Abstract
Peptidoglycan (PGN) recognition proteins (PGRPs) are pattern-recognition receptors of the innate immune system that bind and, in some cases, hydrolyze bacterial PGNs. We determined the crystal structure, at 2.30-Å resolution, of the C-terminal PGN-binding domain of human PGRP-Iα in complex with a muramyl tripeptide representing the core of lysine-type PGNs from Gram-positive bacteria. The peptide stem of the ligand is buried at the deep end of a long binding groove, with N-acetylmuramic acid situated in the middle of the groove, whose shallow end can accommodate a linked N-acetylglucosamine. Although most interactions are with the peptide, the glycan moiety also seems to be essential for specific recognition by PGRPs. Conservation of key PGN-contacting residues shows that all PGRPs employ this basic PGN-binding mode. The structure pinpoints variable residues that likely mediate discrimination between lysine- and diaminopimelic acid-type PGNs. We also propose a mechanism for PGN hydrolysis by Zn2+-containing PGRPs.
Keywords: innate immunity, bacteria, receptor, complex, crystal structure
The innate immune system is the first line of defense against microorganisms in vertebrates and the only defense against microorganisms in invertebrates and plants (1, 2). It recognizes invading microbes by means of pattern-recognition receptors (PRRs) that are highly conserved in evolution to bind unique products of microbial metabolism not produced by the host [pathogen-associated molecular patterns (PAMPs)]. Examples of PAMPs recognized by PRRs such as Toll-like receptors (TLRs), collectins, and peptidoglycan (PGN) recognition proteins (PGRPs) include lipopolysaccharide of Gram-negative bacteria, lipoteichoic acid, mannans, DNA sequences containing unmethylated CpG dinucleotides, flagellin, and PGN, present in both Gram-positive and -negative bacteria (1, 2). However, except in the case of mannans, which are recognized by collectins (3), no structural information is available on how PRRs interact with any of these PAMPs.
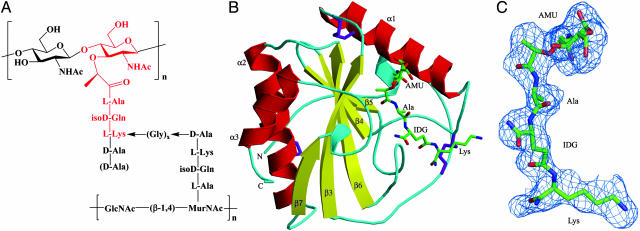
PGNs are located on the surface of virtually all bacteria and, as such, constitute excellent targets for recognition by the innate immune system (1, 2). PGNs are polymers of alternating N-acetylglucosamine (GlcNAc) and N-acetylmuramic acid (MurNAc) in β(1 → 4) linkage, crosslinked by short peptide stems (4, 5) (Fig. 1A). The glycan chains display little variation among different bacterial species. The crosslinking peptides are composed of alternating l and d form amino acids and are similar in all Gram-negative bacteria and in Gram-positive bacilli but vary in length and amino acid composition in Gram-positive cocci. According to the residue at position 3 of the peptide stems, PGNs have been divided into two major types: l-lysine-type (Lys-type) and meso-diaminopimelic acid-type (Dap-type). Dap-type PGN peptides are usually directly crosslinked, whereas Lys-type PGN peptides are interconnected by a peptide bridge that varies in length and amino acid composition in different bacteria (Fig. 1 A). A number of PRRs have been shown to interact with PGNs (2), including CD14 (6), nucleotide-binding oligomerization domain-containing proteins (NODs) (7, 8), and PGRPs (9). However, for none of these innate immune receptors is the basis for PGN recognition known.
Fig. 1.
Structure of Lys-type PGNs and of the PGRP-Iα–MTP complex. (A) The PGN fragment, highlighted in red, corresponds to the MTP ligand used to form the PGRP-Iα–MTP complex. Lys-type PGN peptides are usually crosslinked through a peptide bridge composed of 1–5 glycines. In parentheses is a d-alanine residue at peptide position 5 missing in PGNs from many bacteria. In Dap-type PGNs, l-lysine is replaced by meso-diaminopimelic acid, and the peptide stems are directly connected. (B) Structure of the PGRP-Iα–MTP complex. Helices are shown in red, strands in yellow, and coils in cyan. Disulfide bonds are shown in purple. The labeling of secondary structure elements follows the numbering for unbound PGRP-Iα in ref. 28. The N- and C-termini are indicated. The bound MTP is shown in ball-and-stick representation, with carbon atoms in green, nitrogen atoms in blue, and oxygen atoms in red. (C) σA-weighted Fo – Fc electron density map for the MTP ligand. The contour level is 2σ. NHAc, acetamide; AMU, MurNAc; Ala, l-alanine; IDG, d-isoglutamine; Lys, l-lysine.
PGRPs, which are structurally related to the bacteriophage T7 lysozyme, are highly conserved from insects to mammals (9–14). They bind PGNs with high affinity (15) and are important contributors to host defense against bacterial infections (2). In Drosophila, PGRPs activate two different signaling pathways that induce production of antimicrobial peptides (2). PGRP-SA interacts with Lys-type PGNs from Gram-positive bacteria, which activates the Toll receptor pathway (16). PGRP-LC and PGRP-LE recognize Dap-type PGNs from Gram-negative bacteria and activate the Imd pathway (17–21). Mouse PGRP-S, present in neutrophil tertiary granules, participates in the intracellular killing of bacteria (15). Mice deficient in PGRP-S exhibit increased susceptibility to i.p. infections with low-pathogenicity Gram-positive bacteria (22). Bovine PGRP-S, located in neutrophil and eosinophil granules, inhibits the growth of both Gram-positive and -negative bacteria (23). Certain insect and mammalian PGRPs, termed catalytic PGRPs, hydrolyze the amide bond between the MurNAc and l-alanine moieties of PGNs (24, 25). They are believed to play a scavenger role.
Recently, the crystal structures of Drosophila PGRP-LB (a catalytic PGRP) (26) and Drosophila PGRP-SA (27) and human PGRP-Iα (both noncatalytic PGRPs) (28) in their unliganded forms were reported, showing that the overall conformation of the putative PGN-binding site is maintained across the PGRP family. However, understanding the basis for PGN-binding by PGRPs, for specificity differences among the many family members and for the amidase activity of some PGRPs, requires direct information on PGRP–PGN interactions in the binding site. Accordingly, we determined the crystal structure of the C-terminal PGN-binding domain of human PGRP-Iα (designated PGRP-IαC) in complex with MurNAc-l-Ala-d-isoGln-l-Lys, a muramyl tripeptide (MTP) representing the conserved core of Lys-type PGNs from Gram-positive bacteria (Fig. 1 A).
Methods
PGRP Production. Procedures for expressing recombinant PGRP-IαC (residues 177–341) by in vitro folding from Escherichia coli inclusion bodies have been described (28).
PGN Analog Synthesis. An MTP (MurNAc-l-Ala-d-isoGln-l-Lys) and a muramyl dipeptide (MDP) (MurNAc-l-Ala-d-isoGln) were assembled by using classical fluorenylmethoxycarbonyl (Fmoc) chemistry and standard manual solid-phase peptide synthetic techniques (29). Detailed information is provided in Supporting Methods, which is published as supporting information on the PNAS web site.
Binding Assay. Surface plasmon resonance (SPR) was performed with a Biacore (Uppsala) 3000 biosensor. PGRP-IαC was covalently attached to a Biacore Sensor Chip CM5 by using the amine coupling method. Binding experiments were done at 25°C in PBS (pH 7.4) at a flow rate of 20 μl/min. Graded concentrations of MTP or MDP were then serially injected over the PGRP-IαC surface.
Crystallization and Data Collection. Because extensive attempts to cocrystallize PGRP-IαC and MTP were unsuccessful, the PGRP-IαC–MTP complex was obtained by soaking crystals of the free protein in solutions of the ligand. Unbound PGRP-IαC was crystallized at room temperature in hanging drops by mixing 1 μl of protein solution (6 mg/ml) and 1 μl of reservoir solution containing 20% (wt/vol) polyethylene glycol monomethyl ether 2000 (PEG-MME 2000), 0.01 M NiSO4, and 50 mM Tris·HCl (pH 8.5) (28). Once PGRP-IαC crystals were fully grown, 1 μl of MTP dissolved in water at 4 mg/ml (6.7 mM) was added to the hanging drop. After 2 days, soaked crystals were flash-cooled in a nitrogen stream. X-ray diffraction data to 2.30-Å resolution were recorded at 100 K by using an R-Axis IV++ image plate detector (Rigaku, Tokyo) equipped with Osmic mirrors and mounted on a Rigaku rotating anode generator. The data were processed and scaled by using crystalclear (30) (see Table 1, which is published as supporting information on the PNAS web site).
Structure Determination and Refinement. Crystals of PGRP-IαC soaked in MTP were nearly isomorphous with those of the unliganded protein (28). The free PGRP-IαC structure [Protein Data Bank (PDB) ID code 1SK3] with all water molecules deleted was used as the starting model for refinement of the complex. The side chains of residues lining the binding cleft were removed to avoid phase bias; these were returned during refinement assisted by the Fo – Fc omit map. Refinement was performed with cns (31) by using data in the 30.0- to 2.30-Å resolution range. Rigid body refinement, followed by 50 steps of energy minimization, resulted in Rfactor and Rfree values of 27.2% and 30.0%, respectively. When checking the model and electron density maps in xtalview (32), continuous positive density was observed in the PGN-binding cleft in both 2Fo – Fc and Fo – Fc electron density maps. Based on the Fo – Fc density map, MTP was built manually into PGRP-IαC; σA-weighted 2Fo – Fc and Fo – Fc maps were calculated for further model adjustment. After minimization, Rcryst was reduced to 25.4%, and Rfree was reduced to 27.9%. Group and individual temperature factors (B values) refinements were carried out, and water molecules were added into the final model, which comprises residues 177–341, one nickel ion, 48 waters, and one MTP molecule. The final Rcryst was 22.2%, and Rfree was 25.2% for all data between 30.0 and 2.30 Å (Table 1).
Homology-Assisted Docking of MTP into Drosophila PGRP-LB. PGRP-IαC and Drosophila PGRP-LB (26) share a strongly conserved three-dimensional fold, including a very similar PGN-binding cleft. Based on this high homology, MTP was docked into PGRP-LB by superposing PGRP-IαC onto PGRP-LB and then manually positioning MTP into the binding cleft of the latter; no steric clashes were observed. The manually docked PGRP-LB–MTP model was subjected to 200 cycles of energy minimization in cns (31), during which the MTP ligand was free to move, MTP-contacting residues were restrained, and all other residues were fixed.
Results
Overview of the Complex Structure. The structure of the PGRP-IαC–MTP complex was determined to 2.30-Å resolution by using unbound PGRP-IαC (28) as the starting model for refinement of the complex (Table 1). Superposition of free and MTP-bound PGRP-IαC gave an rms difference of 0.33 Å for 165 Cα atoms, indicating no substantial conformational changes upon complex formation. Minor differences in side-chain orientation were observed for several MTP-contacting residues; however, these differences are comparable with those for surface residues outside the PGN-binding site.
The PGRP-IαC structure contains a central β-sheet composed of five β-strands, four parallel and one (β5) antiparallel, and three α-helices (Fig. 1B). The domain is crosslinked by three disulfide bonds (Cys-178—Cys-300, Cys-194—Cys-238, and Cys-214—Cys-220). The PGN-binding site resides in a long cleft whose walls are formed by helix α1 and five loops (β3–α1, α1–β4, β5–β6, β6–α2, and β7–α3) that project above the β-sheet platform. Located opposite the PGN-binding site is a large hydrophobic groove, formed by residues 177–198 (the PGRP-specific segment), that may serve as a binding site for host effector or signaling proteins (26–28).
Very clear electron density corresponding to the entire MTP molecule was visible in the PGN-binding site, as evident in the Fo – Fc omit map for the ligand (Fig. 1C). The occupancy of MTP is <1.0 according to the electron density levels and temperature factors. In the final refined model, the occupancy was set to 0.6, which gave the best explanation for the electron density levels, and an average atom B value (31.3 Å2) close to the average main-chain atom B value for the protein (32.2 Å2) (Table 1).
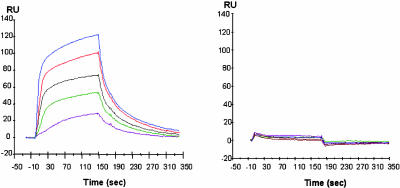
SPR was used to demonstrate specific binding of MTP to PGRP-IαC in solution (Fig. 2). Whereas MTP clearly bound to immobilized PGRP-IαC, the binding of MDP was greatly diminished, revealing a critical role for l-lysine at peptide position 3 (see below).
Fig. 2.
SPR sensograms depicting the binding of MTP versus MDP to PGRP-IαC. Concentrations of 100, 200, 300, 500, and 800 μM MTP (Left) or MDP (Right) in PBS were injected over 14,000 resonance units (RU) of immobilized PGRP-IαC at a flow rate of 20 μl/min for 180 sec. Dissociation was achieved by passing the same buffer for 300 sec.
Interactions in the PGN-Binding Cleft. The PGN-binding cleft of PGRP-IαC is ≈24 Å long, with a shallow (6–7 Å) end flanked by helix α1 and loops β3–α1 and β6–α2 and a deep (12–13 Å) end flanked by loops α1–β4, β5–β6, and β7–α3. The general topology of this groove is maintained in the Drosophila PGRP-LB and PGRP-SA structures (26, 27), as well as in the T7 lysozyme (33). In the complex structure, the tripeptide stem of MTP (l-Ala-d-isoGln-l-Lys) is held in an extended conformation at the deep end of the binding groove, whereas the MurNAc moiety lies in a pocket in the middle of the groove, with its pyranose ring oriented perpendicularly to the base of the pocket (Fig. 1B). The shallow end of the binding cleft is unoccupied by the ligand (see below). The PGRP-IαC–MTP complex buries a total solvent-accessible surface of 1,004 Å2, of which 412 and 592 Å2 are contributed by PGRP-IαC and MTP, respectively. The interfaces with MurNAc and the tripeptide stem account for 42% and 58%, respectively, of the total buried surface. MTP is mostly (62%) buried in the PGN-binding site, with its glycan and peptide portions buried to similar extents. The PGRP-IαC–MTP interface is predominantly hydrophilic yet devoid of buried solvent molecules, an indication of high chemical and shape complementarity.
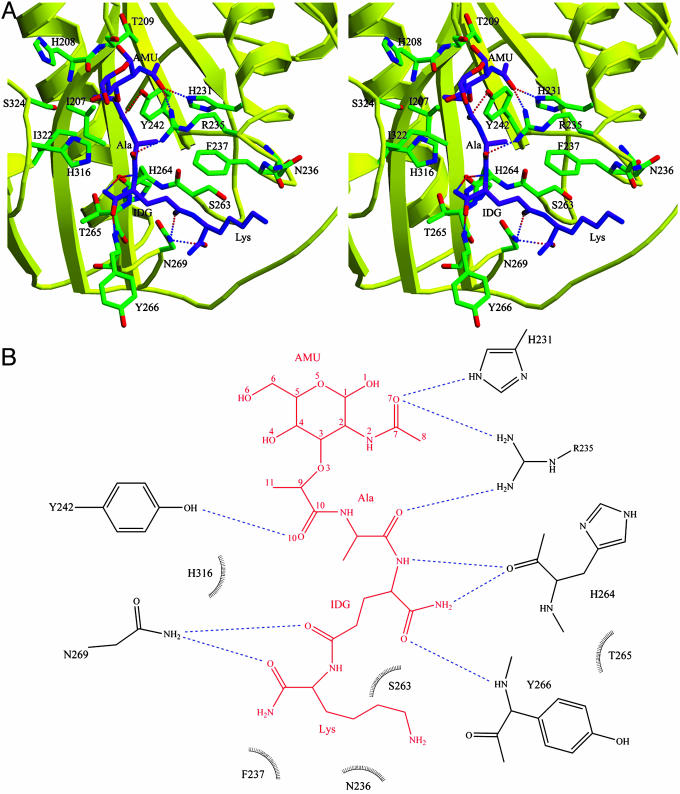
MTP makes extensive contacts with 16 residues lining the binding cleft of PGRP-IαC (Fig. 3A). Most of these interactions (6 of 9 hydrogen bonds and 29 of 43 van der Waals contacts) are with the peptide, rather than glycan, portion of the PGN analog (see Table 2, which is published as supporting information on the PNAS web site). Thus, the MurNAc moiety forms three hydrogen bonds with the side chains of three PGRP-IαC residues, two through atom O7 with Nε2 of His-231 and Nη1 of Arg-235, and another through O10 with Oη of Tyr-242 (Fig. 3B). Most (four of six) of the hydrogen bonds to the tripeptide stem of MTP involve main-chain atoms of the peptide l-Ala O—Arg-235 Nη1, d-isoGln N—His-264 O, d-isoGln Oε1—Asn-269 Nδ2, and l-Lys O—Asn-269 Nδ2. Except for Arg-235, whose imido group makes two hydrogen bonds with MTP, all other residues forming hydrogen bonds to the ligand through their side chains (His-231, Tyr-242, and Asn-269) are highly conserved (>80% identity) across PGRPs (see Fig. 5 which is published as supporting information on the PNAS web site), implying a single PGN-binding mode. No water-mediated hydrogen bonds were observed between PGRP-IαC and MTP, which is atypical for a protein–carbohydrate complex (34).
Fig. 3.
Intermolecular contacts in the PGRP-IαC–MTP complex. (A) Stereoview of interactions between PGRP-IαC and MTP at the PGN-binding site. MTP is shown in purple, PGRP-IαC in yellow, and contacting residues in green. Hydrogen bonds are shown as dashed lines; residues forming van der Waals contacts with MTP are also highlighted. (B) Schematic representation of interactions between MTP and PGRP-IαC. MTP is shown in red; hydrogen bonds are shown as blue dashed lines. Residues making van der Waals contacts with MTP are indicated by arcs with spokes radiating toward the ligand moieties they contact. Only residues making two or more such contacts are shown. No water-mediated interactions were observed. AMU, MurNAc; IDG, d-isoglutamine.
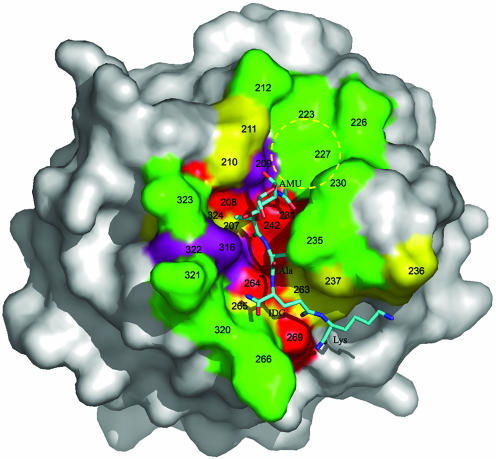
Approximately one-third of the PGN-binding groove of PGRP-Iα, corresponding to its shallow end, is unoccupied in the complex with MTP (Fig. 4). This is readily explained because MTP lacks GlcNAc in β(1 → 4) linkage with MurNAc found in natural PGNs (Fig. 1 A). The empty portion of the binding cleft comprises a pocket that is substantially broader and shallower than the one containing the MurNAc moiety, suggesting an orientation for the pyranose ring of GlcNAc, if bound to PGRP-Iα, parallel to the floor of the pocket. The hexose rings of GlcNAc and MurNAc would then be oriented perpendicularly, the most stable conformation for these linked saccharide units.
Fig. 4.
Surface analysis of the PGN-binding site of PGRP-Iα. The molecular surface is colored according to the percentage identities of residues lining the PGN-binding groove of PGRPs based on the sequence alignments in Fig. 6. Red, >80%; purple, 60–80%; yellow, 40–60%; and green, <40%. The bound MTP is shown in ball-and-stick representation, with carbon atoms in light blue, nitrogen atoms in dark blue, and oxygen atoms in red. A putative binding pocket for the GlcNAc moiety of natural PGNs, not present in the MTP fragment, is circled in yellow. Five of 16 MTP-contacting residues in the PGRP-IαC–MTP complex, at positions 208, 231, 242, 264, and 269, are highly conserved among PGRPs (>80% identity); only two contacting residues, at positions 235 and 266, are <40% conserved. AMU, MurNAc; IDG, d-isoglutamine.
All bacterial PGNs contain d-alanine at peptide position 4, which is absent from MTP, whereas PGNs from some species also include d-alanine at position 5 (Fig. 1 A). If d-alanine were present at position 4 in the PGRP-IαC–MTP structure, it would most likely contact Gln-261, Tyr-266, and Asn-269, the latter two of which are already involved in interactions with the tripeptide stem (Table 2). By contrast, d-alanine at position 5 would extend beyond the binding groove and probably would not contribute to PGN recognition.
Role of Conserved and Variable PGN-Contacting Residues. More than 40 PGRP sequences from insects and mammals have been reported (Fig. 5). Whereas most PGRPs contain only a single PGN-binding domain, some (e.g., human PRGR-Iα and -Iβ and Drosophila PGRP-LF) comprise tandem PGN-binding domains (28). The high sequence homologies among PGRP domains (>45% sequence conservation between any compared pair) indicates that all adopt a very similar fold (Fig. 1B). Although none of the 16 ligand-contacting residues in the PGRP-IαC–MTP complex is invariant in all PGRPs, five of them (His-208, His-231, Tyr-242, His-264, and Asn-269) are >80% identical (Fig. 5). These residues, which form a nearly contiguous patch on the floor of the binding groove (Fig. 4), account for six of nine specific hydrogen bonds to MTP (Fig. 3B). Moreover, if one considers that Tyr-266, which exhibits only 15% identity, hydrogen-bonds to MTP through its main-chain nitrogen, it is apparent that PGRPs most probably use a single PGN-binding mode.
In addition to the highly conserved (>60% identity) PGN-contacting residues at positions 208, 209, 231, 242, 264, 269, 316, and 324 (Fig. 5), PGRP-IαC interacts with MTP through eight other residues that display greater variability. Of particular note are Asn-236 and Phe-237, which form a number of van der Waals contacts with the side chain of l-lysine (Table 2). Sequence variability at these two positions may account for the ability of some PGRPs to discriminate between Lys-type and Dap-type PGNs, as measured in assays of antimicrobial peptide production (16–21). In Drosophila PGRP-SA, which, like PGRP-IαC, preferentially recognizes Lys-type PGNs, the corresponding sequence is Asp-96–Phe-97, very similar (Asn-236–Phe-237) or identical (Asp-242–Phe-243) to those in PGRP-IαC and the C-terminal PGN-binding domain of mouse PGRP-Iα, respectively (Fig. 4). In contrast, the corresponding sequence in Drosophila PGRP-LCx and PGRP-LE, which recognize Dap-type PGNs (2, 21), is Gly–Trp. However, the ability of PGRPs to distinguish between Lys-type and Dap-type PGNs is not absolute. For example, Drosophila PGRP-SC1B and mouse PGRP-L hydrolyze PGN from both Gram-negative and -positive bacteria (24, 35). In addition, Dap-type PGNs activate the Toll pathway, albeit less efficiently than Lys-type PGNs (19). Indeed, a certain degree of crossreactivity would be consistent with the relatively small chemical difference between meso-diaminopimelic acid and l-lysine, which differ by a single carboxyl group attached to the Cε atom of the former. Information on the PGN specificities of other PGRPs will be necessary to refine these structural correlations.
In contrast to the conserved nature of PGRP-IαC residues contacting MurNAc, residues lining the walls of the putative GlcNAc-binding pocket (Thr-209, Gly-211, Val-223, Ile-227, Asn-226, and Phe-230) exhibit much higher variability (Fig. 4). Based on this consideration, and on the relative shallowness of this pocket (7 Å at its deepest point), it appears unlikely that the GlcNAc moiety of PGNs contributes greatly to stabilizing complexes with PGRPs, although this hypothesis will require direct verification.
Proposed Mechanism for PGN Hydrolysis by Catalytic PGRPs. Some PGRPs, including Drosophila PGRP-LB (26) and PGRP-SC1B (24) and human and mouse PGRP-L (25, 35), are Zn2+-dependent amidases that hydrolyze PGNs by cleaving the amide bond between MurNAc and l-alanine. No zinc ion is present in the PGN-binding site of PGRP-IαC (Fig. 3A), which binds but does not hydrolyze PGNs (28). MTP was docked into the PGN-binding cleft of Drosophila PGRP-LB, based on its close similarity to PGRP-IαC (rms difference of 0.75 Å for 136 Cα atoms, excluding the variable PGRP-specific segments). In particular, the main PGN-contacting residues of PGRP-IαC superpose closely onto their counterparts in PGRP-LB (data not shown). In the docked PGRP-LB–MTP complex, MTP forms seven hydrogen bonds with PGRP-LB (Fig. 6A, which is published as supporting information on the PNAS web site), six of which are retained in the PGRP-IαC–MTP structure.
A general mechanism for PGN hydrolysis by catalytic PGRPs, which could also apply to the T7 lysozyme, for which no structure of a complex with PGN has been reported (33), may be proposed based on the Drosophila PGRP-LB–MTP model (Fig. 6B). Acting as an electrophilic catalyst, the bound zinc ion, which is very close (3.5 Å) to the O10 atom of MurNAc, accepts an electron pair from this carbonyl oxygen, polarizing the carbonyl bond and imparting an sp3 character to the C10 atom. This carbonyl carbon then becomes more susceptible to nucleophilic attack by the catalytic water molecule coordinated to the bound Zn2+. In the transition state, decreased delocalization of π electrons on the l-Ala N—C10 amide bond results in bond lengthening and greater basicity of the nitrogen atom, which facilitates transfer of a proton from Tyr-78 to form a leaving group. Another water molecule then attacks the Zn2+ and replaces the carbonyl oxygen and coordination water, releasing the cleavage products (Fig. 6B).
A critical role for Tyr-78 is suggested by its strict conservation in all catalytic PGRPs and the T7 lysozyme. Moreover, superposition of PGRP-LB (or the T7 lysozyme) onto the zinc-dependent metalloproteases carboxypeptidase A and thermolysin (36) through the three zinc-coordinating residues revealed that Tyr-78 is in the position of the catalytic glutamate in the two proteases (data not shown). Finally, mutation of the analogous tyrosine (Tyr-46) of the T7 lysozyme to phenylalanine abolishes amidase activity (33), indicating that the hydroxyl group of this tyrosine, as well as its counterpart in PGRPs, is required for catalysis.
Discussion
The large majority of PGRPs, both insect and mammalian, are soluble proteins localized in intracellular vesicles, where they may encounter phagocytosed bacteria or be secreted upon fusion of these vesicles with the cell membrane (2, 9, 28). Indeed, only Drosophila PGRP-LC has been definitively identified as a cell surface receptor (21). Soluble PGRPs act as adaptors that link PGN recognition to the induction of intracellular signaling or complement cascades. To fulfill this role, it has been proposed that PGRPs have evolved two independent binding sites: a highly conserved site for recognizing PGNs and a topologically variable site (28), located behind the PGN-binding site, for interacting with host effector proteins (37, 38).
Several lines of evidence indicate that MTP represents the minimal PGN motif recognized by PGRPs. Fourier difference maps of PGRP-IαC crystals soaked in high concentrations (10–15 mM) of MDP or MurNAc revealed no electron density corresponding to either compound in the PGN-binding site (data not shown). This is consistent with the lack of binding of MDP to PGRP-IαC by SPR and the finding that MDP is inactive in stimulating the Drosophila Imd pathway (21). Moreover, the minimum PGN fragment hydrolyzed by human PGRP-L is MTP (25). However, based on the PGRP-IαC–MTP structure, addition of d-alanine to MTP at peptide position 4, or of GlcNAc in β(1 → 4) linkage to the MurNAc moiety, might increase affinity for PGRPs, enhancing the efficiency of recognition.
The PGRP-IαC–MTP complex reveals that PGRPs interact with both the peptide stems and glycan chains of PGN. This binding mode, involving differential recognition of variable peptide sequences, provides a structural explanation for the ability of some PGRPs to distinguish among PGNs from different microbes (16–21). In particular, discrimination between Lys-type and Dap-type PGNs is most likely mediated by several variable amino acids located at one extremity of the PGN-binding groove that, in the PGRP-IαC–MTP structure, pack tightly against the side chain of l-lysine in the peptide stem. The importance of both the peptide and glycan portions of PGNs to recognition by PGRPs was demonstrated by using a synthetic lactyl-tetrapeptide corresponding to the peptide stem of tracheal cytotoxin (TCT), a monomeric disaccharide-tetrapeptide fragment of Dap-type PGNs that is a potent activator of the Drosophila Imd pathway (21). Although the peptide alone retained the ability to stimulate the immune response, optimal activation also required the glycan portion of TCT. In the PGRP-IαC–MTP complex, the three-carbon lactyl moiety of MurNAc accounts for 1 of 3 hydrogen bonds and 10 of 14 van der Waals contacts between the sugar and PGRP-IαC (MurNAc consists of GlcNAc in ether linkage with lactic acid). In agreement with the crystal structure, a tetrapeptide lacking the lactyl group lost most of its capacity to stimulate the Imd pathway (21).
PGRPs bind PGN with high (nanomolar) affinity, as measured by the binding of mouse PGRP-S to soluble polymeric PGN immobilized on agarose beads (15). In contrast to collectins such as mannose-binding protein and pulmonary surfactant apoprotein D, which oligomerize through formation of triple-stranded coiled coils (3), PGRPs are monomeric proteins. Hence, unlike collectins, PGRPs cannot use multivalent interactions to augment avidity for carbohydrate-containing ligands. Instead, PGRPs have evolved an alternative strategy to achieve high-affinity binding based on burying both the peptide and MurNAc portions of PGN in a deep (12–13 Å) cleft that completely excludes solvent. It should also be noted that the peptide stems in polymeric PGN are crosslinked, either directly or by a peptide bridge, such that their conformational flexibility should be highly restricted. This situation contrasts sharply with the absence of structural constraints on movements of the peptide stem in monomeric MTP. Assuming that the peptide conformation in crosslinked PGN is similar to the conformation observed in the PGRP-IαC–MTP complex, PGRPs should bind more tightly to polymeric PGN than to its monomeric fragments, because entropic penalties associated with restricting peptide flexibility would be minimized. Indeed, the incomplete occupancy of MTP in the crystal structure, as well as SPR analysis, suggests micromolar binding affinity.
Besides PGRPs, other innate immune receptors, including CD14 (6) and NODs (7, 8), have been shown to recognize PGN. However, these PRRs, which are structurally unrelated to PGRPs (7, 8, 39), are likely to bind PGNs differently than PGRPs do. Whereas NOD2 detects MDP, the minimal PGN motif common to both Gram-positive and -negative bacteria, NOD1 senses the meso-diaminopimelic acid-containing MTP GlcNAc-MurNAc-l-Ala-d-isoGln-(2S,6R)-Dap (Dap-GMTP), a natural product of PGN degradation in Gram-negative bacteria (7, 8, 39, 40). PGRPs, unlike NOD2, do not recognize MDP (21). The tripeptide l-Ala-d-isoGln-(2S,6R)-Dap activates NOD1 as efficiently as Dap-GMTP (40), implying that glycans contribute less to PGN recognition by NOD1 than they do in the case of PGRPs, which require the glycan for maximal immunostimulatory activity (21). The exact PGN structure recognized by CD14 is unknown (6). X-ray crystallographic studies of these other PRRs will be required to define the structural relationship between their PGN-binding modes and those of PGRPs.
Supplementary Material
Acknowledgments
This work was supported by grants from the National Institutes of Health (to G.-J.B. and R.A.M.).
Abbreviations: PRR, pattern-recognition receptor; PGN, peptidoglycan; PGRP, PGN recognition protein; MurNAc, N-acetylmuramic acid; Lys-type, L-lysine-type; Dap-type, meso-diaminopimelic acid-type; NOD, nucleotide-binding oligomerization domain-containing protein; PGRP-IαC, C-terminal PGN-binding domain of human PGRP-Iα; MTP, muramyl tripeptide; MDP, muramyl dipeptide; SPR, surface plasmon resonance.
Data deposition: The atomic coordinates and structure factors have been deposited in the Protein Data Bank, www.pdb.org (PDB ID code 1TWQ).
References
- 1.Medzhitov, R. & Janeway, C. A., Jr. (2002) Science 296, 298–300. [DOI] [PubMed] [Google Scholar]
- 2.Hoffmann, J. A. (2003) Nature 426, 33–38. [DOI] [PubMed] [Google Scholar]
- 3.Weis, W. I., Taylor, M. E. & Drickamer, K. (1998) Immunol. Rev. 163, 19–34. [DOI] [PubMed] [Google Scholar]
- 4.van Heijenoort, J. (2001) Glycobiology 11, 25R–36R. [DOI] [PubMed] [Google Scholar]
- 5.Doyle, R. J. & Dziarski, R. (2001) in Molecular Medical Microbiology, ed. Sussman, M. (Academic, London), pp. 137–153.
- 6.Gupta, D., Kirkland, T. N., Viriyakosol, S. & Dziarski, R. (1996) J. Biol. Chem. 271, 23310–23316. [DOI] [PubMed] [Google Scholar]
- 7.Inohara, N. & Nunez, G. (2003) Nat. Rev. Immunol. 3, 371–382. [DOI] [PubMed] [Google Scholar]
- 8.Chamaillard, M., Girardin, S. E., Viala, J. & Philpott, D. J. (2003) Cell. Microbiol. 5, 581–592. [DOI] [PubMed] [Google Scholar]
- 9.Dziarski, R. (2004) Mol. Immunol. 40, 877–886. [DOI] [PubMed] [Google Scholar]
- 10.Kang, D., Liu, G., Lundstrom, A., Gelius, E. & Steiner, H. A. (1998) Proc. Natl. Acad. Sci. USA 95, 10078–10082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ochiai, M. & Ashida, M. (1999) J. Biol. Chem. 274, 11854–11858. [DOI] [PubMed] [Google Scholar]
- 12.Werner, T., Liu, G., Kang, D., Ekengren, S., Steiner, H. & Hultmark, D. (2000) Proc. Natl. Acad. Sci. USA 97, 13772–13777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Liu, C., Xu, Z., Gupta, D. & Dziarski, R. (2001) J. Biol. Chem. 276, 34686–34694. [DOI] [PubMed] [Google Scholar]
- 14.Christophides, G. K., Zdobnov, E., Barillas-Mury, C., Birney, E., Blandin, S., Blass, C., Brey, P. T., Collins, F. H., Danielli, A., Dimopoulos, G., et al. (2002) Science 298, 159–165. [DOI] [PubMed] [Google Scholar]
- 15.Liu, C., Gelius, E., Liu, G., Steiner, H. & Dziarski, R. (2000) J. Biol. Chem. 275, 24490–24499. [DOI] [PubMed] [Google Scholar]
- 16.Michel, T., Reichhart, J. M., Hoffmann, J. A. & Royet, J. (2001) Nature 414, 756–759. [DOI] [PubMed] [Google Scholar]
- 17.Choe, K. M., Werner, T., Stoven, S., Hultmark, D. & Anderson, K. V. (2002) Science 296, 359–362. [DOI] [PubMed] [Google Scholar]
- 18.Gottar, M., Gobert, V., Michel, T., Belvin, M., Duyk, G., Hoffmann, J. A., Ferrandon, D. & Royet, J. (2002) Nature 416, 640–644. [DOI] [PubMed] [Google Scholar]
- 19.Leulier, F., Parquet, C., Pili-Floury, S., Ryu, J.-H., Caroff, M., Lee, W.-J., Mengin-Lecreulx, D. & Lemaitre, B. (2003) Nat. Immunol. 4, 478–484. [DOI] [PubMed] [Google Scholar]
- 20.Werner, T., Borge-Renberg, K., Mellroth, P., Steiner, H. & Hultmark, D. (2003) J. Biol. Chem. 278, 26319–26322. [DOI] [PubMed] [Google Scholar]
- 21.Kaneko, T., Goldman, W. E., Mellroth, P., Steiner, H., Fukase, K., Kusumoto, S., Harley, W., Fox, A., Golenbock, D. & Silverman, N. (2004) Immunity 20, 637–649. [DOI] [PubMed] [Google Scholar]
- 22.Dziarski, R., Platt, K. A., Gelius, E., Steiner, H. & Gupta, D. (2003) Blood 102, 689–697. [DOI] [PubMed] [Google Scholar]
- 23.Tydell, C. C., Yount, N., Tran, D., Yuan, J. & Selsted, M. E. (2002) J. Biol. Chem. 277, 19658–19664. [DOI] [PubMed] [Google Scholar]
- 24.Mellroth, P., Karlsson, J. & Steiner, H. (2003) J. Biol. Chem. 278, 7059–7064. [DOI] [PubMed] [Google Scholar]
- 25.Wang, Z.-M., Li, X., Cocklin, R. R., Wang, M., Wang, M., Fukase, K., Inamura, S., Kusumoto, S., Gupta, D. & Dziarski, R. (2003) J. Biol. Chem. 278, 49044–49052. [DOI] [PubMed] [Google Scholar]
- 26.Kim, M.-S., Byun, M. & Oh, B.-H. (2003) Nat. Immunol. 4, 787–793. [DOI] [PubMed] [Google Scholar]
- 27.Reiser, J.-B., Teyton, L. & Wilson, I. A. (2004) J. Mol. Biol., 340, 909–917. [DOI] [PubMed] [Google Scholar]
- 28.Guan, R., Malchiodi, E. L., Wang, Q., Schuck, P. & Mariuzza, R. A. (2004) J. Biol. Chem. 279, 31873–31882. [DOI] [PubMed] [Google Scholar]
- 29.Siriwardena, A., Jørgensen, M., Wolfert, M. A., Vandenplas, M. L., Moore, J. N. & Boons, G. J. (2001) J. Am. Chem. Soc. 123, 8145–8146. [DOI] [PubMed] [Google Scholar]
- 30.Pflugrath, J. W. (1999) Acta Crystallogr. D 55, 1718–1725. [DOI] [PubMed] [Google Scholar]
- 31.Brünger, A. T., Adams, P. D., Clore, G. M., DeLano, W. L., Gros, P., Grosse-Kunstleve, R. W., Jiang, J. S., Kuszewski, J., Nilges, M., Pannu, N. S., et al. (1998) Acta Crystallogr. D 54, 905–921. [DOI] [PubMed] [Google Scholar]
- 32.McRee, D. E. (1999) J. Struct. Biol. 125, 156–165. [DOI] [PubMed] [Google Scholar]
- 33.Cheng, X., Zhang, X., Pflugrath, J. W. & Studier, F. W. (1994) Proc. Natl. Acad. Sci. USA 91, 4034–4038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Weis, W. I. & Drickamer, K. (1996) Annu. Rev. Biochem. 65, 441–473. [DOI] [PubMed] [Google Scholar]
- 35.Gelius, E., Person, C., Karlsson, J. & Steiner, H. (2003) Biochem. Biophys. Res. Comm. 306, 988–994. [DOI] [PubMed] [Google Scholar]
- 36.Leung, D., Abbenante, G. & Fairlie, D. P. (2000) J. Med. Chem. 43, 305–341. [DOI] [PubMed] [Google Scholar]
- 37.Gobert, V., Gottar, M., Matskevich, A. A., Rutschmann, S., Royet, J., Belvin, M., Hoffmann, J. & Ferrandon, D. (2003) Science 302, 2126–2130. [DOI] [PubMed] [Google Scholar]
- 38.Pili-Floury, S., Leulier, F., Takahashi, K., Saigo, K., Samain, E., Ueda, R. & Lemaitre, B. (2004) J. Biol. Chem. 279, 12848–12853. [DOI] [PubMed] [Google Scholar]
- 39.Tanabe, T., Chamaillard, M., Ogura, Y., Zhu, L., Qiu, S., Masumoto, J., Ghosh, P., Moran, A., Predergast, M. M., Tromp, G., et al. (2004) EMBO J. 23, 1587–1597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Girardin, S. E., Travassos, L. H., Herve, M., Blanot, D., Boneca, I. G., Philpott, D. J., Sansonetti, P. J. & Mengin-Lecreulx, D. (2003) J. Biol. Chem. 278, 41702–41708. [DOI] [PubMed] [Google Scholar]
- 41.Dziarski, R., Tapping, R. I. & Tobias, P. (1998) J. Biol. Chem. 273, 8680–8690. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.