ABSTRACT
Cytotoxic T-lymphocytes (CTLs) lyse target cells after recognizing the complexes of peptides and MHC class I molecules (pMHC I) on cell surfaces. Tapasin is an essential component of the peptide-loading complex (PLC) and its absence influences the surface repertoire of MHC class I peptides. In the present study, we assessed tapasin expression in 85 primary tumor lesions of non-small cell lung cancer (NSCLC) patients, demonstrating that tapasin expression positively correlated with patient survival. CD8+ T-cell infiltration of tumor lesions was synergistically observed with tapasin expression and correlated positively with survival. To establish a direct link between loss of tapasin and CTL recognition in human cancer models, we targeted the tapasin gene by CRISPR/Cas9 system and generated tapasin-deficient variants of human lung as well as colon cancer cells. We induced the CTLs recognizing endogenous tumor-associated antigens (TAA), survivin or cep55, and they responded to each tapasin-proficient wild type. In contrast, both CTL lines ignored the tapasin-deficient variants despite their antigen expression. Moreover, the adoptive transfer of the cep55-specific CTL line failed to prevent tumor growth in mice bearing the tapasin-deficient variant. Loss of tapasin most likely limited antigen processing of TAAs and led to escape from TAA-specific CTL recognition. Tapasin expression is thus a key for CTL surveillance against human cancers.
KEYWORDS: CD8+ T cells, colon cancer, immune evasion, lung cancer, MHC class I, tapasin
Introduction
Cytotoxic T-lymphocytes (CTLs) recognize endogenously processed peptides presented by MHC class I molecules and lyse the target cells. MHC class I antigen processing consists of multiple steps across the cytosol and the endoplasmic reticulum (ER): The cytosolic proteasomes digest endogenous proteins, thereby generating short fragments of proteins, which are further transported into the ER through the transporter-associated with antigen processing (TAP).1,2 The ER aminopeptidase associated with antigen processing (ERAAP or ERAP1) trims the N-terminal extension of the short protein fragments, optimizing the length of the peptide for binding to MHC class I molecules.3-5 Meanwhile, the MHC I molecules associated with β-2-microglobulin (β2m) form the peptide-loading complex (PLC) consisting of TAP, tapasin, ERp57, and calreticulin, which is necessary for stable pMHC I supply on the cell surface.6,7 Tapasin is an MHC I-specific chaperone tethering MHC I and TAP and plays an indispensable role in shaping the pMHC I repertoire. Many CD8+ T cell responses to tapasin-deficient cells are reduced or diminished in mouse models.8,9
Loss of tapasin expression is a frequent event that has been reported in a wide variety of human cancers, including malignant melanoma, head and neck squamous cell carcinoma (HNSCC), renal cell carcinoma, colorectal carcinoma, glioblastoma, lung carcinoma, and neuroblastoma.10-18 Notably, tapasin expression associated with intratumoral T-cell infiltration has been reported as a prognostic marker of patient survival in ovarian carcinoma, HNSCC, glioblastoma, and colorectal carcinoma.12,13,19-21 It is also reported that loss of tapasin is more frequent among the other antigen-processing machinery (APM) components, strongly suggesting its central role in escape from CTL immune surveillance to tumors.15,22 Impaired CTL responses against tapasin-deficient cells have been well established in mouse models.8,9 In human cases, loss of tapasin alters the peptide repertoire presented by HLA-B*2705 and results in a decrease in alloreactive CTL responses.23 However, a direct evidence showing the processing defect of endogenous tumor-associated antigens (TAA) and following loss of cancer-specific CTL recognitions has been missing due to the lack of appropriate tapasin-deficient models. In the present study, we screened tapasin expression and CD8+ T-cell infiltration of tumor lesions in 85 primary tissue sections from non-small cell lung carcinoma (NSCLC), demonstrating that both tapasin expression and CD8+ T-cell infiltration positively correlate with patient survival. Moreover, we generated the genetically mutated tapasin-deficient human lung and colon cancer cells. Loss of tapasin influences the processing of endogenous TAAs and leads to escape from CTLs recognition.
Results
Tapasin expression and CD8+ T-cell infiltration correlate with prognoses of NSCLC patients
Tapasin expression levels and CD8+ T-cell infiltration in tumor lesions were assessed in 85 cases of human primary NSCLC patients (Table 1 and Table S1). A total of 48.2% of the patients had stage IA disease and stages of the remaining patients varied across IB to IIIB. Most patients were histologically diagnosed as adenocarcinoma (72.9%), followed by squamous cell carcinoma (SCC, 16.5%), and other histological types. Formalin-fixed and paraffin-embedded tumor sections obtained from the patients after surgery were stained with anti-tapasin or anti-CD8+ monoclonal antibodies (mAb) and then scored as indicated in Fig. 1. In summary, the tapasin expression was observed as follows: score 2 (> 75% positive), 27.1%; score 1 (25–75% positive), 37.6%; score 0 (< 25% positive), 35.3%. The results show that abundant tapasin expression is observed only in 27.1% of the tumor lesions and 72.9% cases heterogeneously lack tapasin expression. Meanwhile, CD8+ T-cell infiltration scores were as follows: score 2 (> 75% positive), 63.5%; score 1 (25–75% positive), 18.8%; score 0 (< 25% positive), 17.6%.
Table 1.
Summary of clinical annotations, tapasin scores and CD8+ T-cell infiltration scores of the NSCLC patients
| 66.6±9.42(SD) |
||
|---|---|---|
| Age | n | % total |
| Gender | ||
| Female | 38 | (44.7) |
| Male | 47 | (55.3) |
| Stage | ||
| IA | 41 | (48.2) |
| IB | 16 | (18.8) |
| IIA | 6 | (7.1) |
| IIB | 7 | (8.2) |
| IIIA | 11 | (12.9) |
| IIIB | 4 | (4.7) |
| IV | 0 | (0) |
| Histology | ||
| ADC | 62 | (72.9) |
| SCC | 14 | (16.5) |
| LCC | 4 | (4.7) |
| PLE | 3 | (3.5) |
| ADSQ | 2 | (2.4) |
| Tapasin score | ||
| 2 | 23 | (27.1) |
| 1 | 32 | (37.6) |
| 0 | 30 | (35.3) |
| CD8 score | ||
| 2 | 54 | (63.5) |
| 1 | 16 | (18.8) |
| 0 | 15 | (17.6) |
Note: ADC, adenocarcinoma; SCC, squamous cell carcinoma; LCC, large cell carcinoma; PLE, carcinoma with pleomorphic or sarcomatous elements; ADSQ, adeno-squamous cell carcinoma.
Figure 1.
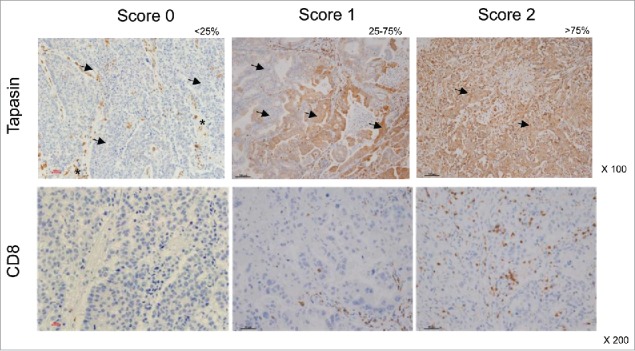
Immunohistochemistry of human primary NSCLC. Representative staining patterns of formalin-fixed, paraffin-embedded tumor lesions with tapasin mAb (upper panels) and CD8+ mAb (lower panels). Tapasin scores and CD8+ scores are determined by percentages of positive tumor cells and by percentages of tumor-infiltrating positive cells in tumor lesions, respectively: 0, <25% positive; 1, 25% to 75% positive; 2, >75% positive. Arrows, tumor cells. Asterisks, stromal (non-tumor) cells. Magnification, x100 and x200.
As expected, the overall survival rate (OS) of stage IA and IB patients was significantly higher than that of stage IIA to IIIB patients (Fig. 2A). We divided the patients into tapasin-positive (score 2) and tapasin-negative (score 1 and 0) groups and compared survival. Notably, the OS of the tapasin-positive group was significantly higher than that of the tapasin-negative group, suggesting that tapasin expression could be a good prognostic marker comparable to clinical stages in NSCLC patients (Fig. 2B). We also divided the patients into CD8+ T-cell infiltration-positive (score 2) and -negative (score 1 and 0) groups and the Kaplan–Meier survival analysis showed a statistically significant higher OS in the CD8+ T-cell infiltration-positive group (Fig. 2C). Tapasin expression and CD8+ T-cell infiltration were statistically correlated (Fig. 2D). Notably, combining tapasin expression and CD8+ T-cell infiltration scores provided further information: prognoses were comparably better among three groups positive to tapasin or CD8+, while only the tapasin and CD8+-double negative group significantly impaired survival (Fig. 2E). These results suggest that losses of tapasin expression and CD8+ T-cell infiltration in tumor lesions synergistically correlate with and significantly influence NSCLC patient prognoses, consistent with the findings of former studies conducted in ovarian carcinoma, HNSCC, glioblastoma, and colorectal carcinoma patients.19-21,24
Figure 2.
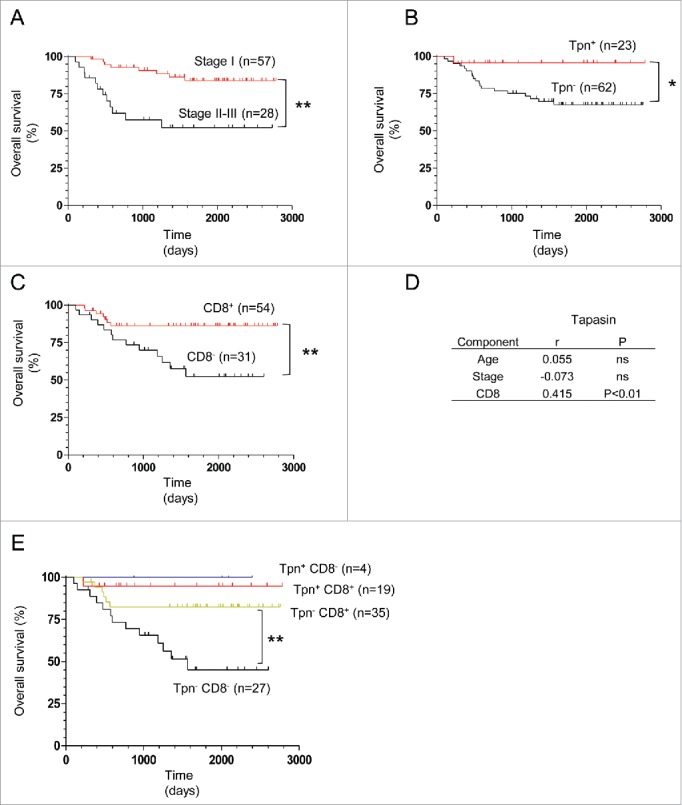
Correlation of tapasin expression or CD8+ T-cell infiltration with NSCLC patient survival. (A, B, C, and E) Kaplan–Meier survival analysis of clinical stages, tapasin expression, CD8+ T-cell infiltration and combined scores of tapasin expression and CD8+ T-cell infiltration (positive, score 2; negative, score 1 or 0). p-values are calculated using the log-rank test. (D) Correlation (r) of tapasin expression scores with ages, stages and CD8+ T-cell infiltration scores. ns, not significant. p-values are calculated using the Spearman rank test.
Generation of a tapasin-deficient human lung cancer line
To establish a direct link between loss of tapasin and cancer-specific CTL evasion, we set about making a pair of human lung cancer lines that express a TAA with or without tapasin expression. Here we chose the LHK2 lung cancer line because it expresses the detectable tapasin protein as well as an endogenous TAA, survivin, which encodes a CTL epitope presented by HLA-A24.25 The CRISPR/Cas9-mediated genome engineering system allowed to introduce random heterozygous mutations at the gRNA targeting site in exon 2 of the tapasin gene, and we established the LHK2 variant, YS3 (Fig. 3A). In contrast to the wild type, which increased tapasin protein expression in an IFNγ dependent manner, the YS3 never expressed tapasin protein both in the absence and presence of IFNγ, most likely due to frameshift mutations taking place in both tapasin alleles (Fig. 3B). The LHK2 line was established from an HLA-A*2402-positive patient and we confirmed the surface expression level, showing that both HLA-A/B/C and HLA-A24 levels significantly decreased on the YS3 variant (Fig. 3C and D). We thus concluded that the introduced tapasin-gene mutations diminished tapasin protein expression and resulted in loss of surface MHC class I expression as observed in tapasin-knockout mouse cases. 8,9
Figure 3.
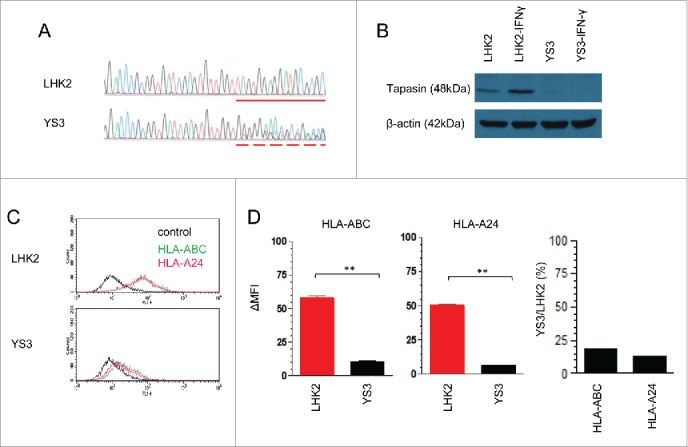
Impaired HLA class I expression on a tapasin-deficient human lung cancer line. (A) Comparison between the tapasin-gene exon 2 sequencing traces of wild-type LHK2 and its variant YS3. The dash line starts with deletion/insertion mutation introduced by the CRISPR/Cas9-mediated genome editing system, followed by frameshift mutation occurred in both alleles. A solid line indicates the corresponding sequence of wild type showing homozygous alleles without mutation. (B) Western blot of LHK2 and YS3 treated with or without IFNγ, stained with tapasin- or actin-mAbs. (C) Flow cytometry of LHK2 and YS3 stained with an HLA-A24-specific c7709A2 mAb (pink), a W6/32 mAb (green), or without a primary antibody (black). (D) Quantification of surface HLA class I levels. ΔMFI values on the y-axis were calculated from three independent experiments shown in Fig. 3C, by subtracting values of negative controls without a primary antibody. Data are shown as mean + SEM (n = 3). p-values were calculated using a two-tailed t-test (**p < 0.01). The mean-percentage changes of YS3 compared with LHK2 in ΔMFI of HLA-A, B, C, and HLA-A24 are also shown.
Loss of tapasin allows lung cancer cells to escape from TAA-specific CTL recognition
A TAA, survivin 2B, is a splicing variant derived from the survivin gene and its expression is widely observed among a variety of cancer lines but not in normal tissues.25 LHK2 and YS3 equally expressed the survivin 2B gene and survivin protein, demonstrating that introducing tapasin-gene mutation did not influence the endogenous expression of the survivin antigen (Fig. 4A and B). We prepared the CTL clone H4 that recognized the survivin 2B epitope presented by HLA-A24 by stimulating cancer-patient PBMC with the synthetic peptide, followed by single-cell sorting as described in methods (Fig. 4C). The clone H4 produced IFNγ in response to T2-A24 cells pulsed with survivin 2B peptides (Fig. 4D). The clone H4 recognized LHK2, suggesting that cancer-patient PBMCs contain survivin 2B-specific CTL precursors (Fig. 4E). By contrast, the CTLs failed to produce IFNγ against YS3. These data indicate that loss of tapasin confers the capability to reduce TAA-specific CTL responses on cancer cells.
Figure 4.
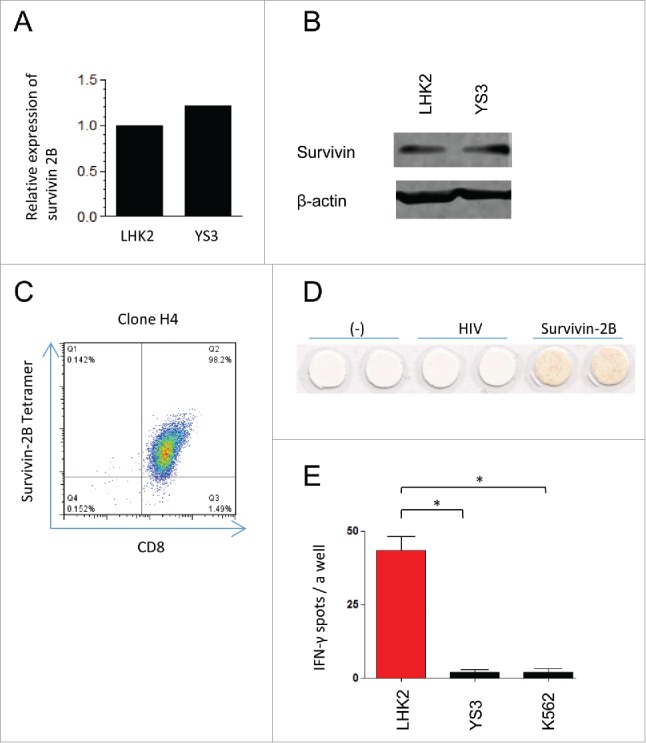
Survivin 2B-specific CTLs failed to recognize the tapasin-deficient lung cancer cells. (A) mRNA expression of survivin 2B. Expression value for YS3 is shown relative to LHK2 (with a value arbitrarily set to 1.0). (B) Western blot of survivin expression in LHK2 and YS3. Actin expression was used as a loading control. (C) Flowcytometry of the CTL clone H4 from cancer-patient PBMC, stained with PE-conjugated survivin 2B tetramers and PC5-conjugated CD8+ mAb. Numbers in plots indicate frequency of corresponding quadrants. (D) ELISPOT IFNγ assay of H4 responding to T2-A24 cells pulsed with synthetic survivin 2B peptides or HIV peptides, which are known to be bound to HLA-A24. Produced IFNγ are detected as red spots. (E) ELISPOT IFNγ assay of H4 responding to LHK2, YS3, and K562. The y-axis indicates the numbers of IFNγ-positive spots per a well. Data are shown as mean + SEM (n = 3). p-values were calculated using a two-tailed t-test (*p < 0.05). Data are representatives of three independent experiments.
Generation of a tapasin-deficient human colon cancer line
To expand the finding over lung cancers and survivin 2B, we next used the human colon cancer line SW480 expressing both survivin 2B and CEPP55 gene, which is another TAA expressed in a wide variety of tumor cell lines including SW480 and encodes a CTL epitope presented by HLA-A24.26 CRISPR/Cas9- system again allowed to introduce random heterozygous mutations in exon 2 of the tapasin gene, and we established the SW480 variant, 3G12 (Fig. 5A). The tapasin-gene mutations resulted in loss of tapasin protein expression, demonstrating that the 3G12 clone was a tapasin-deficient variant (Fig. 5B). The SW480 was originally established from a patient with HLA-A*2402/A*0201 and expressed both HLA class I molecules on the surface (Fig. 5C). Although the surface amount of HLA-A24 was more than two times higher than that of HLA-A2, both amounts were decreased in 3G12 to about 14.4% and 21.7% of the wild type, respectively (Fig. 5D). It is of our interest that the reduction rates were, however, similar between the two HLA-A alleles because HLA-A*0201 is known as a tapasin-independent allele while HLA-*2402 is not.6 We have established another two tapasin-deficient clones, 1D4 and 3A9, and verified comparable trends in loss of tapasin protein and decreased MHC class I surface expression (Fig. S1).
Figure 5.
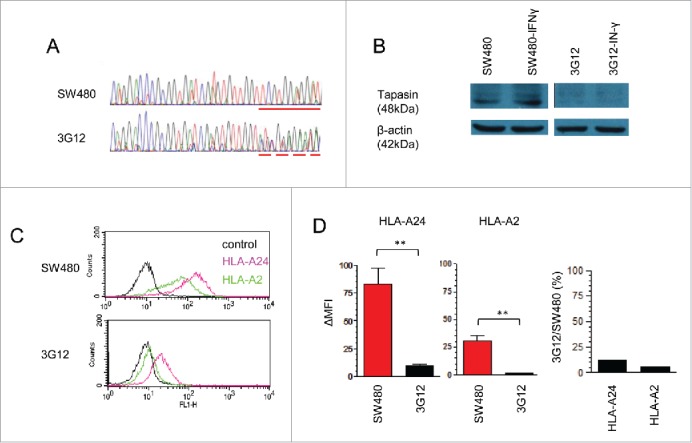
Impaired HLA class I expression on a tapasin-deficient human colon cancer line. (A) Comparison between the tapasin-gene exon 2 sequencing traces of wild-type SW480 and its variant 3G12. The dash line starts with deletion/insertion mutation introduced by the CRISPR/Cas9-mediated genome editing system, followed by frameshift mutation occurred in both alleles. A solid line indicates the corresponding sequence of wild type showing homozygous alleles without mutation. (B) Western blot of SW480 and 3G12 treated with or without IFNγ, stained with tapasin- or actin-mAbs. (C) Flow cytometry of SW480 and 3G12 stained with an HLA-A24-specific c7709A2 mAb (pink), an HLA-A2-specific BB7.2 mAb (green), or without a primary antibody (black). D, Quantification of surface HLA class I levels. ΔMFI values on the y-axis were calculated from three independent experiments shown in Fig. 5D, by subtracting values of negative controls without a primary antibody. Data are shown as mean + SEM (n = 3). p-values were calculated using a two-tailed t-test (**p < 0.01). The mean-percentage changes of 3G12 compared with SW480 in ΔMFI of HLA-24 and HLA-A2 are also shown.
Tapasin-deficient colon cancer line evades TAA-specific CTL lysis
We further investigated the influence of tapasin deficiency in TAA-specific CTL lysis. First, SW480 and 3G12 equally expressed the TAA, survivin 2B, and cep55 genes, which were not observed in a normal colon tissue (Fig. 6A). Second, we subcutaneously injected immunodeficient NSG mice with 1.0 × 105 cells of either SW480 or 3G12, and compared their growth. Injected tumors rapidly developed in both cases and displayed equal tumor growth rates over time (Fig. 6B). These results ensured that the gene mutation introduced by CRISPR/Cas9 yielded no off-target effect on survivin 2B and cep55 gene expression as well as tumor growth rates. Developed SW480 and 3G12 tumors in vivo were dissected and stained with tapasin antibody, showing tapasin-positive and negative staining patterns, respectively (Fig. 6C).
Figure 6.
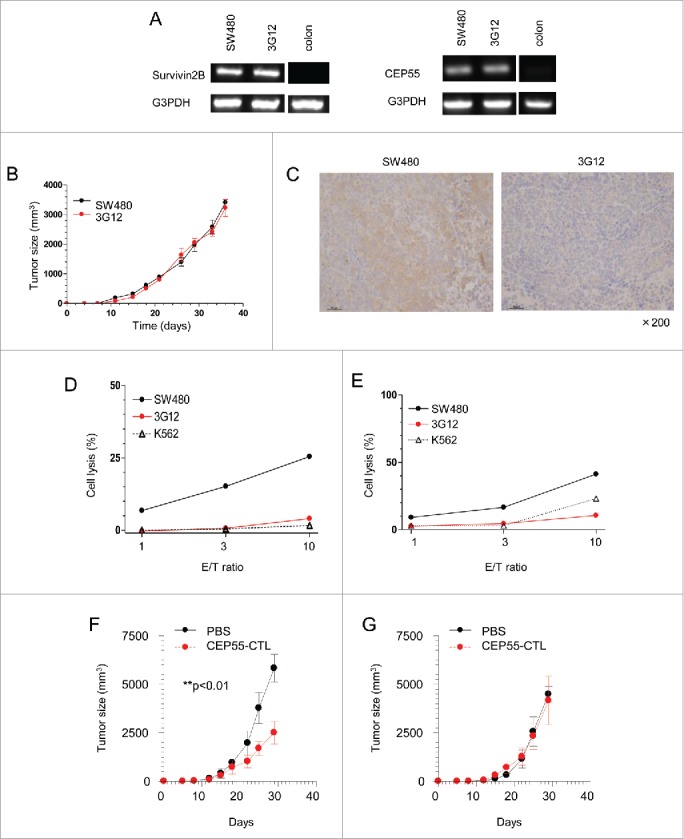
Tapasin deficiency leads to escape from TAA-specific CTL-mediated cytotoxicity. A and B, Survivin and cep55 gene expression in SW480, 3G12 and a normal colon tissue. (B) Tumor growth rates of SW480 and 3G12. 1 × 105 SW480 or 3G12 cells were subcutaneously injected into NSG mice. The x-axis and the y-axis indicate days after injection and the sizes of tumors, respectively. Tumor volume was calculated as follows: volume = xy2/2. Data are shown as mean + SEM (n = 3). (C) Immunohistochemistry of dissected tumors 36 d after injection. Formalin-fixed and paraffin-embedded tissues were stained with tapasin mAb (magnification x200). D and E, LDH cytotoxicity assay of the CTLs specific to the survivin 2B epitope and the cep55 epitope presented by HLA-A24. LDH releases from indicated target cells cultured with survivin 2B-specific CTL clones (D) and cep55-specific CTL clones (E) were measured. The x-axis and y-axis indicate effector/target (E/T) ratios and % target cell lysis, respectively. Data are representatives of three independent experiments. F and G, The adoptive transfer of cep55-specific CTLs and tumor rejection in vivo. SW480 (F) and 3G12 (G) cells were subcutaneously injected into NSG mice. The mice received the adoptive transfer of PBS (black) or 2 × 106 cep55-specific CTLs (red) 10 and 17 d after tumor injection. The x-axis indicates days after tumor injection. Data are shown as mean + SD (n = 3). p-values were calculated using a two-tailed t-test (**p < 0.01).
The survivin 2B-specific CTL clone H4 derived from cancer-patient PBMC in Fig. 4 was used for cytotoxic assay. The H4 lysed SW480 in an E/T ratio dependent manner consistent with clear survivin 2B-gene expression. In contrast, the H4 hardly lysed 3G12 as well as a MHC class I negative-K562 line despite survivin 2B expression equal to SW480 (Fig. 6D). We also used another CTL clone derived from cancer patient PBMC, the clone which specifically recognized the cep55 epitope presented by HLA-A24.26 The CTL clone successfully lysed SW480 expressing the cep55 gene, while failed to recognize 3G12 despite cep55 gene expression equal to SW480 (Fig. 6E). The CTL failed to lyse another tapasin-deficient variants 1D4 and 3A9 as well (Fig. S2). Moreover, we found that loss of tapasin influenced the antitumor effect in vivo. SW480 and 3G12 were subcutaneously injected into NSG mice. The subsequent adoptive transfer of cep55-CTLs significantly prevented the growth of SW480 consistent with the CTL-mediated cell lysis observed in vitro (Fig. 6F). By contrast, the adoptive transfer did not reduce the sizes of tumors in mice bearing tapasin-deficient 3G12 (Fig. 6G).
Loss of tapasin limits the antigen processing of endogenous TAAs
We finally asked whether the impaired CTL responses against endogenous TAAs was caused by a decrease in peptide supply or by a decrease in HLA-A24 on tumor cell surfaces. Because direct CTL responses against SW480 and 3G12 can be influenced by both conditions, we first quantified the amount of CTL epitopes produced in the tumor cells using an exogenous presentation assay (Fig. 7A). Survivin 2B and cep55 CTL clones specifically responded to T2-A24 cells pulsed with survivin 2B and cep55 synthetic peptides, respectively (Fig. 7B). The cell extracts from SW480 and 3G12 were titrated and incubated with T2-A24 cells that serve as APCs. When we used survivin 2B-CTLs as a detector, we found that 2 × 107 cells of SW480, but not the equivalent amount of 3G12, contained the detectable amount of survivin 2B peptide in their extracts (Fig. 7C). And the use of cep55-CTLs demonstrated that the extracts from 2.0 × 105 SW480 and 2.0 × 106 3G12 cells contained the comparable amounts of the cepp55 peptide. These results demonstrate that the amounts of both survivin 2B and cep55 peptides are decreased in 3G12. The amount of HLA-A24 on this surface is decreased in 3G12 compared with SW480, which difference may have contributed to the impaired TAA-specific CTL responses against 3G12 (Fig. 5C). However, the survivin 2B-CTL responses against 5 × 104 cells of SW480 and 3G12 were comparable, when the tumor cells were exogenously pulsed with synthetic survivin 2B peptides (Fig. 7D). Taken together, these results demonstrate that the impaired CTL responses caused by loss of tapasin was mainly due to a decrease in peptide supply but not a decrease in HLA-A24, suggesting that loss of tapasin limits the antigen processing of endogenous TAAs.
Figure 7.
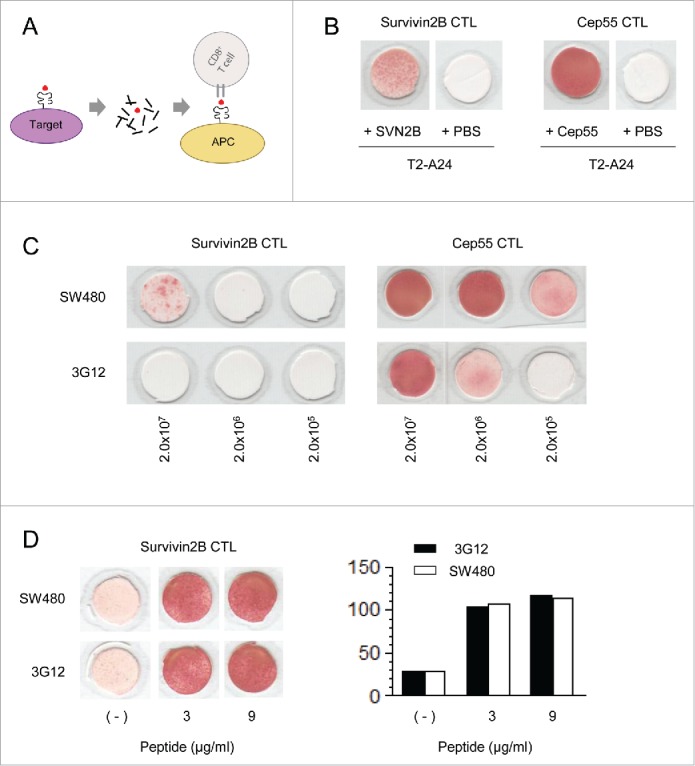
The antigen processing of endogenous TAAs is limited by loss of tapasin. (A) An exogenous presentation assay using antigen-presenting cells (APC) to quantify CTL-epitopes processed in target cells. (B) IFNγ-ELISPOT assay of survivin 2B- or cepp55-CTL clones. T2-A24 cells pulsed with 60 µg of survivin 2B or cepp55 peptides were used as APC. (C) An exogenous presentation assay using T2-A24 cells as APC. IFNγ produced by indicated CTL clones were detected using ELISPOT assay. The <10 kDa cell extracts from SW480 or 3G12 were titrated and incubated with T2-A24 cells and indicated CTL clones as described in methods. The numbers indicate cell equivalents of the extracts. (D) IFNγ-ELISPOT assay of a survivin 2B-CTL clone. The CTL was incubated with 5 × 104 cells of SW480 or 3G12 pulsed with indicated amounts of survivin 2B peptides (left). A right panel shows the quantified numbers of IFNγ spots. Data are representatives of three independent experiments.
Discussion
This study aimed to investigate effect of loss of tapasin on TAA-specific CTL responses that plays a critical role in immune surveillance of human tumors in vivo. Loss of tapasin and its positive correlation to poorer patient prognoses have been reported among a variety of tumors. In a large-cohort study of human colorectal cancers, increased CD8+ T-cell infiltration of tumors was acknowledged as a critical factor positively correlating with patient survival.27 In this study, significant correlation was observed only when cancer tissues lacked tapasin expression as well as CD8+ T-cell infiltration, suggesting that CTL responses to cancer cells influence prognoses in a tapasin-dependent manner, consistent with previous studies in ovarian carcinoma, HNSCC, glioblastoma, and colorectal carcinoma.19-21,24 A former study has demonstrated that restoring tapasin gene expression in a mouse tumor model using viral vector transduction improved survival of model mice bearing lung carcinoma.28 In this study, we generated human lung and colon cancer variants that express TAAs, survivin, or cep55, genetically lacking tapasin protein expression. To our knowledge, this study is first to demonstrate that CTL responses to endogenous TAAs presented by HLA class I molecules are significantly reduced to undetectable levels solely by the absence of tapasin. Our results suggested the antigen processing of HLA-A24 endogenous TAAs is limited in cancer cells. Thus, tapasin expression is a key to regulate cancer-specific CTL surveillance, influencing patient survival.
It is well known that tapasin is the core component tethering MHC class I and TAP molecules and is indispensable to stabilize TAP heterodimers. Loss of tapasin would therefore resulted in disorganized PLC formation and influenced peptide transport into the ER.29 The molecular mechanisms underlying loss of tapasin in tumor cells have been elusive, but it most likely occurs due to several reasons such as deregulation of epigenetic control, transcriptional, and posttranscriptional modulation. Treatment of mouse melanoma and transformed lung cells with a DNA demethylating agent, 5-aza-2′ deoxycytidine (5-AC) or a histone deacetylace inhibitor, trichostatin A (TSA) restored tapasin expression.30-32 An alternative splicing form of tapasin, which failed to restore surface MHC class I levels in tapasin-deficient cells, has been reported in a human melanoma cell line.33 In addition, a recent report has shown a novel germ-line frameshift mutation in a single population of human melanomas, resulting in loss of tapasin, and expression could not be restored by IFNγ treatment.34
Our results raised another intriguing question concerning HLA types and tapasin dependency. It is known that loss of tapasin generally leads to a decrease in HLA surface expression, but the dependency on tapasin differs across HLA-class I allotypes. Because loss of tapasin significantly decreases the quantity of naturally processed peptides that are stably bound to HLA-A2, the majority of peptide-HLA-A2 complexes on tapasin-deficient cell surfaces are potentially unstable.35 Despite that HLA-A*0201 has been reported as a tapasin-independent HLA type, 3G12 clearly showed a decrease in the HLA-A2 surface level in this study.36 The conflicting results between the widely used B-cell line (.220) and the colon cancer line (3G12) may imply an uncharacterized effects of targeting tapasin function and should be further investigated.
Recent clinical successes in immunotherapy using immune-checkpoint inhibitors that improved prognoses of a variety of cancer patients including melanoma, NSCLC, and renal cell carcinoma, strongly emphasize the potential of immune surveillance by endogenous T cells.37-39 NSCLC is indeed a good target for immune checkpoint blockade because of frequent gene mutations yielding neoantigens.40 In contrast, durable clinical effects are not always observed in every case and more than half are yet refractory. Frequent loss of tapasin in NSCLC implies that quite a few proportion of cancer cells are potentially immune-escape variants in vivo. Therefore, counteracting immune-escape variants that are resistant to immunotherapy would be an urgent issue of wide importance. A study with tapasin-deficient mice has showed that loss of tapasin gave rise to quantitative and qualitative change in the surface pMHC repertoire: The repertoire presented by tapasin-deficient cells consists of many lengthy/anchorless peptides, which are not presented by wild-type cells, thereby inducing wild-type CTL responses against tapasin-deficient cells although the antigens remain unknown.41,42 Therefore, endogenous CTL surveillance is potentially able to monitor tapasin deficiency and further investigation of such CTL responses may accelerate the drug development against tumor immune-escape variants.
Materials and methods
Patients
The study was performed with approval of the local ethics committee of Sapporo Medical University. Eigty-five NSCLC patients surgically treated at Sapporo Medical University Hospital, Sapporo, Japan, from 2008 and 2012 were retrospectively included in this study, with informed consent according to the guidelines of the Declaration of Helsinki. Histological diagnoses were determined in accordance with the World Health Organization criteria for histopathology classification. Clinical data were obtained from the patient records and OS was calculated from the day of the operation until October 7, 2015.
Immunohistochemistry
Formalin-fixed paraffin-embedded tumor sections from NSCLC patients or tumor-bearing mice were stained with anti-tapasin TO-3 mAb or anti-CD8+ mAb after epitope retrieval with Novocastra epitope retrieval solution (Leica). Endogenous peroxidase activity was blocked by immersion in 3% peroxidase. The sections were then stained with a corresponding secondary Ab and visualized according to a standard protocol (Leica). All sections were counterstained with hematoxylin-eosin (HE) and cancer lesions were reviewed and verified by pathologists. For scoring CD8+ T-cell infiltration, we randomly assessed 10 high-power fields (HPF, x400) per section and counted the number of fields with >10 CD8+-cells migrating within cancer lesions.
Cells and antibodies
The human lung cancer LHK2, colon carcinoma SW480, HLA class I-null K562, and T2-A24 cells (T2 cells stably expressing HLA-A24) were cultured in RPMI-1640 or Dulbecco's modified Eagle's medium supplemented with 10% fetal bovine serum (Biosera), 1% penicillin-streptomycin mixed solution (Gibco), 1% sodium pyruvate (Gibco), and 50 μM 2-mercaptoethanol (Gibco). The pan-HLA class I, HLA-A24, and HLA-A2 specific mAb were prepared from W6/32 (ATCC), C7709A2 (a gift from Dr. P. G. Coulie), and BB7.2 (ATCC) hybridomas. The anti-tapasin TO-3 mAb (sc-80647; Santa Cruz), anti-human CD8+ mAb (N1592; Daco), anti-actin mAb (clone AC-15; SIGMA), and anti-survivin rabbit polyclonal Ab (novusbiologicals) were obtained commercially. The PE-conjugated tetramer specific to survivin 2B peptide and HLA-A24 was from MBL (Nagoya, Japan).
RT-PCR and qPCR
Total RNA was isolated from cancer cells using TRIzol reagent according to the manufacturer's instructions (Thermo). Total RNA from human adult tissues was purchased (human total RNA master panel; Clontech). cDNA was synthesized from 2 μg of total RNA by reverse transcription with Superscript II and oligo(dT) primer (Life Technologies). PCR reactions were initially incubated at 94°C for 2 min, followed by 35 cycles of denaturation at 94°C for 15 sec, annealing at 58°C for 30 sec, and extension at 68°C for 1 min. Primer pairs used for RT-PCR are as follows: G3PDH, 5′-ACTGTGGCTCCAAACTGCTT-3′ and 5′-GAGCAGCTGTTTCCGTTTTC-3′ (product size 450bp); cep55, 5′- ACTGTGGCTCCAAACTGCTT-3′ and 5′-GAGCAGCTGTTTCCGTTTTC-3′ (product size 181bp); survivin 2B, 5′-TCAAGGACCACCGCATCTCTAC-3′ and 5′-GTGCTGGTATTACAGGCGTAAG-3′ (product size 221bp). Primer pairs used for quantitative PCR are as follows: cep55, 5′-TGAGTTTGCCATCACAGAGC-3′ and 5′-ATTCCACATGGACAAGCAGA-3′; survivin 2B, 5′-TCAAGGACCACCGCATCTCTAC-3′ and 5′-GTGCTGGTATTACAGGCGTAAG-3′.
Western blot
5 × 106 cells were homogenized with buffer containing 20 mM Tris-HCl (pH 7.5), 1% NP40, 150 mM NaCl, and a complete EDTA-free protease inhibitor (Roche) for 30 min at 4˚C. Cell lysates were mixed with an equal volume of 2x sample buffer (125 mM Tris-HCl (pH 6.8), 10% (v/v) 2-mercaptoethanol, 40 mg/mL SDS, 100 mg/ml sucrose, and 4 mg/mL bromophenol blue) and separated by SDS-PAGE. Proteins were then transferred onto polyvinylidene difluoride (PVDF) membranes, which were blocked with 5% skim milk and 2% BSA in 0.1% PBS-Tween at 4˚C overnight. Blots were incubated with anti-human tapasin TO-3 (1:1,000) mAb or anti-actin (1:5,000) mAb overnight at 4˚C, followed by incubation with peroxidase-labeled goat anti-mouse immunoglobulin (1:10,000) for 30 min at room temperature. The membranes were treated with ECL™ Western Blotting Detection Reagents (GE Healthcare) and exposed to blue films (Fuji Photo Film). Odyssey Fc Imaging System 2800 (LI-COR Biosciences) was used for the detection of survivin protein.
Generation of tapasin-deficient human cancer cell lines
Gene targeting with the CRISPR-Cas9 system has been described in detail elsewhere 43 Briefly, we designed guide RNAs (gRNAs; 5′- GGTGATCGAGTGTTGGTTCG-3′) with the online software CRISPRdirect, which predicted a unique site for targeting human tapasin, and introduced it into the GeneArt CRISPR Nuclease vector with orange-fluorescent protein reporter (OFP) (A21174; Thermo). We transfected SW480 cells according to the manufacturer's instructions (Lipofectamine 2000; Thermo) and selected the OFP-positive clones via cell sorting (FACSAria II; BD). The tapasin-deficient clones 3G12, 1D4, and 3A9 were ultimately selected based on their tapasin-gene sequences.
TAA-specific CTL induction and single-cell cloning
PBMCs from an HLA-A24 positive cancer patient were cultured in AIM-V medium (Life Technologies) containing 10% human serum and 50 U/mL IL-2, and repeatedly stimulated with survivin 2B peptide (AYACNTSTL) (40 μg/mL) every 3–4 d. Two weeks later, the cells were stained with a PE-conjugated survivin 2B/HLA-A24 tetramers at 4°C for 20 min, followed by a PC5-conjugated anti-CD8+ mAb (Beckman Coulter) at 4°C for 30 min. A single-cell sorting using FACS (Aria II Special Order, BD, Houston, TX, USA) allowed to isolate the tetramer and CD8+ double-positive clones. The sorted clones were expanded in complete AIM-V medium containing 100 U/mL IL-2, 1 μg/mL PHA, and feeder cells (X-ray irradiated PBMCs) in a U-bottom 96-well plate (Corning). We used the cep55-specific CTL clone that was generated from PBMC of an HLA-A*2402-positive breast cancer patient with stimulation with synthetic cep55 peptide (VYVKGLLAKI) and described elsewhere.26
ELISPOT IFNγ assay
About 1.0 × 105 survivin 2B- or cep55-CTL clones together with 5 × 104 cancer cells (LHK2 or YS3 cells) or 5 × 104 T2-A24 cells pulsed synthetic peptides were added to ELISPOT plates coated with an anti-human IFNγ antibody (BD Biosciences) and incubated in 5% CO2 at 37°C for 24 h. The wells were then incubated with a biotinylated anti-human IFNγ antibody for 2 h at room temperature followed by an ELISPOT streptavidin-HRP antibody for 1 h. IFNγ positive spots were visualized using the ELISPOT AEC Substrate Set according to the manufacture's instruction (BD Biosciences).
LDH cytotoxicity assay
Cytotoxicity against target cells was measured by release of cellular lactate dehydrogenase (LDH) an LDH cytotoxicity detection kit according to the manufacture's instruction (TaKaRa, Japan). The percentage of LDH release was calculated as follows: % release = 100 × (experimental LDH release – spontaneous LDH release)/(maximal LDH release – spontaneous LDH release). Cells treated with 1% Triton X-100 were used as positive controls for maximal LDH releases.
Mice and in vivo tumor models
NSG mice were purchased from the Jackson laboratory. 1 × 105 SW480 or 3G12 cells were subcutaneously injected into NSG mice. The major (x) and minor (y) axes of the tumors were routinely measured. Tumor volume was calculated as follows: volume = xy2/2. In tumor-rejection models, 2.0 × 106 cep55-specific CTLs or PBS was intravenously injected 10 and 17 d after tumor injection. The mice were maintained in the animal facility of Sapporo Medical University and all procedures were done in accordance with the institutional animal care guidelines.
An exogenous presentation assay using cell extracts and APC
Preparation of cell extracts is described in detail elsewhere.44 Briefly, peptide extracts of SW480 or 3G12 cells were prepared by acid extraction using 10% formic acid in the presence of 2 µM of an irrelevant martyr peptide followed by filtration using <10 kDa cut-off spin-column (amicon). The samples were then dried using a vacuum centrifugation for overnight, and assayed for antigenic activities by incubation with 5 × 104 cells of a survivin 2B-CTL clone along with 5 × 104 cells of T2-A24 cells. The produced IFNγ was detected using ELISPOT assay.
Supplementary Material
Disclosure of potential conflicts of interest
No potential conflicts of interest were disclosed.
Funding
This study is supported by Japan Society for the Promotion of Science (T.K.), Suhara Kinen Zaidan (T.K.), Grants-in-Aid for Regional R&D Proposal-Based Program from Northern Advancement Center for Science & Technology of Hokkaido Japan (T.K.), Japan Agency for Medical Research and development (T.T.), Grants-in-Aid of Ono Cancer Research Fund (T.T.), Grant-in-Aid for Scientific Research from the Ministry of Education, Culture, Sports, Science and Technology of Japan (N.S.), and Health and Labor Sciences Research Grants (N.S.).
References
- 1.Neefjes J, Jongsma ML, Paul P, Bakke O. Towards a systems understanding of MHC class I and MHC class II antigen presentation. Nat Rev Immunol 2011; 11:823-36; PMID:22076556; http://dx.doi.org/ 10.1038/nri3084 [DOI] [PubMed] [Google Scholar]
- 2.Blum JS, Wearsch PA, Cresswell P. Pathways of antigen processing. Annu Rev Immunol 2013; 31:443-73; PMID:23298205; http://dx.doi.org/ 10.1146/annurev-immunol-032712-095910 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hammer GE, Kanaseki T, Shastri N. The final touches make perfect the peptide-MHC class I repertoire. Immunity 2007; 26:397-406; PMID:17459809; http://dx.doi.org/ 10.1016/j.immuni.2007.04.003 [DOI] [PubMed] [Google Scholar]
- 4.Rock KL, Farfan-Arribas DJ, Shen L. Proteases in MHC class I presentation and cross-presentation. J Immunol 2010; 184:9-15; PMID:20028659; http://dx.doi.org/ 10.4049/jimmunol.0903399 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Weimershaus M, Evnouchidou I, Saveanu L, van Endert P. Peptidases trimming MHC class I ligands. Curr Opin Immunol 2013; 25:90-6; PMID:23089230; http://dx.doi.org/ 10.1016/j.coi.2012.10.001 [DOI] [PubMed] [Google Scholar]
- 6.Elliott T, Williams A. The optimization of peptide cargo bound to MHC class I molecules by the peptide-loading complex. Immunol Rev 2005; 207:89-99; PMID:16181329; http://dx.doi.org/ 10.1111/j.0105-2896.2005.00311.x [DOI] [PubMed] [Google Scholar]
- 7.Wearsch PA, Cresswell P. The quality control of MHC class I peptide loading. Curr Opin Cell Biol 2008; 20:624-31; PMID:18926908; http://dx.doi.org/ 10.1016/j.ceb.2008.09.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Garbi N, Tan P, Diehl AD, Chambers BJ, Ljunggren HG, Momburg F, Hämmerling GJ. Impaired immune responses and altered peptide repertoire in tapasin-deficient mice. Nat Immunol 2000; 1:234-8; PMID:10973281; http://dx.doi.org/ 10.1038/79775 [DOI] [PubMed] [Google Scholar]
- 9.Grandea AG 3rd, Golovina TN, Hamilton SE, Sriram V, Spies T, Brutkiewicz RR, Harty JT, Eisenlohr LC, Van Kaer L. Impaired assembly yet normal trafficking of MHC class I molecules in Tapasin mutant mice. Immunity 2000; 13:213-22; PMID:10981964; http://dx.doi.org/ 10.1016/S1074-7613(00)00021-2 [DOI] [PubMed] [Google Scholar]
- 10.Dissemond J, Kothen T, Mors J, Weimann TK, Lindeke A, Goos M, Wagner SN. Downregulation of tapasin expression in progressive human malignant melanoma. Arch Dermatol Res 2003; 295:43-9; PMID:12682852; http://dx.doi.org/ 10.1007/s00403-003-0393-8 [DOI] [PubMed] [Google Scholar]
- 11.Krishnakumar S, Abhyankar D, Sundaram AL, Pushparaj V, Shanmugam MP, Biswas J. Major histocompatibility antigens and antigen-processing molecules in uveal melanoma. Clin Cancer Res 2003; 9:4159-64; PMID:14519640 [PubMed] [Google Scholar]
- 12.Ogino T, Bandoh N, Hayashi T, Miyokawa N, Harabuchi Y, Ferrone S. Association of tapasin and HLA class I antigen down-regulation in primary maxillary sinus squamous cell carcinoma lesions with reduced survival of patients. Clin Cancer Res 2003; 9:4043-51; PMID:14519625 [PubMed] [Google Scholar]
- 13.Ogino T, Shigyo H, Ishii H, Katayama A, Miyokawa N, Harabuchi Y, Ferrone S. HLA class I antigen down-regulation in primary laryngeal squamous cell carcinoma lesions as a poor prognostic marker. Cancer Res 2006; 66:9281-9; PMID:16982773; http://dx.doi.org/ 10.1158/0008-5472.CAN-06-0488 [DOI] [PubMed] [Google Scholar]
- 14.Seliger B, Atkins D, Bock M, Ritz U, Ferrone S, Huber C, Störkel S. Characterization of human lymphocyte antigen class I antigen-processing machinery defects in renal cell carcinoma lesions with special emphasis on transporter-associated with antigen-processing down-regulation. Clin Cancer Res 2003; 9:1721-7; PMID:12738726 [PubMed] [Google Scholar]
- 15.Atkins D, Breuckmann A, Schmahl GE, Binner P, Ferrone S, Krummenauer F, Störkel S, Seliger B. MHC class I antigen processing pathway defects, ras mutations and disease stage in colorectal carcinoma. Int J Cancer 2004; 109:265-73; PMID:14750179; http://dx.doi.org/ 10.1002/ijc.11681 [DOI] [PubMed] [Google Scholar]
- 16.Facoetti A, Nano R, Zelini P, Morbini P, Benericetti E, Ceroni M, Campoli M, Ferrone S. Human leukocyte antigen and antigen processing machinery component defects in astrocytic tumors. Clin Cancer Res 2005; 11:8304-11; PMID:16322289; http://dx.doi.org/ 10.1158/1078-0432.CCR-04-2588 [DOI] [PubMed] [Google Scholar]
- 17.Lou Y, Vitalis TZ, Basha G, Cai B, Chen SS, Choi KB, Jeffries AP, Elliott WM, Atkins D, Seliger B et al.. Restoration of the expression of transporters associated with antigen processing in lung carcinoma increases tumor-specific immune responses and survival. Cancer Res 2005; 65:7926-33; PMID:16140964 [DOI] [PubMed] [Google Scholar]
- 18.Raffaghello L, Prigione I, Bocca P, Morandi F, Camoriano M, Gambini C, Wang X, Ferrone S, Pistoia V. Multiple defects of the antigen-processing machinery components in human neuroblastoma: immunotherapeutic implications. Oncogene 2005; 24:4634-44; PMID:15897905; http://dx.doi.org/ 10.1038/sj.onc.1208594 [DOI] [PubMed] [Google Scholar]
- 19.Han LY, Fletcher MS, Urbauer DL, Mueller P, Landen CN, Kamat AA, Lin YG, Merritt WM, Spannuth WA, Deavers MT et al.. HLA class I antigen processing machinery component expression and intratumoral T-Cell infiltrate as independent prognostic markers in ovarian carcinoma. Clin Cancer Res 2008; 14:3372-9; PMID:18519766; http://dx.doi.org/ 10.1158/1078-0432.CCR-07-4433 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Thuring C, Geironson L, Paulsson K. Tapasin and human leukocyte antigen class I dysregulation correlates with survival in glioblastoma multiforme. Anticancer Agents Med Chem 2014; 14:1101-9; PMID:25175688; http://dx.doi.org/ 10.2174/1871520614666140825110402 [DOI] [PubMed] [Google Scholar]
- 21.Sokol L, Koelzer VH, Rau TT, Karamitopoulou E, Zlobec I, Lugli A. Loss of tapasin correlates with diminished CD8(+) T-cell immunity and prognosis in colorectal cancer. J Transl Med 2015; 13:279; PMID:26310568; http://dx.doi.org/ 10.1186/s12967-015-0647-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Thuring C, Follin E, Geironson L, Freyhult E, Junghans V, Harndahl M, Buus S, Paulsson KM. HLA class I is most tightly linked to levels of tapasin compared with other antigen-processing proteins in glioblastoma. Br J Cancer 2015; 113:952-62; PMID:26313662; http://dx.doi.org/ 10.1038/bjc.2015.297 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Purcell AW, Gorman JJ, Garcia-Peydro M, Paradela A, Burrows SR, Talbo GH, Laham N, Peh CA, Reynolds EC, López De Castro JA et al.. Quantitative and qualitative influences of tapasin on the class I peptide repertoire. J Immunol 2001; 166:1016-27; PMID:11145681; http://dx.doi.org/ 10.4049/jimmunol.166.2.1016 [DOI] [PubMed] [Google Scholar]
- 24.Jiang Q, Pan HY, Ye DX, Zhang P, Zhong LP, Zhang ZY. Downregulation of tapasin expression in primary human oral squamous cell carcinoma: association with clinical outcome. Tumour Biol 2010; 31:451-9; PMID:20532727; http://dx.doi.org/ 10.1007/s13277-010-0054-4 [DOI] [PubMed] [Google Scholar]
- 25.Hirohashi Y, Torigoe T, Maeda A, Nabeta Y, Kamiguchi K, Sato T, Yoda J, Ikeda H, Hirata K, Yamanaka N et al.. An HLA-A24-restricted cytotoxic T lymphocyte epitope of a tumor-associated protein, survivin. Clin Cancer Res 2002; 8:1731-9; PMID:12060610 [PubMed] [Google Scholar]
- 26.Inoda S, Hirohashi Y, Torigoe T, Nakatsugawa M, Kiriyama K, Nakazawa E, Harada K, Takasu H, Tamura Y, Kamiguchi K et al.. Cep55/c10orf3, a tumor antigen derived from a centrosome residing protein in breast carcinoma. J Immunother 2009; 32:474-85; PMID:19609239; http://dx.doi.org/ 10.1097/CJI.0b013e3181a1d109 [DOI] [PubMed] [Google Scholar]
- 27.Galon J, Costes A, Sanchez-Cabo F, Kirilovsky A, Mlecnik B, Lagorce-Pages C, Tosolini M, Camus M, Berger A, Wind P et al.. Type, density, and location of immune cells within human colorectal tumors predict clinical outcome. Science 2006; 313:1960-4; PMID:17008531; http://dx.doi.org/ 10.1126/science.1129139 [DOI] [PubMed] [Google Scholar]
- 28.Lou Y, Basha G, Seipp RP, Cai B, Chen SS, Moise AR, Jeffries AP, Gopaul RS, Vitalis TZ, Jefferies WA. Combining the antigen processing components TAP and Tapasin elicits enhanced tumor-free survival. Clin Cancer Res 2008; 14:1494-501; PMID:18316574; http://dx.doi.org/ 10.1158/1078-0432.CCR-07-1066 [DOI] [PubMed] [Google Scholar]
- 29.Garbi N, Tiwari N, Momburg F, Hammerling GJ. A major role for tapasin as a stabilizer of the TAP peptide transporter and consequences for MHC class I expression. Eur J Immunol 2003; 33:264-73; PMID:12594855; http://dx.doi.org/ 10.1002/immu.200390029 [DOI] [PubMed] [Google Scholar]
- 30.Seliger B. Molecular mechanisms of MHC class I abnormalities and APM components in human tumors. Cancer Immunol Immunother 2008; 57:1719-26; PMID:18408926; http://dx.doi.org/ 10.1007/s00262-008-0515-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Khan AN, Gregorie CJ, Tomasi TB. Histone deacetylase inhibitors induce TAP, LMP, Tapasin genes and MHC class I antigen presentation by melanoma cells. Cancer Immunol Immunother 2008; 57:647-54; PMID:18046553; http://dx.doi.org/ 10.1007/s00262-007-0402-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Setiadi AF, Omilusik K, David MD, Seipp RP, Hartikainen J, Gopaul R, Choi KB, Jefferies WA. Epigenetic enhancement of antigen processing and presentation promotes immune recognition of tumors. Cancer Res 2008; 68:9601-7; PMID:19047136; http://dx.doi.org/ 10.1158/0008-5472.CAN-07-5270 [DOI] [PubMed] [Google Scholar]
- 33.Belicha-Villanueva A, Golding M, McEvoy S, Sarvaiya N, Cresswell P, Gollnick SO, Bangia N. Identification of an alternate splice form of tapasin in human melanoma. Hum Immunol 2010; 71:1018-26; PMID:20600451; http://dx.doi.org/ 10.1016/j.humimm.2010.05.019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Chang CC, Pirozzi G, Wen SH, Chung IH, Chiu BL, Errico S, Luongo M, Lombardi ML, Ferrone S. Multiple structural and epigenetic defects in the human leukocyte antigen class I antigen presentation pathway in a recurrent metastatic melanoma following immunotherapy. J Biol Chem 2015; 290:26562-75; PMID:26381407; http://dx.doi.org/ 10.1074/jbc.M115.676130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Barber LD, Howarth M, Bowness P, Elliott T. The quantity of naturally processed peptides stably bound by HLA-A*0201 is significantly reduced in the absence of tapasin. Tissue Antigens 2001; 58:363-8; PMID:11929586; http://dx.doi.org/ 10.1034/j.1399-0039.2001.580604.x [DOI] [PubMed] [Google Scholar]
- 36.Greenwood R, Shimizu Y, Sekhon GS, DeMars R. Novel allele-specific, post-translational reduction in HLA class I surface expression in a mutant human B cell line. J Immunol 1994; 153:5525-36; PMID:7989754 [PubMed] [Google Scholar]
- 37.Robert C, Long GV, Brady B, Dutriaux C, Maio M, Mortier L, Hassel JC, Rutkowski P, McNeil C, Kalinka-Warzocha E et al.. Nivolumab in previously untreated melanoma without BRAF mutation. N Engl J Med 2015; 372:320-30; PMID:25399552; http://dx.doi.org/ 10.1056/NEJMoa1412082 [DOI] [PubMed] [Google Scholar]
- 38.Brahmer J, Reckamp KL, Baas P, Crino L, Eberhardt WE, Poddubskaya E, Antonia S, Pluzanski A, Vokes EE, Holgado E et al.. Nivolumab versus docetaxel in advanced squamous-cell non-small-cell lung cancer. N Engl J Med 2015; 373:123-35; PMID:26028407; http://dx.doi.org/ 10.1056/NEJMoa1504627 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Motzer RJ, Escudier B, McDermott DF, George S, Hammers HJ, Srinivas S, Tykodi SS, Sosman JA, Procopio G, Plimack ER et al.. Nivolumab versus everolimus in advanced renal-cell carcinoma. N Engl J Med 2015; 373:1803-13; PMID:26406148; http://dx.doi.org/ 10.1056/NEJMoa1510665 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Schumacher TN, Schreiber RD. Neoantigens in cancer immunotherapy. Science 2015; 348:69-74; PMID:25838375; http://dx.doi.org/ 10.1126/science.aaa4971 [DOI] [PubMed] [Google Scholar]
- 41.Kanaseki T, Lind KC, Escobar H, Nagarajan N, Reyes-Vargas E, Rudd B, Rockwood AL, Van Kaer L, Sato N, Delgado JC et al.. ERAAP and tapasin independently edit the amino and carboxyl termini of MHC class I peptides. J Immunol 2013; 191(4):1547-55.; PMID:23863903; http://dx.doi.org/ 10.4049/jimmunol.1301043 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Shastri N, Nagarajan N, Lind KC, Kanaseki T. Monitoring peptide processing for MHC class I molecules in the endoplasmic reticulum. Curr Opin Immunol 2014; 26:123-7; PMID:24556408; http://dx.doi.org/ 10.1016/j.coi.2013.11.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Mali P, Yang L, Esvelt KM, Aach J, Guell M, DiCarlo JE, Norville JE, Church GM. RNA-guided human genome engineering via Cas9. Science 2013; 339:823-6; PMID:23287722; http://dx.doi.org/ 10.1126/science.1232033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kanaseki T, Shastri N. Biochemical analysis of naturally processed antigenic peptides presented by MHC class I molecules. Methods Mol Biol 2013; 960:179-85; PMID:23329488; http://dx.doi.org/ 10.1007/978-1-62703-218-6_15 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


