Abstract
Fully conjugated macrocyclic oligothiophenes exhibit a combination of highly attractive structural, optical and electronic properties, and multifunctional molecular thin film architectures thereof are envisioned. However, control over the self-assembly of such systems becomes increasingly challenging, the more complex the target structures are. Here we show a robust self-assembly based on hierarchical non-covalent interactions. A self-assembled monolayer of hydrogen-bonded trimesic acid at the interface between an organic solution and graphite provides host-sites for the epitaxial ordering of Saturn-like complexes of fullerenes with oligothiophene macrocycles in mono- and bilayers. STM tomography verifies the formation of the templated layers. Molecular dynamics simulations corroborate the conformational stability and assign the adsorption sites of the adlayers. Scanning tunnelling spectroscopy determines their rectification characteristics. Current–voltage characteristics reveal the modification of the rectifying properties of the macrocycles by the formation of donor–acceptor complexes in a densely packed all-self-assembled supramolecular nanostructure.
Controlling the self-assembly of oligothiophene complexes that are used in multi-functional thin films can be challenging. Here the authors show a hierarchy of non-covalent interactions for robust self-assembly that orders Saturn-like complexes of fullerenes with oligothiophene macrocycles.
Control over the self-assembly of molecular structures on a solid surface is of particular interest towards three-dimensional (3D) engineering of supramolecular materials1 and designing of molecular electronic devices2,3,4. Hydrogen-bonding is the most versatile, yet specific type of weak interaction to engineer two-dimensional (2D) self-assembled networks5,6 due to the high level of control at the molecular level7,8 and the potential applications9. Among other supramolecular interactions, host–guest complexation on 2D networks provides an interesting platform to create functional multicomponent layers10,11, nanostructures12,13 and prototypical devices14, and establish the first step to tailor 3D architectures15,16. Furthermore, donor (D) - acceptor (A) complexation of common acceptor molecules into the core of giant macrocycles unveils promising building blocks for the fabrication of complex nanostructures15,17,18,19 and self-assembled devices. The design of multifunctional organic materials containing fullerene C60 as an active component has been extensively investigated in organic electronics20,21. The spherical geometry of fullerene C60 and its acceptor properties render a cylindrical or torus-like donor molecule a promising choice to act as a complementary host-site22,23. Similar to its linear counterparts24,25, π-expanded macrocyclic oligothiophenes have attracted attention for their potential applications in molecular electronics and as components in molecular devices15,17,26,27.
Here we show how to create a bilayer of moderately rectifying donor–acceptor complexes via double hosting of C60 molecules. This architecture is achieved by combining the spontaneous formation of a hydrogen-bonded trimesic acid (TMA, 1) as a 2D-template at the interface between its heptanoic acid solution and a highly oriented pyrolytic graphite (HOPG) substrate12,28, the complexation of oligothiophene macrocycles29,30 and C60 in solution, and the site recognition of the cavities in the TMA monolayer by C60 (ref. 31). Moreover, we demonstrate the potential of this multi-step articulated self-assembled construction to engineer multiple epitaxially grown macrocycle layers.
Results
2D-template of trimesic acid
Previous experiments using π-expanded oligothiophenes, that is, macrocycles composed of six32 and eight thiophene units (E,E-8-mer, 2) (ref. 30), have shown that the self-assembly of an ordered monolayer is favourable at the interface between HOPG and a fatty acid solution, where under similar experimental conditions C60 molecules alone do not adsorb33. Here, we found that the attractive interaction between the HOPG substrate and 2 is not sufficient to drive the formation of an ordered monolayer of the Saturn-like complex (E,E-8-mer·C60, 3) at the interface between its heptanoic acid solution and the graphite surface, which may be due to the most favourable energy interaction of the complex. Therefore, we looked for a modification of the substrate to adsorb both components of the complex, macrocycle and C60.
A hydrogen-bonding driven 2D network of TMA has been considered as a template for the complexes due to its capability to host small molecules, such as C60 (ref. 31). Fullerenes C60 can be hosted within the honeycomb-like network of TMA, mainly due to the favourable energetic stabilization of the host–guest interaction, as calculated previously using all force-field methods31. Here we report the recognition of the host-sites in a previously adsorbed self-assembled monolayer of TMA by the in-solution complexed C60 molecules.
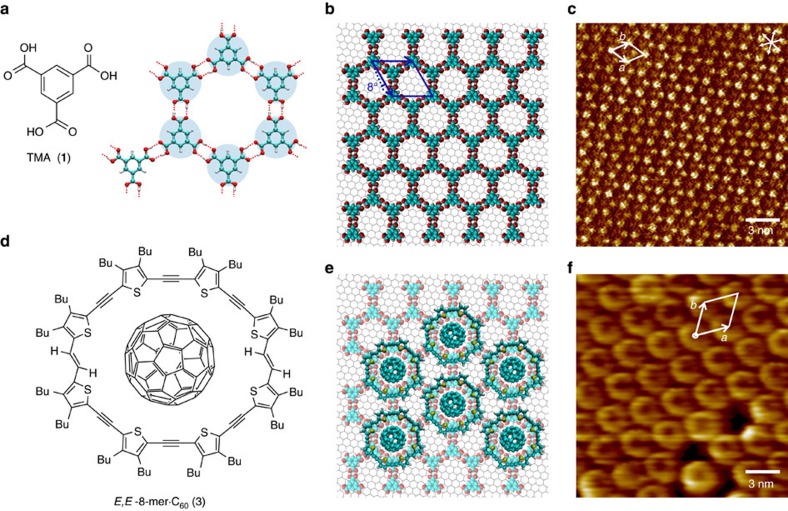
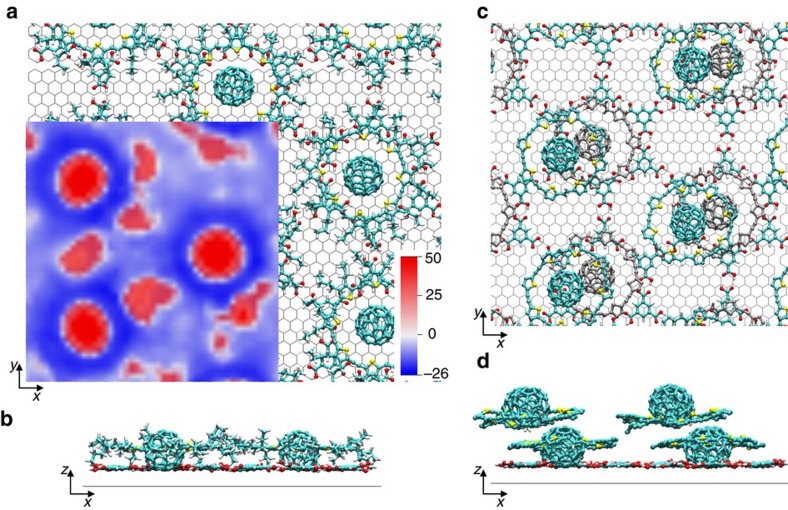
As depicted in Fig. 1a,b, TMA forms an extended honeycomb-like structure when adsorbed at the solid–liquid interface between its solution in heptanoic acid and HOPG. The hexagonal cavities of the periodic arrangement are created by the di-apto hydrogen-bonding of adjacent carboxylic acid groups of six neighbouring molecules, exhibiting a pore diameter of about 12 Å (refs 12, 28). Figure 1c shows a scanning tunnelling microscopy (STM)34 height image of the chicken-wire structure of the 2D TMA-crystal on HOPG upon the addition of a small drop of saturated heptanoic acid solution; the unit cell parameters agree with previous reports12,31.
Figure 1. 2D-template of TMA for a π-expanded macrocycle complex with C60.
(a) Each TMA molecule forms hydrogen-bonding with any three neighbour molecules rotated by 60°, 120° and 180°. (b) Section of the honeycomb-like network formed on the basal plane of HOPG as shown schematically, the size of the host cavity is around 12 Å. The hexagonal unit cell (a=b=1.70 nm, θ=60°) is rotated 8° respect to the HOPG axis. (c) STM height image of a honeycomb-like network of TMA, unit cell a=(1.70±0.09) nm, b=(1.69±0.03) nm, θ=(60±3)°, Us=−1.01 V, It=46 pA. (d) Molecular structure of the π-expanded oligothiophene macrocycles in its E,E form complexed with C60. Fullerenes C60 are coordinated by the electron donor structure of the macrocycle. (e) Molecular schematic of the epitaxially arranged 3 monolayer, where C60 is hosted in the pores of the TMA-template network. (f) STM height image of the 2D network formed upon the addition of a drop of solution of 3 after forming the template in c. Unit cell: a=(2.84±0.14) nm, b=(2.83±0.15) nm, θ=(62±5)°, Us=+0.90 V, It=50 pA.
Self-assembly of E,E-8-mer·C60 on TMA template
We schemed complex 3 (Fig. 1d) to form an epitaxial monolayer with the TMA network as sketched in Fig. 1e, displaying 3 without butyl groups for clarity. The addition of a drop of heptanoic acid solution of 3 to the previously formed honeycomb TMA network causes the spontaneous formation of a hexagonal network with one molecular complex per unit cell (Fig. 1f). Steric hindrance prevents two neighbouring pores to host one complex each; every complex 3 is rather hosted in an alternate pore of the TMA honeycomb, making the unit cell of 3 a (✓3 × ✓3)R30° superstructure of the unit cell of TMA. In the STM image (Fig. 1f) one finds donut-like shaped objects, where the tunnelling current is larger compared with surrounding regions35. This effect is observed at positive sample bias (+0.70 V<Us<+1.20 V), whereas at negative bias the image of the TMA honeycomb-like network is recovered (details in section STM tomography). Contrastingly, when a drop of a heptanoic acid solution of the macrocycle 2 is added to the previously formed honeycomb TMA network, an irregular aggregation of the molecules is observed (Supplementary Fig. 1), suggesting that the formation of an ordered structure depends critically on the presence of the TMA hosting sites along with the C60 molecule of the complex.
Two possible pathways could afford the formation of the templated monolayer of 3: a two-step adsorption, in which non-complexed C60 is adsorbed on TMA vacant sites followed by the formation of the complex 3 again on these TMA sites, and a one-step adsorption, where in-solution complex 3 directly gets adsorbed on the TMA sites. The low solubility of C60 in heptanoic acid and its low density host–guest complexation on a TMA honeycomb network31, favours the one-step adsorption process.
Over large scales the templated monolayer is characterized by single crystalline domains with diameters up to a few 10 nm, separated by grain boundaries, which are mostly slip lines (Supplementary Fig. 2). This observation indicates that the template is not perfectly rigid but somewhat soft. Using the data from the fast fourier transform of Supplementary Fig. 2, we found that the unit cell parameters of the macrocycle network follow the relations: a2=✓3·a1, b2=✓3·b1 and θ1=θ2=60°, where a1, b1 and θ1 are the unit cell parameters of the TMA network. Taking a1=b1=1.7 nm, one gets a2=b2=2.9 nm, which is in perfect accordance with the measured results (including the data presented in Fig. 1f). The maximum length of a complex 3 with fully stretched butyl groups is about 3.2 nm, according to X-ray crystallographic data30. This length is larger than its unit cell parameters on the TMA network and since STM images could not resolve the butyl substituent groups, we assume the side chains of 3 are either back-folded into the supernatant solution or into the empty pores of the template (see molecular dynamics section below), which might be attributed to the incommensurability between the hexagonal TMA-networks and the quasi-eightfold symmetry of 3.
STM tomography
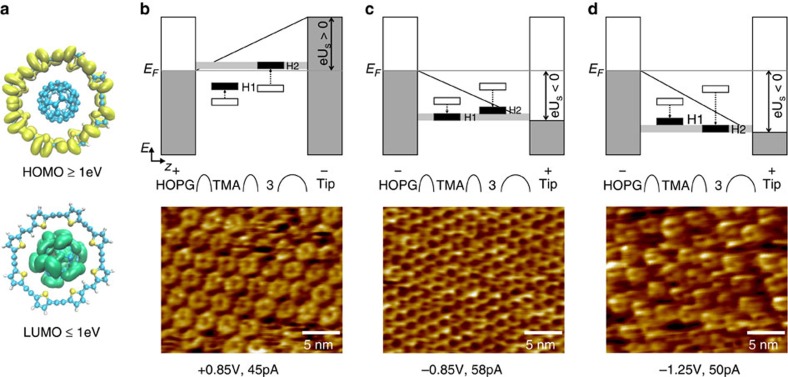
Figures 1f and 2b display STM height images after the formation of the templated monolayer of 3. At currents on the order of 50 pA, stable images could be only recorded at positive sample bias, while at negative sample bias the TMA template was observed (Fig. 2c); at lower negative as well as positive bias (smaller than 0.6 V) no structures were recognized; and at high negative bias (−1.25 V) unstable donut-like molecules were observed (Fig. 2d and Supplementary Fig. 3). Moreover, C60 molecules could not be identified at any substrate bias up to 1.2 V in both polarities, while in similar templated systems (C60 on cyclothiophene17, TMA31 or p-terphenyl-3,5,3′′,5′′-tetracarboxylic acid, TPTC16) they appeared as high-contrast spots. This indicates that in the present case C60 molecules are either absent or not visible. The experimental results reported above can be explained on the basis of a STM tomography model36. The model assumes that the energy frontier orbitals involved in the resonant tunnelling through the molecules within the tip-HOPG gap are shifted proportional to their distance to the surface, mainly due to its asymmetric alignment37. Within the model, the applied bias affects the visibility of the TMA template and the complex 3.
Figure 2. Bias set-point imaging.
(a) Isosurfaces showing the electron probability distribution 1 eV below and above the HOMO and LUMO of the complex 3, respectively. (b) At positive substrate bias, the resonant tunnelling is achieved through the HOMO of 3 (H2), which is symmetrically distributed in the macrocycle. The height image shows below at Us=+0.85 V, It=45 pA. (c) By switching to negative substrate bias, the resonant tunnelling occurs through the HOMO of the underneath TMA (H1), as the image below at Us=−0.85 V, It=58 pA. (d) Lowering the negative substrate bias gives access again to H2, Us=−1.25 V, It=50 pA. The tunnelling does not occur through the LUMO of 3 (localized in the C60) because of the closer proximity of both HOMOs to the Fermi level of the HOPG.
Figure 2a shows the position of the highest occupied molecular orbital (HOMO) of 3, located at the macrocycle, and the lowest unoccupied molecular orbital (LUMO), which lies on the C60, as deduced from gas-phase density functional theory calculations on unsubstituted 3 (Supplementary Fig. 4 and Supplementary Methods).
Figure 2b–d depict energy diagrams of the position of the HOMO (see Supplementary Fig. 5) of the TMA template (H1) and complex 3 (H2) within the tunnelling region for positively biased substrate (Fig. 2b) and negatively biased substrate (Fig. 2c,d), respectively. Empty rectangles show the position of the HOMO when no bias is applied, while filled rectangles are the shifted orbital levels.
As depicted in Fig. 2b, when the substrate is positively biased the resonance effect occurs as the electrons from the tip tunnel through the HOMO of the complex 3 (H2), which is shifted upwards (positive energy) more than the HOMO of the TMA (H1). The STM height image shows the donut-like shape of 3 which is associated with H2. At negative sample bias (Fig. 2c), the lower position (more negative energy) of H1 makes it more accessible at negative imaging bias (between −0.7 and −1.1 V); here the STM height image depicts the honeycomb-like network of TMA. In addition, lowering the voltage even more (Fig. 2d) causes H2 to shift more downwards (negative energy), which makes it accessible again at bias around −1.25 V, but unstable for steady imaging.
Moreover, in the case where the substrate is positively biased, the LUMO of the complex (not shown in Fig. 2) shifts upwards (positive energy), and when it is negatively biased the shift downwards (negative energy) is not sufficient to be in resonance due to the distance of the LUMO to the Fermi-level of HOPG, thus making the C60 invisible in both cases.
Scanning tunnelling spectroscopy
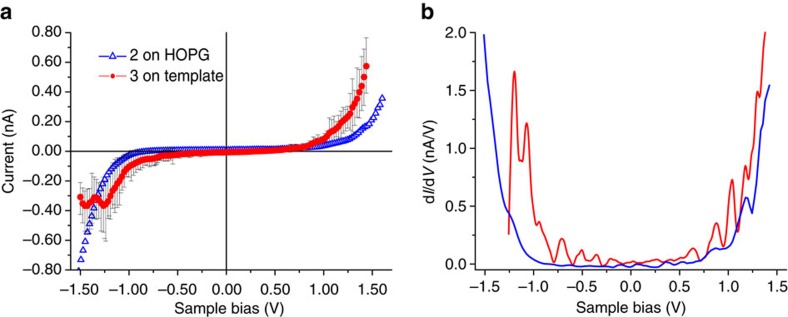
Scanning tunnelling spectroscopy (STS) has been used to determine the local electronic structure of the templated monolayer(s) of 3. Figure 3a depicts the current–voltage (I–V) characteristics across the macrocycle 2 on HOPG and the templated complex 3. The I–V characteristics of 2 exhibits an asymmetric behaviour with a larger tunnelling current at negative sample bias, showing the expected rectification behaviour of a donor molecule38. On the contrary, the I–V characteristics through the donut-like moiety of 3 shows an asymmetric behaviour, with a larger tunnelling current at positive sample bias. The charge transport across the center of the templated complex, where the C60 is located, is symmetric (Supplementary Fig. 6b).
Figure 3. STS of 2 and templated monolayer of 3.
(a) Average I–V characteristics over twenty measurements of a monolayer of templated 3 (red circles, STS conditions Us=+0.94 V and It=50 pA), including error bars (see Supplementary Fig. 6a for scattered data). For comparison the average I–V trace of fifteen measurements of 2 on HOPG was measured too (blue triangles, STS conditions Us=−1.0 V and It=80 pA). For the templated monolayer of 3 the current saturates below −1.2 V, not the case for 2. (b) Differential conductance (dI/dV) versus sample bias, where the saturation effect is evident below −1.2 V.
Moreover, for the case of the transport across the rim of 3, there is a saturation in the current at –(0.35±0.12) nA in a bias range between −1.2 and −1.5 V; similar findings have been previously reported for thiophene macrocycle networks covered with C60 (ref. 17). This tendency is dramatically shown in the dI/dV spectra (Fig. 3b), which reflects more directly the local density of states of the system39. In the range of −1.5 to +1.5 V the charge transport across 2 increases monotonously at both positive and negative bias, while for complex 3 a blockage at −1.2 V is observed, which is attributed to the HOMO. A correspondingly clear feature of the LUMO is, however, not observed in the here accessible bias range up to +1.4 V. The saturation of the current at negative bias values is associated with delocalization of excited states in the infinite π-conjugated system of the macrocycle40 and represents a key spectroscopic characteristic of this type of systems17.
By creating the templated monolayer of 3, in which the presence of the C60 molecules is essential for a regular array of the macrocycles epitaxial to the TMA network, the formation of rectifying monolayer of complexes is accomplished and the rectification behaviour is likewise inversed.
Templated bilayer of Saturn-like complexes
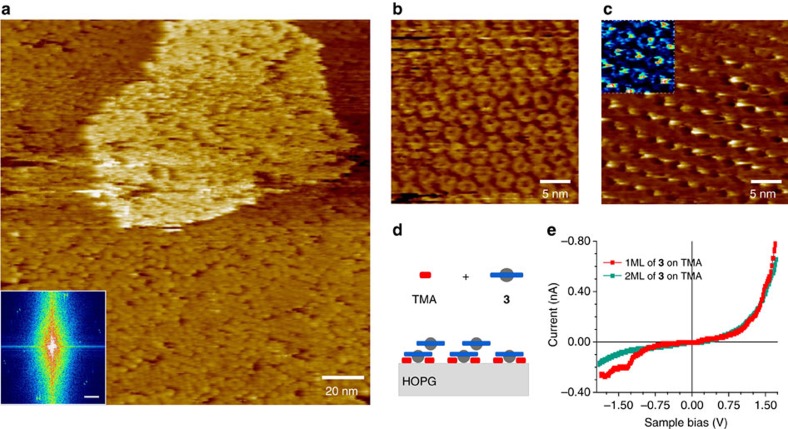
The density of the templated monolayer of 3 increments with time (Supplementary Fig. 7). Saturation of the first monolayer is obtained by increasing the solution concentration. After some minutes, while keeping the imaging conditions, islands similar to the one depicted in Fig. 4a are formed. A histogram of Fig. 4a gives a relative height of 2 Å for the second layer of 3 (Supplementary Fig. 8). This step height is smaller than the typically observed for non-interacting C60 measured by STM41, nevertheless, the apparent height in STM experiments is related to the local density of states and, therefore, is not to taken as an absolute dimension. We attribute these islands to a second monolayer of the complex 3 grown epitaxial to the first monolayer, creating a templated bilayer 3|3. Similar bilayers are known to occur by π–π stacking of polycyclic aromatic hydrocarbons42, metallo-ligand interactions of phthalocyanines43 and guest-induced of two layers of TPTC16.
Figure 4. Growth of a second monolayer of 3.
(a) STM height image of a templated bilayer of 3 in heptanoic acid solution. Measuring conditions Us=+1.05 V, It=42 pA. Two different unit cells can be identified from the fast fourier transform in the inset, the scale bar is 1/10 nm. STM height image at +1.05 V (b) and −1.05 V (c) of a templated bilayer (2ML) of 3 in heptanoic acid solution. The unit cell measures a=(2.85±0.04) nm, b=(2.84±0.04) nm, θ=(61±1)°. An inset in c is showed in a different contrast to highlight the image of the macrocycles of the first templated monolayer (1ML). We attribute the bright spot to the C60 of the layer on top. (d) Scheme of the suggested arrangement of the first and second layer of 3. C60 of the second layer lies on the rim of the first one. (e) I–V characteristic over fifteen measurements through 1ML and 2ML of 3 (STS conditions Us=+0.95 V and It=51 pA) (see Supplementary Fig. 9 for scattering of data).
The arrangement of the second layer relative to the first one is resolved by applying set-point dependent imaging as described earlier for the template monolayer of 3. The image recorded in Fig. 4b shows the top layer of 3 at positive bias. Upon switching the bias to negative values, an arrangement of bright spots and low contrast macrocycles is imaged (Fig. 4c), which we associate with C60 molecules of the top layer of 3 sitting on the rims of the first layer complex. This adsorption site for C60 has been reported to be the most favourable on other types of shape-persistent macrocycles15,17.
STS measurements through one and two monolayers of 3 showed that the forward rectification is kept through the templated bilayer 3|3 (Fig. 4e). However, the saturation point seen across a monolayer, was not reached for values down to −1.8 V in the case of the bilayer. The rectification ratio, defined as R(V0)=|I(V0)/I(−V0)|, where I(V0) is the current at a reference bias V0 (typically the highest bias used), reaches an average value of 4.9±1.2 for the templated bilayer 3|3 at 1.7 V, while for the monolayer of 3 it is 2.7±1.1 at 1.7 V, thus improving the rectification upon addition of a second monolayer. For comparison, the average value of R for a monolayer of 2 on HOPG is 3.5±1.1. Average values of R are the arithmetic mean and standard deviation for each curve in Supplementary Figs 6c and 9.
Molecular dynamics simulation
In order to rationalize the formation of bilayer architectures of 3 on the TMA network in the light of the results in Fig. 4b,c, all force-field calculations were performed using the program NAMD44 with the CHARMM general force field45 (CGenff). The multilayer was simulated in an orthorhombic box with periodic boundary conditions to reproduce an infinite system. First, we placed a twelve pore TMA honeycomb lattice (24 TMA molecules) 6 Å above the 52 Å × 60 Å graphene together with four 3 complexes at 6 Å form the center of every second pore of TMA (12 Å above graphene) (Supplementary Fig. 11), and then energy minimized while keeping the substrate fixed. A two nanosecond molecular dynamic simulation at 300 K shows the stability of the (✓3 × ✓3)R30° superstructure with a TMA-graphene separation of ∼3.45 Å, the fullerenes C60 ∼6.6 Å from graphene and the macrocyclic part of 3 ∼0.6 Å above the complexed C60 molecules. The in-plane position of the complex is stabilized with its center of mass between 0.3 A and 0.9 A to the center of the pore. Moreover, Fig. 5b shows a side view of the templated monolayer, where the alkyl chains are folded up, down into the empty pores of the template and in between complexes, confirming our previous assumption.
Figure 5. Structure of the templated bilayer 3|3|TMA|HOPG.
Snapshot of the self-assembled monolayer of 3 on the TMA template after 2 ns simulation, top (a) and side view (b). A potential mean force map of a templated monolayer of 3 probed with a C60 averaged on 1 ns of molecular dynamic simulation (energy given in kcal mol−1) is also shown. The positions coloured in blue are the most favourable for the molecule in agreement with the STM image in Fig. 4c. (c,d) Top and side view of the templated bilayer of 3 after 2 ns of molecular dynamic simulation at 300 K, the alkyl chains are omitted for clarity.
Furthermore, we exploited implicit ligand sampling (ILS)46 to find the most accessible and energetically favourable position of the second monolayer of molecules. This computational method evaluates the potential mean force (PMF) of placing a probe molecule everywhere close to the system by means of the Gibbs free-energy cost. Figure 5a shows the PMF map of probing with one C60 at 13.5 Å averaged on 1 ns of molecular dynamic simulation at 300 K (25 frames), which locates the minimum energy positions where the probe molecule stands above the rim of the underlying molecule as it was suggested by the STM measurements in Fig. 4. Finally, four 3 complexes were placed on the positions of the ILS map minima forming a second monolayer (Supplementary Fig. 12) and a further two nanoseconds molecular dynamic simulation analysed the behaviour of the templated bilayer layer while keeping the substrate fixed. Figure 5c,d show snapshots after 2 ns of simulation, interestingly the macrocyclic part of 3 of the second layer lowers 1.7 Å with respect to the corresponding C60 (12 Å from HOPG), while the position of the first templated layer is not affected. This positioning decreases the energy of the system in 35 kcal mol−1 per complex 3 in the second layer (Supplementary Fig. 13), which grows linearly with the number of molecules involved in the assembly. The average of the trajectory snapshots of the molecular dynamic simulation under ideal feedback conditions shows that the unit cell for the first and second monolayer is kept within the experimental values (see Supplementary Fig. 14). Full detailed information about the molecular dynamics calculation is provided in Supplementary Note 2.
Discussion
We demonstrated that a conjugated π-expanded macrocyclic oligothiophene hosting fullerenes C60 can be self-assembled into the pores of hexagonally packed TMA to form three-component rectifying bilayers and triple layers with high densities of molecular diodes. The formation of the supramolecular nanostructure depends critically on the presence of both the TMA and the hosted C60. We used bias dependent imaging and all-atom molecular dynamics calculations to establish the structure of the templated mono- and bilayers of complexes and their epitaxial arrangement with respect to the TMA network beneath. STS measurements indicate the inversion of the rectifying characteristics of the macrocycles in the templated monolayer. While the templated monolayer of 3 is a very moderate rectifier, the addition of a second monolayer of 3 improves the rectification ratio. Our findings show that a combination of non-covalent interactions, including hydrogen-bonding and charge transfer in host-guest systems, allows supramolecular engineering of self-assembled functional nanosystems featuring ways to control their electronic properties.
Methods
STM measurements
A home-built beetle-type scanning tunnelling microscope with an Omicron controller was used for all the experimental measurements. Small drops (∼5 μl) of heptanoic acid solutions of the compounds (10−4–10−5 M) were applied to freshly cleavage HOPG substrates. The substrates were glued to a glass holder and electrically contacted using silver paste. STM tips were mechanically cut from a Pt/Ir wire (90/10, 0.25 mm diameter). The STM data were processed using SPIP software (Image Metrology A/S). Thermal drift was corrected with respect to the known hexagonal HOPG lattice underneath; background flattening and despeckling filter were also applied. The HOPG lattice was visualized by lowering the tunnelling voltage to 20 mV and raising the tunnelling current to 1–1.4 nA.
STS measurements
During STM imaging at constant current (feedback on), the STM tip was moved to the point of interest. Hence, the tip position was kept fixed according to the gap voltage and setpoint for the tunnelling current. The feedback loop was then switched off and a bias ramp was swept through and the tunnelling current recorded. In order to decrease the effect of lateral drift while recording STS, the tip was positioned over the region of interest as close as possible to the most recently scanned line. Typically, it takes about 25 ms to perform a single STS curve and the lateral drift velocity is <0.1 nm s−1. The actual tip-sample separation for a single measurement follows a normal distribution around the mean value of the setpoint, thus the average of the data is obtained by a lognormal function to discard inappropriate data which could arise by fluctuations of the tip structure47.
Molecular dynamics calculations
All simulations were performed using the NAMD software (2.10 build for linux), with the CHARMM general force field (CGenff) and parametrization of the molecule 2 (see Supplementary Note 1, Supplementary Fig. 10 and Supplementary Table 1) using the force field toolkit48 implemented in VMD visualization software49. The graphene slab substrate was kept fixed on all simulations, its atoms were assigned atom type CG2R61 and no partial charge. A Langevin thermostat with a damping coefficient of 1 ps−1 and a time step of 2 fs was used in all simulations. A distance cutoff of the size of the periodic box was used to compute non-bonded interactions. Electrostatics interactions were calculated using the Particle mesh Ewald method with a space grid of 0.5 Å. For the ILS calculation, periodic boundary condition were applied, with a resolution (space grid) of 1 Å and a subsampling of 3 points. The trajectory snapshots for PMF map calculation were obtained after extra 1 ns simulation, were both TMA and graphene were kept fix.
Data availability
The authors declare that the data supporting the findings of this study are available within the article and its Supplementary Information file or from the corresponding authors upon reasonable request.
Additional information
How to cite this article: Cojal González, J. D. et al. Templated bilayer self-assembly of fully conjugated π-expanded macrocyclic oligothiophenes complexed with fullerenes. Nat. Commun. 8, 14717 doi: 10.1038/ncomms14717 (2017).
Publisher's note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Material
Supplementary Figures, Supplementary Table, Supplementary Notes, Supplementary Methods and Supplementary References
Acknowledgments
We thank C.-A. Palma (TU München) and N. Severin (HU Berlin) for fruitful discussion. This work was supported by the German Science Foundation (DFG) SFB 765 and Ra482/6-1 and JST, Strategic International Research Cooperative Program (SICP) [Strategic Japanese-German Cooperation Program from JST].
Footnotes
The authors declare no competing financial interests.
Author contributions J.D.C.G. acquired the STM data and performed the molecular dynamic simulations, M.I. developed the synthetic route for the macrocycle 2 and complex 3, J.D.C.G. and J.P.R. analysed the data, J.D.C.G. and J.P.R. conceived and coordinated the experimental work, J.P.R and M.I. coordinated the project and J.D.C.G. and J.P.R. co-wrote the paper with inputs from M.I.
References
- Whitesides G. M., Kriebel J. K. & Mayers B. T. in Nanoscale Assembly (ed.. Huck W. T. S. 217–239Springer (2005). [Google Scholar]
- Müllen K. & Rabe J. P. Nanographenes as active components of single-molecule electronics and how a scanning tunneling microscope puts them to work. Acc. Chem. Res. 41, 511–520 (2008). [DOI] [PubMed] [Google Scholar]
- Palma C.-A. & Samorì P. Blueprinting macromolecular electronics. Nat. Chem. 3, 431–436 (2011). [DOI] [PubMed] [Google Scholar]
- Nirmalraj P. et al. Nanoelectrical analysis of single molecules and atomic-scale materials at the solid/liquid interface. Nat. Mater. 13, 947–953 (2014). [DOI] [PubMed] [Google Scholar]
- Ivasenko O. & Perepichka D. F. Mastering fundamentals of supramolecular design with carboxylic acids. Common lessons from X-ray crystallography and scanning tunneling microscopy. Chem. Soc. Rev. 40, 191–206 (2010). [DOI] [PubMed] [Google Scholar]
- Ciesielski A., Palma C.-A., Bonini M. & Samorì P. Towards supramolecular engineering of functional nanomaterials: pre-programming multi-component 2D self-assembly at solid-liquid interfaces. Adv. Mater. 22, 3506–3520 (2010). [DOI] [PubMed] [Google Scholar]
- Lehn J.-M. in Supramolecular Chemistry Wiley (1995). [Google Scholar]
- Sherrington D. C. & Taskinen K. A. Self-assembly in synthetic macromolecular systems via multiple hydrogen bonding interactions. Chem. Soc. Rev. 30, 83–93 (2001). [Google Scholar]
- Barth J. V., Costantini G. & Kern K. Engineering atomic and molecular nanostructures at surfaces. Nature 437, 671–679 (2005). [DOI] [PubMed] [Google Scholar]
- Lei S. et al. Programmable hierarchical three-component 2D assembly at a liquid−solid interface: recognition, selection, and transformation. Nano Lett. 8, 2541–2546 (2008). [DOI] [PubMed] [Google Scholar]
- Adisoejoso J. et al. Two-dimensional crystal engineering: a four-component architecture at a liquid-solid interface. Angew. Chem. Int. Ed. 48, 7353–7357 (2009). [DOI] [PubMed] [Google Scholar]
- Griessl S., Lackinger M., Edelwirth M., Hietschold M. & Heckl W. M. Self-assembled two-dimensional molecular host-guest architectures from trimesic acid. Single Mol. 3, 25–31 (2002). [Google Scholar]
- Stepanow S. et al. Steering molecular organization and host–guest interactions using two-dimensional nanoporous coordination systems. Nat. Mater. 3, 229–233 (2004). [DOI] [PubMed] [Google Scholar]
- Jäckel F., Watson M. D., Müllen K. & Rabe J. P. Prototypical single-molecule chemical-field-effect transistor with nanometer-sized gates. Phys. Rev. Lett. 92, 188303 (2004). [DOI] [PubMed] [Google Scholar]
- Pan G.-B., Cheng X.-H., Höger S. & Freyland W. 2D supramolecular structures of a shape-persistent macrocycle and Co-deposition with fullerene on HOPG. J. Am. Chem. Soc. 128, 4218–4219 (2006). [DOI] [PubMed] [Google Scholar]
- Blunt M. O. et al. Guest-induced growth of a surface-based supramolecular bilayer. Nat. Chem. 3, 74–78 (2011). [DOI] [PubMed] [Google Scholar]
- Mena-Osteritz E. & Bäuerle P. Complexation of C60 on a cyclothiophene monolayer template. Adv. Mater. 18, 447–451 (2006). [Google Scholar]
- Schmaltz B., Rouhanipour A., Räder H. J., Pisula W. & Müllen K. Filling the cavity of conjugated carbazole macrocycles with graphene molecules: monolayers formed by physisorption serve as a surface for pulsed laser deposition. Angew. Chem. Int. Ed. 48, 720–724 (2009). [DOI] [PubMed] [Google Scholar]
- Jester S.-S., Aggarwal A. V., Kalle D. & Höger S. Mono- and multilayers of molecular spoked carbazole wheels on graphite. Beilstein J. Org. Chem. 10, 2783–2788 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guldi D. M., Illescas B. M., Atienza C. M., Wielopolski M. & Martín N. Fullerene for organic electronics. Chem. Soc. Rev. 38, 1587–1597 (2009). [DOI] [PubMed] [Google Scholar]
- Bendikov M., Wudl F. & Perepichka D. F. Tetrathiafulvalenes, oligoacenenes, and their buckminsterfullerene derivatives: The brick and mortar of organic electronics. Chem. Rev. 104, 4891–4946 (2004). [DOI] [PubMed] [Google Scholar]
- Pan G.-B. et al. Configurations of a Calix[8]arene and a C60/Calix[8]arene complex on a Au(111) surface. Angew. Chem. Int. Ed. 42, 2747–2751 (2003). [DOI] [PubMed] [Google Scholar]
- Song J., Aratani N., Shinokubo H. & Osuka A. A porphyrin nanobarrel that encapsulates C60. J. Am. Chem. Soc. 132, 16356–16357 (2010). [DOI] [PubMed] [Google Scholar]
- Fichou D. Structural order in conjugated oligothiophenes and its implications on opto-electronic devices. J. Mater. Chem. 10, 571–588 (2000). [Google Scholar]
- Otsubo T., Aso Y. & Takimiya K. Functional oligothiophenes as advanced molecular electronic materials. J. Mater. Chem. 12, 2565–2575 (2002). [Google Scholar]
- Krömer J. et al. Synthesis of the first fully α-conjugated macrocyclic oligothiophenes: cyclo[n]thiophenes with tunable cavities in the nanometer regime. Angew. Chem. Int. Ed. 39, 3481–3486 (2000). [PubMed] [Google Scholar]
- Mayor M. & Didschies C. A giant conjugated molecular ring. Angew. Chem. Int. Ed. 42, 3176–3179 (2003). [DOI] [PubMed] [Google Scholar]
- Lackinger M., Griessl S., Heckl W. M., Hietschold M. & Flynn G. W. Self-assembly of trimesic acid at the liquid−solid interfacea study of solvent-induced polymorphism. Langmuir 21, 4984–4988 (2005). [DOI] [PubMed] [Google Scholar]
- Nakao K. et al. Giant macrocycles composed of thiophene, acetylene, and ethylene building blocks. J. Am. Chem. Soc. 128, 16740–16747 (2006). [DOI] [PubMed] [Google Scholar]
- Shimizu H. et al. Synthesis, structures, and photophysical properties of π-expanded oligothiophene 8-mers and their saturn-like C60 complexes. J. Am. Chem. Soc. 137, 3877–3885 (2015). [DOI] [PubMed] [Google Scholar]
- Griessl S. J. H. et al. Room-temperature scanning tunneling microscopy manipulation of single C60 molecules at the liquid−solid interface: playing nanosoccer. J. Phys. Chem. B 108, 11556–11560 (2004). [Google Scholar]
- Iyoda M. et al. Multifunctional π-expanded macrocyclic oligothiophene 6-mers and related macrocyclic oligomers. J. Am. Chem. Soc. 136, 2389–2396 (2014). [DOI] [PubMed] [Google Scholar]
- Uemura S., Samorí P., Kunitake M., Hirayama C. & Rabe J. P. Crystalline C60 monolayers at the solid–organic solution interface. J. Mater. Chem. 12, 3366–3367 (2002). [Google Scholar]
- Rabe J. P. & Buchholz S. Commensurability and mobility in two-dimensional molecular patterns on graphite. Science 253, 424–427 (1991). [DOI] [PubMed] [Google Scholar]
- Lazzaroni R., Calderone A., Brédas J. L. & Rabe J. P. Electronic structure of molecular van der Waals complexes with benzene: Implications for the contrast in scanning tunneling microscopy of molecular adsorbates on graphite. J. Chem. Phys. 107, 99–105 (1997). [Google Scholar]
- Seifert C. et al. Tomography of molecular nanographene double layers using scanning tunneling microscopy. Phys. Rev. B 80, 245429 (2009). [Google Scholar]
- Kornilovitch P. E., Bratkovsky A. M. & Stanley Williams R. Current rectification by molecules with asymmetric tunneling barriers. Phys. Rev. B 66, 165436 (2002). [Google Scholar]
- Jäckel F., Wang Z., Watson M. D., Müllen K. & Rabe J. P. Nanoscale array of inversely biased molecular rectifiers. Chem. Phys. Lett. 387, 372–376 (2004). [Google Scholar]
- Feenstra R. M. Scanning tunneling spectroscopy. Surf. Sci. 299–300, 965–979 (1994). [Google Scholar]
- Iyoda M., Yamakawa J. & Rahman M. J. Conjugated macrocycles: concepts and applications. Angew. Chem. Int. Ed. 50, 10522–10553 (2011). [DOI] [PubMed] [Google Scholar]
- Keeling D. L., Humphry M. J., Moriarty P. & Beton P. H. Attractive mode manipulation of covalently bound molecules. Chem. Phys. Lett. 366, 300–304 (2002). [Google Scholar]
- Watson M. D., Jäckel F., Severin N., Rabe J. P. & Müllen K. A Hexa-peri-hexabenzocoronene cyclophane: An addition to the toolbox for molecular electronics. J. Am. Chem. Soc. 126, 1402–1407 (2004). [DOI] [PubMed] [Google Scholar]
- Klymchenko A. S., Sleven J., Binnemans K. & De Feyter S. Two-dimensional self-assembly and phase behavior of an alkoxylated sandwich-type bisphthalocyanine and its phthalocyanine analogues at the liquid−solid interface. Langmuir 22, 723–728 (2006). [DOI] [PubMed] [Google Scholar]
- Phillips J. C. et al. Scalable molecular dynamics with NAMD. J. Comput. Chem. 26, 1781–1802 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vanommeslaeghe K. et al. CHARMM general force field: a force field for drug-like molecules compatible with the CHARMM all-atom additive biological force fields. J. Comput. Chem. 31, 671–690 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohen J., Arkhipov A., Braun R. & Schulten K. Imaging the migration pathways for O2, CO, NO, and Xe inside myoglobin. Biophys. J. 91, 1844–1857 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Severin N. et al. Data scattering in scanning tunneling spectroscopy. Ultramicroscopy 109, 85–90 (2008). [DOI] [PubMed] [Google Scholar]
- Mayne C. G., Saam J., Schulten K., Tajkhorshid E. & Gumbart J. C. Rapid parameterization of small molecules using the force field toolkit. J. Comput. Chem. 34, 2757–2770 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Humphrey W., Dalke A. & Schulten K. VMD: visual molecular dynamics. J. Mol. Graph. 14, 33–38 (1996). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Figures, Supplementary Table, Supplementary Notes, Supplementary Methods and Supplementary References
Data Availability Statement
The authors declare that the data supporting the findings of this study are available within the article and its Supplementary Information file or from the corresponding authors upon reasonable request.