Abstract
Background:
The aim of this study was to determine the best condition for the production of DT386-BR2 fusion protein, an immunotoxin consisting of catalytic and translocation domains of diphtheria toxin fused to BR2, a cancer specific cell penetrating peptide, for targeted eradication of cancer cells, in terms of the host, cultivation condition, and culture medium.
Materials and Methods:
Recombinant pET28a vector containing the codons optimized for the expression of the DT386-BR2 gene was transformed to different strains of Escherichia coli (E. coli BL21 DE3, E. coli Rosetta DE3 and E. coli Rosetta-gami 2 DE3), followed by the induction of expression using 1 mM IPTG. Then, the strain with the highest ability to produce recombinant protein was selected and used to determine the best expression condition using response surface methodology (RSM). Finally, the best culture medium was selected.
Results:
Densitometry analysis of sodium dodecyl sulfate-polyacrylamide gel electrophoresis of the expressed fusion protein showed that E. coli Rosetta DE3 produced the highest amounts of the recombinant fusion protein when quantified by 1 mg/ml bovine serum albumin (178.07 μg/ml). Results of RSM also showed the best condition for the production of the recombinant fusion protein was induction with 1 mM IPTG for 2 h at 37°C. Finally, it was established that terrific broth could produce higher amounts of the fusion protein when compared to other culture media.
Conclusion:
In this study, we expressed the recombinant DT386-BR2 fusion protein in large amounts by optimizing the expression host, cultivation condition, and culture medium. This fusion protein will be subjected to purification and evaluation of its cytotoxic effects in future studies.
Keywords: DT386-BR2, expression, optimization, response surface methodology
Introduction
Recombinant protein expression is influenced not only by the expression host strain, but also by expression conditions such as temperature, concentration of inducer, pH, incubation time, and composition of the culture medium.[1] The yield of expressed proteins can be increased by optimizing these parameters. Traditionally, changing one variable at a time is used for the optimization of cultivation conditions for overexpression of recombinant proteins.[2] This method is not only time-consuming but also results in data misinterpretation when different variables interact with each other.[3] Response surface methodology (RSM) is the most common multivariate analysis method to determine the optimum culture conditions for protein expression by simultaneously changing several variables based on a minimum number of experiments.[4] RSM has been widely used for studying the interactions of several parameters during bioprocess optimization in different biotechnological processes, based on a reasonable prediction of culture conditions for optimal protein expression.[4,5]
Immunotoxins are recombinant proteins consisting of a toxin moiety conjugated to an antibody or growth factor as the targeting moiety by genetic fusion or chemical ligation, to target a very potent cytotoxic agent to special cells.[6] Various targeting and cytotoxic agents have been used for the production of immunotoxins up to now. Buforin is an antimicrobial peptide (AMP), that forms transient pores in cancer cell membrane and subsequently enters the cells, without any toxic effects on normal cells.[7,8] Therefore, this cell-type-specific cell penetrating peptide is interesting for its potential application in designing new immunotoxins.[8] On the other hand, diphtheria toxin (DT) is the most frequently used bacterial toxin for the construction of fusion proteins by its receptor binding domain replacement with different targeting moieties.[9,10,11,12,13] Hence, in a previous study, we constructed a new fusion protein consisting of BR2 (a short peptide derived from buforin which targets cancer cells more specifically) as a targeting moiety, and catalytic and translocation domains of the DT (DT386) as the cytotoxic moiety. Preliminary studies on specific cytotoxicity of the DT386-BR2 showed reasonable specific cytototxic activities on MCF7 and HeLa tumor cell lines compared to the normal cells, i.e. HUVEC cell line (manuscript is under review).[14] However, to conduct further studies on various aspects of specific and nonspecific toxicity of this fusion protein, in addition to its evaluation through various preclinical studies, a large amount of this protein is required. Therefore, this study was performed to investigate the optimized protein expression conditions for higher DT386-BR2 production.
Materials and Methods
Materials
The recombinant pET28a vector containing DT386-BR2 was obtained from our previous study (manuscript is under review). Kanamycin, chloramphenicol, streptomycin, and tetracycline were purchased from Invivogen (San Diego, California, USA) and used for selection of resistant bacteria. Production of competent cells from different species of Escherichia coli was performed using CaCl2 method. Isopropyl-β-D-thiogalactopyranoside (IPTG) and all other chemicals were obtained from other commercial sources and were of the molecular biology grade. Different growth media were prepared as suggested by the condensed protocols from molecular cloning: A laboratory manual,[15] unless otherwise mentioned.
Methods
Determination of the best host for the expression of DT386-BR2
The coding sequence of the DT386-BR2 fusion protein was optimized for expression in E. coli strains derived from K12 by Biomatik's Proprietary Codon Optimization Service (Canada). The sequence was ordered to be synthesized by GENEray (Hong Kong), and subcloned to the pET28a expression plasmid.
To express DT386-BR2, we used different strains of E. coli including BL21 DE3, Rosetta DE3 and Rosetta-gami 2 DE3 as the hosts. Transformation of the recombinant pET28a vector containing DT386-BR2 insert, with the kanamycin resistance gene, was achieved using heat shock procedure. Five colonies from each bacterium were inoculated into 5 ml of luria-bertani (LB) medium containing appropriate antibiotic and incubated at 37°C for 16 h at 180 rpm. These overnight cultures were used to inoculate (10% v / v) 10 ml of fresh LB medium. The cells were incubated at 37°C until they reached the exponential phase (an OD600 nm of 0.4–0.6). Then the expression of fusion protein was induced by the addition of 1 mM IPTG. At the end of expression time, cells were harvested by centrifugation at 7000 g for 10 min at 4°C. Finally, the pellets of induced bacteria were analyzed by 12% sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE). Protein quantification was performed by densitometry analysis of acrylamide gels using TotalLab Quant software (Nonlinear Inc., Durham NC, USA) and the best host was used for further expression optimization.
Optimization of cultivation condition by response surface methodology
To optimize the protein expression, each experiment was performed under various induction conditions (IPTG concentration, post-induction time, and incubation temperature) as described in experimental design. These variables have shown the greatest effects on induction conditions in previous studies.[16,17] At the end of each experiment, cells were harvested by centrifugation at 7000 g for 10 min.
To systematically evaluate the effects of three independent variables including temperature (factor A), post-induction time (factor B), and IPTG concentration (factor C) on the production of DT386-BR2 in E. coli, an experimental design was developed using Box–Behnken factorial design scheme. Each variable was investigated at levels of-1 (the lower value of the variable), +1 (the higher value of the variable), and 0 (the central point of the variable). As a result, a total of 17 experiments were carried out including one in quintuple at the center point. Surface response methodology was performed for data analyzing of experimental design using Design Expert software (version 8.0.7.1, StatEase Inc., Minneapolis, USA). Protein quantification was performed by densitometry analysis of acrylamide gels using TotalLab Quant software. The band intensities of the expressed DT386-BR2 were analyzed and normalized using albumin with known concentration (1 mg/ml).
Selection of the best culture medium for the expression of DT386-BR2
After determination of the best host and cultivation condition for the overexpression of the fusion protein, the amounts of protein expression of the best host were analyzed in different culture media including LB, terrific broth (TB), and super broth (SB). Finally, the pellets of the cultivated bacteria were analyzed by 12% SDS-PAGE. Again, protein quantification was performed by densitometry analysis of acrylamide gels using TotalLab Quant software. The band intensities of expressed DT386-BR2 samples were analyzed in comparison to each other.
Results
Escherichia coli Rosetta DE3 can be used as the best host for DT386-BR2 expression
The results of analyzing the SDS-PAGE using TotalLab Quant software showed that all five colonies of E. coli BL21 DE3 and Rosetta DE3 can express the desired fusion protein while the expression of this fusion protein in E. coli Rosetta-gami 2 DE3 was not observed. The protein expression in one colony of the E. coli Rosetta DE3 was higher than the other colonies of both E. coli Rosetta DE3 and E. coli BL21 DE3 when compared to albumin at 1 mg/ml concentration (178.07 μg/ml). Hence, this colony was used for determination of the best cultivation condition.
Response surface methodology is a successful method for determining the best condition for DT386-BR2 production
Recombinant E. coli Rosetta DE3 containing pET28a-DT386-BR2 vector (colony 1) was cultured overnight in LB containing 15 and 34 μg/ml of kanamycin and chloramphenicol, respectively, as selective markers, and used for determining the best culture condition according to the design of RSM. Cultivation for the production of DT386-BR2 by changing three variables was performed in shaking flasks so that their individual and synergistic effects on DT386-BR2 expression could be evaluated using a Box–Behnken factorial design. The amounts of protein expression in different culture conditions were analyzed by TotalLab Quant software in comparison to albumin at 1 mg/ml concentration. The measured levels of protein expression for the 17 cultures are summarized in Figure 1 and Table 1.
Figure 1.
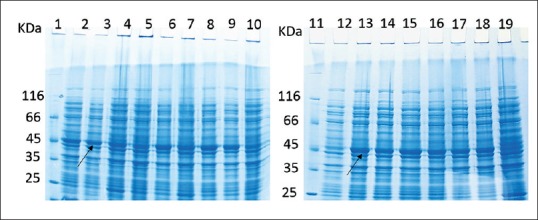
Sodium dodecyl sulfate polyacrylamide gel electrophoresis of 17 different cultivation conditions for the expression of DT386-BR2 fusion protein designed by Design Expert software. Lanes 1 and 11: Protein marker. Lanes 2-10 and 12-19: Different experiment conditions in the terms of temperature, incubation time and IPTG concentration. (Runs 1–17 in Table 1)
Table 1.
Box–Behnken experimental design of three factors and three levels with quintuple at the center point
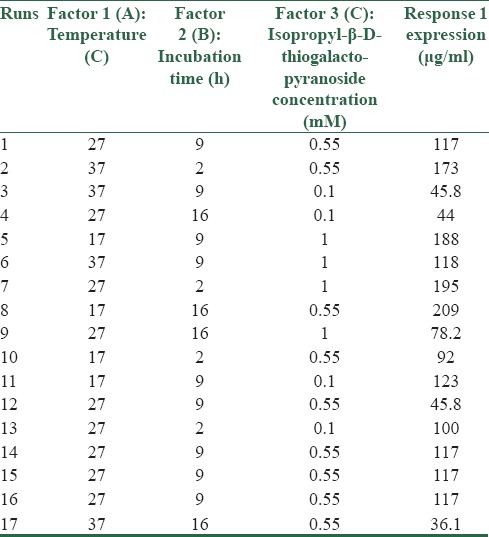
A wide range of DT386-BR2 expression from 36.1 to 209 μg/ml was observed under these investigated culture conditions, indicating the necessity of identifying an optimal condition. To evaluate the experimental error and reproducibility of the model, cultivation was performed in quintuple under the central point conditions (0.55 mM IPTG, 9 h of induction and 27°C).
Correlation of the expressed DT386-BR2 concentration and the investigated variables was determined using the Design Expert software and was represented by the following equation:
Y = 116.89 A – 8.11 B + 41.29 C – 47.5 AB + 1.8 AC – 31.17 BC.
Where Y is the expression (μg/ml), and A, B, and C are temperature (°C), post-induction time (h) and inducer concentration (mM), respectively.
The statistical significance of the above equation was confirmed by F-test and the analysis of variance for response surface 2FI model. The model correlation coefficient (R2) value was 0.7342.
The high value of adequate precision (7.362) indicated that this model had an adequate signal and could be applicable. In this case, C and AB were significant model terms (P < 0.05). The linear IPTG concentration and the term of interaction between induction time and temperature were highly significant.
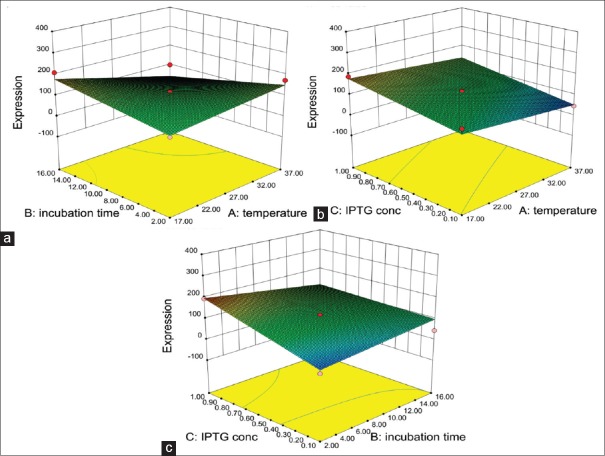
Figure 2a shows the response surface plot of the dependency of temperature and post-induction time on DT386-BR2 expression when the IPTG concentration remains at its mean level (0.55 mM). Based on the graph, DT386-BR2 expression directly correlated to IPTG concentration. In Figure 2b, the dependency of temperature and IPTG concentration has been shown. According to this figure, increasing the incubation time has no effect on the DT386-BR2 expression; on the other hand, the highest level of protein expression was attained after 2 h of incubation and increasing the incubation time did not affect the amount of expression. Finally, Figure 2c shows the dependency of IPTG concentration and incubation time when the temperature is in mean score (27°C). As shown, the highest amount of the protein expression occurred at 37°C. Therefore, it seems that the IPTG concentration has the most effect on protein expression (P < 0.05).
Figure 2.
Response surface of DT386-BR2 expression represents the interaction between two factors in the expression of DT386-BR2 (μg/ml) by keeping other factor constant. (a) interaction between the temperature and incubation time on the amount of fusion protein expression. (b) interaction between the IPTG concentration and temperature on the amount of fusion protein expression. (c) interaction between the IPTG concentration and incubation time on the amount of fusion protein expression
Based on the developed model, the optimal culture conditions for DT386-BR2 expression in E. coli were predicted to be as follows: The temperature of 37°C, postinduction time of 2 h, and 1 mM IPTG. These optimal levels led to a DT386-BR2 concentration of 224.86 μg/ml.
Terrific broth could induce the production of high amounts of the recombinant protein
The overnight culture of colony 1 of the recombinant E. coli Rosetta DE3 containing pET28a-DT386-BR2 was cultured in three different culture media including LB, SB, and TB in the optimum condition (temperature of 37°C, post-induction time of 2 h, and 1 mM IPTG). Figure 3 shows the SDS-PAGE of crude lysate of these three cultures. As shown, the best medium for the expression of DT386-BR2 was TB. The ratios of expressed protein in SB culture medium versus LB and TB culture medium versus LB were 1.93 (equal to 434 μg/ml of the total fusion protein) and 2.54 (equal to 571.14 μg/ml of the total fusion protein), respectively.
Figure 3.
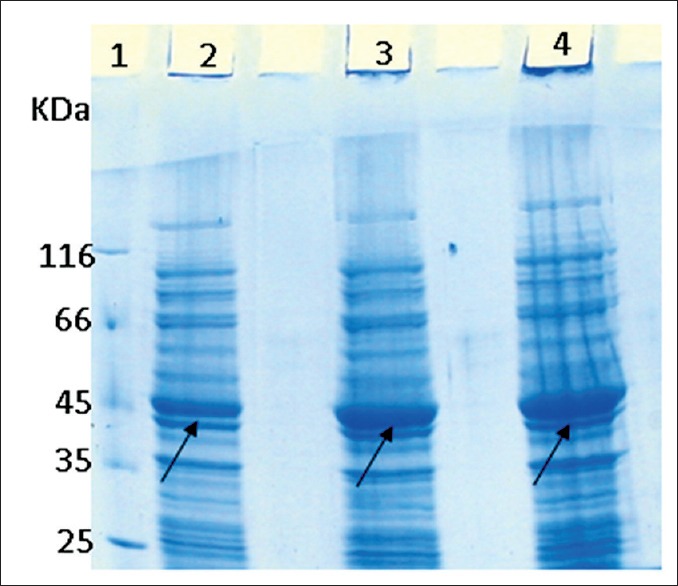
Sodium dodecyl sulfate polyacrylamide gel electrophoresis of different medium culture for expression of DT386-BR2 with 47 KDa molecular weight. Lane 1: Protein marker, lane 2: Expressed DT386-BR2 in luria bertani medium, lane 3: Expressed DT386-BR2 in SB medium, lane 4: expressed DT386-BR2 in terrific broth medium
Discussion
E. coli is the most commonly used host for overexpression of the recombinant proteins because of the several well-known advantages including fast growth kinetics, easily achieved high cell density cultures, possibility to make rich complex media from available and inexpensive components, and fast and easy transformation with exogenous DNA.[18] There are several successfully produced DT-based fusion proteins using E. coli expression system including DT385-p22,[19] DAB389 IL-2,[20] and DTLIL3.[21]
E. coli BL21 is a bacterial strain deficient in the protease coding genes preventing the degradation of foreign and extracellular proteins.[22,23] On the other hand, λDE3 is a prophage that inserts its genome containing the T7 RNA polymerase gene in the BL21 chromosome under the lacUV5 promoter.[24] Consequently, induction of the T7 RNA polymerase expression results in the expression of proteins in this strain (BL21 [DE3]).
Furthermore, Rosetta host strains are BL21 derivatives designed to enhance the expression of eukaryotic proteins containing codons rarely used in E. coli. These strains supply tRNAs for some codons on a compatible chloramphenicol-resistant plasmid.[25] In this study, despite codon optimization of DT386-BR2 gene for expression in E. coli, it has been shown that the expression of this fusion protein in Rosetta is higher than BL21. For E. coli Rosetta-gami DE3, on the other hand, with the abilities of either Rosetta or origami which allows for enhanced expression of eukaryotic proteins containing codons rarely used in E. coli and enhanced disulfide bond formation, respectively,[26] we did not detect the expression of the fusion protein which was attributed to difficulties in growth in the presence of four antibiotics and low yield of cellular mass as well as effort for the expression of antibiotic resistance gene, but not the desired fusion protein.
In the study of Tegel et al., 68 recombinant proteins previously expressed in BL21 (DE3) were retransformed and successfully expressed in Rosetta (DE3).[27] In another study conducted by Bukhtiyarova et al., it was shown that p38α (a type of MAP kinase) expression in Rosetta DE3 cells was significantly higher compared to the wild-type BL21 DE3 strain constituting about 20% of the total cellular protein.[28] The result of this studies showed that the yield of expression significantly improved and the purification procedure was successfully performed. According to the results of the above-mentioned studies and our project, it seems that Rosetta (DE3) is the best host for the expression of recombinant proteins.
Two most important factors affecting protein expression are incubation time and the post-induction temperature.[29] Overall, lowering the post-induction temperature lead to the reduction of the protein synthesis rate; consequently, the formation of inclusion bodies may be reduced. On the other hand, high temperature can promote cell growth resulting in high protein expression, but it is detrimental to protein expression because a higher growth rate would lead to a higher probability of plasmid loss and stimulates mispartition of an expression vector.[30] However, the optimum combination of postinduction temperature and the length of induction is still a trial- and-error matter.[31] As shown in our results, the best temperature for protein expression was determined to be 37°C. As BR2 is an anti-microbial peptide (AMP) that may affect the survival of bacterial host and inclusion bodies are inactive forms of the recombinant proteins, inclusion body formation is a suitable idea for overexpression of this bacteriotoxic fusion protein.
Inducer concentration also affects the protein expression. Low inducer concentration may result in an inefficient induction and consequently, low recombinant protein yields.[32] On the other hand, inducers added in excess can result in toxic effects including reduced cell growth and finally reduced recombinant protein concentration. Thus, inducer concentration should be maintained at slightly higher than the critical concentration. Since the study of Ramirez et al., showed that IPTG concentrations lower than 1 mM did not affect E. coli specific growth rate or maximum cell concentration, we used IPTG at 1 mM final concentration.[32] Regarding our results, it was shown that the effect of inducer concentration is critical for the protein expression.
Another aspect of overexpression of recombinant proteins is its augmentation by growing the culture to higher densities.[33] This can be achieved by changing a few parameters such as medium composition and providing better aeration by vigorous shaking.[34]
LB is the most commonly used culture medium for culturing E. coli, but the cell growth stops at a low density due to the presence of scarce amounts of carbohydrates and divalent cations in this medium culture.[35] Hence, it seems that increasing the amount of peptone or yeast extract and divalent cations supplementation leads to higher cell densities.[33,36] Terrific Broth is a richer medium than LB, resulting in greater cell mass and allowing higher recombinant protein expression levels.[37] For example, high density cultivation in TB medium yielded 56 g/L wet biomass, which was 10-fold higher than that obtained in LB medium[38] and the achieved results in this study confirmed this fact.
Conclusion
In this study, we could express large amounts of the recombinant DT386-BR2 fusion protein by optimizing the host, cultivation condition, and culture medium. Therefore, the expression of this fusion protein will be performed at the obtained optimal expression condition; the protein will be purified and used for evaluation of its cytotoxicities.
Financial support and sponsorship
This study was funded by Research Deputy of Isfahan University of Medical Sciences (grant number 193038).
Conflicts of interest
There are no conflicts of interest.
Acknowledgments
The authors are grateful to thanks Mrs. Fatemeh Moazen for her technical assistance.
References
- 1.Makino T, Skretas G, Georgiou G. Strain engineering for improved expression of recombinant proteins in bacteria. Microb Cell Fact. 2011;10:32. doi: 10.1186/1475-2859-10-32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Czitrom V. One-factor-at-a-time versus designed experiments. Am Stat. 1999;53:126–31. [Google Scholar]
- 3.Araujo PW, Brereton RG. Experimental design II. Optimization. Trends Analyt Chem. 1996;15:63–70. [Google Scholar]
- 4.Bezerra MA, Santelli RE, Oliveira EP, Villar LS, Escaleira LA. Response surface methodology (RSM) as a tool for optimization in analytical chemistry. Talanta. 2008;76:965–77. doi: 10.1016/j.talanta.2008.05.019. [DOI] [PubMed] [Google Scholar]
- 5.Myers RH, Montgomery DC, Anderson-Cook CM. Response Surface Methodology: Process and Product Optimization Using Designed Experiments. 3rd edithion. John Wiley and Sons; 2016. [Google Scholar]
- 6.Reiter Y. Recombinant immunotoxins in targeted cancer cell therapy. Adv Cancer Res. 2001;81:93–124. doi: 10.1016/s0065-230x(01)81003-4. [DOI] [PubMed] [Google Scholar]
- 7.Pushpanathan M, Gunasekaran P, Rajendhran J. Antimicrobial peptides: Versatile biological properties. Int J Pept 2013. 2013:675391. doi: 10.1155/2013/675391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Martín I, Teixidó M, Giralt E. Building cell selectivity into CPP-mediated strategies. Pharmaceuticals. 2010;3:1456–90. doi: 10.3390/ph3051456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Black JH, McCubrey JA, Willingham MC, Ramage J, Hogge DE, Frankel AE. Diphtheria toxin-interleukin-3 fusion protein (DT (388) IL3) prolongs disease-free survival of leukemic immunocompromised mice. Leukemia. 2003;17:155–9. doi: 10.1038/sj.leu.2402744. [DOI] [PubMed] [Google Scholar]
- 10.Frankel AE, Powell BL, Hall PD, Case LD, Kreitman RJ. Phase I trial of a novel diphtheria toxin/granulocyte macrophage colony-stimulating factor fusion protein (DT388GMCSF) for refractory or relapsed acute myeloid leukemia. Clin Cancer Res. 2002;8:1004–13. [PubMed] [Google Scholar]
- 11.Weaver M, Laske DW. Transferrin receptor ligand-targeted toxin conjugate (Tf-CRM107) for therapy of malignant gliomas. J Neurooncol. 2003;65:3–13. doi: 10.1023/a:1026246500788. [DOI] [PubMed] [Google Scholar]
- 12.Woo JH, Lee YJ, Neville DM, Frankel AE. Methods Mol Biol. Springer; 2010. Pharmacology of Anti-CD3 Diphtheria Immunotoxin in CD3 Positive T-Cell Lymphoma Trials. Immunotherapy of Cancer; pp. 157–75. [DOI] [PubMed] [Google Scholar]
- 13.Yang X, Kessler E, Su LJ, Thorburn A, Frankel AE, Li Y, et al. Diphtheria toxin-epidermal growth factor fusion protein DAB389EGF for the treatment of bladder cancer. Clin Cancer Res. 2013;19:148–57. doi: 10.1158/1078-0432.CCR-12-1258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Shafiee F, Rabbani M, Jahanian-Najafabadi A. Production and evaluation of cytotoxic effects of DT386-BR2 fusion protein as a novel anti-cancer agent. J microbial meth. 2016 doi: 10.1016/j.mimet.2016.09.004. In press. [DOI] [PubMed] [Google Scholar]
- 15.Sambrook J, Russell DW, Russell DW. The Condensed Protocols from Molecular Cloning: A Laboratory Manual. New York: Cold Spring Harbor Laboratory Press Cold Spring Harbor; 2006. [Google Scholar]
- 16.Akbari V, Sadeghi HM, Jafarian-Dehkordi A, Chou CP, Abedi D. Optimization of a single-chain antibody fragment overexpression in Escherichia coli using response surface methodology. Res Pharm Sci. 2015;10:75–83. [PMC free article] [PubMed] [Google Scholar]
- 17.Shafiee F, Moazen F, Rabbani M, Mir Mohammad Sadeghi H. Optimization of the Expression of Reteplase in Escherichia coli TOP10 using arabinose promoter. Jundishapur J Nat Pharm Prod. 2015;10:e16676. doi: 10.17795/jjnpp-16676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Rosano GL, Ceccarelli EA. Recombinant protein expression in Escherichia coli: Advances and challenges. Recombinant Protein Expression in Microbial Systems; Front Microbiol. 2014:7. doi: 10.3389/fmicb.2014.00172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zhang Y, Schulte W, Pink D, Phipps K, Zijlstra A, Lewis JD, et al. Sensitivity of cancer cells to truncated diphtheria toxin. PLoS One. 2010;5:e10498. doi: 10.1371/journal.pone.0010498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Liu Y, Wangn J, Qu J, Wang C, Hong T. Cloning and expression of recombinant chimeric toxin DT_(389)-IL-2 and testing of its specific cytotoxicity toward cells which bearing high affinity IL-2 receptor. Chin J Virol. 2000;17:117–21. [Google Scholar]
- 21.Frankel AE, McCubrey JA, Miller MS, Delatte S, Ramage J, Kiser M, et al. Diphtheria toxin fused to human interleukin-3 is toxic to blasts from patients with myeloid leukemias. Leukemia. 2000;14:576–85. doi: 10.1038/sj.leu.2401743. [DOI] [PubMed] [Google Scholar]
- 22.Gottesman S. Proteases and their targets in Escherichia coli. Annu Rev Genet. 1996;30:465–506. doi: 10.1146/annurev.genet.30.1.465. [DOI] [PubMed] [Google Scholar]
- 23.Grodberg J, Dunn JJ. ompT encodes the Escherichia coli outer membrane protease that cleaves T7 RNA polymerase during purification. J Bacteriol. 1988;170:1245–53. doi: 10.1128/jb.170.3.1245-1253.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Studier FW, Moffatt BA. Use of bacteriophage T7 RNA polymerase to direct selective high-level expression of cloned genes. J Mol Biol. 1986;189:113–30. doi: 10.1016/0022-2836(86)90385-2. [DOI] [PubMed] [Google Scholar]
- 25.Kane JF. Effects of rare codon clusters on high-level expression of heterologous proteins in Escherichia coli. Curr Opin Biotechnol. 1995;6:494–500. doi: 10.1016/0958-1669(95)80082-4. [DOI] [PubMed] [Google Scholar]
- 26.Gopal GJ, Kumar A. Strategies for the production of recombinant protein in Escherichia coli. Protein J. 2013;32:419–25. doi: 10.1007/s10930-013-9502-5. [DOI] [PubMed] [Google Scholar]
- 27.Tegel H, Tourle S, Ottosson J, Persson A. Increased levels of recombinant human proteins with the Escherichia coli strain Rosetta (DE3) Protein Expr Purif. 2010;69:159–67. doi: 10.1016/j.pep.2009.08.017. [DOI] [PubMed] [Google Scholar]
- 28.Bukhtiyarova M, Northrop K, Chai X, Casper D, Karpusas M, Springman E. Improved expression, purification, and crystallization of p38alpha MAP kinase. Protein Expr Purif. 2004;37:154–61. doi: 10.1016/j.pep.2004.05.017. [DOI] [PubMed] [Google Scholar]
- 29.Vasina JA, Baneyx F. Expression of aggregation-prone recombinant proteins at low temperatures: A comparative study of the Escherichia coli cspA and tac promoter systems. Protein Expr Purif. 1997;9:211–8. doi: 10.1006/prep.1996.0678. [DOI] [PubMed] [Google Scholar]
- 30.Mosrati R, Nancib N, Boudrant J. Variation and modeling of the probability of plasmid loss as a function of growth rate of plasmid-bearing cells of Escherichia coli during continuous cultures. Biotechnol Bioeng. 1993;41:395–404. doi: 10.1002/bit.260410402. [DOI] [PubMed] [Google Scholar]
- 31.Papaneophytou CP, Kontopidis G. Statistical approaches to maximize recombinant protein expression in Escherichia coli: A general review. Protein Expr Purif. 2014;94:22–32. doi: 10.1016/j.pep.2013.10.016. [DOI] [PubMed] [Google Scholar]
- 32.Ramirez O, Zamora R, Espinosa G, Merino E, Bolivar F, Quintero R. Kinetic study of penicillin acylase production by recombinant E. coli in batch cultures. Process Biochem. 1994;29:197–206. [Google Scholar]
- 33.Atlas RM. Handbook of Microbiological Media. 4th edition. CRC Press; 2010. [Google Scholar]
- 34.Blommel PG, Becker KJ, Duvnjak P, Fox BG. Enhanced bacterial protein expression during auto-induction obtained by alteration of lac repressor dosage and medium composition. Biotechnol Prog. 2007;23:585–98. doi: 10.1021/bp070011x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sezonov G, Joseleau-Petit D, D’Ari R. Escherichia coli physiology in Luria-Bertani broth. J Bacteriol. 2007;189:8746–9. doi: 10.1128/JB.01368-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Studier FW. Protein production by auto-induction in high density shaking cultures. Protein Expr Purif. 2005;41:207–34. doi: 10.1016/j.pep.2005.01.016. [DOI] [PubMed] [Google Scholar]
- 37.Litterer L, Schagat T. Protein expression in less time: A short induction protocol for KRX. Promega Notes. 2007;96:20–1. [Google Scholar]
- 38.Li JF, Zhang J, Song R, Zhang JX, Shen Y, Zhang SQ. Production of a cytotoxic cationic antibacterial peptide in Escherichia coli using SUMO fusion partner. Appl Microbiol Biotechnol. 2009;84:383–8. doi: 10.1007/s00253-009-2109-2. [DOI] [PubMed] [Google Scholar]