Abstract
We have investigated the plasma proteome by using 2D gel electrophoresis and MS from patients with severe acute respiratory syndrome (SARS). A complete proteomic analysis was performed on four patients with SARS in different time courses, and a total of 38 differential spots were selected for protein identification. Most of the proteins identified are acute phase proteins, and their presence represents the consequence of serial cascades initiated by SARS-coronavirus infection. There are several proteins that have never been identified in plasma before using 2D gel electrophoresis, among which peroxiredoxin II was chosen for further study by analyzing additional 20 plasma samples from patients with probable and suspected SARS and patients with fever, respectively. The results showed that the level of plasma peroxiredoxin II in patients with SARS is significantly high and could be secreted by T cells. Taken together, our findings indicate that active innate immune responses, along with the oxidation-associated injuries, may play a major role in the pathogenesis of SARS.
Keywords: proteomic techniques, acute phase proteins, peroxiredoxin II
Severe acute respiratory syndrome (SARS) was first recognized at the end of 2002 in Guangdong, China, and since then the disease has spread to several countries. By late July 2003, >8,000 SARS cases and >700 SARS-related deaths were reported from >25 countries around the world, and no effective treatment has been established so far. Through extensive studies, the causative agent of SARS has been identified as a human SARS-coronavirus (SARS-CoV) (1–3). Although the complete genome sequence of SARS-CoV, the structure of the main protease (3CL protease), and the possible receptor have been determined (4–7), the pathogenesis of SARS is still not fully understood.
Plasma proteins are useful targets for diagnostic, prognostic, and/or therapeutic development. With proteomic tools available recently, profiling of human plasma proteome becomes more feasible in searching for disease-related markers (8). To explore the possible pathogenetic mechanisms involving the progression of SARS, we have analyzed the plasma proteins of 22 different plasma samples from four SARS patients in different time courses by using 2D gel electrophoresis (2DE) in combination with MS. The results showed that most of the plasma proteins found in patients with SARS are acute phase proteins (APPs) generated by inflammatory reactions during SARS-CoV-induced acute lung injuries.
Materials and Methods
Human Subjects. Four SARS patients with a total of 22 plasma samples were selected for complete proteomic analysis. These patients were infected by SARS-CoV through nosocomial transmission during one major SARS outbreak in one municipal hospital in Taipei. For comparison, plasma samples from six healthy individuals were used as controls. The protein concentration of each sample was determined by the standard Bradford method. The use of these samples was approved by the Review Board of the Tri-Service General Hospital, National Defense Medical Center, National Defense University.
2DE and Image Analysis. 2DE was performed with the Amersham Biosciences IPGphor IEF system and Bio-Rad PROTEAN II Multi-Cell electrophoresis unit. Plasma samples were solubilized in 350 μl of lysis buffer containing 7 M urea, 4% 3-[(3-cholamidopropyl)dimethylammonio]-1-propanesulfonate, and 2mM Tri-n-butylphosphine. After sonication, the protein sample (500 μg) was loaded into immobilized pH gradient (IPG) gel strips. Isoelectric focusing was run at 18°C with 6,000 V for a total of 35 kV·h. After isoelectric focusing, the IPG strips were equilibrated for 15 min in a buffer containing 50 mM Tris·HCl (pH 8.8), 6 M urea, 2% SDS, 30% glycerol, and 2% dithioerythritol. A 2DE was run at 45 mA for 5 h. The gels were fixed in 10% methanol and 7% acetic acid for 30 min, stained in 400 ml of Sypro Ruby solution overnight, and soaked in deionized water for 20 min. The 2D images were scanned by using the fluorescence image scanning Typhoon 9200 (Amersham Biosciences) and analyzed by using imagemaster 2d platinum software (Amersham Biosciences). The intensity of each spot was first processed by background subtraction and then normalized between gels as a proportion of the total protein intensity from the entire gel. For those proteins with multiple spots in the gel, such as haptoglobin, the sum of pixel volumes of individual spots from isoforms was used for analysis.
MS and Protein Identification. Protein spots of interest were digested with trypsin by using the Genomic Solutions ProGest workstation. For MALDI-TOF MS analysis, peptide powders were resuspended with 0.1% trifluoroacetic acid in 50% acetonitrile, mixed with matrix (10 mg/ml α-cyano-4-hydroxycinnamic acid), and analyzed by using an Applied Biosystems Voyager-DE-PRO mass spectrometer. Monoisotopic peptide mass values were used for protein identification by searching the Swiss-Prot protein database with MS-Fit (http://prospector.ucsf.edu). For liquid chromatography (LC)-tandem MS (MS/MS) analysis, peptide powders were resuspended with 0.1 μM formic acid containing 5% acetonitrile. LC-MS/MS analyses were performed by using an LC Packings Ultimate capillary LC system coupled to an Applied Biosystems QSTAR Pulsar i quadrupole TOF mass spectrometer. Peptide fragment mass values were searched against the Swiss-Prot protein database by using the mascot search program (www.matrixscience.com).
Western Blot Analysis. Protein samples, including plasma, conditioned media, and cell lysates, were separated by 12% SDS/PAGE and then transferred onto nitrocellulose membranes under semidry conditions by using Transblot (Bio-Rad). Western blot was performed by using a polyclonal antibody against Prx II (LabFrontier, Seoul, South Korea) and peroxidase-conjugated secondary antibody. The reaction was detected by chemiluminescence with an ECL kit (Amersham Biosciences).
Results
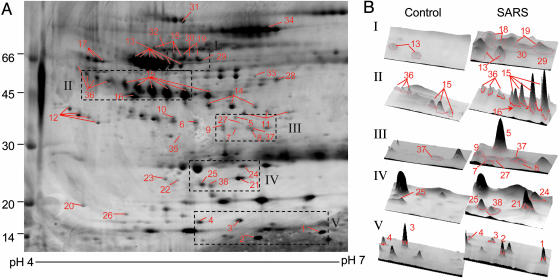
Plasma Protein Profiles of SARS. The protein profiles of 22 plasma samples from four patients with SARS and six samples from health control were analyzed by 2DE with pI values in the range of 4–7 to obtain a greater resolution in protein separation and to eliminate the interference of basic Igs (IgG heavy and light chains). Fig. 1 shows a representative 2DE image of plasma profiles from four patients collected at the day of their admission to Tri-Service General Hospital. About 360 spots in each Sypro Ruby-stained gel can be visualized by imagemaster software. In comparison with the 2DE from the patients with healthy controls, many chains of spots that may represent different degrees of protein modification and/or degradation products were found to be significantly overexpressed.
Fig. 1.
A representative 2D gel of plasma from one SARS patient at his acute stage. (A) Protein spots expressed differently from control plasma were marked on the maps and further identified by MALDI-TOF MS and/or LC-MS/MS. (B) The 3D images of differential spots between control and SARS were analyzed byimagemaster 2d platinum software.
Identification of Differentially Expressed Proteins. Among the 22 SARS-associated 2DE, a total of 385 selected differentially expressed protein spots with >2-fold change were excised and in-gel digested with trypsin, followed by MS analysis. The sequence coverage of the identified proteins by peptide mass fingerprinting ranges from 35% to 68%, depending on the size of proteins and the amount present. For protein spots with insufficient quantities, nanoflow LC-MS/MS was used to confirm protein identification. As shown in Fig. 1, the comparative analysis revealed that 38 protein spots were altered >2-fold with 35 and 3 protein spots showing overexpression and downexpression, respectively. After examination of each gel image, 15 protein spots were overexpressed in all 22 samples, 7 of which were not detectable in normal controls, including proteins of glutathione peroxidase, Prx II, retinol binding protein, vitamin D binding protein (2 spots), and serum amyloid A (SAA) (2 spots) and 8 of which were overexpressed, including pigment epithelium-derived factor, α-2-HS glycoprotein, complement factor H-related protein, α-1-antitrypsin, haptoglobin β chain, inter-α-trypsin inhibitor heavy chain H4 (2 spots), and leucine-rich α-2-glycoprotein. Among these altered plasma proteins, the monomeric transthyretin, spots 3 and 4, showed a 2-fold decrease in patients with SARS, whereas the multimeric transthyretin, spots 5 and 6, revealed overexpression. Other significantly overexpressed protein spots included antithrombin III fragment (19 of 22), angiotensinogen (17 of 22), mannose binding protein (18 of 22), apolipoprotein A-I (19 of 22), apolipoprotein E (15 of 22), and tetranectin (18 of 22). The locations of these 38 differentially expressed protein spots were marked with numbers in one representative gel shown in Fig. 1. A list of the up- or down-regulated proteins as well as their biological functions is summarized in Table 1.
Table 1. List of significant differentially expressed proteins identified in plasma from patients with SARS.
| Spot | Protein name | Swiss-Prot entry | Sequence coverage, % | pl/mass, kDa | Regulation | Average volume*, P/N | Functions† |
|---|---|---|---|---|---|---|---|
| Acute Phase Proteins | |||||||
| 1 | SAA protein | P02735 | 35 | 5.9/12 | ↑ (22/22)‡ | 1.78/ND | a, b, c, d |
| 2 | SAA protein | P02735 | 66 | 5.9/12 | ↑ (22/22) | 1.61/ND | a, b, c, d |
| 3 | Transthyretin | P02766 | 60 | 5.4/14 | ↓ (22/22) | 0.56/5.69 | a, b |
| 4 | Transthyretin | P02766 | 70 | 5.4/14 | ↓ (22/22) | 0.29/0.82 | a, b |
| 5 | Transthyretin (polymer) | P02766 | 40 | 5.4/14 | ↑ (17/22) | 0.34/ND | a, b |
| 6 | Transthyretin (polymer) | P02766 | 68 | 5.4/14 | ↑ (20/22) | 0.28/0.12 | a, b |
| 7 | Mannose-binding protein | P11226 | 16§ | 5.4/24 | ↑ (18/22) | 0.02/ND | a, c |
| 8 | Inter-α-trypsin inhibitor heavy chain H4(35kDa fragment) | Q14624 | 41 | 6.3/27 | ↑ (8/22) | 0.03/ND | a, e, g, l |
| 9 | Inter-α-trypsin inhibitor heavy chain H4(35kDa fragment) | Q14624 | 3§ | 6.3/27 | ↑ (22/22) | 0.08/ND | a, e, g, l |
| 10 | Inter-α-trypsin inhibitor heavy chain H4(35kDa fragment) | Q14624 | 2-3§ | 6.3/27 | ↑ (22/22) | 0.25/0.02 | a, e, g, l |
| 11 | α-1-antitrypsin (fragment) | P01009 | 31-40 | 5.4/44 | ↑ (19/22) | 0.27/ND | a, e, f, o |
| 12 | α-1-antitrypsin (fragment) | P01009 | 9-24§ | 5.4/44 | ↑ (22/22) | 0.82/ND | a, e, f, o |
| 13 | α-1-antitrypsin | P01009 | 35-5§ | 5.4/44 | ↑ (21/22) | 7.96/1.01 | a, e, f, o |
| 14 | Haptoglobin (β chain) (fragment) | P00738 | 19-22§ | 6.3/27 | ↑ (14/22) | 0.39/ND | a, b, c, d |
| 15 | Haptoglobin (β chain) | P00738 | 8-16§ | 6.3/27 | ↑ (22/22) | 4.20/0.26 | a, b, c, d |
| 16 | Complement C3 (fragment) | P01024 | 7§ | 6.0/185 | ↑ (4/22) | 0.50/ND | a, c |
| 17 | α-2-HS glycoprotein | P02765 | 11§ | 4.5/30 | ↑ (22/22) | 0.73/0.52 | a, c |
| 18 | Angiotensinogen | P01019 | 22§ | 5.6/50 | ↑ (17/22) | 0.15/ND | a, e, k, m |
| 19 | Inter-α-trypsin inhibitor heavy chain H4(70kDa fragment) | Q14624 | 7§ | 5.9/71 | ↑ (11/22) | 0.08/ND | a, e, g, l |
| 20 | Complement C4 (fragment) | P01028 | 1§ | 5.7/192 | ↑ (13/22) | 0.06/ND | a, c |
| Antioxidation | |||||||
| 21 | Peroxiredoxin 2 | P32119 | 30§ | 5.7/22 | ↑ (22/22) | 0.32/ND | c, d, l, o |
| 22 | Peroxiredoxin 1 or 2 | P32119 | 6§ | 5.7/22 | ↑ (9/22) | 0.10/ND | c, d, l, o |
| 23 | Peroxiredoxin 1 or 2 | P32119 | 6§ | 5.7/22 | ↑ (5/22) | 0.03/ND | c, d, l, o |
| 24 | Glutathione peroxidase | P22352 | 28§ | 7.8/23 | ↑ (22/22) | 0.22/ND | d |
| Transport Proteins | |||||||
| 25 | Plasma retinol-binding protein | P02766 | 21§ | 5.3/21 | ↑ (22/22) | 0.22/ND | b |
| 26 | Apolipoprotein A-VI (fragment) | P06727 | 11§ | 5.2/43 | ↓ (12/22) | 0.02/0.21 | b |
| 27 | Apolipoprotein E | P02649 | 20§ | 5.5/34 | ↑ (15/22) | 0.01/ND | b, n |
| 28 | Complement factor H-related protein 1 | P02647 | 3§ | 7.5/36 | ↑ (22/22) | 0.29/0.17 | b, c |
| 29 | Vitamin D-binding protein | P02774 | 21§ | 5.2/51 | ↑ (22/22) | 0.14/ND | b |
| 30 | Vitamin D-binding protein | P02774 | 12§ | 5.2/51 | ↑ (22/22) | 0.19/ND | b |
| 31 | Apolipoprotein A-1 (polymer) | P02753 | 44 | 5.3/28 | ↑ (19/22) | 0.04/ND | b, c, g |
| Protease and Protease inhibitors | |||||||
| 32 | Kininogen | P01042 | 5§ | 6.2/70 | ↑ (16/22) | 0.07/ND | c, e, f |
| 33 | Pigment epithelium-derived factor | P36955 | 12§ | 6.0/45 | ↑ (22/22) | 0.02/0.01 | h, i, k |
| 34 | Thrombin | P00734 | 15§ | 8.3/34 | ↑ (2/22) | 0.46/ND | f, i |
| 34 | Antithrombin III | P01008 | 51 | 6.0/49 | ↑ (2/22) | 0.46/ND | e, f, k, o |
| 35 | Antithrombin III (fragment) | P01008 | 5§ | 6.0/ND | ↑ (19/22) | 0.06/ND | e, f, o |
| Others | |||||||
| 36 | Leucine-rich α-2-glycoprotein | P02750 | 49 | 5.7/34 | ↑ (22/22) | 0.58/0.07 | j |
| 37 | Fibrinogen-like protein 1 | Q08830 | 6§ | 5.5/34 | ↑ (10/22) | 0.07/0.03 | h |
| 38 | Tetranectin | P05452 | 5§ | 5.8/20 | ↑ (18/22) | 0.04/ND | g |
The average spot volume (%) in normal controls (N) and in SARS patients (P). ND, not detectable.
a, acute phase protein; b, transport/scavenger proteins; c, immune modulation; d, antioxidant; e, protease inhibitor; f, coagulation; g, tissue remodeling; h, signaling proteins; i, protease; j, hemostasis; k, antiangiogenesis; l, antiapoptosis; m, regulation of blood pressure; n, lipid metabolism; o, antiviral activity (based on the description on http://harvester.embl.de).
Occurrence in 22 gels.
Spots were further confirmed by LC-MS/MS.
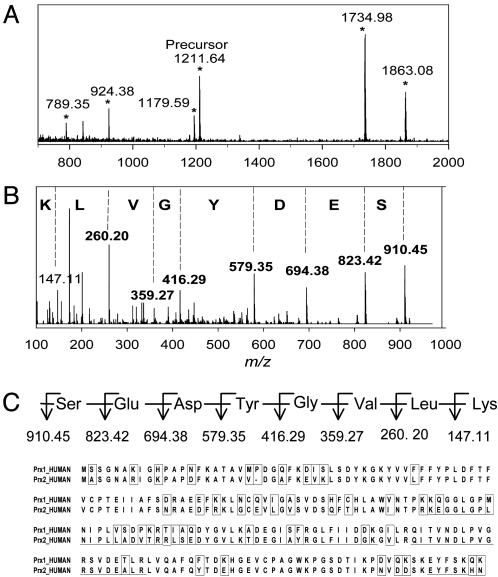
Identification of Prx II. In addition to the known circulating plasma proteins, a surprising observation is the appearance of an intracellular protein Prx II in spot 21 that has not been detected in human plasma in 2DE analysis. Fig. 2A shows the mass map of tryptic peptides from spot 21, identified as Prx II or natural killer enhancing factor (NKEF) B, which was observed exclusively in the plasma of SARS patients but not in healthy individuals. Of the six members of the human Prx protein family, only Prx I (also NKEF-A) is similar to Prx II in both molecular weight and amino acid sequence (9). A database search for the doubly charged ion of 512.29 m/z and its fragment ions revealed the peptide sequence, LSEDYGVLK, unique to Prx II (Fig. 2 B and C). The peptide sequence from spots 22 and 23 were located at the homologous region of Prx I and Prx II; therefore, we could not determine the protein identity from these two spots.
Fig. 2.
Identification of Prx II. (A) MALDI mass spectrum of tryptic peptides from spot 21, identified as Prx II. Peaks marked with stars match the underlined sequence of Prx II shown in C.(B)MS/MS spectrum of peak [M + 2H]2+ at m/z 512.29 from spot 21 matches sequence LSEDYGVLK of Prx II (shaded in C). Only the most intense y-series fragment ions were labeled with single letter codes. (C) Sequence alignment of human Prx I and Prx II. Different amino acid residues between Prx I and Prx II are boxed.
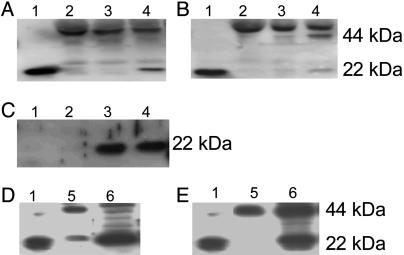
Validation of Plasma Prx II. Western blot analysis of plasma was conducted under reducing and nonreducing (without disrupting the disulfide bonds) conditions by using polyclonal anti-Prx II antibody (10). The result showed that Prx II is present in the monomeric form at 22 kDa in a reducing condition (Fig. 3A) and in the dimeric form at 44 kDa under a nonreducing condition (Fig. 3B). To determine the correlation of Prx II with SARS, 60 plasma samples, including 20 SARS probable cases, 20 SARS suspected cases, and 20 regular fever cases, along with another 20 normal controls were used for further validation. The results showed that 4 of 20 SARS probable cases, 4 of 20 SARS suspected cases, and 3 of 20 simple fever cases revealed the appearance of plasma Prx II. Unexpectedly, 2 of 20 normal controls also revealed the presence of Prx II. When reexamining the samples, we found that these two control samples showed sign of hemolysis, i.e., plasma with red color, which may be caused by mechanical breakage of erythrocytes during blood collection. The plasma Prx II disappeared when the recollection procedure was performed in these two normal controls without hemolysis (Fig. 3C). In addition, to determine whether Prx II could be secreted from T cells as demonstrated in ref. 11, conditioned media and cell lysates from Jurkat T cells were analyzed. The results showed that Prx II could be secreted by T cells in mainly the dimeric form (Fig. 3 D and E). The hemolysis-related Prx II was also observed in samples from SARS patients as shown in Fig. 4. Generally, the more red color that is shown in the plasma, the more Prx II is expressed in Western blot analyses.
Fig. 3.
Western blot analyses of Prx II expression in plasma and T cells. A and B indicate the reducing and nonreducing conditions, respectively. Lane 1, recombinant Prx II; lane 2, normal control; lanes 3 and 4, representative samples from patients with SARS. C shows two normal controls. Lanes 1 and 2 are from the plasma without hemolysis. Lanes 3 and 4 reveal signs of hemolysis. The molecular mass of monomeric and dimeric forms of Prx II are 22 kDa and 44 kDa, respectively. D and E indicate the presence of Prx II in conditioned media (lane 5) and cell lysates (lane 6) from Jurkat T cells under the reducing (D) and nonreducing (E) conditions, respectively. Lane 1 in D and E is recombinant Prx II.
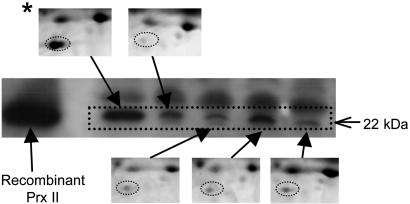
Fig. 4.
2DE gel images of Prx II and their corresponding 1D electrophoresis Western blot analyses. Plasma samples were collected from five different time courses during hospitalization of one representative patient with SARS. *, hemolyzed sample.
After checking all of the samples analyzed, we found that 8 of 22 SARS samples analyzed by 2DE and 16 of 60 samples provided by the Taiwan Center for Disease Control showed sign of hemolysis. Therefore, including 20 normal controls, 24% (24 of 102) of the samples collected in this study showed signs of hemolysis. However, three hemolyzed samples (3 of 24) did not show the presence of Prx II and 16 nonhemolyzed samples, including 14 SARS and 2 fever samples, showed the appearance of Prx II. When excluding the hemolyzed samples, 36% (15 of 42) of samples from patients with SARS and 10% (2 of 20) from patients with fever showed various level of Prx II in plasma, indicating that the presence of plasma Prx II in SARS is not due to contamination of ruptured erythrocytes caused by mechanical damage during blood collection but represents an actual disease-associated marker.
To determine whether endogenous hemolysis could be the reason for plasma Prx II appearance, we analyzed the level of haptoglobin in 2D gel images because it would bind free hemoglobin caused by endogenous hemolysis in the circulation. The decreased level of haptoglobin indicates the presence of hemolysis within the body. After excluding the hemolyzed samples, the statistical analysis showed a slightly negative correlation between Prx II and the haptoglobin level with no statistical significance (r2 = 0.0347), indicating that endogenous hemolysis is not a major source for plasma Prx II.
Discussion
The work presented here demonstrated the use of proteomic analysis to study the nature of an emerging disease. Although analysis of bronchoalveolar lavage fluid (12–14) provides a direct way to investigate lung diseases, it is impractical to use this method in highly contagious diseases such as SARS. Our studies demonstrate that plasma analysis is not only more feasible but safer. In addition, because of the close communication between the alveolar space and blood circulation during lung inflammation, the pathological events occurring in the inflamed lung may be attainable by simply analyzing the patients' plasma. This study has identified a number of overexpressed plasma proteins with different functions in patients with SARS. Selected description of these proteins that may play a role in SARS pathogenesis is discussed as follows.
In this study, we have found many APPs in the plasma of patients with SARS. These proteins may provide a fast and effective control of inflammatory damage until the subsequent defensive mechanisms can begin to operate (15). In the past, there have been very few studies on the response of APPs to bacterial (16) or viral infection (17, 18). Among the APPs found in SARS patients, SAA and haptoglobin have been widely used as inflammatory markers for evaluating the status of infection and inflammatory lesions in veterinary medicine (19). Although SAA is not as commonly used in human medicine as C-reactive protein (CRP), it is more sensitive to acute response than CRP (17, 20, 21).
The mannose-binding protein was identified for the first time through 2DE in the plasma of SARS patients. It is a member of the C-type lectin family closely associated with infectious diseases (22) and host defense (23) against pathogens, such as influenza A virus (24) and HIV (25). The lectin complement pathway is activated upon pathogen exposure to promote chemotactaxis, cytolysis, and inflammation (26). Activation of the complement system in SARS could also be facilitated by the overexpressed complement C4 and C3 fragments and complement factor H-related protein 1 found in this study. We have also identified another C-type lectin, tetranectin, which has been demonstrated to be a specific fibrinolytic marker in patients with rheumatoid arthritis (27). Its participation in the proteolytic process by binding to plasminogen-like hepatocyte growth factor and tissue-type plasminogen activator (28, 29) may be associated with the significant fibrinolytic tissue destruction in SARS.
The proteases and protease inhibitors identified in the plasma of SARS indicate the active status of proteolytic processes and the consequent antiproteolytic responses. The plasma proteins with protease activity were thrombin and pigment epithelium-derived factor, whereas proteins having protease inhibitory effect were antithrombin III, α-1-antitrypsin, inter-α-trypsin inhibitor heavy chain H4 (ITIH4), angiotensinogen, and kininogen. Among these proteins, angiotensinogen and ITIH4 have never been previously identified in 2DE. It is of interest that angiotensin converting enzyme (ACE) 2, a downstream regulator of rennin-angiotensinogen cascade, recently has been shown to be a functional receptor for SARS-CoV (7). In physiological circumstances, angiotensin is cleaved by rennin to give rise to angiotensin I, which is then converted to the active form, angiotensin II, by ACE located in specialized cells in the lung. Patients with severe infection were found to have an increased level of circulation (30, 31), and steroid administration will also increase angiotensinogen and blood pressures (32). Therefore, the increase of angiotensinogen in SARS may not necessarily be related to the enzyme activity of ACE2 but to the inflammatory status and the usage of steroid in patients infected by SARS-CoV.
Among the decreased plasma proteins shown in patients with SARS, transtyretin, also called prealbumin, revealed different expression levels with its different conformation. Because it has been known as a negative APP (33), i.e., a decrease in the plasma concentration during acute inflammation, it is not surprising that the monomeric plasma transtyretin showed downexpression in SARS. The multimeric form of plasma transthyretin, on the contrary, showed an increase in SARS. Transthyretin has been shown to be a valuable prognostic factor of nutritional and inflammatory indices in patients with acute respiratory failure (34). A persistent low level of serum transthyretin is predictive of lethality, whereas increased levels were associated with improved ventilatory performances. Our findings in 2DE analysis suggested the conformational significance of transthyretin in patients with critical respiratory condition, and the development of a means for detection of transthyretin in its monomeric and tetrameric forms is needed to clarify their roles in the disease progression.
Another reported negative APP, α-2-HS glycoprotein (also called fetuin), which was usually expressed at a decreased level in acute inflammatory condition caused by major surgical procedures, trauma, and burns (35), showed overexpression in the plasma from patients with SARS. Recently, fetuin has been shown to play a role in the removal of apoptotic neutrophils through the activation of macrophages during the acute inflammatory response (36), perhaps accounting for the increase of plasma α-2-HS glycoprotein in SARS. Furthermore, inadequate clearance of apoptotic cells has been shown to result in pulmonary fibrosis (37) or autoimmunity (38) that has been demonstrated in some patients with SARS.
The alveolar macrophage acts as the guardian and the modulator at the alveolar–blood interface, serving as the front line of innate defense against respiratory infection (39). The initiation of macrophage activation leads to the inflammatory cascades, including the recruitment of activated neutrophils into the alveolar space, causing further tissue damage and a high degree of oxidative stress. In this study, two cellular antioxidants, glutathione peroxidase and Prx II, were identified in the plasma from patients of SARS that have not been previously reported in 2DE of plasma. Like catalase, glutathione peroxidase and Prx II are responsible for the removal of cytotoxic hydrogen peroxides with the former present in various intracellular compartments (40) and the latter in cytosol (41). In addition, other APPs such as haptoglobin and SAA have also been reported to have antioxidant properties. In animal studies, overexpression of haptoglobin can reduce oxidation-induced lung injury associated with hemolysis in mice (42). As for SAA, studies also showed that it could inhibit oxidative burst response in neutrophils (43). Taken together, the presence of antioxidant enzymes in the plasma of patients with SARS indicated that the antioxidative response plays a major role in SARS-CoV-induced oxidative injury in the lung.
The protein Prx II was found in the plasma of 23% of nontreated HIV-infected patients (detected by using ELISA) and shown to have anti-HIV activity in vitro as well (11). By using multidimensional chromatographic separation followed by MS, Prx II has been identified in normal plasma; however, it could not be detected in plasma from normal health individuals by Western blot analysis (44). By using 2DE followed by MS identification, on the other hand, it could be detected in human erythrocytes (45) and follicular fluid of mature human follicles (46) but not in unfractionated plasma. Unexpectedly, in this study we found that erythrocyte breakage due to mechanical hemolysis during blood collection also contributed to the appearance of Prx II in plasma. About 17–76% of SARS patients are reported to have hemolysis during their hospitalization (47, 48), possibly because of lipid peroxidation on the erythrocyte membrane caused by ribarivin therapy (49). Furthermore, the highly oxidative stress in the inflamed lung may further cause the damage of the erythrocyte membrane. The plasma PRX II may play a role for protecting erythrocytes from further damage by oxidative stress.
After excluding the hemolyzed samples, plasma Prx II was detected in SARS in about one-third of cases, slightly higher than the rate found in the HIV-infected patients. However, the possibility of coexistence of plasma Prx II from SARS and mechanical hemolysis could not be ruled out. The different expression of plasma Prx II between patients from Tri-Service General Hospital and from the Taiwan Center for Disease Control may be due to the discrepancy of disease severity. Patients from Tri-Service General Hospital were all transferred from other hospitals, whereas cases from the Taiwan Center for Disease Control were at the earlier stage, suggesting that the appearance of Prx II in plasma is not an early event of SARS. In terms of the source of plasma Prx II, the finding of an insignificant negative relationship between haptoglobin and Prx II suggested that in addition to endogeneous hemolysis, T lymphocytes may be another source of plasma Prx II. T lymphocytes isolated from patients with HIV infection have been shown to secrete Prx II after stimulation in vitro (11). In this study, although we did not isolate the T lymphocytes from SARS patients to perform the same testing, we did find Prx II in cultured Jurkat T cells in vitro. Because Jurkat cells are immortalized cancer cells, we do not know whether active secretion of Prx II is part of the defense mechanism against the oxidative stress in the culture system or is one of the specific functions of the T lymphocyte. Based on these findings, we suggested that T lymphocytes, at least in part, could secret Prx II into plasma, which is not SARS-specific because we have found that Prx II could be detected in the plasma of patients with pneumonia not caused by SARS-CoV infection and patients with gastric cancer (data not shown).
The function of Prx II is not clear in patients with SARS. Prx II belongs to a ubiquitous Prx family, which has been shown to have multiple functions such as enhancing natural killer cell activity (9, 50), increasing cell resistance to oxidative stress (51, 52), regulating transcription activator protein (53), protecting erythrocytes against oxidative stress (54), and anti-HIV activity (11). In light of its multifaceted biological functions, the plasma Prx II might play a multifunctional role during the acute inflammation induced by SARS-CoV infection or other pathogenic agents and might be considered as a biomarker and a potential therapeutic agent for treating infectious disorders.
In summary, SARS is a complicated disease in that the virus, SARS-CoV, has never infected humans before. In this study we have demonstrated that 2DE in combination with MS is a valuable proteomic tool for observation of the altered plasma proteins. Detailed analysis of these proteins could provide more information of the nature of the disease. The APPs found in patients with SARS suggest that the health status of the liver in patients may affect their defense against SARS-CoV infection. Finally, Prx II identified in this study may be used as one of the disease-associated biomarkers.
Acknowledgments
We thank Academia Sinica and Tri-Service General Hospital for support and the Taiwan Center for Disease Control for providing the SARS plasma samples.
Author contributions: J.-H.C., C.-H.W., and Y.-J.C. designed research; J.-H.C., Y.-W.C., S.-C.H., and K.-Y.C. performed research; J.-H.C., C.-W.Y., T.-S.C., and Y.-J.C. contributed new reagents/analytic tools; J.-H.C., Y.-W.C., C.-W.Y., T.-S.C., S.-C.H., K.-Y.C., A.C., F.-Y.C., C.-H.W., and Y.-J.C. analyzed data; and J.-H.C., Y.-W.C., C.-H.W., and Y.-J.C. wrote the paper.
Abbreviations: APP, acute phase protein; CoV, coronavirus; LC, liquid chromatography; MS/MS, tandem MS; SAA, serum amyloid A; SARS, severe acute respiratory syndrome; 2DE, 2D electrophoresis.
References
- 1.Fleischauer, A. T. (2003) Morb. Mortal. Wkly. Rep. 52, 226–228. [Google Scholar]
- 2.Ksiazek, T. G., Erdman, D., Goldsmith, C. S., Zaki, S. R., Peret, T., Emery, S., Tong, S., Urbani, C., Comer, J. A., Lim, W., et al. (2003) N. Engl. J. Med. 348, 1953–1966. [DOI] [PubMed] [Google Scholar]
- 3.Peiris, J., Lai, S., Poon, L., Guan, Y., Yam, L., Lim, W., Nicholls, J., Yee, W. K., Yan, W. W., Cheung, M. T., et al. (2003) Lancet 361, 1319–1325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Rota, P. A., Oberste, M. S., Monroe, S. S., Nix, W. A., Campagnoli, R., Icenogle, J. P., Peñaranda, S., Bankamp, B., Maher, K., Chen, M. H., et al. (2003) Science 300, 1394–1399. [DOI] [PubMed] [Google Scholar]
- 5.Marra, M. A., Jones, S. J. M., Astell, C. R., Holt, R. A., Brooks-Wilson, A., Butterfield, Y. S. N., Khattra, J., Asano, J. K., Barber, S. A., Chan, S. Y., et al. (2003) Science 300, 1399–1404. [DOI] [PubMed] [Google Scholar]
- 6.Anand, K., Ziebuhr, J., Wadhwani, P., Mesters, J. R. & Hilgenfeld, R. (2003) Science 300, 1763–1767. [DOI] [PubMed] [Google Scholar]
- 7.Li, W., Moore, M. J., Vasilieva, N., Sui, J., Wong, S. K., Berne, M. A., Somasundaran, M., Sullivan, J. L., Luzuriaga, K., Greenough, T. C., et al. (2003) Nature 426, 450–454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Anderson, N. L. & Anderson, N. G. (2002) Mol. Cell. Proteomics 1, 845–867. [DOI] [PubMed] [Google Scholar]
- 9.Shau, H., Butterfield, L. H., Chiu, R. & Kim, A. (1994) Immunogenetics 40, 129–134. [DOI] [PubMed] [Google Scholar]
- 10.Sauri, H., Butterfield, L., Kim, A. & Shau, H. (1995) Biochem. Biophys. Res. Commun. 208, 964–969. [DOI] [PubMed] [Google Scholar]
- 11.Geiben-Lynn, R., Kursar, M., Brown, N. V., Addo, M. M., Shau, H., Lieberman, J., Luster, A. D. & Walker, B. D. (2003) J. Biol. Chem. 278, 1569–1574. [DOI] [PubMed] [Google Scholar]
- 12.Magi, B., Bini, L., Fossi, A., Sanchez, J., Hochstrasser, D., Paesano, S., Raggiaschi, R., Santucci, A., Pallini, V. & Rottoli, P. (2002) Electrophoresis 23, 3434–3444. [DOI] [PubMed] [Google Scholar]
- 13.Wattiez, R., Hermans, C., Cruyt, C., Bernard, A. & Falmagne, P. (2000) Electrophoresis 21, 2703–2712. [DOI] [PubMed] [Google Scholar]
- 14.Lindahl, M., Stahlbom, B., Svartz, J. & Tagesson, C. (1998) Electrophoresis 19, 3222–3229. [DOI] [PubMed] [Google Scholar]
- 15.Moshage, H. (1997) J. Pathol. 181, 257–266. [DOI] [PubMed] [Google Scholar]
- 16.Dowton, S. B. & Colton, H. R. (1988) Semin. Hematol. 25, 84–90. [PubMed] [Google Scholar]
- 17.Miwata, H., Yamada, T., Okada, M., Kudo, T., Kimura, H. & Morishima, T. (1993) Arch. Dis. Child. 68, 210–214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jahoor, F., Gazzard, B., Phillips, G., Sharpstone, D., Delorsario, M., Frazer, M. E., Heird, W., Smith, R. & Jackson, A. (1999) Am. J. Physiol. 276, E1092–E1098. [DOI] [PubMed] [Google Scholar]
- 19.Eckersall, P. D., Duthie, S., Toussaint, M. J., Gruys, E., Heegaard, P., Alava, M., Lipperheide, C. & Madec, F. (1999) Adv. Vet. Med. 41, 643–655. [DOI] [PubMed] [Google Scholar]
- 20.Raynes, J. G. & Cooper, E. H. (1983) J. Clin. Pathol. 36, 798–803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Juan, H. F., Chen, J. H., Hsu, W. T., Huang, S. C., Chen, S. T., Lin, Y. C., Chang, Y. W., Chiang, C. Y., Wen, L. L., Chan, D. C., et al. (2004) Proteomics 4, 2766–2775. [DOI] [PubMed] [Google Scholar]
- 22.Eisen, D. P. & Minchinton, R. M. (2003) Clin. Infect. Dis. 37, 1496–1505. [DOI] [PubMed] [Google Scholar]
- 23.Gordon, S. (2002) Cell 111, 927–930. [DOI] [PubMed] [Google Scholar]
- 24.Hartshorn, K. L., Sastry, K., White, M. R., Anders, E. M., Super, M., Ezekowitz, R. A. & Tauber, A. I. (1993) J. Clin. Invest. 91, 1414–1420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Turner, M. W. (1996) Immunol. Today 17, 532–540. [DOI] [PubMed] [Google Scholar]
- 26.van de Wetering, J. K., van Golde, L. M. G. & Batenburg, J. J. (2004) Eur. J. Biochem. 271, 1229–1249. [DOI] [PubMed] [Google Scholar]
- 27.Kamper, E. F., Kopeikina, L. T., Koutsoukos, V. & Stavridis, J. (1997) J. Rheumatol. 24, 262–268. [PubMed] [Google Scholar]
- 28.Hogdall, C. K. (1998) APMIS 86, 1–31. [Google Scholar]
- 29.Westergaard, U. B., Andersen, M. H., Heegaard, C. W., Fedosov, S. N. & Petersen, T. E. (2003) Eur. J. Biochem. 270, 1850–1854. [DOI] [PubMed] [Google Scholar]
- 30.Hoj Nielsen, A. & Knudsen, F. (1987) Scand. J. Clin. Lab. Invest. 47, 175–178. [PubMed] [Google Scholar]
- 31.Caulfield, M., Lavender, P., Farrall, M., Munroe, P., Lawson, M., Turner, P. & Clark, A. J. (1994) N. Engl. J. Med. 330, 1629–1633. [DOI] [PubMed] [Google Scholar]
- 32.Krakoff, L. R. (1973) J. Clin. Endocrinol. Metab. 37, 110–117. [DOI] [PubMed] [Google Scholar]
- 33.Doherty, N. S., Littman, B. H., Reilly, K., Swindell, A. C. & Buss, J. M. (1998) Electrophoresis 19, 355–363. [DOI] [PubMed] [Google Scholar]
- 34.Schlossmacher, P., Hasselmann, M., Meyer, N., Kara, F., Delabranche, X., Kummerlen, C. & Ingenbleek, Y. (2002) Clin. Chem. Lab. Med. 40, 1339–1343. [DOI] [PubMed] [Google Scholar]
- 35.Lebreton, J. P., Joisel. F., Raoult, J. P., Lannuzel, B., Rogez, J. P. & Humbert, G. (1979) J. Clin. Invest. 64, 1118–1129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Jersmann, H. P. A., Dransfield, I. & Hart, S. P. (2003) Clin. Sci. 105, 273–278. [DOI] [PubMed] [Google Scholar]
- 37.Hagimoto, N., Kuwano, K., Miyazaki, H., Kunitake, R., Fujita, M., Kawasaki, M., Kaneko Y. & Hara, N. (1997) Am. J. Respir. Cell Mol. Biol. 17, 272–278. [DOI] [PubMed] [Google Scholar]
- 38.Botto, M., Dell'Agnola, C., Bygrave, A. E., Thompson, E. M., Cook, H. T., Petry, F., Loos, M., Pandolfi, P. P. & Walport, M. J. (1998) Nat. Genet. 19, 56–59. [DOI] [PubMed] [Google Scholar]
- 39.Sibille, Y. & Reynolds, H. Y. (1990) Am. Rev. Respir. Dis. 141, 471–501. [DOI] [PubMed] [Google Scholar]
- 40.Asayama, K., Yokoda, S., Dobashi, K., Hayashibe, H., Kawaoi, A. & Nakazawa, S. (1994) Histochemistry 102, 213–219. [DOI] [PubMed] [Google Scholar]
- 41.Kang, S. W., Chae, H. Z., Seo, M. S., Kim, K., Baines, I. C. & Rhee, S. G. (1998) J. Biol. Chem. 273, 6297–6302. [DOI] [PubMed] [Google Scholar]
- 42.Yang, F., Haile, D. J., Berger, F. G., Herbert, D. C., Beveren, E. V. & Ghio, A. J. (2003) Am. J. Physiol. 284, L402–L409. [DOI] [PubMed] [Google Scholar]
- 43.Linke, R. P., Bock, V., Valet, G. & Rothe, G. (1991) Biochem. Biophys. Res. Commun. 176, 1100–1105. [DOI] [PubMed] [Google Scholar]
- 44.Pieper, R., Gatlin, C. L., Makusky, A. J., Russo, P. S., Schatz, C. R., Miller, S. S., Su, Q., McGrath, A. M., Estock, M. A., Parmar, P. P., et al. (2003) Proteomics 3, 1345–1364. [DOI] [PubMed] [Google Scholar]
- 45.Low, T. Y., Seow, T. K. & Chung, M. C. M. (2002) Proteomics 2, 1229–1239. [DOI] [PubMed] [Google Scholar]
- 46.Anahory, T., Dechaud, H., Bennes, R., Marin, P., Lamb, N. J. & Laoudj, D. (2002) Electrophoresis 23, 1197–1202. [DOI] [PubMed] [Google Scholar]
- 47.Wong, R. S. M., Wu, A., To, K. F., Lee, N., Lam, C. W. K., Wong, C. K., Chan, P. K. S., Ng, M. H. L., Yu, L. M., Hui, D. S., et al. (2003) Br. Med. J. 326, 1358–1362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Booth, C. M., Matukas, L. M., Tomlinson, G. A., Rachlis, A. R., Rose, D. B., Dwosh, H. A., Walmsley, S. L., Mazzulli, S. L., Avendano, M., Derkach, P., et al. (2003) J. Am. Med. Assoc. 289, 2801–2809. [DOI] [PubMed] [Google Scholar]
- 49.Scharte, M. & Fink, M. P. (2003) Crit. Care Med. 31, Suppl. 12, S651–S657. [DOI] [PubMed] [Google Scholar]
- 50.Shau, H., Gupta, R. K. & Golub, S. H. (1993) Cell. Immunol. 147, 1–11. [DOI] [PubMed] [Google Scholar]
- 51.Shau, H., Kim, A. T., Hedrick, C. C., Lusis, A. J., Tompkins, C., Finney, R., Leung, D. W. & Paglia, D. E. (1997) Free Radical Biol. Med. 22, 497–507. [DOI] [PubMed] [Google Scholar]
- 52.Sarafian, T. A., Rajper, N., Grigorian, B., Kim, A. & Shau, H. (1997) Free Radical Res. 26, 281–289. [DOI] [PubMed] [Google Scholar]
- 53.Shau, H., Huang, A. C. J., Faris, M., Nazarian, R., de Vellis, J. & Chen, W. (1998) Biochem. Biophy. Res. Commun. 249, 683–686. [DOI] [PubMed] [Google Scholar]
- 54.Lee, T. H., Kim, S. U., Yu, S. L., Kim, S. H., Park, D. S., Moon, H. B., Dho, S. H., Kwon, K. S., Kwon, H. J., Han, Y. H., et al. (2003) Blood 101, 5033–5038. [DOI] [PubMed] [Google Scholar]