Abstract
γ-Secretase activity is associated with a presenilin (PS)-containing macromolecular complex. Whether PS contains the active site of γ-secretase has been controversial. One challenge is to find PS that is engaged in the active γ-secretase complex at the cell surface, where some substrates appear to be processed. In this study, we developed an intact cell photolabeling technique that allows the direct visualization of active γ-secretase at the cell surface. We demonstrated that active γ-secretase is present in the plasma membrane. Moreover, the PS1 heterodimer is specifically photolabeled at the cell surface by a potent inhibitor that binds to only the active γ-secretase. We also explored the cellular processing sites of γ-secretase for amyloid precursor protein (APP) and Notch by using small molecular probes. MRL631, a γ-secretase inhibitor that is unable to penetrate the cell membrane, significantly blocks γ-secretase-mediated Notch cleavage but has little effect on APP processing. These results indicate that Notch is processed at the cell surface and that the majority of APP is processed by intracellular γ-secretase. Furthermore, the fact that inhibitors first target γ-secretase in the plasma membrane for Notch processing, and not for APP, will have important implications for drug development to treat Alzheimer's disease and cancer.
Keywords: intact cell photolabeling, intramembrane protease, presenilin
Alzheimer's disease (AD) has become a growing public health crisis with the increase of the elderly population. β-Amyloid (Aβ) peptide, the major constituent of amyloid plaques, is believed to play a central role in the neuropathology of AD according to the “amyloid cascade hypothesis” (1). Aβ is generated from the amyloid precursor protein (APP) by two proteases, β-secretase and γ-secretase. γ-Secretase generates the C termini of Aβ that exists in two major forms consisting of: 40 (Aβ40) and 42 aa (Aβ42). Aβ42 is more prone to aggregation (2). Interventions that reduce Aβ or mitigate plaque formation are promising strategies for AD therapy (3). Because β- and γ-secretases are involved in the earliest steps of the pathogenic pathway of AD, they are prime targets for drug development to treat and/or prevent AD.
β-Secretase has been identified and validated as an aspartyl protease (4–7). However, identification of γ-secretase appears to be much more complicated. First, cleavage of APP by γ-secretase is an extraordinary event because the scissile bond appears to be situated within a transmembrane domain. Second, γ-secretase activity is catalyzed by a macromolecular complex composed of at least four proteins: presenilins (PSs), nicastrin, APH-1, and PEN-2 (8–10). Third, γ-secretase also cleaves Notch (11) and other membrane proteins (12). Therefore, γ-secretase, which serves as a highly effective posttranslational regulator of numerous proteins, controls many biological processes, such as cell differentiation, proliferation, and/or survival. Because PS-1 (PS1) and PS-2 (PS2) familial early-onset AD mutations increase the production of Aβ42, considerable effort has been made to investigate the relationship between PS and γ-secretase (3). The role of PS1 and PS2 in γ-secretase activity has been controversial (13). As a result, two models have been proposed. The first suggests that PS is a chaperone/trafficking protein, which brings the substrate and protease together and/or assembles the γ-secretase complex. There is evidence that PS1 regulates the glycosylation and intracellular trafficking of APP, components of γ-secretase complex, and other membrane proteins (14–18). The second model suggests that PS contains the active site of γ-secretase. Mutagenesis of two Asp residues of PS1 in the transmembrane domains significantly reduces the γ-secretase-mediated APP cleavage (19). More importantly, potent transition-state aspartyl inhibitors directly bind to PS1 and PS2 (20). Furthermore, a PS1-like transmembrane protein displayed signal peptide peptidase activity when it was overexpressed in yeast (21). However, the assignment of γ-secretase activity to the PSs has been challenging because PS1 and the γ-secretase substrates do not appear to be colocalized to the same subcellular site, termed the “spatial paradox” (13, 22, 23). PSs predominantly reside in the endoplasmic reticulum and the early compartments of the secretory apparatus (24–26), whereas the γ-secretase-mediated substrate processing seems to occur in the late secretory compartments and plasma membrane (27–32). Recent studies (33–36) that have discovered low levels of PS1 in the cell surface, lysososome, endosome, and trans-Golgi network seem to resolve the paradox. However, another discrepancy observing that colocalization of the APP substrate and PS1 in the endoplasmic reticulum did not generate the γ-secretase-cleaved products was encountered (23). We previously reported (37) that only a small portion of the PS1 in cellular membranes is engaged in the active γ-secretase complex. Therefore, localization of PS1 itself may not reflect the activity of γ-secretase, and there is no direct evidence to demonstrate that active γ-secretase is present in the cell plasma membrane. In addition to APP, other cell surface proteins are processed by γ-secretase. The cellular action sites of these proteins by γ-secretase have not been established. It is critical to define the localization of γ-secretase and its action sites for different substrates to develop effective therapies for AD, and to understand the molecular basis of regulated intramembrane proteolysis (38). Developing novel functional approaches to detect active γ-secretase or PS1 associated with the active secretase in cells is required to unravel the intricacies of the enigmatic γ-secretase and its substrate processing.
In the present study, we aim to define the γ-secretase-mediated processing of APP and Notch at the cellular level and seek direct evidence that active γ-secretase exists in the cell membrane by using small molecular probes. We discovered a distinctive γ-secretase inhibitor and applied it to examine the processing of APP and Notch. We also developed an intact cell photoaffinity technique by using the active-site targeted inhibitors to localize the active γ-secretase at the cell surface. Moreover, we demonstrated that active γ-secretase is present in the plasma membrane by using activity-based probes.
Materials and Methods
γ-Secretase Inhibitors and Cell-Based Assay for Aβ40 Production. Three γ-secretase inhibitors were used for this study (Fig. 1). L-685,458 was resynthesized in our laboratory (39). MRL631 and MRL505 were kindly provided by Merck Research Laboratories. HEK293 cells were transiently transfected with an expression vector containing the fragment of APP corresponding to the membrane-bound C-terminal stub (APP-C99) produced by β-secretase (37). pGreen-Lantern-1, which contains the gene for the green fluorescence protein (Life Technologies, Grand Island, NY), was also cotransfected for normalization of the transfection efficiency. After transfection for 24 h, fresh media and inhibitors were added to the cells. The conditioned media were removed and assayed for Aβ(X-40) production 24 h later. Aβ40 was detected with biotinylated 4G8 and ruthenylated G2–10 Abs (40).
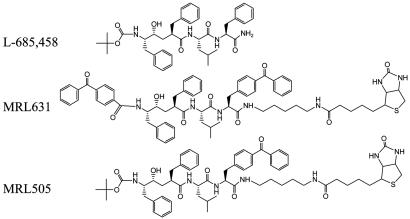
Fig. 1.
Chemical structures of L-685,458, MRL631, and MRL505. Both MRL631 and MRL505 contain a benzophenone moiety that can covalently incorporate into the target proteins after photoactivation.
Notch-Mediated Hes1-Luciferase Reporter Assay. The Notch-Hes1 reporter assay was modified from procedures as described in ref. 41. HEK293 cells were plated in a six-well dish (≈5 × 105 cells) and transiently cotransfected with the following plasmids: luciferase reporter construct Hes1-Luc (0.3 μg), Notch construct pCS2-ΔEMV-6MT (also named Notch ΔE) (3.0 μg), which contains the transmembrane and intracellular domains of Notch1 or control vector pCS2 vector (3.0 μg), and pCH110 (0.1 μg), which expresses β-galactosidase (Amersham Pharmacia) for normalization of the transfection efficiency and luciferase activity. The cells were transfected with these plasmids for 24 h, and then fresh media and inhibitors were added to the cells. The cells were harvested after 24 h and lysed for measuring luciferase activity by using the Luciferase Reporter Gene Assay kit (Roche). The luciferase activity was normalized according to β-galactosidase activity.
Cell Permeabilization with Streptolysin-O (SLO). Cells were permeabilized with SLO (Sigma) as described in ref. 6. SLO was activated by incubation with dithiotreiotol (5 mM) at 37°C for 2 h. HEK293-APP695 cells were washed with PBS without Ca2+ and incubated with the activated SLO (100 ng/ml) plus MRL631 (1 μM) or L-685,458 (1 μM) for 15 min at 37°C. Then fresh medium was added to the cells. The conditioned media were removed and assayed for Aβ (X-40) production 24 h later. The cell viability was determined by AlamarBlue assay.
Immunostaining of Cell-Surface APP. Cells grown in Lab-Tek CC2 chamber slide were incubated for 24 h in the absence or presence of 1 μM inhibitor and then washed in cold PBS supplemented with 1 mM CaCl2 and 0.5 mM MgCl2 (PCM buffer). Live cells without previous fixation or permeabilization were stained with mAb P2–1 (9.6 μg/ml) on ice. After 60 min the cells were washed with cold PCM and incubated with FITC-conjugated anti-mouse IgG (7.5 μg/ml) (Jackson ImmunoResearch) and rhodamine-conjugated wheat germ agglutinin (WGA, 10 μg/ml, Vector Laboratories) on ice for 1 h. Then the cells were washed with PCM and fixed with 4% paraformaldehyde (Sigma) in PBS and mounted by Vectashield-mounting medium with DAPI (Vector Laboratories). Images were examined by Nikon inverted microscope Eclipse TE2000-S with the TE-FM epi-fluorescence attachment.
Intact Cell Photolabeling and Immunostaining. Cells grown in Lab-Tek CC2 chamber slide were washed with PCM and incubated with the photoreactive γ-secretase inhibitor (MRL631 or MRL505) at 25 nM on ice for 1 h in the absence or presence of L-685,458 (2 μM). Then the cells were photolyzed for 45 min with a Rayonet RPR-200 photochemical reactor with PRP-3500A lamps (Southern New England Ultraviolet, Hamden, CT) under cold conditions and washed with ice-cold PCM. The washed cells were incubated with FITC-conjugated antibiotin IgG (10 μg/ml) and rhodamine-conjugated WGA on ice for 1 h. Then the cells were fixed with 4% paraformaldehyde in PBS and mounted by Vectashield with DAPI. Immunofluorescence staining was examined by Zeiss LSM 510 confocal laser scanning microscope.
Western Blotting. The cells were plated in 6-cm Petri dishes and photolabeled with MRL631 as described earlier. The labeled cells were washed with cold PBS three times and lysed in PBS buffer containing 0.5% 3-[(3-cholamidopropyl)dimethylammonio]-2-hydroxy-1-propanesulfonate, 0.5% deoxycholate, and 1% Nonidet P-40. The cell lysates were centrifuged and the resulting supernatant was incubated with Streptavidin beads (Pierce). The beads were washed, and the bound proteins were eluted by boiling for 5 min in SDS sample buffer. The eluted proteins were separated by SDS/PAGE and were analyzed by Western blotting by using anti-PS1-N-terminal fragment (NTF) and anti-PS1-C-terminal fragment (CTF) Abs.
Results
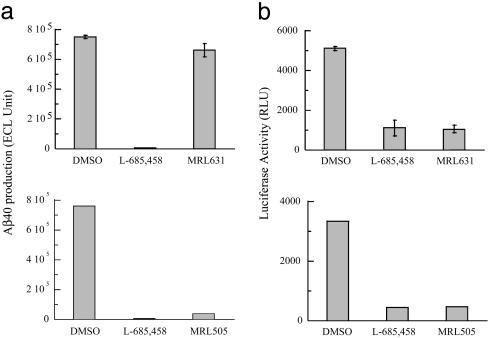
Processing of Notch and APP Can Be Selectively Blocked by MRL631. Our previous study demonstrated that MRL631 and MRL505 (Fig. 1) are potent inhibitors for solubilized γ-secretase and bind directly to PS1 and PS2 (20, 42). The potential application of these compounds for cellular studies was explored. First, their potency to inhibit APP processing was examined. Because cleavage of APP by α-or β-secretase appears to be a prerequisite for γ-secretase-mediated processing, HEK293 cells were transiently transfected with an expression vector encoding the APP fragment (APP-C99) that corresponds to the β-secretase cleavage product. After inhibitor treatment of the transfected cells, secreted Aβ (X-40) was assayed from the conditioned medium. L-685,458 and MRL505 at the concentration of 1 μM inhibited secreted Aβ (X-40) 100% and 95.0%, respectively, as compared with the conditioned medium from cells transfected with controlled vector (Fig. 2a Lower); whereas MRL631 at 1 μM only inhibited 9 ± 4.8% Aβ production (Fig. 2a Upper).
Fig. 2.
Effect of L-685,458, MRL631, and MRL505 on the processing of APP and Notch (a) APP processing. Inhibition of cellular γ-secretase activity for processing APP by MRL631 (Upper) (mean ± SD, n = 3) and MRL505 (Lower) (the average of two experiments) is depicted. HEK293 cells were transfected with the expression vector (APP-C99) (37) and incubated in the absence or presence of γ-secretase inhibitors. The secreted Aβ(X-40) is detected by biotinylated 4G8 and ruthenylated G2–10, which specifically recognizes the C terminus of Aβ40. (b) Notch processing. Inhibition of Notch-induced luciferase reporter activity by MRL631 (Upper) (mean ± SD, n = 3) and MRL505 (Lower) (the average of two experiments) are depicted. HEK293 cells were transiently transfected with Hes1-luc and NotchΔE constructs, and luciferase activity was detected. A plasmid that encodes β-galactosidase was also cotransfected for normalization of the transfection efficiency and luciferase activity.
Second, the effect of the three compounds on Notch cleavage was evaluated. Activated Notch induces expression of the HES1 gene via the Notch intracellular domain. A Hes1-luciferase reporter system was used to investigate γ-secretase-mediated Notch signaling. NotchΔE corresponds to the membrane-bound C-terminal stub produced by S2 site protease and the transduction of the Notch signal is dependent on γ-secretase cleavage. HEK293 cells were transiently cotransfected with the NotchΔE plasmid and the Hes1-luc reporter vectors. After inhibitor treatment, the cells were lysed and assayed for luciferase assay. The background luciferase activity is defined as the activity in the presence of 1 μM L-685,458, which has been shown to inhibit γ-secretase-mediated Notch signaling (41, 43). MRL631 and MRL505 at 1 μM block NotchΔE cleavage-mediated luciferase activity and are as potent as L-685,458 (Fig. 2b). To check whether the loss of reporter activity directly resulted from inhibition of γ-secretase, the Notch intracellular domain vector was used for transfection. This vector contains only the intracellular domain of Notch that is responsible for target gene transactivation whose activation is therefore independent of γ-secretase. We determined that MRL631 has no effect on Notch-mediated luciferase activity in the Notch intracellular domain-transfected cells (data not shown). These results strongly suggest that MRL631 specifically inhibits the Notch-mediated Hes1-luciferase activity via blockage of γ-secretase.
To address whether MRL631 is capable of inhibiting the membrane form of γ-secretase for APP processing, we determined its ability to inhibit the membrane γ-secretase activity by using the C100FLAG substrate and HeLa S3 membrane and demonstrated that it is highly potent with an IC50 of 1.1 nM (data not shown). Hence, the fact that MRL631 has little effect on cellular APP processing may result from a lack of accessibility of MRL631 to γ-secretase inside the cells, rather than an inability to inhibit the protease.
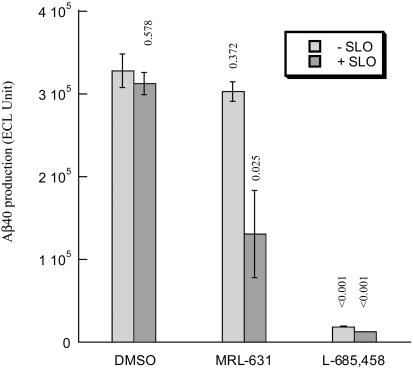
MRL631 Appreciably Inhibits APP Processing in Permeabilized Cells. To test whether ineffective inhibition of cellular APP processing by MRL631 results from its incapability to penetrate the cell membrane, we treated cells with SLO and then examine the effect of MRL631 and L-685,458 on Aβ production in the HEK293-APP695 cells. SLO is a bacterial protein that interacts with cholesterol to form holes in the plasma membrane to permeabilize cells. We found that in the absence of SLO, MRL631 only inhibited ≈8% Aβ production at 1 μM (Fig. 3), which is quite similar to the results observed in the cells transfected with APP-C99. However, MRL631 blocks 60% Aβ production in the permeabilized cells, whereas SLO itself at 100 ng/ml has little or no effect (<3%) on Aβ production. Furthermore, viability assay showed that there is no cytotoxicity with treatment of MRL631 or L-685,458 in the presence and absence of SLO (100 ng/ml) (data not shown). These observations demonstrate that reduction of Aβ production in the MRL631 and SLO-treated cells results from inhibition of γ-secretase, rather than from toxic effect. L-685,458 significantly inhibits Aβ production both in the absence and presence of SLO. These studies revealing that SLO potentiates the cellular inhibitory potency of MRL631 suggest that MRL631 is unable to cross plasma membrane to reach its intracellular targets.
Fig. 3.
Effect of MRL631 and L685,458 on production of Aβ in SLO-permeablized cells (n = 3). The HEK293-APP695 cells were treated with SLO at 100 ng/ml in presence of MRL631 (1 μM) or L-685,458 for 15 min, and then fresh medium was added. The secreted Aβ (X-40) was assayed. The P values calculated from the DMSO control and each treatment is displayed above the bars.
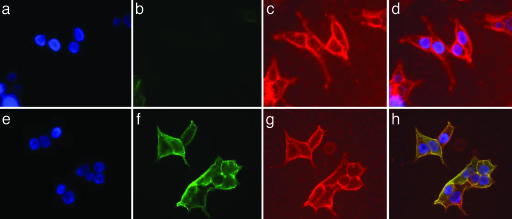
MRL631 and MRL505 Display Different Activities Against the APP Distribution at the Cell Surface. It has been reported that treatment of cells with γ-secretase inhibitors or expression of the loss-of-function PS1 mutant results in accumulation of APP at the cell surface by blocking its internalization (16, 35) and/or increasing APP trafficking from trans-Golgi network to plasma membrane (17). Therefore, we examined the effect of MRL631 and MRL505 on the distribution of APP at the cell surface. The HEK293-APP695 cells were treated with these compounds, and then the APP at the cell surface was probed by anti-APP mAb P2–1 that recognizes the NTF epitope and FITC-conjugated anti-mouse Ab (Fig. 4). The cells were also stained with rhodamine-conjugated WGA that labels proteins containing N-acetylglucosamine. Because the cells were not fixed or permeabilized, these Abs only recognize cell surface antigens. MRL505 increased the level of APP at the cell surface (Fig. 4f), and the APP colocalized with WGA-staining (Fig. 4 e–h). L-685,458 had the same effect on the APP cell surface distribution (data no shown). On the contrary, very little or no APP was detected on the cell surface (Fig. 4B) of the MRL631-treated cells (Fig. 4 a–d) or the control cells (DMSO only, data not shown). These studies clearly establish that MRL631 and MRL505 have different effects on the distribution of APP at the cell surface. Therefore, the regulation of APP distribution at the cell surface is dependent on intracellular γ-secretase, which is accessible by MRL505 and L-685,458 but not by MRL631.
Fig. 4.
Effect on MRL631 and MRL505 on the distribution of APP on the cell surface. The HEK293-APP695-stable cell line was incubated with the γ-secretase inhibitors at 1 μM for 24 h, and cells were probed with anti-APP mAb P2-1 that recognizes an N-terminal epitope of APP on ice for 1 h, and then added a FITC-anti-mouse Ab (green) and rhodamine-WGA (red) for 1 h on ice. The cells were fixed for nuclear staining. Staining of MRL631-treated (a Upper–d Upper) and MRL505-treated (e Lower–h Lower) cells is depicted. (a and e) DAPI staining for nuclei. (b and f) P2-1 staining for APP. (c and g) WGA staining for the cell-surface glycoproteins. (d) Overlay images of a–c.(h) Overlay images of e–g.
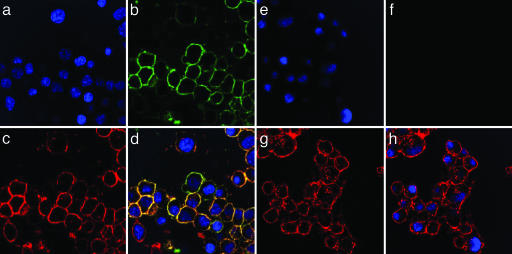
MRL631 Specifically Labels γ-Secretase in the Cell Plasma Membrane. To determine the localization of γ-secretase activity in living cells, we developed an intact cell photoaffinity-labeling technique with MRL631. MRL631 is an active site-directed inhibitor and only binds to the active form of γ-secretase. Photo-activation results in cross-linking of the biotin probes to the target proteins. After photolabeling, the N2a cells were immunostained with FITC-conjugated antibiotin IgG to define the localization of the biotinylated probes. The photolabeled cells were also stained with rhodamine-conjugated WGA. Because the cells were not permeabilized, WGA would only stain the cell surface glycoproteins. Antibiotin Ab detected signal (Green) in the MRL631-treated cells (Fig. 5b), but not in the presence of 2 μM L-685,458 (Fig. 5f). WGA stained both groups of cells (Fig. 5 c and g, red). Furthermore, the antibiotin and WGA staining colocalized (Fig. 5d), which suggests that MRL631 labels the target protein in the plasma membrane. The fact that L-685,458 blocks the MRL631 photoinsertion into the target proteins in the plasma membrane demonstrates that this labeling is specific.
Fig. 5.
Localization of the γ-secretase activity at the cell surface by using the active-site-based probe MRL631. The photolabeled N2a cells with MRL631 in absence (a–d) and presence (e–h) of L-685,458 were stained. (a and e) DAPI (blue) for the nucleus staining. (b and f) FITC-antibiotin IgG (green). (c and g) Rhodamine-WGA (red) for cell-surface staining. (d) Overlay images of a–c.(h) Overlay images of e–g.
To further demonstrate that MRL631 labeling is exclusive to γ-secretase, PS1- and PS2-deficient cells, in which γ-secretase activity for secreted Aβ and Notch processing is abolished, were used. The same intact cell photolabeling technique was applied to the PS1–/–PS2–/– blastocyst-derived cells by using MRL631. We could not detect any antibiotin signal for MRL631 labeling in these cells (data not shown). This result demonstrates that the MRL631 photolabeling is dependent on expression of PS1 and PS2.
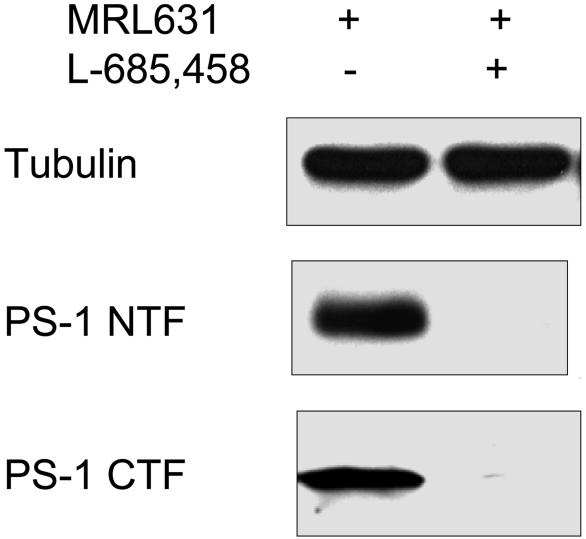
MRL631 Covalently Binds to PS1-NTF and CTF upon Photoactivation. We further analyzed whether PS1 is biotinylated in the plasma membrane at the cell surface by MRL631. After the intact cell labeling with MRL631 in the absence or presence of L-685,458, the cells were lysed, and the cell membrane was prepared and solubilized. To assure the same amount of protein was loaded onto the streptavidin beads in the proceeding step, the solubilized proteins were analyzed by Western analysis by using anti-tubulin Ab (Fig. 6 Top). The biotinylated proteins were captured by the streptavidin beads and released by boiling in SDS-sample buffer. The eluted proteins were separated by SDS/PAGE and probed by Western analysis by using anti-PS1-NTF and anti-PS1-CTF Abs. The MRL631 labeled PS1-NTF and PS1-CTF after photoactivation (Fig. 6 Middle and Bottom, respectively). Moreover, the photoincorporation of MRL631 into PS1 was blocked by an excess of L-685,458 (Fig. 6). We also analyzed nicastrin, which is a component of the γ-secretase complex, and found that it was not photolabeled by MR631 (data not shown).
Fig. 6.
Western analysis of the MRL631-photolabeled cells. After photolysis, the cells were lysed and solubilized. The biotinylated proteins were isolated by streptavidin beads and analyzed by Western blotting. (Top) Anti-β-tubulin. (Middle) Anti-PS1-NTF. (Bottom) Anti-PS1-CTF.
Discussion
Because γ-secretase is an appealing target for drug development to treat AD and because it cleaves multiple transmembrane protein substrates, it is critical to define its cellular action sites including those for APP processing and other substrates. Notch has been identified as another major substrate of γ-secretase (11). We aimed to dissect the APP and Notch cellular processing sites by using small molecule approaches. L-685,458 and MRL505 at 1 μM completely block the γ-secretase-mediated Aβ production and Notch signaling in cells, whereas MRL631 inhibits Notch but not APP processing. These studies distinguished MRL631 from L-685,458 and MRL505 inhibitors in cellular studies and strongly suggest that there are at least two cellular sites for γ-secretase action: one for Notch which is inhibitable by MRL631 and the other for APP which is not blocked by MRL631. Therefore, Notch and APP have distinct γ-secretase pathways. Our conclusion is consistent with earlier studies that showed that APP and Notch do not compete for γ-secretase activity in the cell (44, 45). A lack of competition between the two substrates could arise because the processing of Notch and APP occurs in different subcellular compartments, or because the concentration of both substrates is not saturated. Our studies clearly support the first hypothesis of subcellular compartmentalization of the two substrates. However, others have found that APP and Notch can compete with each other in a similar system (43, 46). These findings suggested that APP and Notch processing occur in the same subcellular compartments or that they significantly overlap, which directly contrasts our conclusion. We argue that the competition results from overproduction of both substrates, thereby altering the subcellular distribution of Notch and APP.
The fact that MRL631 is a potent inhibitor of γ-secretase hydrolysis of APP in vitro, but not in cells, suggests that the action site of γ-secretase for processing APP in cells is not accessible by this compound. This may result from MRL631 failing to get inside cells or from deactivation before reaching the target protease. Potentiation of MRL631 for inhibition of cellular APP processing by SLO strongly suggests that MRL631 is not able to penetrate the plasma membrane. This conclusion is also corroborated by another line of evidence from our current work. Treatment of the HEK293-APP695 cells with MRL631 has no effect on APP distribution, which is in contrast to that of other γ-secretase inhibitors, which are able to penetrate cells, such as MRL505 and L-685,458. It has been reported that a loss of function of PS1 or some γ-secretase inhibitors increase APP distribution on the cell surface (16, 35). Our studies suggest that MRL631 is not able to access intracellular γ-secretase for APP processing and APP trafficking. However, it interacts with γ-secretase residing at the cell surface for Notch processing. Taken together, these studies indicate that the cleavage of Notch by γ-secretase occurs at the cell surface and the majority of APP is processed by intracellular γ-secretase activity. Moreover, the intracellular γ-secretase activity affects APP distribution on the cell surface.
Notch proteolysis (S3 site) by γ-secretase is thought to take place near or in the cell surface (47). However, there is no direct experimental evidence to confirm this action yet. Notch has been shown to interact with PS1 in the endoplasmic reticulum and Golgi and then transport to the plasma membrane as a complex (33). Our inhibitor studies provide strong evidence that the event of γ-secretase-mediated cleavage occurs at the cell surface. These results suggest that the catalytic center of γ-secretase is exposed near or at the cell surface. This observation raises a critical question regarding regulated intramembrane proteolysis, which hypothesizes that proteolysis occurs within the membrane bilayer (38). Recently, reexamination of the topography of the seven-transmembrane model of PS1 suggested that the putative catalytic residues (Asp-257 and Asp-385) is located near or at the cell surface (48).
The finding that Notch is processed on the cell surface raises the question of whether active γ-secretase is present in the plasma membrane. Attempts to search for γ-secretase activity in the plasma membrane were focused on the localization of individual components of the putative γ-secretase complex. A small fraction of total PS1 has been found at the cell surface (33). It has been established that PSs are essential but not sufficient for γ-secretase activity, and only a small portion of PS1 resides in the active γ-secretase complex (37). Therefore, these studies only provide indirect evidence that PS1, engaged in the active γ-secretase, may exist in the plasma membrane. Here, our approach by using transition-state inhibitors that bind only to the active form of the protease directly detects the active γ-secretase. By using this approach, we detected endogenous active γ-secretase in the cell plasma membrane with the following observations: (i) MRL631 labels cell surface proteins that colocalize with WGA staining; (ii) the photolabeling can be blocked by an excess of L-685,458; (iii) MRL631 fails to label PS1 and PS2-deficient cells; and (iv) MRL631 specifically labels PS1-NTF and PS1-CTF at the cell surface. Therefore, our studies provide definitive evidence that PS1, engaged in active γ-secretase, is present in the cell plasma membranes. These studies help to resolve the spatial paradox and supply more evidence that PS contains the active site of γ-secretase. This discovery also provides critical insight into other cell surface proteins, which may be processed by γ-secretase and coupled with other proteases in the plasma membrane for signaling and cell adhesion.
In conclusion, our studies highlight the complexity of γ-secretase-mediated processing of multiple substrates and protein trafficking in cells. Elucidation of the action mechanism of MRL631 and MRL505 inhibitors allows us to distinguish between the cell surface γ-secretase and the intracellular γ-secretase. The former is responsible for Notch cleavage and the latter for APP processing and recycling. We detected in situ active γ-secretase at the cell surface by using transition-state inhibitors. These studies contribute to our understanding of intramembrane proteolysis and have important implications in drug development targeting APP for AD and Notch for cancer therapies.
Acknowledgments
We thank Drs. Elizabeth Chen Dodson, Ming-tain Lai, and Daria Hazuda (Merck) for kindly supplying γ-secretase inhibitors, Abs, cell lines, and other reagents; Dr. Katia Manova (Memorial Sloan-Kettering Cancer Center Molecular Cytology Core Facility) for help with confocal microscopy; Rophael Kopan (Washington University School of Medicine, St. Louis) and Jeffrey Nye (Johnson and Johnson, Skillman, NJ) for all of the Notch constructs; Sam Sisodia (University of Chicago) for the anti-PS1-CTF Ab; and Drs. Dorit Donoviel and Alan Bernstein (Samuel Lunenfeld Research Institute, Mt. Sinai Hospital, Toronto) for providing PS1- and PS2-deficient cells. This work was supported by Mr. William H. Goodwin and Mrs. Alice Goodwin and the Commonwealth Foundation for Cancer Research, the Experimental Therapeutics Center of Memorial Sloan-Kettering Cancer Center, and the William Randolph Hearst Fund in Experimental Therapeutics.
Author contributions: L.T. and Y.-M.L. designed research; L.T., Y.I.Y., and B.B. performed research; L.T. and Y.I.Y. analyzed data; and Y.-M.L. wrote the paper.
Abbreviations: AD, Alzheimer's disease; APP, amyloid precursor protein; PS, presenilin; NTF, N-terminal fragment; CTF, C-terminal fragment; SLO, streptolysin-O; WGA, wheat germ agglutinin; Aβ, β-amyloid.
References
- 1.Hardy, J. & Allsop, D. (1991) Trends Pharmacol. Sci. 12, 383–388. [DOI] [PubMed] [Google Scholar]
- 2.Jarrett, J. T., Berger, E. P. & Lansbury, P. T., Jr. (1993) Ann. N.Y. Acad. Sci. 695, 144–148. [DOI] [PubMed] [Google Scholar]
- 3.Li, Y.-M. (2001) Mol. Interven. 1, 198–207. [PubMed] [Google Scholar]
- 4.Vassar, R., Bennett, B. D., Babu-Khan, S., Kahn, S., Mendiaz, E. A., Denis, P., Teplow, D. B., Ross, S., Amarante, P., Loeloff, R., et al. (1999) Science 286, 735–741. [DOI] [PubMed] [Google Scholar]
- 5.Cai, H., Wang, Y., McCarthy, D., Wen, H., Borchelt, D. R., Price, D. L. & Wong, P. C. (2001) Nat. Neurosci. 4, 233–234. [DOI] [PubMed] [Google Scholar]
- 6.Walev, I., Bhakdi, S. C., Hofmann, F., Djonder, N., Valeva, A., Aktories, K. & Bhakdi, S. (2001) Proc. Natl. Acad. Sci. USA 98, 3185–3190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Luo, Y., Bolon, B., Kahn, S., Bennett, B. D., Babu-Khan, S., Denis, P., Fan, W., Kha, H., Zhang, J., Gong, Y., et al. (2001) Nat. Neurosci. 4, 231–232. [DOI] [PubMed] [Google Scholar]
- 8.De Strooper, B. (2003) Neuron 38, 9–12. [DOI] [PubMed] [Google Scholar]
- 9.Takasugi, N., Tomita, T., Hayashi, I., Tsuruoka, M., Niimura, M., Takahashi, Y., Thinakaran, G. & Iwatsubo, T. (2003) Nature 422, 438–441. [DOI] [PubMed] [Google Scholar]
- 10.Luo, W. J., Wang, H., Li, H., Kim, B. S., Shah, S., Lee, H. J., Thinakaran, G., Kim, T. W., Yu, G. & Xu, H. (2003) J. Biol. Chem. 278, 7850–7854. [DOI] [PubMed] [Google Scholar]
- 11.Kopan, R. & Goate, A. (2000) Genes Dev. 14, 2799–2806. [DOI] [PubMed] [Google Scholar]
- 12.Periz, G. & Fortini, M. E. (2004) J. Neurosci. Res. 77, 309–322. [DOI] [PubMed] [Google Scholar]
- 13.Sisodia, S. S., Annaert, W., Kim, S. H. & De Strooper, B. (2001) Trends Neurosci. 24, S2–S6. [DOI] [PubMed] [Google Scholar]
- 14.Naruse, S., Thinakaran, G., Luo, J. J., Kusiak, J. W., Tomita, T., Iwatsubo, T., Qian, X., Ginty, D. D., Price, D. L., Borchelt, D. R., et al. (1998) Neuron 21, 1213–1221. [DOI] [PubMed] [Google Scholar]
- 15.Nishimura, M., Yu, G., Levesque, G., Zhang, D. M., Ruel, L., Chen, F., Milman, P., Holmes, E., Liang, Y., Kawarai, T., et al. (1999) Nat. Med. 5, 164–169. [DOI] [PubMed] [Google Scholar]
- 16.Leem, J. Y., Saura, C. A., Pietrzik, C., Christianson, J., Wanamaker, C., King, L. T., Veselits, M. L., Tomita, T., Gasparini, L., Iwatsubo, T., et al. (2002) Neurobiol. Dis. 11, 64–82. [DOI] [PubMed] [Google Scholar]
- 17.Cai, D., Leem, J. Y., Greenfield, J. P., Wang, P., Kim, B. S., Wang, R., Lopes, K. O., Kim, S. H., Zheng, H., Greengard, P., et al. (2003) J. Biol. Chem. 278, 3446–3454. [DOI] [PubMed] [Google Scholar]
- 18.Wang, H., Luo, W. J., Zhang, Y. W., Li, Y. M., Thinakaran, G., Greengard, P. & Xu, H. (2004) J. Biol. Chem. 279, 40560–40566. [DOI] [PubMed] [Google Scholar]
- 19.Wolfe, M. S., Xia, W., Ostaszewski, B. L., Diehl, T. S., Kimberly, W. T. & Selkoe, D. J. (1999) Nature 398, 513–517. [DOI] [PubMed] [Google Scholar]
- 20.Li, Y. M., Xu, M., Lai, M. T., Huang, Q., Castro, J. L., DiMuzio-Mower, J., Harrison, T., Lellis, C., Nadin, A., Neduvelil, J. G., et al. (2000) Nature 405, 689–694. [DOI] [PubMed] [Google Scholar]
- 21.Weihofen, A., Binns, K., Lemberg, M. K., Ashman, K. & Martoglio, B. (2002) Science 296, 2215–2218. [DOI] [PubMed] [Google Scholar]
- 22.Annaert, W. & De Strooper, B. (1999) Trends Neurosci. 22, 439–443. [DOI] [PubMed] [Google Scholar]
- 23.Cupers, P., Bentahir, M., Craessaerts, K., Orlans, I., Vanderstichele, H., Saftig, P., De Strooper, B. & Annaert, W. (2001) J. Cell Biol. 154, 731–740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Walter, J., Capell, A., Grunberg, J., Pesold, B., Schindzielorz, A., Prior, R., Podlisny, M. B., Fraser, P., Hyslop, P. S., Selkoe, D. J., et al. (1996) Mol. Med. 2, 673–691. [PMC free article] [PubMed] [Google Scholar]
- 25.Annaert, W. G., Levesque, L., Craessaerts, K., Dierinck, I., Snellings, G., Westaway, D., George-Hyslop, P. S., Cordell, B., Fraser, P. & De Strooper, B. (1999) J. Cell Biol. 147, 277–294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kim, S. H., Lah, J. J., Thinakaran, G., Levey, A. & Sisodia, S. S. (2000) Neurobiol. Dis. 7, 99–117. [DOI] [PubMed] [Google Scholar]
- 27.Haass, C., Koo, E. H., Mellon, A., Hung, A. Y. & Selkoe, D. J. (1992) Nature 357, 500–503. [DOI] [PubMed] [Google Scholar]
- 28.Higaki, J., Quon, D., Zhong, Z. & Cordell, B. (1995) Neuron 14, 651–659. [DOI] [PubMed] [Google Scholar]
- 29.Perez, R. G., Soriano, S., Hayes, J. D., Ostaszewski, B., Xia, W., Selkoe, D. J., Chen, X., Stokin, G. B. & Koo, E. H. (1999) J. Biol. Chem. 274, 18851–18856. [DOI] [PubMed] [Google Scholar]
- 30.Peraus, G. C., Masters, C. L. & Beyreuther, K. (1997) J. Neurosci. 17, 7714–7724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Chyung, J. H. & Selkoe, D. J. (2003) J. Biol. Chem. 278, 51035–51043. [DOI] [PubMed] [Google Scholar]
- 32.Soriano, S., Chyung, A. S., Chen, X., Stokin, G. B., Lee, V. M. & Koo, E. H. (1999) J. Biol. Chem. 274, 32295–32300. [DOI] [PubMed] [Google Scholar]
- 33.Ray, W. J., Yao, M., Mumm, J., Schroeter, E. H., Saftig, P., Wolfe, M., Selkoe, D. J., Kopan, R. & Goate, A. M. (1999) J. Biol. Chem. 274, 36801–36807. [DOI] [PubMed] [Google Scholar]
- 34.Rechards, M., Xia, W., Oorschot, V. M., Selkoe, D. J. & Klumperman, J. (2003) Traffic 4, 553–565. [DOI] [PubMed] [Google Scholar]
- 35.Kaether, C., Lammich, S., Edbauer, D., Ertl, M., Rietdorf, J., Capell, A., Steiner, H. & Haass, C. (2002) J. Cell Biol. 158, 551–561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lah, J. J. & Levey, A. I. (2000) Mol. Cell. Neurosci. 16, 111–126. [DOI] [PubMed] [Google Scholar]
- 37.Lai, M. T., Chen, E., Crouthamel, M. C., DiMuzio-Mower, J., Xu, M., Huang, Q., Price, E., Register, R. B., Shi, X. P., Donoviel, D. B., et al. (2003) J. Biol. Chem. 278, 22475–22481. [DOI] [PubMed] [Google Scholar]
- 38.Brown, M. S., Ye, J., Rawson, R. B. & Goldstein, J. L. (2000) Cell 100, 391–398. [DOI] [PubMed] [Google Scholar]
- 39.Nadin, A., Lopez, J. M. S., Neduvelil, J. G. & Thomas, S. R. (2001) Tetrahedron 57, 1861–1864. [Google Scholar]
- 40.Li, Y. M., Lai, M. T., Xu, M., Huang, Q., DiMuzio-Mower, J., Sardana, M. K., Shi, X. P., Yin, K. C., Shafer, J. A. & Gardell, S. J. (2000) Proc. Natl. Acad. Sci. USA 97, 6138–6143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Berechid, B. E., Kitzmann, M., Foltz, D. R., Roach, A. H., Seiffert, D., Thompson, L. A., Olson, R. E., Bernstein, A., Donoviel, D. B. & Nye, J. S. (2002) J. Biol. Chem. 277, 8154–8165. [DOI] [PubMed] [Google Scholar]
- 42.Xu, M., Lai, M. T., Huang, Q., DiMuzio-Mower, J., Castro, J. L., Harrison, T., Nadin, A., Neduvelil, J. G., Shearman, M. S., Shafer, J. A., et al. (2002) Neurobiol. Aging 23, 1023–1030. [DOI] [PubMed] [Google Scholar]
- 43.Schroeter, E. H., Ilagan, M. X., Brunkan, A. L., Hecimovic, S., Li, Y. M., Xu, M., Lewis, H. D., Saxena, M. T., De Strooper, B., Coonrod, A., et al. (2003) Proc. Natl. Acad. Sci. USA 100, 13075–13080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Chen, F., Yu, G., Arawaka, S., Nishimura, M., Kawarai, T., Yu, H., Tandon, A., Supala, A., Song, Y. Q., Rogaeva, E., et al. (2001) Nat. Cell. Biol. 3, 751–754. [DOI] [PubMed] [Google Scholar]
- 45.Petit, A., St. George-Hyslop, P., Fraser, P. & Checler, F. (2002) Biochem. Biophys. Res. Commun. 290, 1408–1410. [DOI] [PubMed] [Google Scholar]
- 46.Berezovska, O., Jack, C., Deng, A., Gastineau, N., Rebeck, G. W. & Hyman, B. T. (2001) J. Biol. Chem. 276, 30018–30023. [DOI] [PubMed] [Google Scholar]
- 47.Selkoe, D. & Kopan, R. (2003) Annu. Rev. Neurosci. 26, 565–597. [DOI] [PubMed] [Google Scholar]
- 48.Dewji, N. N., Valdez, D. & Singer, S. J. (2004) Proc. Natl. Acad. Sci. USA 101, 1057–1062. [DOI] [PMC free article] [PubMed] [Google Scholar]