Abstract
Background
Almost all hepatic cancer cells have resistance to tumor necrosis factor-related apoptosis-inducing ligand (TRAIL)-induced apoptosis. c-FLIPL and RIP-1 are apoptotic negative regulatory factors. This study investigated the role of c-FLIPL and RIP-1 in hepatic cancer cell resistance to TRAIL-induced apoptosis.
Material/Methods
HepG2 cells were treated by TRAIL, RIP-1 siRNA, and/or BY11-7082. Cell viability was detected by MTT assay. Cell apoptosis was tested by flow cytometry. DISC component proteins, RIP-1, and p-p65 were measured by Western blot. Caspase-8 and caspase-3 were determined by spectrophotometry.
Results
Single TRAIL treatment showed no significant impact on cell proliferation and apoptosis. HepG2 cells expressed high levels of RIP1 and c-FLIPL, while a high concentration of TRAIL upregulated RIP-1 and c-FLIPL expression but not DR4 and DR5. Single TRAIL treatment did not obviously activate caspase-8 and caspase-3. RIP-1 or c-FLIPL siRNA markedly induced cell apoptosis and enhanced caspase-8 and caspase-3 activities. Combined transfection obviously increased apoptotic cells. TRAIL markedly upregulated RIP-1 expression and enhanced p-p65 protein. Downregulating RIP-1 and/or BAY11-7082 significantly reduced NF-κB transcriptional activity, blocked cells in G0/G1 phase, weakened proliferation, elevated caspase-8 and caspase-3 activities, and promoted cell apoptosis.
Conclusions
TRAIL can enhance RIP1 and c-FLIPL expression in HepG2 cells. High expression of RIP1 and c-FLIPL is an important reason for TRAIL resistance. Downregulation of RIP1 and c-FLIPL can relieve caspase-8 suppression, activate caspase-3, and promote cell apoptosis. TRAIL mediates apoptosis resistance through upregulating RIP-1 expression, enhancing NF-κB transcriptional activity, and weakening caspase activity.
MeSH Keywords: Airway Resistance, Apoptosis, Liver Neoplasms, Trail Making Test
Background
Primary hepatic cancer is a common malignant tumor in clinic. In incidence, it is the fifth among malignant tumors, and is the third leading cause of mortality in cancer [1]. Hepatocellular carcinoma (HCC) is the main pathological type, which accounts for more than 85~90% in all primary hepatic cancers [2]. There are numerous hepatitis B virus carriers in our country that account for about 10% of the world, leading to the incidence of up to 52/100,000. There are more than 600,000 new cases around the world every year, while China has more than 340,000, which amounts to more than 55%. Hepatic cancer causes serious harm to the patient’s life and health [3]. Tumor necrosis factor-related apoptosis-inducing ligand (TRAIL) is considered to be one of a new generation of highly selective antineoplastic drugs that can induce tumor cell apoptosis but does not significantly damage normal cells [4]. However, almost all of the HCC cell lines present different degrees of resistance to TRAIL-induced cell apoptosis. After surgical resection, the minimal residual hepatic cancer cells have resistance to TRAIL-induced cell apoptosis as shown by exhibiting drug insensitivity and easy metastasis and recurrence [5]. c-FLIPL and RIP-1 are the two extremely important negative regulatory factors of death-inducing signaling complex (DISC) and apoptosis signaling transduction [6]. It was found that c-FLIPL and/or RIP-1 are highly expressed in hepatic cancer cells [7,8]. Therefore, we hypothesized that RIP-1 and/or c-FLIPL abnormal expression induced DISC formation and apoptosis signaling transduction blockage, which may be the important mechanism of hepatic cancer cell resistance to TRAIL-induced cell apoptosis. This study explored the role and potential mechanism of RIP-1 and/or c-FLIPL in hepatic cancer cell resistance to TRAIL-induced apoptosis.
Material and Methods
Main reagents and materials
HepG2 cells were purchased from the American Type Culture Collection (ATCC). High glucose Dulbecco’s Modified Eagle’s medium (DMEM), fetal bovine serum (FBS), and penicillin-streptomycin were obtained from Gibco. Human recombinant TRAIL was from Peprotech. Trizol and Lipofectamine 2000 were from Invitrogen. PrimeScript RT reagent kit was from Takara. SYBR Green real time PCR kit was from Toyobo. PCR primers were designed and synthetized by Genepharma. Rabbit anti-human c-FLIP antibody, mouse anti-human RIP-1 antibody, and rabbit anti-human phosphor-NF-κB p65 (S536) antibody were from Abcam. Mouse anti-human death receptor 4 (DR4) and DR5 antibodies were from Santa Cruz. Rabbit anti-human cleaved caspase-8 and cleaved caspase-3 antibody were from CST. HRP-coupled goat anti-mouse and goat anti-rabbit secondary antibodies were from Jackson. BAC protein quantification kit, Annexin V/PI apoptosis reagent, and caspase-8 and caspase-3 activities detection kit were from Beyotime. NF-κB specific inhibitor BAY11-7082 was from Selleck.
Cell culture and TRAIL treatment
HepG2 cells were cultured in high glucose DMEM supplemented by 10% FBS, penicillin 100 U/mL, and streptomycin 100 μg/mL, and maintained at 37°C and 5% CO2. The cells in 80% fusion were passaged or used for experiments. The cells were treated by different concentrations of TRAIL (0, 5, 10, 20, 50, and 100 ng/mL) for 24 h.
MTT assay
The cells in logarithmic phase were digested by enzyme and seeded in a 96-well plate at 2000 cells/well. After 24 h, the cells were treated by different concentrations of TRAIL (0, 5, 10, 20, 50, and 100 ng/mL) for 24 h. After addition of 10 μL of MTT for 4 h, the cells were treated with 150 μL of dimethyl sulfoxide (DMSO) for 15 min. At last, the plate was read at 490 nm to test absorbance. Each group was repeated with six replicates. Cell proliferation rate (%)=experimental group mean value/control mean value ×100%.
Cell apoptosis
The cells were collected after digestion and centrifuged at 1000 g for 5 min. The cells were resuspended in 195 μL of Annexin V-FITC buffer and 5 μL of Annexin V-FITC. After mixing, 10 μL of propidium iodide (PI) was added to the cells, and they were incubated at room temperature avoiding light for 10~20 min. At last, the cells were tested by flow cytometry.
siRNA transfection
c-FLIP and RIP-1 siRNA were transfected to the cells. The cells were used for transfection when the density reached 50~60%. siRNA and Lipofectamine 2000 were diluted by Opti-MEM and incubated for 5 min at room temperature, respectively. Then they were gently mixed and added to the cells for 20 min at room temperature. Next, the cells were cultured in an incubator for 6 h, and the mediums were changed. After 48 h, the cells were treated by TRAIL at 100 ng/mL for 24 h and collected for the following experiments. The sequences were listed as follows:
si c-FLIP forward, 5′-GCAGUCUGUUCAAGGAGCATT-3′.
si c-FLIP reverse, 5′-UGCUCCUUGAACAGACUGCTT-3′.
c-FLIP NC forward, 5′-UUCUCCGAACGUGUCACGUTT-3′.
c-FLIP NC reverse, 5′-ACGUGACACGUUCGGAGAATT-3′.
si RIP-1 forward, 5′-GCAAAGACCUUACGAGAAUTT-3′.
si RIP-1 reverse, 5′-AUUCUCGUAAGGUCUUUGCTT-3′.
RIP-1 NC forward, 5′-TTCTCCGAACGTGTCACGTTT-3′.
RIP-1 NC reverse, 5′-ACGTGACACGTTCGGAGAATT-3′.
The cells were divided into five groups, including control: c-FLIP NC group, si c-FLIP group, RIP-1 NC group, si RIP-1 group, and si c-FLIP + si RIP-1 group.
qRT-PCR
Total RNA was extracted using the Trizol method and quantified on an Eppendorf protein nucleic acid detector. Then the RNA was reverse transcripted to cDNA using the ReverTra Ace RT Kit. The reverse transcription system in 20 μL contained 2 μL of total RNA, 1 μL of dNTP (10 mmol/L), 4 μL of RT buffer (5×), 2 μL of RT primer (1 μmol/L), 1.5 μL of reverse transcriptase, 0.5 μL of RNase inhibitor, and ddH2O. Reverse transcription was performed at 16°C for 30 min, 42°C for 15 min, and 85°C for 5 min. The cDNA was stored at −20°C. Then the cDNA was used for the PCR reaction, and the primers used were as follows:
RIP-1PF: 5′-GCACTGTTGTGACTCGTTGG-3′;
RIP-1PR: 5′-GACACCCGACCATACTTTCAG-3′;
c-FLIPLPF: 5′-GTCTGCTGAAGTCATCCATC-3′;
c-FLIPLPR: 5′-ACTACGCCCAGCCTTTTGG-3′;
β-actinPF: 5′-GAACCCTAAGGCCAAC-3′;
β-actinPR: 5′-TGTCACGCACGATTTCC-3′.
The PCR reaction system in 10 μL contained 4.5 μL of 2×SYBR Green Mixture, 0.5 μL of primer (2.5 μm/L), 1 μL of cDNA, and 3.5 μL of ddH2O. The PCR reaction was performed on an ABI ViiA7 amplifier at 40 cycles of 95°C for 15 s, 60°C for 30 s, and 74°C for 30 s. U6 and β-actin were adopted as internal references for miRNA and mRNA. Each sample was repeated three times. The comparative Ct method (2−ΔΔCT) was applied for quantitative analysis.
Western blot
Total protein was extracted and quantified by the BCA method. A total of 40 μg of protein was separated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and transferred to PVDF membrane. After blocking in 5% skim milk at room temperature for 60 min, the membrane was incubated in primary antibody at 4°C overnight. After washing with phosphate buffered saline with Tween 20 (PBST) three times, the membrane was further incubated in HRP-tagged secondary antibody at room temperature for 60 min. At last, the membrane was treated with ECL chemiluminiscence and scanned on Epson to collect data. The band was analyzed by Image J. Relative protein level = target band gray value/β-actin band gray value.
Spectrophotometry detection of caspase-8 and caspase-3 activity
Caspase-8 and caspase-3 activity was detected using the kit according to the manual. pNA (10 mM) provided by the kit was diluted to 0, 10, 20, 50, 100, and 200 μM as the standard substance. The standard substance was tested at 405 nm to prepare the standard curve. The cells were digested by enzyme and centrifuged at 600 g and 4°C for 5 min for collection. After washing with PBS, the cells were treated by lysis at 100 μL/2,000,000 cells on ice for 15 min. After centrifugation at 18,000 g and 4°C for 15 min, the supernatant was moved to the pre-cooled Ep tube to test caspase-3 activity. The cells were treated with 10 μL of Ac-DEVD-pNA (2 mM) and incubated at 37°C for 120 min. At last, the cells were read at 405 nm to measure the concentration.
Cell cycle
The cells were digested by 0.25% enzyme and 70% ethanol was added at 4°C overnight. After washing twice with PBS, the cells were resuspended in 500 μL of PBS and treated with Rnase A 50 μg/mL at 37°C for 30 min. Next, the cells were incubated on ice for 5 min to stop the reaction. Then the cells were treated by 0.1% Triton X-100 for 30 min, and PI 100 μg/mL was added for staining at 4°C for 30 min. At last, the cells were detected by flow cytometry to calculate G0/G1, S, and G2/M cell ratio.
Statistical analysis
All data analysis was performed with SPSS 18.0 software. Measurement data are presented as mean ± standard deviation. One-way analysis of variance (ANOVA) was applied for group comparison. P<0.05 was considered as statistical significance.
Results
TRAIL showed no significant impact on cell apoptosis and proliferation
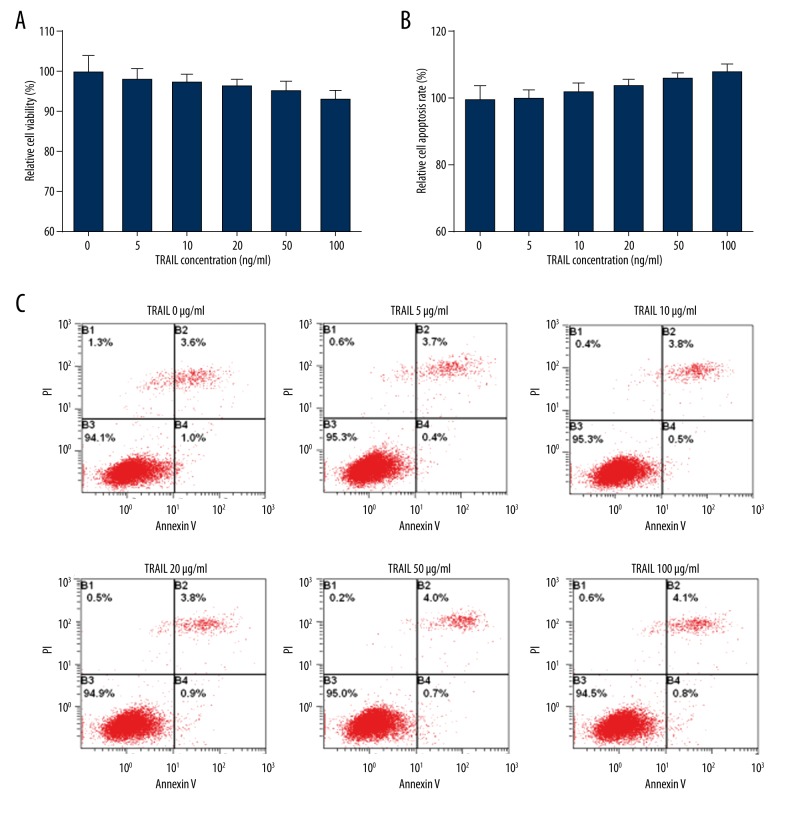
MTT assay showed that single TRAIL treatment slightly reduced the HepG2 cell survival rate following concentration increase (P>0.05), suggesting that HepG2 cells have resistance to TRAIL (Figure 1A). Flow cytometry revealed that cell apoptosis did not present obvious changes (P>0.05), indicating that HepG2 cells show resistance to TRAIL-induced cell apoptosis (Figure 1B, 1C).
Figure 1.
TRAIL showed no significant impact on cell apoptosis and proliferation. (A) MTT assay detection of cell proliferation. (B) Relative cell apoptosis rate. (C) Flow cytometry detection of cell apoptosis.
TRAIL induced RIP-1 and c-FLIPL elevation in DISC
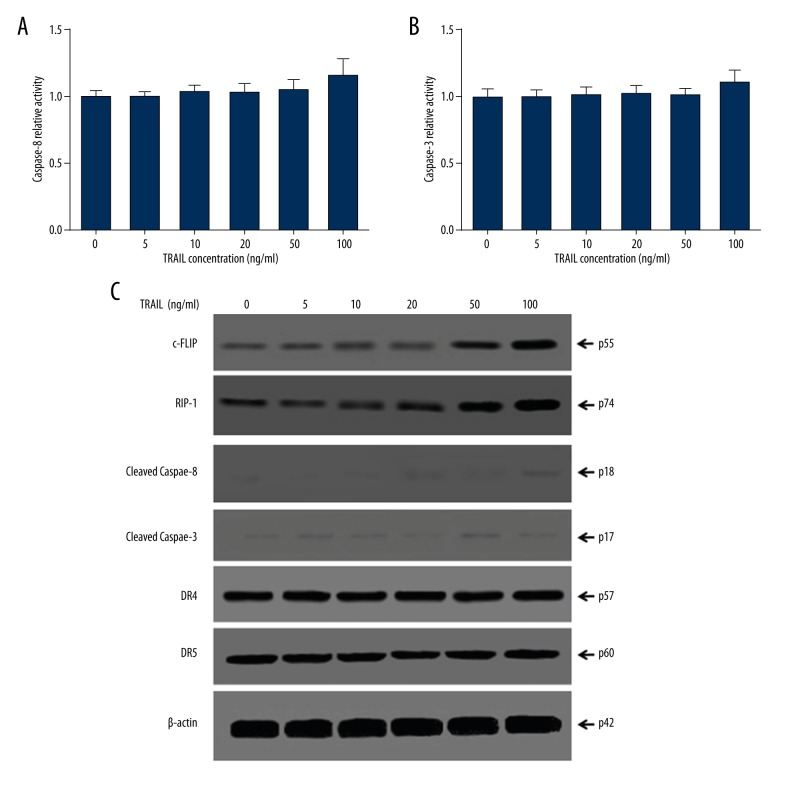
Caspase-8 and caspase-3 activity in DISC was observed after TRAIL treatment. Spectrophotometry demonstrated that compared with control, different concentrations of TRAIL failed to effectively enhance caspase-8 and caspase-3 activity in HepG2 cells (P>0.05), suggesting that TRAIL did not produce a significant impact on caspase-3 activation and triggering apoptosis (Figure 2A, 2B). HepG2 cells expressed a high abundance of RIP-1 and c-FLIPL proteins. Low concentrations (5, 10, 20 μg/mL) of TRAIL did not affect protein expression, while high concentrations (>50 μg/mL) of TRAIL markedly upregulated RIP-1 and c-FLIPL protein expression in cells and reached the top at 100 μg/mL (Figure 2C). We failed to detect cleaved caspase-8 and cleaved caspase-3 protein expression, which may be related to high abundance of RIP-1 and c-FLIPL protein expression in HepG2 cells. TRAIL treatment showed no significant impact on DR4 and DR5 expression.
Figure 2.
TRAIL-induced RIP-1 and c-FLIPL elevation in DISC. (A) Spectrophotometry detection of caspase-8 activity. (B) Spectrophotometry detection of caspase-3 activity. (C) Western blot detection of proteins in DISC.
si RIP-1 and si c-FLIP obviously downregulated RIP-1 and c-FLIPL protein expression
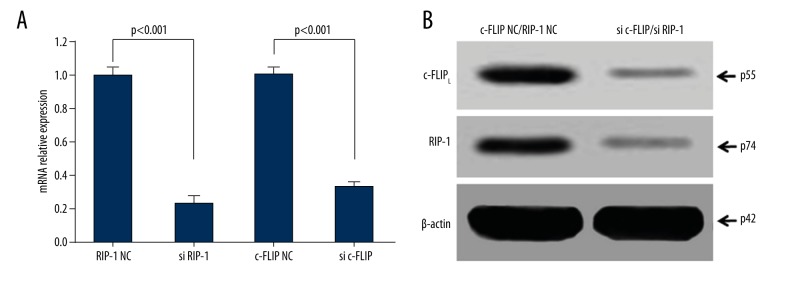
Western blot showed that RIP-1 and c-FLIPL mRNA and protein expression markedly declined after si RIP-1 and si c-FLIP transfection (Figure 3A, 3B), suggesting good transfection efficacy.
Figure 3.
si RIP-1 and si c-FLIP obviously downregulated RIP-1 and c-FLIPL protein expression. (A) qRT-PCR detection of RIP-1 and c-FLIPL mRNA expression. (B) Western blot detection of RIP-1 and c-FLIPL protein expression.
RIP-1 and c-FLIPL interference promoted cell apoptosis
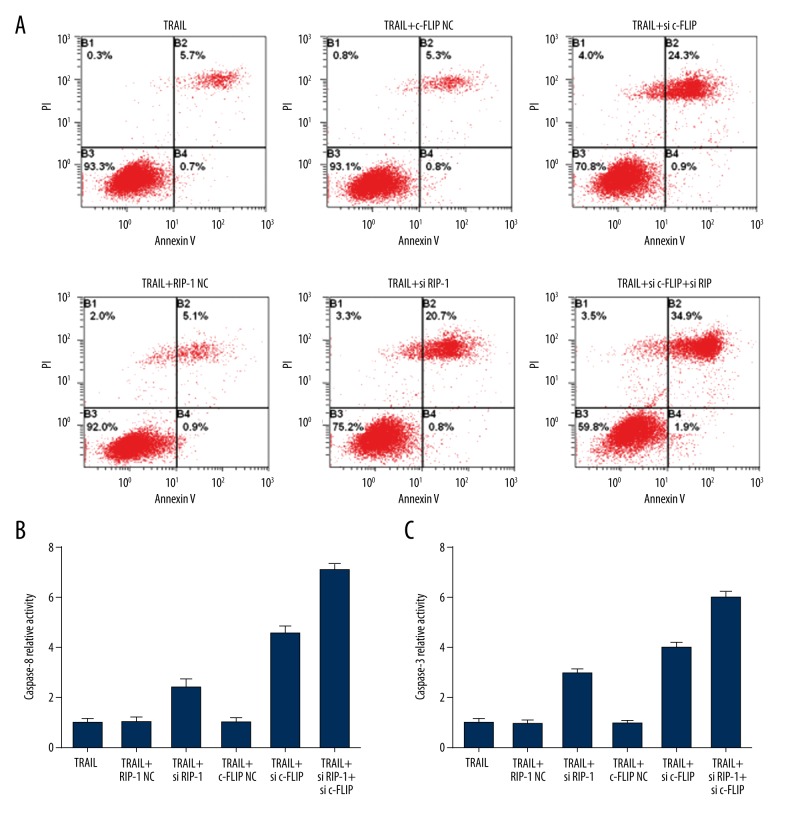
HepG2 cells showed a high abundance of RIP-1 and c-FLIPL, while a high concentration of TRAIL obviously induced RIP-1 and c-FLIPL expression. Therefore, this study discusses the role of RIP-1 and c-FLIPL in HepG2 cell resistance to TRAIL-induced apoptosis. Compared with a single 100 ng/mL TRAIL treatment, RIP-1 NC or c-FLIP NC transfection failed to enhance cell apoptosis. si RIP-1 or si c-FLIP transfection obviously elevated cell apoptosis, and combined transfection further increased cell apoptosis, indicating that interfering with RIP-1 and c-FLIP expression can reverse HepG2 resistance to apoptosis and enhance sensitivity to TRAIL-induced apoptosis (Figure 4A). RIP-1 or c-FLIP knockdown significantly enhanced caspase-8 and caspase-3 activity, while combined transfection further enhanced their activities (Figure 4B, 4C). The results demonstrated that high expression of RIP-1 and c-FLIPL in DISC was an important reason for HepG2 cell resistance to TRAIL. Downregulating RIP-1 and c-FLIPL can relieve caspase-8 suppression and activate caspase-3, thus triggering the apoptosis signaling pathway and promoting cell apoptosis.
Figure 4.
RIP-1 and c-FLIPL interference promoted cell apoptosis. (A) Flow cytometry detection of cell apoptosis. (B) Spectrophotometry detection of caspase-8 activity. (C) Spectrophotometry detection of caspase-3 activity.
RIP-1 knockdown reduced NF-κB activity, activated caspase, and induced apoptosis
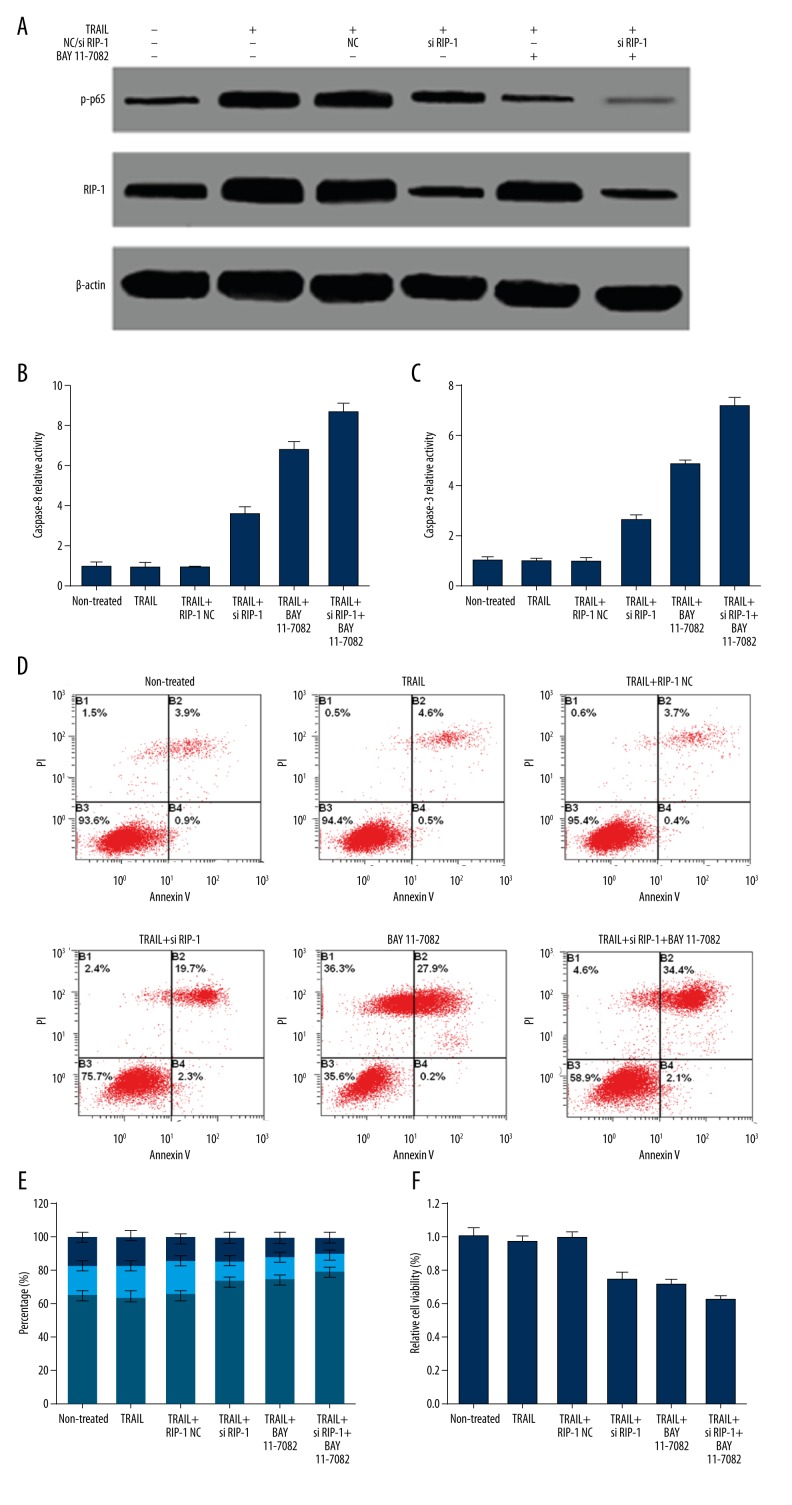
Except for binding with Fas-associated death domain (FADD) and procaspase-8 to form DISC and activate the apoptosis signaling pathway, after binding with DR4 or DR5, TRAIL can recruit and bind with TRADD and RIP-1 to activate the NF-κB signaling pathway, thus promoting cell proliferation and survival. This study explored whether high abundance of RIP-1 expression was related to NF-κB activation and apoptosis resistance. The results showed that phospho-NF-κB p65 protein was expressed in HepG2 cells, suggesting NF-κB activation. TRAIL treatment at 100 ng/mL significantly upregulated p-p65 protein expression and NF-κB transcriptional activity (Figure 5A). si RIP-1 transfection obviously reduced the RIP-1 protein level and NF-κB transcriptional activity, while the NF-κB specific inhibitor BAY11-7082 markedly downregulated the p-p65 level and NF-κB transcriptional activity but not RIP-1 expression (Figure 5A). Both RIP-1 interference and BAY11-7082 significantly suppressed caspase-8 and caspase-3 activity (Figure 5B, 5C), and promoted cell apoptosis (Figure 5D). Their simultaneous treatment showed a stronger effect on caspase-3 and cell apoptosis. Moreover, suppressing RIP-1 expression and/or weakening NF-κB transcriptional activity obviously reduced cell rate in S phase and G2/M phase, blocked cells in G0/G1 phase (Figure 5E), and restrained cell proliferation (Figure 5F), indicating that RIP-1 promoted HepG2 cell survival through upregulating NF-κB transcriptional activity and enhanced cell resistance to apoptosis.
Figure 5.
RIP-1 knockdown reduced NF-κB activity, activated caspase, and induced apoptosis. (A) Western blot detection of protein expression. (B) Spectrophotometry detection of caspase-8 activity. (C) Spectrophotometry detection of caspase-3 activity. (D) Flow cytometry detection of cell apoptosis. (E) Flow cytometry detection of cell cycle. (F) MTT assay detection of cell proliferation.
Discussion
Primary hepatic cancer is one of the common malignant tumors in our country with high morbidity and mortality [9]. It usually occurs in young male patients, featuring a high degree of malignancy and rapid development. Most patients are in an advanced stage when diagnosed, resulting in poor curative effect and prognosis [10]. TRAIL, belonging to tumor necrosis factor (TNF) superfamily, is an endogenous protein molecule targeting death receptor (DR) [11]. TRAIL can highly selectively induce tumor cell, transformed cell, and virus-infected cell apoptosis, and effectively avoid killing normal cells [4]. Apoptosis mainly includes the exogenous mitochondrial independent pathway mediated by DR and the endogenous mitochondrial dependent pathway [12]. Death receptor-mediated apoptosis signaling pathway suppression leads to imbalance in a variety of apoptosis regulatory factors, resulting in tumor cell insensitivity to apoptosis and drug resistance. Apoptosis resistance not only can improve the survival ability of tumor cells and their ability to escape immune surveillance, but also can enhance tumor cells’ resistance to chemotherapy and other treatments. It is considered to be one of the most common and important reasons for malignant tumor occurrence and progression [13]. The unique biological characteristic of TRAIL brings new hope for tumor treatment, and it is considered to be a safe and effective new-generation antitumor drug. It is now in phase II clinical trials [14]. Although TRAIL shows a significant apoptosis-inducing effect on numerous tumor cells, almost all HCC cell lines present resistance to TRAIL-induced apoptosis, leading to a limited effect of single TRAIL therapy [15]. Even after radical resection, the minimal residual hepatic cancer cells still can escape immunological surveillance and cause rapid recurrence. This is mainly caused by hepatic cancer cells’ resistance to TRAIL-induced cell apoptosis, though the specific mechanism is still unclear [5].
After binding with DR4 or DR5 on the cell membrane, TRAIL forms a ligand-receptor trimerization compound that induces the death domain (DD) on DR cytoplasmic domain binding with the DD on the C terminal of Fas-associated death domain (FADD). The death effector domain (DED) on the N-terminal of FADD combines with procaspase-8 to form DR4/DR5-FADD-procaspase-8 death induced signal complex, which triggers procaspase-8 self-splicing into the active apoptosis starter caspase-8. The latter activates the caspase cascade reaction, including caspase-3, -6, and -7, to degrade structural protein and functional protein in the cells directly, eventually triggering apoptosis [16]. DISC is a key regulatory factor in the TRAIL apoptosis signaling pathway, while c-FLIPL and RIP-1 are the two important negative regulatory proteins on DISC and the apoptosis signaling pathway [6]. Anti-apoptotic protein c-FLIPL is a key regulatory factor in the TNF family to induce apoptosis tolerances [17]. c-FLIPL is a natural inhibitor of caspase-8 protein (containing a structure and sequence similar to those of caspase-8), but it lacks the cysteine structure necessary for enzyme catalytic activity [18]. c-FLIPL competitively combines with FADD and/or caspase-8 and -10 through its two DEDs on the N-terminal, leading to blockage of the caspases’ cascade reaction and TNF, TRAIL, FasL, and corresponding DR-mediated apoptosis [19]. c-FLIPL and/or RIP-1 are overexpressed in various tumors, such as prostate cancer [20], colorectal cancer [21], and melanoma [22]. It was also found that c-FLIPL and/or RIP-1 overexpression existed in hepatic cancer cells [7,8]. Thus, we hypothesized that RIP-1 and/or c-FLIPL abnormal expression induced DISC formation and apoptosis signaling transduction blockage, which may be the important mechanism of hepatic cancer cell resistance to TRAIL-induced cell apoptosis. This study explored the role of RIP-1 and/or c-FLIPL in hepatic cancer cell resistance to TRAIL-induced apoptosis.
MTT and apoptosis detection showed that different concentrations of TRAIL had no obvious impact on HepG2 cell proliferation activity and apoptosis, confirming that HepG2 cells had strong resistance to apoptosis induced by TRAIL [23]. In view of the negative regulatory effect of RIP-1 and c-FLIPL within DISC on the apoptosis signaling pathway, we examined DISC component protein expression in HepG2 cells treated by TRAIL. The results showed that HepG2 presented a high abundance of RIP-1 and c-FLIPL, and high concentrations of TRAIL treatment can significantly increase RIP-1 and c-FLIPL expression. It might be one of the reasons for hepatic cancer cells acquiring TRAIL resistance [24]. TRAIL failed to cause obvious caspase-8 and caspase-3 activation, or induce obvious changes in DR4 and DR5 expression, suggesting that RIP-1 and c-FLIPL overexpression may play a role in caspase-8 suppression and TRAIL resistance. Ganten et al. [25] observed the inhibitory effect of TRAIL on caspase-8 activity in hepatic cancer cells, and considered that caspase activity suppression by c-FLIP overexpression in DISC was the reason for hepatic cancer cell resistance to TRAIL. However, in the Shin et al. [15] study, c-FLIP expression showed no linear relationship with hepatic cancer cell resistance to TRAIL; thus, the relationship between c-FLIP and TRAIL resistance was still unclear. Moreover, we discuss that high expression of RIP-1 and c-FLIPL played a role in HepG2 cells’ resistance to apoptosis induced by TRAIL. With use of the siRNA technique to silence endogenously RIP-1 and c-FLIPL expression, HepG2 cells’ apoptosis obviously increased, and caspase-8 and caspase-3 enzyme activities also were significantly enhanced. They reached the top when both RIP-1 and c-FLIPL were suppressed, indicating that high expression of RIP-1 and c-FLIPL plays a critical role in HepG2 TRAIL resistance.
Except for binding with FADD and procaspase-8 to form DISC and activate the apoptosis signaling pathway, after binding with DR4 or DR5, TRAIL can recruit and bind with TNF receptor type 1-associated DEATH domain protein (TRADD) and RIP-1 to activate the NF-κB signaling pathway. RIP-1 recruitment is the key in NF-κB activation induced by TRAIL. RIP-1 recruitment and NF-κB activation are extremely important in promoting cell survive and proliferation. The latter mediates cell resistance to TRAIL-induced cell apoptosis [26]. At present, most studies considered that RIP expression can activate NF-κB, whereas the relationship between its elevation and caspase activity suppression is controversial. This study found that TRAIL upregulated RIP-1 expression and enhanced phospho-NF-κB p65 protein expression to elevate NF-κB transcriptional activity in HepG2 cells, revealing that RIP-1 elevation and NF-κB activation may play a role in apoptosis resistance. RIP-1 downregulation reduced NF-κB transcriptional activity, blocked the cell cycle in G0/G1 phase, weakened cell proliferation, enhanced caspase-8 and caspase-3 activity, and increased cell apoptosis. As the specific inhibitor of NF-κB, BAY11-7082 showed a similar effect with RIP-1 interference. Their simultaneous treatment presented a stronger effect, suggesting that TRAIL enhanced NF-κB transcriptional activity, reduced caspase activity, and mediated apoptosis resistance through upregulating RIP-1 expression.
Conclusions
TRAIL can enhance RIP1 and c-FLIPL expression in HepG2 cells. High expression of RIP1 and c-FLIPL is an important reason for TRAIL resistance. Downregulation of RIP1 and c-FLIPL can relieve caspase-8 suppression, activate caspase-3, and promote cell apoptosis. TRAIL mediates apoptosis resistance through upregulating RIP-1 expression, enhancing NF-κB transcriptional activity, and weakening caspase activity.
Footnotes
Disclosure of conflict of interest
The authors declare no competing financial or commercial interests in this manuscript.
Source of support: Departmental sources
References
- 1.Wallace MC, Preen D, Jeffrey GP, Adams LA. The evolving epidemiology of hepatocellular carcinoma: A global perspective. Expert Rev Gastroenterol Hepatol. 2015;9:765–79. doi: 10.1586/17474124.2015.1028363. [DOI] [PubMed] [Google Scholar]
- 2.Lafaro KJ, Demirjian AN, Pawlik TM. Epidemiology of hepatocellular carcinoma. Surg Oncol Clin N Am. 2015;24:1–17. doi: 10.1016/j.soc.2014.09.001. [DOI] [PubMed] [Google Scholar]
- 3.Tanaka M, Katayama F, Kato H, et al. Hepatitis B and C virus infection and hepatocellular carcinoma in China: A review of epidemiology and control measures. J Epidemiol. 2011;21:401–16. doi: 10.2188/jea.JE20100190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sophonnithiprasert T, Nilwarangkoon S, Nakamura Y, Watanapokasin R. Goniothalamin enhances TRAIL-induced apoptosis in colorectal cancer cells through DR5 upregulation and cFLIP downregulation. Int J Oncol. 2015;47:2188–96. doi: 10.3892/ijo.2015.3204. [DOI] [PubMed] [Google Scholar]
- 5.Li J, Sun RR, Yu ZJ, et al. Galectin-1 modulates the survival and tumor necrosis factor-related apoptosis-inducing ligand (TRAIL) sensitivity in human hepatocellular carcinoma cells. Cancer Biother Radiopharm. 2015;30:336–41. doi: 10.1089/cbr.2015.1857. [DOI] [PubMed] [Google Scholar]
- 6.Matsuda I, Matsuo K, Matsushita Y, et al. The C-terminal domain of the long form of cellular FLICE-inhibitory protein (c-FLIPL) inhibits the interaction of the caspase 8 prodomain with the receptor-interacting protein 1 (RIP1) death domain and regulates caspase 8-dependent nuclear factor kappaB (NF-kappaB) activation. J Biol Chem. 2014;289:3876–87. doi: 10.1074/jbc.M113.506485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chen F, Zhu HH, Zhou LF, et al. Inhibition of c-FLIP expression by miR-512-3p contributes to taxol-induced apoptosis in hepatocellular carcinoma cells. Oncol Rep. 2010;23:1457–62. doi: 10.3892/or_00000784. [DOI] [PubMed] [Google Scholar]
- 8.Sun J, Luo H, Nie W, et al. Protective effect of RIP and c-FLIP in preventing liver cancer cell apoptosis induced by TRAIL. Int J Clin Exp Pathol. 2015;8:6519–25. [PMC free article] [PubMed] [Google Scholar]
- 9.Yao H, Bian X, Mao L, et al. Preoperative enteral nutritional support in patients undergoing hepatectomy for hepatocellular carcinoma: A strengthening the reporting of observational studies in epidemiology article. Medicine (Baltimore) 2015;94:e2006. doi: 10.1097/MD.0000000000002006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Liu PH, Huo TI. Prognosis of hepatocellular carcinoma: Can scores guide treatment decisions? J Hepatol. 2016;64(3):601–8. doi: 10.1016/j.jhep.2015.10.029. [DOI] [PubMed] [Google Scholar]
- 11.Huang K, Zhang J, O’Neill KL, et al. Cleavage by caspase 8 and mitochondrial membrane association activate bid during TRAIL-induced apoptosis. J Biol Chem. 2016;291(22):11843–51. doi: 10.1074/jbc.M115.711051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sinha K, Das J, Pal PB, Sil PC. Oxidative stress: the mitochondria-dependent and mitochondria-independent pathways of apoptosis. Arch Toxicol. 2013;87:1157–80. doi: 10.1007/s00204-013-1034-4. [DOI] [PubMed] [Google Scholar]
- 13.Lee YJ, Lee JH, Moon JH, Park SY. Overcoming hypoxic-resistance of tumor cells to TRAIL-induced apoptosis through melatonin. Int J Mol Sci. 2014;15:11941–56. doi: 10.3390/ijms150711941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bellail AC, Qi L, Mulligan P, et al. TRAIL agonists on clinical trials for cancer therapy: The promises and the challenges. Rev Recent Clin Trials. 2009;4:34–41. doi: 10.2174/157488709787047530. [DOI] [PubMed] [Google Scholar]
- 15.Shin EC, Seong YR, Kim CH, et al. Human hepatocellular carcinoma cells resist to TRAIL-induced apoptosis, and the resistance is abolished by cisplatin. Exp Mol Med. 2002;34:114–22. doi: 10.1038/emm.2002.17. [DOI] [PubMed] [Google Scholar]
- 16.Ramamurthy V, Yamniuk AP, Lawrence EJ, et al. The structure of the death receptor 4-TNF-related apoptosis-inducing ligand (DR4-TRAIL) complex. Acta Crystallogr F Struct Biol Commun. 2015;71:1273–81. doi: 10.1107/S2053230X15016416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Tsuchiya Y, Nakabayashi O, Nakano H. FLIP the switch: Regulation of apoptosis and necroptosis by cFLIP. Int J Mol Sci. 2015;16:30321–41. doi: 10.3390/ijms161226232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kataoka T. The caspase-8 modulator c-FLIP. Crit Rev Immunol. 2005;25:31–58. doi: 10.1615/critrevimmunol.v25.i1.30. [DOI] [PubMed] [Google Scholar]
- 19.Jarvinen K, Hotti A, Santos L, et al. Caspase-8, c-FLIP, and caspase-9 in c-Myc-induced apoptosis of fibroblasts. Exp Cell Res. 2011;317:2602–15. doi: 10.1016/j.yexcr.2011.08.014. [DOI] [PubMed] [Google Scholar]
- 20.Lee SW, Cho JM, Cho HJ, et al. Expression levels of heat shock protein 27 and cellular FLICE-like inhibitory protein in prostate cancer correlate with Gleason score sum and pathologic stage. Korean J Urol. 2015;56:505–14. doi: 10.4111/kju.2015.56.7.505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Carson R, Celtikci B, Fenning C, et al. HDAC inhibition overcomes acute resistance to MEK inhibition in BRAF-mutant colorectal cancer by downregulation of c-FLIPL. Clin Cancer Res. 2015;21:3230–40. doi: 10.1158/1078-0432.CCR-14-2701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Shao Y, Le K, Cheng H, Aplin AE. NF-kappaB regulation of c-FLIP promotes TNFalpha-mediated RAF inhibitor resistance in melanoma. J Invest Dermatol. 2015;135:1839–48. doi: 10.1038/jid.2015.91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lu G, Liu Y, Ji B, et al. Synergistic effect of celecoxib on TRAIL-induced apoptosis in hepatocellular carcinoma cells. Cancer Invest. 2010;28:629–34. doi: 10.3109/07357900903095631. [DOI] [PubMed] [Google Scholar]
- 24.Thorburn A, Behbakht K, Ford H. TRAIL receptor-targeted therapeutics: Resistance mechanisms and strategies to avoid them. Drug Resist Updat. 2008;11:17–24. doi: 10.1016/j.drup.2008.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ganten TM, Haas TL, Sykora J, et al. Enhanced caspase-8 recruitment to and activation at the DISC is critical for sensitisation of human hepatocellular carcinoma cells to TRAIL-induced apoptosis by chemotherapeutic drugs. Cell Death Differ. 2004;11(Suppl 1):S86–96. doi: 10.1038/sj.cdd.4401437. [DOI] [PubMed] [Google Scholar]
- 26.Bellail AC, Tse MC, Song JH, et al. DR5-mediated DISC controls caspase-8 cleavage and initiation of apoptosis in human glioblastomas. J Cell Mol Med. 2010;14:1303–17. doi: 10.1111/j.1582-4934.2009.00777.x. [DOI] [PMC free article] [PubMed] [Google Scholar]