Abstract
Plasminogen activator inhibitor-1 (PAI-1) plays an important role in the pathogenesis of obesity-driven type 2 diabetes mellitus and associated cardiovascular complications. Here, we show that perturbation of caveolar microdomains leads to insulin resistance and concomitant up-regulation of PAI-1 in 3T3L1 adipocytes. We present several lines of evidence showing that the phosphatidylinositol 3-kinase (PI3K) pathway negatively regulates PAI-1 gene expression. Insulin-induced PAI-1 gene expression is up-regulated by a specific inhibitor of PI3K. In addition, serum PAI-1 level is elevated in protein kinase Bα-deficient mice, whereas it is reduced in p70 ribosomal S6 kinase 1-deficient mice. The PI3K pathway phosphorylates retinoblastoma protein (pRB), known to release free E2 (adenoviral protein) factor (E2F), which we have previously demonstrated to be a transcriptional repressor of PAI-1 gene expression. Accordingly, cell-penetrating peptides that disrupt pRB–E2F interaction, and thereby release free E2F, are able to suppress PAI-1 levels that are elevated during insulin-resistant conditions. This study identifies a caveolar-dependent signal pathway that up-regulates PAI-1 in insulin-resistant adipocytes and proposes a previously undescribed pharmacological paradigm of disrupting pRB–E2F interaction to suppress PAI-1 levels.
Keywords: diabetes
Type 2 diabetes mellitus (T2DM) is characterized by insulin resistance, where the insulin receptor (IR) fails to elicit the metabolic signaling that is required for glucose metabolism and energy homeostasis. In insulin-sensitive tissues, the IR transduces two main signaling cascades: a metabolic signaling that is responsible for glucose uptake and glycogen synthesis and a mitogenic signaling that is responsible for cell proliferation and growth. The IR substrate (IRS)–phosphatidylinositol 3-kinase (PI3K)–protein kinase B (PKB) and Cbl-CAP-Flotillin pathways represents the major metabolic signaling, whereas the Shc-Ras-extracellular regulated kinase (Erk) pathway represents the major mitogenic signaling (1). Both in animal models and clinical T2DM subjects, a selective impairment of metabolic signaling has been observed, whereas mitogenic signaling is more or less unaffected (2–4).
Obesity is prominent among the plethora of factors that leads to the development of T2DM, although the molecular mechanism underlying the pathogenesis of obesity-driven T2DM is not well understood. Comparative analysis of large and small fat cells within the same fat pad reveals a 2-fold reduction in the levels of plasma membrane cholesterol in large fat cells, suggesting that a decrease in membrane cholesterol is characteristic of adipocyte hypertrophy per se (5). Recently, it has been proposed that the protein levels of caveolin-1 and -3 are inversely correlated to the body-mass index.§ Plasma membrane cholesterol (6) and caveolins (7) are indispensable for the structural and functional integrity of caveolar microdomains. We therefore reasoned that obesity might lead to caveolar dysfunction. Because the IR and several of its downstream signal transducers are localized in caveolae (8), it was intriguing to investigate whether caveolar dysfunction could lead to insulin resistance.
There is compelling evidence that plasminogen activator inhibitor-1 (PAI-1), whose levels are elevated in both obesity and T2DM, plays an important role in the development of cardiovascular disorders (9). PAI-1, a primary physiological inhibitor of plasminogen activators (uPA and tPA), inhibits both fibrinolysis and proteolysis and plays an important role in mediating the cardiovascular complications associated with T2DM such as nephropathy (10), retinopathy (11), coronary artery disorders (12), and hypertension (13). Consequently, it is believed that normalizing plasma PAI-1 levels will retard the progression of cardiovascular complications (14). Insulin-induced Erk phosphorylation, followed by the activation of transcription factors of the AP-1 family, is considered to be partly, if not wholly, responsible for insulin-induced PAI-1 up-regulation in insulin-sensitive tissues (15, 16). However, because the mitogenic mitogen-activated protein kinase pathway is not affected during hyperinsulinemic conditions, such as insulin resistance or T2DM, this pathway is unlikely to be the primary cause of PAI-1 elevation during these pathological conditions. It is, therefore, intriguing to investigate whether insulin induction of PAI-1 during T2DM can be explained by directly linking the compromised PI3K pathway to PAI-1 up-regulation. Interestingly, it has been shown that IR-mediated activation of metabolic signaling (PI3K pathway) induces the phosphorylation of retinoblastoma protein (pRB) in adipocytes (17). pRB phosphorylation is known to lead to the release of free E2 (adenoviral protein) factor (E2F) (18). We have demonstrated (19) that E2F transcription factors can negatively regulate PAI-1 gene expression by repressing PAI-1 promoter activity independently of its binding to pocket proteins, revealing a novel mechanism for the E2F-mediated repression of gene expression. In this study, we investigate whether caveolar dysfunction can lead to insulin resistance, and whether the resulting impairment of the PI3K-PKB-E2F pathway can by itself lead to the up-regulation of PAI-1. We also explore the pharmacological disruption of the E2F–pRB interaction to release free E2F, which we hypothesize will attenuate PAI-1 transcription and hence its plasma level. Adipocytes are chosen for this study, because they are responsible for obesity, abundant in caveolae (20), highly sensitive to insulin (>200,000 receptors per cell) (21), the primary site for insulin resistance (22), and a major contributor of plasma PAI-1 in the obese (23).
Materials and Methods
Reagents. Methyl-β-cyclodextrin (MBCD) and filipin III were obtained from Sigma. Monoclonal antibodies against E2F1 (KH-95) and E2F2 (TFE-25) and rabbit polyclonal antibodies against E2F3 (C-18), E2F4 (C-20), E2F5 (C-20), pRB (M-153), p130 (C-20), p107 (C-18), IRS-1 (C-20), Erk, and PAI-1 (H-135) were from Santa Cruz Biotechnology. Rabbit polyclonal antibodies against phospho-pRB (Ser-795) and phospho-Erk were from Cell Signaling Technology (Beverly, MA). Rabbit polyclonal antibodies against Shc and phospho-tyrosine (G410) were from Transduction Laboratories (Lexington, KY). Sheep polyclonal antibody against PAI-1 was from American Diagnostics (Greenwich, CT). All reagents for real-time PCR were from Applied Biosystems. The oligonucleotide E2pro, the sequence of which corresponded to nucleotides –72 to –32 of the adenovirus E2 promoter and contained E2F-binding sites, had the following sequence (only the upper strand is given) and was used for gel-shift assays: 5′-GAT CAG TTT TCG CGC TTA AAT TTG AGA AAG GGC GCG AAA CTA G-3′.
Adipocyte Differentiation. 3T3-L1 preadipocytes were cultured in DMEM containing 10% FCS, and 2 days after cells reached confluency, the medium was changed to DMEM containing 10% FCS, 10 μg/ml insulin, 1 μM dexamethasone, and 0.5 mM isobutylmethylxanthine. Two to three days later, this medium was replaced with DMEM supplemented only with 10 μg/ml insulin, and cells were kept for 2 days. The medium was then replaced with DMEM containing 10% FCS every 2 days. The cells were serum-starved overnight before experiments.
Immunoprecipitation and Western Blotting. Immunoprecipitation and Western blotting were performed as described (24).
Glucose Uptake Assay. Measurements of 2-deoxyglucose uptake into adipocytes were carried out as described (25).
RNA Isolation and Northern Blot Analysis. Total RNA (12 μg) was isolated and subjected to Northern blot analysis as described (26). The cDNA clone for mouse PAI-1 was provided by A. Riccio (University of Naples, Naples).
Quantitative Real-Time PCR. One microgram of total RNA was reverse transcribed and 1 μl of RT reaction was added to 24 μl of PAI-1 PCR reaction (1× universal master mix/900 nM forward primer [5′-CCTGGCCGACTTCACAAGTC-3′]/900 nM reverse primer [5′-TTGCAGTGCCTGTGCTACAGA-3′]/200 nM TaqMan probe [5′-FAM-TCCGACCAAGAGC-MGB-3′]). Thermal cycling was done as follows: 50°C for 2 min, followed by 95°C for 10 min and then 45 cycles of 95°C for 1 min and 60°C for 1 min. The fluorophor dyes for the PAI-1 probe and 18S rRNA (internal control) probe were 6-carboxyfluorescein and VIC, respectively. The quencher in both probes was tetra-methylrhodamine. The reaction was carried out in an ABI Prism 7700. The output raw data were normalized with internal control and statistically analyzed by using ms excel (Microsoft).
Small Interfering RNA (siRNA) Nucleofection. siRNA used for targeting caveolin-1 mRNA has the following sequence: sense, 5′-GAGCUUCCUGAUUGAGAUU-3′ and antisense, 5′-AAUCUCAAUCAGGAAGCUC-3′. Control siRNA sequences: sense, 5′-GUACCUGACUAGUCGCAGAAG-3′ and antisense, 5′-UCUGCGACUAGUCAGGUACGG-3′. These sequences contain 3′ UU overhangs. Specificities of these sequences were confirmed by performing a blast search against the GenBank/European Molecular Biology Laboratory database. Each siRNA (final concentration 1 μM) was mixed with NIH 3T3 cells overexpressing the human IR (NIH-IR) cell suspension (2 × 106 cells in 0.1 ml of buffer-T/transfection), transferred to a 2-mm electroporation cuvette, and electroporated by using an Amaxa Nucleofector (Amaxa, Cologne, Germany) by using the program a-23. After electroporation, cells were immediately transferred to 1 ml of growth medium, and cultured in six-well plates at 37°C until analysis.
p70 Ribosomal S6 Kinase 1 (S6K1)–/– and PKBβ–/– Mice. PKBα knockout mice were generated as described (27). S6K1 knockout mice (28) were kindly provided by G. Thomas (Friedrich Miescher Institute). The S6K1–/– mice and their wild-type counterparts were fed with high-fat diet for 5–6 months (required for hyperactivity of PI3K pathway) before their blood was taken by using tail punctures. PKBα–/– mice were fed with a normal chow diet.
Cell-Penetrating Peptide Treatment. The sequence of the interfering peptide of 18-aa length was derived from the pRB-binding region of E2F1 (amino acids 402–419: LDYHFGLEEGE-GIRDLFD) (29). A control peptide with the same amino acid composition, GEELEGFHDGLLDFDIR, was prepared by randomly shuffling the sequence of this peptide. Cell-penetrating peptides were prepared by coupling these peptides to the carboxy terminal of the cell-penetrating region of the HIV tat protein (amino acid 47–57: RRRQRRKKR) (30) via hinge peptide G. Differentiated adipocytes were separated from undifferentiated cells by using a Percoll density gradient as described (31) with a slight modification. Adipocytes were pretreated with collagenase (2 mg/ml) for 30 min, centrifuged at 1,500 rpm (Sorvall H4000, Sorvall) for 5 min at 4°C, and mixed with Percoll solution (1.025 g/ml) to form a homogenous suspension. The cell suspension was then layered on preformed Percoll solution (1.035 g/ml) and centrifuged at 3,000 rpm (Sorvall H4000, Sorvall) for 20 min at 4°C. The cells collected from the upper layer were resuspended in media and reseeded for the experiment. The penetrating peptide was added to cells, incubated for 16 h, and then subjected to various treatments.
Nuclear Extracts and Electromobility-Shift Assays. Nuclear extracts (5 μg) were first incubated at room temperature for 15 min in 20 μl of binding reaction mixture containing 50 mM KCl, 20 mM Hepes (pH 7.9), 0.2 mM EDTA, 6% glycerol, 0.5% Ficoll 400, 1 μg of salmon sperm DNA, 6 μg of BSA, and 1 mM DTT with or without penetrating peptide and antibodies, followed by a further 15-min incubation after addition of 0.3 ng of radiolabeled oligonucleotide probes. Oligonucleotide probes were radiolabeled by using Escherichia coli polynucleotide kinase and [γ-32P]ATP. Aliquots (5 μl) of reaction mixture were separated in a 4.5% polyacrylamide gel run in 0.25× TBE buffer (90 mM Tris/64.6 mM boric acid/2.5 mM EDTA, pH 8.3) at room temperature. The gel was dried and analyzed in a PhosphorImager.
Results
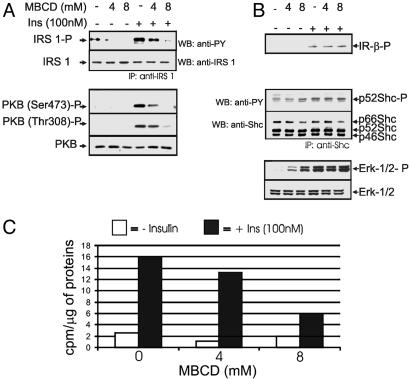
Perturbation of Caveolar Function Mimics Insulin Resistance in 3T3L1 Adipocytes. To find out whether caveolar dysfunction can cause insulin resistance, we perturbed the integrity of caveolar microdomains and examined the subsequent effects on insulin signaling. Cholesterol depletion using MBCD, a reagent widely used to perturb the structural integrity of caveolae, was used. MBCD pretreatment dose-dependently inhibited insulin-induced IRS-1 phosphorylation, PKB phosphorylation (Fig. 1A), and 2-deoxyglucose uptake (Fig. 1C). On the other hand, MBCD pretreatment did not affect insulin-induced phosphorylation of the β-subunit of the IR, Shc, or Erk (Fig. 1B). MBCD treatment alone (in the absence of insulin) was also found to increase the phosphorylation of p52 Shc and Erk-1/2, although to a lesser extent (Fig. 1B).
Fig. 1.
Effect of cholesterol depletion on insulin signaling. 3T3L1 adipocytes were treated with 0, 4, or 8 mM MBCD for 40 min, followed by insulin treatment (100 nM) for 10 min, and then whole-cell extracts were prepared. The cell lysates were fractionated by SDS/PAGE, followed by Western blotting analysis using the specific antibodies indicated. For the analysis of IRS-1 and Shc, the lysates were first immunoprecipitated by using IRS-1 and Shc antibodies, respectively, before Western blotting. (A) Analysis of PKB and IRS-1, molecules involved in the metabolic signal pathway. (B) Analysis of the IR (IR) and its mitogenic signal transducers Shc and Erk. (C) Effect of MBCD on insulin-induced glucose uptake. Adipocytes were treated similarly as above and then subjected to a glucose uptake assay. All data shown here are representative of at least three independent experiments.
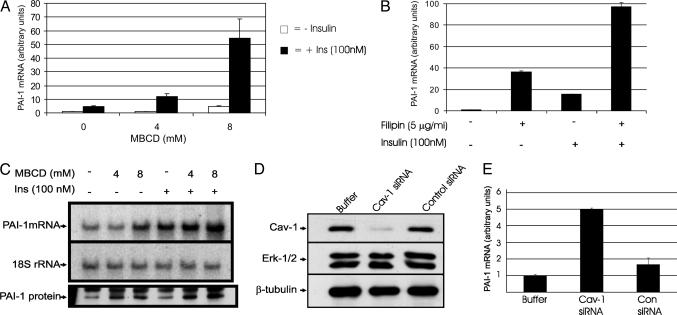
Induction of Insulin Resistance Leads to Concomitant Increase in PAI-1 Gene Expression. To determine whether MBCD-induced insulin resistance leads to the up-regulation of insulin-induced PAI-1 gene expression, we measured PAI-1 mRNA levels after insulin treatment with or without MBCD pretreatment. Results from both real-time PCR (Fig. 2A) and Northern blot hybridization (Fig. 2C) show that MBCD dose-dependently increased insulin-induced PAI-1 levels. To find out whether this increase at the mRNA level is reflected at the protein level, PAI-1 protein levels were measured in the media. Corresponding to the mRNA levels, insulin-induced PAI-1 protein levels were dose-dependently up-regulated by MBCD treatment. Filipin, a structurally distinct sterol-binding compound, also augmented the insulin-induced PAI-1 levels (Fig. 2B), suggesting that the observed MBCD effects were through perturbation of caveolae function per se. Fig. 2 A and B clearly show that the effect of cholesterol depletion synergistically up-regulated insulin-induced PAI-1 gene expression. To ascertain that the up-regulation of PAI-1 observed here is due to caveolar dysfunction and not due to an unspecific effect of cholesterol depletion, we depleted the caveolin-1 protein using siRNA directed against caveolin-1 mRNA. In this experiment, NIH-IR were used instead of adipocytes, which proved difficult to transfect efficiently with siRNA. NIH-IR cells were chosen as the suitable alternative to adipocytes, because they are insulin-sensitive, rich in caveolae, share a common cell lineage with adipocytes, and can be differentiated into adipocytes (32). This siRNA showed specific effects, because it significantly lowered the caveolin-1 protein levels but did not affect the levels of other proteins such as Erk and β-tubulin (Fig. 2D). Furthermore, control siRNA had no effect on any of these proteins. Caveolin-1 siRNA treatment enhanced the levels of insulin-induced PAI-1 5-fold, whereas control siRNA had no significant effect (Fig. 2E).
Fig. 2.
Effect of caveolar dysfunction on insulin-induced PAI-1 mRNA levels. (A) Effect of MBCD on insulin-induced PAI-1 mRNA levels. 3T3L1 adipocytes were treated with 0, 4, or 8 mM MBCD for 40 min, with or without subsequent insulin treatment (100 nM) for 2 h. The total RNA was prepared and PAI-1 mRNA levels were measured by using real-time PCR with 18S rRNA as an internal control. (B) Effect of filipin on insulin-induced PAI-1 mRNA levels. Adipocytes were treated with filipin (5 μg/ml) for 40 min, with or without subsequent 2 h insulin (100 nM) treatment. Total RNA was analyzed as above. (C) Northern and Western blotting analysis. The same RNA samples used in A were subjected to Northern blot hybridization analysis for PAI-1 mRNA levels. For the analysis of PAI-1 protein levels, cells were treated with 0, 4, or 8 mM MBCD for 40 min, followed by insulin (100 nM) treatment for 6 h. Then, PAI-1 protein was immunoprecipitated from the conditioned media (because PAI-1 is a secreted protein) by using protein-G beads coupled to sheep anti-PAI-1 antibodies, and analyzed by Western blotting by using rabbit anti-PAI-1 antibodies. (D and E) Effects of caveolin down-regulation. NIH-IR cells were electroporated by using buffer without siRNA or with Cav-1 or control siRNA. After 24 h, the cells were treated with insulin (100 nM) for 2 h. Protein levels of caveolin-1, Erk, and β-tubulin (D) and mRNA levels of PAI-1 (E) were examined by Western blotting and RT-PCR, respectively.
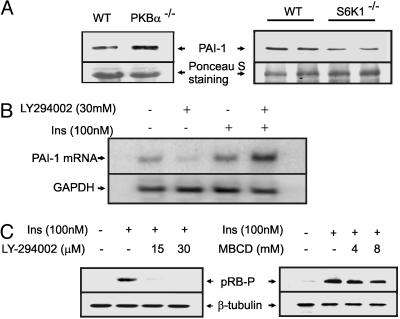
Impairment of PI3K Pathway Leads to Transcriptional Up-Regulation of PAI-1. To see whether the up-regulation of insulin-induced PAI-1 by caveolar dysfunction was a direct consequence of impaired metabolic signaling (PI3K pathway), we examined the levels of plasma PAI-1 in two different mouse models that are hallmarked by augmented (S6K1–/–) and attenuated (PKBα–/–) metabolic signaling. Activated S6K1 phosphorylates IRS-1 at serine residues and suppresses its tyrosine phosphorylation by IR. Thus, deletion of S6K1 augments PI3K signaling in these mice (33). On the other hand, PKB is a critical mediator of PI3K signaling. Thus, deletion of PKB would attenuate the PI3K signal pathway. As shown in Fig. 3A, plasma PAI-1 levels were up-regulated in PKBα–/– mice, whereas they were down-regulated in S6K1–/– mice. The negative regulation of PAI-1 gene expression by the PI3K pathway was further confirmed in adipocyte cell culture, where PI3K inhibitor (LY294002) enhanced insulin-induced PAI-1 mRNA levels (Fig. 3B). Treatment with LY294002 and MBCD, both shown to inhibit the PI3K pathway, dose-dependently inhibited insulin-induced pRB phosphorylation (Fig. 3C), thus providing a possible explanation of why PI3K pathway activation leads to the down-regulation of PAI-1 gene expression (see below).
Fig. 3.
The role of the PI3K pathway in PAI-1 gene expression. (A) Serum PAI-1 levels of PKBα–/–, S6K1–/– mice, and their wild-type counterparts were analyzed by PAI-1 immunoprecipitation followed by Western blotting. The membranes were stained with Ponceau S for loading control. (B) 3T3L1 adipocytes were treated with LY294002 (30 μM) for 45 min, followed by insulin (100 nM) for 2 h. Total RNA was prepared and analyzed for levels of PAI-1 and GAPDH (loading control) mRNAs by Northern blot hybridization. (C) 3T3L1 adipocytes were treated with increasing concentrations of either LY294002 or MBCD for 45 min, followed by insulin (100 nM) for 2 h, and the phosphorylation status of pRB and the protein levels of β-tubulin (loading control) were measured by Western blotting.
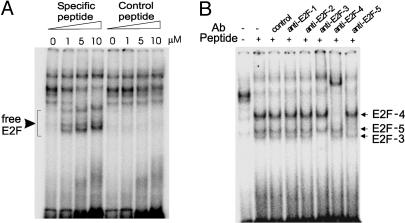
Disruption of pRB–E2F Interaction Using a Cell-Penetrating Peptide. Hypophosphorylated pRB is bound to E2F forming an inactive complex, whereas hyperphosphorylation of pRB leads to the release of free E2F (18). We have previously shown that overexpression of E2F isoforms can down-regulate PAI-1 gene expression (19). It has also been shown that active pRB, which can bind to E2F, reverses this down-regulation (26). Taken together, these results suggest that free E2F acts as a transcriptional repressor of the PAI-1 gene. We have observed that pRB phosphorylation is increased, whereas E2F-1 protein levels are reduced during adipogenesis (data not shown), suggesting that E2F activity (corresponding to free E2F) may be compromised in differentiated adipocytes. To restore the potential of E2F-mediated transcriptional repression of the PAI-1 gene in adipocytes, we sought to release free E2F by disrupting the E2F–pRB complex using an interfering peptide that corresponds to amino acids 402–419 of E2F1, the domain interacting with the pocket proteins (29). A blast search revealed that this domain is fairly conserved among members of the E2F family (excluding E2F-6) and, therefore, the peptide that we designed may disrupt the pocket protein–E2F interaction in general. As a control, we used a peptide with a randomly shuffled sequence. To render these peptides cell-penetrable, an HIV-1 Tat-derived peptide sequence was tagged to these peptides (30). The avidity of these peptides to disrupt the E2F–pRB complex was confirmed by DNA gel-shift assays using adipocyte nuclear extracts and a radioactive oligonucleotide, the sequence of which corresponds to the E2F-binding site of the adenovirus E2 promoter. As shown in Fig. 4A, DNA–protein complexes shifted to low-molecularweight forms when increasing concentrations of the specific peptide were added to binding mixtures; the control peptide had no effect. Supershift assays using specific antibodies against E2F1–5 revealed that the main isoforms in the freed E2F fractions were E2F3, -4, and -5 (Fig. 4B). These peptides thus serve as an effective tool to disrupt endogenous E2F–pRB interactions.
Fig. 4.
Effect of cell-penetrating interfering peptide on free E2F levels. (A) In DNA gel-shift assays using 32P-labeled E2pro oligonucleotide and nuclear extracts from adipocytes, cell-penetrating peptide (specific and control peptides) was added to binding reactions at increasing concentrations. (B) Nuclear extracts (day 8) were preincubated with the cell-penetrating interfering peptide together with specific antibodies against different E2F members for 15 min and then analyzed for E2F DNA-binding activity by gel-shift assays as above.
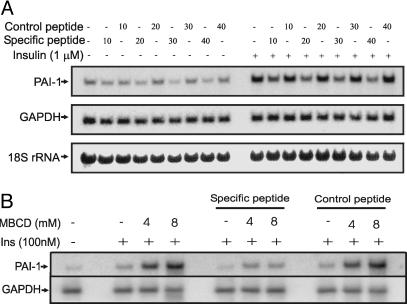
Elevation of PAI-1 Levels in Insulin Resistance Is Suppressed by the Cell-Penetrating Peptide. To examine whether interfering peptide treatment can compromise induction of PAI-1 by hyperinsulinemia and insulin resistance, we performed the following experiments. Adipocytes were prepared according to Materials and Methods and were treated with either the specific or control peptide for 12 h followed by 1 μM insulin for 2 h. As shown in Fig. 5A, the specific peptide, but not the control peptide, suppressed both basal and insulin-induced PAI-1 levels in a dose-dependent manner, the latter being more potently affected (Fig. 5A). Furthermore, the specific peptide also suppressed PAI-1 expression that was up-regulated by MBCD-induced insulin resistance (Fig. 5B). Again, the control peptide had no effect.
Fig. 5.
Effect of cell-penetrating interfering peptide on PAI-1 mRNA levels. (A) Dose-response of the peptides. 3T3L1 adipocytes (day 8) were treated with increasing concentrations (0–40 μM) of either specific or control peptide for 6 h, followed by treatment with or without 1 μM insulin for 2 h. Total RNA was prepared, and levels of PAI-1 and GAPDH (loading control) mRNAs were measured by Northern blot hybridization. Ribosomal RNAs were stained with methylene blue to serve as an additional control for equal loading and blotting. (B) Effect of the peptides on PAI-1 mRNA levels, which are elevated under insulin-resistant conditions. Cells were serum-starved and then treated with 40 μM of specific or control peptides for 12 h. Cells were then treated with MBCD for 40 min, followed by insulin (100 nM) for 2 h. Total RNA was prepared and analyzed for PAI-1 mRNA as above.
Discussion
Obesity is characterized by increased adipocyte mass and altered adipocyte physiology. These traits contribute to the progression from obesity to insulin resistance (34). Reductions in plasma membrane cholesterol and protein levels of caveolin-1/3 are associated with adipocyte hypertrophy and obesity, respectively (5, 6). It is thought that this could lead to caveolar dysfunction, because plasma membrane cholesterol and caveolin are essential for the structural and functional integrity of caveolae (6, 7). To study the acute effects of caveolar dysfunction in adipocytes, we used cholesterol-scavenging reagents, such as MBCD, which are known to perturb caveolae in adipocytes (35). Kinetic studies have suggested that the vast majority of cholesterol scavenged by cyclodextrins is from the plasma membrane (36), where >90% of cellular cholesterol is known to reside (37). Treatment with MBCD did not affect insulin-induced tyrosine phosphorylation of the IR and its downstream mitogenic signaling, but impaired the IRS-PKB signal pathway that leads to glucose uptake (Fig. 1 A and B). This suggests that cholesterol depletion and, presumably, consequent caveolar dysfunction would lead to insulin resistance. Consistently, it was recently reported that caveolin-1–/– and caveolin-3–/– mice, both known to have a dramatic reduction in caveolae, exhibit insulin resistance (38, 39). Moreover, in adipocytes treated with TNF-α, the IR accumulated less in the detergent-insoluble low-density membrane fractions (microdomains) and shifted to the high-density fractions, suggesting a translocation of IR from caveolar to noncaveolar fractions (40). TNF-α is a potent inducer of insulin resistance (41) and PAI-1 gene expression (42). It is chronically elevated in conditions of obesity and is a major culprit in the pathogenesis of obesity-driven insulin resistance (41). Thus, denying the caveolar platform for IR signaling could be a plausible mechanism through which obesity per se and its secondary effectors elicit insulin resistance. Our hypothesis warrants further investigations into the structural and functional integrity of caveolae in obese and T2DM patients.
Treatment of adipocytes with two structurally distinct cholesterol-depleting reagents (MBCD and filipin) leads to up-regulation of insulin-induced expression of PAI-1 mRNA and protein levels (Fig. 2 A–C). Similar up-regulation is also obtained in NIH-IR cells after RNAi-mediated caveolin-1 down-regulation (Fig. 2E). Thus, perturbation of caveolae by depleting their integral components, either cholesterol or caveolin-1, leads to significant up-regulation of insulin-induced PAI-1 gene expression. MBCD alone induces PAI-1 mRNA to a certain extent (Fig. 2), but this level of induction can be explained by a slight activation of Erk, because Erk has been reported to up-regulate the PAI-1 gene by activating the AP-1 transcription factor (43). However, the MBCD-mediated synergistic increase in insulin-induced PAI-1 gene expression cannot be explained by Erk activation, because the extent of insulin-induced Erk phosphorylation (Fig. 1B) was not affected by MBCD cotreatment. We therefore postulate an alternative mechanism for the up-regulation of insulin-induced PAI-1 gene expression mediated by caveolar dysfunction (see below).
Insulin-induced PAI-1 mRNA levels are augmented by treatment with a specific PI3K inhibitor, suggesting that the PI3K pathway inhibits PAI-1 gene expression (Fig. 3B). We show that the level of serum PAI-1 is increased in PKBα–/– mice, whereas it is reduced in S6K1–/– mice (Fig. 3A). The whole blood protein concentration in both PKBα and S6K1–/– is similar to that of their wild-type littermates. These results suggest that the functional state of the PI3K pathway plays an important role in the regulation of PAI-1 gene expression.
There are several reports of the PI3K pathway regulating pRB–E2F interactions in a cell-type-independent manner. Activation of PKB by inhibition of its phosphatases reduced the pRB phosphorylation and E2F1 release in C141 cells (44). In T lymphocytes, expression of active PKB is sufficient to induce E2F activity (45). In C33A cells, both wortmannin (PI3K inhibitor) treatment and overexpression of PTEN (a negative regulator of the PI3K-PKB pathway) inhibited pRB phosphorylation, and this was reversed by coexpression of a catalytically active subunit of PI3K (46). In NIH 3T3 fibroblasts, epidermal growth factor-induced pRB phosphorylation and hence G1 to S phase cell cycle progression were inhibited by both LY294002 and wortmannin (47). In agreement with the finding of Usui et al. (17), we show that LY294002 inhibits insulin-induced phosphorylation of pRB in 3T3L1 adipocytes. MBCD also shows similar effects (Fig. 3C), implying that impaired metabolic signaling, characteristic of insulin resistance, may reduce the phosphorylation of pRB. Hypophosphorylated pRB is known to bind E2F1–3 proteins and inhibits their activity either by directly binding to and masking the transactivation domain of E2F or by recruiting histone deacetylases (18). Reduction in pRB phosphorylation, therefore, will result in the reduction of free E2F levels. We have previously shown (26) that free E2F can act as a repressor of PAI-1 gene expression in a variety of cell lines, including U2OS, T98G, SAOS2, LLC-PK1, and MEF cells, suggesting that the E2F-mediated negative regulation of PAI-1 gene expression is a general phenomenon independent of cell type. These observations taken together, we propose that the PI3K pathway in insulin signaling negatively regulates the PAI-1 gene through phosphorylation of pRB and subsequent release of free E2F. This, in essence, is suggestive of differential regulation of PAI-1 gene expression by insulin; positive regulation through the mitogenic pathway, and negative regulation through the metabolic pathway (see Fig. 6, which is published as supporting information on the PNAS web site). The compromised metabolic pathway may explain why insulin-induced PAI-1 levels are up-regulated in states of caveolar dysfunction.
It is widely accepted that PAI-1 levels are raised in conditions of obesity and insulin resistance (9) and play a key role in the development of cardiovascular complications in these patients. Recent studies show that PAI-1 knockout mice are resistant to high-fat-diet-induced obesity and insulin resistance, although the underlying molecular mechanisms are not well understood (48, 49). In line with the data from Vuori et al. (50) showing that vitronectin–αvβ3 integrin interaction facilitates insulin-induced IRS-1 activation, we had earlier proposed a model in which PAI-1 can induce insulin resistance by binding to vitronectin and inhibiting its interaction with αvβ3 integrin (51). This suggests that PAI-1 can be a cause as well as a consequence of insulin resistance, and reducing its levels may offer immense therapeutic value. Both basal and insulin-induced PAI-1 levels could be dramatically reduced by treating adipocytes with a cell-penetrating peptide that physically disrupts the pRB–E2F interaction (Fig. 5A). Furthermore, the interfering peptide was also able to suppress the elevation of PAI-1 mRNA levels in insulin-resistant adipocytes (Fig. 5B). These results strengthen our hypothesis that insulin negatively suppresses PAI-1 gene expression through E2F proteins.
Conclusion
We have demonstrated that caveolar dysfunction leads to the selective impairment of the PI3K pathway in adipocytes. This compromises E2F-mediated suppression of PAI-1 gene expression, resulting in the concomitant up-regulation of PAI-1 levels. Our work thus establishes a direct link between impaired PI3K pathway and elevated PAI-1 gene expression, both characteristics of insulin-resistant conditions. We also propose a pharmacological paradigm of disrupting pRB–E2F interaction to suppress PAI-1 levels that are elevated during insulin resistance (see Fig. 6). Recently, the crystal structure of E2F bound to pRB was solved (52), and this should facilitate the development of small molecule inhibitors of E2F–pRB interaction.
Supplementary Material
Acknowledgments
We are grateful to Sung Hee Um and Francesca Frigerio (Friedrich Miescher Institute) for help with the S6K1–/– mice and Marco Falasca (University College London, London) for the NIH-IR cells. We thank Timothy Garvey (Medical University of South Carolina, Charleston, SC), Wilhelm Krek (Eidgenössische Technische Hochschule, Zurich) and Sandra Kleiner (Friedrich Miescher Institute) for helpful discussions. Derek Brazil (Conway Institute, Dublin) and Hoanh Tran (Medical Research Council–Laboratory of Molecular Biology, Cambridge, U.K.) are duly acknowledged for critical reading of the manuscript. This work was partly supported by the Roche Research Foundation (fellowship to K.H.).
This paper was submitted directly (Track II) to the PNAS office.
Abbreviations: PAI-1, plasminogen activator inhibitor-1; PI3K, phosphatidylinositol 3-kinase; PKB, protein kinase B; S6K1, p70 ribosomal S6 kinase 1; pRB, retinoblastoma protein; E2F, E2 (adenoviral protein) factor; MBCD, methyl-β-cyclodextrin; IR, insulin receptor; IRS, IR substrate; Erk, extracellular regulated kinase; T2DM, type 2 diabetes mellitus; siRNA, small interfering RNA; NIH-IR, NIH 3T3 cells overexpressing the human IR.
Footnotes
Maianu, L., Chen, Y., Simmons, A., Wallace, P., Hutto, A., Shaughnessy, S., Fernandes, J. & Garvey, W. (2003) Diabetes 52, A330 (abstr).
References
- 1.Saltiel, A. R. & Kahn, C. R. (2001) Nature 414, 799–806. [DOI] [PubMed] [Google Scholar]
- 2.Shao, J., Yamashita, H., Qiao, L. & Friedman, J. E. (2000) J. Endocrinol. 167, 107–115. [DOI] [PubMed] [Google Scholar]
- 3.Cusi, K., Maezono, K., Osman, A., Pendergrass, M., Patti, M. E., Pratipanawatr, T., DeFronzo, R. A., Kahn, C. R. & Mandarino, L. J. (2000) J. Clin. Invest. 105, 311–320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Krook, A., Bjornholm, M., Galuska, D., Jiang, X. J., Fahlman, R., Myers, M. G., Jr., Wallberg-Henriksson, H. & Zierath, J. R. (2000) Diabetes 49, 284–292. [DOI] [PubMed] [Google Scholar]
- 5.Le Lay, S., Krief, S., Farnier, C., Lefrere, I., Le Liepvre, X., Bazin, R., Ferre, P. & Dugail, I. (2001) J. Biol. Chem. 276, 16904–16910. [DOI] [PubMed] [Google Scholar]
- 6.Dreja, K., Voldstedlund, M., Vinten, J., Tranum-Jensen, J., Hellstrand, P. & Sward, K. (2002) Arterioscler. Thromb. Vasc. Biol. 22, 1267–1272. [DOI] [PubMed] [Google Scholar]
- 7.Drab, M., Verkade, P., Elger, M., Kasper, M., Lohn, M., Lauterbach, B., Menne, J., Lindschau, C., Mende, F., Luft, F. C., et al. (2001) Science 293, 2449–2452. [DOI] [PubMed] [Google Scholar]
- 8.Anderson, R. G. (1998) Annu. Rev. Biochem. 67, 199–225. [DOI] [PubMed] [Google Scholar]
- 9.McGill, J. B., Schneider, D. J., Arfken, C. L., Lucore, C. L. & Sobel, B. E. (1994) Diabetes 43, 104–109. [DOI] [PubMed] [Google Scholar]
- 10.Eddy, A. A. (2002) Am. J. Physiol. 283, F209–F220. [DOI] [PubMed] [Google Scholar]
- 11.Lambert, V., Munaut, C., Noel, A., Frankenne, F., Bajou, K., Gerard, R., Carmeliet, P., Defresne, M. P., Foidart, J. M. & Rakic, J. M. (2001) FASEB J. 15, 1021–1027. [DOI] [PubMed] [Google Scholar]
- 12.Vaughan, D. E. (2003) in Diabetes mellitus, eds. Porte, D., Sherwin, R. S. & Baron, A. (McGraw–Hill, New York), pp. 175–178.
- 13.Kaikita, K., Fogo, A. B., Ma, L., Schoenhard, J. A., Brown, N. J. & Vaughan, D. E. (2001) Circulation 104, 839–844. [DOI] [PubMed] [Google Scholar]
- 14.Lyon, C. J. & Hsueh, W. A. (2003) Am. J. Med. 115 Suppl 8A, 62S–68S. [DOI] [PubMed] [Google Scholar]
- 15.Samad, F., Pandey, M., Bell, P. A. & Loskutoff, D. J. (2000) Mol. Med. 6, 680–692. [PMC free article] [PubMed] [Google Scholar]
- 16.Griffiths, M. R., Black, E. J., Culbert, A. A., Dickens, M., Shaw, P. E., Gillespie, D. A. & Tavare, J. M. (1998) Biochem. J. 335, 19–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Usui, I., Haruta, T., Iwata, M., Takano, A., Uno, T., Kawahara, J., Ueno, E., Sasaoka, T. & Kobayashi, M. (2000) Biochem. Biophys. Res. Commun. 275, 115–120. [DOI] [PubMed] [Google Scholar]
- 18.Trimarchi, J. M. & Lees, J. A. (2002) Nat. Rev. Mol. Cell. Biol. 3, 11–20. [DOI] [PubMed] [Google Scholar]
- 19.Koziczak, M., Krek, W. & Nagamine, Y. (2000) Mol. Cell. Biol. 20, 2014–2022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Parton, R. G. (2003) Nat. Rev. Mol. Cell. Biol. 4, 162–167. [DOI] [PubMed] [Google Scholar]
- 21.White, M. F. & Kahn, C. R. (1994) J. Biol. Chem. 269, 1–4. [PubMed] [Google Scholar]
- 22.Hotamisligil, G. S. (2000) Int. J. Obes. Relat. Metab. Disord. 24 Suppl 4, S23–27. [DOI] [PubMed] [Google Scholar]
- 23.Mavri, A., Alessi, M. C., Bastelica, D., Geel-Georgelin, O., Fina, F., Sentocnik, J. T., Stegnar, M. & Juhan-Vague, I. (2001) Diabetologia 44, 2025–2031. [DOI] [PubMed] [Google Scholar]
- 24.Faisal, A., Kleiner, S. & Nagamine, Y. (2004) J. Biol. Chem. 279, 3202–3211. [DOI] [PubMed] [Google Scholar]
- 25.Sweeney, G., Somwar, R., Ramlal, T., Volchuk, A., Ueyama, A. & Klip, A. (1999) J. Biol. Chem. 274, 10071–10078. [DOI] [PubMed] [Google Scholar]
- 26.Koziczak, M., Muller, H., Helin, K. & Nagamine, Y. (2001) Eur. J. Biochem. 268, 4969–4978. [DOI] [PubMed] [Google Scholar]
- 27.Yang, Z. Z., Tschopp, O., Hemmings-Mieszczak, M., Feng, J., Brodbeck, D., Perentes, E. & Hemmings, B. A. (2003) J. Biol. Chem. 278, 32124–32131. [DOI] [PubMed] [Google Scholar]
- 28.Shima, H., Pende, M., Chen, Y., Fumagalli, S., Thomas, G. & Kozma, S. C. (1998) EMBO J. 17, 6649–6659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Helin, K., Lees, J. A., Vidal, M., Dyson, N., Harlow, E. & Fattaey, A. (1992) Cell 70, 337–350. [DOI] [PubMed] [Google Scholar]
- 30.Vives, E. & Lebleu, B. (2002) in Cell-Penetrating Peptides, ed. Langel, U. (CRC, Boca Raton, FL), Vol. 1, pp. 3–23. [Google Scholar]
- 31.Ramsay, T. G., Hausman, G. J. & Martin, R. J. (1987) J. Anim. Sci. 64, 735–744. [DOI] [PubMed] [Google Scholar]
- 32.Hwang, C. S., Loftus, T. M., Mandrup, S. & Lane, M. D. (1997) Annu. Rev. Cell Dev. Biol. 13, 231–259. [DOI] [PubMed] [Google Scholar]
- 33.Um, S. H., Frigerio, F., Watanabe, M., Picard, F., Joaquin, M., Sticker, M., Fumagalli, S., Allegrini, P. R., Kozma, S. C., Auwerx, J. & Thomas, G. (2004) Nature 431, 200–205. [DOI] [PubMed] [Google Scholar]
- 34.Kahn, S. E. & Porte, D. (2003) in Ellenberg and Rifkin's Diabetes Mellitus, eds. Porte, D., Sherwin, R. S. & Baron, A. (McGraw–Hill, New York), pp. 331–367.
- 35.Gustavsson, J., Parpal, S., Karlsson, M., Ramsing, C., Thorn, H., Borg, M., Lindroth, M., Peterson, K. H., Magnusson, K. E. & Stralfors, P. (1999) FASEB J. 13, 1961–1971. [PubMed] [Google Scholar]
- 36.Yancey, P. G., Rodrigueza, W. V., Kilsdonk, E. P., Stoudt, G. W., Johnson, W. J., Phillips, M. C. & Rothblat, G. H. (1996) J. Biol. Chem. 271, 16026–16034. [DOI] [PubMed] [Google Scholar]
- 37.Lange, Y., Swaisgood, M. H., Ramos, B. V. & Steck, T. L. (1989) J. Biol. Chem. 264, 3786–3793. [PubMed] [Google Scholar]
- 38.Cohen, A. W., Razani, B., Wang, X. B., Combs, T. P., Williams, T. M., Scherer, P. E. & Lisanti, M. P. (2003) Am. J. Physiol. 285, C222–C235. [DOI] [PubMed] [Google Scholar]
- 39.Oshikawa, J., Otsu, K., Toya, Y., Tsunematsu, T., Hankins, R., Kawabe, J., Minamisawa, S., Umemura, S., Hagiwara, Y. & Ishikawa, Y. (2004) Proc. Natl. Acad. Sci. USA 101, 12670–12675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kabayama, K., Sato, T., Kitamura, F., Uemura, S., Kang, B. W., Igarashi, Y. & Inokuchi, J. I. (2004) Glycobiology, in press. [DOI] [PubMed]
- 41.Hotamisligil, G. S., Shargill, N. S. & Spiegelman, B. M. (1993) Science 259, 87–91. [DOI] [PubMed] [Google Scholar]
- 42.Samad, F., Uysal, K. T., Wiesbrock, S. M., Pandey, M., Hotamisligil, G. S. & Loskutoff, D. J. (1999) Proc. Natl. Acad. Sci. USA 96, 6902–6907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Hsueh, W. A. & Law, R. E. (1998) Am. J. Med. 105, 4S–14S. [DOI] [PubMed] [Google Scholar]
- 44.Zhang, Z., Gao, N., He, H., Huang, C., Luo, J. & Shi, X. (2004) Mol. Cell Biochem. 255, 227–237. [DOI] [PubMed] [Google Scholar]
- 45.Brennan, P., Babbage, J. W., Burgering, B. M., Groner, B., Reif, K. & Cantrell, D. A. (1997) Immunity 7, 679–689. [DOI] [PubMed] [Google Scholar]
- 46.Paramio, J. M., Navarro, M., Segrelles, C., Gomez-Casero, E. & Jorcano, J. L. (1999) Oncogene 18, 7462–7468. [DOI] [PubMed] [Google Scholar]
- 47.Takuwa, N., Fukui, Y. & Takuwa, Y. (1999) Mol. Cell. Biol. 19, 1346–1358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ma, L. J., Mao, S. L., Taylor, K. L., Kanjanabuch, T., Guan, Y., Zhang, Y., Brown, N. J., Swift, L. L., McGuinness, O. P., Wasserman, D. H., Vaughan, D. E. & Fogo, A. B. (2004) Diabetes 53, 336–346. [DOI] [PubMed] [Google Scholar]
- 49.Schafer, K., Fujisawa, K., Konstantinides, S. & Loskutoff, D. J. (2001) FASEB J. 15, 1840–1842. [DOI] [PubMed] [Google Scholar]
- 50.Vuori, K. & Ruoslahti, E. (1994) Science 266, 1576–1578. [DOI] [PubMed] [Google Scholar]
- 51.Lopez-Alemany, R., Redondo, J. M., Nagamine, Y. & Munoz-Canoves, P. (2003) Eur. J. Biochem. 270, 814–821. [DOI] [PubMed] [Google Scholar]
- 52.Xiao, B., Spencer, J., Clements, A., Ali-Khan, N., Mittnacht, S., Broceno, C., Burghammer, M., Perrakis, A., Marmorstein, R. & Gamblin, S. J. (2003) Proc. Natl. Acad. Sci. USA 100, 2363–2368. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.