Abstract
Several lines of indirect evidence suggest that hominoids (apes and humans) and cercopithecoids (Old World monkeys) diverged around 23–25 Mya. Importantly, although this range of dates has been used as both an initial assumption and as a confirmation of results in many molecular-clock analyses, it has not been critically assessed on its own merits. In this article we test the robusticity of the 23- to 25-Mya estimate with ≈150,000 base pairs of orthologous DNA sequence data from two cercopithecoids and two hominoids by using quartet analysis. This method is an improvement over other estimates of the hominoid–cercopithecoid divergence because it incorporates two calibration points, one each within cercopithecoids and hominoids, and tests for a statistically appropriate model of molecular evolution. Most comparisons reject rate constancy in favor of a model incorporating two rates of evolution, supporting the “hominoid slowdown” hypothesis. By using this model of molecular evolution, the hominoid–cercopithecoid divergence is estimated to range from 29.2 to 34.5 Mya, significantly older than most previous analyses. Hominoid–cercopithecoid divergence dates of 23–25 Mya fall outside of the confidence intervals estimated, suggesting that as much as one-third of ape evolution has not been paleontologically sampled. Identifying stem cercopithecoids or hominoids from this period will be difficult because derived features that define crown catarrhines need not be present in early members of these lineages. More sites that sample primate habitats from the Oligocene of Africa are needed to better understand early ape and Old World monkey evolution.
Keywords: evolution, molecular clock, primates
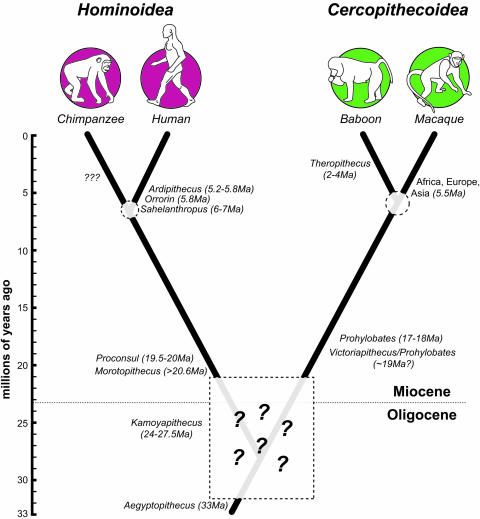
Both the fossil record and analyses of molecular data have been used to infer a hominoid–cercopithecoid divergence of ≈23–25 Mya (1–4). Fossil-based dates are typically the result of rounding up the oldest hominoid or cercopithecoid (Fig. 1). Proconsul is often cited as the oldest hominoid at ≈20 Mya (5, 6), although there is some evidence of earlier apes: Morotopithecus is dated to >20.6 Mya (7), and the dentally hominoid-like catarrhine Kamoyapithecus is radiometrically dated to 24–27.5 Mya (8). Fossil cercopithecoids are found at a number of Early Miocene African sites but are not found in any context that predates the oldest hominoids (9–11). Because the hominoid status of Kamoyapithecus is debatable at this time (12), Morotopithecus is the oldest undoubted hominoid or cercopithecoid at ≈21 Mya (13, 14). Molecular-clock analyses yield interpolated hominoid–cercopithecoid divergence dates that are largely congruent with the fossil record. Kumar and Hedges (3) interpolated a divergence time of 23 Mya by using a combination of calibration points for birds and mammals (310 Mya) and primates and rodents (110 Mya). Glazko and Nei (4) also calculated a date of 23 Mya by calibrating the divergence of primates and artiodactyls at 90 Mya. The congruence of fossil and genetic estimates is important because a number of molecular-clock analyses have used a 23- to 25-Mya estimate either as an initial assumption (15–17) or as a confirmation of results (3, 18). If this estimate were found to be inaccurate, it would have consequent effects on how we interpret crown catarrhine evolution. However, until now the robusticity of the estimate has not been directly tested by any analysis. A more critical assessment of the fossil and molecular evidence indicates the 23- to 25-Mya date is not as well supported as would first appear, suggesting that a more specific test of the estimate is warranted.
Fig. 1.
Phylogeny of the four taxa analyzed by using the quartet method, and the dates and names of fossil taxa relevant to the divergences among these species.
First, whereas it is possible to estimate from fossils when a split has occurred, it is more difficult to say when a split has not occurred. In other words, one can infer from the earliest hominoid fossil (≈21 Mya) that the divergence must have occurred by this time, but it is difficult to know just how much earlier the divergence might have occurred (1, 5, or 10 million years prior?). The Oligocene sediments from the Fayum are well located in both time (36–33 Mya) and space (Africa) to address this question. Primates from the Fayum include Aegyptopithecus, a taxon plausibly inferred as ancestral to all living catarrhines, and neither hominoids nor cercopithecoids appear among the Fayum fauna, suggesting that 33 Mya is a reasonable upper limit to their divergence date (19, 20). Sadly, much of the rest of this 33- to 21-Mya time span is so fossil poor in Africa, particularly for primates (21, 22), that it is impossible to pinpoint when the divergence had not occurred. Given this scarcity of evidence, the fossil record is equally supportive of any divergence time between 32 and 21 Mya.
Second, data sets that either interpolated a 23–25 Mya hominoid–cercopithecoid divergence date from an older calibration (e.g., refs. 18 and 23) or were calibrated by using this date (e.g., refs. 16, 24, and 25) underestimate portions of the hominoid tree for which we have good fossil evidence (14). For example, Kumar and Hedges (3) estimate the human–chimpanzee divergence at 5.5 ± 0.2 Mya, a date that is at odds with recent discoveries of hominin fossil material dating to as early as 7 Mya (26). When the afore-mentioned data sets are instead calibrated by using a 7-Mya hominin date, the hominoid–cercopithecoid split is pushed back between 29–31 Mya (mean = 30.1 Mya), significantly older than 23–25 Mya (1, 14). Reliance on a single fossil calibration point can also be problematic if it is not well supported. For example, Kumar and Hedges (3) have been criticized for the use of questionable fossil calibrations for the divergence of both birds/mammals and rodents/primates as well as their methodology (27). This criticism casts doubt on their estimates for other nodes, including hominoid–cercopithecoid.
Third, rate heterogeneity among different lineages is problematic for molecular clock analyses. Within mammals there is considerable evidence for evolutionary rate heterogeneity, with different mammal groups evolving at different rates (28–37), although this finding has been questioned (38). While some studies run contrary to this finding (39, 40), there is also considerable evidence for rate heterogeneity within primates, particularly that hominoids are evolving slower than cercopithecoids (the “hominoid slowdown”) (15, 32, 34, 41–47). These findings conflict with the proposition of a global molecular clock for mammals (23). Divergence dates estimated with a constant rate of molecular change among all lineages are likely complicated by such rate heterogeneity. Local molecular clocks have been used to control for the rate variations to date primate divergences (35, 48), but the heterogeneity within catarrhine primates (i.e., the hominoid slowdown) has not yet been incorporated into analyses specifically to estimate the hominoid–cercopithecoid divergence.
Together these caveats suggest that from both molecular and paleontological perspectives, the 23- to 25-Mya date for the hominoid–cercopithecoid divergence is not as strongly supported as is frequently supposed. There is reason to believe that (i) fossil data from the Oligocene and Early Miocene are much more equivocal, (ii) interpolated molecular clock dates based on single fossil calibrations are not entirely consistent with the more robustly supported hominin fossil dates, and (iii) rate heterogeneity within primates is problematic for dating primate divergences and must be taken into account. Here we estimate the hominoid–cercopithecoid divergence using a method that has four major improvements over previous studies. First, this method incorporates two fossil calibration points, one from within hominoids and one from within cercopithecoids, to date a single divergence point. Second, >150,000 base pairs of orthologous DNA sequence data are analyzed to estimate the divergence time, a large increase over previous studies. Third, this method integrates a statistically defined model of molecular evolution within the hominoids and cercopithecoids, controlling for rate heterogeneity. Fourth, through the use of maximum likelihood analyses, confidence intervals can be placed around the date estimates, enabling a statistical test of the 23- to 25-Mya divergence of hominoids and cercopithecoids.
Methods
The divergence date for the hominoid–cercopithecoid split is estimated by using a maximum likelihood-based quartet analysis (49), a method that has found widespread taxonomic utility in calculating divergence times (33, 49–56). Here this method is expanded to large genomic contigs sequenced in catarrhine primates. Quartet analyses use DNA sequence data from four species, two species each from two clades that are monophyletic with respect to one another. Within each pair, an independent, paleontologically derived divergence date is used to calibrate the overall rate of molecular evolution, which is then used to date the divergence between the two pairs. In this case the two pairs are hominoids (human and chimpanzee) and cercopithecoids (macaque and baboon), and the divergence date estimated is the hominoid–cercopithecoid split (Fig. 1). These four species were chosen for two reasons: Within both pairs, fossil data are available to estimate the divergence of the two species pairs with reasonable confidence, and within each of these four species a number of genomic contigs have been sequenced, allowing alignment and analysis of large regions of orthologous DNA.
Divergence Dates. The split between the hominoid pair (human–chimpanzee divergence) is calibrated by using the earliest known hominin, Sahelanthropus tchadensis, which has been dated between 6 and 7 Mya by using faunal comparisons (26). This date is supported by two other early hominins, Orrorin tugenensis (≈5.8 Mya) (57) and Ardipithecus kadabba (5.2–5.8 Mya) (58). Although there are no known fossil chimpanzees to date, this evidence strongly suggests that hominins and chimpanzee ancestors diverged by at least 6 Mya, and possibly as early as 7 Mya. However, the lack of an African ape fossil record in the late Miocene does not preclude earlier dates. To reflect this uncertainty, divergence dates of 6 and 7 Mya will be used as the human–chimpanzee divergence date. The split between the cercopithecoid species is calibrated by using an estimate for the split between macaques and baboons/mangabeys (together known as papionins). According to Delson et al. (9), the earliest fossil evidence of papionins is from teeth found in North Africa and Kenya of late Miocene age (6–8 Mya). Macaques are known from North Africa (77) and Europe (78) by 5.5 Mya, and there is evidence of their arrival in Asia at about the same time.†† Papionins other than macaques are well sampled from the middle to late Pliocene: ≈4–2 Mya. Delson (10) and Delson et al. (9) estimate the split at ≈7–8 Mya. Conservatively, we use macaque–baboon divergence dates of 5 and 7 Mya.
Assembly of Genomic Contigs. We designed a method to identify and align regions of orthologous DNA from genomic contigs sequenced in humans (Homo sapiens), chimpanzees (Pan troglodytes), rhesus macaques (Macaca mulatta), and anubis baboons (Papio anubis). The data assembly method had five steps. First, a database was constructed of all of the completely sequenced genomic contigs from both Papio and Macaca available in GenBank. All of the repeat elements (e.g., Alu insertions, LINEs) were removed from these contigs by using repeatmasker (available from Smit and Green at http://repeatmasker.org), resulting in a database of “masked” contigs. Second, each Papio contig was compared to the masked Macaca database by using blast (59) to find contigs that were orthologous between the two species. A reciprocal blast search, from Macaca to Papio, confirmed the matches. From this list of corresponding contigs, five nonoverlapping pairs were chosen. Third, blast was used to identify the contigs available in GenBank from humans and chimpanzees that were orthologous to the five cercopithecoid contig pairs. Once identified, the repeat elements were removed from the hominoid contigs. In total, 39.51% of the contig base pairs were identified as repeat elements. Fourth, these five sets, each containing one contig each from human, chimpanzee, baboon, and macaque, were trimmed to include only base pairs common to all four species. These five sets were aligned by using clustalw and clustalx (60, 61) and reviewed to remove any remaining regions of nonhomology. Finally, all non-point mutations, such as mutated CpG sites, mononucleotide repeat stretches, and other complex mutations, were removed from the alignments. These types of mutations can bias mutation models, especially in the case of CpG sites (62). Omission of these sites, which totaled 1.5% of the total data set, had no significant impact on the results (when all sites are included, the difference in the global average is ≈1 Mya greater than the estimate presented here). This method resulted in five alignments of orthologous, nonrepeat DNA sequences, each referred to as a “contig set” (Table 1).
Table 1. Summary contig set information.
| Contig set | Contigs | Base pairs | Ti/Tv ratio* | α-Parameter† | |
|---|---|---|---|---|---|
| H. sapiens | P. troglodytes | ||||
| A | AC007568.1 | AC144512.1 | 27,591 | 2.11 | 0.76 |
| P. anubis | M. mulatta | ||||
| AC087103.3 | AC123537.3 | ||||
| H. sapiens | P. troglodytes | ||||
| B | AC002066.2 | AC087512.2 | 23,374 | 1.87 | 1.16 |
| P. anubis | M. mulatta | ||||
| AC084730.2 | AC124158.3 | ||||
| H. sapiens | P. troglodytes | ||||
| C | AC106873.3 | AC142297.1 | 11,856 | 1.66 | 3.03 |
| P. anubis | M. mulatta | ||||
| AC087215.5 | AC130184.4 | ||||
| H. sapiens | P. troglodytes | ||||
| D | AC002542.1 | AC144752.3 | 63,965 | 2.19 | 0.55 |
| P. anubis | M. mulatta | ||||
| AC087252.3 | AC124159.3 | ||||
| H. sapiens | P. troglodytes | ||||
| E | AC000061.1 | AC087835.2 | 23,434 | 1.94 | 0.65 |
| P. anubis | M. mulatta | ||||
| AC091381.3 | AC123966.4 | ||||
Transition-to-transversion ratio.
Parameter for the gamma rate heterogeneity correction.
Likelihood Ratio Testing and Quartet Dating Analysis. These contig sets, in conjunction with the two divergence dates, were used to estimate the rates of evolution within these species lineages and date the hominoid split by using the quartet method implemented by the qdate program (Version 1.1) (49). The method calculated likelihoods assuming the HKY model of molecular evolution (63) with a gamma correction for site-specific rate heterogeneity (64) (HKY+Γ) and a user-defined transition/transversion ratio [both parameters estimated by using maximum likelihood in paup* (65)] (Table 1). Although the choice of model has been shown to have an impact on estimation of divergence dates (66), in the present case estimated divergence dates were robust to the use of different models (data not shown). Likelihood values were calculated under three different conditions of lineage specific rate heterogeneity. First, the likelihoods of the “one-rate” condition were calculated. In this condition, a single rate of molecular evolution was assumed for all of the branches of the tree, a constant (or global) molecular clock. Second, the likelihoods of the “two-rate” condition were calculated, assuming one rate of molecular evolution for the hominoid branches and a second rate for the cercopithecoid branches, allowing an estimation of the hominoid slowdown. A third “free-rate” condition was examined, where each branch was allowed to evolve at its own rate and no molecular clock is assumed. Comparing the likelihoods under these different conditions, rate constancy can be examined with a likelihood ratio test (67) using a χ2 approximation to test the significance of the difference (Δ) in log likelihood values between the given conditions, as implemented in qdate. First, the one-rate condition is tested against the free-rate condition, to test whether one rate of molecular evolution characterizes the data. Second, the two-rate condition is tested against the free-rate condition, to test whether a model where cercopithecoids and hominoids are evolving at two different rates is a statistically better fit than a model with each branch allowed to evolve at a different rate (essentially a test of the hominoid slowdown hypothesis). Subsequently, when using comparisons where rate constancy holds (one-rate or two-rate), the branch lengths will be converted to ordinal dates, using the calibration points discussed above. The statistically defined confidence intervals on these dates are calculated as described in Rambaut and Bromham (49). Here, an additional degree of uncertainty in the date estimates of the hominoid and cercopithecoid divergence is incorporated in the analysis by examining a narrow range of plausible fossil divergences within hominoids and cercopithecoids (discussed above).
Results and Discussion
Likelihood ratio tests were constructed to assess whether one or more molecular clocks characterized the evolution of the hominoid and cercopithecoid lineages with each of the four different fossil divergence date estimates (Table 2). In none of the contig sets did all four comparisons support rate constancy among the hominoids and cercopithecoids. In three contig sets (A, C, and E), two of the four comparisons were consistent with rate constancy, and in the remaining two contig sets (B and D) there were no comparisons consistent with rate constancy. In total, 6 of 20 comparisons were evolving in a clock-like fashion. Overall, these data do not support a model of rate constancy between hominoids and cercopithecoids. An alternative possibility is that the calibration points are incorrect. However, decreasing the hominoid calibration to dates younger than 6 Mya would be inconsistent with the hominid fossil record, and increasing the cercopithecoid calibration substantially past 7 Mya has no fossil justification.
Table 2. Likelihood ratio tests and estimated divergence dates for the hominoid–cercopithecoid split.
| One-rate tests
|
Two-rate tests
|
|||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Divergence dates,* Mya
|
One-rate vs. free-rate
|
One-rate date estimates, Mya
|
Two-rate vs. free-rate
|
Two-rate date estimates, Mya
|
||||||||||
| Contig set | Rate† | Δ‡ | Sig.§ | Date¶ | Low∥ | High** | Rate 1†† | Rate 2‡‡ | Δ | Sig. | Date | Low | High | |
| A | (6,5) | 8.79 | 1.1 | n.s. | 24.0 | 22.1 | 26.1 | 8.57 | 1.06 | 23.9 | 22.0 | 26.0 | ||
| (6,7) | 7.44 | 6.0 | ** | 8.57 | 0.76 | 0.9 | n.s. | 28.0 | 25.8 | 30.5 | ||||
| (7,5) | 8.06 | 3.8 | * | 7.34 | 1.23 | 25.7 | 23.6 | 28.0 | ||||||
| (7,7) | 6.91 | 1.9 | n.s. | 30.5 | 28.1 | 33.2 | 7.34 | 0.88 | 30.5 | 28.1 | 33.2 | |||
| Average | 7.80 | 27.2 | 25.1 | 29.6 | 8.73 | 0.98 | 27.0 | 24.9 | 29.4 | |||||
| B | (6,5) | 9.83 | 26.8 | ** | 6.98 | 1.90 | 24.5 | 22.5 | 26.8 | |||||
| (6,7) | 8.32 | 7.3 | ** | 6.98 | 1.36 | 1.6 | n.s. | 30.2 | 27.7 | 33.0 | ||||
| (7,5) | 9.02 | 40.4 | ** | 5.98 | 2.22 | 25.8 | 23.7 | 28.2 | ||||||
| (7,7) | 7.72 | 14.4 | ** | 5.98 | 1.58 | 32.1 | 29.5 | 35.1 | ||||||
| Average | 8.72 | 6.48 | 1.76 | 28.1 | 25.8 | 30.8 | ||||||||
| C | (6,5) | 6.83 | 4.9 | ** | 5.74 | 1.42 | 28.6 | 24.8 | 33.3 | |||||
| (6,7) | 5.78 | 2.2 | n.s. | 34.3 | 29.8 | 39.9 | 5.74 | 1.02 | 2.2 | n.s. | 34.4 | 29.8 | 39.9 | |
| (7,5) | 6.27 | 7.8 | ** | 4.92 | 1.66 | 30.4 | 26.3 | 35.4 | ||||||
| (7,7) | 5.37 | 2.8 | n.s. | 37.0 | 32.1 | 43.0 | 4.92 | 1.18 | 37.0 | 32.1 | 43.0 | |||
| Average | 6.06 | 35.7 | 30.9 | 41.4 | 5.33 | 1.32 | 32.6 | 28.3 | 37.9 | |||||
| D | (6,5) | 7.25 | 39.3 | ** | 5.40 | 1.75 | 28.9 | 27.2 | 30.7 | |||||
| (6,7) | 6.14 | 6.4 | ** | 5.40 | 1.25 | 0.2 | n.s. | 35.3 | 33.2 | 37.6 | ||||
| (7,5) | 6.65 | 63.8 | ** | 4.63 | 2.04 | 30.5 | 28.6 | 32.4 | ||||||
| (7,7) | 5.70 | 17.9 | ** | 4.63 | 1.46 | 37.7 | 35.5 | 40.1 | ||||||
| Average | 6.44 | 5.02 | 1.63 | 33.1 | 31.1 | 35.2 | ||||||||
| E | (6,5) | 6.62 | 3.6 | ** | 5.75 | 1.40 | 31.9 | 28.7 | 35.5 | |||||
| (6,7) | 5.60 | 0.7 | n.s. | 38.1 | 34.4 | 42.5 | 5.75 | 1.00 | 0.5 | n.s. | 38.0 | 34.3 | 42.4 | |
| (7,5) | 6.07 | 8.1 | ** | 4.93 | 1.63 | 34.0 | 30.6 | 37.9 | ||||||
| (7,7) | 5.20 | 0.8 | n.s. | 41.1 | 37.0 | 45.8 | 4.93 | 1.16 | 41.1 | 37.0 | 45.8 | |||
| Average | 5.87 | 39.6 | 35.7 | 44.1 | 5.34 | 1.30 | 36.2 | 32.6 | 40.4 | |||||
| Weighted average | (6,5) | 7.80 | 1 n.s./4 | 24.0 | 22.1 | 26.1 | 6.31 | 1.57 | 27.7 | 25.6 | 30.2 | |||
| (6,7) | 6.61 | 2 n.s./4 | 36.9 | 32.8 | 41.6 | 6.31 | 1.12 | all n.s. | 33.5 | 30.9 | 36.5 | |||
| (7,5) | 7.16 | 0 n.s./4 | 5.41 | 1.83 | 29.4 | 27.1 | 32.0 | |||||||
| (7,7) | 6.13 | 3 n.s./4 | 35.7 | 32.2 | 39.7 | 5.41 | 1.30 | 36.0 | 33.2 | 39.2 | ||||
| Global average | 6.92 | 32.2 | 29.0 | 35.8 | 5.86 | 1.45 | 31.6 | 29.2 | 34.5 | |||||
(hominoid, cercopithecoid).
Substitution rate per site per million years × 10-10.
Differences between likelihood values in the two models.
Significance as determined by a likelihood ratio test (*, P < 0.5; **, P < 0.01). n.s., not significant.
Maximum likelihood estimate of the divergence date for hominoids and cercopithecoids.
Lower 95% boundary.
Upper 95% boundary.
Substitution rate in hominoids per site per million years × 10-10.
Substitution rate in cercopithecoids, expressed as a ratio in terms of rate 1.
The comparisons that exhibited rate constancy allow an estimate of the hominoid–cercopithecoid divergence with a constant molecular clock. Within each contig set, only contig set A had a confidence interval overlapping a 23- to 25-Mya divergence. In no cases did the average estimated confidence intervals overlap a 23- to 25-Mya hominoid–cercopithecoid divergence. The global average of the hominoid–cercopithecoid divergence dates consistent with rate constancy was 32.2 Mya, with an average lower bound of 29.0 Mya. This estimate is not consistent with either a 23- or 25-Mya divergence date for hominoids and cercopithecoids.
Under a constant molecular clock, rate estimates for all five contig sets ranged from 5.20 × 10–10 to 9.83 × 10–10 substitutions per site per year. This estimate not only incorporates estimates from five distinct contigs, but also incorporates paleontological uncertainty for both the hominoid and cercopithecoid divergence dates. However, as shown, the assumption of a constant molecular clock is not valid for most of the comparisons, and these estimates are therefore not preferred. When restricted to the comparisons fitting the one-rate model, the global average is 6.28 × 10–10 substitutions per site per year. This estimate is ≈3 times slower than a global nuclear molecular clock estimated for placental mammals (22.2 × 10–10) (23), estimated in part by using a 5.5-Mya divergence for humans and chimpanzees and a 23-Mya divergence for hominoids and cercopithecoids. The rates derived here are more similar to those estimated exclusively from humans and chimpanzees, 7.9 × 10–10, which used a 7.5-Mya divergence for these taxa (34). When both cercopithecoids and hominoids are analyzed, Yi et al. (34) estimated a rate of 15 × 10–10 substitutions per site per year when using a 23-Mya divergence between these taxa and 11.7 × 10–10 when using a 30-Mya divergence date. Rates estimated here with a one-rate model are more similar to those estimated from only primates, suggesting that within mammals, different groups evolve at different rates.
Unlike the above cases, there were no significant differences found between the two- and the free-rate models for any comparisons in the five contig sets (Table 2). Together, these two tests show that when each branch is allowed to evolve at a different rate, they fall into two rate categories, with one rate for hominoids and a second rate for cercopithecoids. Of the five contig sets, four show cercopithecoids evolving more quickly than hominoids. When partitioned, hominoids are evolving at an average rate of 5.86 × 10–10 substitutions per site per year, whereas cercopithecoids are evolving 1.45 times faster at 8.52 × 10–10. These tests and the rates of molecular evolution derived for these species strongly support the hominoid slowdown hypothesis as first proposed by Goodman (41).
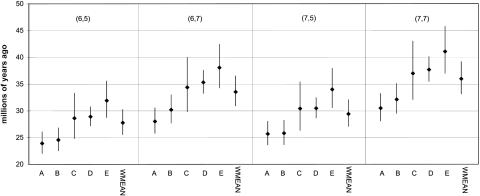
By using the two rates estimated for these taxon pairs, a date for the hominoid–cercopithecoid divergence can be estimated. Within each contig set, the average divergence dates yield different estimates for the hominoid–cercopithecoid divergence. By examining the within-contig set averages, the uncertainty of the paleontologically derived divergences within hominoids and cercopithecoids can be incorporated into the estimates of the hominoid–cercopithecoid split. Contig A has the youngest average for the maximum likelihood estimates (27.0 Mya), and contig E has the oldest (36.2 Mya). The youngest average lower bound was 24.9 Mya, and the highest average upper bound was 40.4 Mya, essentially bracketing the entire Oligocene. However, of the 20 comparisons, only 2 had confidence intervals that overlap a 23-Mya divergence with 3 additional comparisons overlapping a 25-Mya divergence (Fig. 2). When incorporating the paleontological uncertainty in the divergence within each pair, these estimates underscore the heterogeneity in divergence estimates between each of the contig sets.
Fig. 2.
Divergence date estimates for the hominoid–cercopithecoid split based on the two-rate model for each contig set (A–E) and the weighted mean (WMEAN) under four different divergence dates within hominoids and cercopithecoids. Divergence date estimates are in millions of years and denote (hominoid, cercopithecoid).
The average dates within each contig set were further averaged to reduce the heterogeneity due to differences among the contig sets, while still incorporating the uncertainty of the divergence dates within the hominoids and cercopithecoids. The youngest average date of the hominoid–cercopithecoid divergence, 27.7 Mya, was found by using the youngest hominoid and cercopithecoid calibration points (6 Mya and 5 Mya, respectively). The average lower bound for this estimate was 25.6 Mya. Divergence dates of 7 Mya for hominoids and 7 Mya for cercopithecoids produced the oldest average estimate, 36.0 Mya, with an average lower bound of 33.2 Mya. In averaging across the contig sets, none of the divergence dates have average confidence intervals that encompassed a 23- to 25-Mya divergence for hominoids and cercopithecoids. Instead, the global average yields a substantially older date for the divergence of hominoids and cercopithecoids: 31.6 Mya with confidence intervals from 29.2 to 34.5 Mya. This range reflects both statistical and paleontological uncertainty, as discussed in Methods. With additional fossil evidence, the fossil calibration points within cercopithecoids and hominoids may be revised, refining this estimate further. For example, better resolution of the age of Sahelanthropus may preclude a human–chimpanzee divergence younger than 7 Mya.
It is interesting to note that the only contig set that was evolving close to a constant rate, set A, yielded the youngest divergences for the hominoid–cercopithecoid split. However, as shown from the remaining contigs where rate heterogeneity is considerable, the estimated divergence estimates are significantly older. This confirms the finding that removing data sets that do not evolve with rate constancy, a common practice (e.g., ref. 23), is a source of bias (34).
Two findings suggest possible biases for our estimate of the hominoid–cercopithecoid divergence. First, simulations show that early single calibration points can bias clock estimates of older divergence dates upwards (68). In these simulations, the estimated dates begin to converge on the actual dates when the length of the simulated data sets approached 500 aa. Our study likely overcomes this bias, because each of the five contig sets analyzed is >12,000 base pairs. A second potential for bias is in the sample of contigs analyzed, which are all homologous to human chromosome 7. Evidence shows that the substitution rate differs among hominoid chromosomes (69). However, these substitution regimes are conserved over large phylogenetic distances (69) and therefore it is not clear whether this pattern could bias molecular clock estimates. The correspondence of the present data to two other studies further suggests that the present study is not biased. A study of mitochondrial genomes (35) suggested a 30- to 40-Mya cercopithecoid–hominoid divergence when employing local molecular clocks. DNA–DNA hybridization studies (24, 70) when recalibrated with a 7-Mya human–chimpanzee divergence, date the hominoid–cercopithecoid divergence to ≈30 Mya (14). These findings are within the confidence intervals presented here.
A 29.2- to 34.5-Mya range for the hominoid–cercopithecoid split is significantly older than is usually estimated, implying that ≈10 million years of ape and Old World monkey evolution are largely unsampled. The most obvious reason for this underestimation is that the lack of definitive hominoid or cercopithecoid fossils from the Early and Late Oligocene has been erroneously viewed as evidence of absence. But there are only a handful of known African sites in the period between 33 and 21 Mya on which to make this assessment (the Fayum, Lothidok, and Chilga). Of these, only Lothidok (24–27.5 Mya) has produced a possible hominoid or cercopithecoid primate (Kamoyapithecus), although it cannot be definitively linked to either catarrhine group. Our estimate of the divergence time suggests that the absence of hominoid or cercopithecoid fossils is likely due to other factors: current sites sample inappropriate fauna/environments, are taphonomically biased (only large-bodied fauna), or are incompatible with the preservation of primate fossils (low densities of hominoids/cercopithecoids compared with other mammals).
In addition, identifying stem hominoids or cercopithecoids from this 33- to 21-Mya gap is potentially problematic. Derived features used to characterize these groups may not be present in early members, making it difficult to determine whether a catarrhine from the Early or Late Oligocene is a stem catarrhine, stem hominoid, or stem cercopithecoid. Kamoyapithecus is an example of this problem: dental similarities of hominoids are primitive for catarrhines (71–74), complicating any assessment of its phylogenetic position. Even Proconsul, which is often considered to be a stem hominoid ancestral to all other known apes, has been suggested by some to be a stem catarrhine (e.g., refs. 71 and 75) because it has few derived features linking it to hominoids other than the absence of a tail and large body size (72, 76). Compared with Aegyptopithecus, cercopithecoid taxa are more postcranially and dentally derived than hominoids (2), but it is possible that diagnostic features such as bilophodonty are not present in the earliest members of this lineage. Indeed, at the time of their divergence hominoid and cercopithecoid lineages would be as morphologically similar as sister species and virtually unrecognizable as precursors to more distinct groups, raising the possibility that easily recognizable traits such as the absence of a tail or the presence of bilophodonty may not be especially diagnostic of early hominoids or cercopithecoids. In this context, supposed stem catarrhines such as Dendropithecus or Limnopithecus (72) may be more closely related to either hominoids or cercopithecoids than previously thought.
Ultimately, it may be difficult to confidently recognize the earliest members of two closely related lineages, but a reassessment of hominoid and cercopithecoid diagnostic features is warranted given the distance between our estimated divergence and what is known from the fossil record. This gap in our knowledge underscores how poorly we understand the earliest portions of ape and Old World monkey evolution and indicates that finding new sites within Afro-Arabia or reinvestigating old sites from within this time period will be critical to reconstructing the earliest portions of their evolution.
Acknowledgments
We thank W.-G. Qiu, C. Roos, J. Rossie, and two anonymous reviewers for comments on the manuscript. T.Y.S. thanks W.-G. Qiu and the Evolutionary Bioinformatics Laboratory at Hunter College for computational support. Some of the sequence data used were generated by the National Institutes of Health (NIH) Intramural Sequencing Center (www.nisc.nih.gov). This work was supported by Research Centers in Minority Institutions Award RR-03037 (to M.E.S.) from the National Center for Research Resources (NCRR) of the NIH, which supports the infrastructure of the Anthropological Genetics Laboratory at Hunter College, and by Cora du Bois Charitable Trust and American School for Prehistoric Research graduate fellowships (to N.M.Y.).
Author contributions: M.E.S. and N.M.Y. designed research; M.E.S., N.M.Y., and T.Y.S. performed research; M.E.S., N.M.Y., and T.Y.S. analyzed data; and M.E.S. and N.M.Y. wrote the paper.
This paper was submitted directly (Track II) to the PNAS office.
Abbreviation: Mya, million years ago.
Footnotes
Delson, E. (1996) in Abstracts, International Symposium: Evolution of Asian Primates (Primate Research Institute, Inuyama, Japan), p. 40.
References
- 1.Pilbeam, D. R. & Young, N. M. (2004) C. R. Palevol. 3, 303–319. [Google Scholar]
- 2.Gingerich, P. (1984) Yrbk. Phys. Anthrop. 27, 57–72. [Google Scholar]
- 3.Kumar, S. & Hedges, S. B. (1998) Nature 392, 917–920. [DOI] [PubMed] [Google Scholar]
- 4.Glazko, G. V. & Nei, M. (2003) Mol. Biol. Evol. 20, 424–434. [DOI] [PubMed] [Google Scholar]
- 5.Andrews, P., Harrison, T., Martin, L. & Pickford, M. (1981) J. Hum. Evol. 10, 123–128. [Google Scholar]
- 6.Pickford, M. & Andrews, P. (1981) J. Hum. Evol. 10, 1–33. [Google Scholar]
- 7.Gebo, D. L., MacLatchy, L., Kityo, R., Deino, A., Kingston, J. & Pilbeam, D. (1997) Science 276, 401–404. [DOI] [PubMed] [Google Scholar]
- 8.Boschetto, H. B., Brown, F. H. & McDougall, I. (1992) J. Hum. Evol. 22, 47–71. [Google Scholar]
- 9.Delson, E., Tattersall, I., Van Couvering, J. A. & Brooks, A. S. (2000) in Encyclopedia of Human Evolution and Prehistory, eds. Delson, E., Tattersall, I., Van Couvering, J. A. & Brooks, A. S. (Garland, New York), 2nd Ed., pp. 166–171.
- 10.Delson, E. (1992) in Cambridge Encyclopedia of Human Evolution, eds. Jones, S., Martin, R. D. & Pilbeam, D. R. (Press Syndicate of the University of Cambridge, Cambridge, U.K.), pp. 217–222.
- 11.Jablonski, N. J. (2002) in The Primate Fossil Record, ed. Hartwig, W. (Cambridge Univ. Press, Cambridge, U.K.), pp. 255–300.
- 12.Leakey, M. G., Ungar, P. S. & Walker, A. (1995) J. Hum. Evol. 28, 519–531. [Google Scholar]
- 13.MacLatchy, L. (2004) Evol. Anthropol. 13, 90–103. [Google Scholar]
- 14.Young, N. M. & MacLatchy, L. (2004) J. Hum. Evol. 46, 163–184. [DOI] [PubMed] [Google Scholar]
- 15.Bailey, W. J., Fitch, D. H., Tagle, D. A., Czelusniak, J., Slightom, J. L. & Goodman, M. (1991) Mol. Biol. Evol. 8, 155–184. [DOI] [PubMed] [Google Scholar]
- 16.Stauffer, R. L., Walker, A., Ryder, O. A., Lyons-Weiler, M. & Hedges, S. B. (2001) J. Hered. 92, 469–474. [DOI] [PubMed] [Google Scholar]
- 17.Schrago, C. G. & Russo, C. A. (2003) Mol. Biol. Evol. 20, 1620–1625. [DOI] [PubMed] [Google Scholar]
- 18.Goodman, M., Porter, C. A., Czelusniak, J., Page, S. L., Schneider, H., Shoshani, J., Gunnell, G. & Groves, C. P. (1998) Mol. Phylogenet. Evol. 9, 585–598. [DOI] [PubMed] [Google Scholar]
- 19.Simons, E. L. & Rasmussen, D. T. (1994) Evol. Anthropol. 3, 128–139. [Google Scholar]
- 20.Rasmussen, D. T. (2002) in The Primate Fossil Record, ed. Hartwig, W. (Cambridge Univ. Press, Cambridge, U.K.), pp. 203–220.
- 21.Sanders, W. J., Kappelman, J. & Rasmussen, D. T. (2004) Acta Palaeontol. Pol. 49, 365–392. [Google Scholar]
- 22.Kappelman, J., Rasmussen, D. T., Sanders, W. J., Feseha, M., Bown, T., Copeland, P., Crabaugh, J., Fleagle, J., Glantz, M., Gordon, A., et al. (2003) Nature 426, 549–552. [DOI] [PubMed] [Google Scholar]
- 23.Kumar, S. & Subramanian, S. (2002) Proc. Natl. Acad. Sci. USA 99, 803–808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Caccone, A. & Powell, J. R. (1989) Evolution (Lawrence, Kans.) 43, 925–942. [DOI] [PubMed] [Google Scholar]
- 25.Sibley, C. G. & Ahlquist, J. E. (1987) J. Mol. Evol. 26, 99–121. [DOI] [PubMed] [Google Scholar]
- 26.Brunet, M., Guy, F., Pilbeam, D., Mackaye, H. T., Likius, A., Ahounta, D., Beauvilain, A., Blondel, C., Bocherens, H., Boisserie, J. R., et al. (2002) Nature 418, 145–151. [DOI] [PubMed] [Google Scholar]
- 27.Graur, D. & Martin, W. (2004) Trends Genet. 20, 80–86. [DOI] [PubMed] [Google Scholar]
- 28.Laird, C. D., McConaughy, B. L. & McCarthy, B. J. (1969) Nature 224, 149–154. [DOI] [PubMed] [Google Scholar]
- 29.Wu, C. I. & Li, W. H. (1985) Proc. Natl. Acad. Sci. USA 82, 1741–1745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Britten, R. J. (1986) Science 231, 1393–1398. [DOI] [PubMed] [Google Scholar]
- 31.Gu, X. & Li, W. H. (1992) Mol. Phylogenet. Evol. 1, 211–214. [DOI] [PubMed] [Google Scholar]
- 32.Li, W. H., Ellsworth, D. L., Krushkal, J., Chang, B. H. & Hewett-Emmett, D. (1996) Mol. Phylogenet. Evol. 5, 182–187. [DOI] [PubMed] [Google Scholar]
- 33.Douzery, E. J., Delsuc, F., Stanhope, M. J. & Huchon, D. (2003) J. Mol. Evol. 57, Suppl. 1, S201–S213. [DOI] [PubMed] [Google Scholar]
- 34.Yi, S., Ellsworth, D. L. & Li, W. H. (2002) Mol. Biol. Evol. 19, 2191–2198. [DOI] [PubMed] [Google Scholar]
- 35.Yoder, A. D. & Yang, Z. (2000) Mol. Biol. Evol. 17, 1081–1090. [DOI] [PubMed] [Google Scholar]
- 36.Adkins, R. M., Honeycutt, R. L. & Disotell, T. R. (1996) Mol. Biol. Evol. 13, 1393–1404. [DOI] [PubMed] [Google Scholar]
- 37.Gissi, C., Reyes, A., Pesole, G. & Saccone, C. (2000) Mol. Biol. Evol. 17, 1022–1031. [DOI] [PubMed] [Google Scholar]
- 38.Easteal, S. & Collet, C. (1994) Mol. Biol. Evol. 11, 643–647. [DOI] [PubMed] [Google Scholar]
- 39.Easteal, S. (1991) Mol. Biol. Evol. 8, 115–127. [DOI] [PubMed] [Google Scholar]
- 40.Herbert, G. & Easteal, S. (1996) Mol. Biol. Evol. 13, 1054–1057. [DOI] [PubMed] [Google Scholar]
- 41.Goodman, M. (1961) Hum. Biol. 33, 131–162. [PubMed] [Google Scholar]
- 42.Goodman, M., Barnabas, J., Matsuda, G. & Moore, G. W. (1971) Nature 233, 604–613. [DOI] [PubMed] [Google Scholar]
- 43.Koop, B. F., Goodman, M., Xu, P., Chan, K. & Slightom, J. L. (1986) Nature 319, 234–238. [DOI] [PubMed] [Google Scholar]
- 44.Li, W. H. & Tanimura, M. (1987) Nature 326, 93–96. [DOI] [PubMed] [Google Scholar]
- 45.Li, W. H., Tanimura, M. & Sharp, P. M. (1987) J. Mol. Evol. 25, 330–342. [DOI] [PubMed] [Google Scholar]
- 46.Ellsworth, D. L., Hewett-Emmett, D. & Li, W. H. (1993) Mol. Phylogenet. Evol. 2, 315–321. [DOI] [PubMed] [Google Scholar]
- 47.Seino, S., Bell, G. I. & Li, W. H. (1992) Mol. Biol. Evol. 9, 193–203. [DOI] [PubMed] [Google Scholar]
- 48.Poux, C. & Douzery, E. J. (2004) Am. J. Phys. Anthropol. 124, 1–16. [DOI] [PubMed] [Google Scholar]
- 49.Rambaut, A. & Bromham, L. (1998) Mol. Biol. Evol. 15, 442–448. [DOI] [PubMed] [Google Scholar]
- 50.Bromham, L., Rambaut, A., Fortey, R., Cooper, A. & Penny, D. (1998) Proc. Natl. Acad. Sci. USA 95, 12386–12389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Delgado, S., Casane, D., Bonnaud, L., Laurin, M., Sire, J. Y. & Girondot, M. (2001) Mol. Biol. Evol. 18, 2146–2153. [DOI] [PubMed] [Google Scholar]
- 52.Huchon, D., Catzeflis, F. M. & Douzery, E. J. (2000) Proc. R. Soc. London B 267, 393–402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Huchon, D., Madsen, O., Sibbald, M. J., Ament, K., Stanhope, M. J., Catzeflis, F., de Jong, W. W. & Douzery, E. J. (2002) Mol. Biol. Evol. 19, 1053–1065. [DOI] [PubMed] [Google Scholar]
- 54.Kawashita, S. Y., Sanson, G. F., Fernandes, O., Zingales, B. & Briones, M. R. (2001) Mol. Biol. Evol. 18, 2250–2259. [DOI] [PubMed] [Google Scholar]
- 55.Pitra, C., Lieckfeldt, D., Frahnert, S. & Fickel, J. (2002) Mol. Phylogenet. Evol. 23, 63–74. [DOI] [PubMed] [Google Scholar]
- 56.Springer, M. S., Murphy, W. J., Eizirik, E. & O'Brien, S. J. (2003) Proc. Natl. Acad. Sci. USA 100, 1056–1061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Senut, B., Pickford, M., Gommery, D., Mein, P., Chebol, K. & Coppens, Y. (2001) C. R. Acad. Sci. Ser. IIa 332, 137–144. [Google Scholar]
- 58.Haile-Selassie, Y., Suwa, G. & White, T. D. (2004) Science 303, 1503–1505. [DOI] [PubMed] [Google Scholar]
- 59.Altschul, S. F., Gish, W., Miller, W., Myers, E. W. & Lipman, D. J. (1990) J. Mol. Biol. 215, 403–410. [DOI] [PubMed] [Google Scholar]
- 60.Thompson, J. D., Higgins, D. G. & Gibson, T. J. (1994) Nucleic Acids Res. 22, 4673–4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Thompson, J. D., Gibson, T. J., Plewniak, F., Jeanmougin, F. & Higgins, D. G. (1997) Nucleic Acids Res. 25, 4876–4882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Rosenberg, M. S. & Kumar, S. (2003) Mol. Biol. Evol. 20, 610–621. [DOI] [PubMed] [Google Scholar]
- 63.Hasegawa, M., Kishino, H. & Yano, T. (1985) J. Mol. Evol. 22, 160–174. [DOI] [PubMed] [Google Scholar]
- 64.Yang, Z. (1994) J. Mol. Evol. 39, 306–314. [DOI] [PubMed] [Google Scholar]
- 65.Swofford, D. L. (2001) paup*: Phylogenetic Analysis Using Parsimony (*and Other Methods) (Sinauer, Sunderland, MA), Version 4.
- 66.Yang, Z. & Yoder, A. (2003) Syst. Biol. 52, 705–716. [DOI] [PubMed] [Google Scholar]
- 67.Felsenstein, J. (1981) J. Mol. Evol. 17, 368–376. [DOI] [PubMed] [Google Scholar]
- 68.Rodriguez-Trelles, F., Tarrio, R. & Ayala, F. J. (2002) Proc. Natl. Acad. Sci. USA 99, 8112–8115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Ebersberger, I., Metzler, D., Schwarz, C. & Paabo, S. (2002) Am. J. Hum. Genet. 70, 1490–1497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Sibley, C. G., Comstock, J. A. & Ahlquist, J. E. (1990) J. Mol. Evol. 30, 202–236. [DOI] [PubMed] [Google Scholar]
- 71.Harrison, T. (1987) J. Hum. Evol. 16, 41–80. [Google Scholar]
- 72.Harrison, T. (2002) in The Primate Fossil Record, ed. Hartwig, W. (Cambridge Univ. Press, Cambridge), pp. 311–338.
- 73.Fleagle, J. G. & Kay, R. (1983) in New Interpretations of Ape and Human Ancestry, eds. Ciochon, R. & Corruccini, R. (Plenum, New York), pp. 181–210.
- 74.Corruccini, R., Ciochon, R. & McHenry, H. (1976) Primates 17, 205–223. [Google Scholar]
- 75.Rossie, J. B., Simons, E. L., Gauld, S. C. & Rasmussen, D. T. (2002) Proc. Natl. Acad. Sci. USA 99, 8454–8456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Ward, C. V., Walker, A. & Teaford, M. F. (1991) J. Hum. Evol. 21, 215–220. [Google Scholar]
- 77.Delson, E. (1980) in The Macaques: Studies in Ecology, Behavior, and Evolution, ed. Lindburg, D. E. (Van Nostrand, New York),
- 78.Köhler, M., Moyà-Solà, S. & Alba, D. M. (2000) J. Hum. Evol. 38, 447–452. [DOI] [PubMed] [Google Scholar]