ABSTRACT
Despite the success of immune checkpoint blockade in melanoma, the majority of patients do not respond. We hypothesized that the T and NK cell subset frequencies and expression levels of their receptors may predict responses and clinical outcome of anti-CTLA-4 treatment. We thus characterized the NK and T cell phenotype, as well as serum levels of several cytokines in 67 melanoma patients recruited in Italy and Sweden, using samples drawn prior to and during treatment. Survival correlated with low expression of the inhibitory receptor TIM-3 on circulating T and NK cells prior to and during treatment and with the increased frequency of mature circulating NK cells (defined as CD3−CD56dim CD16+) during treatment. Survival also correlated with low levels of IL-15 in the serum. Functional experiments in vitro demonstrated that sustained exposure to IL-15 enhanced the expression of PD-1 and TIM-3 on both T and NK cells, indicating a causative link between high IL-15 levels and enhanced expression of TIM-3 on these cells. Receptor blockade of TIM-3 improved NK cell-mediated elimination of melanoma metastasis cell lines in vitro. These observations may lead to the development of novel biomarkers to predict patient response to checkpoint blockade treatment. They also suggest that induction of additional checkpoints is a possibility that needs to be considered when treating melanoma patients with IL-15.
KEYWORDS: Anti-CTLA-4, IL-15, melanoma, NK cells, PD-1, TIM-3
Introduction
Fundamental research in immunology is being successfully translated to improve cancer therapy.1-3 For example, treatment with anti-CTLA-4 and anti-PD-1 monoclonal antibodies—also referred to as “immune checkpoint blockade”—unleashes patient immune responses to tumors by blocking inhibitory receptors on immune cells.4 Initial response rates to anti-CTLA-4 (ipilimumab) were in the range of 20%5,6 of patients. Although these are higher with anti-PD-1 (nivolumab and pembrolizumab) and even higher with a combination of the two, it is unclear why certain patients respond and others do not. The evaluation of responses to immune checkpoint blockade therapy has been based mainly on conventional clinical biomarkers, while cellular and molecular immune responses have only been studied to a limited extent.7-14 There is ample evidence for T cell involvement in immunity to melanoma,15,16 and CTLA-4 blockade could influence T cell activity by different mechanisms, e.g., (a) expanding the pool of antigen specific of CD8+ T cells and CD4+ T helper cells, (b) depleting CD4+ regulatory T cells (Tregs).17-20
NK cells display potent cytotoxicity against tumor cells expressing low levels of MHC class I molecules21-23 and/or increased levels of tumor or stress associated activating ligands, but their potential in cancer immunotherapy is yet to be fully exploited. We and others have recently demonstrated that (a) NK cells selectively target freshly explanted tumor cells, (b) activation and frequency are related to the clinical outcome of melanoma and (c) specific NK cell subsets found in tumor-infiltrated lymph nodes of melanoma patients can eliminate autologous tumor cells.24-29
We hypothesized that the in-depth characterization of T and NK cells in the peripheral blood of melanoma patients before and during anti-CTLA-4 treatment could help develop a better integrated understanding of the complexity of immune responses and ultimately contribute to a more efficient immune therapy. Moreover, comparative analysis of T and NK cells in patients stratified into groups based on survival in response to the immune therapy may reveal an association of specific variables with clinical outcome after anti-CTLA-4 treatment. This could lead to the identification of new biomarkers able to predict responses to immune checkpoint blockade.
Results
Multivariate analysis of T, NK cells and sera of melanoma patients before ipilimumab treatment distinguish long- and short-term survival patients
To evaluate the putative association between immune variables and treatment outcome, 67 melanoma patients treated with ipilimumab were stratified into two groups according to survival; 12 mo or longer (long-term survivors, n = 33) and shorter than 12 mo (short-term survivors, n = 34). Peripheral blood lymphocytes (PBL) and sera were collected from patients before the administration of the first dose of ipilimumab (withdrawal 0, W0); at week 3 (withdrawal, W1) before the second administration; at week 6 (withdrawal, W2), before the third administration; and at week 9 (withdrawal, W3) before the fourth and last administration. At each time point, a total of 134 parameters consisting of subset frequencies, the receptor repertoire of T and NK cells, as well as serum concentrations of cytokines and chemokines, were analyzed in multivariate as well as in univariate mode. The same biological parameters were analyzed in healthy donor controls.
To establish potential correlations between the immune variables analyzed and survival, multivariate projection models were built. At W0, 65 patients were included in the analysis (long-term survivors = 32, short-term survivors = 33) and at W3 51 patients (long-term survivors = 27, short-term survivors = 24). The modeling approach, Orthogonal Projections to Latent Structures, Discriminant Analysis (OPLS-DA) is based on Principal Component Analysis, taking all variables into account simultaneously with equal importance independent of the range of values. Thus, MFI and percentages can be analyzed together and co-variation between two or more variables, as well as the correlation to a specific group of patients (positive or negative, significance and magnitude of correlation) can easily be assessed and displayed graphically. OPLS-DA in particular helps identifying variables correlated with group separation since the components are orthogonal, meaning that the first principal component (here: horizontal axis) only represents the difference between the two most distinct groups, while variation unrelated to survival is represented as orthogonal components (here: vertical axis). The advantage in a clinical setting is that fewer patients are needed for biomarker identification than for traditional statistical methods, since general patterns can more easily be discerned when all variables are allowed to contribute.
At the start of the treatment, the OPLS-DA model yielded a good separation between patients (Fig. 1A) that could explain 89.7% of the difference between long- and short-term survival patients. The cross-validated predictive capacity was 63.8%. In summary, the constellation of immune variables applied in the multivariate analysis led to a clear separation of the two discrete patient groups and identified the most promising biomarkers correlating with a good prognosis.
Figure 1.
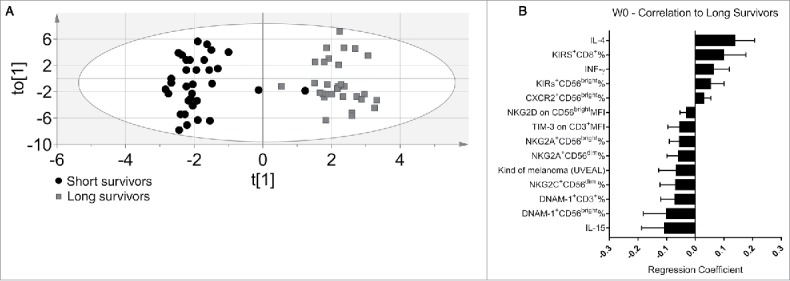
Discriminant analysis and immunoprofile of melanoma patients before the treatment (W0). (A) Discriminant analysis: Gray squares = long survivors (31), 12 mo or more. Black circles = short survivors, <12 mo (33). Horizontal axis = predictive component. Vertical axis = Orthogonal component not related to difference between groups. Ellipse = Hotelling's T2 95% confidence interval limit. (B) The 14 most significant variables correlated with long survival at the start of treatment. Error bars = 95% confidence intervals. Positive correlation to long survival means negative correlation to short survival, and vice versa.
Immunological parameters associated with survival time
We next studied in detail which individual parameters were associated with disease outcome. Eighteen variables were significantly different between long- and short-term survivors. The most significant variables are shown in Fig. 1B. We analyzed T and NK cell subsets in blood by flow cytometry, using the gating strategy depicted in Fig. S1. Variables correlated with long survival were increased frequencies of the CD56bright NK subsets expressing respectively KIRs receptors as well as CXCR2 chemokines receptors (Fig. 1B). Moreover, high IL-4 and IFN-γ serum levels and increased frequencies of KIRs+ CD8+ T cells also correlated with the long-survival endpoint. It should be noted that high CXCR2 frequencies in circulating NK cell subsets in melanoma patients confirm our previous study.29
Other variables that contribute to the molecular signature of long-survival patients in the OPLS-DA model were: low IL-15 serum levels, low frequencies of NKG2A+, DNAM-1+ CD56bright NK cells, NKG2C+ and NKG2A+CD56dim NK cells, DNAM-1+ on T cells and low expression of NKG2D on CD56bright and TIM-3 on T cells (Fig. 1B).
The most surprising finding was the association of lower serum IL-15 concentration with long-term survival, since IL-15 it is known to boost the antitumor cytotoxic immune response.30 It was significantly higher for short-term survivors than for long-term survivors before the first dose (W0) and elevated in most patients before treatment when compared with healthy donors. Since this observation was counterintuitive, additional patients were included in the study for this individual parameter. The IL-15 measurement has thus been performed on 129 (W0) and 109 (W3) sera in total. The result on this larger cohort of patients confirmed this finding and further reinforced the statistical significance (Table 1).
Table 1.
Immunoprofile of melanoma patients before the treatment (W0).
| Variable | Association with survival | Short survivors mean value ± S.D. | Long survivors mean value ± S.D. | p-value |
|---|---|---|---|---|
| % of CD3−CD56brightTIM-3+ cells | Negative | 72.77 ± 11.76 | 63.05 ± 15.47 | 0.01581 |
| MFI of TIM-3 on CD3−CD56bright | Negative | 1589 ± 801.4 | 1049 ± 549.4 | 0.00221 |
| MFI of TIM-3 on CD3−CD56dim | Negative | 1320 ± 602.4 | 1038 ± 448.9 | 0.04261 |
| MFI of KIRs on CD3−CD56dim | Negative | 6690 ± 3598 | 4687 ± 2323 | 0.02361 |
| MFI of TIM-3 on CD3+CD56− | Negative | 1259 ± 406.7 | 1007 ± 314.6 | 0.00762 |
| IL-15 serum concentration | Negative | 3.862 ± 2.285 | 2.849 ± 1.392 | 0.01211 |
According to the distribution, data were analyzed with Mann–Whitney test (1), for non-normally-distributed data, or unpaired t test (2), for normally-distributed data.
The differences in TIM-3 expression on T cells, and the IL-15 serum concentrations were statistically significant between long and short-term survivors also after univariate analysis. Moreover, the univariate analysis observed reduced frequencies of circulating CD56bright TIM-3+ and CD56dim KIR+ NK cells subsets in long survival patients (Table 1).
In conclusion, IL-15 and TIM-3 were the individual parameters that correlated most strongly with survival prior to treatment start, and which were also confirmed by univariate analysis. The fact that the expression of TIM-3 was associated with poor survival suggests that this inhibitory receptor may play a role as new immune checkpoint.
Analysis of T, NK cells and sera of melanoma patients during ipilimumab treatment
The first dose of ipilimumab did not induce broad modifications in the immune profile of NK and T cells between short- and long-term survivors. A change, however, occurred after the first (W1) and second dose (W2) (Fig. S2), when the average of CXCR2+ CD56bright NK cells percentage increased in the long-term survivors (Fig. S2C and F).
A new pattern emerged in the immune profile of the last withdrawal (Fig. 2A). Here, the adverse side effects colitis, hipophysitis and skin rash were also included in the model. The multivariate OPLS-DA model could explain 83.8% of the variation in the data at this time-point, and the cross-validated predictive capacity for new data was 63.5%. Forty-three variables were significantly different between long- and short-term survivors. The most relevant are shown in Fig. 2B. Among the variables that positively correlated with long-term survival were: percentages of circulating CXCR2+ CD56bright, CD56dim, CD16+CD56dim NK cells, DNAM-1+ CD56dim and NKG2D+ CD56dim. The T cell compartment was characterized by high frequencies of CCR2+ and NKG2D+ cells. Finally, higher serum levels of IL-4 and IFNγ correlated with long-term survival (Fig. 2B). The most significant variables that correlated with long-term survival were the reduced concentration of IL-15 in the patients' sera and a lower expression of KIRs on the CD56dim NK cells subset. These two parameters also correlated with each other, meaning that the same long-term survivors often displayed both reduced levels of IL-15 and low expression of KIRs on NK cells. The T cell compartment of long-term survival patients was dominated by a low expression TIM-3 and CCR7 and a reduced frequency of PD1+ T cells (Fig. 2B).
Figure 2.
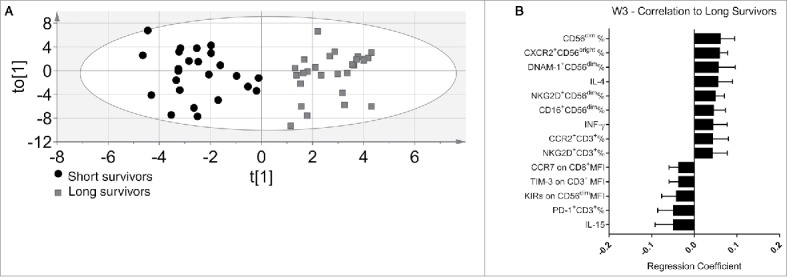
Discriminant analysis and immunoprofile of melanoma patients after the third treatment (W3). (A) Discriminant analysis: Gray squares = long survivors (28), 12 m or more. Black circles = short survivors, <12 mo (24). Horizontal axis = predictive component, vertical axis = order of patients, not related to differences between groups. (B) The 14 most significant variables correlated with long survival at the end of treatment. Error bars = 95% confidence intervals. Positive correlation to long survival means negative correlation to short survival, and vice versa.
In Table 2, we summarized the variables confirmed by univariate analysis that associated with the patients survival after the third ipilimumab treatment. Two variables were confirmed to positively correlate with long survival in univariate analysis: the frequencies of circulating CD56dim NK cells having a higher proportion of CD16+CD56dim cells. While five variables inversely correlate with long survival: KIRs on CD56dim and CCR7 expression on CD56bright NK cells, IL-15 serum levels, TIM-3 levels on CD3+ T cells and PD-1 expression levels on CD8+ T cells, respectively. Notably, there were no changes in the absolute number of lymphocytes expressed as number of cells/μL among long and short survivors groups before and after ipilimumab treatment (data not show). Thus, the univariate analysis confirmed that low serum IL-15 concentrations at W0 as well as at W3 characterize long-term survival patients.
Table 2.
Immunoprofile of melanoma patients after the third treatment univariate analysis (W3).
| Variable | Association with survival | Short survivors mean value ± S.D. | Long survivors mean value ± S.D. | p-value |
|---|---|---|---|---|
| % of CD3−CD56dim | Positive | 12.90 ± 5.868 | 20.55 ± 9.099 | 0.0005 |
| % of CD3−CD56dimCD16+ cells | Positive | 80.81 ± 16.41 | 89.87 ± 7.074 | 0.02201 |
| MFI of KIRs on CD3−CD56dim | Negative | 6337 ± 3334 | 4324 ± 1909 | 0.00961 |
| MFI of CCR7 on CD3−CD56bright | Negative | 2655 ± 1367 | 1872 ± 918.3 | 0.0198 |
| % of CD3+CD56− | Negative | 60.28 ± 13.63 | 53.16 ± 11.60 | 0.02201 |
| MFI of TIM-3 on CD3+CD56− | Negative | 1203 ± 370.5 | 980.3 ± 267.8 | 0.02821 |
| MFI of PD-1 on CD3+CD8+ | Negative | 2459 ± 1424 | 1698 ± 751.7 | 0.03571 |
| IL-15 serum concentration | Negative | 3.850 ± 2.357 | 2.937 ± 1.951 | 0.02571 |
According to the distribution, data were analyzed with Mann–Whitney test (1), for non-normally-distributed data, or unpaired t test (2), for normally-distributed data.
In conclusion, IL-15, TIM-3, PD-1 and KIRs were the parameters that correlated with survival after the ipilimumab treatment which were also confirmed by univariate analysis.
Patterns emerging during the treatments
A longitudinal analysis of treatment effects on the clinical and immune variables was performed. All the measured parameters were analyzed in univariate analysis and correlated with patient survival. To understand whether clusters of variables correlate to survival, a longitudinal multivariate analysis was also performed. A clear decline of the percentages of DNAM-1+ CD56bright NK cells was observed in short-term survival patients. Interestingly, in long-term survival patients a switch of the CD4+/CD8+ frequencies occurred between W1 and W2 in the peripheral blood, but this pattern was not sustained in the blood in the next withdrawal. At W2, the CD8+ T cells percentage prevailed over the CD4+ T cells for these patients (Table 3). To further investigate changes in the NK and T cell compartments in response to treatment, we longitudinally investigated lymphocyte activation in smaller subset of patients (Fig. 3). NK cells in PBL from both patient groups (six long and six short survivors) displayed a more robust degranulating activity at W0, in comparison with the healthy donors. Moreover, at W1 the long-term survivors showed higher proportion of activated CD107a+ NK cells than was found in short-term survivors. These differences were even more evident after exogenous stimulation with the highly NK susceptible K562 cell line (Fig. 3A, right panel). Strikingly, while circulating NK cells from long-term survivor patients promptly responded to the exogenous K562 stimulation, as measured by CD107a degranulation assays, NK cells from the short-term survival patients did not. The increased activation of NK cells degranulation in long-term survivors was mostly evident at W0 and W1 while differences in CD107a+ percentage of T cells was observed at W3 (Fig. 3B). Thus, while NK cells in long-term survival melanoma patients responded already after the first ipilimumab treatment (W1) in short-term survival melanoma patients did not. Moreover, the exogenous pulsing with K562 reveals that circulating NK cells isolated from short-term survival patients did not respond. These observations may indicate that NK cells of short survival patients could be exhausted or anergic.
Table 3.
Changes in NK and T cells phenotype in short and long survivors during anti-CTLA-4 treatment.
| Variable | Patients subgroup | Samples withdrawal | p-value | Mean ± S.D. |
|---|---|---|---|---|
| % of CD3−CD56brightDNAM-1+ cells | Long survivors | n.s. | ||
| Short survivors2 | W0 vs. W3 | ** | 97.34 ± 2.21 vs.91.49 ± 8.27 | |
| W1 vs. W3 | * | 96.38 ± 3.39 vs. 91.49 ± 8.27 | ||
| Long survivors2 | W1 vs. W2 | * | 59.12 ± 11.36 vs. 47.18 ± 13.37 | |
| % of CD3+CD4+ cells | Short survivors2 | n.s. | ||
| % of CD3+CD8+ cells | Long survivors2 | W1 vs. W2 | * | 37.67 ± 10 23 vs. 47.97 ± 11.78 |
| Short survivors2 | n.s. |
According to the distribution, data were analyzed with Kruskal–Wallis test followed by Dunn's correction (1), for non-normally-distributed data, and ANOVA followed by Bonferroni's correction (2), for normally-distributed data.
p-value < 0.001;
p-value < 0.01;
p-value < 0.05.
Figure 3.
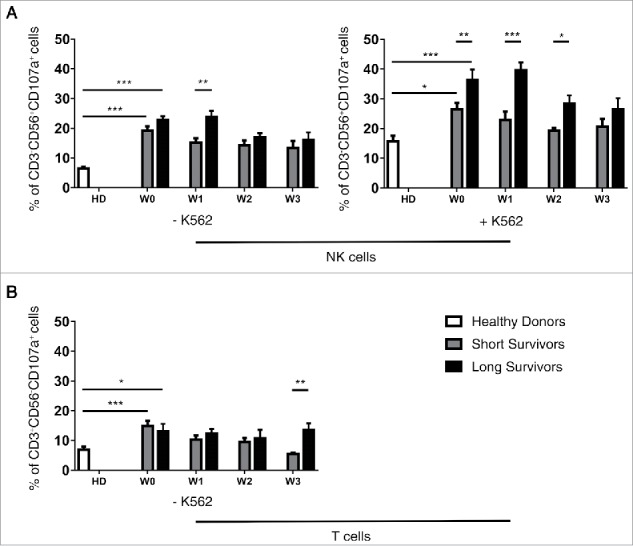
NK and T cells function analysis in short and long survivors during anti CTLA-4 treatment. Long survivors (black bars), short survivors (gray bars) and healthy donors (white bars). CD107a degranulation by NK (A) and T (B) cells on six healthy donors, six long and six short survival patients. Statistical analyses were performed with ANOVA followed by Bonferroni's correction; ***p-value < 0.001; **p-value < 0.01; *p-value < 0.05.
IL-15 upregulates TIM-3 and PD-1 on NK and T cells in vitro
The data presented above suggested a correlation between IL-15 serum levels and expression of PD-1 on T cells, KIRs on NK cells and TIM-3 on both. We therefore tested the hypothesis that IL-15 modulates the expression of these inhibitory receptors on NK and T cells. We stimulated PBL of healthy donor with recombinant human IL-15 (Fig. 4) in vitro. After a sustained exposure to IL-15 (5 d), a clear increase of both frequencies and expression levels of TIM-3 and PD-1 on mature NK and T cells was observed (Figs. 4A–C). Sustained stimulation with IL-15 also increased the KIR expression selectively on NK cells (Fig. 4A). Similar experiments conducted with 10-fold lower dose of IL-15 gave the same trend, although with less impressive induction of TIM-3, PD-1 and KIRs (Fig. S3).
Figure 4.
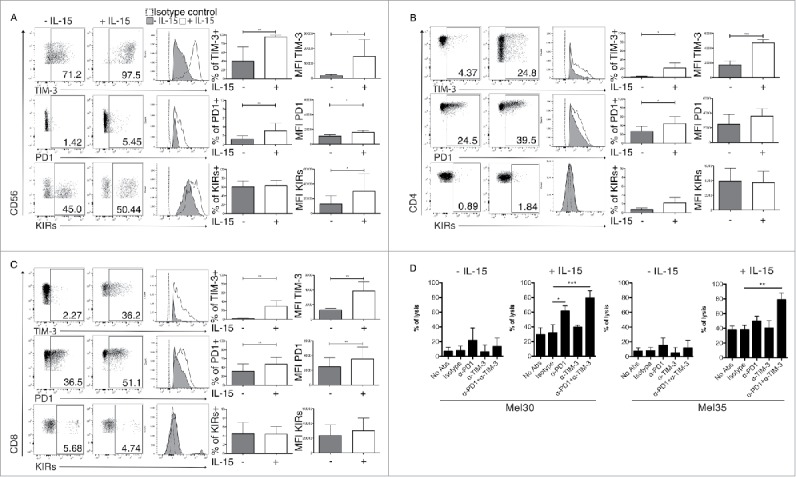
In vitro effects of IL-15 stimulation on NK and T cells phenotype and function. FACS analysis of TIM-3, PD-1 and KIRs expression (mean fluorescent intensities MFI) and frequencies: (A) NK cells (CD3-CD56+), (B) CD4+ T cells (CD3+CD4+) and (C) CD8+ T cells (CD3+CD8+) before and after 120 h of stimulation with IL-15. The data are showed as representative dot plots where the percentage of cells in each quadrant is indicated, and as representative histogram plots. Columns represent the statistical analysis from six independent experiments, ***p-value < 0.001; **p-value < 0.01; *p-value < 0.05, by Student's t test. Error bars represent standard deviations. (D) Effect either single or combined PD-1, TIM-3 antibodies blockade on resting or 120 h IL-15 activated NK cells cytotoxicity against primary metastatic melanoma cells; Mel30 and Mel35. Statistical analysis from three independent experiments made in single. ***p-value < 0.001; **p-value < 0.01; *p-value < 0.05 by one-way ANOVA with Bonferroni's post-test. Error bars, S.D.
Next, the possible role of TIM-3 and PD-1 in the modulation of NK cell cytotoxicity against melanoma cells was evaluated with receptor blockade experiments. While PD-1 blocking alone or in combination with TIM-3 did not significantly alter the cytotoxicity of unstimulated NK cells, these treatments significantly increased the cytotoxicity of IL-15 stimulated NK cells (Fig. 4D). These results may suggest that the short-term survival of melanoma patients associated with high IL-15 levels could be due to its induction of additional immune checkpoints on NK and T cells that can interfere with the anti-CTLA-4 treatment.
Prediction of survival based on immune biomarkers before and during treatment
To explore whether TIM-3, KIRs, IL-15 and NK subsets could work as predictive markers for long or short survival, an indicator variable was used to build and compare two Kaplan–Meier survival (KM) curves. Medians of the selected variables obtained from the whole patient cohort demonstrated to be a meaningful threshold with a good discriminating power. The log rank test was used to compare the KM curves and calculate the p-value.
Four predictors were identified that differentiated patients before the treatment had started: (i) CD16+CD56dim NK cell frequency; (ii and iii) the expression level of TIM-3 on CD56bright NK cells and this subset frequency, (iv) the expression of TIM-3 on CD56dim NK cells.
Patients with low frequencies of circulating CD16+CD56dim NK cells, with high frequency of TIM-3+ CD56bright NK cells and high expression levels of TIM-3 on both CD56dim and CD56bright NK cells, displayed a statistically significant shorter survival (Fig. 5A).
Figure 5.
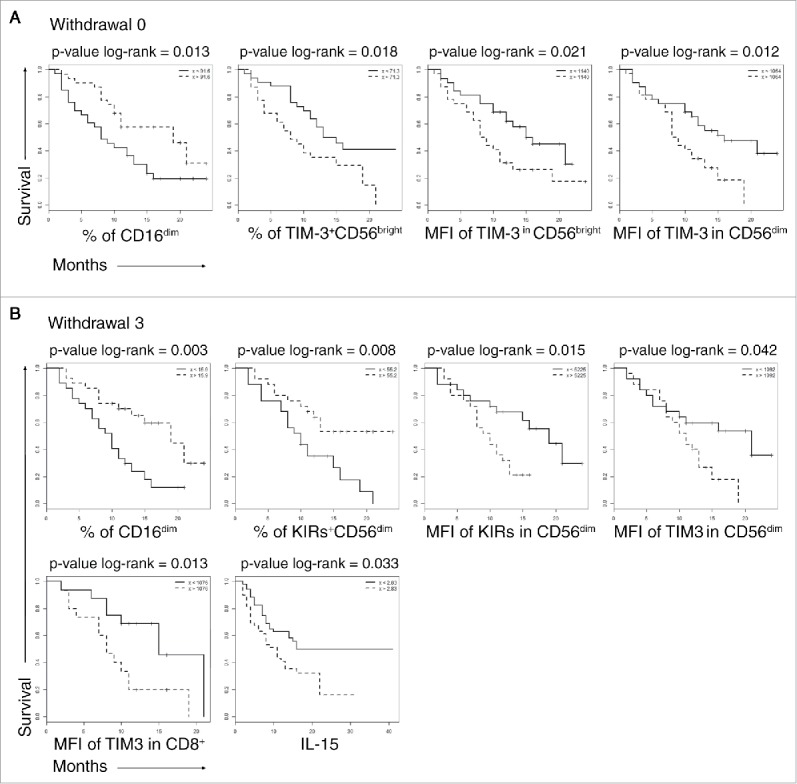
Overall survival of patients stratifying by selected biomarkers. Kaplan–Meier survival curves in long survival dotted lines and short survival solid lines patients. (A), W0, biomarkers measured before the anti-CTLA-4 treatment, (B) W3, biomarkers measured after the third anti CTLA-4 treatment. Variables are reported on the x-axis. The curves were compared with log-rank test the p-values are reported on the top of each panel.
After the third administration of ipilimumab, parameters inversely correlated with survival were high TIM-3 expression on CD8+T cells and on CD56dim NK cells. The increased frequency of KIR+ CD56dim circulating NK cells was associated with adverse prognosis. On the other hand, increased frequency of CD16+ CD56dim NK cells at this time point strongly correlated with good prognosis and long survival (Fig. 5B).
To adjust for potential confounding factors, separate Cox models were fitted for each variable independently (TIM-3 expression, IL-15 concentration, CD56dim frequency). The models were adjusted for age, sex, kind of melanoma = UVEAL and disease stage (RECIST PD)
The analysis at W0 showed that the expression of TIM-3 on CD56bright and CD56dim, and the IL-15 sera concentration were associated with increased risk. All estimated hazard ratios (hr) were significantly different from one. However, those associated with TIM-3 and CD56 were very close to one, indicating a small effect on the risk of death. Instead, the hazard ratio of IL-15 was relatively large (hr = 1.26, 95% CI: 1.10, 1.43, p-value < 0.001). At W3, IL-15 was associated with an increased risk (hr = 1.22, 95% CI: 1.07, 1.401, p-value = 0.002), whereas CD56dim was associated with a reduced risk (hr = 0.92, 95% CI = 0.86, 0.99, p-value = 0.020) (Table S6).
Discussion
In this explorative study, we identified a number of parameters that are associated either positively or negatively with the prognosis of ipilimumab-treated melanoma patients. Here, we will focus on the serum concentration of IL-15, frequency and function of NK cells, and expression of TIM-3, PD-1 and KIRs on NK and T cells.
High IL-15 serum levels, before as well as during the treatment, was associated with poor prognosis in patients with NK/T cell lymphoma.31 IL-15 is usually considered favorably in the context of cancer immunotherapy because it induces expansion and maturation of NK cells, as well as expansion of memory CD8+ T-cells, without amplifying the Tregs cell compartment.30 Clinical trials are ongoing in melanoma patients with infusion of IL-15 over 3 weeks.32 Indeed, our own in vitro data confirmed that IL-15 could activate NK cells.
There are several potential explanations for our findings in ipilimumab-treated melanoma patients. One possibility is that high IL-15 is just a downstream effect of another disease feature associated with poor prognosis, e.g., cancer dissemination or chronic inflammation. However, a more interesting possibility is that high IL-15 levels actually contribute to the poor outcome. The multivariate analysis showed that IL-15 serum concentration was associated with increased expression and frequency of TIM-3 and PD-1 on both NK and T cells, which were factors associated with shorter survival. We also found that exposure to IL-15 for 5 d in vitro indeed led to enhanced expression of TIM-3 and PD-1 on both NK and T cells, and of KIRs selectively in NK cells. Our data confirm and expand previous observations indicating that IL-15 may induce TIM-3 expression on human NK cells.33 It should be noted that TIM-3 recognizes ligands expressed on the melanoma cell surface that can inhibit the NK cells cytotoxic recognition. We demonstrated, in vitro, that antibody blockade of TIM-3 and PD-1 reverts NK cell exhaustion leading to melanoma metastasis killing.34-35
To the best of our knowledge, it has not previously been shown that IL-15 stimulation upregulates PD-1 on NK cells. These data support the possibility that the association between high IL-15 levels and TIM-3/PD-1 expression on NK cells and T cells may represent a causative link. Our results further suggest that IL-15 may prevent the beneficial effect of anti-CTLA-4 blockade by inducing additional immune check points, leading to a dampened immune response and as a consequence more rapid tumor progression.
Previous studies in mice have shown that administration of IL-15 over 3 weeks induces NK cell senescence and imbalance toward expression of inhibitory receptors, whereas administration of IL-15 over 2 d leads to NK cells maturation, proliferation and activation of cytotoxicity.36,37 Based on this and our own results, we suggest that a more effective strategy for IL-15 therapy might be short and frequent administrations, possibly intervening with a second line of checkpoint blockade targeting PD-1, TIM-3 and/or KIR.
The multivariate analysis showed that higher levels of IL-4 and IFN-γ before ipilimumab treatment were associated with long survival. The reason for this association is unknown. As a speculation, we suggest that this may be related to the influence of the microbiome. In previous studies in mice and in patients, antibiotics treatment weakened the antitumor response after anti-CTLA-4 blockade.38 This was correlated with how certain bacteria in the normal gut flora influenced production of cytokines in co-cultures of T-cells and dendritic cells, e.g., IL-10 and IFN-γ. However, IL-4 was not measured in those studies, nor was IL-15. Moreover, these data may suggest a role for the NKT population during anti CTLA-4 treatment.
Several observations from our study implicate that patient NK cells are activated during anti-CTLA-4 treatment and that this may correlate to its therapeutic effect. For example, upon one or two doses of anti-CTLA-4, many patients in the long-term survival group displayed NK cells with increased capacity to de-granulate, whereas patients in the short-term survival group tended to show weaker response as well as increased expression of TIM-3 and KIR. Moreover, once the treatment had been completed, overall survival correlated with high frequency of NK cells in the lymphocyte population, and within the NK compartment, particularly the mature CD56dimCD16+ NK cells. Again, there are several possible explanations. A reduced NK cells frequency could be a consequence of other factors that also lead to poor prognosis, e.g., disseminated disease or immunomodulation. The other general possibility is that this reflects a response to ipilimumab which could be either indirect via sustained secretion of cytokines by T cells or deletion of Treg cells, or via a direct effect on NK cells. Indeed, reports suggest that ipilimumab may activate cytokine production in NK cells through CD16.39,40 We observed that CXCR2+CD56bright NK cells frequency was associated with good prognosis before treatment and after the first ipilimumab administration. CXCR2 recognizes IL-8 that is produced by metastatic melanoma cells acting as a tumor autocrine growth factor increasing the melanoma cells migration in metastatic lesions.41 We found it conceivable to speculate that the CXCR2+ NK cells subsets may be able to migrate in the melanoma metastatic foci.
Regardless of the mechanism of action of anti-CTLA-4, our results in this exploratory study clearly highlight the NK cell population as a source of variables that may offer predictive biomarkers in the context of checkpoint blockade. This should be investigated and possibly validated in larger study. It should be noted that such new biomarkers may be easily detected in the patient's peripheral blood allowing the development of a new simple and affordable diagnostic approach to be used in real time before and during therapy. Our data also suggest that NK cells are worth considering in the diagnosis and therapy of melanoma patients. NK cells are indeed already used in the experimental therapeutic trials of other malignancies, for example, in AML and Multiple Myeloma. Therefore, NK cells could open new avenues for the next generation of immune intervention in melanoma patients, i.e., KIR blockade, and/or NK cell-based adoptive immune therapy.42
Methods
Study design
For the study, 115 stage IV melanoma patients were enrolled in Italy at the Naples National Cancer Institute and 24 patients were recruited at the Oncology clinic, Karolinska University Hospital, Stockholm, Sweden. Ethical Committees associated with Naples Cancer Institutet, Naples and Karolinska University Hospital, Stockholm granted ethical permission for each patient cohort. Written informed consent was obtained from all patients in accordance with the Declaration of Helsinki to the use of human biological samples for research purpose. Each patient received four infusions of ipilimumab with 21 d between infusions. PBL and sera were collected from patients at withdrawal 0 (W0) before the administration of the first dose of ipilimumab; at week 3 (withdrawal, W1) before the second administration; at week 6 (withdrawal, W2), before the third administration; and at week 9 (withdrawal, W3) before the fourth and last administration. We isolated PBMCs from 67 patients to analyze the lymphocyte compartment and 12 were used in functional assays. Based on their survival, we divided patients in two groups: long survival ≥ 12 mo, short survival < 12 mo after treatment. We also collected blood and sera respectively from 37 and 28, sex and age matched, healthy donors as controls at the Pugliese-Ciaccio Hospital and University Magna Graecia of Catanzaro, Catanzaro, Italy. The experiments were performed once per patient.
Isolation of peripheral blood lymphocytes and tumor cell lines
Blood from 70 melanoma patients and 37 healthy donors were isolated by Biocoll separating solution (Biochrom AG, Berlin, Germany) density gradient centrifugation. Cells were counted and put in culture with RPMI-1640 medium (Life Technology, Milan, Italy) supplemented with penicillin (100 IU/mL) and streptomycin (100 μg/mL) and 10% FBS. Cells were frozen the next day. Ten Swedish patients were recruited at the Oncology clinic, Karolinska University Hospital, Stockholm, Sweden.
Blood samples were collected in 10 mL heparinized vacutainer tubes (BD diagnostics, NJ, USA). Peripheral mononuclear cells were isolated by density gradient centrifugation (Ficoll-Paque, GE Healthcare) within 2 h of sample collection and frozen in 90% FCS + 10% DMSO freezing medium.
K562 cell line was obtained from ATCC. Melanoma cell lines (Mel30 and Mel35) were obtained, based on informed consent, from surgical specimens of melanoma metastases from patients admitted at the Fondazione IRCCS Istituto Nazionale dei Tumori, Milan. The cell lines were cultured in RPMI 1640 medium (Life Technology, Milan, Italy) supplemented with penicillin (100 IU/mL) and streptomycin (100μg/mL) and 10% FBS.
Immunofluorescence staining
After thawing, PBL from metastatic melanoma patients and healthy donors were processed for an immunofluorescence staining. Cells were washed, counted and divided in 96-well plates and incubated for 15 min at room temperature with 50µL of inactivated human serum to achieve not specific binding. Then, cells were washed once with phosphate buffered saline (PBS) 1X and were incubated at 4°C for 30 min, with the following antibodies: PE or APC CD56 (clone REA196)/IgG1, FITC CD3 (clone BW264/56)/IgG2a, PeCy7 CD4 (clone VIT4)/IgG2a, APCCy7 CD8 (clone BW135/80)/IgG2a, PE or PeCy7 CD279 (PD-1, clone PD1.3.1.3)/IgG2b, PeCy7 CCR7 (clone REA108)/IgG1, PE CD158a/h (KIR2DL1/S1, clone 11BP6)/IgG1, PE CD158b (KIR2DL2/DL3, clone DX27)/IgG2aκ, PE CD158e (KIR3DL1, clone DX9)/IgG1κ, PE CD57 (clone TB03)/IgM, PE CD69 (clone FN50)/IgG1κ, PE CD314 (NKG2D, clone BAT221)/IgG1, PE CD226 (DNAM-1, clone DX11)/IgG1, PE CD337 (NKp30, clone AF29–4D12)/IgG1, PE CD336 (NKp44, clone 2.29)/IgG1, PE CD335 (NKp46, clone 9E2)/IgG1κ were from Miltenyi Biotech. APCCy7 CD16 (clone 3G8)/IgG1κ, AlexaFluor 647 CD192 (CCR2, clone 6C6)/IgG1γ, APC CXCR2 (IL8RB, clone 48607)/IgG2bκ, were from BD PharMingen. PE TIM-3 (clone F382E2)/IgG1, were from Biolegend, PE NKG2C (clone 134)/IgG1 were from R&D Systems. After that, cells were washed twice with PBS 1X and acquired by FACS CANTO II. 7-AAD Staining Solution (BD Italy) was added before each acquisition. In all experiments, the isotype-matched controls were used to set up the negative values. Data were analyzed using Flow-Jo version 9.3.1 software analysis. The analysis of T and NK cell subsets in peripheral blood by flow cytometry was performed using the gating strategy depicted in Fig. S1.
CD107a mobilization assay after K562 pulsing
For degranulation assays quantifying cell surface CD107a expression, thawed lymphocytes derived from melanoma patients (six long and six short survivors) and healthy donors (six healthy donors), were cultured at 37°C in 5% CO2 at 1:1 effector:target ratio with K562 (ATCC) in the presence of 8µL of PE-conjugated CD107a/IgG1 antibody (BD PharMingen) in U-bottom 96-well plates. After 1 h, Brefeldin A (10µg/mL) (Sigma Aldrich Saint Louis, MO, United States) was added to cultures for an additional 3 h of incubation. At this time, cells were collected, washed with PBS and stained with anti-CD56APC and anti-CD3FITC (Miltenyi Biotec San Diego, California) mAbs for 30 min at 4°C in the dark. The cells were washed two times with PBS 1X and then analyzed by flow cytometry. To detect cell viability, we used 7AAD (BD).
Cytotoxicity assay
Human NK cells were purified from 120 h IL-15 (10 ng/mL) treated PBMCs by negative selection (Miltenyi Biotech). Purified NK cells were incubated for 1 h at room temperature with the following blocking antibodies at 10μg/mL: Purified mouse IgG1 Isotype control (DAKO), Ultra-LEAFTM Purified anti-human TIM-3 (Clone F38–2E2, Biolegend), Ultra-LEAFTM Purified anti-human PD-1 (Clone EH12–2H7, Biolegend) or the combination of TIM-3 and PD-1. Target cells Mel30 or Mel35, previously labeled with Na[51Cr]O4 at 37°C for 1 h, were added at Effector:Target ratio of 6:1 and the co-culture was allowed to proceed for 4 h at 37°C. The 51Cr activity of the supernatant was measured with a gamma-counter. Percentage of specific lysis was calculated as ([cpm experimental release − cpm spontaneous release]/[cpm maximum release − cpm spontaneous release]) × 100. Spontaneous release represents the 51Cr release from target cells alone. Experimental release represents the release from target cells incubated with effector cells and maximum release represents the 51Cr content of re-suspended target cells.
Microarray cytokine assay
For cytokine profile analysis, the serum of patients was analyzed using the biochip analyzer Evidence Investigator (Randox Labs, Antrim, UK) and the “Cytokine Array I” kit (Randox, Antrim, UK), according to the manufacturer's instructions for the simultaneous quantification of interleukin-2 (IL-2), IL-4, IL-6, IL-8, IL-10, IL-1α, IL-1β, vascular endothelial growth factor (VEGF), interferon-γ (IFNγ), monocyte chemotactic protein-1 (MCP1), tumor necrosis factor-α (TNF-α), Epidermal Growth Factor (EGF). This assay was performed following the manufacturer's recommended procedure.
Samples were thawed before the analysis. Briefly, the principle of this multianalyte testing relies on a sandwich ELISA, in which the analytes of interest are captured by specific antibodies bound to discrete regions of the surface chemistry of the biochip; horseradish peroxidase (HRP) labeled secondary antibodies, which specifically recognize the analytes, trigger a luminol-based electrochemiluminescent signal emission, registered by a charge-coupled device (CCD) camera and quantified by a software.
IL-15 and IL-21 were evaluated by ELISA kit following manufacturer's instructions (R&D Systems and BioVendor).
Statistical analysis
Functional data obtained from multiple experiments were calculated as mean ± S.D. and analyzed for statistical significance using the two-tailed Student's t-test for independent groups, or analysis of variance followed by Bonferroni correction for multiple comparisons. For immunoprofiling data with non-parametric distributions, statistical univariate analyses were performed with Mann–Whitney test. p-values ≤ 0.05 were considered statistically significant. Statistical computations were done using the Graph Pad Prism 5.0 software.
To estimate survival curves, we used the standard product-limit estimator (Kaplan–Meier). We created Kaplan–Meier curves to compare patients below/above the median of each of the variables validated in the univariate and multivariate analysis. The log-rank test was used to compare the KM curves and calculate the p-value. We used the R statistical software, version 3.3.1 (www.r-project.org), and in particular the “survival” package. To compute survival curves and compare them, we use the Kaplan–Meier estimator. To compute hazard ratios, and adjust for potential confundings we used the Cox model, testing for proportionality using Schoenfeld's test.
Multivariate analysis
SIMCA, version 14 (MKS Data Analytics Solutions, Umeå Sweden) was applied for multivariate analysis.43 In total, 165 variables were applied in the analysis. The biographical, immune and clinical variables analyzed are reported in Table S1–5. The majority of variables originated from flow cytometry. Fourteen different cytokines, as well as variables characterizing the patients (such as age, gender, site of melanoma and counts of various types of blood cells) were also included in the analysis. Orthogonal Projections to Latent Structures and Discriminant Analysis (OPLS-DA) was used to distinguish groups and identify biomarkers for healthy donors, long survivors and short survivors.44 OPLS-DA has recently been employed as a tool for biomarker discovery in endometriosis research.45
OPLS is a projection method developed from principal component analysis where systematic variation in the data are summarized into scores (T). The scores are latent variables, where t[1] represents the systematic variation in the N-dimensional variable space related to an Y-variable outcome.44 Discriminant analysis, OPLS-DA, models are created with the aim of finding maximal separation between pre-defined groups. Our groups were long survivors, short survivors and healthy donors. All variation related to separation between groups is present in the predictive component(s) t[1]. More predictive components can be calculated if there are several groups in the data, which also are distinct enough to separate. Variation unrelated to separation between groups is visualized as “orthogonal components” to[1], to[2] and so on. The quality of OPLS-DA models are assessed by internal cross-validation, and presented as percentage of data that can be explained and predicted. A good biological model is defined as having > 40% predictive capacity.43
The models were constructed with the following workflow. First, biologically irrelevant variables were excluded, for example subsets with no signal, or side effects at time point W0 where the side effects had not yet occurred. Second, the models were screened for outliers. A patient was only considered an outlier if it deviated significantly both from its own group and the global OPLS-DA model. At W0, the model was built on 127 variables, and no consistent outliers were observed. The model could explain 89.7% of the difference between long-term and short-term survivors and predict with 63.8% accuracy the survival of potential new patients screened for the key biomarkers. At W3, the model was built on 129 variables. One consistent patient outlier was observed, displaying very high values for most of the MFIs. After excluding this outlier, the model could explain 83.8% of the difference between long-term and short-term survivors and predict 63.5% accuracy the survival of potential new patients screened for the key biomarkers.
To estimate survival curves, we used the standard product-limit estimator (Kaplan–Meier). We created Kaplan–Meier curves to compare patient below/above the median of each of the variables validated in the univariate and multivariate analysis. The log rank test was used to compare the KM curves and calculate the p-value.
Supplementary Material
Disclosure of potential conflicts of interest
Paolo A. Ascierto has/had a consultant/advisory role for Bristol Myers Squibb, Roche-Genentech, Merck Sharp & Dohme, Ventana, Novartis, Amgen, and Array. He also received research funds from Bristol Myers Squibb, Roche-Genentech, Ventana, and Array.
Acknowledgments
We thank Jeffrey Miller, Minnesota University (Minneapolis, MN), and Julie Djeu Moffet Cancer Center, (Tampa, FL) for the suggestions and discussion.
Funding
E.C. was supported by Associazione Italiana Ricerca Cancro AIRC-IG 15521, Fondazione Melanoma, Naples and Wenner-Gren Stiftelserna, Sweden. Ministry of Health grant “Progetto ricerca finalizzata 2011–2012” grant CO-2011–02348049 co-funded by Bristol Myers Squibb, Fondazione Melanoma Onlus, Naples. R.K. was supported by The Swedish Cancer Society (2013/379), The Cancer Society in Stockholm and The King Gustaf V's Jubilee Foundation (144102), The Swedish Medical Research Council (521–2013–4100), Stockholm City Council Project Grant 20140036 (ALF Medicin 2015) and the “Knut and Alice Wallenberg Foundations.” R.T. was supported by FIRC Fellowship “Luciana Selce.” K.K was supported by The Swedish Cancer Society (Cancerfonden). CAN 2015/1365. S.J. and E.S. were supported by the Swedish Research Council (Vetenskapsrådet) Medicine. R.S. is a recipient of a fellowship from the Foundation Blanceflor Boncompagni Ludovisi, née Bildt.
Author contributions
R.T. performed the experiments, analyses and manuscript writing. C.M.C performed experiments and analysis, E.S. constructed and interpreted discriminant analysis performed data analysis and mathematical models, C.G. performed experiments, E.P, R.S., V.C. performed experiments, M.C., G.M., D.M., E.S., A.M.G. sampling and clinical data collection, Y.P.C sampling collection manuscript writing, S.J. performed data analysis and mathematical models, E.G. data analysis, A.A. and F.C. were involved in the analyses, and manuscript writing, G.C, R.K., K.K., P.A.A. were involved in data analyses, interpretation and manuscript writing. P.A.A. was involved in sample collection, clinical data collection, data analyses, interpretation, manuscript writing. E.C. conceived the study, analyzed the data, wrote the manuscript and designed the experiments. All the authors critically read and approved the manuscript.
Ethical statements
The study has been approved in accordance with the Helsinki Declaration of 1975 by the Ethics Committee of Human Experimentation of the: (a) National Cancer Institute of Naples “G Pascale,” Naples, nr.49 29.01.2007, (b) the Ethics Committee of Human Experimentation of the University Magna Graecia of Catanzaro, Catanzaro, nr. 2014.33 16.04.2014, (c) the Ethics Committee of the Karolinska University Hospital, Stockholm, nr. 2011/143–32/1.
References
- 1.Krummel MF, Allison JP. CD28 and CTLA-4 have opposing effects on the response of T cells to stimulation. J Exp Med 1995; 182:459-65; PMID:7543139; http://dx.doi.org/ 10.1084/jem.182.2.459 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Leach DR, Krummel MF, Allison JP. Enhancement of antitumor immunity by CTLA-4 blockade. Science 1996; 271:1734-36; PMID:8596936; http://dx.doi.org/ 10.1126/science.271.5256.1734 [DOI] [PubMed] [Google Scholar]
- 3.Freeman GJ, Long AJ, Iwai Y, Bourque K, Chernova T, Nishimura H, Fitz LJ, Malenkovich N, Okazaki T, Byrne MC et al.. Engagement of the PD-1 immunoinhibitory receptor by a novel B7 family member leads to negative regulation of lymphocyte activation. J Exp Med 2000; 192:1027-34; PMID:11015443; http://dx.doi.org/ 10.1084/jem.192.7.1027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Postow MA, Callahan MK, Wolchok JD. Immune checkpoint blockade in cancer therapy. J Clin Oncol 2015; 33:1974-82; PMID:25605845; http://dx.doi.org/ 10.1200/JCO.2014.59.4358 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Postow MA, Chesney J, Pavlick AC, Robert C, Grossmann K, McDermott D, Linette GP, Meyer N, Giguere JK, Agarwala SS et al.. Nivolumab and ipilimumab versus ipilimumab in untreated melanoma. N Engl J Med 2015; 372:2006-17; PMID:25891304; http://dx.doi.org/ 10.1056/NEJMoa1414428 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hodi FS, O'Day SJ, McDermott DF, Weber RW, Sosman JA, Haanen JB, Gonzalez R, Robert C, Schadendorf D, Hassel JC et al.. Improved survival with ipilimumab in patients with metastatic melanoma. N Engl J Med 2010; 363:711-23; PMID:20525992; http://dx.doi.org/ 10.1056/NEJMoa1003466 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Weide B, Di Giacomo AM, Fonsatti E, Zitvogel L. Immunologic correlates in the course of treatment with immunomodulating antibodies. Semin Oncol 2015; 42:448-58; PMID:25965363; http://dx.doi.org/ 10.1053/j.seminoncol.2015.02.016 [DOI] [PubMed] [Google Scholar]
- 8.Ascierto PA, Kalos M, Schaer DA, Callahan MK, Wolchok JD. Biomarkers for immunostimulatory monoclonal antibodies in combination strategies for melanoma and other tumor types. Clin Cancer Res 2013; 19:1009-20; PMID:23460532; http://dx.doi.org/ 10.1158/1078-0432.CCR-12-2982 [DOI] [PubMed] [Google Scholar]
- 9.Kitano S, Tsuji T, Liu C, Hirschhorn-Cymerman D, Kyi C, Mu Z, Allison JP, Gnjatic S, Yuan JD, Wolchok JD. Enhancement of tumor-reactive cytotoxic CD4+ T cell responses after ipilimumab treatment in four advanced melanoma patients. Cancer Immunol Res 2013; 1:235-44; PMID:24396833; http://dx.doi.org/ 10.1158/2326-6066.CIR-13-0068 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wang W, Yu D, Sarnaik AA, Yu B, Hall M, Morelli D, Zhang Y, Zhao X, Weber JS. Biomarkers on melanoma patient T cells associated with ipilimumab treatment. J Transl Med 2012; 10:146; PMID:22788688; http://dx.doi.org/ 10.1186/1479-5876-10-146 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Simeone E, Gentilcore G, Giannarelli D, Grimaldi AM, C Caracò, Curvietto M, Esposito A, Paone M, Palla M, Cavalcanti E et al.. Immunological and biological changes during ipilimumab treatment and their potential correlation with clinical response and survival in patients with advanced melanoma. Cancer Immunol Immunother 2014; 63:675-83; PMID:24695951; http://dx.doi.org/ 10.1007/s00262-014-1545-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Weide B, Martens A, Hassel JC, Berking C, Postow MA, Bisschop K Simeone E, Mangana J, Schilling B, Di Giacomo AM et al.. Baseline biomarkers for outcome of melanoma patients treated with pembrolizumab. Clin Cancer Res 2016; clincanres 0127.2016; PMID:27185375; http://dx.doi.org/26787752 10.1158/1078-0432.CCR-16-0127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Martens A, Wistuba-Hamprecht K, Geukes Foppen M, Yuan J, Postow MA, Wong P, Romano E, Khammari A, Dreno B, Capone M et al.. Baseline Peripheral Blood Biomarkers Associated with Clinical Outcome of Advanced Melanoma Patients Treated with Ipilimumab. Clin Cancer Res 2016; 22:2908-18; PMID:26787752; http://dx.doi.org/ 10.1158/1078-0432.CCR-15-2412 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Martens A, Wistuba-Hamprecht K, Yuan J, Postow MA, Wong P, Capone M, Madonna G, Khammari A, Schilling B, Sucker A et al.. Increases in absolute lymphocytes and circulating CD4+ and CD8+ T cells are associated with positive clinical outcome of melanoma patients treated with ipilimumab. Clin Cancer Res 2016; 22(19):4848-4858; pii: clincanres.0249.2016; PMID:27169993; http://dx.doi.org/22130166 10.1158/1078-0432.CCR-16-0249 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kvistborg P, Philips D, Kelderman S, Hageman L, Ottensmeier C, Joseph-Pietras D, Welters MJ, van der Burg S, Kapiteijn E, Michielin O et al.. Anti-CTLA-4 therapy broadens the melanoma-reactive CD8+ T cell response. Sci Transl Med 2014; 6:254ra128; http://dx.doi.org/ 10.1126/scitranslmed.3008918 [DOI] [PubMed] [Google Scholar]
- 16.Weber JS, Hamid O, Chasalow SD, Wu DY, Parker SM, Galbraith S, Gnjatic S, Berman D. Ipilimumab increases activated T cells and enhances humoral immunity in patients with advanced melanoma. J Immunother 2012; 35:89-97; PMID:22130166; http://dx.doi.org/ 10.1097/CJI.0b013e31823aa41c [DOI] [PubMed] [Google Scholar]
- 17.Romano E, Kusio-Kobialka M, Foukas PG, Baumgaertner P, Meyer C, Ballabeni P, Michielin O, Weide B, Romero P, Speiser DE. Ipilimumab-dependent cell-mediated cytotoxicity of regulatory T cells ex vivo by nonclassical monocytes in melanoma patients. Proc Natl Acad Sci U S A 2015; 112:6140-45; PMID:25918390; http://dx.doi.org/ 10.1073/pnas.1417320112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wing K, Onishi Y, Prieto-Martin P, Yamaguchi T, Miyara M, Fehervari Z, Nomura T, Sakaguchi S. CTLA-4 control over Foxp3+ regulatory T cell function. Science 2008; 322:271-5; PMID:18845758; http://dx.doi.org/ 10.1126/science.1160062 [DOI] [PubMed] [Google Scholar]
- 19.Suarez N, Alfaro C, Dubrot J, Palazon A, Bolaños E, Erro L, Hervas-Stubbs S, Martinez-Forero I, Morales-Kastresana A, Martin-Algarra S et al.. Synergistic effects of CTLA-4 blockade with tremelimumab and elimination of regulatory T lymphocytes in vitro and in vivo. Int J Cancer 2011; 129:374-86; PMID:20853321; http://dx.doi.org/ 10.1002/ijc.25681 [DOI] [PubMed] [Google Scholar]
- 20.Simpson TR, Li F, Montalvo-Ortiz W, Sepulveda MA, Bergerhoff K, Arce F, Roddie C, Henry JY, Yagita H, Wolchok JD et al.. Fc-dependent depletion of tumor-infiltrating regulatory T cells co-defines the efficacy of anti-CTLA-4 therapy against melanoma. J Exp Med 2013; 210:1695-710; PMID:23897981; http://dx.doi.org/ 10.1084/jem.20130579 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kiessling R, Klein E, Wigzell H. “Natural” killer cells in the mouse. I. Cytotoxic cells with specificity for mouse Moloney leukemia cells. Specificity and distribution according to genotype. Eur J Immunol 1975; 5:112-7; PMID:1234049; http://dx.doi.org/ 10.1002/eji.1830050208 [DOI] [PubMed] [Google Scholar]
- 22.Kiessling R, Klein E, Pross H, Wigzell H. “Natural” killer cells in the mouse. II. Cytotoxic cells with specificity for mouse Moloney leukemia cells. Characteristics of the killer cell. Eur J Immunol 1975; 5:117-21; PMID:1086218; http://dx.doi.org/ 10.1002/eji.1830050209 [DOI] [PubMed] [Google Scholar]
- 23.Kärre K, Ljunggren HG, Piontek G, Kiessling R. Selective rejection of H-2-deficient lymphoma variants suggests alternative immune defence strategy. Nature 1986; 319:675-8; PMID:3951539; http://dx.doi.org/19349689 10.1038/319675a0 [DOI] [PubMed] [Google Scholar]
- 24.Lakshmikanth T, Burke S, Ali TH, Kimpfler S, Ursini F, Ruggeri L, Capanni M, Umansky V, Paschen A, Sucker A et al.. NCRs and DNAM-1 mediate NK cell recognition and lysis of human and mouse melanoma cell lines in vitro and in vivo. J Clin Invest 2009; 119:1251-63; PMID:19349689; http://dx.doi.org/ 10.1172/JCI36022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ferrari de Andrade L, Ngiow SF, Stannard K, Rusakiewicz S, Kalimutho M, Khanna KK, Tey SK, Takeda K, Zitvogel L, Martinet L et al.. Natural killer cells are essential for the ability of BRAF inhibitors to control BRAFV600E-mutant metastatic melanoma. Cancer Res 2014; 74:7298-308; PMID:25351955; http://dx.doi.org/ 10.1158/0008-5472.CAN-14-1339 [DOI] [PubMed] [Google Scholar]
- 26.Mamessier E, Sylvain A, Thibult ML, Houvenaeghel G, Jacquemier J, Castellano R, Gonçalves A, P André, F Romagné, Thibault G et al.. Human breast cancer cells enhance self tolerance by promoting evasion from NK cell antitumor immunity. J Clin Invest 2011; 121:3609-22; PMID:21841316; http://dx.doi.org/ 10.1172/JCI45816 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Delahaye NF, Rusakiewicz S, Martins I, Ménard C, Roux S, Lyonnet L, Paul P, Sarabi M, Chaput N, Semeraro M et al.. Alternatively spliced NKp30 isoforms affect the prognosis of gastrointestinal stromal tumors. Nat Med 2011; 17:700-7; PMID:21552268; http://dx.doi.org/ 10.1038/nm.2366 [DOI] [PubMed] [Google Scholar]
- 28.Takeuchi H, Maehara Y, Tokunaga E, Koga T, Kakeji Y, Sugimachi K. Prognostic significance of natural killer cell activity in patients with gastric carcinoma: a multivariate analysis. Am J Gastroenterol 2001; 96:574-8; PMID:11232710; http://dx.doi.org/ 10.1111/j.1572-0241.2001.03535.x [DOI] [PubMed] [Google Scholar]
- 29.Ali TH, Pisanti S, Ciaglia E, Mortarini R, Anichini A, Garofalo C, Tallerico R, Santinami M, Gulletta E, Ietto C et al.. Enrichment of CD56(dim)KIR + CD57 + highly cytotoxic NK cells in tumour-infiltrated lymph nodes of melanoma patients. Nat Commun 2014; 5:5639; PMID:25472612; http://dx.doi.org/ 10.1038/ncomms6639 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Conlon KC, Lugli E, Welles HC, Rosenberg SA, Fojo AT, Morris JC, Fleisher TA, Dubois SP, Perera LP, Stewart DM et al.. Redistribution, hyperproliferation, activation of natural killer cells and CD8 T cells, and cytokine production during first-in-human clinical trial of recombinant human interleukin-15 in patients with cancer. J Clin Oncol 2015; 33:74-82; PMID:25403209; http://dx.doi.org/ 10.1200/JCO.2014.57.3329 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wang H, Zhu JY, Liu CC, Zhu MY, Wang JH, Geng QR, Lu Y. Increased serum levels of interleukin 15 correlate with negative prognostic factors in extra nodal NK / T cell lymphoma. Med Oncol 2015; 32:370; PMID:25428383; http://dx.doi.org/ 10.1007/s12032-014-0370-4 [DOI] [PubMed] [Google Scholar]
- 32.Waldmann TA. Interleukin-15 in the treatment of cancer. Expert Rev Clin Immunol 2014; 10:1689-701; PMID:25359408; http://dx.doi.org/ 10.1586/1744666X.2014.973856 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ndhlovu LC, Lopez-Vergès S, Barbour JD, Jones RB, Jha AR, Long BR, Schoeffler EC, Fujita T, Nixon DF, Lanier LL. Tim-3 marks human natural killer cell maturation and suppresses cell-mediated cytotoxicity. Blood 2012; 119:3734-43; PMID:22383801; http://dx.doi.org/ 10.1182/blood-2011-11-392951 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.da Silva IP, Gallois A, Jimenez-Baranda S, Khan S, Anderson AC, Kuchroo VK, Osman I, Bhardwaj N. Reversal of NK-cell exhaustion in advanced melanoma by Tim-3 blockade. Cancer Immunol Res 2014; 2:410-22; PMID:24795354; http://dx.doi.org/ 10.1158/2326-6066.CIR-13-0171 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Gallois A, Silva I, Osman I, Bhardwaj N. Reversal of natural killer cell exhaustion by TIM-3 blockade. Oncoimmunology 2015; 3:e946365; PMID:25964857; http://dx.doi.org/ 10.4161/21624011.2014.946365 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Elpek KG, Rubinstein MP, Bellemare-Pelletier A, Goldrath AW, Turley SJ. Mature natural killer cells with phenotypic and functional alterations accumulate upon sustained stimulation with IL-15/IL-15Ralpha complexes. Proc Natl Acad Sci U S A 2010; 107:21647-52; PMID:21098276; http://dx.doi.org/ 10.1073/pnas.1012128107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Gleason MK, Lenvik TR, McCullar V, Felices M, O'Brien MS, Cooley SA, Verneris MR, Cichocki F, Holman CJ, Panoskaltsis-Mortari A et al.. Tim-3 is an inducible human natural killer cell receptor that enhances interferon gamma production in response to galectin-9. Blood 2012; 119:3064-72; PMID:22323453; http://dx.doi.org/ 10.1182/blood-2011-06-360321 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Vétizou M, Pitt JM, Daillère R, Lepage P, Waldschmitt N, Flament C, Rusakiewicz S, Routy B, Roberti MP, Duong CP et al.. Anticancer immunotherapy by CTLA-4 blockade relies on the gut microbiota. Science 2015; 350:1079-84; PMID:2654161; http://dx.doi.org/17100888 10.1126/science.aad1329 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ghiringhelli F, Ménard C, Martin F, Zitvogel L. The role of regulatory T cells in the control of natural killer cells: relevance during tumor progression. Immunol Rev 2006; 214:229-38; PMID:17100888; http://dx.doi.org/ 10.1111/j.1600-065X.2006.00445.x [DOI] [PubMed] [Google Scholar]
- 40.Laurent S, Queirolo P, Boero S, Salvi S, Piccioli P, Boccardo S, Minghelli S, Morabito A, Fontana V, Pietra G et al.. The engagement of CTLA-4 on primary melanoma cell lines induces antibody-dependent cellular cytotoxicity and TNF-α production. J Transl Med 2013; 11:108; PMID:23634660; http://dx.doi.org/ 10.1186/1479-5876-11-108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Uen WC, Hsieh CH, Tseng TT, Jiang SS, Tseng JC, Lee SC. Anchorage independency promoted tumor malignancy of melanoma cells under reattachment through elevated interleukin-8 and CXC chemokine receptor 1 expression. Melanoma Res 2015; 25:35-46; PMID:25426644; http://dx.doi.org/ 10.1097/CMR.0000000000000134 [DOI] [PubMed] [Google Scholar]
- 42.Miller JS. Therapeutic applications: natural killer cells in clinic. Hematology Am Soc Hematol Educ Program 2013; 2013:247-53; PMID:24319187; http://dx.doi.org/ 10.1182/asheducation-2013.1.247 [DOI] [PubMed] [Google Scholar]
- 43.Eriksson L, Johansson E. Multi- and Megavariate Data Analysis: Principles and application. Umetrics A B 2001; Umetrics Academics Publishing 533 pp. 2001. ISBN 919737301X, 9789197373012. [Google Scholar]
- 44.Trygg J, Wold S. Orthogonal Projections to Latent Structures (O-PLS). Chemometrics 2002; 16:119-28; http://dx.doi.org/ 10.1002/cem.695 [DOI] [Google Scholar]
- 45.Dutta M, Subramani E, Taunk K, Gajbhiye A, Seal S, Pendharkar N, Dhali S, Ray CD, Lodh I, Chakravarty B et al.. Investigation of serum proteome alterations in human endometriosis. J Proteomics 2015; 114:182-96; PMID:25449831; http://dx.doi.org/ 10.1016/j.jprot.2014.10.021 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


