Abstract
Growth hormone regulates its biological properties via a sequential hormone-induced receptor homodimerization mechanism. Using a mutagenesis-scanning analysis of 81 single and 32 pairwise double mutations, we show that the hormone's two spatially distal receptor binding sites (Site1 and Site2) are allosterically coupled. These allosteric effects are focused among a relatively few residues centered around the interaction between Asp-116 of the hormone and Trp-169 of the receptor in Site2. A rearrangement of this interaction triggered by mutations in Site1 produces both a major conformation and energetic reorganization of Site2, surprisingly without a reduction in overall binding affinity. Additionally, the data suggest a change in the conformational dynamics of several groups in Site2 that appear to be important in defining the Site2 interaction. Changes in binding energy of the affected Site2 residues usually range in magnitude from 3- to 60-fold, but in one case are as large as 104.
Within the cytokine super family, the growth hormone (GH)/prolactin family of hormones and receptors is arguably the most extensively characterized system focused on establishing the structure–function basis for protein–protein interactions (1–5). These studies and those of related cytokine systems have been instrumental in defining structural similarities within the family that are deterministic for hormone action and regulation (6–12). The structure-based mechanisms by which these systems activate are similar (3, 5, 9, 13). However, although these mechanisms are conceptually simple (hormone-induced receptor aggregation), it is becoming more appreciated that the molecular strategies that are used by each particular system are complex and hardly predictable (4, 6, 7, 14).
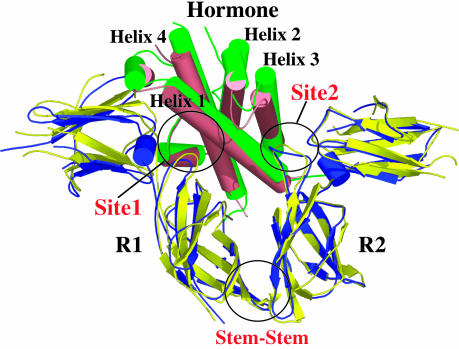
A hallmark of the GH-induced biological activity is a receptor aggregation process involving a receptor homodimerization through a mechanism that requires the two receptor extracellular domains (ECDs) to bind to the hormone in a highly regulated sequential order (13). The sequential order of binding is a consequence of the difference in binding affinities of the hormone's two spatially separated receptor binding sites. A high-affinity site, Site1, is always occupied first by the receptor ECD (R1) to form a stable 1:1 hormone-receptor intermediate complex [human GH (hGH)/R1] (15). This 1:1 complex provides the structural platform for binding the second receptor ECD (R2) through two spatially distinct subsites, which are independently weak but together produce a tight R2 association. One subsite is located on the opposite face of the hormone to Site1 and is referred to as Site2 (Fig. 1). The second subsite, the so-called stem–stem contact, is formed by an extensive set of interactions between the C-terminal portions of the ECDs of the two receptor ECDs. Although it is clear that the two receptor ECDs are structurally integrated through this stem–stem contact, there is no direct evidence to suggest that the Site1 and Site2 binding sites on the hormone have any structural linkage.
Fig. 1.
Diagram of the hGHv/R1/R2 receptor (hGHR) ternary complex superimposed onto the WT hGH/R1/R2 complex (6, 16). The hGHv complex is shown in salmon (hormone) and blue (R1/R2), and the WT hGH complex is shown in green (hormone) and yellow (R1/R2). The structures were superimposed by using the β-sheets in both the N- and C-terminal domains of the hGH receptor (hGHR). The Site1, Site2, and Stem–Stem binding interfaces between both hGHv and WT hGH and the hGHR R1 and R2 are circled. All structural figures were generated by using the program pymol (DeLano Scientific, San Carlos, CA, www.pymol.org).
The supposition that the two hormone binding sites are structurally independent has been recently called into question by our structural study of a ternary complex involving a high-affinity Site1 variant of hGH bound to R1 and R2 (16). This variant (hGHv), which was produced by phage display mutagenesis and had 15 mutations located exclusively in its Site1, binds to R1 ≈400-fold tighter than the WT hormone (17).
Not surprisingly, crystallographic analysis showed that the introduction of multiple Site1 mutations resulted in major structural changes in that interface. Most unexpectedly, however, the structure also showed that the Site2 hormone-receptor interface undergoes a significant conformational reorganization, even though no Site2 mutations had been introduced. In addition, it is noteworthy that this highly modified Site2 interface results in a virtually identical affinity for R2 binding as is found in the WT complex even though six of the seven hydrogen bonds in the interface are not conserved between these complexes (16).
The fact that Site1 mutations have such profound structural consequences at the distal Site2 interface indicates that there is measurable cross-talk between these binding sites at some level. However, it is not clear whether these effects are triggered only when all or a majority of the 15 mutations in hGHv are included, or whether there is a much smaller subset of critically disposed residues that when mutated propagate the majority of the effect. If this is the case, what is the spatial organization of this subset of residues and what are the possible structural linkages between them and Site2? Additionally, it is an important general molecular recognition question to determine how the energy is distributed in the reorganized Site2 interface in the hGHv complex. This “synthetically evolved” interface has such a different hydrogen-bonding structure that it is almost inconceivable that it would have the same energetic organization as the WT Site2 interface. The overriding importance of characterizing this interface is that it provides a window to evaluate systematically the inherent interaction potential of cytokine hormone-receptor interfaces and provides some insight into the structure–function parameters for how cross-reactivity between different cytokines might have evolved.
We describe here how the binding energetics of R2 to Site2 of hGH are affected by the allosteric coupling to Site1 and identify which residues are responsible for this effect. We find that the effect on equilibrium dissociation constant (Kd) of this coupling can be as large as 104 on R2 binding. The alanine scanning mutagenesis of Site2 residues in the hGHv-receptor complex clearly identified two spatially proximal residues, D116H of the hormone and W169R2 of the receptor R2 (superscript H indicates a hormone residue and the subscripts R1 and R2 indicate the ECD1 and ECD2 residues, respectively), as groups being most affected by the allosteric coupling of the binding sites. Although these two groups form a hydrogen-bond interaction in the hGHv complex, a series of homologous substitutions at each of these sites indicates that the most important contribution to the observed allosteric effects is through their local van der Waals packing arrangement, not their hydrogen-bond interaction. Based on structure and mutagenesis data it appears that R19 in Site1 of the hGHv has a role in promoting cross-talk between receptor binding sites even though it was not mutated itself. We find that the allosteric coupling effects in Site2 can be triggered by mutations in either helix 1 or helix 4 in Site1, although these effects are smaller than those observed when the mutations in both helices are combined.
Materials and Methods
Sample Preparation. hGHv contains the following 15 residue mutations in hGH: F10A, M14W, H18D, H21N, K41I, Y42H, L45W, Q46W, F54P, R64K, R167N, D171S, E174S, F176Y, and I179T, whereas phage-optimized helix (POH) 4 and POH1 variants contain the following mutations: R167N, D171S, E174S, F176Y, and I179T and F10A, M14W, H18D, and H21N, respectively. hGHvs were expressed and purified as described (18). All variants of the ECD (residues 29–238) of the human growth receptor were expressed and purified as described (19, 20). All mutations were made by using Kunkel mutagenesis (21) and confirmed by DNA sequencing. All protein purity and molecular weights were analyzed by analytical HPLC and MS, respectively.
Surface Plasmon Resonance (SPR). SPR experiments were performed by using a Biacore (Neuchatel, Switzerland) 2000 instrument at 25°C. The sensor chip preparation has been described (22, 23). Trimolecular SPR experiments [in 10 mM Hepes (pH 7.4), 150 mM NaCl, 3 mM EDTA, and 0.005% Tween 20 with a flow rate of 50 μl·min–1] were begun with a saturating concentration injection of 100 μl of 250 nM of a hGHv, followed by a 250-μl injection of varying concentration of free R2 variant, followed by a 700-s dissociation period. Typically, 2-fold serial dilutions of concentrations of 0, 3.75, 7.5, 15, 30, and 60 nM for the second R2 concentrations were used. The surface was regenerated for subsequent runs with a 5-μl injection of 4.5 M MgCl2. Typically, 200–300 injections were made on a sensor chip surface.
The six sensorgrams for each binding experiment were globally fit to the decaying surface model (24) in the program clamp99 (25) to determine R2 kon and koff binding rate constants to the 1:1 hormone/R1 complex. To be consistent with the previous kinetic results of R2 binding to the WT hGH/R1 complex (23), the hGHv and POH4 reactions were also fit to a decaying surface model. Each protein variant was measured in triplicate, and the average rate constants were reported. The rate constants showed differences of <5% in replicated measurements.
Results
Binding of High-Affinity GH Variants. Using trimolecular SPR we determined that the ECD R2 binds to the 1:1 hormone/R1 intermediates through moderately fast on and slow off kinetics. The resulting affinity for R2 binding to the hGHv/R1 complex is 4.4 nM, which compares closely with the value determined for the WT hGH complex (Kd = 3.8 nM) (23). R2 binds to the POH4 variant, which contains the helix 4 mutations R167N, D171S, E174S, F176Y, and I179T (17) complex (POH4/R1) with a Kd of 2.2 nM. Although this is a relatively small increase in binding affinity compared with the hGHv or WT complexes, the accuracy of the measured kon and koff values by trimolecular SPR is high enough to say confidently that the POH4 variant binds R2 slightly tighter than the other two complexes.
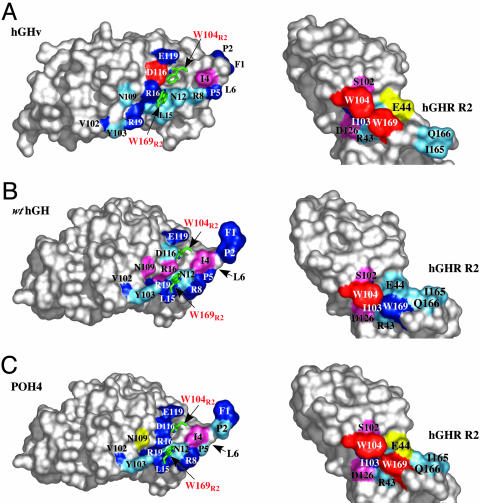
hGHv Ala Scan. The binding energetics of the hGHv's Site2 binding interface with R2 were determined by using a 15-residue Ala scan covering contact residues contained in the molecule's N terminus (residues 1–6), helix 1 (residues 7–32), and helix 3 (residues 108–127). The residues scanned contribute 460 Å2 to the binding interface and form six hydrogen bonds with receptor residues. The Ala scan showed that no single hormone residue in the interface by itself is essential for R2 binding (Fig. 2A). The majority (11 of 15) of the hGHv Ala-scanned mutants showed a <5-fold decrease in binding affinity compared to WT hGH (Table 1) (23). These differences arise almost exclusively from changes in koff rate constants; the kon rate constants for all of the mutants are almost invariant. The most significant decreases in binding among the Ala mutants are found in I4HA (24-fold), R16HA (8-fold), D116HA (61-fold), and E119HA (9-fold). These groups are spatially dispersed in the interface; residues 116H and 119H are located in helix 3, residue 16H is in helix 1, and residue I4H is in the N-terminal segment of the hormone (Fig. 2), and the observed decreases can be assigned to van der Waals and H-bonding factors.
Fig. 2.
Site2 Ala scans determined by using trimolecular SPR for hGHv, POH4, WT hGH, and their corresponding R2s. The results are color-coded as a function of fold decrease or increase [Kd(Ala)/Kd(Ref)]: red, >50-fold decrease; pink, 13- to 50-fold decrease; dark blue, 3- to 13-fold decrease; light blue, 0.5- to 3-fold decrease; and yellow, >2-fold increase. (A) Peeled-away surface diagram of the Site2 Ala scan of hGHv and R2 shown as a function of fold decrease or increase. (B) Peeled-away surface diagram depicted for the Site2 Ala scan of WT hGH and R2 (23). Leu-6 on WT hGH is located behind P5 in this orientation (depicted by the arrow). (C) Similar surface diagram of the Site2 Ala scan of POH4 and R2 results plotted on the same scale. The crystal structure of the WT hGH ternary complex (Protein Data Bank ID code 3HHR) was used to map the Site2 binding energetics of POH4 and R2 surfaces. Also depicted on the surface of each hormone are the Site2 R2 residues, W104R2 and W169R2 (red letters).
Table 1. Normalized Kd values for GH mutants binding to R2.
| Mutation | hGHv | POH4 | WT hGH* |
|---|---|---|---|
| F1A | 5 | 10.0 | 9.0 |
| P2A | 3 | 2.0 | 5.0 |
| I4A | 24 | 17.0 | 23.0 |
| P5A | 5 | 2.0 | 5.0 |
| L6A | 4 | 2.0 | 2.0 |
| R8A | 2 | 3.0 | 6.0 |
| N12A | 1 | 0.9 | 0.7 |
| L15A | 1 | 3.0 | 5.0 |
| R16A | 8 | 5.0 | 13.0 |
| R19A | 3 | 3.0 | 3.0 |
| V102A | 3 | 3.0 | 3.0 |
| Y103A | 2 | 1.0 | 2.0 |
| N109A | 2 | 0.3 | 19.0 |
| D116A | 61 | 12.0 | 0.8 |
| E119A | 9 | 3.0 | 3.0 |
| D116N | 6 | 3.0 | 3.0 |
| D116V | 9 | 7.0 | 2.0 |
| L6A, D116A | 49 (244) | 33 (2.0) | |
| N12A, D116A | 35 (61) | 35 (0.6) |
Equilibrium dissociation constants were calculated from the ratios of the measured kon and koff rate constant values. The normalized Kd values are relative to the respective GH variant: Kd (Mutant)/Kd. The R2 Kd values were 4.4, 2.1, and 3.8 nM for hGHv, POH4, and WT hGH, respectively. hGHv contains the following mutations: F10A, M14W, H18D, H21N, K41I, Y42H, L45W, Q46W, F54P, R64K, R167N, D171S, E174S, F176Y, and I179T. POH4 contains the following mutations: R167N, D171S, E174S, F176Y, and I179T. Values in parentheses are calculated by binding energy additivities based on the single point mutations: Kd(Mutant1)/Kd·Kd(Mutant2)/Kd.
Values taken from Walsh et al. (23).
In contrast, for N12H and R19H, both of which form intermolecular hydrogen bonds to R2 and bury significant amounts of surface area on binding (≈80 and 70 Å2, respectively), the alanine mutations result in surprisingly little change in binding considering the loss of hydrogen bonding and packing they produce (Table 1 and Fig. 3). Additionally, N109H, which makes an intermolecular hydrogen bond to R2 in the hGHv complex and buries ≈25 Å2 of its side chain in the interface, exhibits no substantial decrease in binding when mutated to alanine. On the other hand, in the WT hGH complex, where no hydrogen bond is made or surface area buried, the N109HA mutation decreases binding by 19-fold (23).
Fig. 3.
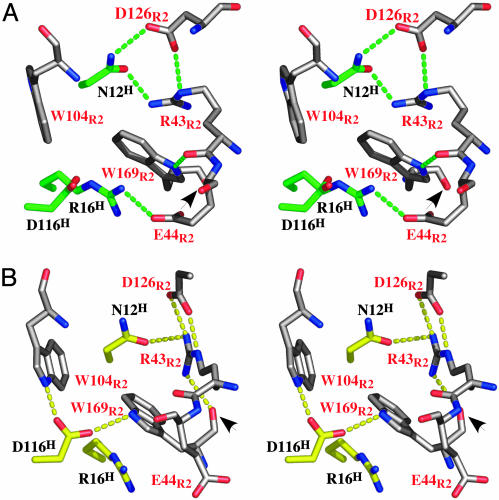
Site2 interfaces of hGHv and WT hGH with their respective R2s. (A) Expanded stereoview of the WT hGH (green)/R2 (white) complex also around D116. (B) Expanded stereoview of the Site2 interface in the hGHv (yellow)/R2 (white) complex around D116. The hormone and receptor residues are labeled in black and red, respectively. The intermolecular H bonds and salt bridges between the hormone and the receptor are depicted as broken lines. The arrows point to the peptide bond flip observed for W169E2 between the two crystal structures.
A notably different pattern is seen for the D116HA mutation. In WT hGH this mutation has essentially no effect, whereas in hGHv it produces a 61-fold decrease in binding (Table 1). In this case, however, there is a strong structural rationale for this observed difference in behavior. In Site2 of the WT hGH complex the side chain of D116H plays a bystander role, burying only ≈32 Å2 of surface area at the R2 interface. In contrast, the structure of the hGHv complex shows that in the reorganized Site2 interface, D116H makes two hydrogen bonds to the indole nitrogens of the important W104R2 and W169R2 residues of the receptor (Fig. 3).
R2 Ala Scan. Overall, the nine-residue Ala scan of R2 assayed against the WT hGH and hGHv hormones showed a similar effect on binding for the majority of the Ala-substituted residues (Table 2 and Fig. 2). For instance, the W104R2A mutation eliminates binding to both hormones. A notable outlier is the W169R2A mutation, which is highly hormone-dependent. In the context of the WT hGH complex the mutant produces an 11-fold decrease in binding affinity, whereas in the context of the hGHv complex, the W169R2A mutation completely inhibits R2 binding.
Table 2. Normalized Kd values for R2 mutants binding to GH/R1 complexes.
| Mutation | hGHv | POH4 | POH1 | WT hGH* |
|---|---|---|---|---|
| R43A-R2 | 1.0 | 0.5 | 2 | |
| E44A-R2 | 0.3 | 0.2 | 1 | |
| S102A-R2 | 15.0 | 15.0 | 13 | |
| I103A-R2 | 8.0 | 4.0 | 5 | |
| W104A-R2 | NB | NB | NB | NB |
| D126A-R2 | 33.0 | 30.0 | 37 | |
| I165A-R2 | 1.0 | 1.0 | 2 | |
| Q166A-R2 | 1.0 | 1.0 | 1 | |
| W169A-R2 | NB | 61.0 | 64 | 11 |
| W104F-R2 | 125.0 | 37.0 | 56 | 81 |
| W169F-R2 | 3.0 | 6.0 | 3 | 6 |
Values are normalized to Kd values for hGHv, POH4, POH1, and WT hGH of 4.4, 2.1, 4.1, and 3.8 nM, respectively. POH1 contains the following mutations: F10A, M14W, H18D, and H21N. NB, no binding (Kd > 1,000 nM).
Values taken from Walsh et al. (23).
Surprisingly, the Ala scan shows that the R43R2 side chain, which makes several hydrogen bonds (three intramolecular and one intermolecular) in the hGHv complex and its aliphatic groups form extensive van der Waals packing contact with the important W169R2 side chain (Fig. 3), produces no measurable effect on binding. This mutation not only eliminates these contacts, but its absence also presumably creates a hydrophobic hole in the Site2 interface that would not be energetically favorable to be filled by a water molecule without a major rearrangement of the local structure. In comparison, the hormone side chain of R16H performs a similar packing role on the opposite face of the W169R2 side chain in the hGHv complex. In this case, however, the alanine mutation produces a marked decrease in binding (8-fold). Interestingly, removing the side chain of E44R2, which forms a salt bridge with the R16H side chain in the WT hGH complex, does not change the affinity, yet removing this side chain appears to actually promote a slightly higher affinity interaction between R2 and the hormone variants (Table 2). These findings indicate that whereas removing the W169R2 side chain impacts the binding significantly, removing the side chains that form its hydrophobic packing environment surprisingly has little effect on R2 binding.
D126R2A also displays some anomalous binding behavior. The ≈35-fold decrease in binding that it produces in all hormone complexes is consistent with the fact that it forms two hydrogen bonds: one to N12H of the hormone and the other to R43R2 of the R2 (Fig. 3). However, these effects are not mirrored in the complementary mutations (N12HA and R43R2A) (see above), which eliminate the hydrogen bonds to D126R2 individually but show no effect themselves.
To differentiate between the hydrogen-bonding and van der Waals contributions in producing the large binding energies of the buried side chains of W104R2 and W169R2, a homologous mutation (Trp to Phe) was individually introduced at each of the two sites. Although it is not possible to quantify fully the effect of the W104R2F mutant, because the baseline (W104R2A) shows no binding whatsoever, nevertheless, it is clear that this mutation rescues binding by at least 103 (Table 2). The effects are even more dramatic in the case of the W169R2F mutant. Although it produces only a 2-fold enhancement over the alanine in the case of the WT hGH complex, in the hGHv complex this mutation essentially replaces all of the binding energy provided by the Trp side chain, close to a 105 enhancement in binding over the W169R2A mutant (Table 2).
Origin of the Conformational Effects at Site2. To establish the source of the allosteric coupling between binding sites, we wanted to determine whether there is a smaller subset of the Site1 mutations in hGHv that will themselves produce a large effect on the binding energetics of Site2. To address this, we performed the identical 15-residue Site2 Ala scan in the context of only five mutations in helix 4 of Site1. These mutations, referred to as POH4 mutations (17), were chosen because they encompassed the major portion of the Site1 “hot spot” region and when introduced as a group they increased the Site1 binding by ≈20-fold over WT hGH (26, 27).
We found that the energy distribution determined from the Ala scan of the POH4 variant paralleled that of the WT hGH and hGHv with several notable exceptions (Table 1 and Fig. 2). The D116HA and N109HA mutations, the two Site2 residues that produced the largest differences between WT hGH and hGHv, were also the largest outliers in the POH4 scan, albeit showing a different pattern. Although N109HA produced little change in binding to R2 in the context of hGHv and a 19-fold decrease in affinity in WT hGH, surprisingly the POH4 variant actually produced a 3-fold increase in binding affinity. For D116HA, which produced no effect in WT hGH and a 61-fold decrease in binding for hGHv, combining it with the POH4 mutations produced a 12-fold decrease in affinity, which is intermediate between the WT hGH and hGHv values.
For the nine-residue Ala scan of the R2 residues comparable patterns of binding of POH4 and the other two hormones were observed, with the exception of W169R2A (Table 2 and Fig. 2). Although this mutant does not bind to hGHv and shows an ≈11-fold reduction in binding to WT hGH, it binds to the POH4 variant with a Kd of 128 nM, a 61-fold reduction in binding compared with the WT R2.
The finding that the binding affinity of W169R2A to the POH4 variant is intermediate between WT hGH and hGHv suggests that other features in the hGHv molecule, in addition to the POH4 mutations, contribute to the observed binding effects. To test this notion, a third hormone variant, containing only the four mutations in helix 1 of hGH (POH1: F10A, M14W, H18D, and H21N) (Table 1) (17), was made and assayed against the W104R2A and W169R2A ECD variants (Table 2). Similar to the other hormone variants, the POH1 variant did not bind to W104R2A; however, it bound to W169R2A with about the same affinity observed for the POH4 variant. Although these results are open to several interpretations, we believe that the POH1 and POH4 variants work through the same general mechanism in influencing the binding of W169R2 (see below).
D116H and W169R2: The Epicenter for Structural Effects from Site1–Site2 Coupling. For the following discussion it is noteworthy that the hGHv–receptor crystal structure contained two independent hGHv–receptor complexes in the asymmetric unit and the extensive set of interactions described below are generally recapitulated in each independent copy of the complex.
In the WT hGH complex the side chain of D116H plays a bystander role with R2; however, in the rearranged hGHv–R2 interface, the carboxylate oxygens make hydrogen bonds to the indole nitrogens of W104R2 and W169R2 (Fig. 3). The importance of these hydrogen bonds is not clear, particularly the one to the indole of W169R2 in the hGHv complex, because the W169R2F mutant, where no hydrogen bond can be made to the indole, results in only an ≈3- to 5-fold decrease in binding (compared to no binding for the W169R2A mutant). This finding suggests that the most important feature of the W169R2 side chain's contributing to binding is through its extensive van der Waals interactions.
Hydrogen-Bonding Role of D116H in the Site2 Conformational Switch. To evaluate the hydrogen-bonding role of the D116H side chain in determining the Site2 conformational switch produced by hGHv, two isosteric mutants (Asp to Asn and Asp to Val) were made and evaluated (Table 3). The D116HN mutant limits hydrogen-bonding to one of the side-chain indole nitrogens, and D116HV eliminates all hydrogen-bonding capability and calibrates the van der Waals contributions between this side chain and tryptophan side chains. As expected based on structural arguments, neither of these mutations produced significant effects (≈2- to 3-fold) on R2 binding in the WT hGH complex, reflecting the fact that residue 116H contributes minimally for R2 binding. On the other hand, these mutations generate more substantial decreases in binding in the context of the hGHv–R2 interface (6- to 9-fold), albeit much smaller than is seen with the D116HA mutant. These data suggest that the packing void created by removing the 116H side chain is the most dominant effect in this set of mutations and that the loss of the hydrogen bonds to the tryptophans produces an ≈3-fold effect.
Table 3. Normalized pairwise Kd values for GH 116 mutants and R2 Trp mutants.
| Variants | WT R2 | W104F-R2 | W169F-R2 |
|---|---|---|---|
| D116A-HGHv | 61 | NB (7,600) | NB (183) |
| D116A-POH4 | 12 | NB (444) | 68 (72) |
| D116A-hGH* | 1 | NB (66) | 3 (4) |
| D116N-hGHv | 6 | 28 (750) | 17 (18) |
| D116N-POH4 | 3 | 68 (11) | 21 (18) |
| D116N-hGH* | 3 | NB (243) | 15 (14) |
| D116V-hGHV | 9 | 76 (1,100) | 28 (27) |
| D116V-POH4 | 7 | NB (259) | 39 (42) |
| D116V-hGH* | 2 | NB (162) | 24 (9) |
Values are normalized to the Kd values for hGHv, POH4, and WT hGH of 4.4, 2.1, and 3.8 nM, respectively. NB, no binding (Kd > 1,000 nM). Values in parentheses are calculated binding energy additivities based on the single-point mutations: Kd(Mutant1)/Kd(Ref)·Kd(Mutant2)/Kd.
Values taken from Walsh et al. (23).
To assess the pairwise relationships between hormone residue 116H and the two tryptophans, double mutants were evaluated between combinations of the three 116H mutants (Ala, Asn, and Val) and the phenylalanine substitutions at W104R2F and W169R2F. Overall, the effects on binding produced by these sets of double mutations were generally additive (Table 3). In the framework of the WT hGH–R2 interface, none of the hGH 116H mutants bound to the ECD containing W104R2F. In the context of hGHv, Asn and Val bound, but not Ala. For the POH4 variant, only D116HV bound to the W104R2F mutant. In contrast, W169R2F bound to all of the hGHvs with the exception of the hGHv D116HA mutant, which completely abolished binding (Table 3).
Taken together, these findings suggest certain distinguishing properties of the Site2 interfaces produced by the coupling to the Site1 mutations. First, in the WT hGH complex, W104R2 dominates all other Site2 residues in conferring R2 binding. W169R2 plays a secondary role and its Trp side-chain moiety can be changed to a Phe without significant effect. However, it is essential that W169R2F has some form of van der Waals contact with the 116H side chain in the hGHv complex. It is less important whether the side chain is hydrophilic or hydrophobic because D116HN and D116HV bind with reasonable affinity; however, D116HA, which would presumably lead to a packing void, does not bind.
Discussion
Structural Basis of the Allosteric Trigger in Site1. Although the mutagenesis data indicate that in Site2 D116H and W169R2 are the most significantly affected residues in the allosteric coupling with Site1, it is less clear whether there might be a particular set of a few residues in Site1 that influences this phenomenon. The different sets of mutations in the POH4 and POH1 variants lead to changes in binding of the D116HA and W169R2A mutants that lie approximately intermediate between those observed for WT hGH and the high-affinity hGHv, which contains 15 Site1 mutations. Two different models can explain this behavior. It could be that both sets of mutations lead to an altogether different structural solution in Site2 than is seen for the WT hGH and hGHv complexes, or that they both produce a weighted average between the WT hGH and hGHv interactions.
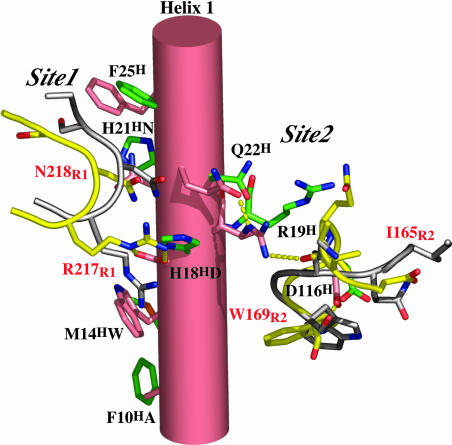
Although the Site1 and Site2 receptor binding sites are on approximately opposite faces of the hGH molecule, with their centroids separated through the molecule by >15 Å, their peripheries come close to overlapping at a point of intersection between helix 1 and helix 4 (Fig. 1). Three residues that form new interactions to R1 in hGHv are spatially clustered in this region, two in helix 1 (H18HN and H21HD) and one in helix 4 (E174HS). In the hGHv Site1 interaction S174H forms a hydrogen bond with the indole nitrogen of W104R1, whereas N18H and D21H form hydrogen bonds with the receptor side chains of R217R1 and N218R1, respectively. The adjacent residue to N18H (R19H) is a binding determinant for R2 binding, as are the proximal helix 1 residues R16H and N12H. This region of helix 1, which contains an interspersed set of Site1 and Site2 determinants, is one candidate for having a direct influence on the observed Site1–Site2 allosteric effects (Fig. 4).
Fig. 4.
Structural depiction of the potential allosteric coupling trigger going from Site1 to Site2 for the WT hGH to hGHv complexes. The hGHv complex residues are shown in salmon and yellow, respectively. The WT hGH complex residues are shown in green and white, respectively. The hormone and receptor residues are labeled in black and red, respectively. Hydrogen bonds for the hGHv complex are depicted as broken yellow dashes.
Additionally, the rather subtle global changes seen in the packing of helix 1 and helix 3 (≈5° rotation and 2.5-Å translation of helix 1) produce other significant structural changes in the hormone, R1, and R2 in this region. At the center of these changes is the new hydrogen-bonding arrangement of the side chain of R19H. In the WT hGH complex this plays an indirect role, whereas in the hGHv complex it moves ≈5 Å to hydrogen-bond to the carbonyl oxygen of I165R2 (Fig. 4). This change requires a significant reorganization of the main chain between residues 163 and 169 of R2, with the Cα positions of 165 and 166 moving by ≈5 Å (Fig. 4). We suggest that this change is responsible for the flipping of the peptide bond and side chain of residue W169R2 that is the central element of the rearrangement of Site2. In this position the carbonyl is positioned to form an H-bond with R43R. These changes in the backbone conformation of the receptor are structurally complemented by a large conformational change (up to 6 Å in Cαs between the unbound and bound form) in the hGHv molecule between residues 102 and 112. Besides the new hydrogen bond involving R19H, a hydrogen bond between N109HNδ2 and Q166R2O is formed.
The Conformational Switch. The positioning of the segment of residues around N109H appears to be important in stabilizing the receptor loop containing W169R2. Although there are significant differences in the global conformation of the structured loop containing N109H in the unbound and bound hGHv molecules (16), these differences are almost exclusively produced by the flip of a single peptide bond at positions 107–108. Superposition of five residues before and after this peptide flip shows similar local conformations (rms deviation <0.5 Å). Thus, the transition between the unbound and bound conformations, while resulting in a significant global change of several residues in the loop, is a simple low-energy event that should not have any large accompanying entropic penalty.
The conformational change in this loop that occurs on binding in the WT hGH complex is similarly governed by a single peptide bond flip. However, in this case the flip occurs between residues 102 and 103, resulting in a quite different orientation and composition of the groups that interact with the receptor. Thus, it appears that this binding loop (residues 102–112) exists in several conformations connected by low-energy barriers, and that it is the relative energetic partitioning between the populations of these conformations that is affected by the Site1 mutations. Interestingly, its binding “partner” in R2, the binding loop of the receptor containing W169R2, probably also shares similar properties because the conformational transition between its binding orientation in the WT hGH complex and the hGHv counterpart is produced by a single peptide flip at position 169. However, because the conformation of this loop seems to be equally adept at forming an interaction with either conformation of the 102–112 loop, the implication is that the energetic preference of the 102–112 loop in hGHv or WT hGH is the overriding factor in determining the conformation of the 169 loop and thus, the resulting structural relationships in the Site2 interfaces.
Acknowledgments
This work was supported by National Institutes of Health Grant DK-61602 (to A.A.K.). S.T.R.W. was a Burroughs Wellcome Fund Fellow of the Life Sciences Research Foundation.
Author contributions: S.T.R.W. and A.A.K. designed research; S.T.R.W. and J.E.S. performed research; S.T.R.W. contributed new reagents/analytic tools; S.T.R.W. and A.A.K. analyzed data; and S.T.R.W. and A.A.K. wrote the paper.
This paper was submitted directly (Track II) to the PNAS office.
Abbreviations: GH, growth hormone; hGH, human GH; hGHv, hGH variant; hGHR, hGH receptor; ECD, extracellular domain; POH, phage-optimized helix; SPR, surface plasmon resonance.
References
- 1.Bazan, J. F. (1990) Proc. Natl. Acad. Sci. USA 87, 6934–6938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Wells, J. A. (1991) Methods Enzymol. 202, 390–411. [DOI] [PubMed] [Google Scholar]
- 3.Sprang, S. R. & Bazan, J. F. (1993) Curr. Opin. Struct. Biol. 3, 815–827. [Google Scholar]
- 4.Kossiakoff, A. A., Somers, W., Ultsch, M., Andow, K., Muller, Y. A. & de Vos, A. M. (1994) Protein Sci. 3, 1697–1705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wells, J. A. & de Vos, A. M. (1996) Annu. Rev. Biochem. 65, 609–634. [DOI] [PubMed] [Google Scholar]
- 6.de Vos, A. M., Ultsch, M. & Kossiakoff, A. A. (1992) Science 255, 306–312. [DOI] [PubMed] [Google Scholar]
- 7.Somers, W., Ultsch, M., de Vos, A. M. & Kossiakoff, A. A. (1994) Nature 372, 478–481. [DOI] [PubMed] [Google Scholar]
- 8.Walter, M. R., Windsor, W. T., Nagabhushan, T. L., Lundell, D. J., Lunn, C. A., Zauodny, P. J. & Narula, S. K. (1995) Nature 376, 230–235. [DOI] [PubMed] [Google Scholar]
- 9.Banner, D. W., D'Arcy, A. D., Chene, C., Winkler, F. K., Guha, A., Konigsberg, W. H., Nermerson, Y. & Kirchhofer, D. (1996) Nature 380, 41–46. [DOI] [PubMed] [Google Scholar]
- 10.Wiesmann, C., Fuh, G., Christinger, H. W., Eigenbrot, C., Wells, J. A. & de Vos, A. M. (1997) Cell 91, 695–704. [DOI] [PubMed] [Google Scholar]
- 11.Syed, R. S., Reid, S. W., Li, C., Cheetham, J. C., Aoki, K. H., Liu, B., Zhan, H., Osslund, T. D., Chirino, A. J., Zhang, J., et al. (1998) Nature 395, 511–516. [DOI] [PubMed] [Google Scholar]
- 12.Boulanger, M. J., Bankovich, A. J., Kortemme, T., Baker, D. & Garcia, K. C. (2003) Mol. Cell 12, 577–589. [DOI] [PubMed] [Google Scholar]
- 13.Kossiakoff, A. A. & de Vos, A. M. (1998) Adv. Protein Chem. 52, 67–108. [DOI] [PubMed] [Google Scholar]
- 14.Atwell, S., Ultsch, M., de Vos, A. M. & Wells, J. A. (1997) Science 278, 1125–1128. [DOI] [PubMed] [Google Scholar]
- 15.Fuh, G., Colosi, P., Wood, W. I. & Wells, J. A. (1992) Science 268, 5376–5381. [PubMed] [Google Scholar]
- 16.Schiffer, C., Ultsch, M., Walsh, S., Somers, W., de Vos, A. M. & Kossiakoff, A. (2002) J. Mol. Biol. 316, 277–289. [DOI] [PubMed] [Google Scholar]
- 17.Lowman, H. B. & Wells, J. A. (1993) J. Mol. Biol. 234, 564–578. [DOI] [PubMed] [Google Scholar]
- 18.Chang, C. N., Rey, M., Bochner, B., Heyneker, H. & Gray, G. (1987) Gene 55, 189–196. [DOI] [PubMed] [Google Scholar]
- 19.Fuh, G., Mulkerrin, M. G., Bass, S., McFarland, N., Brochier, M., Bourell, J. H., Light, D. R. & Wells, J. A. (1990) J. Biol. Chem. 265, 3111–3115. [PubMed] [Google Scholar]
- 20.Clackson, T., Ultsch, M. H., Wells, J. A. & de Vos, A. M. (1998) J. Mol. Biol. 277, 1111–1128. [DOI] [PubMed] [Google Scholar]
- 21.Kunkel, T. A., Roberts, J. D. & Zakour, R. A. (1987) Methods Enzymol. 154, 367–382. [DOI] [PubMed] [Google Scholar]
- 22.Bernat, B., Pal, G., Sun, M. & Kossiakoff, A. A. (2003) Proc. Natl. Acad. Sci. USA 100, 952–957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Walsh, S. T., Jevitts, L. M., Sylvester, J. E. & Kossiakoff, A. A. (2003) Protein Sci. 12, 1960–1970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Joss, L., Morton, T. A., Doyle, M. L. & Myszka, D. G. (1998) Anal. Biochem. 261, 203–210. [DOI] [PubMed] [Google Scholar]
- 25.Myszka, D. G. & Morton, T. A. (1998) Trends Biochem. Sci. 23, 149–150. [DOI] [PubMed] [Google Scholar]
- 26.Pal, G., Kossiakoff, A. A. & Sidhu, S. S. (2003) J. Mol. Biol. 332, 195–204. [DOI] [PubMed] [Google Scholar]
- 27.Bernat, B., Sun, M., Dwyer, M., Feldkamp, M. & Kossiakoff, A. A. (2004) Biochemistry 43, 6076–6084. [DOI] [PubMed] [Google Scholar]