Abstract
Bacteria use type IV secretion systems (T4SS) to translocate DNA and protein substrates to target cells of phylogenetically diverse taxa. Recently, by use of an assay termed transfer DNA immunoprecipitation (TrIP), we described the translocation route for a DNA substrate [T-DNA, portion of the Ti (tumor-inducing) plasmid that is transferred to plant cells] of the Agrobacterium tumefaciens VirB/D4 T4SS in terms of a series of temporally and spatially ordered substrate contacts with subunits of the secretion channel. Here, we report that the bitopic inner membrane protein VirB10 undergoes a structural transition in response to ATP utilization by the VirD4 and VirB11 ATP-binding subunits, as monitored by protease susceptibility. VirB10 interacts with inner membrane VirD4 independently of cellular energetic status, whereas the energy-induced conformational change is required for VirB10 complex formation with an outer membrane-associated heterodimer of VirB7 lipoprotein and VirB9, as shown by coimmunoprecipitation. Under these conditions, the T-DNA substrate is delivered from the inner membrane channel components VirB6 and VirB8 to periplasmic and outer membrane-associated VirB2 pilin and VirB9. We propose that VirD4 and VirB11 coordinate the ATP-dependent formation of a VirB10 “bridge” between inner and outer membrane subassemblies of the VirB/D4 T4SS, and that this morphogenetic event is required for T-DNA translocation across the A. tumefaciens cell envelope.
Keywords: conjugation, energy coupling, pathogenesis, membrane complex, translocation
Bacteria use type IV secretion systems (T4SS) to deliver macromolecular substrates to target cells, often for purposes associated with pathogenesis (1, 2). The T4SS share a common ancestry with bacterial conjugation systems, surface organelles recognized for their widespread phylogenetic distribution and capacity to mediate horizontal gene transfer among many species of Gram-negative and -positive bacteria. The Agrobacterium tumefaciens T-DNA transfer system is an archetypal T4SS, assembled from 11 VirB mating-pair-formation (Mpf) subunits and the VirD4 coupling protein (T4CP) (1). To better understand the mechanism of action of the VirB/D4 T4SS, we recently developed a transfer DNA immunoprecipitation (TrIP) assay to monitor formation of DNA substrate [T-DNA, portion of the Ti (tumor-inducing) plasmid that is transferred to plant cells] contacts with channel components during translocation (3). Studies of WT and T4SS mutant strains led to formulation of a T-DNA translocation pathway described as a series of temporally and spatially ordered interactions with channel subunits. In the postulated pathway, the T-DNA first binds the VirD4 receptor and then, in turn, forms close contacts with the VirB11 ATPase, the VirB6 and VirB8 inner membrane (IM) subunits, and, finally, VirB2 and VirB9 localized in the periplasm and outer membrane (OM). Among the remaining VirB subunits that do not form detectable contacts with the translocating substrate, VirB4 coordinates substrate transfer to the VirB6 and VirB8 subunits, whereas VirB3, VirB5, and VirB10 promote transfer from VirB6 and VirB8 to the VirB2 and VirB9 subunits (3).
As with other bacterial translocation systems (4), ATP provides a source of energy for substrate transfer through the T4SS. Previous studies established that VirD4, VirB11, and VirB4 are the energizing components for the VirB/D4 T4SS, each bearing conserved nucleoside triphosphate binding sites (Walker A motifs) required for translocation of T-DNA to target cells (5, 6). Recently, we used the TrIP assay to characterize effects of the Walker A mutations on specific transfer reactions required for formation of substrate–channel subunit contacts. These studies supplied evidence that VirD4 and VirB11 retain WT substrate binding activities in the absence of ATP binding or hydrolysis. VirD4, VirB11, and VirB4 physically interact with each other and, by a mechanism dependent on utilization of ATP, catalyze substrate transfer to the VirB6 and VirB8 IM channel subunits (3, 7).
Here, we present evidence that ATP also regulates T-DNA substrate passage through the portion of the secretion channel extending through the periplasm to the cell exterior. These studies originated from an observation that several structural features of the bitopic IM subunit VirB10 resemble those of the energy coupling protein TonB (8). We thus tested a model wherein VirB10 functions as an energy sensor for the VirB/D4 T4SS. We present evidence that the VirD4 and VirB11 ATP-binding subunits induce a structural transition in VirB10 that most probably is necessary for a late stage of machine biogenesis and, in turn, passage of substrate from the IM to the cell surface. We discuss our findings in the context of accumulating evidence that ATP-sensing by TonB-like subunits might contribute to the biogenesis and function of diverse macromolecular trafficking systems in bacteria.
Experimental Procedures
Strains and Plasmids. A. tumefaciens A136 is strain C58 cured of the nopaline-type pTiC58 plasmid. WT A348 is strain A136 bearing the octopine-type pTi6NC plasmid (9). All of the mutants used in this study are A348 derivatives: Mx355 (virD4::Tn3HoHo1), PC1000 (ΔvirB operon), and PC1001 to PC1011 (ΔvirB1 to ΔvirB11, nonpolar null mutants) (9). Plasmids expressing WT virB11, virB11ΔGKT174–176, virB11K175Q, WT virD4, or virD4K152Q under the control of the virB promoter have been described (7, 10).
Media and Chemicals. A. tumefaciens strains were grown routinely in LB media supplemented with glutamate and mannitol (MG/L broth) at 28°C. Virulence (vir) gene expression was induced with acetosyringone (AS) as described (9). Sources of other reagents were as follows: carbonyl cyanide m-chlorophenylhydrazone (CCCP, Sigma), sodium arsenate (Allied Chemical, Morristown, NJ), unlabeled tetraphenylphosphonium bromide (TPP+, Aldrich). Tritiated TPP+ (Amersham Pharmacia Bioscience) was a gift from William Dowhan (University of Texas-Houston Medical School).
Cell Treatments. A. tumefaciens cultures were induced with AS for 12–14 h and directly treated with CCCP (10 μM) for 10 min or sodium arsenate (25 mM) for 30 min (11). Cells were harvested by centrifugation and subjected to spheroplast formation, immunoprecipitation, or measurements of ATP cellular levels or transmembrane potential (Δψ), as described below.
Spheroplast Formation. Pelleted AS-induced cells were resuspended at 1/30 vol of the inducing culture in 20 mM Tris·HCl (pH 8.0), 20% (wt/vol) sucrose. Cells were converted to spheroplasts by addition of 100 μg/ml of lysozyme (Sigma), incubation for 5 min on ice, addition of 2 mM EDTA, and further incubation for 45 min on ice. Cells were then centrifuged at 5,000 × g for 5 min, and the supernatant, containing the periplasmic material, was discarded. Spheroplasts were retained intact by gentle suspension in 20 mM Tris·HCl (pH 8.0), 10 mM MgSO4, and 20% sucrose.
Protease Accessibility Assay. Spheroplasts were incubated with Streptomyces griseus protease (Sigma) at a final concentration of 100 μg/ml for 15 min on ice, harvested by mixing with an equal volume of 2× Laemmli's buffer, and then immediately boiled for 10 min. Protein samples were analyzed by SDS/PAGE and immunostaining of Western blots.
Transembrane Potential (Δψ) Measurements. Transmembrane potential (Δψ) was measured as described (12) with minor modifications. Five milliliters of AS-induced cells was harvested by centrifugation, resuspended in 250 μl of 100 mM Tris·HCl, pH 7.8/1 mM EDTA, and then incubated for 3 min at 37°C. Permeabilized cells were then centrifuged and resuspended in 1 ml of 100 mM sodium phosphate (pH 7.8), 1 mM KCl, and 0.4% glycerol. TPP+ (tritiated probe diluted 40-fold in 0.5 mM unlabeled TPP+) was added to 200 μl of the mixture at a final concentration of 10 μM, and the cells were incubated for 10 min at 37°C. Cells were recovered by filtration on GF/F filters (Whatman) and washed twice with 3 ml of 100 mM sodium phosphate buffer (pH 7.8). As a control for nonspecific TPP+ binding to cells, aliquots of cells were first incubated with 10 μM CCCP for 5 min at room temperature before addition of the TPP+ solution, incubation, and filtration.
ATP Measurements. Untreated or CCCP- or arsenate-treated AS-induced cells were concentrated 10- to 20-fold before ATP measurements. The ATP levels were measured by using an ATP bioluminescence assay kit (CLSII, Boehringer Mannheim, Indianapolis) as described (11). Briefly, 20 μl of concentrated cells were mixed with 80 μl of DMSO (Sigma) and further diluted in 400 μl of ice-cold water. Luminescence was measured with 20–50 μl of the sample mixed with 200 μl of luciferase reagent, by using a fluorimeter (Jobin Yvon, Longjumeau, France) equipped with a cooled detector.
Immunoprecipitation. AS-induced cells (10 ml) were pelleted, resuspended in 500 μl of 50 mM Hepes (pH 8.0), 5 mM EDTA, and 100 μg/ml lysozyme, and incubated for 45 min on ice. After sonication with 4× 10-s cycles (80% duty cycle, output 7) by using a Branson sonifier 250 microtip, the cell extract was brought to 1-ml final volume by addition of 50 mM Hepes (pH 8.0) containing 69 μl of 2-mercaptoethanol (2-ME, 1% final concentration), 67 μl of 15% lauryldimethylamine oxide (LDAO, 1% final concentration; Calbiochem), 200 μl of 2.5% deoxycholate (DOC, 0.5% final concentration; Sigma), and 33 μl of 30× solution of protease inhibitors (complete EDTA free; Boehringer Mannheim). The suspension was incubated for 4 h at 4°C on a wheel, and centrifuged for 15 min at 14,000 × g to remove unsolubilized material. The supernatant used for the immunoprecipitation was first incubated with 20 μl of bed volume of Protein A-Sepharose CL-4B (Pharmacia Biotech, Piscataway, NJ) to remove nonspecifically bound proteins. The supernatant was then incubated for 14 h at 4°C with 2.5 μl of anti-VirB antibodies and 40 μl of bed vol of Protein A-Sepharose CL-4B. Beads were washed twice with Hepes 50 mM (pH 8.0) supplemented with 2% LDAO and 0.5% DOC and once in Hepes buffer supplemented with 0.2% LDAO. Beads were then resuspended in loading buffer and boiled for 15 min, and samples were analyzed by Western blotting and immunostaining. VirD4 is insoluble and aggregates upon heat treatment, and thus the above protocol was modified as described (7) for recovery of VirD4-containing complexes.
TrIP. TrIP and quantitative TrIP (QTrIP) experiments were performed as described (3).
Miscellaneous. Standard methods were used for protein and DNA manipulations, SDS/PAGE, and Western blotting. Goat anti-rabbit antibodies conjugated to alkaline phosphatase were used for immunodetection with bromo-chloro-indolyl phosphate (BCIP, Sigma) and nitroblue tetrazolium (NBT, Sigma).
Results
VirB10 Shares Structural Similarities with TonB-Like Proteins. The VirB10 components of T4SS possess an overall domain structure resembling that of the IM energy sensor protein TonB (see Fig. 6, which is published as supporting information on the PNAS web site). Common features include a bitopic membrane topology and a region enriched in Pro residues shown for TonB to form an elongated structure in the periplasm (13, 14). VirB10 proteins also possess protein–protein interaction motifs, such as coiled coils near the transmembrane segment (TMS) and clusters of highly conserved hydrophobic residues near the C terminus, that might be important for establishment of interactions with known partner proteins at the IM and OM, respectively (refs. 15 and 16; see below). Similarly, TonB possesses motifs enabling contacts with its partner proteins at both membranes. For TonB, these transenvelope protein contacts are thought to form simultaneously or through a shuttle mechanism to mediate coupling of IM transmembrane potential to the gating of OM transporters (17–19). Thus, we tested whether VirB10 might also contribute to T4SS machine assembly or function through an energy-sensing mechanism.
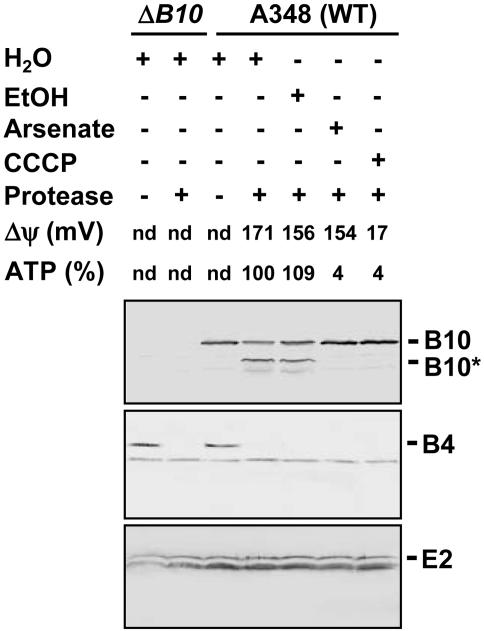
VirB10 Protease Susceptibility as a Function of Cellular ATP Level. Upon sensing of proton motive force (pmf) through partner protein interactions at the IM, TonB undergoes a structural transition as monitored by a change in protease susceptibility (19). Similarly, we found that VirB10 adopts alternative protease-susceptible and -resistant conformations as a function of cellular energetic status. As shown in Fig. 1, treatment of spheroplasts from the WT strain A348 with the S. griseus serine protease degraded the 48-kDa native VirB10 to an ≈40-kDa species. This species, designated VirB10*, was not detected in extracts of the similarly treated ΔvirB10 mutant, confirming its identity as a proteolytic product of VirB10. As controls for spheroplast integrity, the serine protease degraded VirB4 in agreement with previous evidence that this ATPase spans the IM, but had no effect on VirE2, a substrate of the VirB/D4 T4SS that accumulates in the cytoplasm before export (see ref. 5).
Fig. 1.
Identification of an energy-dependent VirB10 conformational switch. A348 (WT) or a virB10-null mutant (ΔB10) were treated with the energy poisons arsenate (Ars) or CCCP, or H2O or EtOH as controls. Spheroplasts from these cells were incubated in the presence (+) or absence (–) of the S. griseus protease. Immunoblots were probed with the antibodies to VirB10 (Top) or, as controls for spheroplast integrity, the VirB4 integral membrane subunit (Middle), or cytosolic VirE2 (Bottom). The positions of full-length VirB10 (B10) and the degradation product (B10*) are indicated at right. Transmembrane potential (Δψ) in millivolts (mV) and ATP levels in percent relative to the H2O-treated control were quantitated as described in the text. nd, Not determined.
We treated A. tumefaciens cells with the protonophore CCCP, which collapses the transmembrane potential (Δψ) and also dissipates cellular ATP levels. Cells were also treated with arsenate, which dissipates cellular ATP without affecting Δψ. The values reported in Fig. 1 for Δψ and cellular ATP measured in the absence and presence of CCCP or arsenate agree with values reported previously for other Gram-negative respiring bacteria (12, 20, 21). Of considerable interest, the protease degraded VirB10 to VirB10* from the H20- or EtOH-treated control cells, but failed to degrade VirB10 from the CCCP- or arsenate-treated cells (Fig. 1). Neither energy poison affected the VirB4 or VirE2 protease susceptibility patterns. We cannot exclude a possible contribution of Δψ to the conformational status of VirB10, but the demonstrated effects of arsenate treatment indicate that VirB10 undergoes a structural transition in response to depletion of cellular ATP levels, as monitored by a change in protease susceptibility.
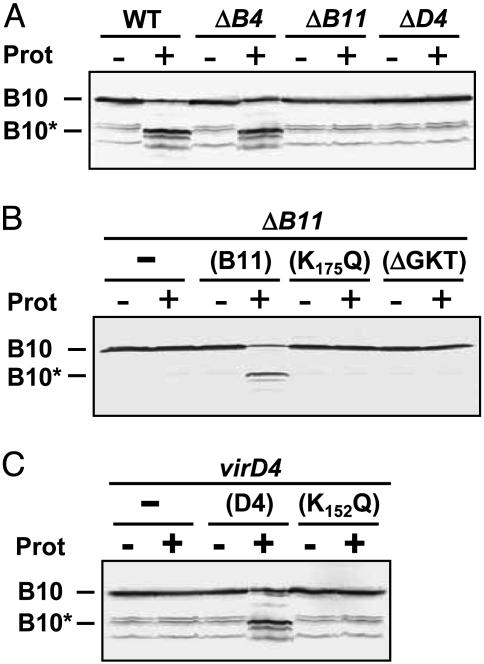
VirD4 T4CP and VirB11 ATPase Modulate the VirB10 Conformation. Next, we tested whether VirB10 senses ATP through one or more of the three ATP-binding subunits of the VirB/D4 T4SS. VirB10 displayed a WT pattern of protease susceptibility upon treatment of spheroplasts from a mutant lacking VirB4, but a protease-resistant conformation in mutants lacking VirD4 or VirB11 (Fig. 2A). In parallel studies, VirB10 displayed a WT pattern of protease susceptibility in the remaining virB gene deletion mutants, establishing that the conversion to protease resistance was specifically correlated with the absence of VirD4 and VirB11 (data not shown). Complementation studies further confirmed that VirB10 was degraded to VirB10* in the virD4 and ΔvirB11 mutants, producing native VirD4 and VirB11 from an IncP replicon (Fig. 2 B and C).
Fig. 2.
Dependence of the VirB10 conformation on energetic subunits. Protease susceptibility of VirB10 in the WT or virB4, virB11 or virD4 mutant strains (A); the virB11 strain (ΔB11) producing native VirB11 (B11) or Walker A mutant proteins (K175Q and ΔGKT) (B); or the virD4 strain producing native VirD4 (D4) or a Walker A mutant protein (K152Q) (C). Spheroplasts of strains indicated were incubated in the presence (+) or absence (–) of protease. Immunoblots were developed with anti-VirB10 antibodies. Positions of the intact VirB10 protein and the degradation product (B10*) are indicated at the left. In A and C, immunoblots show the presence of nonspecific VirB10 degradation products of ≤40-kDa detected previously (42).
Fig. 2 B and C further shows that VirB10 adopted a protease-resistant conformation in strains producing the Walker A substitution mutant proteins VirD4K152Q or VirB11K175Q or the deletion mutant VirB11ΔGKT. These strains fail to deliver substrates to recipient cells (6, 10), and, furthermore, purified forms of VirD4 and VirB11 homologs bearing similar Walker A mutations display defects in ATP utilization in vitro (22–24). Thus, our results suggest that the VirD4 and VirB11 channel subunits induce a structural transition in VirB10 through an ATP-binding-dependent mechanism.
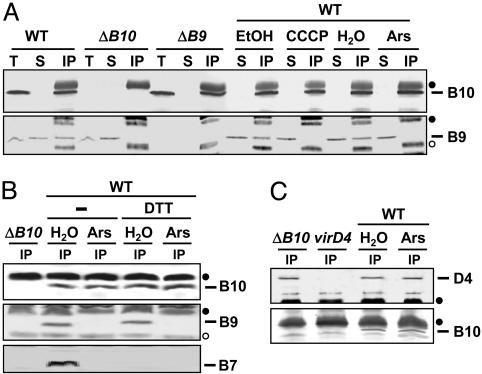
The VirB10 Conformational Change Is Required for Complex Formation with VirB9 but Not VirD4. VirB10 interacts with the OM-associated subunit VirB9, as shown by yeast two-hybrid analysis (25). To determine whether the VirB10 structural transition is necessary for this interaction in A. tumefaciens, we assayed for formation of an immunoprecipitable VirB10–VirB9 complex in untreated and energy-depleted cells. As shown in Fig. 3A, anti-VirB10 antibodies coprecipitated VirB10 and VirB9 from extracts of untreated or H20- or EtOH-treated WT cells. In striking contrast, the antibodies precipitated VirB10 but no detectable VirB9 from extracts of CCCP- or arsenate-treated cells, indicating that the ATP-dependent conformation is required for VirB10 complex formation with VirB9 in vivo. In the control experiments, the anti-VirB10 antibodies precipitated only VirB10 from a virB9-null mutant and neither VirB10 nor VirB9 from a virB10 mutant.
Fig. 3.
Effects of cellular energy on formation of VirB10 complexes. WT or mutant strains were treated with CCCP, arsenate (Ars), or the corresponding EtOH or H2O controls, then detergent-solubilized as described in Experimental Procedures. (A) Material precipitated with anti-VirB10 antibodies from extracts of energized (EtOH- or H2O-treated) or energy-depleted (Ars- or CCCP-treated) WT cells, or the ΔvirB9 (ΔB9) or ΔvirB10 (ΔB10) control strains. (B) Material precipitated with anti-VirB10 antibodies from extracts of energized (H2O) or energy-depleted (Ars) WT cells in the absence (–) or presence of reducing agent (DTT). (C) Material precipitated with anti-VirD4 antibodies from extracts of energized and energy-depleted WT cells or the ΔvirB10 or virD4 control strains. Immunoblots were developed with antibodies to the T4SS subunits listed at the right. Crossreactive material was immunoreactive heavy (filled circle) and light (open circle) chain IgG. T, total cell extract; S, material in supernatant after the immunoprecipitation; IP, immunoprecipitated material.
In A. tumefaciens, VirB9 assembles as a disulfide-crosslinked heterodimer with the OM-associated lipoprotein VirB7 (see ref. 5). As expected, in the absence of reducing agent, the anti-VirB10 antibodies coprecipitated a presumptive complex of VirB10, VirB9, and VirB7 from untreated WT cells (Fig. 3B). From the arsenate-treated cells, these antibodies precipitated VirB10 but no VirB9 or VirB7 regardless of the postlysis addition of the reducing agent DTT (Fig. 3B). Therefore, the ATP-induced conformational switch is required for VirB10 complex formation with the VirB7–VirB9 heterodimer through the VirB9 contact.
Recently, we showed that VirB10 also interacts with the IM VirD4 T4CP (7). Interestingly, in contrast to the above findings, the anti-VirD4 antibodies coprecipitated VirD4 and VirB10 from extracts of arsenate-treated WT cells, suggesting that cellular energy is not required for VirD4–VirB10 complex formation (Fig. 3C). We also tested for formation of a VirB10–VirB11 complex but were unable to gain evidence for such an interaction by coimmunoprecipitation (data not shown).
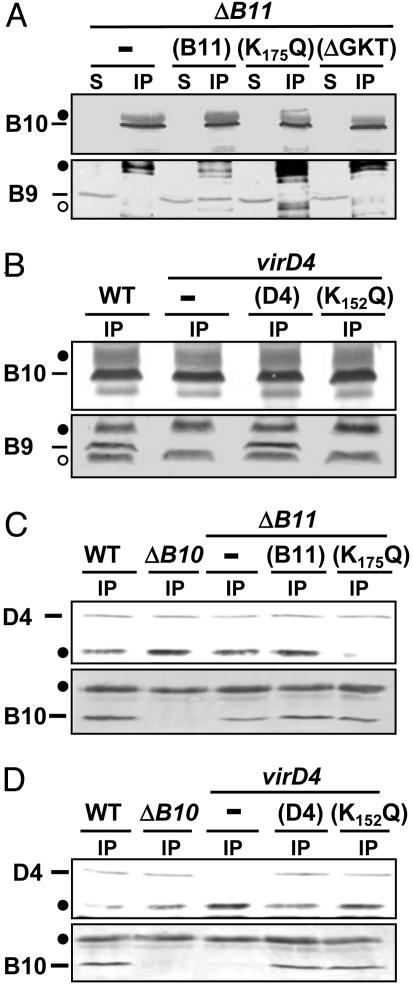
We further asked whether the VirD4 and VirB11 subunits contribute to VirB10 multimerization. Consistent with data presented in Fig. 3, the anti-VirB10 antibodies precipitated VirB10 but no VirB9 from extracts of the virD4- or virB11-null mutants (Figs. 4 A and B). These antibodies also precipitated VirB10 but no VirB9 from extracts of the isogenic strains producing the VirD4K152Q or VirB11K175Q mutant proteins (Fig. 4 A and B). In contrast, the anti-VirD4 antibodies coprecipitated VirD4K152Q and VirB10 from extracts of a strain producing this Walker A substitution mutant (Fig. 4C), and also coprecipitated VirD4 and VirB10 from extracts of strains lacking VirB11 or producing VirB11K175Q (Fig. 4D). Together, these data indicate that the ATP-binding-dependent activities of VirD4 and VirB11 are required for interaction of VirB10 with the VirB7–VirB9 heterodimer at the OM, but dispensable for interaction of VirB10 with VirD4 at the IM.
Fig. 4.
Effects of virD4- and virB11-null and Walker A mutations on VirB10 complex formation. (A) Material precipitated with anti-VirB10 antibodies from extracts of the ΔvirB11 strain producing native VirB11 or the VirB11 Walker A mutants (K175Q and ΔGKT). (B) Material precipitated with anti-VirB10 antibodies from extracts of the virD4 mutant producing native VirD4 or the VirD4 Walker A mutant (K152Q). (C) Material precipitated with anti-VirD4 antibodies from extracts of the ΔvirB10 mutant or the ΔvirB11 mutant producing VirB11 or VirB11K175Q. (D) Material precipitated with anti-VirD4 antibodies from extracts of the virD4 mutant producing VirD4 or VirD4K152Q. Immunoblots were developed with antibodies to the T4SS subunits listed at the left. Crossreactive material was immunoreactive heavy (filled circle) and light (open circle) chain IgG. S, material in supernatant after the immunoprecipitation; IP, immunoprecipitated material.
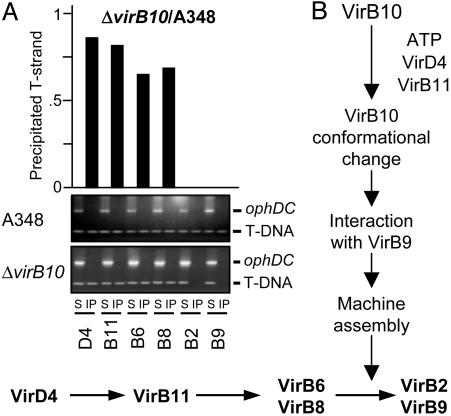
VirB10 Is Required for T-Strand Transfer Across the Cell Envelope. Previously, by use of the TrIP assay, we reported that a virB10-null mutation blocks T-DNA substrate transfer from the VirB6 and VirB8 IM channel subunits to VirB2 and VirB9 located in the periplasm and OM (3). However, this assay cannot be used to detect quantitative effects of VirB10 on early substrate transfer to the IM components, or sensitive enough to detect possible low-level substrate transfer to VirB2 and/or VirB9 in the absence of VirB10. To further explore the physiological importance of VirB10 for substrate transfer across the cell envelope, we used a quantitative version of the TrIP assay (QTrIP) to compare substrate transfer efficiencies in isogenic strains producing and lacking VirB10.
Results yielded two findings of interest (Fig. 5A). First, the IM channel components VirD4 and VirB11 reproducibly bound the T-DNA substrate less efficiently in the virB10 mutant compared with the WT strain. This difference in substrate binding was more pronounced for VirB6 and VirB8; these subunits bound <65–70% of the amount of T-DNA in the virB10 mutant vs. the WT strain. Previous studies have shown that VirB10 production does not affect steady-state accumulation of these IM components, excluding problems of protein instability as the basis for diminished substrate binding in the virB10 mutant (7, 9). Thus, our findings indicate that VirB10 contributes to the efficiency of substrate translocation through the inner membrane channel. The importance of VirB10 for the early substrate transfer reactions is further illustrated by results of a recent in vivo reconstitution study. In that study, we determined that a strain producing a subset of T4SS subunits including the IM components VirD4, VirB11, VirB4, VirB6, and VirB8, together with the VirB7 lipoprotein and VirB10, successfully delivers substrate to VirB6 and VirB8. In contrast, the isogenic strain lacking VirB10 fails to transfer substrate beyond VirD4 (7).
Fig. 5.
TrIP studies with the virB10 strain. (A) TrIP and QTrIP measurements of the T-strand interaction with VirB proteins from the WT or the ΔvirB10 strain. S, supernatant after immunoprecipitation with the anti-VirB antibodies listed on bottom; IP, immunoprecipitated material. For TrIP, the T-strand substrate (T-DNA) and the Ti control fragment (ophDC) were detected by PCR amplification and gel electrophoresis. For QTrIP, quantitative data (Upper) are presented as cpm of incorporated radionucleotide during one cycle in the logarithmic phase of PCR amplication and reported as a ratio of cpm recovered with a given antibody from the virB10 mutant vs. the WT strain. QTrIP data are reported for a single experiment; several repetitions of these experiments showed <5% deviation of the values shown for a given T4SS subunit. (B) Schematic showing the proposed contribution of VirB10 energy coupling to assembly and function of the VirB/D4 T4SS.
VirB10 synthesis is absolutely essential for substrate transfer from VirB6 and VirB8 to VirB2 and VirB9, as shown by QTrIP (Fig. 5A). Thus, our findings indicate that three sequential reactions [(i) ATP utilization by VirD4 and VirB11, (ii) a VirB10 structural transition, and (iii) VirB10 complex formation with the OM VirB7–VirB9 heterodimer] mediate a late stage of machine biogenesis required for substrate passage to and through the periplasmic portion of the secretion channel (Fig. 5B).
Discussion
In this study, we identify a mechanism by which ATP energy mediates assembly and function of the A. tumefaciens VirB/D4 T4SS. Specifically, through sensing of ATP utilization by the VirD4 substrate receptor and the VirB11 ATPase, VirB10 undergoes a structural transition that is required for stable complex formation with the VirB7–VirB9 heterodimer. Under these conditions, VirB10 interacts both with VirD4 at the IM and VirB7–VirB9 complex associated with the OM, thus likely forming a bridge between T4SS subassemblies at the two membranes. That this transenvelope bridge is important for machine function is supported by two lines of evidence. First, our TrIP studies show a strict correlation between the ATP-dependent VirB10 conformational switch and substrate transfer from VirB6 and VirB8 at the IM to VirB2 and VirB9, the postulated periplasmic and OM channel components. Second, previously we presented evidence that the VirB7–VirB9 heterodimer plays a critical role in stabilizing VirB7 and VirB9 as well as several IM components including VirB4, VirB10, and VirB11 (see ref. 5). Therefore, reactions involving VirB10 both at the IM (ATP utilization by VirD4 and VirB11 and activation of a VirB10 conformational switch) and the OM (assembly of the VirB7–VirB9 heterodimer and complex formation with the ATP-altered form of VirB10) together mediate assembly of a stable and functional secretory apparatus.
Molecular details of the interactions among VirB10 and VirD4 and VirB11 at the IM await further study. The available data suggest that VirB10 senses energy at least in part through transmembrane interactions with the T4CP. In addition to our finding that VirB10 stably interacts with VirD4 independently of ATP utilization by either the T4CP or VirB11 (Figs. 3 and 4), recent studies showed that two VirB10 homologs, TrwE and TrhB, interact with cognate (16, 26) as well as heterologous T4CPs (26). Moreover, interaction domains were mapped to the N-terminal membrane-spanning regions of both sets of partner proteins by two-hybrid analyses (16, 26). VirB10 is also dependent on VirB11 ATP utilization for the structural transition, yet so far there is no evidence for complex formation between VirB10 and VirB11. In contrast, a VirD4–VirB11 interaction is supported by genetic suppression (27), TrIP data (3, 7), and results of coimmunoprecipitation studies indicating that VirD4 and VirB11 stably interact independently of other T4SS subunits (7). VirB11 might induce the VirB10 conformational switch indirectly through its interaction with VirD4, although at this time a direct transient or weak affinity contact between VirB11 and VirB10 cannot be excluded. Crystallographic studies have shown that homologs of both VirB11 and VirD4 form homo-hexameric ring structures and undergo ATP-dependent conformational changes (28–30). These conformational changes might activate the VirB10 structural transition detected in this study.
The finding that ATP energy mediates formation of the VirB10 contact with the VirB7–VirB9 heterodimer is of interest in view of biochemical evidence that the VirB7 lipoprotein and VirB9 partition predominantly with the OM (5). Additionally, sequence analyses of VirB9 and its homologs have identified multiple potential β-strands (ref. 15; S. Buchanan, personal communication), prompting a proposal that these subunits might form a β-pleated sheet structure in the OM possibly comprising part or all of an OM pore complex (15). Whether the VirB7–VirB9 complex indeed forms an OM pore, or instead comprises a portion of the secretion channel in the periplasm, awaits further study. Nevertheless, our findings suggest that the VirD4 and VirB11 IM channel subunits induce a conformational change in VirB10 required for stable complex formation with VirB9 in the periplasm. Although we favor a model whereby this energy-dependent interaction corresponds to a late stage of channel assembly (Fig. 5B), an alternative possibility is that the VirB10–VirB9 interaction stimulates the opening of a channel gate to allow substrate passage through the periplasm.
Interestingly, a TonB paradigm based on utilization of ATP energy as opposed to proton motive force has been invoked as a possible mechanism for activating assembly or function of other bacterial macromolecular trafficking systems. For example, in Aeromonas hydrophila, the ExeA ATP-binding and the ExeB bitopic subunits of a type II secretion system (T2SS) form a complex that is altered in its conformation by mutation of nucleotide triphosphate binding motifs (31, 32). Moreover, formation of the ExeAB complex was shown to promote multimerization of the secretin ExeD, leading to the suggestion that the ExeAB complex energizes assembly of the secretin ring required for aerolysin secretion across the OM (33). In other T2SS, the TonB-like GspC proteins have been shown to interact both with an IM platform composed of VirB11-like GspE and proteins GspF, -L, and -M, and an OM complex composed of GspS lipoprotein and GspD secretin (34–37). Studies of chimeric T2SS suggest that the GspC subunits participate in gating of the OM secretin complexes, although the contribution of ATP energy to GspC complex formation or function has not yet been demonstrated (38). Finally, in the well characterized bacterio-phage f1 extrusion system, protein pI possesses an ATP-binding domain near its N terminus, and the rest of the protein extends through the periplasm to establish contact with the pIV secretin protein (39). Both cellular ATP and an intact Walker A motif of pI are required for phage secretion, again consistent with a proposal that pI uses the energy of ATP to gate an OM secretin (40). These findings, together with results of the present study, support a general proposal that ATP energy sensing by a bitopic IM subunit contributes to the morphogenesis and function of evolutionarily diverse trafficking pathways of Gram-negative bacteria (see Fig. 6 and Fig. 7, which are published as supporting information on the PNAS web site).
In summary, we propose that VirD4 and VirB11 induce a structural change in VirB10 to stimulate formation of a “bridge” linking inner and outer membrane-associated subassemblies of the VirB/D4 T4SS (Fig. 5B). It is interesting to note that VirD4 and VirB11 most probably repetitively bind and hydrolyze ATP during the course of machine assembly or substrate transfer, raising the possibility that this transenvelope bridge forms only transiently. In this context, it is intriguing that ABC transporters, and possibly also the TonB-dependent systems, form transient transenvelope structures specifically in response to energy and ligand binding (19, 41). Studies examining the possible contribution of substrate binding to biogenesis of the VirB/D4 T4SS are thus warranted.
Supplementary Material
Acknowledgments
We thank William Dowhan for providing the tritiated TPP+, Lucienne Letellier for advice for membrane potential measurements, Roland Lloubès and James Sturgis for the use of the spectrofluorimeter, and Sophie Bleves and members of the Christie laboratory for helpful comments and critical reading of the manuscript.
Author contributions: E.C. and P.J.C. designed research, performed research, analyzed data, and wrote the paper.
This paper was submitted directly (Track II) to the PNAS office.
Abbreviations: T4SS, type IV secretion system; TrIP, transfer DNA immunoprecipitation; QTrIP, quantitative TrIP; T-DNA, portion of the Ti (tumor-inducing) plasmid that is transferred to plant cells; IM, inner membrane; OM, outer membrane; vir, virulence; AS, acetosyringone; CCCP, carbonyl cyanide m-chlorophenylhydrazone; TPP+, tetraphenylphosphonium bromide.
References
- 1.Cascales, E. & Christie, P. J. (2003) Nat. Rev. Microbiol. 1, 137–150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Llosa, M. & O'Callaghan, D. (2004) Mol. Microbiol. 53, 1–8. [DOI] [PubMed] [Google Scholar]
- 3.Cascales, E. & Christie, P. J. (2004) Science 304, 1170–1173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Holland, B. (2004) Biochim. Biophys. Acta, in press.
- 5.Christie, P. J. (2004) Biochim. Biophys. Acta, in press.
- 6.Kumar, R. B. & Das, A. (2002) Mol. Microbiol. 43, 1523–1532. [DOI] [PubMed] [Google Scholar]
- 7.Atmakuri, K., Cascales, E. & Christie, P. J. (2004) Mol. Microbiol., in press. [DOI] [PMC free article] [PubMed]
- 8.Postle, K. & Kadner, R. J. (2003) Mol. Microbiol. 49, 869–882. [DOI] [PubMed] [Google Scholar]
- 9.Berger, B. R. & Christie, P. J. (1994) J. Bacteriol. 176, 3646–3660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Rashkova, S., Spudich, G. M. & Christie, P. J. (1997) J. Bacteriol. 179, 583–589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cascales, E., Gavioli, M., Sturgis, J. N. & Lloubes, R. (2000) Mol. Microbiol. 38, 904–915. [DOI] [PubMed] [Google Scholar]
- 12.Letellier, L., Howard, S. P. & Buckley, J. T. (1997) J. Biol. Chem. 272, 11109–11113. [DOI] [PubMed] [Google Scholar]
- 13.Evans, J. S., Levine, B. A., Trayer, I. P., Dorman, C. J. & Higgins, C. F. (1986) FEBS Lett. 208, 211–216. [DOI] [PubMed] [Google Scholar]
- 14.Williamson, M. P. (1994) Biochem. J. 297, 249–260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cao, T. B. & Saier, M. H., Jr. (2001) Microbiology 147, 3201–3214. [DOI] [PubMed] [Google Scholar]
- 16.Gilmour, M. W., Gunton, J. E., Lawley, T. D. & Taylor, D. E. (2003) Mol. Microbiol. 49, 105–116. [DOI] [PubMed] [Google Scholar]
- 17.Letain, T. E. & Postle, K. (1997) Mol. Microbiol. 24, 271–283. [DOI] [PubMed] [Google Scholar]
- 18.Larsen, R. A., Foster-Harnett, D., McIntosh, M. A. & Postle, K. (1997) J. Bacteriol. 179, 3213–3221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Larsen, R. A., Thomas, M. G. & Postle, K. (1999) Mol. Microbiol. 31, 1809–1824. [DOI] [PubMed] [Google Scholar]
- 20.Ahmed, S. & Booth, I. R. (1981) Biochem. J. 200, 573–581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Possot, O. M., Letellier, L. & Pugsley, A. P. (1997) Mol. Microbiol. 24, 457–464. [DOI] [PubMed] [Google Scholar]
- 22.Krause, S., Pansegrau, W., Lurz, R., de la Cruz, F. & Lanka, E. (2000) J. Bacteriol. 182, 2761–2770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hormaeche, I., Alkorta, I., Moro, F., Valpuesta, J. M., Goni, F. M. & De La Cruz, F. (2002) J. Biol. Chem. 277, 46456–46462. [DOI] [PubMed] [Google Scholar]
- 24.Schroder, G. & Lanka, E. (2003) J. Bacteriol. 185, 4371–4381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Das, A. & Xie, Y.-H. (2000) J. Bacteriol. 182, 758–763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Llosa, M., Zunzunegui, S. & de la Cruz, F. (2003) Proc. Natl. Acad. Sci. USA 100, 10465–10470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zhou, X.-R. & Christie, P. J. (1997) J. Bacteriol. 179, 5835–5842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Yeo, H.-J., Savvides, S. N., Herr, A. B., Lanka, E. & Waksman, G. (2000) Mol. Cell 6, 1461–1472. [DOI] [PubMed] [Google Scholar]
- 29.Savvides, S. N., Yeo, H. J., Beck, M. R., Blaesing, F., Lurz, R., Lanka, E., Buhrdorf, R., Fischer, W., Haas, R. & Waksman, G. (2003) EMBO J. 22, 1969–1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gomis-Ruth, F. X., Moncalian, G., Perez-Luque, R., Gonzalez, A., Cabezon, E., de la Cruz, F. & Coll, M. (2001) Nature 409, 637–641. [DOI] [PubMed] [Google Scholar]
- 31.Howard, S. P., Meiklejohn, H. G., Shivak, D. & Jahagirdar, R. (1996) Mol. Microbiol. 22, 595–604. [DOI] [PubMed] [Google Scholar]
- 32.Schoenhofen, I. C., Stratilo, C. & Howard, S. P. (1998) Mol. Microbiol. 29, 1237–1247. [DOI] [PubMed] [Google Scholar]
- 33.Ast, V. M., Schoenhofen, I. C., Langen, G. R., Stratilo, C. W., Chamberlain, M. D. & Howard, S. P. (2002) Mol. Microbiol. 44, 217–231. [DOI] [PubMed] [Google Scholar]
- 34.Bleves, S., Gerard-Vincent, M., Lazdunski, A. & Filloux, A. (1999) J. Bacteriol. 181, 4012–4019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Possot, O. M., Vignon, G., Bomchil, N., Ebel, F. & Pugsley, A. P. (2000) J. Bacteriol. 182, 2142–2152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sandkvist, M. (2001) Mol. Microbiol. 40, 271–283. [DOI] [PubMed] [Google Scholar]
- 37.Py, B., Loiseau, L. & Barras, F. (2001) EMBO Reports 2, 244–248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lindeberg, M., Salmond, G. P. & Collmer, A. (1996) Mol. Microbiol. 20, 175–190. [DOI] [PubMed] [Google Scholar]
- 39.Russel, M. (1993) J. Mol. Biol. 231, 689–697. [DOI] [PubMed] [Google Scholar]
- 40.Feng, J. N., Model, P. & Russel, M. (1999) Mol. Microbiol. 34, 745–755. [DOI] [PubMed] [Google Scholar]
- 41.Letoffe, S., Delepelaire, P. & Wandersman, C. (1996) EMBO J. 15, 5804–5811. [PMC free article] [PubMed] [Google Scholar]
- 42.Ward, J. E., Jr., Dale, E. M., Nester, E. W. & Binns, A. N. (1990) J. Bacteriol. 172, 5200–5210. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.