ABSTRACT
Evidence of cancer immunosurveillance and immunoediting processes has been primarily demonstrated in mouse models of chemically induced oncogenesis. Although these models are very tractable, they are characterized by high mutational loads that represent a minority of human cancers. In this study, we sought to determine whether cancer immunosurveillance and immunoediting could be demonstrated in a more clinically relevant oncogene-induced model of carcinogenesis, the MMTV-PyMT (PyMT) mammary carcinoma model. This model system in the FVB/NJ strain background was previously used to demonstrate that adaptive immunity had no role in limiting primary cancer formation and in fact promoted metastasis, thus calling into question whether cancer immunosurveillance operated in preventing the development of breast cancer. Our current study in the C57BL/6 strain backgrounds provides a different conclusion, as we report here the existence of an adaptive immunosurveillance of PyMT mammary carcinomas using two independent models of immune deficiency. PyMT mice bred onto a Rag1−/− background or immune suppressed by chronic tacrolimus therapy both demonstrated accelerated development of mammary carcinomas. By generating a bank of cell lines from these animals, we further show that a subset of PyMT cell lines had delayed growth after transplantation into wild-type (WT) syngeneic, but not immune-deficient hosts. This reduced growth rate in immunocompetent animals was characterized by an increase in immune cell infiltration and tissue differentiation. Furthermore, loss of the immune cell infiltration that characterized immunoediting of slow growing cell lines, changed them into fast growing variants capable of progressing in the immunocompetent model. In conclusion, our study provides evidence that immunosurveillance and immunoediting of PyMT-derived cell lines modulate tumor progression in this oncogene-induced model of cancer.
KEYWORDS: Growers and fast growers, immune cell infiltration, immune-mediated slow, immunosurveillance, immunoediting, mammary cancer, MMTV-PyMT, oncogene-induced model
Abbreviations
- BFA
brefeldin A
- HPF
high-power field
- MCA
methylchloranthrene
- MMTV-PyMT
mouse mammary tumor virus-polyoma virus middle T antigen
- vs.
versus
- WT
wild type
- d
days
Introduction
Evidence for cancer immunosurveillance comes from studies monitoring differences in cancer incidence in immunocompetent versus (vs.) immunodeficient animals. These studies utilize chemical carcinogenesis models, spontaneous models, and genetic cancer models. Using the well-studied 3-methylcholanthrene (MCA) chemically induced tumorigenesis model, several groups showed a requirement for adaptive immunity, natural killer (NK) cells, and NK-T cells in the surveillance of murine sarcomas.1-7 These studies have shown that immunosurveillance of cancer cells occurs as part of an elimination step in the cancer immunoediting process.8 Animals that fail to completely eradicate MCA-induced sarcoma cells can undergo an equilibrium phase,1 in which a small number of persisting tumor cells is held in check by an active antitumor immune response. This stage can be followed by the emergence of immune “edited,” escaped cancers that have acquired or abolished the expression of certain genes to avoid immune recognition.3,9-12 In addition to the MCA model, other studies using spontaneous or genetic models of cancer have found a role for perforin, NKT cells, NKG2D, and CD226 in the surveillance of hematologic malignancies.7,13-17 Furthermore, genetic models showed that hepatocarcinoma and prostate cancer are surveilled by adaptive immune responses.18-20
Although considerable support for cancer immunoediting has emerged, there is a paucity of studies and lack of consensus on the role of immunity in oncogene-induced non-hematologic cancers. In fact, recent studies using oncogene-induced sarcomas have shown that there is no cancer surveillance and immunoediting unless specific antigens were introduced into these cancers,21 in contrast to the robust cancer surveillance and immunoediting seen with MCA-induced sarcoma models.2,12,22 The basis for this difference could lie in the scarcity of mutations in genetic vs. carcinogen-induced models of cancer.12
Breast cancer has a large inflammatory component that typically promotes cancer cell growth and metastasis.23-26 Mouse models of breast cancer are abundant,27 but perhaps the most studied model is the MMTV-PyMT (PyMT) model,28 in which the middle T antigen from the polyoma virus is expressed as an oncogene in mammary tissues to drive tumor formation. Studies using this model have come to differing conclusions on the role of immunity in tumor formation and progression. One group found that genetic depletion of the main immune components of the adaptive immune system performed by generating PyMT mice with a homozygous null mutation in Rag1, or mice more specifically lacking CD4+, CD8+, or B cells had no impact on primary tumor latency or progression, but were resistant to metastasis.25 In contrast, recent studies have found that deficiency in IL-15 led to an increase in cancer development in PyMT animals, suggesting that innate lymphocytes could inhibit breast cancer formation in this model.29
Earlier studies investigating the effect of immunity on PyMT tumor formation had been performed in the FVB/NJ strain background, which has a tumor latency 6 weeks shorter than the C57BL/6J strain.30,25 We sought to determine if this discrepancy was due to the extremely short tumor latency and rapid tumor progression observed in FVB/NJ PyMT mice. Indeed, we hypothesized that immunosurveillance would be insufficient to control the rapid and aggressive oncogene-driven tumor progression in the FVB/NJ background.
In the current study, we show that the adaptive immune system can delay cancer formation in PyMT mice. In addition, by generating a bank of cell lines with varying growth kinetics that depend on the presence of adaptive immune cells, we further demonstrate the occurrence of immunosurveillance and immunoediting in breast cancer. This study provides much-needed evidence that immunosurveillance and immunoediting of cancer cells can occur outside of carcinogen-induced cancer models and moreover suggest that breast cancer patients may benefit from cancer immune therapy.
Materials and methods
Experimental procedures
All experiments involving mice were conducted under an animal protocol approved by the University of California, San Diego Institutional Animal Care and Use Committee (IACUC protocol #S06201).
Mice
C57BL/6 (PyMT) MMTV-PyMT30 and FVB/NJ (PyMT) MMTV-PyMT28 mice were used in these studies. C57BL/6 PyMT mice were bred with recombinase active gene 1 (Rag1)−/− mice to generate PyMT/Rag1−/−. C57BL/6 WT, C57BL/6 Rag1−/−, and Rag2−/− x γc−/− mice were used for tumor transplantation experiments. All mice were bred in-house and therefore exposed to similar microbiota. In syngeneic model systems, tumor cell lines were transplanted into sex-matched animals. In spontaneous tumor formation experiments, tumor sites were palpated biweekly to monitor tumor latency and progression.
Tumor transplantation
Orthotopic engraftment of mammary tumor cell lines was performed by injections of 20 µL of 105 PyMT cells in PBS mixed 1:1 with matrigel (#E1270, Sigma) into the #4 and #9 fat pads of C57BL/6 WT, C57BL/6 Rag1−/−, and Rag2−/− x γc−/− mice anesthetized by a combination of ketamine (Ketaset, Zoetis) and xylazine (AnaSed, Lloyd) delivered intraperitoneally (i.p.). Tumor sites were palpated biweekly to determine the initiation of tumor formation.
Tacrolimus-induced immune suppression
13 mg biodegradable 90 d-releasable tacrolimus pellets or placebo-containing pellets (Innovative Research of America, FL, USA and Tacropellet, MD, USA) were implanted subcutaneously in the interscapular region of the back in C57BL/6 PyMTand FVB/NJ PyMT mice anesthetized by a combination of ketamine and xylazine, as described previously.31 C57BL/6 WT control animals were implanted with 13 mg biodegradable 90 d-releasable tacrolimus pellets or placebo-containing pellets.
Generation of PyMTcell lines
C57BL/6 PyMT and C57BL/6 PyMT/Rag1−/− mammary tumor cell lines were established from spontaneous mammary tumors arising in C57BL/6 PyMT females as described previously.32 Tumors were minced and incubated at 37°C with 250 rpm agitation for at least an hour in 4 mL of Ham's F12K medium containing 1 mg/mL collagenase type II (#LS004174, Worthington Biochemicals, Lakewood, NJ, USA), 2 mg/mL soybean trypsin inhibitor (#T6522, Sigma-Aldrich, St. Louis, MO, USA), and 2% BSA (Sigma-Aldrich, St. Louis, MO, USA). Minced and digested tissues were sequentially re-suspended in red blood cell lysis solution (Sigma), then 0.25% trypsin/EDTA solution (Gibco) and dispase (#07923, Stem Cell technologies).
After multiple washes with a FCS-containing medium, the suspension was passed through a 70 μm nylon filter (Fisher Scientific, Pittsburg, PA, USA) and the single cells were pelleted by centrifugation and cultured in Ham's F12K medium (Gibco) containing 5% FCS, 2.5 μg/mL fungizone (#FG-70, Omega Scientific), 50 μg/mL gentamycin (Gibco), and MITO+ (#355006, Corning). The cells were grown in flasks for several weeks. Typically, after the first week, a small population of immortalized cancer cells emerges. These are expanded and frozen. All cell lines were derived in an identical manner and are considered representative of subpopulations of tumor cells within the original tumor mass.
Histology and immunostaining
Tumors were fixed for 24 h in 10% neutral buffered formalin (BDH) and stored in 70% ethanol before embedding in paraffin. Sections were stained with H&E for pathological analysis and adjacent sections were evaluated by immunohistochemistry (IHC) with an anti-CD45 antibody (#ab10558, Abcam). For IHC, the slides were de-paraffinized and blocked for endogenous peroxidase and endogenous biotin and overlayed with the antibody at 5 µg/mL in a humidified chamber overnight at 4°C. Slides were washed with tris buffered saline with 0.1% tween 20 (TBST) after every incubation. Separate slides received irrelevant IgG control. Bound antibody was detected using biotinylated anti-mouse antibody, followed by HRP labeled Streptavidin and substrate color development used AEC (Amino-ethyl carbazole, Vector Labs Burlingame CA), following the manufacturer's recommendations. Nuclei were counterstained using Mayer's hematoxylin and the slides were mounted in aqueous mounting media for visualization using an Olympus BH2 light microscope with digital photomicrography using Olympus Magnafire software. The number of CD45+ events per 40× high-power field were counted using a Leica DM2500 microscope.
Splenocyte isolation
Spleens were dissected from placebo or tacrolimus pellet implanted animals and homogenized by mechanical disruption. To obtain a single-cell suspension, the homogenized tissues were first subjected to red blood cell lysis, washed with PBS and then filtered through a 70 μm nylon filter (Fisher Scientific, Pittsburg, PA, USA).
Antibodies and flow cytometry
Splenocytes were vigorously re-suspended and washed in FACS buffer (PBS + 1% FCS + 0.05% NaN3; Sigma-Aldrich). The following anti-mouse antibodies were used anti-: CD45 (#103116), CD4+ (GK1.5, #103116) (Biolegend) and CD8+ (53–6.7, 11–0081–85) (eBioscience). Cell surface staining was conducted for 15–20 min at 4°C in FACS tubes containing 1–2×106 total cells, 0.1–0.2 µg of antibody, 1 µg of Fc blocking anti-CD16/32 antibody (for tumor transplant analysis), and 100 µL of FACS buffer. 7AAD (EMD Millipore) was added at 1 µg/mL immediately before FACS analysis.
For intracellular staining, stimulated and unstimulated splenocytes were incubated with or without 1 μg/mL Brefeldin A (BFA, BD Biosciences) for 5 h and then prepared for intracellular FACS staining. Briefly, the cells were washed with PBS, incubated with Cytofix for 15 min at 4°C and washed twice with Perm/Wash solution (BD biosciences). Fixed cells were then incubated for 30 min in the dark with antibodies against the intracellular proteins IFNγ and IL-2 (eBioscience) diluted in the Perm/Wash solution. Cells were analyzed on a BD FACSCanto.
T cell stimulation
Splenocytes were cultured in 48-well plates pre-coated with 1 µg/mL anti-CD3 (Clone: 17A2, Catalog#: BE0002, BioXcell) and anti-CD28 antibodies (Clone: 37.51 Catalog#: BE0015–1, BioXcell) for either 5 h with BFA or 3 d with 50 U/mL of IL-2 (#589102, Biolegend). Stimulated splenocytes were then processed for FACS staining.
Statistical analysis
Statistical significance between two groups at defined time points was determined by the Student's t-test using two-tailed analysis to obtain p-values. The Log-Rank (Mantel-Cox) test was used to compare the survival of mice across tumor transplantation or primary oncogenesis and * reflects the p values obtained in between groups at a given time point calculated by the Student's t-test. Error bars are depicted using the SEM, mean is represented as center values and *p <0.05, **p <0.01, ***p <0.001. All experiments were done at least twice and representative data are shown. In primary transplant experiments, non-growing tumors in the Rag2−/− x γc−/− environment were excluded, as the fat pad morphology in this immune background did not allow a 100% rate of success for the mammary fat pad injections in contrast to WT mice.
Results
Early immune suppression and adaptive immunodeficiency accelerates mammary tumor latency in C57BL/6 PyMT mice
To study the impact of acute immunosuppression on tumor latency, 6- or 11-week-old PyMT C57BL/6 mice were implanted with a 13 mg slow-release tacrolimus pellet in the interscapular region of the back (Figs. 1A and B).31 CD4+ and CD8+ T cells are activated by IL-2 produced by a calcium-dependent Il-2 gene transcription that is blocked by tacrolimus.33 We confirmed that the slow release of tacrolimus led to sustained immune suppression as CD8+ mediated-IL-2 production under basal conditions as well as after CD3/CD28 stimulation was blocked (Fig. S1A). Moreover, IL-2 production by CD4+ cells after CD3/CD28 stimulation (Fig. S1B) as well as the number and percentages of CD4+ cells with or without CD3/CD28/IL-2 stimulation (Figs. S1F and H) was diminished. NK cells, however, were not impacted by tacrolimus-mediated immune suppression (data not shown).
Figure 1.
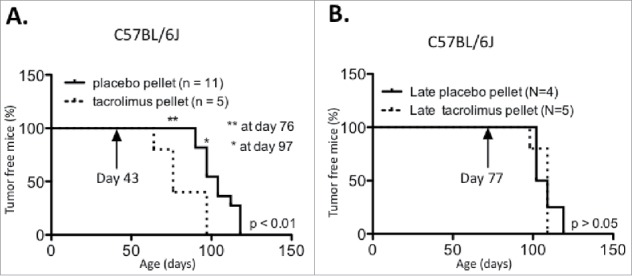
Tacromilus immune suppression can impact PyMT tumor latency in the C56BL/6 background. PyMT C56BL/6 mice were implanted at age (A) 43 d or (B) 77 d with a 13 mg biodegradable 90 d-releasable tacrolimus or placebo pellet in the interscapular region of the back, and tumor onset was recorded over time.
Having confirmed that tacrolimus caused immune suppression, we compared tumor formation in cohorts of C57BL/6J PyMT mice implanted with tacrolimus vs. placebo pellets. The results showed that in mice implanted at 6 weeks, tumor latency was accelerated in the presence of tacrolimus, with an average tumor onset of 82 d vs. 104 d for mice implanted with the placebo (Fig. 1A, p < 0.01). In concordance, the number of tumor-bearing mice was significantly increased with tacrolimus treatment at days 76 and 97. However, when the pellet was inserted at 11 weeks of age, tacrolimus-mediated immune suppression had no impact on tumor latency (Fig. 1B).
We tested our hypothesis that strain background had a significant effect on the efficacy of immunosurveillance by implanting 13 mg biodegradable 90 d-releasable tacrolimus pellets at age 30 d in the interscapular region of the backs of FVB/NJ PyMT mice. Immune suppression had no impact on tumor latency in the FVB/NJ background (Fig. 2A), suggesting a strain-specific immunosurveillance of tumorigenesis that could be attributed to the early and aggressive tumor progression that characterizes PyMT mice generated in the FVB/NJ background.
Figure 2.
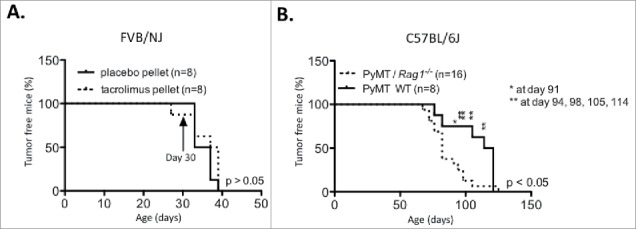
Impact of adaptive immune deficiency in the C56BL/6 PyMT background and strain specific tumor latency in the PyMT FVB/NJ background. (A) PyMT FVB/NJ mice were implanted at 30 d of age with a 13 mg biodegradable 90 d-releasable tacrolimus or placebo pellet in the interscapular region of the back. (B) Tumor onset was compared between C57BL/6 PyMT/Rag1−/− and PyMT WT mice.
We next sought to confirm the tacrolimus results using a genetic approach similar to that used in previous studies.25 Fig. 2B shows that PyMT/Rag1−/− mice in the C57BL/6J background displayed a significantly earlier tumor onset than PyMT WT mice (86.5 vs. 107.62 d, p < 0.05). These data contradict previously published results obtained on the FVB/NJ background25,34 and provide further support for our hypothesis.
Immune-dependent growth kinetics of PyMT cell lines
We next hypothesized that the earlier cancer formation occurring in PyMT/Rag1−/− mice resulted in unedited cancers. To test this hypothesis, 12 cell lines were generated from primary mammary tumors arising in either PyMT/Rag1−/− hosts (n = 6) or PyMT WT hosts (n = 6) by producing a single-cell suspension from the tumor masses and cultivating the cells in vitro for a week (Fig. 3A). The immunogenicity of each cell line was determined by injecting 105 cells into the mammary fat pads of severely immunodeficient Rag2−/− x γc−/− or WT mice (Fig. 3B), as performed previously to define regressor and progressor cells in the MCA-sarcoma system.2,3
Figure 3.
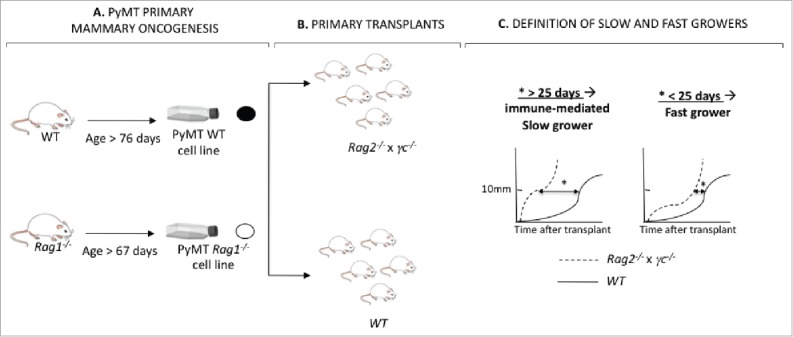
Generation of PyMT cell lines from Rag1−/− and WT hosts. (A) PyMT mice in the Rag1−/− or WT immune background developed mammary tumors over time. Tumors from each animal were harvested into a single-cell suspension and a cell line was generated. (B) Each cell line was transplanted into five Rag2−/− x γc−/− and five WT mice. (C) The kinetic of each cell line in immune deficient Rag2−/− x γc−/− vs. immune competent WT mice was compared by determining the difference in time to reach 10×10 mm in size in Rag2−/− x γc−/− vs. WT mice. When this value (*) was inferior to 25 d, the cell lines were identified as immune-mediated slow growers. Cell lines for which this value was superior to 25 d, the cell lines were identified as fast growers.
The influence of the immune system on tumor growth was examined by comparing the growth of transplanted cell lines in severely immunodeficient Rag2−/− x γc−/− hosts vs. immunocompetent WT host (Fig. 3C). Classic regressor growth was not observed in this model system. Rather, we observed that some cell lines exhibited delayed growth in WT but not Rag2−/− x γc−/− hosts. To demonstrate the growth delay caused by host immunity, we calculated the average time to reach a tumor size of 10×10 mm in WT vs. Rag2−/− x γc−/− for all the cell lines (Table 1: Time10×10(WT) − Time10×10(Rag2−/− x γc−/−)). Based on this measure, cell lines were subsequently assigned either a slow grower (Time10×10(WT) − Time10×10 (Rag2−/− x γc−/−) < 25 d) or fast grower (Time10×10(WT) − Time10×10 (Rag2−/− x γc−/−) > 25 d) phenotype (Figs. 3C, 4A and B, Table 1). Surprisingly, the number of slow growers and fast growers were almost identical for both genotypes from which the cell lines originated. Four out of the six PyMT/Rag1−/− cell lines (#53, #29, #31, #32) and PyMT WT cell lines (#252, #234, #256, #235) showed a fast grower phenotype.
Table 1.
Characteristics of the PyMT cell lines generated from tumors arising in a WT or a Rag1−/− background.
| PRIMARY ONCOGENESIS |
PRIMARY TRANSPLANT |
||||||
|---|---|---|---|---|---|---|---|
| # | Genotype | Age onset (days) | Age at tumor harvest (days) | Average time: 10×10 mm in WT host1(days) | Average time: 10×10 mm in Rag2−/− x γc−/− host2(days) | Time1– Time2 | Growth phenotype |
| 53 | PyMT/Rag1−/− | 79 | 32 | 32 | 33 | −1 | FAST |
| 29 | PyMT/Rag1−/− | 69 | 40 | 40 | 35 | 5 | FAST |
| 31 | PyMT/Rag1−/− | 97 | 62 | 62 | 56 | 6 | FAST |
| 32 | PyMT/Rag1−/− | 97 | 47 | 47 | 26 | 21 | FAST |
| 55 | PyMT/Rag1−/− | 91 | 90 | 90 | 50 | 40 | SLOW |
| 69 | PyMT/Rag1−/− | 80 | 92 | 92 | 25 | 67 | SLOW |
| 252 | PyMT WT | 74 | 26 | 26 | 29 | −3 | FAST |
| 234 | PyMT WT | 117 | 37 | 37 | 33 | 4 | FAST |
| 256 | PyMT WT | 115 | 74 | 74 | 74 | 0 | FAST |
| 235 | PyMT WT | 93 | 36 | 36 | 14 | 22 | FAST |
| 276 | PyMT WT | 78 | 96 | 96 | 59 | 37 | SLOW |
| 238 | PyMT WT | 113 | 108 | 108 | 53 | 55 | SLOW |
Figure 4.
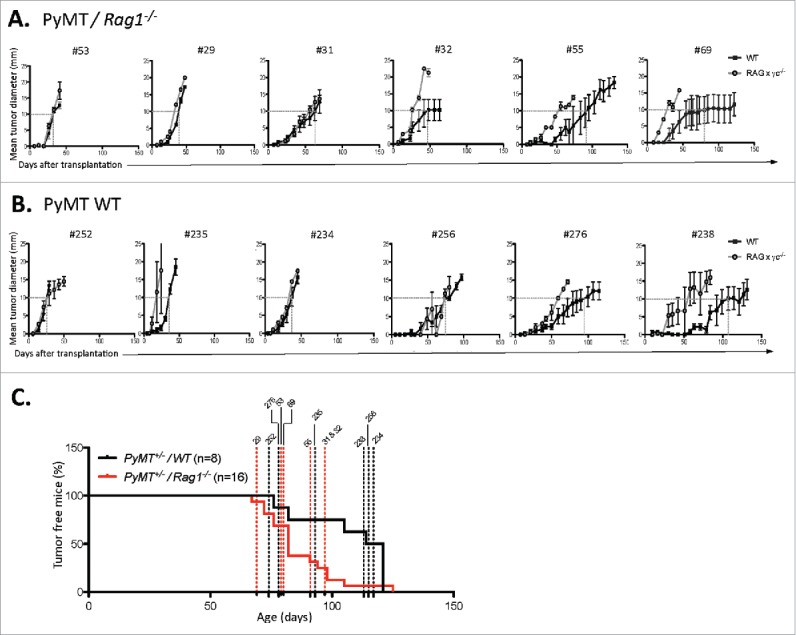
Immune-dependent growth kinetics of PyMT cell lines. PyMT cell lines were generated from primary tumors from PyMT mice (A) Rag1−/− or (B) WT mice. Each cell line was subsequently passaged in either severely immune deficient Rag2−/− x γc−/− (open circle) or WT mice (closed square). For each cell line, a dotted line shows the age at which tumor size reached a 10×10 mm mean tumor diameter. (C) The primary tumor latency of each cell lines was reported on the graph showing the primary tumor onset of PyMT/Rag1−/− mice vs. PyMT WT mice.
Two cell lines from PyMT/Rag1−/− hosts (#69 and #55) and two cell lines from PyMT WT hosts (#276 and #238) displayed a slow grower phenotype in the presence of complete immunity, taking more than 90 d to form tumors in WT hosts. No correlations were found regarding age of tumor onset, age of tumor harvest, and tumor burden at end point between slow and fast growers (Fig. 4C, Table 1).
In accordance with the identical number of slow and fast growers in each of the genotypes tested, no significant differences were found when comparing the genotype of the cell lines for their survival after primary transplants in immunocompetent (Fig. 5A) vs. immunodeficient hosts (Fig. 5B). Similarly, there were no significant differences in duration to reach 10×10 mm of diameter in WT vs. Rag2−/− x γc−/− hosts (Fig. 5C).
Figure 5.
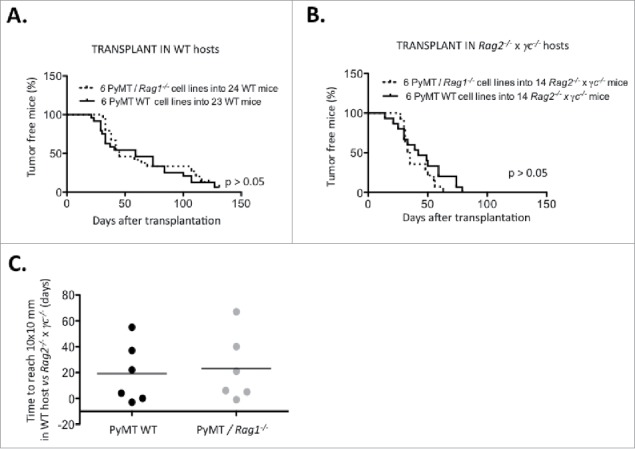
Immune-dependent growth kinetics of PyMT cell lines, Using a cut-off of 10×10 mm mean tumor diameter, survival of (A) 24 WT mice and (B) 14 Rag2−/− x γc−/− mice transplanted with either six PyMT/Rag1−/− cell lines or six PyMT WT cell lines was analyzed. (C) The time difference to reach 10×10 mm in WT vs. Rag2−/− x γc−/− hosts is plotted.
Nevertheless, several cell lines (#55, #69, #238, #276) displayed a clear delay in growth after transplantation into WT vs. immune deficient Rag2−/− x γc−/−. These cell lines were defined as immunogenic and selected for further study.
Slow growers are characterized by increased immune cell infiltration and a more differentiated tumor histology
To determine the impact of infiltrating immune cells on the immunogenicity of cancer cell lines subsequently derived from a primary tumor, sections from primary PyMT tumors (Fig. 3A) were analyzed for their CD45 immune infiltration content using IHC. Primary tumors from which slow growing cell lines (#69, #238, #55) were derived showed massive CD45+ cell infiltration within the tumor mass. In contrast, primary tumors from which fast growers (#252, #234, #29) were derived showed the presence of CD45+ cells confined to the periphery of the tumor tissue (Fig. 6A). Quantitative analysis of immune cell infiltration within the H&E positive tumor tissues showed a significant increase in immune cell infiltration within the primary slow grower tumor mass (Fig. 6B). Interestingly, the number of CD45+ infiltrating cells correlated with the genotype of the mouse from which the tumor originated: PyMT/Rag1−/− were more infiltrated with immune cells compared with PyMT WT (Fig. 6C). Histological analysis of primary tumors for dedifferentiation, necrosis, and cystic changes revealed a significant increase in cystic changes in slow growers, indicative of tissue differentiation (Fig. 6D).35 The amount of necrosis and the area of poorly differentiated tissue were not significantly altered between slow growers and fast growers (Fig. S2).
Figure 6.
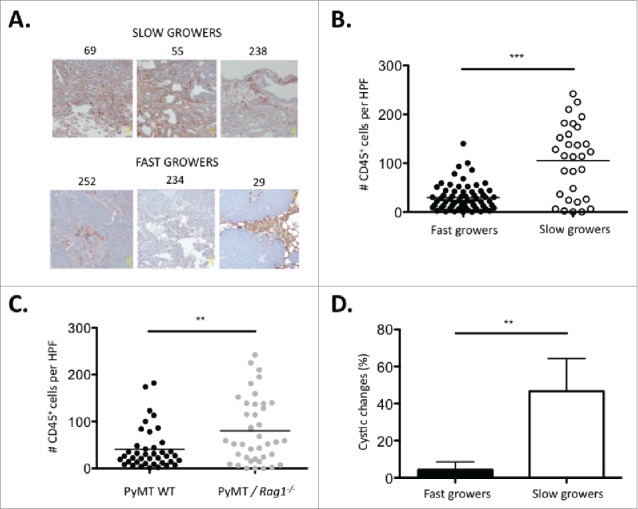
Slow growing tumors are characterized by increased immune cell infiltration and a more differentiated tumor histology. (A) Representative micrographs of primary tumors from three immune-mediated slow growers (#69, #55, #238) and fast growers (#252, #234, #29) sectioned and stained with H&E and anti-CD45 antibody. (B) Number of tumor-infiltrating CD45+ cells per high-power field (HPF) in primary tumors from three immune-mediated slow growers (#69, #55, #238) vs. five fast growers (#252, #234, #29, #235, #256). Ten HPF were counted per tumor sample. (C) Number of tumor infiltrating CD45+ cells per HPF in primary tumors from four PyMT/Rag1−/− (#69, #55, #29, #31) vs. four PyMT WT (#252, #234, #256, #238) cell lines. Ten HPF were counted per tumor sample. The percent of cystic changes as a morphological indication of tumor differentiation was examined on H&E stained sections in the primary tumor of three immune-mediated slow growers (#69, #55, #238) vs. five fast growers (#252, #234, #29, #235, #256).
Immune pressure in WT hosts converts slow grower tumors into fast growers—evidence of immunoediting in the PyMT model
To determine if cell lines were edited during their growth in WT recipients, passaged cell lines were generated after transplantation of the primary cell lines and tested for immunogenicity (Figs. 7A and B). Cell lines #69 (slow grower) and #252 (fast grower) (Fig. 7A) were transplanted into either WT or Rag2−/− x γc−/− hosts. Cell lines generated from the resulting tumor masses (Fig. 7B) were then re-transplanted into syngeneic WT or Rag2−/− x γc−/− hosts (Fig. 7C). The tumorigenicity of the WT- or Rag2−/− x γc−/−-passaged-cell lines after secondary transplantations into immune deficient or WT hosts is shown in Fig. 8. Notably, the slow growing cell line #69, when passaged through WT hosts, was converted from a slow grower to a fast grower (Figs. 7B, C and 8B), whereas passaging through severely immunodeficient Rag2−/− x γc−/− hosts did not change the slow growing profile (Figs. 7B, C and 8A). Indeed, the passaged cell line failed to grow in three out of four WT mice transplanted (Fig. 8A). Fig. 8C shows the combined survival curves of the secondary-transplanted WT- and Rag2−/− x γc−/− -passaged #69 cell lines in WT recipients, demonstrating a significant difference (p < 0.01) in tumorigenicity of cell lines passaged through WT vs Rag2−/− x γc−/− mice. In contrast, the growth kinetics of the fast grower #252 cell line in secondary transplants remained unchanged after passage in WT hosts compared its Rag2−/− x γc−/−-passaged counterpart (Figs. 7C, 8E and F).
Figure 7.
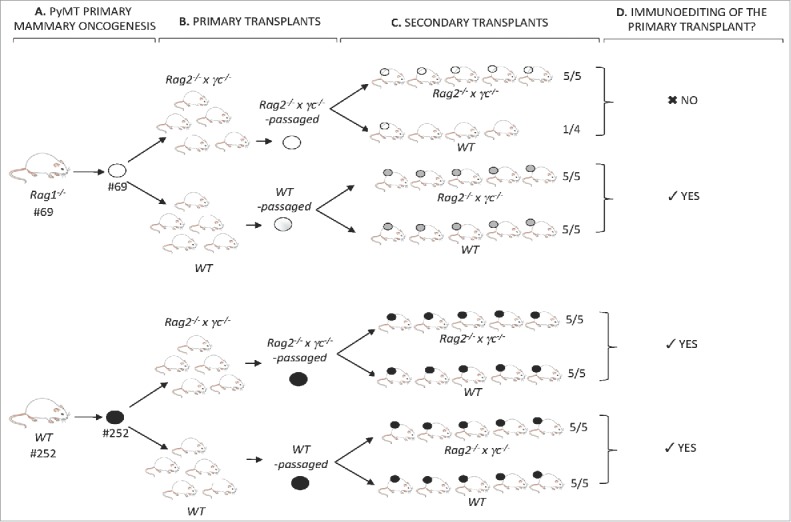
Identification of immunoediting in the PyMT model. (A) PyMT mice #69 and #252, respectively, of the Rag1−/− or WT immune background, developed mammary tumors over time. Tumors from each animal were harvested into a single-cell suspension and a cell line was generated. (B) Each cell line was transplanted into five Rag2−/− x γc−/− and five WT mice. Tumors were harvested into a single-cell suspension and Rag2−/− x γc−/−-passaged and WT-passaged cell lines of both #69 and #252 primary cell lines were generated. (C) Rag2−/− x γc−/− and WT-passaged primary transplants of #69 and #252 were transplanted as secondary transplants in either Rag2−/− x γc−/− or WT mice. (D) Secondary transplant kinetics in immune deficient Rag2−/− x γc−/− and immune competent WT mice revealed the occurrence of immunoediting of the primary-passaged transplant.
Figure 8.
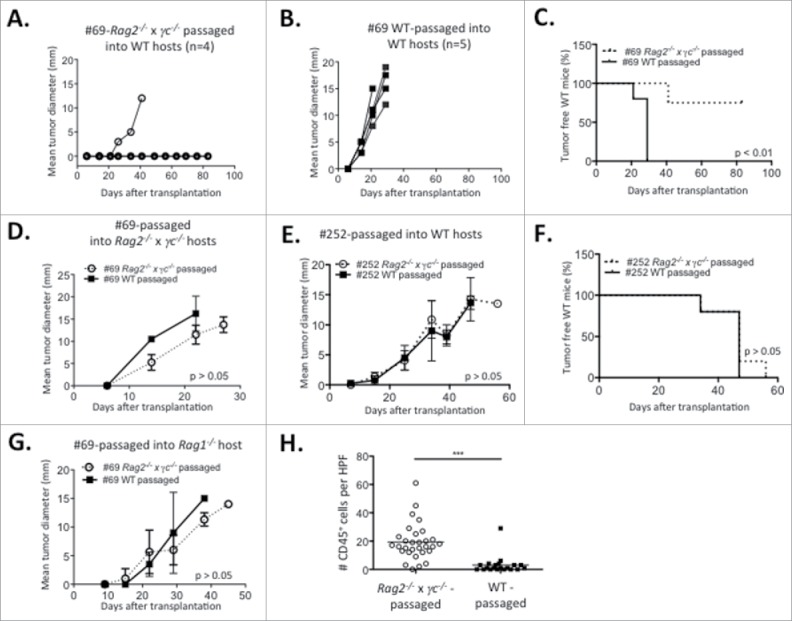
Immunoediting in the PyMT model. Tumor progression over time of (A) Rag2−/− x γc−/− and (B) WT- passaged slow growing cell line #69 transplanted in WT recipients. Rag2−/− x γc−/− or WT-passaged slow growing cell line #69 were transplanted into (C) WT or (D) Rag2−/− x γc−/− recipients and (C) survival or (D) tumor progression was assessed. (E) Tumor progression of the Rag2−/− x γc−/− and WT-passaged cell lines originating from the fast grower #252 in WT recipients. (F) Survival of WT mice transplanted with Rag2−/− x γc−/− and WT-passaged cell lines originating from the fast grower #252. (G) Tumor progression of Rag1−/− mice transplanted with either Rag2−/− x γc−/− or WT-passaged slow growing cell line #69. (H) Number of tumor-infiltrating CD45+ cells per high-power field (HPF) in the Rag2−/− x γc−/− and WT-passaged #69 cell lines secondarily transplanted in Rag1−/− host. Experiments were repeated twice.
The difference in tumorigenicity between the secondary Rag2−/− x γc−/−-passaged vs. the WT-passaged #69 cell line could be due to intrinsic differences in growth rate. To eliminate this possibility, the growth of the primary-passaged #69 cell lines was compared in vitro and after transplantation into immune deficient animals. Fig. S3 shows that the passaged cell lines had equivalent growth in vitro and Figs. 8D and 8G show that the passaged cell lines also had equivalent growth in immune deficient Rag1−/− and Rag2−/− x γc−/− mice. Interestingly, despite having identical growth kinetics, there was still a heavy infiltration of CD45+ cells in the Rag2−/− x γc−/− -passaged vs. the WT-passaged #69 cell line after secondary transplant into Rag1−/− mice (Fig. 8H).
Discussion
In this study, we show that adaptive immunosurveillance occurs in the oncogene-induced PyMT C57BL/6 mammary carcinoma model, where genetic ablation of adaptive immune responses and early acute immune suppression with tacrolimus accelerated tumor latency. We also generated a bank of PyMT tumor cell lines, with differing degrees of growth capacity in immune competent vs. immune deficient hosts. Using this novel set of cell lines, we show that immune infiltration into the primary tumor mass correlated with tumor cystic changes and could impact the immunogenicity of cell lines subsequently generated from the tumor. Overall, these results extend the cancer immunoediting paradigm to breast cancer and an oncogene-induced model system, while providing a useful set of reagents to model immune cell infiltration into breast cancer.
Our results are consistent with recent reports demonstrating that immunosurveillance of PyMT tumors was mediated in part by tissue resident innate lymphocytes that depended on IL-15 and perforin for their development and activity.29 Among these novel actors of immunosurveillance, a population of T cell receptor-expressing innate lymphoid cells, termed ILTC1, was identified. Since all T cells require RAG to develop, our mice also lacked this population of cells; however, it is not known whether the lack of immunosurveillance we observed is due to lack of ILTC1 cells or another cell type that requires RAG for its development. Our possession of cell lines derived from these mice will allow us to determine whether the slow growing cell lines are enriched in genes that stimulate ILTC1 cells or other cell types that could mediate cancer immunoediting.
We found that PyMT/Rag1−/− mice had significantly reduced tumor latency compared with PyMT WT mice in the C57BL/6 background, a result that was not seen when similar animals were studied in the FVB/NJ background.25 This strain specificity is likely to be due to the rapid oncogenesis occurring in the FVB/NJ strain where the oncogene-driven rapid primary tumor progression does not allow for an environmental immune cell influence. Alternatively, it is possible that the Th1 tendencies in C57BL/6 animals36,37 promotes better antitumor immunity than the Th2 bias in FVB/NJ animals.38,39 Other groups have also observed a strain specificity in tumor progression in this animal model. In studying the role of nitric oxide in PyMT tumor progression, Davie et al. found that PyMT/iNOS−/− mice had delayed tumor formation in the C57BL/6 but not in the FVB/NJ background.30
Genetic models of oncogenesis have been used extensively to document immune-mediated tumor promotion in solid cancers.25,30,40-42 However, evidence of immunosurveillance and immunoediting in genetic models of “solid,” non-lymphoid, oncogenesis have been sparse and have yielded conflicting results.34,43 For example, in the p53−/+ model of oncogenesis, knockout of Ifngr1 accelerated tumor onset and the spectrum of tumors of non-lymphoid origins.34 In the TRAMP model, deficiencies in NKG2D or TCRδ resulted in higher incidence, higher grade, or extensive prostate carcinomas.17,44 Conversely, deficiencies in TRAIL and TRAIL receptor DR5 did not affect tumor onset in colon adenocarcinoma and mammary tumors.45,46 In genetic models of sarcoma and lung cancer, immunosurveillance and immunoediting were shown to be mediated through the lentiviral-enforced expression of exogenous antigens by the primary tumor, therefore positing the expression of tumor-specific antigen as key for the occurrence of immunoediting.21,47 In addition, naturally occurring immunosurveillance against a self-antigen was documented in a genetic model of prostate cancer.20 Relevant to breast cancer, using the Her2/Neu model of murine breast cancer Street et al. showed that perforin could diminish mammary cancer progression but had no effect on incidence.48 Stat1−/− mice also had a higher incidence of breast cancer, although the effect was attributed to a role for STAT1 as a cell intrinsic tumor suppressor rather than an immune deficiency.49 Our results provide evidence that adaptive immunity is required for optimal breast cancer surveillance.
In addition, we show here that innate immune cells, found in Rag1−/− mice, could be recruited to slow growing tumors and correlated with the cystic changes in the tumor histology, as reported in an oncogene-induced model of carcinogenesis in a perforin-deficient model.50 We speculate that the ability to recruit innate immune cells is a pre-requisite to create an environment that will in turn recruit adaptive immune cells. Additionally, this increase in immune cell infiltration in Rag1−/− primary and transplant recipients suggests that this infiltration is independent of the antigenicity of the tumor, as adaptive immune cells are absent. Whether this increased immune recruitment is due to a chemokine or cytokine secreted by immunogenic tumor cells3 is not known but will be studied in the future. Importantly, identification of these key molecular mediators of innate cell recruitment, as we have done previously, may open new avenues for cancer therapy of tumors with a low level of antigenicity such as breast cancer.3,51
Supplementary Material
Disclosure of potential conflicts of interest
No potential conflicts of interest were disclosed.
Acknowledgments
We thank Dr Nissi Varki for assistance in interpreting tumor histology and Calvin Lee for technical support. We thank members of the Bui laboratory for their meticulous analysis of the manuscript.
Funding
J.D.B. is supported by the NIH grant CA157885 and The Hartwell Foundation.
References
- 1.Koebel CM, Vermi W, Swann JB, Zerafa N, Rodig SJ, Old LJ, Smyth MJ, Schreiber RD. Adaptive immunity maintains occult cancer in an equilibrium state. Nature 2007; 450:903-7; PMID:18026089; http://dx.doi.org/ 10.1038/nature06309 [DOI] [PubMed] [Google Scholar]
- 2.Shankaran V, Ikeda H, Bruce AT, White JM, Swanson PE, Old LJ, Schreiber RD. IFNgamma and lymphocytes prevent primary tumour development and shape tumour immunogenicity. Nature 2001; 410:1107-11; PMID:11323675; http://dx.doi.org/ 10.1038/35074122 [DOI] [PubMed] [Google Scholar]
- 3.O'Sullivan T, Saddawi-Konefka R, Gross E, Tran M, Mayfield SP, Ikeda H, Bui JD. Interleukin-17D mediates tumor rejection through recruitment of natural killer cells. Cell Rep 2014; 7:989-98; PMID:24794441; http://dx.doi.org/ 10.1016/j.celrep.2014.03.073 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Crowe NY, Smyth MJ, Godfrey DI. A critical role for natural killer T cells in immunosurveillance of methylcholanthrene-induced sarcomas. J Exp Med 2002; 196:119-27; PMID:12093876; http://dx.doi.org/ 10.1084/jem.20020092 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Crowe NY, Coquet JM, Berzins SP, Kyparissoudis K, Keating R, Pellicci DG, Hayakawa Y, Godfrey DI, Smyth MJ. Differential antitumor immunity mediated by NKT cell subsets in vivo. J Exp Med 2005; 202:1279-88; PMID:16275765; http://dx.doi.org/ 10.1084/jem.20050953 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Smyth MJ, Swann J, Cretney E, Zerafa N, Yokoyama WM, Hayakawa Y. NKG2D function protects the host from tumor initiation. J Exp Med 2005; 202:583-8; PMID:16129707; http://dx.doi.org/ 10.1084/jem.20050994 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zitvogel L, Pitt JM, Daillere R, Smyth MJ, Kroemer G. Mouse models in oncoimmunology. Nat Rev Cancer 2016; PMID:27687979; http://dx.doi.org/ 10.1038/nrc.2016.91 [DOI] [PubMed] [Google Scholar]
- 8.Dunn GP, Old LJ, Schreiber RD. The three Es of cancer immunoediting. Annu Rev Immunol 2004; 22:329-60; PMID:15032581; http://dx.doi.org/ 10.1146/annurev.immunol.22.012703.104803 [DOI] [PubMed] [Google Scholar]
- 9.Kretz-Rommel A, Qin F, Dakappagari N, Ravey EP, McWhirter J, Oltean D, Frederickson S, Maruyama T, Wild MA, Nolan MJ et al.. CD200 expression on tumor cells suppresses antitumor immunity: new approaches to cancer immunotherapy. J Immunol 2007; 178:5595-605; PMID:17442942; http://dx.doi.org/ 10.4049/jimmunol.178.9.5595 [DOI] [PubMed] [Google Scholar]
- 10.Dorand RD, Nthale J, Myers JT, Barkauskas DS, Avril S, Chirieleison SM, Pareek TK, Abbott DW, Stearns DS, Letterio JJ et al.. Cdk5 disruption attenuates tumor PD-L1 expression and promotes antitumor immunity. Science 2016; 353:399-403; PMID:27463676; http://dx.doi.org/ 10.1126/science.aae0477 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Saddawi-Konefka R, Seelige R, Gross ET, Levy E, Searles SC, Washington A Jr., Santosa EK, Liu B, O'Sullivan TE, Harismendy O et al.. Nrf2 Induces IL-17D to Mediate Tumor and Virus Surveillance. Cell Rep 2016; 16:2348-58; PMID:27545889; http://dx.doi.org/ 10.1016/j.celrep.2016.07.075 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Matsushita H, Vesely MD, Koboldt DC, Rickert CG, Uppaluri R, Magrini VJ, Arthur CD, White JM, Chen YS, Shea LK et al.. Cancer exome analysis reveals a T-cell-dependent mechanism of cancer immunoediting. Nature 2012; 482:400-4; PMID:22318521; http://dx.doi.org/ 10.1038/nature10755 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bolitho P, Street SE, Westwood JA, Edelmann W, Macgregor D, Waring P, Murray WK, Godfrey DI, Trapani JA, Johnstone RW et al.. Perforin-mediated suppression of B-cell lymphoma. Proc Natl Acad Sci U S A 2009; 106:2723-8; PMID:19196996; http://dx.doi.org/ 10.1073/pnas.0809008106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Smyth MJ, Thia KY, Street SE, MacGregor D, Godfrey DI, Trapani JA. Perforin-mediated cytotoxicity is critical for surveillance of spontaneous lymphoma. J Exp Med 2000; 192:755-60; PMID:10974040; http://dx.doi.org/ 10.1084/jem.192.5.755 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Guillerey C, Ferrari de Andrade L, Vuckovic S, Miles K, Ngiow SF, Yong MC, Teng MW, Colonna M, Ritchie DS, Chesi M et al.. Immunosurveillance and therapy of multiple myeloma are CD226 dependent. J Clin Invest 2015; 125:2904; PMID:26075821; http://dx.doi.org/ 10.1172/JCI82646 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Swann JB, Uldrich AP, van Dommelen S, Sharkey J, Murray WK, Godfrey DI, Smyth MJ. Type I natural killer T cells suppress tumors caused by p53 loss in mice. Blood 2009; 113:6382-5; PMID:19234138; http://dx.doi.org/ 10.1182/blood-2009-01-198564 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Guerra N, Tan YX, Joncker NT, Choy A, Gallardo F, Xiong N, Knoblaugh S, Cado D, Greenberg NM, Raulet DH. NKG2D-deficient mice are defective in tumor surveillance in models of spontaneous malignancy. Immunity 2008; 28:571-80; PMID:18394936; http://dx.doi.org/ 10.1016/j.immuni.2008.02.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Donkor MK, Sarkar A, Savage PA, Franklin RA, Johnson LK, Jungbluth AA, Allison JP, Li MO. T cell surveillance of oncogene-induced prostate cancer is impeded by T cell-derived TGF-beta1 cytokine. Immunity 2011; 35:123-34; PMID:21757379; http://dx.doi.org/ 10.1016/j.immuni.2011.04.019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kang TW, Yevsa T, Woller N, Hoenicke L, Wuestefeld T, Dauch D, Hohmeyer A, Gereke M, Rudalska R, Potapova A. Senescence surveillance of pre-malignant hepatocytes limits liver cancer development. Nature 2011; 479:547-51; PMID:22080947; http://dx.doi.org/ 10.1038/nature10599 [DOI] [PubMed] [Google Scholar]
- 20.Savage PA, Vosseller K, Kang C, Larimore K, Riedel E, Wojnoonski K, Jungbluth AA, Allison JP. Recognition of a ubiquitous self antigen by prostate cancer-infiltrating CD8+ T lymphocytes. Science 2008; 319:215-20; PMID:18187659; http://dx.doi.org/ 10.1126/science.1148886 [DOI] [PubMed] [Google Scholar]
- 21.DuPage M, Mazumdar C, Schmidt LM, Cheung AF, Jacks T. Expression of tumour-specific antigens underlies cancer immunoediting. Nature 2012; 482:405-9; PMID:22318517; http://dx.doi.org/ 10.1038/nature10803 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.O'Sullivan T, Saddawi-Konefka R, Vermi W, Koebel CM, Arthur C, White JM, Uppaluri R, Andrews DM, Ngiow SF, Teng MW et al.. Cancer immunoediting by the innate immune system in the absence of adaptive immunity. J Exp Med 2012; 209:1869-82; PMID:22927549; http://dx.doi.org/ 10.1084/jem.20112738 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kitamura T, Qian BZ, Soong D, Cassetta L, Noy R, Sugano G, Kato Y, Li J, Pollard JW. CCL2-induced chemokine cascade promotes breast cancer metastasis by enhancing retention of metastasis-associated macrophages. J Exp Med 2015; 212:1043-59; PMID:26056232; http://dx.doi.org/ 10.1084/jem.20141836 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wculek SK, Malanchi I. Neutrophils support lung colonization of metastasis-initiating breast cancer cells. Nature 2015; 528:413-7; PMID:26649828; http://dx.doi.org/ 10.1038/nature16140 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.DeNardo DG, Barreto JB, Andreu P, Vasquez L, Tawfik D, Kolhatkar N, Coussens LM. CD4(+) T cells regulate pulmonary metastasis of mammary carcinomas by enhancing protumor properties of macrophages. Cancer Cell 2009; 16:91-102; PMID:19647220; http://dx.doi.org/ 10.1016/j.ccr.2009.06.018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bos PD, Plitas G, Rudra D, Lee SY, Rudensky AY. Transient regulatory T cell ablation deters oncogene-driven breast cancer and enhances radiotherapy. J Exp Med 2013; 210:2435-66; PMID:24127486; http://dx.doi.org/ 10.1084/jem.20130762 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Fantozzi A, Christofori G. Mouse models of breast cancer metastasis. Breast Cancer Res 2006; 8:212; PMID:16887003; http://dx.doi.org/ 10.1186/bcr1530 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Guy CT, Cardiff RD, Muller WJ. Induction of mammary tumors by expression of polyomavirus middle T oncogene: a transgenic mouse model for metastatic disease. Mol Cell Biol 1992; 12:954-61; PMID:1312220; http://dx.doi.org/ 10.1128/MCB.12.3.954 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Dadi S, Chhangawala S, Whitlock BM, Franklin RA, Luo CT, Oh SA, Toure A, Pritykin Y, Huse M, Leslie CS et al.. Cancer immunosurveillance by tissue-resident innate lymphoid cells and innate-like T Cells. Cell 2016; 164:365-77; PMID:26806130; http://dx.doi.org/ 10.1016/j.cell.2016.01.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Davie SA, Maglione JE, Manner CK, Young D, Cardiff RD, MacLeod CL, Ellies LG. Effects of FVB/NJ and C57Bl/6J strain backgrounds on mammary tumor phenotype in inducible nitric oxide synthase deficient mice. Transgenic Res 2007; 16:193-201; PMID:17206489; http://dx.doi.org/ 10.1007/s11248-006-9056-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sevc J, Goldberg D, van Gorp S, Leerink M, Juhas S, Juhasova J, Marsala S, Hruska-Plochan M, Hefferan MP, Motlik J et al.. Effective long-term immunosuppression in rats by subcutaneously implanted sustained-release tacrolimus pellet: effect on spinally grafted human neural precursor survival. Exp Neurol 2013; 248:85-99; PMID:23748136; http://dx.doi.org/ 10.1016/j.expneurol.2013.05.017 [DOI] [PubMed] [Google Scholar]
- 32.Gibby K, You WK, Kadoya K, Helgadottir H, Young LJ, Ellies LG, Chang Y, Cardiff RD, Stallcup WB. Early vascular deficits are correlated with delayed mammary tumorigenesis in the MMTV-PyMT transgenic mouse following genetic ablation of the NG2 proteoglycan. Breast Cancer Res 2012; 14:R67; PMID:22531600; http://dx.doi.org/ 10.1186/bcr3174 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kiani A, Rao A, Aramburu J. Manipulating immune responses with immunosuppressive agents that target NFAT. Immunity 2000; 12:359-72; PMID:10795734; http://dx.doi.org/ 10.1016/S1074-7613(00)80188-0 [DOI] [PubMed] [Google Scholar]
- 34.Swann JB, Smyth MJ. Immune surveillance of tumors. J Clin Invest 2007; 117:1137-46; PMID:17476343; http://dx.doi.org/ 10.1172/JCI31405 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hedlund M, Ng E, Varki A, Varki NM. alpha 2–6-Linked sialic acids on N-glycans modulate carcinoma differentiation in vivo. Cancer Res 2008; 68:388-94; PMID:18199532; http://dx.doi.org/ 10.1158/0008-5472.CAN-07-1340 [DOI] [PubMed] [Google Scholar]
- 36.Watanabe H, Numata K, Ito T, Takagi K, Matsukawa A. Innate immune response in Th1- and Th2-dominant mouse strains. Shock 2004; 22:460-6; PMID:15489639; http://dx.doi.org/ 10.1097/01.shk.0000142249.08135.e9 [DOI] [PubMed] [Google Scholar]
- 37.Schulte S, Sukhova GK, Libby P. Genetically programmed biases in Th1 and Th2 immune responses modulate atherogenesis. Am J Pathol 2008; 172:1500-8; PMID:18467709; http://dx.doi.org/ 10.2353/ajpath.2008.070776 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kim EM, Bae YM, Choi MH, Hong ST. Cyst formation, increased anti-inflammatory cytokines and expression of chemokines support for Clonorchis sinensis infection in FVB mice. Parasitol Int 2012; 61:124-9; PMID:21820080; http://dx.doi.org/ 10.1016/j.parint.2011.07.001 [DOI] [PubMed] [Google Scholar]
- 39.Disis ML, Palucka K. Evaluation of cancer immunity in mice. Cold Spring Harb Protoc 2014; 2014:231-4; PMID:24173315; http://dx.doi.org/ 10.1101/pdb.top078816 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Boyle ST, Faulkner JW, McColl SR, Kochetkova M. The chemokine receptor CCR6 facilitates the onset of mammary neoplasia in the MMTV-PyMT mouse model via recruitment of tumor-promoting macrophages. Mol Cancer 2015; 14:115; PMID:26047945; http://dx.doi.org/ 10.1186/s12943-015-0394-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Pylayeva-Gupta Y, Das S, Handler JS, Hajdu CH, Coffre M, Koralov SB, Bar-Sagi D. IL35-producing B cells promote the development of pancreatic neoplasia. Cancer Discov 2016; 6:247-55; PMID:26715643; http://dx.doi.org/ 10.1158/2159-8290.CD-15-0843 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Pylayeva-Gupta Y, Lee KE, Hajdu CH, Miller G, Bar-Sagi D. Oncogenic Kras-induced GM-CSF production promotes the development of pancreatic neoplasia. Cancer Cell 2012; 21:836-47; PMID:22698407; http://dx.doi.org/ 10.1016/j.ccr.2012.04.024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Raulet DH, Guerra N. Oncogenic stress sensed by the immune system: role of natural killer cell receptors. Nat Rev Immunol 2009; 9:568-80; PMID:19629084; http://dx.doi.org/ 10.1038/nri2604 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kaplan DH, Shankaran V, Dighe AS, Stockert E, Aguet M, Old LJ, Schreiber RD. Demonstration of an interferon gamma-dependent tumor surveillance system in immunocompetent mice. Proc Natl Acad Sci U S A 1998; 95:7556-61; PMID:9636188; http://dx.doi.org/ 10.1073/pnas.95.13.7556 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Yue HH, Diehl GE, Winoto A. Loss of TRAIL-R does not affect thymic or intestinal tumor development in p53 and adenomatous polyposis coli mutant mice. Cell Death Differ 2005; 12:94-7; PMID:15514675; http://dx.doi.org/ 10.1038/sj.cdd.4401523 [DOI] [PubMed] [Google Scholar]
- 46.Zerafa N, Westwood JA, Cretney E, Mitchell S, Waring P, Iezzi M, Smyth MJ. Cutting edge: TRAIL deficiency accelerates hematological malignancies. J Immunol 2005; 175:5586-90; PMID:16237043; http://dx.doi.org/ 10.4049/jimmunol.175.9.5586 [DOI] [PubMed] [Google Scholar]
- 47.DuPage M, Cheung AF, Mazumdar C, Winslow MM, Bronson R, Schmidt LM, Crowley D, Chen J, Jacks T. Endogenous T cell responses to antigens expressed in lung adenocarcinomas delay malignant tumor progression. Cancer Cell 2011; 19:72-85; PMID:21251614; http://dx.doi.org/ 10.1016/j.ccr.2010.11.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Street SE, Zerafa N, Iezzi M, Westwood JA, Stagg J, Musiani P, Smyth MJ. Host perforin reduces tumor number but does not increase survival in oncogene-driven mammary adenocarcinoma. Cancer Res 2007; 67:5454-60; PMID:17545627; http://dx.doi.org/ 10.1158/0008-5472.CAN-06-4084 [DOI] [PubMed] [Google Scholar]
- 49.Chan SR, Vermi W, Luo J, Lucini L, Rickert C, Fowler AM, Lonardi S, Arthur C, Young LJ, Levy DE et al.. STAT1-deficient mice spontaneously develop estrogen receptor alpha-positive luminal mammary carcinomas. Breast Cancer Res 2012; 14:R16; PMID:22264274; http://dx.doi.org/ 10.1186/bcr3100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Macagno M, Bandini S, Stramucci L, Quaglino E, Conti L, Balmas E, Smyth MJ, Lollini PL, Musiani P, Forni G et al.. Multiple roles of perforin in hampering ERBB-2 (Her-2/neu) carcinogenesis in transgenic male mice. J Immunol 2014; 192:5434-41; PMID:24790144; http://dx.doi.org/ 10.4049/jimmunol.1301248 [DOI] [PubMed] [Google Scholar]
- 51.Cimino-Mathews A, Foote JB, Emens LA. Immune targeting in breast cancer. Oncology (Williston Park) 2015; 29:375-85; PMID:25979549 [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


