potential biological weapon) and the insect pathogen Bacillus thuringiensis (a pesticide) (15). Recent availability of the genome sequences of B. cereus 14579 (American Type Culture Collection) and B. anthracis A2012 have revealed a large core set of genes (75–80%) conserved (with 80–100% identity) between these two Bacilli species, indicating a common ancestor (16, 17).
Genes that are highly homologous to E. coli ppk1 and ppx and to P. aeruginosa pap have been found in B. cereus. In this study, these homologues were cloned and overexpressed in E. coli, and they were confirmed in their enzymatic activities. Each of the ppk, ppx, and pap genes was knocked out by deletion mutagenesis, and the phenotypes of these mutants are reported here.
Materials and Methods
Reagents. The following reagents were used: ATP, creatine kinase, DNase I, and RNase A (Roche Molecular Biochemicals); [γ-32P]ATP and [γ-32P]Pi (Amersham Biosciences); polyethyleneimine-cellulose F TLC plates (Merck); restriction enzymes (New England Biolabs); and common chemicals (Sigma).
Strains and Plasmids. B. cereus 14579 was obtained from the American Type Culture Collection; B. cereus suicide vector pSPUC was a gift from Jo Handelsman (University of Wisconsin, Madison); B. cereus/E. coli shuttle vector pHT304 was kindly provided by Didier Lereclus (Institut National de la Recherche Agronomique, Paris). The full list of strains and plasmids is given in Table 1, which is published as supporting information on the PNAS web site.
Media and Growth Conditions. Cultures were grown in LB or brain–heart infusion (BHI) media; CCY medium (casein hydrolysate/yeast-containing medium) (18) was used to assay sporulation. For poly-P accumulation, a Mops-buffered minimal medium contained glucose (4 mg/ml) as a carbon source and K2HPO4 as the Pi source. The low-Pi medium contained 0.1 mM Pi, and the high-Pi medium contained 2 mM Pi (11). Media were supplemented with ampicillin (250 μg/ml), kanamycin (250 μg/ml), spectinomycin (300 μg/ml), tetracycline (10 μg/ml), chlorophenicol (10 μg/ml), or erythromycin (10 μg/ml) for selection of antibiotic-resistant transformants of B. cereus.
Plasmid Construction. B. cereus ppk, ppx, and pap coding sequences were obtained by PCR using genomic DNA as a template. Each pair of primers yielded a 2.1-kb (for ppk), 1.6-kb (for ppx), or 0.8-kb (for pap) PCR product. For expression in E. coli, the PCR fragments were digested with NcoI and BamHI and cloned into pTrc99A predigested with the same restriction enzymes. The resulting plasmids were designated as pTrc-BcPPK, pTrc-BcPPX, and pTrc-BcPAP, respectively.
For complementation of null mutants, a low-copy-number plasmid, pHT304, was used (19). The PCR products were digested with BamHI and HindIII and cloned into pHT304. The sequences of the primers used are given in Table 2, which is published as supporting information on the PNAS web site.
Construction of the B. cereus ppk, ppx, and pap Null Mutants. Null mutant alleles of ppk, ppx, or pap were obtained by using a two-step PCR strategy. A 0.9-kb 5′-flanking region and a 0.9-kb 3′-flanking region of ppk were obtained by PCR using the following primers: K1 (with a BamHI restriction site at the 5′ end) and K2 for the upstream region of ppk; and K3 and K4 (with a HindIII site at the 5′ end) for downstream of ppk. A tetracycline-resistance gene was also amplified from vector pDG1515 with primers K5 and K6. The 5′ of K5 has a tail on it which is complementary to the 3′ sequence of upstream region of ppk, and similarly, the 5′ of primer K6 is complementary to the 5′ sequence downstream of ppk. The three PCR products were then mixed together and used as templates for the second round of PCR. By using K1 and K4 as primers, the second-round PCR product (ppk::Tet fragment), which has 5′- and 3′-flanking regions of ppk and an insertion of the tet resistance gene instead of the ORF of ppk, was amplified. This sequence was then digested with BamHI/HindIII and ligated into the B. cereus suicide vector pSPUC, which was precut with the same restriction enzymes. The plasmid constructed as described for allelic displacement of native ppk in the B. cereus genome was designated as pSPUC-PPK-KO. The plasmids for allelic displacement of native ppx and pap, designated as pSPUC-PPX-KO and pSPUC-PAP-KO, were constructed by following the same procedure. The sequences of the primers that we used are given in Table 2. DNA fragments were sequenced by the Protein and Nucleic Acid (PAN) Facility at Stanford University.
pSPUC-derivative plasmids for making the knockouts of ppk, ppx, and pap were transformed into B. cereus by electroporation (20). Transformants of single crossovers were selected on tet/spectinomycin plates. These transformants were then grown in tet/LB medium for 4–6 days with regular subculturing to allow for the excision and loss of the plasmid. Cultures were properly diluted and plated onto tet/TSA plates, and the knockout mutants (double-crossover recombinants) were obtained as tetracycline-resistant but streptomycin-sensitive colonies. Chromosomal DNA was isolated from selected mutants, and PCR analysis was performed to confirm the replacements of the WT genes by double-homologous recombination events in the ppk, ppx, and pap loci. The mutants were further verified by enzymatic assays of PPK, PPX, or poly-P/AMP phosphotransferase (PAP).
Biochemical Assays. Poly-P was extracted and measured by both radioactive and nonradioactive methods (9, 12). PPK, PPX, and PAP enzyme activities were assayed as described in refs. 5 and 6.
Surface-Attachment Assay. Cells grown overnight at 30°C in LB were diluted 1:100 into 200 μl of LB in 96-well polystyrene plates (Falcon 3911 Microtest III flexible assay plate, Dickinson, Oxnard, CA) and incubated at 30°C without shaking for 6–18 h. Biofilm initiation was monitored and quantified by microtiterplate assay, as described (21).
Motility Assays. Media for swimming and swarming assays were LB or BHI containing 0.3% (wt/vol) agarose (GIBCO/BRL) or 0.5% (wt/vol) Bacto-agar (Difco). Plates were inoculated with bacteria from an overnight LB culture on agar plates with a sterile toothpick and incubated at 30°C for 8–10 h for swimming assays and 12–16 h for swarming assays (13).
Sporulation Assay. Cultures were grown in CCY sporulation medium (19). Samples were taken at intervals at 6–36 h after inoculation, diluted in PPM buffer (10 mM phosphate saline, pH 7.4/50 mM KCl/1 mM MgSO4) and plated on LB agar before and after heat treatment at 70°C for 15 min. The sporulation efficiency was the proportion of the population that survived the heat treatment that was counted as colonies the next day (19).
Phenotype MicroArray Tests. Phenotype MicroArray (Biolog, Hayward, CA) analysis was performed. Two independent mutants with mutations in each gene of ppk, ppx, and pap were examined. Mutants were compared with the isogenic control strain.
Results
The ppk, ppx, and pap Genes in B. cereus. The genes for the poly-P-metabolizing enzymes PPK, PPX, and PAP in B. cereus 14579 (18) are highly homologous to those in E. coli and P. aeruginosa. In B. cereus, the ppk and ppx genes are adjacent (110 bp apart); pap is remote from ppk and ppx. The sequences homologous to ppk, ppx, or pap were amplified by PCR, cloned, and overexpressed in the E. coli ppk–ppx– mutant CF5802; enzyme assays confirmed the identity of these enzymes (data not shown).
The deduced amino acid sequence of B. cereus PPK, with 702 aa and a calculated molecular mass of 80.8 kDa, is 33% identical and 55% similar to that of E. coli and 42% identical and 61% similar to that of P. aeruginosa PPK1. The deduced amino acid sequence of B. cereus PPX, with 512 residues and a calculated molecular mass of 59.1 kDa, is 24% identical and 42% similar to that of E. coli PPX. The B. cereus PAP, to which no homologue has been found in E. coli, is presumed to be 267 aa in length with a molecular mass of 32.2 kDa and 51% identical and 69% similar to PAP of P. aeruginosa.
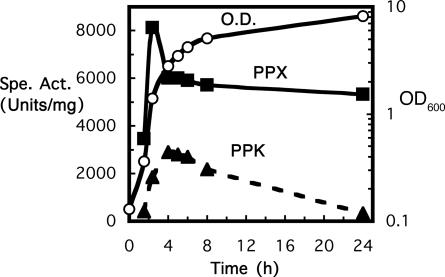
Purification and Characterization of PPK. PPK-assay reaction mixtures that omitted salt and included poly-P (2 mM poly-P type 15; Sigma) raised the PPK activity in the cell lysates 10-fold to 3,000 units/mg, which is a level that is comparable with that of PPK1 in E. coli and P. aeruginosa. Expressions of PPK and PPX in B. cereus are growth-phase-dependent (Fig. 1). During vegetative growth, PPK reached its maximum in the early stationary phase and then decreased; PPX peaked earlier and then dropped in the exponential phase, after which it remained stable.
Fig. 1.
Growth-phase dependency of PPK and PPX in B. cereus. With a 1% inoculum of an overnight culture, B. cereus was grown in LB media at 37°C with vigorous shaking. At the indicated time points (corresponding to middle-log, late-log, early-stationary, and late-stationary phases), cells were harvested, suspended in TED buffer (50 mM Tris·HCl, pH 7.4/1 mM EDTA/1 mM DTT), and treated with lysozyme and sonication to give crude lysates for enzyme assays (see Materials and Methods).
B. cereus PPK is unique among the bacterial PPKs in its low pI value of 6 (compared with 8 in the other bacterial PPKs), lability at low and at high pH, and low affinity to heparin and phosphocellulose columns. PPK was purified 500-fold from lysates of the B. cereus Δppx mutant by lack of adsorption to heparin and phosphocellulose and by chromatography on DEAE–Sepharose. The specific activity is 2 × 106 units/mg protein, which is a value that is comparable with the PPK1 of E. coli and P. aeruginosa. The poly-P-synthesizing reaction of B. cereus PPK is processive, producing poly-P with a uniform length (≈700), similar to the product of E. coli PPK1 (Fig. 2).
Fig. 2.
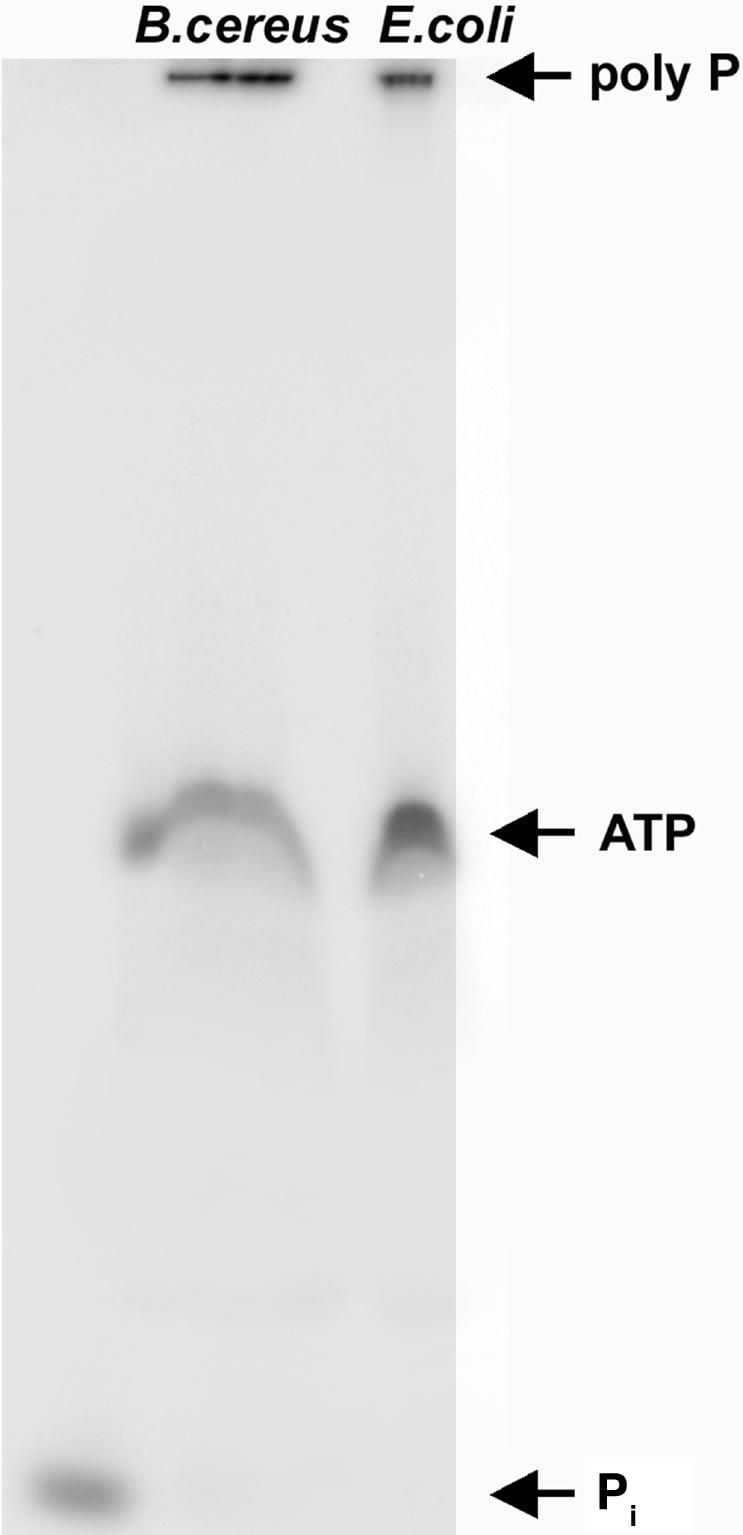
PAGE analysis of poly-P synthesized by B. cereus PPK. The products of incubation of [γ-32P]ATP with 40 ng of purified B. cereus PPK or 20 ng of E. coli PPK at 37°C for 30 min were separated by 20% PAGE containing 7 M urea, and visualized with a PhosphorImager (Molecular Dynamics).
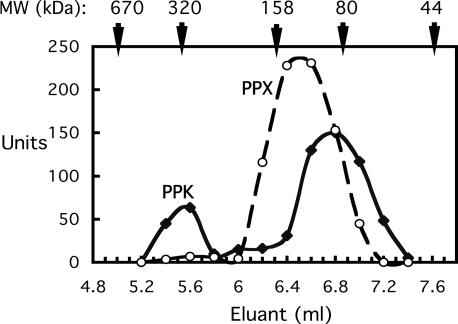
Null Mutants of ppk, ppx, and pap. Null mutants were obtained by replacement of the ORFs of ppk, ppx, and pap with a tetracycline-resistance gene (see Materials and Methods). The enzymatic activities of PPK in the Δppk and PPX in the Δppx mutant were undetectable (<0.1 units/mg protein). The PPX level in the Δppk mutant was similar to that in the WT, but remarkably, the PPK level in the Δppx mutant was ≈10-fold higher than that of the WT. However, an inhibitory complex of PPX with PPK was not observed in the WT cell lysates on size-exclusion chromatography. On a Tosoh G2000SW-XL HPLC column (Amersham Biosciences), PPK behaved as a monomer of 80 kDa, but in the presence of P15 or P65 (2 mM in Pi residues; Sigma), approximately one-fifth of the PPK migrated as a tetramer of 320 kDa. PPX appeared to be a dimer of 130 kDa, whether or not poly-P was present (Fig. 3).
Fig. 3.
Size-exclusion chromatography of PPK and PPX. Cell lysates of WT cells were prepared as described in HEDSP running buffer (50 mM Hepes·KOH, pH 7.6/150 mM Na2SO4/2 mM MgCl2/1 mM EDTA/1mMDTT/2 mM poly-P15) (Sigma). After incubation for 30 min on ice, 200 μl of lysates were applied to a Tosoh G2000SW-XL HPLC column, eluted with HEDSP buffer, and assayed for enzyme activities. Positions of molecular mass standards were thyroglobulin (670 kDa), bovine γ globulin (158 kDa), and chicken ovalbumin (44 kDa). Arrows at 80 and 320 kDa are judged to mark the PPK monomer and tetramer, respectively.
When E. coli is downshifted from a high-Pi (2 mM)-Mopsbuffered minimal medium to low-Pi (0.1 mM), poly-P accumulated to a level of ≈50 nmol/mg protein (11). Such a poly-P accumulation was not observed in B. cereus. Under the same conditions, WT cells accumulated poly-P to a level of only ≈2 nmol/mg protein; the Δppk mutant had <5% the WT poly-P level, and the Δpap mutant contained poly-P at a level similar to the WT. Remarkably, the Δppx mutant accumulated poly-P to a level of 2,600 nmol/mg protein, which is ≈1,000-fold that of the WT.
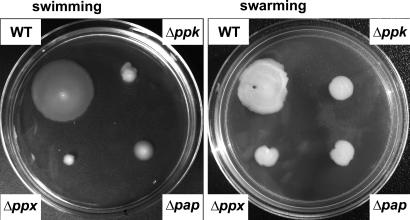
Impaired Motility in the Mutants. Deletion of ppk1 in many Gram-negative strains, such as E. coli, P. aeruginosa, and Vibrio cholerae, impairs their motility (14, 22). In B. cereus, flagellar motility observed as swimming and swarming was impaired not only in the Δppk mutant but even more so in the Δppx mutant, despite a massive accumulation of poly-P in this mutant (Fig. 4). Deficiency in motility was similar in the Δpap and Δppk mutants. The motility defects were complemented by introducing the corresponding gene on a low-copy-number plasmid, pHT304 (data not shown).
Fig. 4.
Swimming and swarming motility of WT and the Δppk, Δppx, and Δpap mutants. Cells were inoculated on swimming plates [BHI containing 0.3% (wt/vol) agarose] or swarming plates [BHI containing 0.5% (wt/vol) agar] with an overnight culture (LB agar plates) with a sterile toothpick, and they were photographed after an 8-h incubation for swimming plates or a 14-h incubation for swarming plates at 30°C.
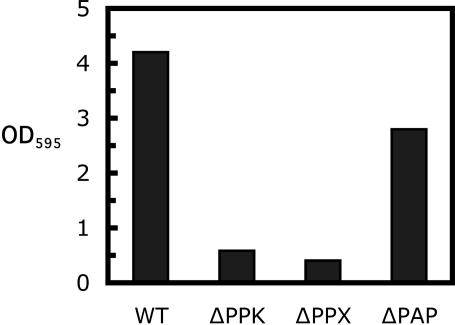
Impaired Biofilm Formation in the Mutants. As in the P. aeruginosa ppk1 mutant (13), mutants of ppk, ppx, and pap in B. cereus are each defective in swimming and swarming, as well as in the initiation of biofilm formation. Static biofilm formation was measured by crystal violet staining of attachment to an abiotic (polystyrene) surface. After a 12-h static incubation, only ≈14% of the WT level remained in the Δppk mutant, 10% in the Δppx mutant, and 67% in the Δpap mutant (Fig. 5). The deficiencies were not attributable to impairment of growth, the rates of which were indistinguishable from the WT.
Fig. 5.
Abiotic surface attachment of WT and mutants. An overnight LB culture (2 μl) was inoculated into 200 μl of LB in 96-well polystyrene plates and incubated at 30°C without shaking. After a 12-h incubation, the media were removed, each well was rinsed with sterile water, and the surface attachments were stained with crystal violet, extracted, and measured as described in Materials and Methods.
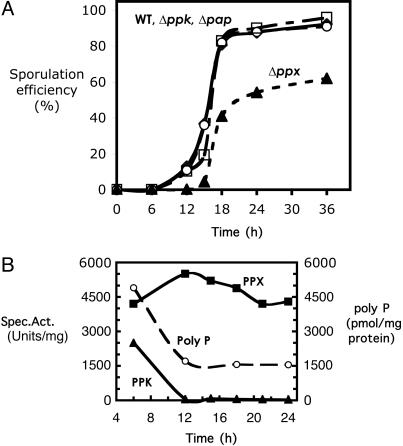
Impaired Sporulation in the ppx Null Mutant. Confronted by environmental stress as in nutrient depletion, the spore-forming bacteria Bacillus and Clostridium produce dormant spores by undergoing sporulation. As shown in Fig. 6A, B. cereus cells started to sporulate in CCY medium after 9–12 h of growth, and sporulation efficiency reached ≈90% after 24 h and almost 100% after 36 h. The Δppk and Δpap mutants showed similar sporulation efficiency as the WT. Remarkably, the Δppx mutant showed a 3-h delay in sporulation, and the efficiency was only ≈60%, even after a 36-h incubation. In the WT cells, PPK dropped immediately with the start of sporulation, and remained at ≈5% of the level of vegetative cells, whereas PPX was unchanged during sporulation (Fig. 6B). Consistent with the PPK activity, the poly-P level in WT cells fell during sporulation and remained at a level of 1.8 nmol/mg protein in the spores (Fig. 6B), which is approximately one-third that of the vegetative cells. The Δppx mutant accumulated poly-P to ≈60 nmol/mg protein, which is a level similar to that of Δppx vegetative cells.
Fig. 6.
Sporulation of WT and mutants. (A) Sporulation efficiency. Cultures were grown in CCY sporulation medium. Samples at the indicated times were diluted, heated at 70°C for 15 min, and plated on LB agar. Spore frequency was determined by the population that survived the heat treatment and was counted as colonies the next day. (B) PPK, PPX enzymatic activities, and the poly-P level in the sporulating WT cells. Enzyme assays and poly-P determination were performed as described in Materials and Methods.
Discussion
Previous studies of the enzymes and genes in the generation and use of poly-P in Gram-negative bacteria (e.g., E. coli and P. aeruginosa) demonstrated the essential roles that poly-P plays in responses to stresses and for survival (2, 3, 9–12). In this study, we have explored the distribution of poly-P and the poly-P enzymes, genes, and metabolic responses in the Gram-positive, spore-forming B. cereus, which is related closely to the serious pathogen B. anthracis. PPK, PAP, and PPX and their encoding genes have each been identified and characterized. Null mutants of each gene display phenotypic features that interfere with their behavior and development, such as defective swimming and swarming motilities (Fig. 4) and diminished attachment to an abiotic surface (Fig. 5), which is indicative of poor biofilm development.
Defects in motility are commonly associated with a decrease in virulence (13) as is poor biofilm formation (13, 22). B. cereus has alternative forms of flagella-driven motility, such as swimming by individual cells with short and few flagella and swarming by collective swarm cells with long and many flagellae (23). Each of the mutants (ppk, ppx, and pap) showed impairment in both types (Fig. 4). Surface attachment was greatly reduced in the ppk and ppx mutants but only moderately reduced in the pap mutant (Fig. 5). The fact that the ppx null mutant showed a more severe defect than the ppk mutant indicates that these impairments are likely caused by the fluctuation of the poly-P level in the cell, rather than the alteration of the PPK activity. Maintenance of the intracellular concentration of poly-P within a narrow range may be essential for effectiveness.
The mechanisms by which large fluctuations in poly-P and related enzymes levels produce the various phenotypic features require further study. Phenotype arrays showed that all three mutants had increased sensitivity to the antibiotic reagents miltefosine and alexidine (Table 3, which is published as supporting information on the PNAS web site). Miltefosine is an inhibitor of phosphocholine cytidylyl transferase (CTP), which is a key regulator in phosphatidylcholine biosynthesis (24, 25). Alexidine is a cationic antiseptic that inhibits glucosyltransferases which catalyze the formation of glucan from sucrose (28). Electron microscopy showed that mutants grown in a low-phosphate, Mops-buffered medium accumulated phase bright clusters (data not shown), perhaps the result of membrane invaginations during fixation due to the changes in the cytoplasmic membranes or the cytoskelons in the mutants (A. Makhov, personal communication).
Of the several PPKs isolated from E. coli (5), P. aeruginosa (13), Salmonella sp. (29), Shigella flexneri (29), V. cholerae (22), and Helicobacter pylori (3), that of B. cereus is the most distinctive. The PPK of B. cereus has a low pI, fails to adsorb to anion exchangers, and is stimulated 10-fold in activity in lysates by the presence of poly-P. This stimulation may be due to facilitating the formation or stabilization of a more active oligomer of PPK, as observed with the PPK2 of P. aeruginosa (26, 27), or counteracting a high level of PPX activity. In the ppx null mutant, PPK activity in the lysates is increased 10-fold, and the accumulation of poly-P in the cell increased 1,000-fold. No such effects have been observed with other ppx mutants. The adjacency of ppk and ppx genes may constitute an operon encoding a PPK–PPX complex. Inhibition of PPK activity by PPX in such a complex might account for the heightened PPK activity and the accumulation of poly-P in the ppx mutant. Colocalization of PPK and PPX has been observed in the E. coli nucleoid (A. Kuroda, personal communication).
Another distinctive feature of poly-P metabolism in B. cereus is the lack of its accumulation in response to a nutritional stringency. Unlike E. coli and P. aeruginosa in which a switch from a high to a low Pi level in the medium results in a massive accumulation of poly-P (i.e., 50–100 nmol/mg protein), the response in B. cereus is feeble (i.e., ≈2 nmol/mg protein). In E. coli, the ppk mutant is deficient in adapting to stresses and fails to survive in stationary phase (11). B. cereus possesses both a rapid, reversible adaptive stress response that is governed by σB and an ability to differentiate into dormant spores under extreme conditions (30–32). Although sporulation did not appear to be defective in the ppk mutant with the assay used, it may prove to be defective under other conditions. For example, sporulation efficiency in the Myxococcus xanthus ppk mutant is decreased when induced by glycerol, and in Dictyostelium discoideum, the ppk1 mutant shows a marked delay in fruiting body formation that precedes sporulation (H. Zhang, personal communication). Remarkably, the ppx mutant of B. cereus showed a decreased sporulation efficiency; in spores of the mutant, poly-P accumulated up to 60 nmol/mg protein, >30-fold than in the WT spores. The lack of poly-P accumulation in response to a nutritional stringency in WT B. cereus and the defects in sporulation of the ppx mutant accompanied with poly-P accumulation in its spores indicate important roles that poly-P plays in development. In the WT cells, PPK and poly-P levels dropped immediately with the start of sporulation, whereas the PPX level remained unchanged (Fig. 6). Loss of poly-P in the cell triggers the start of sporulation, where Spo0A, the key regulator for initiation of sporulation may be the target of poly-P regulation in the nucleoid.
The fact that PPK can serve as a target for screening antibiotics has already been demonstrated for PPK1 in P. aeruginosa (S. Lee, personal communication); PPX and PAP now provide additional targets. Recent availability of genome sequences of B. cereus and B. anthracis has revealed that a large core set of genes is conserved between these two species (17, 18). The defects in the virulence-related phenotypes of null mutants of ppk, ppx, and pap genes and the very high homologies of the corresponding proteins in B. anthracis (97–98% similarity) are attractive targets for discovery of antibiotics for the treatment of anthrax.
Supplementary Material
Acknowledgments
We thank Dr. Michael Brown and Leroy Bertsch for advice and assistance in preparation of this manuscript, Dr. Jo Handelsman for plasmid pSPUC and technical suggestions for making knockout mutants, Dr. Didier Lereclus for plasmid pHT304, and Dr. Bill Burkholder (Stanford University) for plasmid pDG1515. This work was supported by the National Institutes of Health.
Abbreviations: poly-P, inorganic polyphosphate; PPK, poly-P kinase; PPX, exopolyphosphatase; PAP, poly-P/AMP phosphotransferase; BHI, brain–heart infusion.
Data deposition: The sequence reported in this paper has been deposited in the GenBank database [accession no. BK005415 (pap)].
References
- 1.Kulaev, I. S. (1979) in The Biochemistry of Inorganic Polyphosphate (Wiley, New York).
- 2.Kornberg, A. (1995) J. Bacteriol. 177, 491–496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kornberg, A., Rao, N. N. & Ault-Riché, D. (1999) Annu. Rev. Biochem. 68, 89–125. [DOI] [PubMed] [Google Scholar]
- 4.Wood, H. G. & Clark, J. E. (1988) Annu. Rev. Biochem. 57, 235–260. [DOI] [PubMed] [Google Scholar]
- 5.Ahn, K. & Kornberg, A. (1990) J. Biol. Chem. 265, 11734–11739. [PubMed] [Google Scholar]
- 6.Akiyama, M., Crooke, E. & Kornberg, A. (1993) J. Biol. Chem. 268, 633–639. [PubMed] [Google Scholar]
- 7.Kumble, K. & Kornberg, A. (1996) J. Biol. Chem. 271, 27146–27151. [DOI] [PubMed] [Google Scholar]
- 8.Sethuraman, A., Rao, N. N. & Kornberg, A. (2001) Proc. Natl. Acad. Sci. USA 98, 8542–8547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Rao, N. N. & Kornberg, A. (1996) J. Bacteriol. 178, 1394–1400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Shiba, T., Tsutsumi, K., Yano, H., Ihara, Y., Kameda, A., Tanaka, K., Takahashi, H., Munekata, M., Rao, N. N. & Kornberg, A. (1997) Proc. Natl. Acad. Sci. USA 94, 11210–11215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Rao, N. N., Liu, S. & Kornberg, A. (1998) J. Bacteriol. 180, 2186–2193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ault-Riché, D., Fraley, C. D., Tzeng, C. M. & Kornberg, A. (1998) J. Bacteriol. 180, 1841–1847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Rashid, H., Rumbaugh, K., Passador, L., Davies, D. G., Hamood, A. N., Iglewski, B. H. & Kornberg, A. (2000) Proc. Natl. Acad. Sci. USA 97, 9636–9641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wang, L., Fraley, C. D., Faridi, J., Kornberg, A. & Roth, R. A. (2003) Proc. Natl. Acad. Sci. USA 100, 11249–11254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Helgason, E., Okstad, O. A., Caugant, D. A., Johansen, H. A., Fouet, A., Mock, M., Hegna, I. & Kolsto, A. B. (2000) Appl. Environ. Microbiol. 66, 2627–2630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ivanova, N., Sorokin, A., Anderson, I., Galleron, N., Candelon, B., Kapatral, V., Bhattacharyya, A., Reznik, G., Mikhailova, N., Lapidus, A., et al. (2003) Nature 423, 87–91. [DOI] [PubMed] [Google Scholar]
- 17.Read, T. D., Peterson, S. N., Tourasse, N., Baillie, L. W., Paulsen, I. T., Nelson, K. E., Tettelin, H., Fouts, D. E., Eisen, J. A., Gill, S. R., et al. (2003) Nature 423, 81–86. [DOI] [PubMed] [Google Scholar]
- 18.Stewart, G. S., Johnstone, K., Hagelberg, E. & Ellar, D. J. (1981) Biochem. J. 198, 101–106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Arantes, O. & Lereclus, D. (1991) Gene 108, 115–119. [DOI] [PubMed] [Google Scholar]
- 20.Dunn, A. K., Klimowicz, A. K. & Handelsman J. (2003) Appl. Environ. Microbiol. 69, 1197–1205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.O'Toole, G. A. & Kolter, R. (1998) Mol. Microbiol. 28, 449–461. [DOI] [PubMed] [Google Scholar]
- 22.Ogawa, N., Tzeng, C.-M., Fraley, C. D. & Kornberg, A. (2000) J. Bacteriol. 182, 6687–6693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Senesi, S., Celandroni, F., Salvetti, S., Beecher, D. J., Wong, A. C. & Ghelardi E. (2002) Microbiology 148, 1785–1794. [DOI] [PubMed] [Google Scholar]
- 24.Martinez-Morales, F., Schobert, M., Lopez-Lara, I. M. & Geiger, O. (2003) Microbiology 149, 3461–3471. [DOI] [PubMed] [Google Scholar]
- 25.Sohlenkamp, C., Lopez-Lara, I. M. & Geiger, O. (2003) Prog. Lipid Res. 42, 115–162. [DOI] [PubMed] [Google Scholar]
- 26.Zhang, H., Ishige, K. & Kornberg, A. (2002) Proc. Natl. Acad. Sci. USA 99, 16678–16683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ishige, K., Zhang, H. & Kornberg, A. (2002) Proc. Natl. Acad. Sci. USA 99, 16684–16688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Venkitaraman, A. R., Vacca-Smith, A. M., Kopec, L. K. & Bowen, W. H. (1995) J. Dent. Res. 74, 1695–1701. [DOI] [PubMed] [Google Scholar]
- 29.Kim, K. S., Rao, N. N., Fraley, C. D. & Kornberg, A. (2002) Proc. Natl. Acad. Sci. USA 99, 7675–7680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Petersohn, A., Brigulla, M., Haas, S., Hoheisel, J. D., Völker, U. & Hecker, M. (2001) J. Bacteriol. 183, 5617–5631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ireton, K., Rudner, D. Z., Siranosian, K. J. & Grossman, A. (1993) Genes Dev. 7, 283–294. [DOI] [PubMed] [Google Scholar]
- 32.Hamon, M. A. & Lazazzera, B. A. (2001) Mol. Microbiol. 42, 1199–1209. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.