Abstract
Gene expression analysis implicates an increasing number of novel genes in the brain as potential targets for the treatment of neurological and psychiatric disorders. Frequently, these genes are ubiquitously expressed in the brain and, thus, may contribute to a pathophysiological state through actions in several brain nuclei. Current strategies employing genetically modified animals for in vivo validation of such targets are time-consuming and often limited by developmental adaptations. Somatic gene manipulation using viral-mediated RNA interference (RNAi) has emerged recently, although restricting the target validation to specific brain nuclei. We investigated whether nonviral infusion of short interfering RNA (siRNA) into the ventricular system would enable a sequence-specific gene knockdown. The temporality and extent of siRNA-induced down-regulation were analyzed by targeting a transgene, EGFP, in mice overexpressing EGFP. Extensive knockdown of EGFP was observed, especially in regions adjacent or dorsoventrally and mediolaterally distant to the infusion site (dorsal third ventricle), with lesser knockdown in more distal regions. We challenged our RNAi approach to generate a specific knockdown of an endogenous gene, encoding the dopamine transporter (DAT) in regions (ventral midbrain) far distal to the infusion site. DAT-siRNA infusion in adult mice produced a significant down-regulation of DAT mRNA and protein in the brain and also elicited a temporal hyperlocomotor response similar to that (but delayed) obtained upon infusion of GBR-12909, a pharmacologically selective DAT inhibitor. Application of this nonviral RNAi approach may accelerate target validation for neuropsychiatric disorders that involve a complex interplay of gene(s) from various brain regions.
Keywords: in vivo, target validation, EGFP, dopamine transporter
The burgeoning use of microarray analyses to detect potential target genes relevant to neuropsychiatric disorders necessitates the validation of such targets in vivo (1–3). The approach of genetically modifying animals (knockouts or transgenics) for target validation often is limited by developmental adaptations and genetic compensation that may mask the establishment of a clear phenotype. Further, such methodology is laborious and time-consuming and not applicable for high-throughput in vivo validation of the large number of hits generated from modern microarray and proteomic target-identification strategies. Additional approaches using ribozymes and antisense oligodeoxynucleotides for somatic gene manipulation also suffer from drawbacks of eliciting nonspecific actions. RNA interference (RNAi) recently has emerged as a potentially superior alternative to the traditional approaches for assessing gene function in adult animals (4).
RNAi is a cellular surveillance mechanism that responds to double-stranded RNA (dsRNA) by destroying cytoplasmic mRNAs containing sequences homologous to the dsRNA trigger (5). dsRNAs longer than 30 nt introduced in mammalian cells can induce an undesirable IFN response to produce cell death (6, 7). Conversely, short interfering RNAs (siRNAs; 21–23 nt) that mimic the cleavage products of dsRNA in mammalian cells elude the IFN response and produce sequence-specific gene silencing in these cells (6, 7). siRNAs have been used successfully to investigate gene function in neuronal cultures; however, their delivery into the mammalian brain to achieve the same knockdown in vivo still remains a challenge (8, 9). Recent studies describe the use of viral vectors to attain stable RNAi in the brain, albeit only in specific brain regions (10–13). This method may be pertinent for phenotyping of genes expressed in a few distinct loci, but not of genes that are broadly expressed and frequently contribute to a pathophysiological state by influencing a complex circuitry involving neurons from more than one brain region (14–18). Application of a regionally selective approach also would be difficult for investigating the function of novel genes with lack of knowledge of their mRNA-processing events or of brain regions relevant to their (patho)physiological actions. Therefore, to facilitate the functional assessment of such ubiquitously expressed genes and also to enable a rapid and regionally unbiased phenotyping of novel genes, we assessed whether siRNA could be delivered efficiently to produce gene silencing in the whole brain.
We first used mice overexpressing EGFP under the control of β-actin promoter (19) as an in vivo model to test the gene-silencing efficiency of our nonviral RNAi method. EGFP expression in these mice is ubiquitous and nearly uniform throughout the brain, thus enabling the determination of the extent of siRNA-induced knockdown of target (EGFP) mRNA and protein levels in the brain. The specificity, temporality, and magnitude of EGFP knockdown were evaluated after infusion of an EGFP-targeting siRNA in the dorsal third ventricle. Further, we tested the validity of this RNAi approach by targeting an endogenous gene that codes for the dopamine transporter (DAT) within the ventral midbrain neurons, far distal from the siRNA infusion site (20). DAT controls the temporal and spatial activity of dopamine released into the synapse by facilitating a rapid uptake of the neurotransmitter into presynpatic terminals. DAT is, therefore, a key regulator of dopamine actions on locomotion, emotion, and cognition, and it is implicated in the etiology of hyperkinetic disorders, such as attention-deficit hyperactivity disorder (21–24). Here, we compare the behavioral as well as molecular consequences of infusing the DAT-siRNA vs. those of a selective pharmacological DAT inhibitor in the brain.
Methods
Animals. Male EGFP-expressing mice were generated on a BALB/c background as described in ref. 19. Briefly, the plasmids pEGFP-N1 (Clontech) and pRAY-2 (25), and a 5.9-kbp KpnI genomic subclone of β-actin were assembled to produce a construct, wherein a promoterless EGFP gene was inserted at the ATG of β-actin and driven from the β-actin promoter. Homologous recombination was performed in BALB/c-I embryonic stem cells. Modification of the β-actin locus was confirmed by PCR and Southern blotting. Male BALB/c mice (21–29 g; Iffa Credo) were used for all other experiments. All mice were housed two per cage before surgery and one per cage thereafter in a humidity- and temperature-controlled room with a 12-h/12-h light/dark cycle (lights on at 6 a.m.). Food pellets and tap water were available ad libitum, except during behavioral testing. Animals were acclimatized to these housing conditions for at least 1 week after their arrival before starting any procedure. Animal experimentation was performed during the light cycle, in accordance with the Veterinary Authority of Basel-Stadt, Switzerland.
Drug/siRNA Administration in Mouse Brain. We used an in vitro validated siRNA, targeting EGFP (antisense, 5′-AUGAACUUCAGGGUCAGCUTG-3′, and sense, 5′-AGCUGACCCUGAAGUUCAUCT-3′) (6), and a siRNA of scrambled sequence (scrRNA) (antisense, 5′-UCGUCAUAACGUUCAUAGGCG-3′, and sense, 5′-CCUAUGAACGUUAUGACGATT-3′). They were 21-mer double-stranded oligonucleotides with a 19-bp oligoribonucleotide region and dinucleotide overhangs on the 3′ end of each strand consisting of either 2′-O-(2-methoxyethyl)-modified nucleotide residues linked by means of a phosphorothioate group (26) in the case of EGFP-targeting siRNA or deoxynucleotide residues linked by means of a phosphodiester group in the case of DAT-targeting siRNA. A standardized mRNA fusion-construct assay (27) was used to screen 12 different siRNAs with antisense strands targeting DAT at nucleotides 283–303, 335–355, 607–627, 612–632, 670–690, 682–702, 716–736, 1042–1062, 1168–1188, 1192–1212, 1469–1489, and 1858–1878 (GenBank accession no. BC054119). DAT-siRNA, targeting nucleotides 682–702, revealed a maximum knockdown activity (80%; data not shown) in this assay and was chosen for all in vivo experiments. A corresponding 3-nt mismatch siRNA (mmRNA) (antisense, 5′-UUGUGGCAUUGGAGCCAUGGG-3′, and sense, 5′-CAUGGCUCCAAUGCCACAATT-3′) was used as a control. Antisense and sense strands of each siRNA were annealed in an isotonic RNAi buffer (100 mM potassium acetate/30 mM Hepes-KOH/2 mM magnesium acetate/26 mM NaCl, pH 7.4, at 37°C) as described in ref. 26. GBR-12909.2HCl (Sigma), a selective DAT inhibitor, was dissolved in the isotonic RNAi vehicle maintained at 37°C. Osmotic minipumps were filled with the vehicle for infusion at a rate of 12 or 6 μl/day for 1 week (Alzet model 1007D, Durect, Cupertino, CA) or 2 weeks (Alzet model 1002), respectively. Minipumps also were filled to infuse 0.4 mg of siRNA or the indicated dose of GBR-12909 per day. A relatively high dose of siRNA was chosen because preliminary studies indicated that administration of siRNA up to this dose was well tolerated, and no signs of neurotoxicity (hind-limb paralysis, vocalization, food intake, or neuroanatomical damage) were present. Further, it corresponds to the dose that was maximally effective in rat studies upon intrathecal administration (26). A brain-infusion cannula (Plastics One, Roanoke, VA) was stereotaxically placed, as described by the manufacturer, for infusion from the s.c.-implanted minipump into the dorsal third ventricle (anteroposterior, –0.5 mm; mediolateral, 0 mm; dorsoventral, –3 mm relative to bregma; ref. 28). All mice received Temgesic (0.5 mg/kg, i.p.; Essex Chemicals, Lucerne, Switzerland) and, 45 min later, an anesthetic (100 mg/kg ketamine and 10 mg/kg xylazine i.p.; Sigma) before surgery, followed by sterile saline (1 ml i.p.) after surgery.
Locomotor Activity. Locomotor activity was analyzed between 8 and 10 a.m. as described in ref. 29. Activity was assessed on days –1 and 0 to habituate the animals to the environment and to obtain a baseline, where day 0 is the day of implanting the cannula-minipump assembly in mice. Thereafter, mice were assessed for activity levels on days 3, 6, 9, 12, and 14.
Processing of the Brain for mRNA and Protein Analysis. Mice involved in the EGFP-siRNA study were decapitated on day 8 or 15, depending on the model of osmotic minipump implanted. For the DAT-siRNA experiment, mice were decapitated ≈24 h after their final locomotor activity trial. Brains were removed, and serial coronal sections of 10-μm thickness were obtained for each brain region at the following anteroposterior coordinates in mm relative to bregma (28): 3.56 (olfactory bulb), 2.46 (prefrontal cortex), 1.18 (caudate putamen and nucleus accumbens), –0.22 (globus pallidus), –0.46 (to confirm the site of injection), –1.58 (cerebral cortex, hippocampus, habenula, thalamus, hypothalamus, and amygdala), –3.08 (substantia nigra and ventral tegmental area), –4.36 (periaqueductal gray and dorsal raphe), –5.34 (locus coeruleus), and –7.2 (spinal trigeminal nucleus). Sections were thaw-mounted onto poly(l-lysine)-coated slides, coded for a blind analysis of mRNA and protein, and stored at –80°C until use in <1 week.
In Situ Hybridization Analysis. Brain sections were subjected to an in situ hybridization protocol as described in ref. 30, by using [35S]-labeled antisense riboprobes for detecting EGFP, GABA type A (GABAA) receptor subunit α2 (GABAAα2), DAT, or tyrosine hydroxylase (TH) mRNA. Hybridizing adjacent brain sections with the corresponding 35S-labeled sense riboprobes did not reveal any mRNA signal and served as a control for each probe. The DNA templates for riboprobe synthesis were generated from cDNA fragments of EGFP (GenBank accession no. U55762; nucleotides 679–1398), mouse GABAAα2 (RefSeq accession no. NM_008066; nucleotides 293–898), DAT (RefSeq accession no. NM_010020; nucleotides 1507–1941), or TH (GenBank accession no. M69200; nucleotides 62–1654). At the end of the in situ hybridization procedure, slides were dipped in liquid nuclear emulsion, and the optical density (OD) of silver grains, positive for the probed mRNA, was quantified as described in ref. 30. Cresyl violet staining (0.5%) facilitated the localization and subsequent quantification of mRNA levels in different brain regions from each section. EGFP mRNA is expressed throughout the brain of EGFP-expressing mice and was quantified in the above-mentioned brain regions. GABAAα2 mRNA was measured in the same regions of adjacent brain sections. DAT mRNA in the substantia nigra compacta and ventral tegmental area was collectively quantified as expression in ventral midbrain nuclei. Adjacent brain sections were probed similarly for TH mRNA expression in the ventral midbrain neurons.
Quantification of EGFP Fluorescence. Images of EGFP fluorescence, from microscopic fields of sectioned brain regions, were recorded with equal exposure times by using an Olympus BX51 fluorescence microscope outfitted with a SIS ColorView III camera (Soft Imaging System, Münster, Germany). Specific nuclei imaged as a representative of each brain region are given in parentheses as follows: olfactory bulb (granular cell layer), prefrontal cortex (secondary motor cortex), caudate putamen (central area), nucleus accumbens (core and shell), globus pallidus (lateral), cerebral cortex (secondary and trunk region of the primary somatosensory cortex), hippocampus (CA1–CA3 and dentate gyrus), habenula (medial and lateral), thalamus (mediorostral lateral posterior and ventrolateral), hypothalamus (dorsomedial and ventrolateral ventromedial), amygdala (basolateral and anterior basomedial), substantia nigra (reticular and compacta), periaqueductal gray (dorsolateral and dorsomedial), dorsal raphe (dorsal and ventral), and locus coeruleus and spinal trigeminal nucleus (interpolar part of the spinal 5 nucleus). analysis 3.2 software (Soft Imaging System) was used for image processing and analysis of fluorescence intensity as a measure of EGFP levels in each region. The integral fluorescence intensity of each image was measured in a preset ellipsoidal area of 0.1 mm2, except for the CA1–CA3 neurons and dentate gyrus, which were marked with boundaries of fixed areas of 0.02 and 0.1 mm2, respectively.
Quantitative Autoradiography. Protein levels of DAT or serotonin transporter (SERT) in the caudate putamen, nucleus accumbens (core and shell), and olfactory tubercles of adjacent brain sections were quantified by using [125I]RTI-55 autoradiography as described in ref. 31 with minor modifications. Briefly, a nonsaturating concentration of [125I]RTI-55 [10 pM; 2,200 Ci/mmol (1 Ci = 37 GBq), PerkinElmer] selectively labeled DAT, provided the binding to SERT was occluded with 50 nM citalopram HBr. The same radioligand was used to label SERT upon inhibition of DAT binding with 10 μM GBR-12909. By using tissue-calibrated data from the coexposed radioactive standards, OD values of autoradiograms were transformed to levels of radioactivity bound (nCi/mg tissue protein) to specific brain regions in tissue sections. Nonspecific binding was determined in adjacent brain sections by using 10 μM GBR-12909 and 50 nM citalopram, in addition to [125I]RTI-55, and was equivalent to the autoradiographic film background.
Results
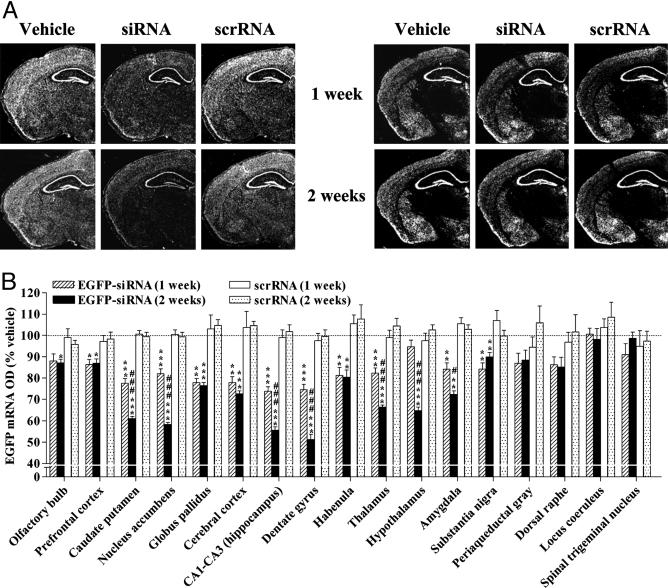
siRNA-Mediated Widespread, Temporal, and Specific Knockdown of a Broadly Expressed Transgene in the Brain. EGFP-targeting siRNA, scrRNA, or vehicle was intracerebroventricularly infused in mice that exhibited a ubiquitous and nearly uniform expression of EGFP in the brain. We chose the dorsal third ventricle as the site of infusion with an aim of achieving a bilateral and widespread distribution of siRNA in the brain. siRNA infusion for 1 week produced a significant down-regulation of EGFP mRNA in 11 of 17 regions tested along the rostrocaudal, dorsoventral, and mediolateral axes of the brain (Fig. 1). The prefrontal cortex was the anterior-most region (to the infusion site) that revealed a significant reduction of EGFP mRNA in siRNA-treated mice as compared with levels in vehicle- or scrRNA-treated mice (Fig. 1B). Higher-magnitude reductions in EGFP mRNA were observed in regions more proximal to the infusion site, including the caudate putamen, nucleus accumbens, and globus pallidus. Among the regions tested on the posterior end of the siRNA infusion site, substantia nigra was the most distal that exhibited a significant down-regulation of EGFP mRNA. With the exception of the hypothalamus, a significant decline in EGFP mRNA levels was noted in all other regions examined between the siRNA infusion site and substantia nigra. Maximal decrease in EGFP mRNA was seen in the hippocampus (CA1–CA3, dentate gyrus; Fig. 1).
Fig. 1.
siRNA-induced specific, temporal, and widespread knockdown of EGFP mRNA in the brain. EGFP-expressing mice received an infusion of vehicle, EGFP-targeting siRNA, or scrRNA into the dorsal third ventricle for the indicated time. EGFP mRNA levels were reduced, in a temporal fashion, in several brain regions of mice infused with siRNA as compared with levels in vehicle- and scrRNA-treated mice. Levels of GABAAα2 mRNA were unaffected. (A) Low-magnification (×10), dark-field photomicrographs of representatives of in situ hybridization with EGFP (Left) and GABAAα2 (Right) riboprobes in adjacent coronal brain sections, after emulsion-dipping of slides. (B) Densitometric quantification of mRNA-positive grains in each brain region is expressed as percent OD values of those in corresponding regions from vehicle-treated mice. Bars represent means ± SEM of 16–36 observations (four to six observations per animal and four to six animals per group). *, P < 0.05; **, P < 0.01; ***, P < 0.001, significantly different from vehicle- and scrRNA-treated mice in each brain region. #, P < 0.05; ###, P < 0.001, significantly different from siRNA treatment for 1 week in the same region, two-way ANOVA followed by Tukey's post hoc test.
siRNA-induced knockdown of hippocampal EGFP mRNA was substantiated upon 2-week siRNA infusion (Fig. 1). A temporal decrease in EGFP mRNA levels also was observed in the caudate putamen, nucleus accumbens, thalamus, and amygdala after siRNA treatment for 2 weeks (Fig. 1B). Moreover, a significant RNAi response was noted in the olfactory bulbs and hypothalamus that were initially resistant to the first week of siRNA infusion. Other regions, such as the globus pallidus, habenula, substantia nigra, and the prefrontal and cerebral cortices, showed no further reduction in EGFP mRNA levels after 2 weeks compared with those seen after 1 week of siRNA treatment.
We also assessed changes in mRNA levels of GABAAα2 after 1- and 2-week siRNA, scrRNA, or vehicle infusions in mice. The expression pattern of GABAAα2 closely parallels that of EGFP, except for the thalamus where GABAAα2 mRNA is present only in the central and mediodorsal thalamic nuclei (Fig. 1A). EGFP siRNA failed to alter the levels of GABAAα2 mRNA in any of the brain regions tested at either time point, indicative of an EGFP-specific silencing action of siRNA (Fig. 1A and Fig. 6, which is published as supporting information on the PNAS web site). Surprisingly, scrRNA infusion for 1 week, but not 2, produced a small decrease in GABAAα2 mRNA in the substantia nigra. The significance of this finding is difficult to ascertain, especially considering that 2-week scrRNA infusion failed to influence GABAAα2 mRNA levels in this or any other region (Fig. 6). Nevertheless, unaltered levels of EGFP mRNA after 1- or 2-week scrRNA infusion (Fig. 1) provide further evidence for specificity of the RNAi response observed with siRNA.
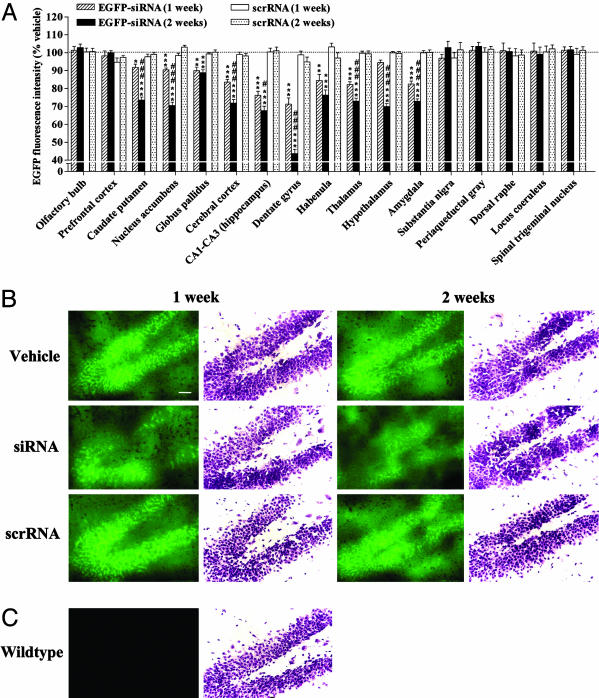
siRNA infusion significantly and time-dependently reduced the protein levels of EGFP in several brain regions (Fig. 2A). siRNA-induced changes in EGFP protein correlated well with those in mRNA, with a maximal effect obtained in the hippocampus (Fig. 2). However, the magnitude of decrease in protein was, in most cases, less than that in mRNA, such that the regions demonstrating <15% decrease in EGFP mRNA showed no changes in EGFP protein with siRNA treatment (Figs. 1B and 2A). Treatment with scrRNA did not alter the protein levels of EGFP in any of the regions tested throughout the brain (Fig. 2).
Fig. 2.
siRNA-mediated temporal and extensive down-regulation of EGFP in the mouse brain. (A) Green fluorescence images from microscopic fields of sectioned brain regions were analyzed for quantifying fluorescence intensity as an indicator of EGFP levels in each region. Bars represent mean fluorescence intensity units ± SEM of 20–40 observations (five to eight observations per animal and four to six animals per group). siRNA infusion in the mouse brain time-dependently down-regulated the EGFP levels in several brain regions tested. *, P < 0.05; **, P < 0.01; ***, P < 0.001, significantly different from vehicle- and scrRNA-treated mice in each brain region. #, P < 0.05; ###, P < 0.001, significantly different from siRNA treatment for 1 week in the same region, two-way ANOVA followed by Tukey's post hoc test. (B) Representative microscopic fluorescence images of an area of the dentate gyrus demonstrating a temporal reduction of EGFP levels in siRNA-treated mice as compared with those in vehicle- or scrRNA-treated mice. Nissl-stained adjacent brain sections also are shown for each treatment. (C) Representative microscopic fluorescence image indicating lack of any fluorescence emitted from the same area of dentate gyrus of wild-type mice that do not express EGFP. Nissl staining of adjacent section is shown. (Scale bar, 50 μm.)
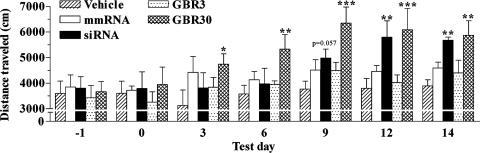
siRNA-Induced Knockdown of an Endogenous Gene in the Brain. Having demonstrated that siRNA infusion in the dorsal third ventricle produced a significant knockdown of an exogenously expressed gene in the brain and that this effect caudally extended up to the ventral midbrain neurons of the substantia nigra, we tested the efficiency of this RNAi approach in producing gene knockdown of an endogenous gene, DAT. DAT is selectively expressed in the dopaminergic ventral midbrain neurons of the ventral tegmental area and substantia nigra compacta (20). Mice received one of the following: a 2-week infusion of an in vitro validated DAT-targeting siRNA, a corresponding mmRNA, a selective DAT inhibitor (3 and 30 μg/day GBR-12909), or vehicle. Locomotor activity was measured every third day, from the start of infusion to day 14. Vehicle-treated mice did not habituate to this behavioral assessment and exhibited a constant locomotor activity response during the entire testing period (Fig. 3). Repeated-measures ANOVA revealed a significant effect of treatment (F4,120 = 3.56, P = 0.024) and time (F6,120 = 13.42, P < 0.001) on locomotor activity, and there was a significant treatment × time interaction (F24,120 = 1.86, P = 0.016). Post hoc analyses illustrated a significant hyperlocomotor response in mice receiving GBR-12909 (30 μg/day) on all test days, starting from day 3 onward (Fig. 3). This increase in locomotor activity, particularly on day 9, was significantly different from that on days 3 (P < 0.001) and 6 (P = 0.033), indicative of a behavioral sensitization response to the higher dose of GBR-12909. Thereafter, the hyperlocomotor response of these mice plateaued up to the last test day (Fig. 3). DAT siRNA infusion in mice also produced a similar, although delayed, hyperlocomotor effect, where there was a strong trend toward an increased locomotor activity on day 9 (P = 0.057); this effect was highly significant on days 12 and 14 (Fig. 3). Infusion of mmRNA had no significant effect on the locomotor activity of mice throughout the test period (Fig. 3).
Fig. 3.
Effects of infusing a DAT-targeting siRNA or the pharmacological inhibitor GBR-12909 on mouse locomotor activity. Locomotor activity was assessed for 30 min on the indicated days, 0 being the day of surgical implantation of the minipump-cannula assembly for infusing vehicle, DAT-targeting siRNA, mmRNA, or GBR-12909 (GBR3, 3 μg/day; GBR30, 30 μg/day) in the dorsal third ventricle of adult mice. Bars represent mean distance traveled (cm) ± SEM from six animals per group. Infusion of GBR-12909 (30 μg/day) resulted in a significant hyperlocomotor activity in mice on all test days. siRNA elicited a trend toward a significant hyperlocomotion effect on day 9 that was significant on days 12 and 14. *, P < 0.05; **, P < 0.01; ***, P < 0.001, significantly different from vehicle, using repeated-measure ANOVA with treatment as the between factor and time as the within-subject variable, followed by Fisher's post hoc test.
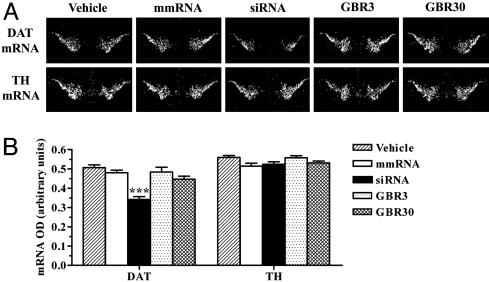
DAT mRNA and protein levels in the brain were quantified to correlate with changes in behavior after various treatments. A significant down-regulation of DAT mRNA, by 33%, was observed in the ventral midbrain of mice infused with siRNA (Fig. 4). siRNA-induced knockdown of DAT mRNA was greater than that of EGFP mRNA in the ventral midbrain. EGFP mRNA levels in the transgenic mice used in our study were much higher than the endogenous levels of DAT mRNA. Therefore, levels of target transcript possibly may play a role in determining the knockdown potency of EGFP-siRNA vs. DAT-siRNA, despite the fact that a chemically modified siRNA was used for targeting EGFP to increase its resistance against nuclease degradation in vivo. DAT mRNA levels were unchanged with infusion of vehicle, mmRNA, or GBR-12909. TH, a rate-limiting enzyme for dopamine biosynthesis, is expressed in all of the dopaminergic ventral midbrain neurons (32). Therefore, we probed adjacent brain sections for detecting any changes in the expression of this dopaminergic marker. None of the treatments, including siRNA, produced a significant alteration in midbrain levels of TH mRNA (Fig. 4).
Fig. 4.
siRNA-induced specific knockdown of DAT mRNA in the substantia nigra and ventral tegmental area. Mice received a 2-week infusion of vehicle, DAT-targeting siRNA, mmRNA, or GBR-12909 (3 or 30 μg/day) into the dorsal third ventricle. DAT mRNA levels were reduced significantly in the ventral midbrain regions of mice infused with siRNA. Levels of TH mRNA in the same regions were unaffected by any treatment. (A) Low-magnification (×20), dark-field photomicrographs of representatives of in situ hybridization with DAT (Upper) and TH (Lower) riboprobes in adjacent brain sections, after emulsion-dipping of slides. (B) Densitometric quantification of mRNA positive grains is presented as mean OD values ± SEM of 20–24 observations (two to four observations per animal and six animals per group). ***, P < 0.001, significantly different from all other treatments, using one-way ANOVA followed by Tukey's post hoc test.
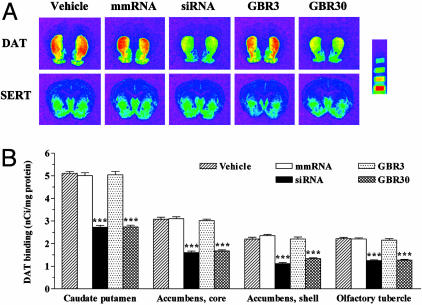
The processing of DAT mRNA for protein synthesis takes place in the perikarya of neurons originating in the substantia nigra compacta and ventral tegmental area. The resulting DAT protein then is transported down the axons to the dopaminergic projection areas, where it is found abundantly in the caudate putamen, accumbens core and shell, and the olfactory tubercle (33). Quantitative autoradiographic analysis demonstrated up to a 49% reduction in protein levels of DAT in all of these dopaminergic projection areas upon infusion of either DAT siRNA or the higher dose of GBR-12909 (Fig. 5). We quantified levels of SERT in adjacent brain sections and found that they were unaffected by any treatment in all of the brain regions tested (Fig. 5A and Fig. 7, which is published as supporting information on the PNAS web site).
Fig. 5.
Down-regulation of the DAT protein after infusion of siRNA or GBR-12909. (A) Representative autoradiograms of [125I]RTI-55 binding to DAT (Upper) or SERT (Lower) in adjacent brain sections of mice infused for 2 weeks with vehicle, DAT-targeting siRNA, mmRNA, or GBR-12909 (3 or 30 μg/day). siRNA or GBR-12909 (30 μg/day) produced a significant down-regulation of DAT in all of the dopaminergic projection areas of the forebrain. None of the treatments produced a significant alteration of SERT protein levels in the same brain regions. (Microscale bars, from top to bottom, represent binding values of 1.52, 2.58, 5.16, and 10.96 nCi/mg.) (B) Quantification of DAT binding in the caudate putamen, accumbens core and shell, and olfactory tubercles. Bars represent mean nCi/mg tissue protein ± SEM of 36–40 observations (six to eight observations per animal and six animals per group). ***, P < 0.001, significantly different from all other treatments, using one-way ANOVA followed by Tukey's post hoc test.
Discussion
The ability to achieve a rapid, inexpensive, and specific knockdown of neuronal genes for in vivo target validation is one of the prime ambitions of modern neuroscience, particularly with the immensity of targets being identified in gene-expression analysis of brain disorders. Frequently, these targets exhibit an expression and functional profile that encompasses a complex neuronal circuitry involving several brain regions. In the present study, we characterize a nonviral in vivo method of somatic gene manipulation by using siRNA to attain a widespread sequence-specific gene knockdown in the brain. This method would accelerate the functional investigation of broadly expressed target genes implicated in neurological and psychiatric disorders.
We evaluated the efficiency of an intracerebroventricularly infused siRNA in knocking down a transgene, EGFP, that was overexpressed in the mouse genome. siRNA produced a significant, time-dependent, and specific reduction of transgene mRNA in the brain. This effect was pronounced in regions adjacent to the site of infusion and spread as far as the prefrontal cortex and substantia nigra (rostrocaudal axis), cerebral cortex and hypothalamus (dorsoventral axis), and amygdala (mediolateral axis). Although siRNA infusion appears to have a general spread of action in the brain, its efficacy is clearly determined by the nature of neurons constituting each region. This concept is apparent because a time-dependent silencing effect of siRNA was not observed in all brain regions near the infusion site. In fact, regions such as the globus pallidus (closest to the infusion site) and habenula (with relatively ready access to the dorsal third ventricle) showed no further decline in EGFP mRNA after 2 weeks as compared with 1 week of siRNA infusion. On the contrary, the hypothalamus, which was resistant to the gene-silencing effect of siRNA in the first week, revealed a potent RNAi response after the second week of siRNA treatment. The magnitude of decrease in EGFP mRNA also varied among brain regions, with the hippocampal neurons demonstrating a maximal siRNA response.
We quantified changes in EGFP protein levels based on a correlation with changes in the intensity of fluorescence emitted from EGFP in microscopic fields of each brain region. A siRNA-induced decline in protein levels of EGFP closely paralleled the mRNA reduction in each brain region, with a maximal effect obtained in the hippocampus as well. The magnitude of decrease in protein was, in most cases, less than that in EGFP mRNA, such that regions demonstrating <15% decrease in EGFP mRNA showed no changes in protein levels with siRNA treatment. Such a trend could suggest that not all changes in EGFP mRNA precisely translate to those at the protein level. However, brain sections of mice overexpressing EGFP, under the control of β-actin promoter, emit an intense “carpet-like” fluorescent signal that is likely to underestimate the inhibiting potency of siRNA on EGFP. Despite the elevated expression and stability of EGFP in these mice, siRNA produced a specific and temporal knockdown (up to 50%) of this transgene with an efficiency comparable with that reported by using viral-mediated RNAi (25–30%; refs. 11 and 13). Moreover, the spread of RNAi (≈5.7 mm anteroposterior) obtained in the brain with our nonviral approach is much more extensive than that demonstrated by viral methods (≈0.8 mm anteroposterior; ref. 11).
We next validated the efficacy of our RNAi protocol in producing a widespread gene knockdown by targeting an endogenous gene expressed far caudal to the infusion site. We chose to investigate DAT knockdown, because the behavioral effects of pharmacological or genetic inhibition of this protein have been well characterized, particularly hyperactivity (34, 35). Infusion of an in vitro validated siRNA into the dorsal third ventricle led to a significant down-regulation of DAT mRNA in the ventral midbrain neurons. Levels of DAT protein also were down-regulated in the midbrain projection areas, such as the caudate putamen, nucleus accumbens, and olfactory tubercle. The behavioral consequence of down-regulating the DAT gene, by using siRNA in adult mice, was a marked hyperlocomotor response, as has been reported in DAT-knockout mice (34). This increase in locomotor activity was not observed up to day 6 of siRNA infusion but was observed on subsequent test days, which is consistent with the ≈7-day turnover rate of DAT protein in vivo (36, 37). In contrast, pharmacological inhibition of the DAT protein with GBR-12909 produced an almost immediate hyperlocomotor response as observed on the first test day after infusion. GBR-12909 also produced a significant down-regulation of DAT protein without affecting the mRNA levels. Similar effects of GBR-12909 are described in refs. 35 and 38.
siRNA infusion into the dorsal third ventricle takes advantage of a direct access to many brain regions simultaneously that contribute to a complex neuronal circuitry underlying neuropsychiatric psychopathologies, thus enabling a widespread and bilateral gene knockdown in the brain. Furthermore, a rapid clearance from the ventricular system reduces the likelihood of any nonspecific effects. In the present study, mRNA or protein levels of two other genes (Th and SERT) were unaltered in the corresponding brain regions, suggestive of a sequence-specific effect of the DAT siRNA. Moreover, no signs of general toxicity (neuroanatomical damage and loss of body weight) were evident in mice after infusion of siRNA at the specified dose. In fact, all mice receiving DAT siRNA survived throughout the experimental period as opposed to the DAT-knockout mice that exhibit a high propensity to premature death (34).
Here, we demonstrate the previously undescribed potential utility of siRNA in achieving a widespread sequence-specific silencing of exogenous and, more interestingly, endogenous neuronal genes in vivo in the brain. Our results highlight the temporal effects of siRNA, requiring a constant minipump-mediated infusion in the dorsal third ventricle for a stable and bilateral knockdown of gene expression in the brain. This finding is in agreement with previous studies demonstrating a transient or no RNAi effect after a single or short-term application of siRNA in a specific brain region (39, 40). Further modifications of the RNAi protocol, presented in the current study, also will allow for reversible gene knockdown or simultaneous knockdown of multiple genes in the adult brain. Our nonviral RNAi approach is efficient in producing a distinct behavioral phenotype similar to that obtained either in knockout animals or after pharmacological inhibition of the gene product. Pump-mediated infusion of siRNA, therefore, provides a rapid means of in vivo validation of novel targets implicated in neurological and psychiatric disorders, where ligands interacting with the protein products of these targets are unavailable.
Supplementary Material
Acknowledgments
We thank Sabine Leonhard, Hugo Bürki, Dominique Fehlmann, Dora Khar, David Kirk, Jose Crespo, Gilles Sansig, and Cedric Mombereau for excellent technical assistance; Dr. Christian Chatenay-Rivauday, Dr. Anna Ottlecz, and Hugues Ryckelynck for help in quantifying the fluorescent signal; Drs. Kumlesh K. Dev and Klemens Kaupmann for critical review of the manuscript; and Dr. Graeme Bilbe (Head of Neuroscience Research, Novartis Institutes for Biomedial Research) for continuous support of our RNAi projects.
This paper was submitted directly (Track II) to the PNAS office.
Abbreviations: RNAi, RNA interference; siRNA, short interfering RNA; scrRNA, scrambled sequence RNA; mmRNA, mismatch siRNA; TH, tyrosine hydroxylase; SERT, serotonin transporter; DAT, dopamine transporter; GABAA, GABA type A; GABAAα2, GABAA receptor subunit 2.
References
- 1.Mirnics, K., Middleton, F. A., Marques, A., Lewis, D. & Levitt, P. (2000) Neuron 28, 53–67. [DOI] [PubMed] [Google Scholar]
- 2.Dickey, C. A., Loring, J. F., Montgomery, J., Gordon, M. N., Eastman, P. S. & Morgan, D. (2003) J. Neurosci. 23, 5219–5226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.McClung, C. A. & Nestler, E. J. (2003) Nat. Neurosci. 6, 1208–1215. [DOI] [PubMed] [Google Scholar]
- 4.Dorsett, Y. & Tuschl, T. (2004) Nat. Rev. Drug Discov. 3, 318–329. [DOI] [PubMed] [Google Scholar]
- 5.Fire, A., Xu, S., Montgomery, M. K., Kostas, S. A., Driver, S. E. & Mello, C. C. (1998) Nature 391, 806–811. [DOI] [PubMed] [Google Scholar]
- 6.Caplen, N. J., Parrish, S., Imani, F., Fire, A. & Morgan, R. A. (2001) Proc. Natl. Acad. Sci. USA 98, 9742–9747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Elbashir, S. M., Harborth, J., Lendeckel, W., Yalcin, A., Weber, K. & Tuschl, T. (2001) Nature 411, 494–498. [DOI] [PubMed] [Google Scholar]
- 8.Davidson, B. L. & Paulson, H. L. (2004) Lancet Neurol. 3, 145–149. [DOI] [PubMed] [Google Scholar]
- 9.Holen, T. & Mobbs, C. V. (2004) Neuroscience 126, 1–7. [DOI] [PubMed] [Google Scholar]
- 10.Xia, H., Mao, Q., Paulson, H. L. & Davidson, B. L. (2002) Nat. Biotechnol. 20, 1006–1010. [DOI] [PubMed] [Google Scholar]
- 11.Hommel, J. D., Sears, R. M., Georgescu, D., Simmons, D. L. & DiLeone, R. J. (2003) Nat. Med. 9, 1539–1544. [DOI] [PubMed] [Google Scholar]
- 12.van den Haute, C., Eggermont, K., Nuttin, B., Debyser, Z. & Baekelandt, V. (2003) Hum. Gene Ther. 14, 1799–1807. [DOI] [PubMed] [Google Scholar]
- 13.Xia, H., Mao, Q., Eliason, S. L., Harper, S. Q., Martins, I. H., Orr, H. T., Paulson, H. L., Yang, L., Kotin, R. M. & Davidson, B. L. (2004) Nat. Med. 10, 816–820. [DOI] [PubMed] [Google Scholar]
- 14.Lewis, D. A. & Lieberman, J. A. (2000) Neuron 28, 325–334. [DOI] [PubMed] [Google Scholar]
- 15.Nestler, E. J., Gould, E., Manji, H., Bucan, M., Duman, R. S., Greshenfeld, H. K., Hen, R., Koester, S., Lederhendler, I., Meaney, M., et al. (2002) Biol. Psychiatry 52, 503–528. [DOI] [PubMed] [Google Scholar]
- 16.Sisodia, S. S. & St George-Hyslop, P. H. (2002) Nat. Rev. Neurosci. 3, 281–290. [DOI] [PubMed] [Google Scholar]
- 17.Millan, M. J. (2003) Prog. Neurobiol. 70, 83–244. [DOI] [PubMed] [Google Scholar]
- 18.Chao, J. & Nestler, E. J. (2004) Annu. Rev. Med. 55, 113–132. [DOI] [PubMed] [Google Scholar]
- 19.Matsumoto, Y., Hof, A., Baumlin, Y., Müller, M., Prescott, M. F. & Hof, R. P. (2003) Transplantation 76, 1569–1572. [DOI] [PubMed] [Google Scholar]
- 20.Giros, B., El Mestikawy, S., Bertrand, L. & Caron, M. G. (1991) FEBS Lett. 295, 149–154. [DOI] [PubMed] [Google Scholar]
- 21.Amara, S. G. & Kuhar, M. J. (1993) Annu. Rev. Neurosci. 16, 73–93. [DOI] [PubMed] [Google Scholar]
- 22.Giros, B. & Caron, M. G. (1993) Trends Pharmacol. Sci. 14, 43–49. [DOI] [PubMed] [Google Scholar]
- 23.Cook, E. H. Jr., Stein, M. A., Krasowski, M. D., Cox, N. J., Olkon, D. M., Kieffer, J. E. & Leventhal, B. L. (1995) Am. J. Hum. Genet. 56, 993–998. [PMC free article] [PubMed] [Google Scholar]
- 24.Gill, M., Daly, G., Heron, S., Hawi, Z. & Fitzgerald, M. (1997) Mol. Psychiatry 2, 311–313. [DOI] [PubMed] [Google Scholar]
- 25.Storck, T., Kruth, U., Kolhekar, R., Sprengel, R. & Seeburg, P. H. (1996) Nucleic Acids Res. 24, 4594–4596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Dorn, G., Patel, S., Wotherspoon, G., Hemmings-Mieszczak, M., Barclay, J., Natt, F. J. C., Martin, P., Bevan, S., Fox, A., Ganju, P., et al. (2004) Nucleic Acids Res. 32, e49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hüsken, D., Asselbergs, F., Kinzel, B., Natt, F., Weiler, J., Martin, P., Häner, R. & Hall, J. (2003) Nucleic Acids Res. 31, e102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Paxinos, G. & Franklin, K. B. J. (2001) The Mouse Brain in Stereotaxic Coordinates (Academic, London), 2nd Ed.
- 29.Cryan, J. F., Kelly, P. H., Chaperon, F., Gentsch, C., Mombereau, C., Lingenhoehl, K., Froestl, W., Bettler, B., Kaupmann, K. & Spooren, W. P. (2004) J. Pharmacol. Exp. Ther. 310, 952–963. [DOI] [PubMed] [Google Scholar]
- 30.Bischoff, S., Barhanin, J., Bettler, B., Mulle, C. & Heinemann, S. (1997) J. Comp. Neurol. 379, 541–562. [DOI] [PubMed] [Google Scholar]
- 31.Sellings, L. H. L. & Clarke, P. B. S. (2003) J. Neurosci. 23, 6295–6303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lorang, D., Amara, S. G. & Simerly, R. B. (1994) J. Neurosci. 14, 4903–4914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ciliax, B. J., Heilman, C., Demchyshyn, L. L., Pristupa, Z. B., Ince, E., Hersch, S. M., Niznik, H. B. & Levey, A. I. (1995) J. Neurosci. 15, 1714–1723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Giros, B., Jaber, M., Jones, S. R., Wightman, R. M. & Caron, M. G. (1996) Nature 379, 606–612. [DOI] [PubMed] [Google Scholar]
- 35.Izenwasser, S., French, D., Carroll, F. I. & Kunko, P. M. (1999) Behav. Brain Res. 99, 201–208. [DOI] [PubMed] [Google Scholar]
- 36.Kimmel, H. L., Carroll, F. I. & Kuhar, M. J. (2000) Neuropharmacology 39, 578–585. [DOI] [PubMed] [Google Scholar]
- 37.Fleckenstein, A. E., Pogun, S., Carroll, F. I. & Kuhar, M. J. (1996) J. Pharmacol. Exp. Ther. 279, 200–206. [PubMed] [Google Scholar]
- 38.Kunko, P. M., Loeloff, R. J. & Izenwasser, S. (1997) Naunyn-Schmiedeberg's Arch. Pharmacol. 356, 562–569. [DOI] [PubMed] [Google Scholar]
- 39.Isacson, R., Kull, B., Salmi, P. & Wahlestedt, C. (2003) Acta Physiol. Scand. 179, 173–177. [DOI] [PubMed] [Google Scholar]
- 40.Makimura, H., Mizuno, T. M., Mastaitis, J. W., Agami, R. & Mobbs, C. V. (2002) BMC Neurosci. 3, 18. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.