Abstract
Most paramyxoviruses circumvent the IFN response by blocking IFN signaling and limiting the production of IFN by virus-infected cells. Here we report that the highly conserved cysteine-rich C-terminal domain of the V proteins of a wide variety of paramyxoviruses binds melanoma differentiation-associated gene 5 (mda-5) product. mda-5 is an IFN-inducible host cell DExD/H box helicase that contains a caspase recruitment domain at its N terminus. Overexpression of mda-5 stimulated the basal activity of the IFN-β promoter in reporter gene assays and significantly enhanced the activation of the IFN-β promoter by intracellular dsRNA. Both these activities were repressed by coexpression of the V proteins of simian virus 5, human parainfluenza virus 2, mumps virus, Sendai virus, and Hendra virus. Similar results to the reporter assays were obtained by measuring IFN production. Inhibition of mda-5 by RNA interference or by dominant interfering forms of mda-5 significantly inhibited the activation of the IFN-β promoter by dsRNA. It thus appears that mda-5 plays a central role in an intracellular signal transduction pathway that can lead to the activation of the IFN-β promoter, and that the V proteins of paramyxoviruses interact with mda-5 to block its activity.
Keywords: interferon, NF-κB, innate immunity
The Paramyxoviridae family is divided into two subfamilies, the Paramyxovirinae and Pneumovirinae (reviewed in ref. 1). Members of the Paramyxovirinae subfamily include viruses such as measles, mumps, parainfluenza viruses (PIV) of humans, Newcastle Disease virus of birds, Sendai virus (SeV) of rodents, and simian virus 5 (SV5), which has been isolated from monkeys, dogs, pigs, and humans. Paramyxoviruses also have zoonotic potential, as has been observed with the newly emergent Hendra (HeV) and Nipah viruses, which naturally infect fruit bats but can cause serious, often fatal infections when transmitted to farm and domestic animals and to humans (reviewed in ref. 2). Like all viruses, upon infection of cells, paramyxoviruses are subjected to a variety of intracellular antiviral responses, including the IFN response (reviewed in refs. 3–5). Over the last few years, it has become clear that protein products of the P/V/C gene of viruses within the Paramyxovirinae subfamily (for review of the molecular biology of paramyxoviruses, see ref. 1) specifically reduce the effectiveness of the IFN response. For example, the V protein of SV5 targets signal transducer and activator of transcription 1 (STAT1) for degradation, thereby blocking both IFN-α/β and IFN-γ signaling within infected cells (6), whereas the C proteins of SeV block IFN signaling by interfering with STAT phosphorylation or stability (reviewed in refs. 7–9). As well as blocking IFN signaling, these viruses also specifically limit the production of IFN by virus-infected cells (10–12). The block on IFN-β production is at the level of transcription, because very little IFN-β mRNA is induced in cells infected with SV5. In contrast, large amounts of IFN-β mRNA (and thus IFN-β) are produced by cells infected with a recombinant of SV5 (SV5VΔC) that produces a truncated V protein lacking the cysteine-rich C terminus (which is dispensable for virus replication), suggesting that the V protein is responsible for the block on IFN production. This conclusion is supported by the observation that in gene reporter assays, the V proteins of SV5, PIV2, and SeV inhibit the activation of the IFN-β promoter in response to intracellular dsRNA (11).
Initial transcription from the IFN-β promoter requires the activation of a number of cellular transcription factors, including IFN regulatory factor (IRF)-3 and NF-κB, leading to the formation of an enhanceosome complex that associates with the basal transcriptional machinery to recruit RNA polymerase II to the IFN-β promoter (reviewed in refs. 3 and 13). The molecular details of how the V proteins of paramyxoviruses block IFN production are not known, but the block affects the signal transduction pathway that activates both NF-κB and IRF-3 in response to dsRNA. Thus, these transcription factors are not activated in cells infected with wild-type SV5 but are activated in cells infected with SV5VΔC. Furthermore, ectopic expression of SV5 V inhibits the activation of IRF-3 and NF-κB by both dsRNA and infection with SV5VΔC (10, 11). Unlike the targeted degradation of signal transducer and activator of transcription 1 (STAT1), which requires both the N- and C-terminal domains of V (14, 15), only the cysteine-rich C-terminal domain of V is required to inhibit dsRNA activation of the IFN-β promoter (11).
Our current understanding of signaling in response to dsRNA is incomplete. Extracellular dsRNA is thought to activate transcription through binding TLR3, which recruits an adaptor called TRIF, leading in turn to the activation of a kinase complex (virally activated kinase, or VAK) that contains TRAF-family-member-associated NF-κB activator 1 (TBK1) and inhibitor of NF-κB kinase ε (IKKε) (16, 17), and which leads to the activation of IRF-3. However, recent observations suggest that intracellular dsRNA, such as that generated during virus replication, does not require TLR3 (18, 19) to activate VAK (17). Indeed, in our hands, the SV5 V protein does not inhibit extracellular dsRNA or TLR3-initiated signals (unpublished results), suggesting that its target represents a component of an as-yet-uncharacterized intracellular dsRNA signaling pathway. Thus, by further defining how SV5 V inhibits IFN production, greater insights into these signaling pathways may be gained. Here we report that the C-terminal domain of the V protein of SV5, and of other paramyxoviruses, binds specifically to the melanoma differentiation-associated gene 5 (mda-5) product. mda-5 is a host cell IFN-inducible protein, which is an RNA helicase with a caspase recruitment domain (CARD) at its N terminus. Exogenous expression of mda-5 activates the IFN-β promoter (a property of many signaling intermediates involved in activation of the IFN-β promoter) and enhances its activation by dsRNA; both these activities are blocked by coexpression of the V proteins of a diverse group of paramyxoviruses.
Materials and Methods
Cells, Viruses, and IFN. SV5 strains CPI- and W3A were propagated, and Vero, 293, HeLa, 2fTGH, and 2fTGH/SV5-V(CPI-) cells were maintained as described (14). Transfections were carried out by using Lipofectamine (Invitrogen), Fugene (Roche, Diagnostics), or Polyfect (Qiagen, Valencia, CA) by using standard procedures. For induction by synthetic dsRNA, poly(I)-poly(C) (Amersham Pharmacia Biosciences) was transfected into cells by using Lipofectamine (Invitrogen) under conditions specified by the manufacturer. Induction by IFN was as described (6). The amount of IFN secreted by cells was estimated by using a biological assay in Vero cells. Cells were pretreated with dilutions of culture media containing IFN for 24 h before infection with SV5VΔC [which is sensitive to IFN (10)] and the development of a cytopathic effect (cpe) monitored 36 h postinfection. The end point (1 unit of IFN) was the dilution of media, which reduced the cpe by ≈50%.
Plasmids. The following plasmids have been described (see refs. 6 and 11): the mammalian expression vector pEFplink2 and the virus V-expressing plasmids, pEF.SV5-V and V deletions, pEF.Se-V, pEF.hPIV2-V, and the SV5 P expression plasmid, pEF.SV5-P; the reporter plasmids for IFN-α/β [p(9–27ISRE)4tkΔ(-39)lucter] and IFN-γ [p(GAS)2tkΔ(-39)lucter]; the reporter plasmid for the IFN-β promoter [pIFΔ(-116)lucter]; the reporter plasmid for NF-κB [p[PRD2]5tkΔ(-39)lucter]; the reporter plasmid for lexA fusions and the lexA.IRF-3 fusion plasmid, pEF.mlexA.IRF-3; the constitutive β-galactosidase reporter plasmid, pJATlacZ. pEF.HeV-V, pEF.mumps-V, and pEF.mumps-V(c) containing the cDNA of the respective virus V proteins is described elsewhere (N.S., S.G., and R.E.R., unpublished work). The yeast two-hybrid GAL4 activation domain–SV5 V fusion plasmid, pGADT7.SV5V, was constructed by inserting the full-length SV5 V cDNA into pGADT7 (Clontech).
The full-length cDNA for mda-5 was prepared from total RNA from 293 cells treated for 16 h with 1,000 units/ml of IFN-α, by sequential reverse transcription and PCR reactions by using SuperScript III (Invitrogen) and AccuPrime Pfx polymerase (Invitrogen), respectively, with gene-specific primers under the conditions specified by the manufacturer. The integrity of the cDNA sequence was confirmed by sequencing. The full-length cDNA was cloned between the NcoI and BamHI sites of pEFplink2, pEFTag and pGBKT7 to create pEF.mda-5, pEF.mda-5.c-myc, and pGBKT7.mda5, respectively. Mammalian expression vectors for the CARD-only domain of mda-5 (amino acids 1–196; pEF.mda-5CARD) and mda-5 lacking the CARD domain (amino acids 197–1025; pEF.mda-5ΔN) were generated from pEF.mda-5 by standard methods.
RNA Interference (RNAi). Short RNA molecules for RNAi were generated by in vitro transcription and annealed to produce dsRNA, as described (20). For mda-5, the T7 promoter oligonucleotide (5′-TAATACGACTCACTATAG-3′) was annealed to 5′-AAGACGTCTCTAACTAGAAGCTATAGTGAGTCGTATTA-3′ and 5′-AAGCTTCTAGTTAGAGACGTCTATAGTGAGTCGTATTA-3′, respectively, and the 21 base transcripts annealed to generate a duplex with two base 3′ overhangs (spanning mda-5 nucleotides 382–403 with respect to the initiator AUG). Control oligonucleotides incorporated base changes at the underlined positions. Equal amounts of test and control RNAi duplexes were cotransfected with reporter genes as described (20) and transfections left for 96 h before assay. pRetroSuper-derived RNAi to mda-5 was constructed by inserting the annealed oligonucleotide pair 5′-GATCCCCGTGCATGGAGGAGGAACTGTTCAAGAGACAGTTCCTCCTCCATGCACTTTTTGGAAA-3′ and 5′-AGCTTTTCCAAAAAGTGCATGGAGGAGGAACTGTCTCTTGAACAGTTCCTCCTC CATGCACGGG-3′ between the BglII and HindIII sites of pRetroSuper, with pRetroSuper-derived RNAi to CD2 being used as a negative control (21).
Immunoprecipitation and Immunoblot Analysis. SV5-infected or -transfected cells were metabolically labeled for 1 h with l-[35S]methionine and subjected to immunoprecipitation with a pool of mAbs to the NP, P, V, M, and HN proteins of SV5, a polyclonal antiserum to the P/V proteins of SV5, or the 9E10 mAb to the myc tag, as described (14). Labeled polypeptides, after separation by SDS/PAGE, were visualized by PhosphorImager analysis. Myc-tagged mda-5 was detected by immunoblot analysis by using the myc tag antibody (9E10), also as described (14).
Results
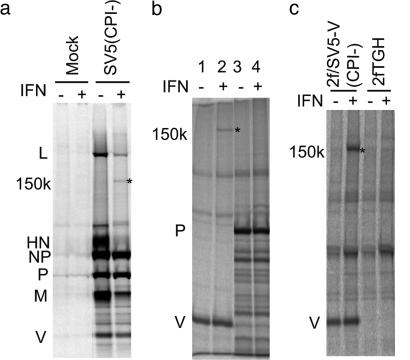
The V Protein of SV5 Binds mda-5. We have previously observed that the host cell protein DDB1 [which has an essential role in the targeted degradation of signal transducer and activator of transcription 1 (STAT1) by the V protein of SV5 (14, 22, 23)] and a less prominent unidentified protein of 150 kDa interact with the V protein of SV5 (14, 24). While examining the replication of an isolate of SV5 termed CPI- that fails to bind to DDB1 and to block IFN signaling (25), but which remains a poor inducer of IFN (11), we noted a substantial increase (compared with wild-type SV5 isolates) in the levels of the 150-kDa protein coimmunoprecipitated with virus proteins from cells treated with exogenous IFN (Fig. 1a). We confirmed that the 150-kDa protein specifically associates with the V protein and is IFN-inducible by transiently transfecting Vero cells with plasmids that expressed either the V or P proteins of CPI- followed by immunoprecipitation of the P, V, and associated host cell proteins (Fig. 1b). Because we had already isolated 2fTGH cells that constitutively express the V protein of CPI- [2f/SV5-V(CPI-) cells (14)], we examined whether increased amounts of the 150-kDa protein could also be coprecipitated with V from these cells by treating them with IFN. Fig. 1c shows this, and we therefore used 2f/SV5-V(CPI-) cells treated with IFN to purify sufficient material to enable identification of the 150-kDa protein as mda-5 by an in-house MALDI–TOF MS service, with a probability-based Mowse score of 260 (the next-nearest match had a score of 73, with only scores over 75 being considered significant). mda-5 is a host cell IFN-inducible protein and an RNA helicase with a CARD at its N terminus (26). The mouse equivalent reflects these properties in its designation as HELI-CARD (27).
Fig. 1.
An IFN-inducible 150-kDa protein interacts with the SV5 V protein. (a) Vero cells were mock-infected or infected with SV5 (CPI-) at a high multiplicity of infection (10 plaque-forming units per cell) for 12 h, incubated in the presence (+) or absence (–) of rHuIFN-αA/D for 6 h, and metabolically labeled with 35[S]methionine for 1 h. Virus proteins (and associated cellular proteins) were immuno-precipitated with a pool of mAbs to the NP, P, V, M, and HN proteins. (b) Vero cells were transfected with plasmids that express either the V (lanes 1 and 2) or P (lanes 3 and 4) proteins of SV5 (CPI-). Thirty-six hours posttransfection, the cells were, or were not, treated with IFN-β for 6 h (as shown); the cells metabolically labeled with 35[S]methionine for 1 h; and the P and V proteins immunoprecipitated. (c) Confluent monolayers of 2f/SV5-V(CPI-) cells were, or were not, treated with IFN-β for 6 h before metabolic labeling with 35[S]methionine for 1 h and subjected to immunoprecipitation by using polyclonal anti-P/V antisera. Precipitated proteins were separated by electrophoresis through 4–12% (a) or 10% (b and c) polyacrylamide gels and radioactively labeled proteins visualized by PhosphorImager analysis. The 150-kDa IFN-inducible protein is highlighted with *.
The identification of mda-5 was based on its interaction with the V protein of the IFN-sensitive SV5 (CPI-) strain (25). To ascertain whether the V proteins of IFN-resistant isolates of SV5 also interacted with mda-5, Vero cells were cotransfected with a plasmid that expressed mda-5 together with a plasmid expressing the V protein of SV5(W3A) that blocks IFN signaling. mda-5 was clearly coprecipitated with the V protein of W3A (Fig. 2 and Fig. 6, which is published as supporting information on the PNAS web site). This interaction is likely to be direct, because we also demonstrated a specific interaction between mda-5 and SV5 V by using the yeast two-hybrid assay (Fig. 6).
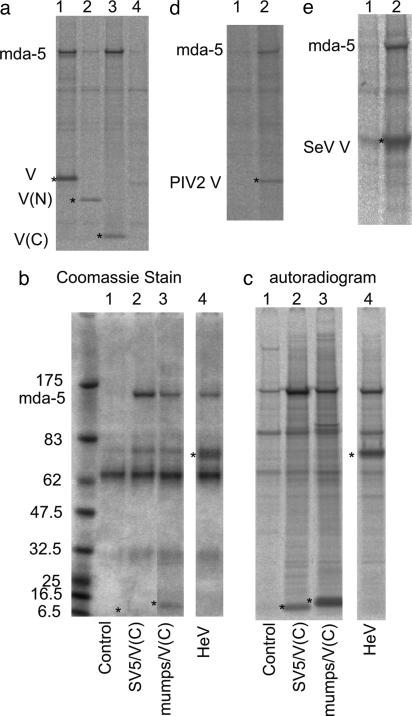
Fig. 2.
mda-5 interacts with the C terminus of the SV5 V protein and with other paramyxovirus V proteins. (a) Cells were transfected for 36 h with vectors expressing nontagged mda-5 (lane 4), together with myc-tagged versions of SV5 V(W3A) (lane 1), the N terminus of V (1–174) [V(N); lane 2], or the C terminus of V (126–222) [V(C); lane 3], or a control “empty vector” (lane 4) and immunoprecipitated with anti-myc antibody. (b and c) Coomassie stain and autoradiogram, respectively, of immunoprecipitates, by using anti-myc antibody, of cells transfected for 36 h with plasmids expressing nontagged mda-5, together with a control “empty vector” (lane 1) or plasmids expressing myctagged versions of the C terminus of SV5 [SV5/V(C); lane 2] and of mumps [mumps/V(C); lane 3] or myc-tagged HeV V (lane 4). (d and e) Autoradiograms of immunoprecipitates from cells transfected with a plasmid that express the nontagged mda-5, together with a control plasmid (lane 1), a plasmid expressing PIV 2/V(d, lane 2), or a plasmid expressing myc-tagged SeV V (e, lane 2). Cells were metabolically labeled with 35[S]methionine for 1 h before the myc-tagged proteins being immunoprecipitated with the 9E10 mAb and the V protein of hPIV-2 immunoprecipitated with the mAb Pk2 (32). *, the bands corresponding to the V proteins or derivatives. Proteins were separated on 4–12% polyacrylamide gradient gels (Invitrogen).
The V Proteins of a Diverse Group of Paramyxoviruses Bind mda-5 via Their Highly Conserved Cysteine-Rich C-Terminal Domains. The V protein of SV5 consists of an N-terminal domain, which is common to the P protein, and a unique cysteine-rich C-terminal domain, which is highly conserved among different paramyxoviruses (1). To ascertain whether mda-5 could bind to either of these domains alone, cells were transfected with a plasmid that expresses untagged mda-5, together with plasmids that express N-terminal myc-tagged SV5 V, the N terminus of V, or the C terminus of V. From these experiments, it was clear that mda-5 binds to the highly conserved C-terminal domain of V but not to the N-terminal domain (Fig. 2a).
We next examined whether the V proteins of other paramyxoviruses also bound mda-5. Cells were cotransfected with a plasmid that expresses untagged mda-5 together with plasmids expressing PIV2-V, myc-tagged V proteins from SeV or HeV, or with a plasmid that expresses the cysteine-rich C-terminal domain only of mumps virus. The V proteins were then immunoprecipitated with the appropriate antibody (Fig. 2). These experiments clearly demonstrated that the V proteins of all these viruses interact with mda-5. Because mda-5 was also clearly coimmunoprecipitated with the C-terminal domain of mumps virus, this confirmed that binding was via the highly conserved C terminus of V.
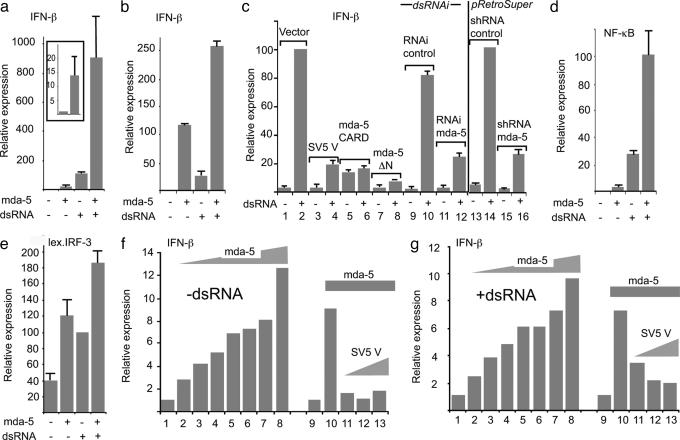
mda-5 Significantly Enhances the Activity of dsRNA-Responsive Promoters, Including the IFN-β Promoter, and Is a Target for Paramyxovirus V Proteins. Given the known ability of SV5 V to block IFN signaling and IFN production, we next investigated whether mda-5 may have a role to play in either of these cellular responses. In reporter assays, it was clear that mda-5 does not have a major effect on either type I or II IFN signaling (see Fig. 7, which is published as supporting information on the PNAS web site). To determine whether mda-5 plays a role in stimulation of the IFN-β promoter, we cotransfected Vero cells with mda-5 and a luciferase-reporter plasmid under the control of the human IFN-β promoter. Fig. 3a shows that the basal level of this promoter is strongly stimulated by mda-5 expression (≈10-fold in these cells; see Fig. 3a Inset). In equivalent experiments in HeLa cells, the basal stimulation was in excess of 100-fold (Fig. 3b). (Note: The activation of basal transcription of the IFN-β promoter by signaling intermediates, including IRF-1, IRF-3, IRF-7, p65, TRIF, IKKε, TBK1, PKR, ATF-2, and c-Jun, has frequently been reported in transient transfections; see, for example, ref. 17). This dramatic enhancement by mda-5 was also magnified when either cell type was transfected with synthetic dsRNA, a potent inducer of IFN-β expression (Fig. 3 a and b). The conclusion of these reporter assays was supported by measuring the release of IFN. 293 cells transfected with mda-5 secreted significantly more IFN than control cells when stimulated with dsRNA, and this could be blocked by coexpression of SV5 V but not SV5 P (Fig. 4). We then examined what effect fragments of mda-5 had on the activation of IFN-β promoter in Vero cells. Fig. 3c shows that a fragment of mda-5 (mda-5ΔN) lacking the CARD domain is able to dominantly inhibit dsRNA induction of the IFN-β promoter (compare lanes 1 and 2 with lanes 7 and 8). However, expression of the CARD domain alone led to low-level constitutive activation of the IFN-β promoter but an inhibition of additional response to dsRNA (Fig. 3c, lanes 5 and 6). Again, these reporter assays were supported by studies measuring the release of IFN. Transient transfection of 293 cells with mda-5, but not with mda-5ΔN, increased the amounts of IFN released by these cells in response to dsRNA. However, although expression of the CARD domain led to constitutive production of IFN, this response was not increased by treating the cells with dsRNA (data not shown).
Fig. 3.
mda-5 is an activator of dsRNA-responsive IFN-β transcription and is a target for inhibition by the SV5 virus V protein. Vero cells (a and c–g) or HeLaE cells (b) were cotransfected with the indicated luciferase reporter constructs; the β-galactosidase expression vector, pJATlacZ; and mammalian expression plasmids driving the overexpression of either mda-5 or fragments thereof, SV5-V, or the control “empty vector.” Transfected cells were either mock-treated or treated with dsRNA and cell extracts prepared for reporter gene assays. In each case, luciferase activity was corrected to the β-galactosidase activity to normalize for variations in the transfection efficiency. Transfection experiments were repeated at least twice, and averages and error bars are shown. Expression levels are compared with the basal level of the empty vector (=1) in each case except c and e, where the reference point is the induced level of the empty vector (=100%). (a and b) mda-5 stimulates basal and dsRNA-induced activity of the IFN-β promoter. Note that a Inset shows a magnified scale of the effects of mda-5 on basal expression. (c) mda-5 is essential for dsRNA response of the IFN-β promoter. Lanes 1–8 show the effect of dsRNA on IFN-β induction in the presence of the empty vector (lanes 1 and 2), the SV5 V protein (lanes 3 and 4), the mda-5 CARD domain (lanes 5 and 6), and mda-5 lacking the CARD domain (mda-5ΔN, lanes 7 and 8). Lanes 9–12 show the effect on induction of short dsRNAi molecules directed against mda-5 (lanes 11 and 12) compared with a control dsRNAi with two nucleotide mismatches (lanes 9 and 10). Lanes 13–16 show the same effect by using pRetroSuper-derived RNAi against mda-5 (lanes 15 and 16) compared with a control against CD2 (lanes 13 and 14). (d and e) mda-5 stimulates basal and dsRNA-induced activity of NF-κB(d) and IRF-3 (e) reporters. In the latter case, the reporter is a dimerized lexA operator sequence placed upstream of the herpes simplex virus 1 (HSV-1) thymidine kinase promoter, and transcriptional activity is conferred by a cotransfected Lex-IRF-3 fusion. (f and g) The basal (f) and dsRNA-induced (g) activation of the IFN-β promoter by mda-5 is inhibited by the SV5 V protein. Lanes 1 and 9 show the activity of the IFN-β promoter in the absence of mda-5, whereas lanes 2–8 show the effect of an increasing level of mda-5 expression plasmid (4, 8, 16, 40, 80, 320, and 468 ng). Lanes 10–13 show the effect of increasing levels of SV5 V expression plasmid (40, 80, and 160 ng) in the presence of 40 ng of mda-5 expression plasmid.
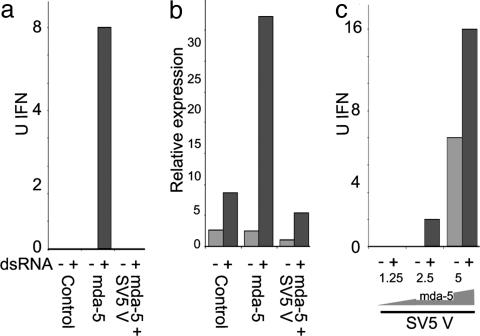
Fig. 4.
mda-5 stimulates the production of endogenous IFN, and this activity can be blocked by coexpression of the SV5 V protein. (a and b): 293 cells were transfected with control “empty vector” (5 μg), mda-5 (1.25 μg and “empty vector” 3.75 μg), or mda-5 (1.25 μg) and SV5-V (3.75 μg) expression vectors. After 24 h, cells were either mock-treated or treated with dsRNA for a further 18 h. The amount of IFN present in the culture media was estimated by either a virus reduction assay (a) or an IFN-stimulated regulatory element reporter assay (b, as described in ref. 6) in Vero cells. (c) As a above, but increasing amounts (1.25–5 μg) of the mda-5 expression vector were transfected with 3.75 μg of the SV5 V plasmid.
Although IFN-inducible, mda-5 is constitutively expressed at low levels in cells in the absence of IFN stimulation (ref. 28; unpublished observations). Therefore, to determine whether mda-5 can influence dsRNA signaling to the IFN-β promoter in the absence of ectopically expressed mda-5, we examined the effects of transfecting into Vero cells a synthetic RNAi molecule directed against mda-5. Fig. 3c (lanes 9–12) shows that induction in response to transfected dsRNA is impaired by RNAi to mda-5, but not by a control synthetic RNAi with a two-nucleotide mismatch to mda-5. Similar results were obtained both (i) by in vivo transcription of a short hairpin (sh)RNA fragment of mda-5 from a pRetroSuper vector but not with a control shRNA (pRetroSuper vector to CD2; Fig. 4c, lanes 13–16) and (ii) by using RNAi generated by RNase III digestion of full-length mda-5 dsRNA (data not shown).
The induction of IFN-β by dsRNA proceeds through the activation of a number of cellular transcription factors, the most important of which are NF-κB and IRF-3. We have previously shown that the SV5 V protein is able to inhibit the activation of both of these factors by dsRNA (10, 11). To determine whether mda-5 effects the activation of these factors, the mda-5 expression plasmid was cotransfected with reporters that are under the control of NF-κB or a lexA-IRF-3 fusion (necessary because cells contain multiple IRF family members that compete for binding to an IRF site). Fig. 3 d and e show that mda-5 stimulates both the basal and dsRNA-induced activities of NF-κB and IRF-3, respectively, again a property shared by several cellular signaling intermediates involved in the activation of the IFN-β promoter.
To determine whether these activities of mda-5 could be inhibited by the expression of the SV5 V protein, the dose-responsiveness of the IFN-β promoter to mda-5 was initially analyzed. Fig. 3 f and g show that both basal and dsRNA-induced activities responded to mda-5 in a dose-dependent manner, and that activation could be seen at the lowest level tested in both cases. We next determined whether coexpression of SV5 V could block the mda-5-dependent stimulation of the IFN-β promoter at a concentration of mda-5 in the midrange of activation. Fig. 3 f and g show that the SV5 V protein efficiently blocks the mda-5-dependent activation of the IFN-β promoter in these reporter assays.
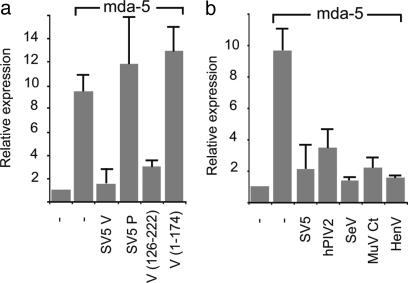
Given that the cysteine-rich C terminus of SV5 V alone both inhibits the activation of the IFN-β promoter by dsRNA (10, 11) and interacts with mda-5, we next tested whether this region of the protein alone was sufficient to inhibit IFN-β promoter activation by mda-5. Fig. 5a shows that this is indeed the case, with a fragment containing only the C-terminal 96 aa (V[126–222] that interacts with mda-5; see Fig. 3) being sufficient to block activation. By contrast, neither an N-terminal fragment (V[1–174]) of V (that does not interact with mda-5; see Fig. 2) nor the SV5 P protein blocked the activation of the IFN-β promoter by mda-5. However, as expected because they interact with mda-5 (Fig. 2), the V proteins of PIV2, mumps, SeV, and HeV blocked the mda-5-dependent transactivation of the IFN-β promoter (Fig. 5b).
Fig. 5.
mda-5 activity is inhibited by the C terminus of SV5 V and is a property of other paramyxovirus V proteins. Vero cells were cotransfected with the IFN-β luciferase reporter, the β-galactosidase expression vector, pJATlacZ, and mammalian expression plasmids driving the overexpression of mda-5 or the control “empty vector,” and either the V or P proteins of SV5 or truncations of the SV5 V protein (a) or the V proteins of other paramyxoviruses (b). In each case, luciferase activity was corrected to the β-galactosidase activity to normalize for variations in the transfection efficiency. Transfection experiments were repeated at least twice, and averages and error bars are shown. Expression levels are compared with the basal level of the empty vector (=1) in each case.
Discussion
In this work, we report that the V proteins of a diverse group of paramyxoviruses (including representatives of the Rubulavirus, Respirovirus, and Henipavirus genera of the subfamily Paramyxovirinae) all bind mda-5 via their highly conserved C-terminal domain and present evidence that links this property to the ability of these viruses to reduce the production of IFN by infected cells. We show in reporter assays that mda-5 stimulated the basal activity of the IFN-β promoter, a property shared by many signaling intermediates for the IFN-β promoter, and that overexpression of mda-5 enhances the activation of the IFN-β promoter in response to intracellular dsRNA. Both of these activities are blocked by coexpression of the V proteins of SV5, PIV2, mumps, HeV, and SeV, and for SV5 and mumps, only the C-terminal domain is required. The results obtained with the reporter gene assays are supported by measurements of endogenous IFN production by 293 cells; cells transfected with mda-5 secrete detectable amounts of IFN that are further increased by treatment of cells with dsRNA. Again, both these activities were blocked by coexpression of the V, but not P, protein of SV5. Because mda-5 also activated both NF-κB and IRF-3 in reporter gene assays, it appears that mda-5 plays an important role upstream of the activation of these transcription factors in response to dsRNA. The inhibition by the V protein of both IRF-3 and NF-κB activation would explain why these factors are not activated in SV5-infected cells but are activated in cells infected with SV5VΔC. Thus it seems reasonable to suggest that paramyxoviruses specifically interact with mda-5 to reduce the amount of IFN released by infected cells.
We have previously reported that paramyxovirus V proteins are unable to completely block IFN production by dsRNA (11). Interestingly, although treatment of cells with RNAi to mda-5 significantly reduced their ability to activate the IFN-β promoter in response to transfected dsRNA, it also did not completely abolish it. These results are consistent with the existence of an mda-5-independent dsRNA response pathway(s) that is insensitive to paramyxovirus V proteins. Given that SV5 is such a poor inducer of IFN, it is possible that SV5 fails to activate this additional pathway(s) or has an additional blocking mechanism.
Interestingly, while this manuscript was in preparation, it was reported that retinoic acid inducible gene I (RIG-I) has similar properties to mda-5 in dsRNA-induced production of IFN-β (19). Like mda-5, RIG-I is IFN-inducible and has a DExD/H box RNA helicase C-terminal domain and a CARD domain at its N terminus. The binding of dsRNA to the helicase domain of RIG-I has been hypothesized to result in the activation of the ATPase, leading to conformational changes to the substrate dsRNA, the helicase domain, and the CARD domain; the “activated” CARD then transmits a signal to downstream molecules that results in the activation of IRF-3 and NF-κB (19). Given our observations on the ability of mda-5 to activate the IFN-β promoter and on the activity of the isolated mda-5 CARD domain, a similar model may apply for mda-5. Furthermore, the properties of mda-5 and RIG-I are reminiscent of those exhibited by nucleotide-binding oligmerization domain (NOD)-1 and NOD2, proteins that recognize intracellular peptidoglycans with the consequent activation of their N-terminal CARD domains and subsequent NF-κB activation (reviewed in ref. 29 and 30). Thus mda-5, RIG-I, NOD1, and NOD2 may be representative of a wider class of intracellular pattern recognition molecules.
Because mda-5 and RIG-I are IFN-inducible, induction of these proteins by IFN may help explain the phenomenon of IFN priming, in which the amount of IFN produced by cells in response to dsRNA or virus infection is significantly increased if cells are first pretreated with low levels of IFN (31). However, the relative importance of mda-5 and RIG-I, and indeed other potential members of this family, in stimulating the IFN-β promoter remains to be established. We have no evidence that the V protein of SV5, or other paramyxoviruses, interacts with RIG-I even in IFN-treated cells, and it is tempting to suggest that RIG-I comprises part of the mda-5-independent dsRNA response pathway(s) discussed above. It will be of great interest to identify the upstream and downstream signaling molecules with which mda-5 and RIG-I interact, to determine whether they are differentially expressed and to ascertain whether groups of viruses, other than paramyxoviruses, also specifically inhibit the activity of mda-5, RIG-I, or any of the molecules with which they interact.
Supplementary Material
Acknowledgments
We thank Linfa Wang (Commonwealth Scientific and Industrial Research Organization, Melbourne) and Bert Rima (Queen's University of Belfast, Belfast, U.K.) for providing the V genes of HeV and mumps virus, respectively. This work was supported by the Wellcome Trust and Biotechnology and Biological Sciences Research Council.
Author contributions: J.A., S.G., K.S.C., and R.E.R. designed research; J.A., K.S.C., D.F.Y., T.S.C., N.S., and S.G. performed research; S.G. and R.E.R. analyzed data; and R.E.R. and S.G. wrote the paper.
Abbreviations: PIV, parainfluenza viruses; SeV, Sendai virus; SV5, simian virus 5; HeV, Hendra virus; mda-5, melanoma differentiation-associated gene 5; CARD, caspase recruitment domain; RNAi, RNA interference; RIG-I, retinoic acid inducible gene I; IRF, IFN regulatory factor.
References
- 1.Lamb, R. A. & Kolakofsky, D. (2001) in Fields' Virology, eds. Fields, B. N., Knipe, D. M., Howley, P. M. & Griffin, D. E. (Lippincott Williams & Wilkins, Philadelphia), Vol. 1, pp. 1305–1340. [Google Scholar]
- 2.Wang, L.-F. & Eaton, B. (2001) Infect. Dis. Rev. 3, 52–69. [Google Scholar]
- 3.Goodbourn, S., Didcock, L. & Randall, R. E. (2000) J. Gen. Virol. 81, 2341–2364. [DOI] [PubMed] [Google Scholar]
- 4.Sen, G. C. (2001) Annu. Rev. Microbiol. 55, 255–281. [DOI] [PubMed] [Google Scholar]
- 5.Grandvaux, N., tenOever, B. R., Servant, M. J. & Hiscott, J. (2002) Curr. Opin. Infect. Dis. 15, 259–267. [DOI] [PubMed] [Google Scholar]
- 6.Didcock, L., Young, D. F., Goodbourn, S. & Randall, R. E. (1999) J. Virol. 73, 9928–9933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Nagai, Y. & Kato, A. (2004) in Biology of Negative Strand RNA Viruses, ed. Kawaoka, Y. (Springer, Berlin), Vol. 283, pp. 197–248. [DOI] [PubMed] [Google Scholar]
- 8.Garcia-Sastre, A. (2004) in Biology of Negative Strand RNA Viruses, ed. Kawaoka, Y. (Springer, Berlin), Vol. 283, pp. 249–280. [DOI] [PubMed] [Google Scholar]
- 9.Horvath, C. M. (2004) Cytokine Growth Factor Rev. 15, 117–127. [DOI] [PubMed] [Google Scholar]
- 10.He, B., Paterson, R. G., Stock, N., Durbin, J. E., Durbin, R. K., Goodbourn, S., Randall, R. E. & Lamb, R. A. (2002) Virology 303, 15–32. [DOI] [PubMed] [Google Scholar]
- 11.Poole, E., He, B., Lamb, R. A., Randall, R. E. & Goodbourn, S. (2002) Virology 303, 33–46. [DOI] [PubMed] [Google Scholar]
- 12.Komatsu, T., Takeuchi, K., Yokoo, J. & Gotoh, B. (2004) Virology 325, 137–148. [DOI] [PubMed] [Google Scholar]
- 13.Servant, M. J., Grandvaux, N. & Hiscott, J. (2002) Biochem. Pharmacol. 64, 985–992. [DOI] [PubMed] [Google Scholar]
- 14.Andrejeva, J., Poole, E., Young, D. F., Goodbourn, S. & Randall, R. E. (2002) J. Virol. 76, 11379–11386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Young, D. F., Chatziandreou, N., He, B., Goodbourn, S., Lamb, R. A. & Randall, R. E. (2001) J. Virol. 75, 3363–3370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sharma, S., tenOever, B. R., Grandvaux, N., Zhou, G. P., Lin, R. & Hiscott, J. (2003) Science 300, 1148–1151. [DOI] [PubMed] [Google Scholar]
- 17.Fitzgerald, K. A., McWhirter, S. M., Faia, K. L., Rowe, D. C., Latz, E., Golenbock, D. T., Coyle, A. J., Liao, S. M. & Maniatis, T. (2003) Nat. Immunol. 4, 491–496. [DOI] [PubMed] [Google Scholar]
- 18.Diebold, S. S., Montoya, M., Unger, H., Alexopoulou, L., Roy, P., Haswell, L. E., Al-Shamkhani, A., Flavell, R., Borrow, P. & Reis e Sousa, C. (2003) Nature 424, 324–328. [DOI] [PubMed] [Google Scholar]
- 19.Yoneyama, M., Kikuchi, M., Natsukawa, T., Shinobu, N., Imaizumi, T., Miyagishi, M., Taira, K., Akira, S. & Fujita, T. (2004) Nat. Immunol. 5, 730–737. [DOI] [PubMed] [Google Scholar]
- 20.Donze, O. & Picard, D. (2002) Nucleic Acids Res. 30, e46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Brummelkamp, T. R., Bernards, R. & Agami, R. (2002) Science 296, 550–553. [DOI] [PubMed] [Google Scholar]
- 22.Ulane, C. M. & Horvath, C. M. (2002) Virology 304, 160–166. [DOI] [PubMed] [Google Scholar]
- 23.Leupin, O., Bontron, S. & Strubin, M. (2003) J. Virol. 77, 6274–6283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Andrejeva, J., Young, D. F., Goodbourn, S. & Randall, R. E. (2002) J. Virol. 76, 2159–2167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chatziandreou, N., Young, D., Andrejeva, J., Goodbourn, S. & Randall, R. E. (2002) Virology 293, 234–242. [DOI] [PubMed] [Google Scholar]
- 26.Kang, D. C., Gopalkrishnan, R. V., Wu, Q., Jankowsky, E., Pyle, A. M. & Fisher, P. B. (2002) Proc. Natl. Acad. Sci. USA 99, 637–642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kovacsovics, M., Martinon, F., Micheau, O., Bodmer, J. L., Hofmann, K. & Tschopp, J. (2002) Curr. Biol. 12, 838–843. [DOI] [PubMed] [Google Scholar]
- 28.Kang, D. C., Gopalkrishnan, R. V., Lin, L., Randolph, A., Valerie, K., Pestka, S. & Fisher, P. B. (2004) Oncogene 23, 1789–1800. [DOI] [PubMed] [Google Scholar]
- 29.Bouchier-Hayes, L. & Martin, S. J. (2002) EMBO Rep. 3, 616–621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Inohara, N. & Nunez, G. (2003) Nat. Rev. Immunol. 3, 371–382. [DOI] [PubMed] [Google Scholar]
- 31.King, P. & Goodbourn, S. (1994) J. Biol. Chem. 269, 30609–30615. [PubMed] [Google Scholar]
- 32.Dunn, C., O'Dowd, A. & Randall, R. E. (1999) J. Immunol. Methods 224, 141–150. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.