Abstract
Ethylene biosynthesis and the ethylene signaling pathway regulate plant salt tolerance by activating the expression of downstream target genes such as those related to ROS and Na+/K+ homeostasis. The Salt Overly Sensitive (SOS) pathway regulates Na+/K+ homeostasis in Arabidopsis under salt stress. However, the connection between these two pathways is unclear. Through genetic screening, we identified two sos2 alleles as salt sensitive mutants in the ein3-1 background. Neither Ethylene-Insensitive 2 (EIN2) nor EIN3 changed the expression patterns of SOS genes including SOS1, SOS2, SOS3 and SOS3-like Calcium Binding Protein 8 (SCaBP8), but SOS2 activated the expression of one target gene of EIN3, Ethylene and Salt-inducible ERF 1 (ESE1). Moreover, Ser/Thr protein kinase SOS2 phosphorylated EIN3 in vitro mainly at the S325 site and weakly at the S35, T42 and S606 sites. EIN3 S325A mutation reduced its transcriptional activating activity on ESE1 promoter:GUS in a transient GUS assay, and impaired its ability to rescue ein3-1 salt hypersensitivity. Furthermore, SOS2 activated salt-responsive ESE1 target gene expression under salt stress. Therefore, EIN3-SOS2 might link the ethylene signaling pathway and the SOS pathway in Arabidopsis salt responses.
Salt stress is one of the major factors reducing crop yield. Plant salt stress response pathways consist of ionic and osmotic homeostasis signaling pathways, detoxification response pathways, and pathways for growth regulation1,2,3. Plant hormone signaling, ROS production, transcription regulation, and ionic homeostasis are interconnected and constitute a complex network modulating plant growth and survival under high salt stress2,4,5.
Ethylene is one of the hormones regulating plant development and stress responses, and both ethylene biosynthesis and ethylene signaling affect plant salt responses6. The enzyme 1-aminocyclopropane-1-carboxylate synthase (ACS) is crucial in ethylene biosynthesis7. Moreover, ethylene-overproducer 1 (eto1), an Arabidopsis mutation, which negatively regulates ACS activity and ethylene production, is salt tolerant8. In ethylene signaling, ethylene is perceived by receptor proteins, Ethylene Response 1/2 (ETR1/2), Ethylene Response Sensor 1/2 (ERS1/2) and Ethylene-INsensitive 4 (EIN4), which physically interact with the kinase Constitutive Triple Response 1 (CTR1) and the metal transporter-like protein EIN29. Ethylene binding leads to CTR1 inactivation, EIN2 dephosphorylation and proteolytic cleavage. Subsequently, the split EIN2 C-terminal fragment is transported into the nucleus and stabilizes EIN3, thus inducing the transcription of Ethylene-Response-Factor 1 (ERF1) and other ethylene-responsive genes10,11,12. Constitutive activation of ethylene signaling in ctr1 leads to salt tolerance, whereas the inactivation of ethylene signaling in ein2 and ein3 results in salt sensitivity in Arabidopsis4,13,14. The overexpression of either EIN3 or Ethylene and Salt-inducible ERF 1 (ESE1) enhances the expression of salt-responsive genes and salt tolerance in Arabidopsis15.
Ionic homeostasis in Arabidopsis is regulated by the Salt Overly Sensitive (SOS) pathway, consisting of SOS116, SOS217, SOS318 and SOS3-like Calcium Binding Protein 8 (SCaBP8)19,20. Under salt stress, the calcium binding protein SOS3 senses the increase in cytosolic calcium concentration and then the SOS3-SOS2 protein kinase complex activates the SOS1 ion transporter, which pumps excess cytosolic sodium out of plant cells1,21,22. SCaBP8, an SOS3 homolog, is primarily expressed in shoots, but SOS3 is primarily expressed in roots19.
Crosstalk might exist between different salt response pathways4,23,24. SOS signaling is mainly considered to be sodium homeostasis regulation under salt stress, but the transcription of several hundred genes, including ERF genes, changes under salt stress in sos2 and sos3 mutants25. Ethylene biosynthesis and the ethylene signaling pathway participate in salt stress signaling6. Furthermore, ethylene signaling and gibberellin signaling coordinately regulate plant survival and growth under salt stress14. Although the mechanism by which both ethylene signaling and SOS pathways regulating plant salt tolerance has been discovered, the connection between ethylene signaling and SOS pathway in salt stress responses remains unknown.
In this study, we isolated two new sos2 alleles through screening for salt sensitive mutants in the ein3-1 background and found that SOS2 activates the expression of ESE1, which encodes an ERF transcription factor, possibly by phosphorylation and activation of EIN3.
Results
EIN3 and SOS2 together regulate salt tolerance in Arabidopsis
To isolate salt sensitive mutants in the ein3-1 background, we constructed an Arabidopsis mutation pool by ethyl methane sulfonate (EMS)-mutagenesis of ein3-1. And from salt tolerance screening in the M2 seedlings of mutagenized ein3-1, we found that two seedlings, 453 and 751, were more sensitive to salt stress than ein3-1 (Supplementary Fig. S1). A detailed analysis of 453 and 751 mutants using M3 seeds showed that the two mutants had similar salt-sensitive phenotypes (Supplementary Fig. S2). Through map-based cloning, we found that the 453 and 751 mutations were two new alleles of sos2 mutants17.
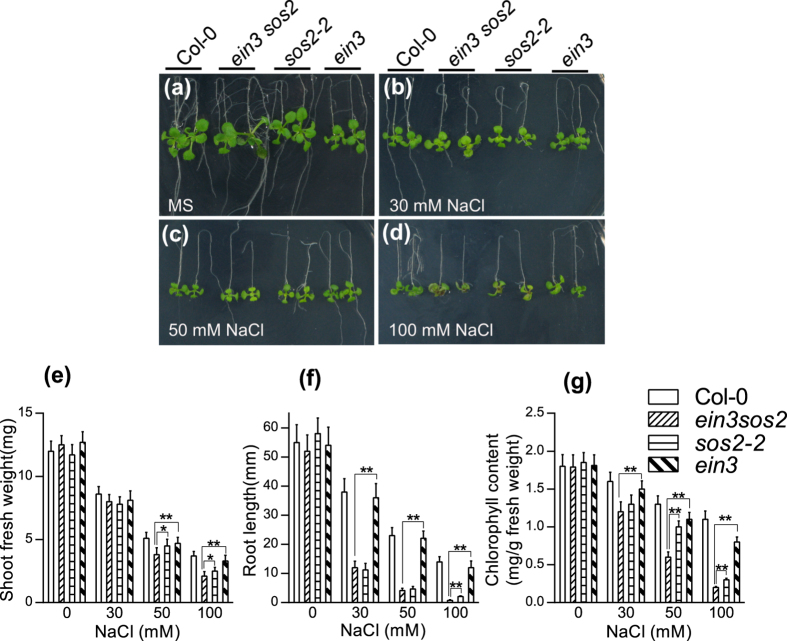
SOS2, a protein kinase, is required to activate the plasma membrane Na+/H+ antiporter SOS1 under salt stress16,17, and the overexpression of SOS2 to improve kinase activity conferred increased plant salt tolerance26. We previously showed that the overexpression of either EIN3 or ESE1 to enhance the expression of salt-responsive genes improved salt tolerance in Arabidopsis15. To dissect the salt tolerance relationship between EIN3 and SOS2, we analyzed seedling growth of Col-0, ein3-1, sos2-2 and ein3-1 sos2 (751) in MS medium containing 0, 30, 50 and 100 mM NaCl (Fig. 1). As shown in Fig. 1a–d, the seedling growth of Col-0, ein3-1, sos2-2 and ein3-1 sos2 showed no difference on MS medium (Fig. 1a), but under salt stress with 30 (Fig. 1b), 50 (Fig. 1c) and 100 mM NaCl (Fig. 1d), the shoot fresh weight and root growth of sos2-2 and ein3-1 sos2 were less than those in Col-0 and ein3-1 (Fig. 1e,f). Under salt stress, the leaves of sos2-2 and ein3-1 sos2 turned yellow-green, thus indicating less chlorophyll content. Therefore, we determined the chlorophyll content by using a spectrophotometric method after pigment extraction with 80% acetone. As shown in Fig. 1g, under salt stress, the chlorophyll content in ein3-1 sos2 was less than that in sos2-2. Under 100 mM NaCl stress, the euphylla of ein3-1 sos2 turned brown as a result of chlorophyll degradation and anthocyanin accumulation19.
Figure 1. The Arabidopsis ein3 sos2 double mutant is more sensitive to salt stress than the ein3-1/sos2 single mutants at seedling stage.
(a–d) Arabidopsis seedlings at 4-day stage were transferred to MS medium containing 0 (a), 30 (b), 50 (c) and 100 mM NaCl (d), respectively. (e–g) Shoot fresh weight (e), root growth (f) and chlorophyll content (g) were determined 5 days after transfer. Values are means ± SD. *Indicates significant difference at p = 0.05 by t-test; **indicates significant difference at p = 0.01 by t-test.
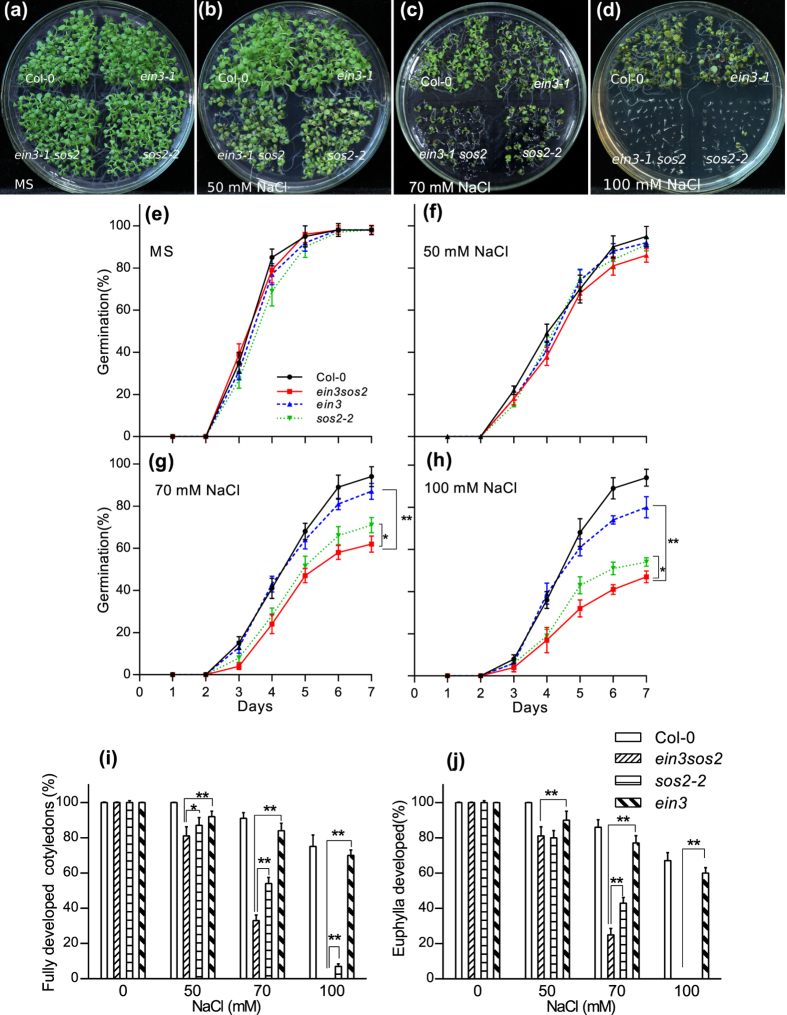
To investigate salt responses regulated by EIN3 and SOS2 in germination, we germinated Col-0, ein3-1, sos2-2 and ein3-1 sos2 (751) seeds in MS medium containing different concentrations of NaCl (Fig. 2). The germination rates showed no significant differences among Col-0, ein3-1, sos2-2 and ein3-1 sos2 seeds (Fig. 2a,e). On MS medium containing 50 mM NaCl, the germination rate of ein3-1 was similar to that of sos2-2 but was slower than that of Col-0, whereas the germination rate of the ein3-1 sos2 double mutant was the lowest (Fig. 2b,f). On MS medium containing 70 or 100 mM NaCl, the germination rate of sos2 was less than that of ein3-1 but more than that of the ein3-1 sos2 double mutant (Fig. 2c,d,g,h).
Figure 2. The Arabidopsis ein3 sos2 double mutant is more sensitive to salt stress than the ein3-1/sos2 single mutants during germination.
(a–h) Germination of Col-0, ein3-1, sos2-2, and ein3 sos2 double mutant seeds on MS medium containing 0 (a,e), 50 (b,f), 70 (c,g) and 100 mM NaCl (d,h). (i) Percentage of seedlings with fully developed cotyledons 7 days after germination. (j) Percentage of seedlings with euphylla 7 days after germination. Values are means ± SD. *Indicates significant difference at p = 0.05 by t-test; **indicates significant difference at p = 0.01 by t-test.
Seven days after germination, all seedlings on MS medium had well developed cotyledons and several euphylla, but on MS medium containing 50 mM NaCl, the euphylla of sos2 and ein3-1 sos2 were similar but much smaller than those of Col-0 and ein3-1 (Fig. 2b,i,j). On MS medium containing 70 mM NaCl, the percentages of seedlings with fully developed cotyledons and euphylla of sos2-2 were more than that of ein3-1 sos2 but less than that of ein3-1 (Fig. 2c,i,j). On MS medium containing 100 mM NaCl, about 80% of Col-0 developed euphylla, but almost all of sos2 and ein3-1 sos2 had a bleached appearance and died despite a 40% rate of germination (Fig. 2d,h,i,j).
The above results indicate that ein3-1 sos2 double mutant is more sensitive to salt stress than ein3-1 or sos2-2 single mutant during the germination process and seedling growth stage. Therefore, EIN3 and SOS2 might synergistically regulate the salt stress response in Arabidopsis.
EIN3 does not influence the transcription of SOS genes, but SOS2 positively regulates the expression of the EIN3 target gene ESE1
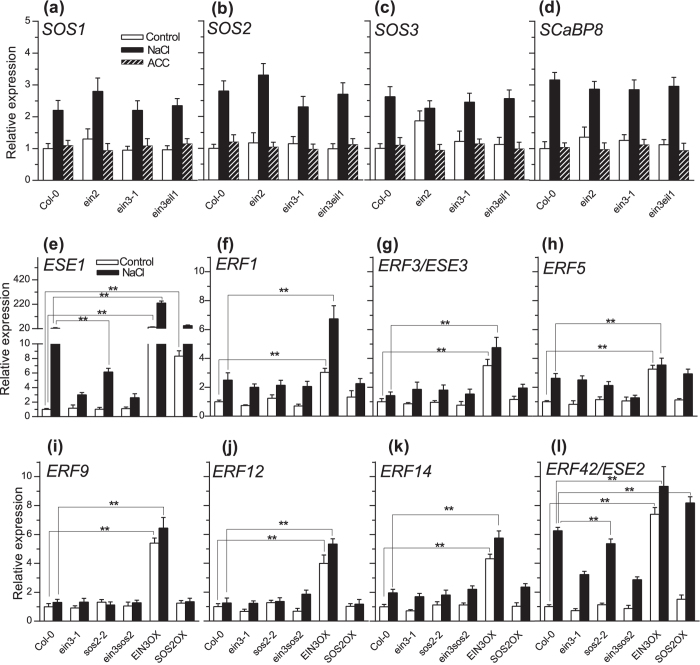
Next, we explored the regulatory relationship between EIN3 and SOS2 in the salt response. Because EIN3 is a transcription factor, we first analyzed SOS genes (SOS1, SOS2, SOS3, SCaBP8) expression in the ein3 mutant via qPCR. As shown in Fig. 3a–d, we found that the expression of all four SOS genes increased 2-3 times after 150 mM NaCl treatment for 2 hours, but was not different between ein3 and Col-0. The induction of SOS genes was almost similar between ein3 eil1 double mutant and ein3 after salt treatment. Furthermore, the expression patterns of SOS genes were not changed in ein2 mutant compared with Col-0 (Fig. 3a–d). In addition, the treatment with ethylene biosynthesis precursor 1-aminocyclopropane-1-carboxylic acid (ACC) at 10 μM for 3 hours did not change the expression levels of SOS genes in wild type plants and ethylene signaling mutants ein2, ein3 and ein3 eil1 plants (Fig. 3a–d).
Figure 3. SOS2 positively regulates the expression of ESE1, but the expression levels of SOS genes are not influenced by EIN3 or EIN2.
(a–d) The expression of SOS genes SOS1 (a), SOS2 (b), SOS3 (c) and SCaBP8 (d) in Col-0, ein2, ein3 and ein3 eil1 in the control, after the treatment by 150 mM NaCl for 2 hours, or by 10 μM ACC for 3 hours. (e–l) The expression of ESE1 (e), ERF1 (f), ERF3 (g), ERF5 (h), ERF9 (i), ERF12 (j), ERF14 (k) and ERF42 (l) in Col-0, ein3, sos2, ein3 sos2 mutants and EIN3/SOS2 overexpression lines before and 2 hours after treatment by 150 mM NaCl. Values are means ± SD (n = 3). **Indicates significant difference at p = 0.01 by t-test. EIN3OX and SOS2OX are EIN3 and SOS2 overexpression lines, respectively.
Previously, we identified 3 ERF genes that were strongly induced by both salt and ACC, and we also found 5 ERF genes were slightly induced by salt and ACC by examining the expression data of 122 putative ERF genes in Geneinvestigator databases followed by qPCR confirmation15. The salt and ethylene/ACC induced genes might participate in ethylene and salt responses, and therefore we determined whether SOS2 affects the expression of these putative EIN3 target ERF genes in the salt response (Fig. 3e–l). Under normal conditions, the basal expression level of ESE1 was similar among Col-0, ein3-1, sos2-2, and ein3 sos2. Two hours after treatment with 150 mM NaCl, the ESE1 induction was lower in the ein3 sos2 double mutant than in the ein3 or sos2 single mutant. Constitutive expression of SOS2 also increased the ESE1 expression level under normal or salt stress conditions (Fig. 3e). The expression pattern of ERF42 (ESE2) was similar to that of ESE1, but induced to a lesser extent (Fig. 3l).
The expression of ERF1, ERF3, ERF5 and ERF14 was induced after salt stress treatment, but the expression patterns were similar in Col-0, sos2-2, and SOS2 overexpression lines, whereas their expression was enhanced in EIN3 overexpression lines (Fig. 3f–h,k). Although the transcription of ERF9 and ERF12 was activated by EIN3, it was not altered by SOS2 (Fig. 3e,j).
The salt induction of ESE1 and ERF42 (ESE2) in ein3 sos2 double mutant was similar to the level in ein3-1 but less than that of sos2, suggesting that salt induction of ethylene-responsive genes might depend more on ethylene signaling than on SOS pathway. Therefore, SOS2 might regulate the transcription of ERFs by modulating ethylene signaling components.
SOS2 phosphorylates EIN3 in vitro
SOS2, a Ser/Thr protein kinase in Arabidopsis, phosphorylates SOS1, thereby activating its Na+ pump activity under salt stress17,26,27. EIN3 is a transcription factor in the ethylene signaling pathway. EIN3 is constitutively expressed, but EIN3 protein interacts with two F-box proteins (EBF1 and EBF2) and is degraded by the 26S proteasome without ethylene. T592 phosphorylation promotes EIN3 degradation, whereas T174 phosphorylation stabilizes EIN3, thus indicating that the stability of EIN3 is regulated by its phosphorylation status28.
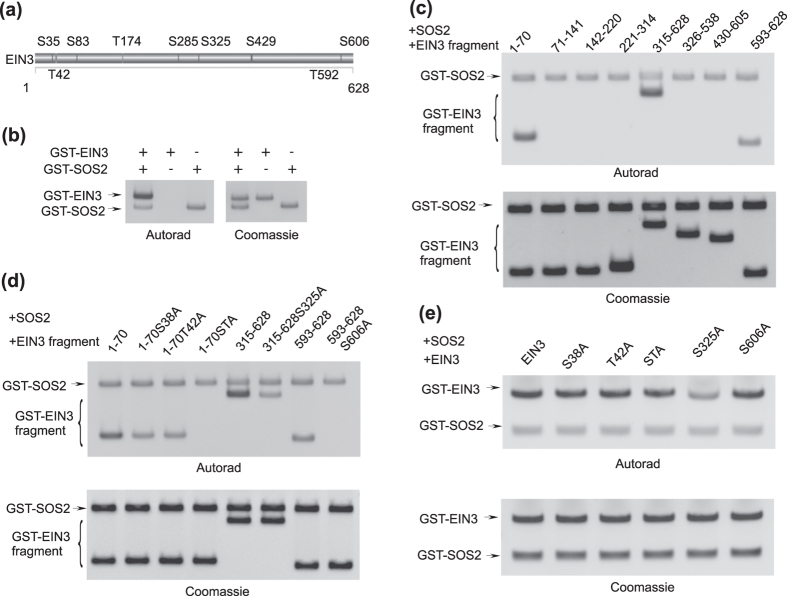
EIN3 consists of 10 putative Ser/Thr phosphorylation sites, as predicted by a bioinformatics search (Fig. 4a). To determine whether the Ser/Thr kinase SOS2 phosphorylates EIN3 and regulates downstream gene expression, we conducted an in vitro phosphorylation assay by incubating GST-SOS2 and GST-EIN3 recombinant proteins with γ-32P-ATP. As shown in Fig. 4b, full length EIN3 was phosphorylated by SOS2 in vitro.
Figure 4. SOS2 phosphorylates EIN3 in vitro.
(a) Putative sites in EIN3 phosphorylated by Ser/Thr kinase, as predicted by NetPhos (http://www.cbs.dtu.dk/ services/NetPhos/). In this prediction, nine amino acids (Ser35, Thr42, Ser83, Thr174, Ser285, Ser325, Ser429, Thr592, Ser606) are potential sites phosphorylated by Ser/Thr protein kinases. (b) Phosphorylation of EIN3 by protein kinase SOS2 in vitro. (c–e) Phosphorylation EIN3 fragments (c), EIN3 fragments with point mutations (d) and full length EIN3 with point mutations (e) by protein kinase SOS2 in vitro. Numbers (1–70, 71–141, 142–220, 221–314, 315–628, 326–538, 430–605, 593–628) indicate GST-EIN3 fragments with corresponding EIN3 sequences. S35A, T42A, S325A and S606A indicate mutation of Ser/Thr to Ala. STA indicates double mutation of Ser35 and Thr42 to Ala. The uncropped images are shown in Supplementary Fig. S4.
To determine the sites of EIN3 phosphorylated by SOS2, we divided EIN3 into small fragments, each of which had 1–3 putative phosphorylation sites, and then conducted in vitro phosphorylation experiments. As shown in Fig. 4c, fragments 1–70, 315–628, and 593–628 were phosphorylated by SOS2.
Next, we analyzed the phosphorylation of EIN3 with point mutations (Fig. 4d). Fragment 1–70 consisted of two putative phosphorylation sites S35 and T42, and an in vitro kinase assay showed that fragment 1–70 with S35A or T42A single mutation was phosphorylated by SOS2, but the signal was much less than that of wild-type fragment 1–70. Furthermore, fragment 1–70 with the S35A T42A double mutation (1-70STA) was not phosphorylated by SOS2. Together, these results suggest that both S35 and T42 in EIN3 may be phosphorylated by SOS2 in vitro. Fragment 315–628 displayed the highest signal in the in vitro kinase assay, but fragment 315–628 with S325A mutation displayed a weak phosphorylation signal. Fragment 593–628, consisting of only one putative phosphorylation site S606, was phosphorylated, thus suggesting that S606 may be one phosphorylation site. SOS2 did not phosphorylate fragment 593–628 with the S606A mutation, thus confirming that S606 is one site in EIN3 that is phosphorylated by SOS2 in vitro (Fig. 4d). These results indicate that S325 might be the main phosphorylation site, and S606 might be a weak phosphorylation site. In addition, phosporylation analysis of full length EIN3 with point mutations by SOS2 revealed that S325A greatly reduced EIN3 phosphorylation, and S35A, T42A or S606A only slightly reduced EIN3 phosphorylation (Fig. 3e). Therefore, S325 may be the main phosphorylation site, whereas S35, T42 and S606 might be weak sites in EIN3 that are phosphorylated by SOS2 in vitro.
In Arabidopsis, EIN3 has 5 homologs, EIL1~EIL5. To determine whether the phosphorylation sites of EIN3 are conserved among EIN3/EIL proteins, we performed multiple sequence alignments using CLUSTAL 2.0.1229 (Supplementary Fig. S3). Among the four phosphorylation sites of EIN3, S325 is the most conserved amino acid, followed by T42, whereas the positions of S35 and S606 are highly diverse. Because S325, the main EIN3 site phosphorylated by SOS2 in vitro is conserved among the EIN3/EIL family, S325 might be crucial for the function of EIN3/EILs.
EIN3 S325A mutation reduces its transcriptional activating activity on ESE1, and impairs its ability to rescue ein3-1 salt hypersensitivity
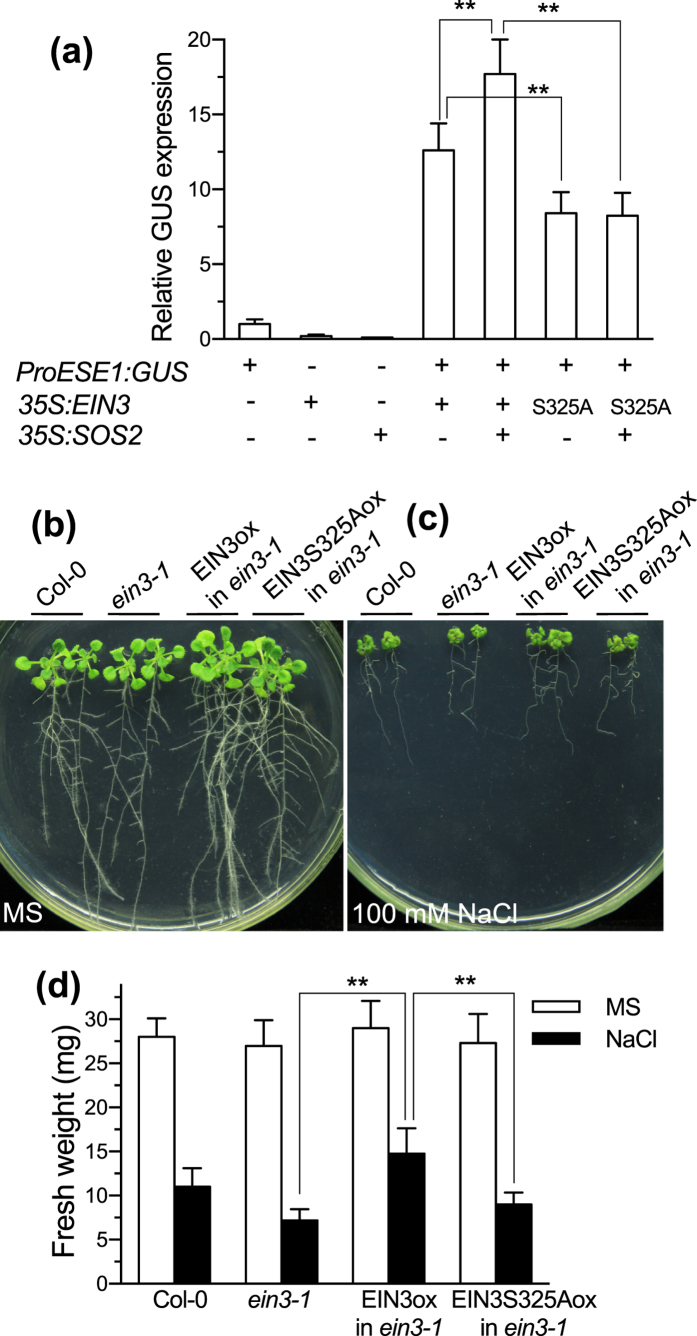
To determine the effect of S325 phosphorylation in EIN3 to its transcriptional activating activity on ESE1, we performed a transient assay by co-transformation of 35S:EIN3, 35S:SOS2 and the reporter ESE1 promoter:GUS with 35S:LUC as an internal control (Fig. 5a). The results showed that the transcriptional activity of ESE1 promoter was reduced at about 30% by S325A mutation in EIN3. Furthermore, SOS2 increased the transcriptional activating activity of EIN3, but not EIN3S325A, on ESE1 promoter at about 25%. These results suggest that S325 phosphorylation is critical for the transcriptional activity of ESE1 regulated by EIN3 and SOS2.
Figure 5. EIN3 S325A mutation reduces its transcriptional activation on ESE1 and impairs its ability to rescue ein3–1 salt hypersensitivity.
(a) Transient GUS assay for ESE1 promoter transcriptional activity by EIN3 and its mutated protein in tobacco leaves. 35S:LUC is an internal control. Values are means ± SD (n = 5). **Indicates significant difference at p = 0.01 by t-test. (b–d) Seedling at 4-day stage were transferred to MS medium with 0 (a) and 100 mM NaCl. Ten days after transfer, photos were captured (b,c) and shoot fresh weight per plant was determined (d). Pictures are representative photos of 3 independent transgenic lines with similar results. Values are means ± SD with 3 independent transgenic lines in 3 repetitions. **Indicates significant difference at p = 0.01 by t-test.
Next, we investigated the salt tolerance of EIN3 or EIN3S325A overexpression lines in ein3-1 background (Fig. 5b–d). The results showed that the overexpression of EIN3 improved ein3-1 salt tolerance in root growth and shoot fresh weight, whereas the overexpression of EIN3S325A only partially rescued ein3-1 salt hypersensitivity.
The above results indicate that the phosphorylation of EIN3S325 by SOS2 is critical for ESE1 transcriptional activation in vivo and plant salt tolerance, at least partially.
EIN3 and SOS2 coactivate downstream salt-responsive gene expression
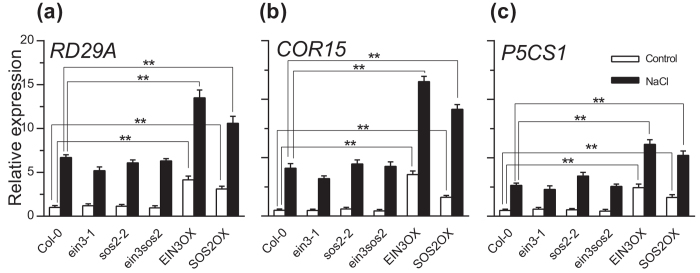
As shown above, SOS2 activated the expression of the EIN3 target gene ESE1, and SOS2 phosphorylated EIN3 in vitro; therefore, the phosphorylation of EIN3 by SOS2 under salt stress might upregulate ESE1 expression. Previously, we have found that ese1 is salt sensitive (Supplementary Fig. S5)15, and EIN3 and ESE1 activate the expression of salt-responsive genes, including RD29A30, COR1531 and P5CS32, by binding to their promoter regions and activating gene expression15. Previous studies showed that the loss-of-function mutant of p5cs1 is salt sensitive32. In salt tolerance test, seedling growth was inhibited much more by salt stress in rd19a and p5cs1 than in wild type plants, but the growth of cor15 was almost similar to wild type plants in salt stress (Supplementary Fig. 6a–f). To investigate whether EIN3 and SOS2 coactivated salt-responsive gene expression, we analyzed the expression of RD29A, COR15 and P5CS1 in ein3-1, sos2-2, ein3 sos2, EIN3OX and SOS2OX in control and salt-stressed plants (Fig. 6a–c). Under normal conditions, the expression of RD29A, COR15 and P5CS1 was not significantly different in ein3-1, sos2-2, ein3 sos2 mutants and wild type but was upregulated in EIN3OX and SOS2OX plants. After salt stress, the expression of RD29A, COR15 and P5CS1 was induced by salt treatment in all plants, but the expression was much higher in EIN3OX and SOS2OX than in the other plants. These results suggest that both EIN3 and SOS2 positively regulate salt-responsive gene expression.
Figure 6. EIN3 and SOS2 activate stress-responsive genes under salt stress.
(a–c) The expression of RD29A (a), COR15 (b) and P5CS1 (c) in Col-0, ein3, sos2, ein3 sos2 mutants and EIN3/SOS2 overexpression lines before and 2 hours after treatment by 150 mM NaCl. Values are means ± SD (n = 3). **Indicates significant difference at p = 0.01 by t-test. EIN3OX and SOS2OX are EIN3 and SOS2 overexpression lines, respectively.
Discussion
Both ethylene biosynthesis8 and the ethylene signaling4,13,14 play positive roles in Arabidopsis salt tolerance. In Arabidopsis, the SOS pathway, consisting of SOS116, SOS217, SOS318 and SCaBP819, is crucial for plant salt tolerance by regulating iron homeostasis under conditions of salt stress. In this study, we isolated two new sos2 alleles as salt sensitive mutants in the ein3-1 background, and found SOS2 phosphorylated and activated EIN3, which suggests a connection between the ethylene signaling component EIN3 and the SOS pathway component protein kinase SOS2 in plant salt response regulation.
EIN3 is a transcription factor, but neither EIN2 nor EIN3 changes the expression pattern of SOS genes under normal or salt stress conditions. However, SOS2 activates the expression of ESE1, which encodes an ERF transcription factor downstream of EIN3. Microarray analysis in sos2 mutant has demonstrated that some EREBP genes and ACS gene are upregulated25, thus implying a crosstalk between the SOS pathway and ethylene signaling in plant salt responses. In vitro phosphorylation of EIN3 by SOS2 and the consequent upregulation of the ERF gene downstream of EIN3 indicate that SOS2-EIN3 might serve as junction between the SOS pathway and the ethylene signaling pathway in plant salt response regulation.
Protein phosphorylation is one means of regulation of EIN3 in ethylene signaling28. T174 and T592 are two phosphorylation sites of EIN3 that affect EIN3 stability in opposite ways. Without ethylene, CTR1, a Raf-like MAPK kinase kinase, inactivates MKK9–MPK3/6 and activates downstream MAPKs, which in turn phosphorylate T592 and promote EIN3 degradation. Ethylene inactivates CTR1, thus resulting in MKK9-MPK3/6 activation and T174 phosphorylation and the consequent stabilization of EIN3. In this study, we found that SOS2 phosphorylates S325, but not T174 or T592 of EIN3 in kinase assays in vitro. These results suggest that phosphorylation of EIN3 at different positions might have various functions: T174 and T592 may act in ethylene signaling, and S325 in salt responses.
Previous studies have shown that the expression of many stress-responsive genes changes in sos mutants16,25,33. However, it is unknown how protein kinase SOS2 regulates gene expression. In this study, we found that the mutation of EIN3 S325A, a phosphorylation site by SOS2, reduces its transcriptional activating activity on ESE1 promoter:GUS, and weakens its ability to rescue ein3-1 salt hypersensitivity, revealing that SOS2 might regulate plant salt response by phosphorylating EIN3 to transcriptionally activate downstream salt stress responsive genes. Therefore, EIN3 and SOS2 might be linked together to modulate plant salt stress response via the phosphorylation of EIN3 by SOS2.
Salt stress increases EIN3 protein accumulation by promoting the proteasomal degradation of EBF1 and EBF234. Meanwhile, salt stress induces cytosolic calcium accumulation, which is sensed by the calcium binding proteins SOS3 and SCaBP818,19,20. Then SOS2, which is activated by SOS3 or SCaBP8, phosphorylates and activates SOS11,16,17,18,19,20,21,22. In this study, we found SOS2 phosphorylates EIN3 protein, which expands the range of proteins phosphorylated by SOS2 in addition to SOS1 and SCaBP820,21. Thus the phosphorylation of EIN3 by SOS2 might increase EIN3 protein stability or directly enhance EIN3 transcriptional activity. Therefore, EIN3 and SOS2 synergistically modulate plant salt tolerance.
Since either the overexpression of EIN315 or the overexpression of SOS2 to improve kinase activity26 increases plant salt tolerance, and the expression of SOS genes is not changed in ein3 and ein2 mutants or by ACC treatment, future experiments to test the salt tolerance of EIN3 overexpresser in sos2 background and SOS2 overexpresser in ein3 background would provide more cues for the genetic interaction between SOS2 and EIN3 in plant salt response regulation. In addition, the function of the other putative sites in EIN3 phosphorylated by SOS2 in salt response regulation needs to be addressed in future investigations. And whether the phosphorylation of EIN3 by SOS2 is regulated by other factors is unknown yet. Furthermore, to dissect additional components connecting the SOS and ethylene pathways is an interesting topic.
In summary, through genetic screening, we identified sos2 as a salt sensitive mutant in the ein3-1 background. SOS2 phosphorylates EIN3 and consequently activates downstream ERF gene expression and induces stress responsive gene expression under salt stress conditions. Therefore, SOS2-EIN3 might link the SOS and ethylene pathways in plant salt responses.
Methods
Isolation of salt sensitive mutants in an ein3-1 background
Approximately 20,000 ein3-1 seeds were treated with 25 volumes of 0.2% (v/v) ethyl methane sulfonate (EMS) for 15 hours. The seeds were washed 10 times with water after removal of the EMS. Seeds were suspended in 0.1% agarose and sown on soil. M2 seeds were collected in pools after plants had matured35.
M2 seeds of ein3-1 were sterilized in a solution containing 20% sodium hypochlorite and 0.1% Triton X-100 for 10 min, washed five times with sterilized water, and sown on MS medium with 0.6% Phytagel (Sigma-Aldrich, P8169). The plates were placed at 4 °C for 2 days, and then the seeds were germinated vertically at 23 °C under 16/8 (light/dark) hours illumination. Four-day-old seedlings with a root length of 1.5 cm were transferred onto MS medium supplemented with 100 mM NaCl and cultured vertically with the roots upward and shoots downward. The root growth and shoot growth was checked after cultivation for 4 days19.
Seedlings with a hyper salt sensitive phenotype were planted in soil to collect M3 seeds. After confirmation of the salt sensitive phenotype in M3 seedlings, M3 plants were crossed with Ler-0. Then, the mutation sites were determined by map-based cloning using F2 segregating population.
Salt sensitivity analysis in Arabidopsis
The salt sensitivity analysis of Arabidopsis seedlings was carried out as described above. For salt sensitivity analysis of Arabidopsis in germination, the seeds were sterilized in a solution containing 20% sodium hypochlorite and 0.1% Triton X-100 for 10 min, washed five times with sterilized water, and sown on MS medium (0.2% Phytagel, Sigma-Aldrich, P8169) with 0, 50, 70 or 100 mM NaCl added. The plates were incubated at 4 °C for 2 days, and the seeds were germinated at 23 °C under 16/8 (light/dark) hours illumination. The germination rate was measured daily.
Chlorophyll content determination
Arabidopsis leaves were weighed and fractured in 80% acetone in water to extract chlorophyll. The debris was removed by centrifugation, and the absorbance of the supernatant at 646 and 663 nm was determined with a spectrometer; the chlorophyll concentration was calculated as described previously36.
Gene expression analysis via real-time quantitative PCR (qPCR)
For Arabidopsis, 7-day seedlings were treated by 150 mM NaCl for 2 hours, and leaves were collected and stored in liquid nitrogen. For rice, 2-week seedlings were stressed with 100 mM NaCl for 2 hours or 150 mM NaCl for 3 hours, and leaves were collected and stored in liquid nitrogen. Total RNA from leaves was extracted via the TRIzol method (ThermoFisher)37,38 and digested with DNase free RNase to remove genomic DNA. Reverse transcription was performed using 1 mg of total RNA and MLV reverse transcriptase (Promega). The cDNA was then used for PCR amplification with gene specific primers in Table S1 on a real-time PCR machine (Bio-rad IQ5). The gene expression level was calculated with Bio-rad IQ5 software (version 2.1).
Recombinant proteins expression and purification
Recombinant SOS2 and EIN3 proteins were expressed as GST fusions. GST-SOS2 has been described previously19,20. For GST-EIN3, an EIN3 full-length coding region was amplified using primers with BamHI and SalI sites (Table S1) from Arabidopsis cDNA. The PCR product was digested with BamHI and SalI and cloned into a pGEX-6P-1 vector. For GST-EIN3 fragments, EIN3 fragment coding region was amplified with the corresponding primers in Table S1 by using a GST-EIN3 construct as a template, digested with BamHI and SalI, and cloned into the pGEX-6P-1 vector (GE Healthcare 28-9546-48). To generate EIN3 with a Ser/Thr to Ala point mutation, we introduced the mutation sites by PCR in vitro via site-directed mutagenesis using primers in Table S139. The EIN3 S35A, T42A double mutant was generated by introducing T42A in EIN3 S35A through the method described above.
The GST fusion constructs were transformed into Escherichia coli strain BL21 (DE3). The expression of recombinant proteins was induced with 0.5 mM IPTG at 18 °C for 6 hours. The cells were then collected by centrifugation and lysed by sonication. The recombinant proteins were purified with glutathione Sepharose 4B (GE Healthcare 17075601) according to the manufacturer’s protocol.
In vitro kinase assays
In vitro kinase assays were performed in kinase buffer including 20 mM Tris-HCl, pH 8.0, 5 mM MgCl2, 10 μM ATP, and 1 mM DTT. In a total volume of 20 μL, 1 μg protein and 0.5 μL of [γ-32P] ATP (5 μCi) were added to the kinase buffer and incubated at 30 °C for 30 min. The reactions were terminated by the addition of 4 μL 6 × SDS loading buffer, then incubated at 95 °C for 5 min. The proteins were separated via 12% (w/v) SDS-PAGE and stained with Coomassie brilliant blue R250, and then the signals were captured with an X-film (Kodak) or a phosphor screen (Amersham Biosciences)19,20.
Sequence alignments
Protein sequences of EIN3 and EIL1~5 were downloaded from The Arabidopsis Information Resource (TAIR) (http://www.arabidopsis.org/), and multiple sequence alignments were performed with CLUSTAL 2.0.1229. The alignment output was printed out with BoxShade (http://www.ch.embnet.org/software/BOX%5Fform. html).
Transient GUS assay
A transient GUS assay was performed by transient expression in tobacco leaves as described previously15,40. The constructs ProESE1:GUS, 35S:EIN3(/S325A), 35S:SOS2 and 35S:LUC were transferred into Agrobacterium strain GV3101 separately by electroporation. Agrobacteria were harvested and incubated with induction medium containing 100 μM acetosyringone. Then the bacteria were injected into tobacco leaves that still attached to the intact plant. Two days after agroinfiltration, GUS activity was determined using 4-methylumbelliferyl-D-glucuronide as a substrate, and luciferase activity was used as an internal control.
Additional Information
How to cite this article: Quan, R. et al. EIN3 and SOS2 synergistically modulate plant salt tolerance. Sci. Rep. 7, 44637; doi: 10.1038/srep44637 (2017).
Publisher's note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Material
Acknowledgments
This work was supported by National Science Foundation of China (31670280), National Key Program of Transgenic Biology (2016ZX08009-003-005) and the National Basic Research Program of China (2012CB114204).
Footnotes
The authors declare no competing financial interests.
Author Contributions R.H. conceived and designed the project. D.Y. isolated the ein3 sos2 double mutant. R.Q. performed the other experiments. J.W., H.Z., R.Q. and Z.Z. analyzed the data. R.Q. wrote the manuscript.
References
- Zhu J.-K. Salt and drought stress signal transduction in plants. Annu. Rev. Plant Biol. 53, 247–273 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Julkowska M. M. & Testerink C. Tuning plant signaling and growth to survive salt. Trends Plant Sci. 20, 586–594 (2015). [DOI] [PubMed] [Google Scholar]
- Munns R. & Gilliham M. Salinity tolerance of crops - what is the cost? New Phytol. 208, 668–673 (2015). [DOI] [PubMed] [Google Scholar]
- Kazan K. Diverse roles of jasmonates and ethylene in abiotic stress tolerance. Trends Plant Sci. 20, 219–229 (2015). [DOI] [PubMed] [Google Scholar]
- Dinneny J. R. Traversing organizational scales in plant salt-stress responses. Curr. Opin. Plant Biol. 23, 70–75 (2015). [DOI] [PubMed] [Google Scholar]
- Tao J.-J. et al. The role of ethylene in plants under salinity stress. Front. Plant Sci. 6, 1059 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang K. L.-C., Li H. & Ecker J. R. Ethylene biosynthesis and signaling networks. Plant Cell 14 Suppl, S131–S151 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang C., Belfield E. J., Cao Y., Smith J. A. C. & Harberd N. P. An Arabidopsis soil-salinity-tolerance mutation confers ethylene-mediated enhancement of sodium/potassium homeostasis. Plant Cell 25, 3535–3552 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merchante C., Alonso J. M. & Stepanova A. N. Ethylene signaling: simple ligand, complex regulation. Curr. Opin. Plant Biol. 16, 554–560 (2013). [DOI] [PubMed] [Google Scholar]
- Qiao H. et al. Processing and subcellular trafficking of ER-tethered EIN2 control response to ethylene gas. Science 338, 390–393 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ju C. et al. CTR1 phosphorylates the central regulator EIN2 to control ethylene hormone signaling from the ER membrane to the nucleus in Arabidopsis. Proc. Natl. Acad. Sci. USA 109, 19486–19491 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Solano R., Stepanova A., Chao Q. & Ecker J. R. Nuclear events in ethylene signaling: a transcriptional cascade mediated by ETHYLENE-INSENSITIVE3 and ETHYLENE-RESPONSE-FACTOR1. Genes Dev. 12, 3703–3714 (1998). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao W.-H. et al. Modulation of ethylene responses affects plant salt-stress responses. Plant Physiol. 143, 707–719 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Achard P. et al. Integration of plant responses to environmentally activated phytohormonal signals. Science 311, 91–94 (2006). [DOI] [PubMed] [Google Scholar]
- Zhang L. et al. An AP2 domain-containing gene, ESE1, targeted by the ethylene signaling component EIN3 is important for the salt response in Arabidopsis. Plant Physiol. 157, 854–865 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi H., Ishitani M., Kim C. & Zhu J. K. The Arabidopsis thaliana salt tolerance gene SOS1 encodes a putative Na+/H+ antiporter. Proc. Natl. Acad. Sci. USA 97, 6896–6901 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J., Ishitani M., Halfter U., Kim C. S. & Zhu J. K. The Arabidopsis thaliana SOS2 gene encodes a protein kinase that is required for salt tolerance. Proc. Natl. Acad. Sci. USA 97, 3730–3734 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J. & Zhu J. K. A calcium sensor homolog required for plant salt tolerance. Science 280, 1943–1945 (1998). [DOI] [PubMed] [Google Scholar]
- Quan R. et al. SCABP8/CBL10, a putative calcium sensor, interacts with the protein kinase SOS2 to protect Arabidopsis shoots from salt stress. Plant Cell 19, 1415–1431 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin H. et al. Phosphorylation of SOS3-LIKE CALCIUM BINDING PROTEIN8 by SOS2 protein kinase stabilizes their protein complex and regulates salt tolerance in Arabidopsis. Plant Cell 21, 1607–1619 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ji H. et al. The Salt Overly Sensitive (SOS) pathway: established and emerging roles. Mol. Plant 6, 275–286 (2013). [DOI] [PubMed] [Google Scholar]
- Zhu J.-K. Cell signaling under salt, water and cold stresses. Curr. Opin. Plant Biol. 4, 401–406 (2001). [DOI] [PubMed] [Google Scholar]
- Hartmann L. et al. Crosstalk between two bZIP signaling pathways orchestrates salt-induced metabolic reprogramming in Arabidopsis roots. Plant Cell 27, 2244–2260 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dar T. A., Uddin M., Khan M. M. A., Hakeem K. & Jaleel H. Jasmonates counter plant stress: a review. Environ. Exp. Bot. 115, 49–57 (2015). [Google Scholar]
- Kamei A. et al. Analysis of gene expression profiles in Arabidopsis salt overly sensitive mutants sos2-1 and sos3-1. Plant Cell Environ. 28, 1267–1275 (2005). [Google Scholar]
- Guo Y. et al. Transgenic evaluation of activated mutant alleles of SOS2 reveals a critical requirement for its kinase activity and C-terminal regulatory domain for salt tolerance in Arabidopsis thaliana. Plant Cell 16, 435–449 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiu Q.-S. et al. Regulation of vacuolar Na+/H+ exchange in Arabidopsis thaliana by the Salt-Overly-Sensitive (SOS) pathway. J. Biol. Chem. 279, 207–215 (2004). [DOI] [PubMed] [Google Scholar]
- Yoo S.-D., Cho Y.-H., Tena G., Xiong Y. & Sheen J. Dual control of nuclear EIN3 by bifurcate MAPK cascades in C2H4 signalling. Nature 451, 789–795 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larkin M. et al. Clustal W and Clustal X version 2.0. Bioinformatics 23, 2947–2948 (2007). [DOI] [PubMed] [Google Scholar]
- Kasuga M., Liu Q., Miura S., Yamaguchi-Shinozaki K. & Shinozaki K. Improving plant drought, salt, and freezing tolerance by gene transfer of a single stress-inducible transcription factor. Nat. Biotechnol. 17, 287–291 (1999). [DOI] [PubMed] [Google Scholar]
- Jaglo-Ottosen K. R., Gilmour S. J., Zarka D. G., Schabenberger O. & Thomashow M. F. Arabidopsis CBF1 overexpression induces COR genes and enhances freezing tolerance. Science 280, 104–106 (1998). [DOI] [PubMed] [Google Scholar]
- Székely G. et al. Duplicated P5CS genes of Arabidopsis play distinct roles in stress regulation and developmental control of proline biosynthesis. Plant J. 53, 11–28 (2008). [DOI] [PubMed] [Google Scholar]
- Gong Z. et al. Genes that are uniquely stress regulated in salt overly sensitive (sos) mutants. Plant Physiol. 126, 363–375 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng J. et al. Salt-induced stabilization of EIN3/EIL1 confers salinity tolerance by deterring ROS accumulation in Arabidopsis. PLoS Genet. 10, e1004664, doi: 10.1371/journal.pgen.1004664 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weigel D. & Glazebrook J. Arabidopsis: A Laboratory Manual (Cold Spring Harbor Laboratory Press, 2002). [Google Scholar]
- Porra R. J. The chequered history of the development and use of simultaneous equations for the accurate determination of chlorophylls a and b. Photosynth. Res. 73, 149–156 (2002). [DOI] [PubMed] [Google Scholar]
- Chomczynski P. & Sacchi N. Single-step method of RNA isolation by acid guanidinium thiocyanate-phenol-chloroform extraction. Anal. Biochem. 162, 156–159 (1987). [DOI] [PubMed] [Google Scholar]
- Chomczynski P. & Sacchi N. The single-step method of RNA isolation by acid guanidinium thiocyanate-phenol-chloroform extraction: twenty-something years on. Nat. Protoc. 1, 581–585 (2006). [DOI] [PubMed] [Google Scholar]
- Ho S. N., Hunt H. D., Horton R. M., Pullen J. K. & Pease L. R. Site-directed mutagenesis by overlap extension using the polymerase chain reaction. Gene 77, 51–59 (1989). [DOI] [PubMed] [Google Scholar]
- Yang Y., Li R. & Qi M. In vivo analysis of plant promoters and transcription factors by agroinfiltration of tobacco leaves. Plant J. 22, 543–551 (2000). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.