Abstract
Herpes simplex virus type 1 (HSV-1) can enter cells expressing any one of multiple entry receptors, including the herpesvirus entry mediator (HVEM), nectin-1, and sites in heparan sulfate generated by specific 3-O-sulfotransferases. The viral ligand for these receptors is glycoprotein D (gD). To define structural requirements for functional interactions of gD with its receptors and to obtain viral mutants altered for receptor usage, we generated a library of HSV-1 mutants with random mutations in the gD gene. Viral isolates selected on a monkey cell line (Vero) were screened for the loss of ability to infect cells expressing each of the HSV-1 receptors. The 10 HSV-1 mutants obtained had 12 mutations in gD, affecting 11 amino acids. All mutations reduced or abrogated viral entry through HVEM and 3-O-sulfated heparan sulfate, indicating that similar features of gD are critical for functional interactions with both these receptors. None of the mutations reduced viral entry through nectin-1, whereas a subset of the mutations conferred ability to use nectin-2 as an entry receptor. These and other results show that features of gD, including conformation of the N terminus, critical for functional interactions with HVEM/3-O-sulfated heparan sulfate, differ from those critical for interactions with nectin-1.
Keywords: glycoprotein D, herpes simplex virus
Many viruses can use multiple alternative receptors for cell entry. The viral ligands for the receptors, either envelope glycoproteins or capsid determinants, can recognize several members of the same receptor family or even different families of receptors. Usage of alternative receptors can enable a virus to infect multiple cell types that differ in receptor expression. Selective pressures on the virus can permit the emergence of variants with enhanced use of one receptor and reduced ability to use another, altering cell tropism and pathogenesis. Also, if viral engagement of cell surface receptors results in activation of signaling pathways, then use of different receptors might also have different consequences for the infected cell.
Herpes simplex virus (HSV) can bind to cells by interaction with heparan sulfate but entry requires interaction of envelope glycoprotein D (gD) with any one of three classes of cell-surface receptors (1, 2). These receptors include the herpesvirus entry mediator (HVEM), a member of the tumor necrosis factor receptor family; nectin-1 and nectin-2, cell adhesion molecules in the Ig superfamily; and specific sites in heparan sulfate generated by certain isoforms of 3-O-sulfotransferase (the product is referred to as 3-O-sulfated heparan sulfate, 3-O-S HS). Binding of gD to one of these receptors triggers fusion of the viral envelope with a cell membrane, an event that requires also the action of envelope glycoprotein B, glycoprotein H, and glycoprotein L (1). We and others have engaged in targeted mutagenesis studies to define the residues in gD that are critical for physical and functional interactions with the several HSV entry receptors (3–6). These studies have been guided in part by serendipity and in part by an x-ray structure of a truncated form of HSV type 1 (HSV-1) gD in complex with a truncated form of HVEM (7).
An approach for random mutagenesis of gD and screening of viral mutants for altered receptor preferences, unbiased except for the requirement that the mutants retain ability to use at least one receptor, was needed to obtain the range of mutants desired and to define the structural determinants of gD critical for interactions with each receptor. We adapted existing technology to develop a system for precise gene replacement at the gD locus in the HSV-1 genome and to generate a library of viral mutants altered in the gD gene, a library that could be screened for the desired phenotypes. This system relied on (i) insertion of an FLP recombination target (FRT) for the site-specific yeast recombinase FLP (8) in place of the gD ORF in the HSV-1 genome; (ii) PCR-induced mutagenesis of the cloned gD ORF bounded by FRT sites; (iii) use of FLP recombinase to insert mutagenized copies of the circularized gD ORF (the two ends linked by one FRT site) into the viral genome; (iv) screening of virus isolates for the ability to infect cells expressing each of the known entry receptors; and (v) plaque purification and further analysis of virus isolates exhibiting receptor usage different from that of WT HSV-1. For WT HSV-1, HVEM and nectin-1 are excellent entry receptors, 3-O-S HS is moderately active, and nectin-2 has very little activity (1). Nectin-2 was originally identified as an entry receptor for a particular HSV-1 mutant and for HSV type 2 (HSV-2) (9).
We present here the results of our first large screening of a library of HSV-1 viruses enriched for mutants carrying altered gD genes. The virus isolates screened were first selected for their ability to form plaques on Vero (African green monkey kidney) cells, which are known to express primate forms of HVEM and nectin-1 (10, 11). Sequencing of the gD genes identified mutations that significantly reduced or eliminated functional interactions with both HVEM and 3-O-S HS, whereas entry activity with nectin-1 was left intact. A subset of these mutations also enhanced functional interactions with nectin-2. Whereas most of the mutations that altered interactions with HVEM were in the interface with HVEM, as could be predicted, three were not but could be explained by effects on conformation of the N-terminal hairpin or by postbinding effects on gD function with HVEM. The results presented here and elsewhere indicate that certain features of gD critical for functional interactions with HVEM and 3-O-S HS are distinct from those critical for interactions with nectin-1.
Materials and Methods
Cells and Viruses. Vero cells and VD60 cells (Vero cells inducible for HSV-1 gD expression by infection with HSV) (12) were grown in DMEM supplemented with 10% FBS. Viral infections were performed in DMEM supplemented with 1% heat-inactivated FBS. CHO-K1 cells and CHO-K1 cells stably expressing human HVEM (13), nectin-1 (14), or nectin-2 (9) were grown in Ham's F12 medium supplemented with 10% FBS. CHO-K1 cells were transiently transfected with a plasmid expressing human 3-O-sulfotransferase-3B as described in ref. 15. HSV-1(KOS)tk12 is a recombinant virus that carries the lacZ gene in the viral thymidine kinase locus and expresses β-galactosidase on viral entry (9).
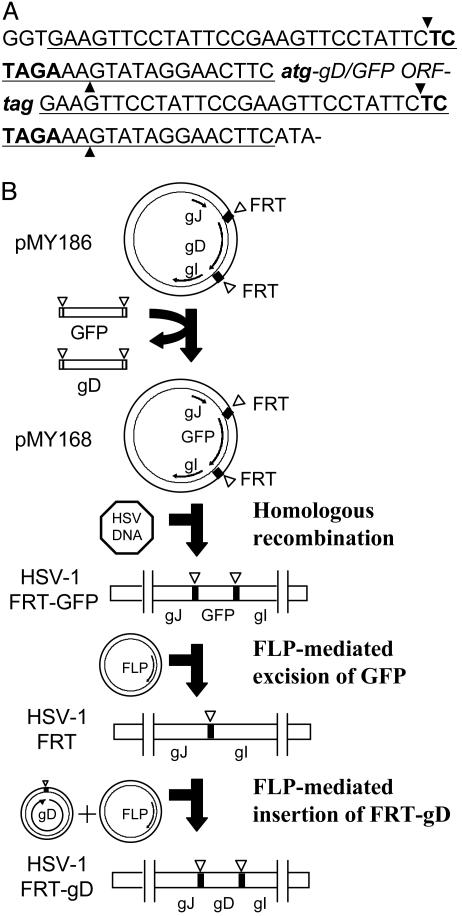
Plasmids. A segment of the HSV-1(KOS) genome, including the genes for glycoprotein J, gD, and glycoprotein I, was amplified by PCR and cloned into pUC19 between EcoRI and PstI sites to generate pMY165. Plasmid pMY186 (Fig. 1), containing two 47-bp FRT sites (8) immediately upstream and downstream of the gD ORF, was generated from pMY165 by using the QuikChange II site-directed mutagenesis kit (Stratagene). A GFP ORF, flanked by two 34-bp minimal FRT sites, was also generated by using pEGFP-N1 (Clontech) as a template to obtain pMY151. An XbaI site is present in both the 34- and 47-bp FRT sites. Plasmid pMY168 was generated by replacing the XbaI fragment containing the gD ORF by the XbaI fragment containing the GFP ORF. The XbaI fragment, containing the gD ORF flanked by partial FRT sites, was excised from pMY186 and subcloned into pUC19 to generate pMY171. Targeted mutations in gD were generated by using pMY171 as a template and the QuikChange II kit. All of the plasmids produced for this study were verified by DNA sequencing of the inserts. The plasmid encoding yeast FLP recombinase, pOG44, was purchased from Invitrogen.
Fig. 1.
Use of FLP recombinase and FRT sites to perform precise gene replacements in the HSV-1 genome and to generate HSV-1/FRT-gD. (A) Sequence of the FRT sites in pMY186 and pMY168. The 47-bp FRT site (underlined) contains an XbaI site (bold type). Strand cleavage by FLP recombinase occurs at the positions marked by arrowheads. The FRT sites are located immediately upstream and downstream of the start and stop codons (lowercase italic bold type) of the gD or GFP ORFs. (B) Plasmids containing the gD ORF (pMY186) or GFP ORF (pMY168) bounded by FRT sites and by regions of the HSV-1 genome normally found upstream and downstream of the gD ORF were constructed as described in Materials and Methods. The gD ORF in the HSV-1 genome was replaced by the GFP ORF, bounded by FRT sites by homologous recombination, to generate HSV-1/FRT-GFP. HSV-1/FRT was obtained after cotransfection of cells with the HSV-1/FRT-GFP genome and a plasmid expressing FLP recombinase, which excised the GFP ORF. HSV-1/FRT-gD was obtained by site-directed recombination after cotransfection of VD60 cells with the HSV-1/FRT genome, a plasmid expressing FLP recombinase and the WT gD ORF with ends joined by a single 47-bp FRT site.
Virus Construction. Recombinant viruses were obtained from cells cultured in six-well plates that had been cotransfected with HSV-1 genomic DNA and the appropriate plasmid DNAs by using Lipofectamine 2000 (Invitrogen) in serum-free medium for 6 h with a change to medium containing 10% FBS overnight. Cells were then incubated with medium containing 1% heatinactivated FBS for 5 days and harvested and sonicated to obtain virus stocks. HSV-1/FRT-GFP (Fig. 1) was isolated from VD60 cells cotransfected with HSV-1(KOS)tk12 DNA and pMY168. The desired recombinant virus obtained by homologous recombination was plaque-purified on the basis of GFP expression in complementing VD60 cells and the inability to form plaques on Vero cells. HSV-1/FRT was isolated from VD60 cells cotransfected with HSV-1/FRT-GFP DNA and pOG44 expressing FLP recombinase by screening for nonfluorescent plaques on VD60 cells. HSV-1/FRT-gD was isolated by site-specific recombination in Vero cells cotransfected with HSV-1/FRT DNA, pOG44, and the WT gD ORF excised from pMY186 by XbaI and then ligated to circularize the gD ORF and regenerate one 47-bp FRT site. Similarly, viruses containing defined mutations at positions 26, 28, and 29 in gD were isolated from Vero cells cotransfected with HSV-1/FRT DNA, pOG44, and the appropriate excised and circularized mutant gD ORF. The recombinant viruses were plaque-purified on Vero cells. Sequences of the gD ORFs were determined from four independent clones obtained by PCR amplification and TA cloning.
Construction and Screening of a Library of HSV-1 Recombinants Containing Random Mutations in gD. Random mutagenesis of the gD ORF was carried out by using pMY171 as a template and the Genemorph random mutagenesis kit (Stratagene). The primer pairs used were 5′-CATTTCGACTCTAGAAAGTATAGGAACTTCATG-3′ and 5′-GGTCTAGAGAATAGGAACTTCGGAATAGGAACTTCCTA-3′. The first and last codons of the gD ORF are shown in boldface, and the XbaI sites are underlined. The PCR products were digested with XbaI and circularized with T4 DNA ligase (New England Biolabs). The library of mutant viruses was generated by site-specific recombination in VD60 cells cotransfected with HSV-1/FRT DNA, pOG44, and circularized FRT-gD ORFs containing random mutations. A portion of the virus stock was amplified by passage on Vero cells to select for recombinants that had acquired a gD gene encoding a product capable of mediating entry into these cells. Dilutions of the virus stock obtained from Vero cells were plated on Vero cells in 96-well plates. Virus isolates were obtained from the wells of plates on which ≈50% of the wells remained uninfected. Culture supernatants from each well were passed at 1:10 dilution to a new well of Vero cells and then at 1:100 dilution to another new well. Virus isolates from 1,000 wells were amplified by passage on six-well plates of Vero cells and then screened on CHO-K1 cells expressing HVEM, nectin-1, nectin-2, or 3-O-sulfotransferase-3B. These cells in 96-well plates were inoculated with a 1:100 dilution of each virus isolate. After 6 h of incubation, the cells were fixed, permeabilized, and stained with X-Gal to identify infected cells. Virus isolates exhibiting enhanced or reduced usage of any of the entry receptors, in comparison with the WT virus, were subjected to two additional rounds of plaque purification. Sequences of the gD ORFs were determined from four independent clones obtained by PCR amplification and TA cloning.
Viral Entry Assay. The assay for viral entry is described in ref. 13. Briefly, CHO cells expressing one of the entry receptors and grown in 96-well plates were inoculated with serial dilutions of the recombinant viruses and incubated for 6 h. The cells were then washed, permeabilized, and incubated with the β-galactosidase substrate o-nitrophenyl β-d-galactopyranoside (ONPG, Sigma). The reaction was monitored at 410 nm to quantitate viral entry by using a Spectra Max250 plate reader (Molecular Devices).
Single-Step Replication Kinetics. Vero cells were inoculated at 5 plaque-forming units (pfu) per cell. After 2 h, the cells were washed and treated with 0.1 M sodium citrate buffer (pH 3.0) for 1 min to inactivate unpenetrated viruses, washed again, and incubated with fresh medium containing 1% heat-inactivated FBS. At different times after inoculation, the cells were scraped into the medium containing released viruses and lysed by sonication. Then virus titers were determined by plaque assay on Vero cells.
Results
Replacement of the HSV-1 gD ORF with an FRT Site and Site-Specific Recombination to Reinsert a WT gD ORF. Strain HSV-1(KOS)tk12, which expresses β-galactosidase as a reporter of viral entry, was the parental virus used for this study. The gD ORF in this strain was replaced by a single FRT site in a two-stage process (Fig. 1 and Materials and Methods). The gD-null virus obtained, HSV-1/FRT, was able to replicate in VD60 cells, which can express gD to complement the deficiency of a gD-negative virus, but not in Vero cells. The complemented virus stock used here had a titer of ≈108 pfu/ml on VD60 cells and <103 pfu/ml on Vero cells.
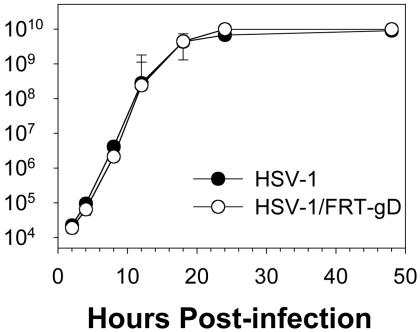
Reinsertion of a gD ORF by site-specific recombination results in the presence of two complete FRT sites flanking the gD ORF. To determine whether the FRT sites influenced viral replication, we isolated HSV-1/FRT-gD, which contains the WT form of the gD gene flanked by FRT sites (Fig. 1). HSV-1/FRT-gD replicated in Vero cells with kinetics and at levels indistinguishable from those of the parental virus strain without any FRT sites (Fig. 2).
Fig. 2.
Replication kinetics of HSV-1/FRT-gD and parental HSV-1 in Vero cells. Replicate cultures of cells were inoculated with 5 pfu per cell of each virus. Cell lysates were prepared at the times indicated after viral inoculation. Virus yields as a function of time after inoculation were determined by plaque assay on Vero cells with duplicate determinations. Similar results were obtained in two additional experiments.
Construction and Screening of a Library of HSV-1 Recombinants with Random Mutations in gD. PCR-generated random mutagenesis of the gD gene was carried out as described in Materials and Methods by using the GeneMorph kit (Stratagene) with conditions set to yield 1–2 mutations per 1,000 bp. The gD ORF is 1,185 bp. The mutagenized PCR products were digested with XbaI and ligated to obtain circularized gD ORFs with ends joined by a single complete FRT site. A library of viral recombinants was produced by site-specific recombination in VD60 cells cotransfected with HSV-1/FRT DNA, the FLP recombinase expression plasmid, and the library of mutagenized and circularized gD ORFs. Samples of the library were plated on Vero cells and on VD60 cells to determine what fraction of the progeny obtained had acquired a gD gene capable of mediating entry into Vero cells. This fraction was 0.33% of the library compared with 1.2% for the WT gD gene transferred to the HSV-1/FRT genome under similar conditions, indicating that ≈75% of gD-positive viral progeny in the library had mutations that precluded infection of Vero cells. A portion of the mutagenized library was passaged once on Vero cells. Then virus isolates (n = 1,000) were purified on Vero cells by limiting dilution and screened for mutants altered with respect to viral entry mediated by HVEM, nectin-1, nectin-2, or 3-O-S HS. Ten such mutants, 1% of the viruses screened, were identified, further plaque-purified, and characterized.
Sequencing of their gD genes revealed that nine mutants had nucleotide substitutions causing a single amino acid change, whereas the 10th had mutations causing two amino acid substitutions and a deletion (Table 1). All of the mutants were essentially unaltered with respect to use of nectin-1 as an entry receptor (Fig. 3 and Table 1), suggesting that selection of the mutants on Vero cells depended on their ability to use nectin-1 for entry, despite the evidence that HVEM is also expressed. The mutants fell into three categories on the basis of their phenotypes with other receptors (Fig. 3 and Table 1). Class I mutants with amino acid substitutions N15E, Q132P, or K190P had reduced ability to use HVEM and 3-O-S HS for entry but were not significantly different from WT with respect to entry by nectin-1 or nectin-2. Class II mutants (M11P, P14R, D30P, or G43P) were similar to class I mutants except that no entry by HVEM could be detected. Class III mutants (Q27P, Q27A, or D26R/ΔL28/T29P) differed from the class II mutants only in that entry through nectin-2 was significantly enhanced. Remarkably, all of the mutations reduced or abolished entry through both HVEM and 3-O-S HS, indicating that similar features of gD are critical for functional interactions with both these receptors but not with nectin-1.
Table 1. Summary of HSV-1 gD mutations and their effects on entry through various HSV entry receptors.
| Viral entry*
|
||||||
|---|---|---|---|---|---|---|
| Mutant class | Codon changes | Amino acid changes | HVEM | Nectin-1 | Nectin-2 | 3-O-S HS |
| None | None | +++ | +++ | +/- | ++ | |
| I | AAT to GAA | N15E | ++ | +++ | +/- | +/- |
| CAG to CCG | Q132P | + | +++ | - | - | |
| AAG to CCG | K190P | + | +++ | - | - | |
| GAC to CGC | D26R† | ++ | +++ | +/- | - | |
| CTG to CGG | L28R† | + | +++ | - | - | |
| II | ATG to CCG | M11P | - | +++ | +/- | + |
| CCC to CGC | P14R | - | +++ | +/- | + | |
| GAC to CCC | D30P | - | +++ | - | - | |
| GGC to CCC | G43P | - | +++ | - | - | |
| CTG to - - - | ΔL28† | - | +++ | - | - | |
| CTG to GGG | L28G† | - | +++ | - | - | |
| CTG to CCG | L28P† | - | +++ | - | - | |
| ACC to CCC | T29P† | - | +++ | - | - | |
| III | CAG to CCG | Q27P | - | +++ | +++ | - |
| CAG to GCG | Q27A | - | +++ | +++ | - | |
| GAC CAG | D26R/Δ28/T29P | - | +++ | ++ | - | |
| CTG ACC to CGC CAG - - - CCC | ||||||
| CTG to GCG | L28A† | - | +++ | ++ | - | |
β-Galactosidase activity was quantitated as a measure of viral entry, and virus dose was plotted against β-galactosidase activity (Figs. 3 and 4). The values for the symbols used are as follows, with OD at 410 nm of 1.0-2.5, +++; 0.5-1.0, ++; 0.3-0.5, +; 0.3, +/-; and <0.3, - at input virus doses of 106-107 pfu per well.
Recombinant HSV-1 viruses were generated after site-directed mutagenesis at positions 26, 28, or 29 in gD.
Fig. 3.
Entry of parental HSV-1 and the HSV-1 mutants obtained by random mutagenesis into CHO-K1 cells expressing (from left to right) HVEM, nectin-1, nectin-2, or 3-O-sulfotransferase-3B (which generates 3-O-S HS). Cells in 96-well plates (5 × 104 cells per well) were inoculated with serial dilutions of the β-galactosidase-expressing viruses. Input doses of virus are indicated. Effective viral doses (amount of virus that actually makes contact with a cell) are much less than the input because of the necessity of using a relatively large inoculum per monolayer surface area in the small wells. After 6 h, the cells were washed and lysed for the quantitation of β-galactosidase activity as a measure of viral entry. Values plotted are OD at 410 nm of the o-nitrophenyl β-d-galactoside reaction product as a function of viral dose. The means of triplicate determinations with SDs for one representative experiment are shown. Similar results were obtained in two additional experiments. Mutants belonging to class I, II, or III (Table 1) are shown in Top, Middle, and Bottom, respectively.
None of these particular mutations has been described previously except Q27P and Q27A (3, 4, 9). The phenotypes reported here are consistent with results presented in these earlier reports. Mutations at L25 (L25A and L25P) were shown to reduce, but not abolish, activity with HVEM (3, 4). This is the only position at which mutations were previously shown to affect activity with HVEM but not altered among the mutants described here.
Separation of the Mutations in the Triple Mutant. To determine which mutations in D26R/ΔL28/T29P were responsible for the phenotype observed, each mutation was separately introduced into the gD gene. In addition, several different amino acid substitutions at position 28 were engineered. The mutant gD genes were inserted into the HSV-1/FRT genome as described above. Fig. 4 and Table 1 show that, whereas all three single mutants (D26R, ΔL28, and T29P) differed from WT HSV-1, none had the profile of entry activities characteristic of the triple mutant. The various substitutions at position L28 also affected entry through one or more receptors. These mutants fell into the three already described classes as follows: class I, D26R and L28R; class II, Δ28, L28G, L28P, and T29P; and class III, L28A. L28A was described as retaining some activity with HVEM (4), which result differs from ours.
Fig. 4.
Entry of parental HSV-1, and the HSV-1 mutants generated by targeted mutagenesis and site-specific gene transfer, into CHO cells expressing HVEM (Upper Left), nectin-1 (Upper Right), nectin-2 (Lower Left), and 3-O-sulfotransferase-3B (Lower Right). Methods and data presentation are as described in the Fig. 3 legend.
Discussion
The HSV-1 library, randomly mutagenized in the gD gene, was subjected first to selection on Vero cells to obtain virus isolates that were capable of using at least one viral entry receptor and that retained functions of gD, aside from receptor binding, required for viral entry (such as interactions with the other viral glycoproteins required for viral entry). The isolates were then screened for the loss of ability to use each of the known HSV entry receptors. Although Vero cells express monkey versions of nectin-1 and HVEM and both are functional HSV-1 entry receptors (10, 11), all of the mutants obtained retained full entry activity with nectin-1 but not HVEM. This finding suggests that HVEM is not a functional entry receptor in Vero cells or that HSV-1 mutants capable of using HVEM for entry, but not nectin-1, may be rarer than the converse. In fact, the only previously described HSV-1 mutants that were fully functional for interactions with HVEM but not nectin-1 had specific double or triple amino acid substitutions in gD (5, 6) and will be discussed further below.
The phenotypes resulting from the randomly generated gD mutations will be considered here in the context of x-ray structures providing evidence that the N terminus of HSV-1 gD is conformationally flexible (7). In the structure of gD crystallized alone, the N terminus was extended, with the first 15 aa disordered. In the structure of gD cocrystallized with HVEM, the entire N terminus of gD was ordered and folded into a hairpin. All contacts with HVEM were within this hairpin. Arguments can be made, as outlined below, that the N-terminal hairpin conformation is critical for functional interactions of gD with HVEM (and perhaps 3-O-S HS), whereas a different conformation of the N terminus is critical for functional interactions of gD with nectin-1 (and perhaps nectin-2).
All of the mutants described here had little or no ability to enter cells through HVEM. It is not surprising that eight of the amino acids substituted or deleted (M11, P14, N15, D26, Q27, L28, T29, and D30) are in the N-terminal HVEM contact regions (shown in yellow or red in Fig. 5). In most cases, the random mutations described here resulted in nonconservative amino acid replacements. Targeted Ala substitutions at these positions also reduced physical or functional interactions with HVEM for all positions listed above except P14 and D26 (4). D26 has numerous side-chain contacts with HVEM, whereas P14 does not (7). The D26R substitution described here would undoubtedly disrupt these contacts, whereas the P14R substitution may have larger effects on stability of the hairpin. In the gD–HVEM complex, this hairpin is sandwiched between HVEM and an α-helix downstream in gD. Stability of the hairpin is probably dependent on intramolecular interactions within gD, involving both arms of the hairpin and the α-helix, and on interactions with HVEM. Four of the gD residues replaced in the mutants described here (M11, N15, D26, and T29) make numerous side-chain contacts with HVEM (7).
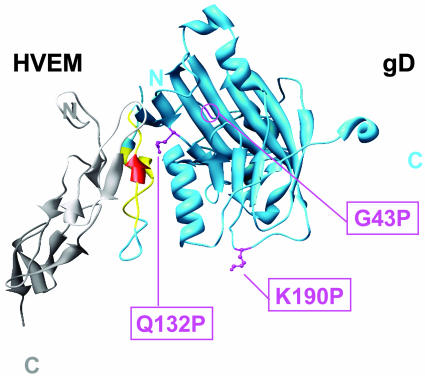
Fig. 5.
Locations of the critical mutations on the structure of gD (blue) cocrystallized with HVEM (gray). Most of the mutations that reduced or abolished entry activity with HVEM and 3-O-S HS are in the N-terminal hairpin of gD (yellow or red). The mutations that also enhanced entry activity with nectin-2 are at positions 27 or 28 (red). Three additional mutations (G43P, Q132P, and K190P; pink) also reduced entry activity with HVEM and 3-O-S HS. The structure shown is based on coordinates deposited in the Protein Data Bank (21) for entry 1JMA (7). This molecular graphic image was produced by using the chimera package (22) from the Computer Graphics Laboratory, University of California, San Francisco (supported by National Institutes of Health Grant P41 RR-01081).
Three of the mutations described here (G43P, Q132P, and K190P) are not in a known contact region with HVEM and yet reduced or abolished entry activity with HVEM without affecting entry through nectin-1 (shown in pink in Fig. 5). Two of these mutations, G43P and Q132P, may indirectly affect the conformation of the N-terminal HVEM-binding hairpin. The G43P substitution could introduce a kink in the loop between the N-terminal hairpin and an Ig-like fold. An insertion of 5 aa after G43 was shown to cause a phenotype similar to that of the G43P substitution (16). The Q132 side chain, extending from the Ig fold, is buried under the N-terminal hairpin. The Q132P substitution could be expected to alter the conformation of neighboring regions of the molecule and also would not provide a side chain perhaps needed to stabilize the N-terminal hairpin. The third mutation (K190P) is in a loop downstream of the Ig fold and distant from the N-terminal hairpin. It is unclear why this mutation would reduce functional interactions with HVEM. Possibly, there are long-distance effects on conformation of the N terminus, perhaps involving a large region of the gD ectodomain (amino acids 260–316) whose structure has yet to be resolved. Alternatively, binding to HVEM occurs but subsequent conformational changes necessary to initiate membrane fusion are blocked by the K190P mutation. If the latter is true, this finding implies that K190 is not critical for such conformational changes after nectin-1 binding.
It is of interest that all of the mutants described here also have reduced or no activity with 3-O-S HS, consistent with previous analyses of other gD mutants having substitutions or deletions of the HVEM contact residues (3). From the properties of heparan sulfate, it could be predicted that basic residues in gD would be critical for interaction with 3-O-S HS, but only one of the mutations (K190P) altered a basic residue. When the N terminus of gD assumes its hairpin conformation, however, there are clusters of basic residues positioned at both ends (7) that could interact with 3-O-S HS. We propose that the N-terminal hairpin conformation permits 3-O-S HS to interact with its contact residues, which have not yet been identified but are probably different from the critical HVEM contact residues.
None of the mutations described here impaired functional interactions of gD with nectin-1. Deletions of amino acids 7–32 (or subsets thereof) in either HSV-1 or HSV-2 gD also permitted full retention of functional interactions with nectin-1 (3). Clearly, the N terminus of gD is dispensable for entry through nectin-1. Double and triple amino acid substitutions at positions D215, R222, and F223 in gD abrogated functional interactions with nectin-1 and nectin-2 but did not diminish function with HVEM and 3-O-S HS (5). Amino acid substitutions at Y38 also abrogated function with nectin-1 and reduced function with HVEM; interestingly, a double mutation (A3C/Y38C), designed to clamp the N terminus into its hairpin conformation by disulfide bonding, abrogated function with nectin-1 but permitted full activity with HVEM (6). Amino acids Y38, D215, R222, and F223 are located on one surface of gD that could serve as an interface for the nectins, provided the N terminus assumes some conformation other than the hairpin found in the gD–HVEM complex (5, 6).
Nectin-2 is an entry receptor for HSV-2, but not WT HSV-1, because of several amino acid differences within the first 53 aa of gD (17). Also, certain amino acid substitutions in HSV-1 gD, as described here and elsewhere (3, 4, 9), conferred the ability to use nectin-2 as an entry receptor. Nectin-2 is a less active entry receptor than the related nectin-1, but equivalent regions are critical for functional interactions with gD (18–20). It seems likely that, for both, the major contacts with gD are as proposed in the preceding paragraph. However, functional interactions with nectin-2, but not nectin-1, require the first 32 aa of gD (3) and depend on the sequence of the N terminus. We propose that the N terminus of gD can assume a conformation permitting interactions with other regions of gD necessary to enable a stable association with nectin-2 or that the N terminus itself makes secondary contacts with nectin-2 necessary to stabilize the primary contacts.
The method of precise gene replacement described here, coupled with PCR-generated mutagenesis of the gD ORF, has provided a means to identify new mutations that reduce or abolish entry of HSV-1 through HVEM or 3-O-S HS or to enhance entry through nectin-2 without demonstrable effect on entry through nectin-1. By selecting for viral mutants on cell lines offering only HVEM, nectin-2, or 3-O-S HS as entry receptors, it should be possible to obtain additional panels of mutants with different constellations of receptor preferences. These mutants will be invaluable for defining more precisely the interactions of gD with each of the HSV entry receptors and for assessing the roles of each in viral entry and viral pathogenesis.
Acknowledgments
We thank Nanette Susmarski (Northwestern University) for excellent technical assistance and the authors of ref. 6 for permission to cite their article prior to its publication. This work was supported by U.S. Public Health Service Grants AI-31494, AI-36293, and AI-53774.
Author contributions: M.Y. and P.G.S. designed research; M.Y. performed research; M.Y. and P.G.S. analyzed data; and M.Y. and P.G.S. wrote the paper.
Abbreviations: FRT, FLP recombination target; gD, glycoprotein D; HSV-1, herpes simplex virus type 1; HVEM, herpesvirus entry mediator; 3-O-S HS, 3-O-sulfated heparan sulfate; pfu, plaque-forming units.
References
- 1.Spear, P. G., Eisenberg, R. J. & Cohen, G. H. (2000) Virology 275, 1–8. [DOI] [PubMed] [Google Scholar]
- 2.Spear, P. G. & Longnecker, R. (2003) J. Virol. 77, 10179–10185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Yoon, M., Zago, A., Shukla, D. & Spear, P. G. (2003) J. Virol. 77, 9221–9231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Connolly, S. A., Landsburg, D. J., Carfi, A., Wiley, D. C., Cohen, G. H. & Eisenberg, R. J. (2003) J. Virol. 77, 8127–8140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Manoj, S., Jogger, C. R., Myscofski, D., Yoon, M. & Spear, P. G. (2004) Proc. Natl. Acad. Sci. USA 101, 12414–12421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Connolly, S. A., Landsburg, D. J., Carfi, A., Whitbeck, J. C., Zuo, Y., Wiley, D. C., Cohen, G. H. & Eisenberg, R. J. (2005) J. Virol., in press. [DOI] [PMC free article] [PubMed]
- 7.Carfi, A., Willis, S. H., Whitbeck, J. C., Krummenacher, C., Cohen, G. H., Eisenberg, R. J. & Wiley, D. C. (2001) Mol. Cell 8, 169–179. [DOI] [PubMed] [Google Scholar]
- 8.Andrews, B. J., Proteau, G. A., Beatty, L. G. & Sadowski, P. D. (1985) Cell 40, 795–803. [DOI] [PubMed] [Google Scholar]
- 9.Warner, M. S., Geraghty, R. J., Martinez, W. M., Montgomery, R. I., Whitbeck, J. C., Xu, R., Eisenberg, R. J., Cohen, G. H. & Spear, P. G. (1998) Virology 246, 179–189. [DOI] [PubMed] [Google Scholar]
- 10.Foster, T. P., Chouljenko, V. N. & Kousoulas, K. G. (1999) Virology 258, 365–374. [DOI] [PubMed] [Google Scholar]
- 11.Milne, R. S. B., Connolly, S. A., Krummenacher, C., Eisenberg, R. J. & Cohen, G. H. (2001) Virology 281, 315–328. [DOI] [PubMed] [Google Scholar]
- 12.Ligas, M. W. & Johnson, D. C. (1988) J. Virol. 62, 1486–1494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Montgomery, R. I., Warner, M. S., Lum, B. J. & Spear, P. G. (1996) Cell 87, 427–436. [DOI] [PubMed] [Google Scholar]
- 14.Geraghty, R. J., Krummenacher, C., Cohen, G. H., Eisenberg, R. J. & Spear, P. G. (1998) Science 280, 1618–1620. [DOI] [PubMed] [Google Scholar]
- 15.Shukla, D., Liu, J., Blaiklock, P., Shworak, N. W., Bai, X., Esko, J. D., Cohen, G. H., Eisenberg, R. J., Rosenberg, R. D. & Spear, P. G. (1999) Cell 99, 13–22. [DOI] [PubMed] [Google Scholar]
- 16.Jogger, C. R., Montgomery, R. I. & Spear, P. G. (2004) Virology 318, 318–326. [DOI] [PubMed] [Google Scholar]
- 17.Zago, A. & Spear, P. G. (2003) J. Virol. 77, 9695–9699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Martinez, W. M. & Spear, P. G. (2001) J. Virol. 75, 11185–11195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Martinez, W. M. & Spear, P. G. (2002) J. Virol. 76, 7255–7262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Struyf, F., Martinez, W. M. & Spear, P. G. (2002) J. Virol. 76, 12940–12950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Berman, H. M., Westbrook, J., Feng, Z., Gilliland, G., Bhat, T. M., Weissig, H., Shindyalov, I. N. & Bourne, P. E. (2000) Nucleic Acids Res. 28, 235–242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Huang, C. C., Couch, G. S., Pettersen, E. F. & Ferrin, T. E. (1996) chimera, An Extensible Molecular Modeling Application Constructed Using Standard Components (Computer Graphics Lab., Univ. of California, San Francisco).