Abstract
The inflammatory middle ear disease known as otitis media can become chronic or recurrent in some cases due to failure of the antibiotic treatment to clear the bacterial etiological agent. Biofilms are known culprits of antibiotic-resistant infections; however, the mechanisms of resistance for non-typeable Haemophilus influenzae biofilms have not been completely elucidated. In this study, we utilized in vitro static biofilm assays to characterize clinical strain biofilms and addressed the hypothesis that biofilms with greater biomass and/or thickness would be more resistant to antimicrobial-mediated eradication than thinner and/or lower biomass biofilms. Consistent with previous studies, antibiotic concentrations required to eliminate biofilm bacteria tended to be drastically higher than concentrations required to kill planktonic bacteria. The size characterizations of the biofilms formed by the clinical isolates were compared to their minimum biofilm eradication concentrations for four antibiotics. This revealed no correlation between biofilm thickness or biomass and the ability to resist eradication by antibiotics. Therefore, we concluded that biofilm size does not play a role in antibiotic resistance, suggesting that reduction of antibiotic penetration may not be a significant mechanism for antibiotic resistance for this bacterial opportunist.
Keywords: otitis media, Haemophilus, biofilm, antibiotic
Surprisingly, thickness, density and biomass of biofilms formed by non-typeable Haemophilus influenzae were not significantly correlated with resistance properties.
INTRODUCTION
Otitis media (OM) is a highly burdensome disease that affects most children by the age of 3 (Klein 2000; Pichichero 2013; Dickson 2014). The Gram-negative bacterium non-typeable Haemophilus influenzae (NTHi) is one of the leading causes of OM. While NTHi is able to live commensally in the nasopharynx, inflammation and dysfunction of the Eustachian tube may allow the bacteria to enter the middle ear space and establish an infection (Pichichero 2013). NTHi is a highly genetically diverse species, with most clinical isolates being unique from one another (Erwin et al.2008; Lacross et al.2008; Kaur et al.2011).
Along with NTHi, Streptococcus pneumoniae and Moraxella catarrhalis are the other most common bacterial causes of OM, and the disease can often be polymicrobial in nature (Murphy et al.2009; Holder et al.2012). Identification of the etiological agent of a case of OM typically remains undetermined. Therefore, OM is treated empirically with antibiotics, with high-dose amoxicillin being the first-line recommendation (Lieberthal et al.2013). However, treatment failure and relapse can occur, resulting in chronic or recurrent infections that may require multiple courses of antibiotics (Sox et al.2008; Pichichero 2013; Dickson 2014).
A significant reason antimicrobial therapy is often unsuccessful is that NTHi is able to form biofilms in the middle ear, which have been observed in both the chinchilla model of OM and in the middle ears of children with OM (Ehrlich et al.2002; Hall-Stoodley et al.2006). Enhanced tolerance to antimicrobials and evasion of the host immune defenses are attributes of biofilm communities. Several mechanisms have been described that could contribute to the increased antibiotic resistance, including a reduced ability of the drug to penetrate the biofilm structure (Costerton, Stewart and Greenberg 1999; Mah and O'Toole 2001; Fux et al.2005; Slinger et al.2006).
In this study, we have isolated clinical strains of NTHi from children undergoing tympanostomy tube placement in order to characterize their biofilms and antibiotic susceptibility profiles. We hypothesized that NTHi strains forming larger (in terms of biomass or thickness) biofilms would be more resistant to eradication by antibiotics than strains forming smaller biofilms.
METHODS
Bacterial strains and growth conditions
Eight NTHi clinical strains were isolated and used in this study: Otis1, Otis2, Otis3, Otis4, Otis6, Otis7, Otis8 and Otis10. NTHi strains were grown on brain heart infusion (BHI) agar (BD) supplemented with hemin (10 μg mL−1; MP Biomedicals) and NAD (10 μg mL−1; Sigma); this media is referred to as supplemented BHI (sBHI). For in vitro assays, NTHi strains were grown in sBHI broth.
In order to isolate clinical NTHi strains, middle ear fluid samples from children undergoing placement of tympanostomy tubes at Wake Forest Baptist Hospital were collected as discard tissue samples with no patient identifiers. This work was reviewed and approved as an exempt study by the Wake Forest Internal Review Board. To isolate clinical strains, middle ear fluids were plated on sBHI supplemented with 3 mg mL−1 vancomycin (Sigma) and incubated overnight at 37°C and 5% CO2. Individual colonies were selected based on colony morphology and confirmed to be NTHi by Gram stain and requirement for factors X (hemin) and V (NAD), and then passed onto sBHI agar plates. Overnight plates of the isolates were swabbed, and the bacteria were resuspended, and frozen for future use in 2 × BHI broth and 50% glycerol (1:1) at −80°C.
Crystal violet assay
NTHi strains were inoculated to ∼1 × 108 colony forming units (CFU) mL−1 in sBHI broth and seeded into 96-well plates (Corning). After incubation at 37°C and 5% CO2 for 4, 12 or 24 h, biofilm supernatants were removed and the biofilms were washed with water twice, and allowed to dry. The biofilms were then stained with 0.1% crystal violet (Sigma) for 30 min at room temperature, the stain was removed and the biofilms were washed four times with water. The biofilms were solubilized in 100% ethanol and the OD540 was measured using a BioTek ELx800 plate reader.
Viability assay
NTHi strains were seeded into 24-well plates (Corning) at a concentration of ∼1 × 108 CFU mL−1 and incubated at 37°C and 5% CO2. At 4, 12 and 24 h time points, the supernatants were removed and the biofilms were resuspended in phosphate-buffered saline (PBS; pH = 7.2), serially diluted and plated on sBHI agar. Plates were incubated for ∼24 h at 37°C and 5% CO2 before colonies were enumerated in order to determine viable bacterial counts.
Confocal laser scanning microscopy
Biofilms for confocal laser scanning microscopy (CLSM) were established by seeding 1 mL of bacterial suspension at ∼1 × 108 CFU mL−1 in Lab-TekII two-chamber no. 1.5 German coverglass slides (Nunc) and incubating for 24 h at 37°C and 5% CO2. The supernatants were removed and the biofilms were washed once with PBS and stained with LIVE/DEAD BacLight Viability Kit (Invitrogen) for 30 min in the dark at room temperature. The stain was removed and 1 mL PBS was added to the biofilms. A Nikon Eclipse C1 confocal laser scanning microscope was used to acquire z-stack images of the biofilms (experiment repeated three times per strain, with six to nine frames of view each). These images were converted using Nikon Elements software and analyzed using COMSTAT v.1 in MATLAB R2014a (Heydorn et al.2000).
Minimum inhibitory concentration assay
Minimum inhibitory concentrations (MICs) were determined by broth microdilution assay in sBHI broth (Andrews 2001). Briefly, antibiotics at 2× concentrations were diluted 2-fold up a microtiter plate. NTHi suspension was added to the antibiotic at a 1:1 ratio (final concentration ∼105 CFU mL−1). The following antibiotics were used: amoxicillin (Sigma), ceftriaxone (Sigma), clarithromycin (Sigma) or azithromycin (USP Reference Standard). After incubation (37°C and 5% CO2) for 20 h, the MIC was determined visually and by measurement of the OD600.
Minimum biofilm eradication concentration assay
Static biofilms were established as described for the crystal violet assay. After 24 h of incubation, supernatants were removed and replaced with either the vehicle control in sBHI or various concentrations of amoxicillin, ceftriaxone, clarithromycin or azithromycin. Following additional 24 h of incubation, supernatants were removed, and biofilms were resuspended in PBS, serially diluted and plated onto sBHI agar. Limit of detection (LOD) = 1.5 × 103 CFU mL−1.
Statistics
Statistical analyses were carried out using GraphPad Prism 5 (GraphPad Software, San Diego, CA). Significance was determined using the Kruskal-Wallis non-parametric ANOVA, followed by Dunn's multiple comparisons test. Spearman's rank correlation test was used to measure correlation coefficients between biofilm size and minimum biofilm eradication concentration (MBECs).
RESULTS
NTHi clinical strains vary in biofilm size and structure
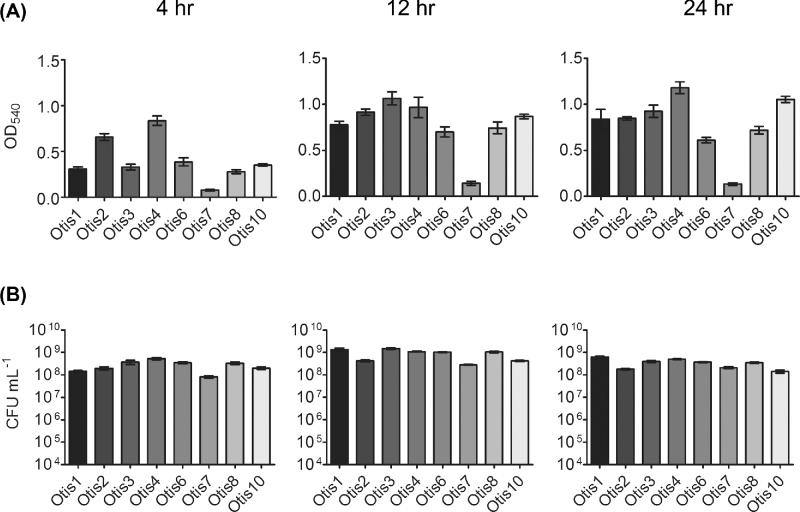
To characterize the biofilms formed by clinical isolates of NTHi, we measured total biomass and viability by crystal violet staining and enumeration of viable bacteria, respectively. Total biomass (including bacteria and extracellular matrix components) was assessed for static biofilms at 4, 12 and 24 h post-inoculation. Biomass varied significantly (P < 0.0001) among the eight clinical strains at all three time points (Fig. 1a). Initial attachment was represented at 4 h, with strains such as Otis2 and Otis4 already establishing dense formations. Biomass increased, to varying degrees among the strains, by 12 h; at 24 h, biofilms were well established. Significant variability (P < 0.0001) was also observed for the viability of the biofilms (Fig. 1b). However, bacterial counts ranged from approximately 108–109 CFU mL−1 (not falling below the initial inocula counts); therefore, we concluded that the biofilms were viable.
Figure 1.
Variability of biofilms formed by NTHi clinical isolates. (A) Biomass in static biofilms was measured by crystal violet staining. (B) Viable counts of biofilms as assessed by plate count.
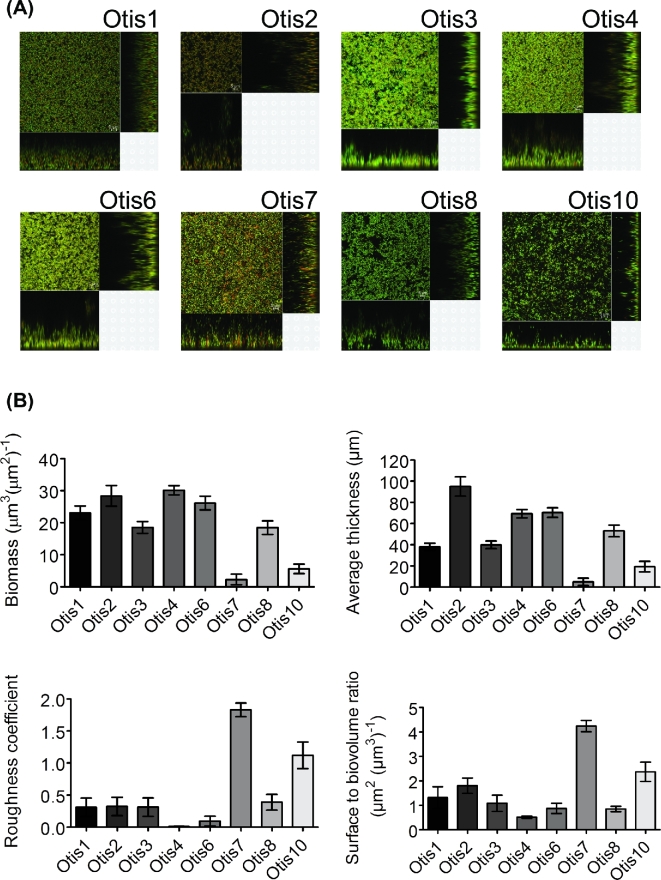
Further characterization was performed utilizing CLSM to visualize the biofilms. Static 24 h biofilms were labeled with BacLight LIVE/DEAD viability stain for imaging of the live bacteria, stained in green with SYTO9, and dead bacteria, stained in red with propidium iodide, within the biofilm. Figure 2a shows overview and cross-sectional images of representative biofilm sections of each NTHi strain. Differences in thickness, structure and areas of viable and dead bacteria were readily observed. Size and structural parameters were quantified from the z-stack images by COMSTAT analysis (Heydorn et al.2000) (Fig. 2b). Significant differences (P < 0.0001) in biofilm size, as measured by biomass and average thickness, were detected. Structural variation was also observed (P < 0.0001). Biofilms like those of Otis7 and Otis10 had higher roughness coefficients, indicating less smooth biofilm surfaces with more peaks and valleys. Otis7 and Otis10 also had higher surface to biovolume ratios suggesting increased surface exposed bacteria that could potentially encounter more nutrient flow (Heydorn et al.2000).
Figure 2.
(A) NTHi clinical strain biofilms vary in size and structure. Representative images are shown for the eight strain biofilms. (B) Z-stack images were acquired and analyzed by COMSTAT to quantify biomass, average thickness, roughness coefficient and surface to biovolume ratio.
These results demonstrate that the NTHi clinical isolates form biofilms to varying degrees in vitro. Based on our hypothesis, the larger biofilms like those of Otis2 or Otis4 should be less susceptible to antibiotic eradication than the smaller biofilms like that of Otis7.
MICs of NTHi clinical strains
The MIC assay is routinely used in clinical settings to determine bacterial susceptibility to antibiotics, and assist in informing physicians on how to treat infections (Andrews 2001). The results are a determination of susceptibility of bacteria in planktonic culture, which is often not a reflection of the state of the bacteria causing the infection. However, in order to compare the degree of antibiotic tolerance of biofilms to planktonic cultures of the clinical NTHi strains, we first needed to determine the concentration of antibiotic required to inhibit bacterial growth in the planktonic state. MICs were determined for four antibiotics that have been used in the treatment of OM (amoxicillin, ceftriaxone, clarithromycin and azithromycin) (Table 1). An important mechanism of β-lactam resistance seen in many NTHi strains is the ability to produce a β-lactamase (Tristram, Jacobs and Appelbaum 2007). All eight clinical NTHi isolates tested negative for β-lactamase production by nitrocefinase disk test (data not shown). Knowing the sensitivities of the strains in the planktonic state, as presented in Table 1, then allowed us to determine antibiotic concentrations with which to treat pre-established biofilms.
Table 1.
Minimal inhibitory concentrations for clinical isolates of NTHi (μg ml−1).
| Strain | Amoxicillin | Ceftriaxone | Clarithromycin | Azithromycin |
|---|---|---|---|---|
| OTIS1 | 1 | 0.004 | 4 | 1 |
| OTIS2 | 1 | 0.016 | 4 | 0.5 |
| OTIS3 | 1 | 0.008 | 4 | 1 |
| OTIS4 | 1 | 0.008 | 2 | 0.25 |
| OTIS6 | 0.5 | 0.008 | 4 | 1 |
| OTIS7 | 2 | 0.032 | 2 | 0.25 |
| OTIS8 | 2 | 0.008 | 8 | 2 |
| OTIS10 | 2 | 0.016 | 8 | 1 |
The ability to resist eradication by antibiotics is not dependent on biofilm size
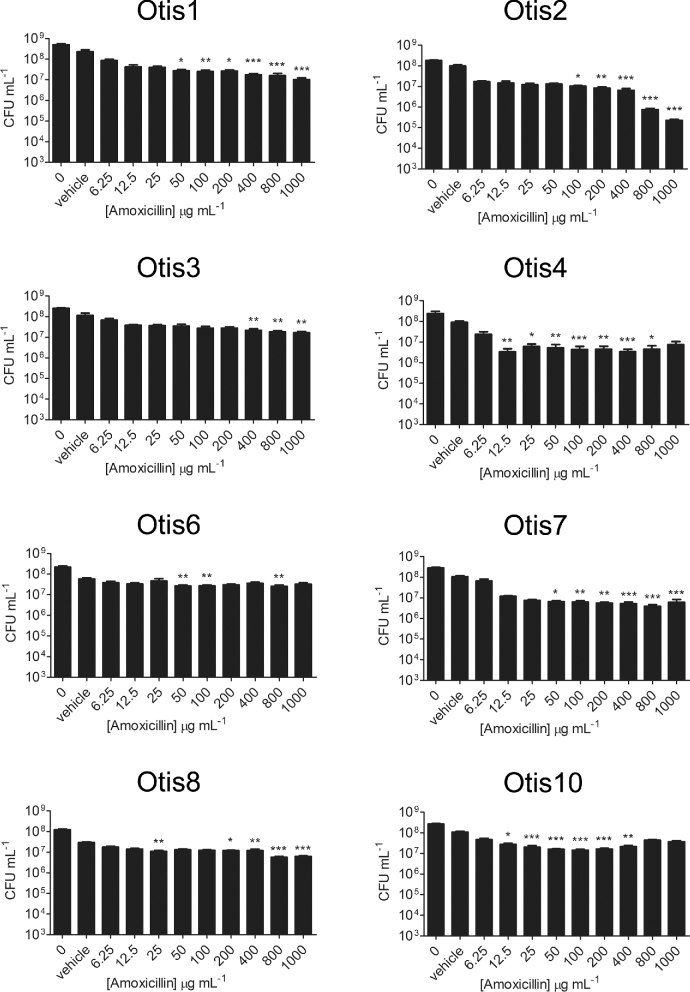
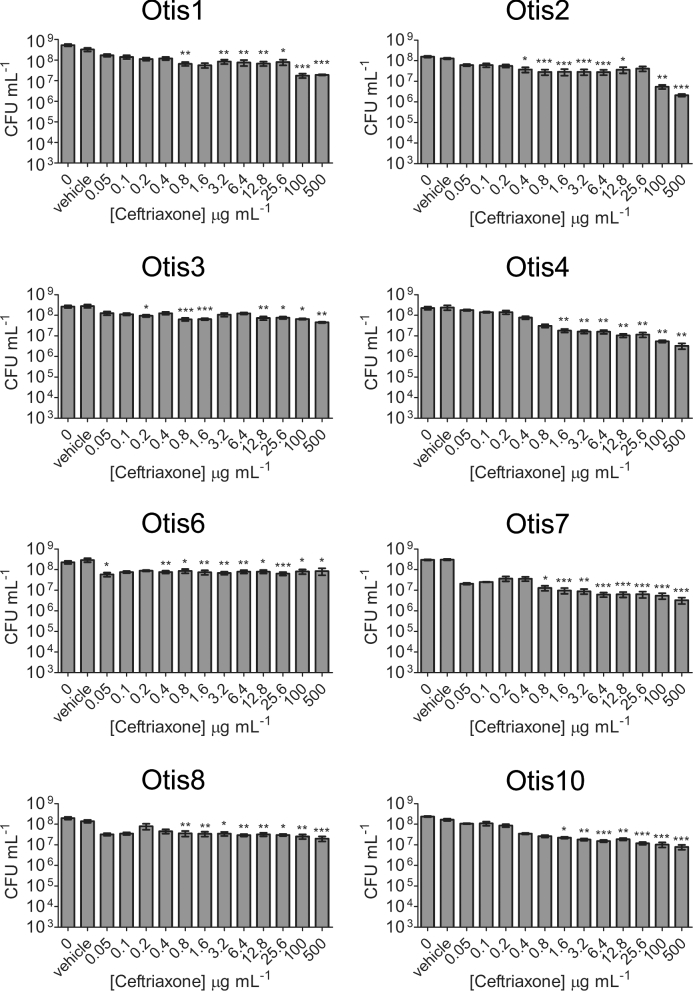
A well-described quality of biofilms is their increased tolerance to antimicrobial treatment compared to the bacteria in their planktonic states. We therefore tested a higher range of antibiotic concentrations against the biofilms formed by the NTHi clinical isolates in order to determine the concentration of antibiotic required to eradicate the biofilm bacteria (i.e. decrease viable counts to the LOD). Established 24 h static biofilms were treated with amoxicillin ranging from 6.25 to 1000 μg mL−1or ceftriaxone ranging from 0.05 to 500 μg mL−1, and viable bacteria were quantified after 24 h of exposure. Compared to the vehicle controls, statistically significant decreases in viability were detected with some β-lactam antibiotic concentrations (Figs 3 and 4). However, the most bacterial killing observed with either antibiotic was of Otis2, which was decreased by ∼3 logs post treatment with 1000 μg mL−1 amoxicillin (Fig. 3). This concentration of amoxicillin was 1000× higher than the MIC for Otis2. Additionally, the biofilm failed to be eradicated, leaving viable bacteria that may continue proliferating. These results demonstrated that the NTHi clinical isolate biofilms were all highly resistant to amoxicillin and ceftriaxone treatment, and that the MBECs of the β-lactam antibiotics were higher than the greatest concentrations tested.
Figure 3.
NTHi clinical strain biofilms are highly resistant to amoxicillin treatment. Static biofilms were treated with a range of concentrations of amoxicillin and then viable bacteria quantified by plate count. Results represent the means with standard error of the mean (SEM) of three experiments each done in triplicate. One-way ANOVA measured statistical significance of antibiotic-treated biofilms compared to the vehicle control (*P < 0.05; **P < 0.01; ***P < 0.001).
Figure 4.
NTHi clinical strain biofilms are resistant to high ceftriaxone concentrations. Static biofilms were treated with a range of ceftriaxone concentrations. Biofilm bacteria were plated for viability 24 h later. The results show the means with SEM of three experiments each done in triplicate. One-way ANOVA measured statistical significance of antibiotic treated biofilms compared to the vehicle control (*P < 0.05; **P < 0.01; ***P < 0.001).
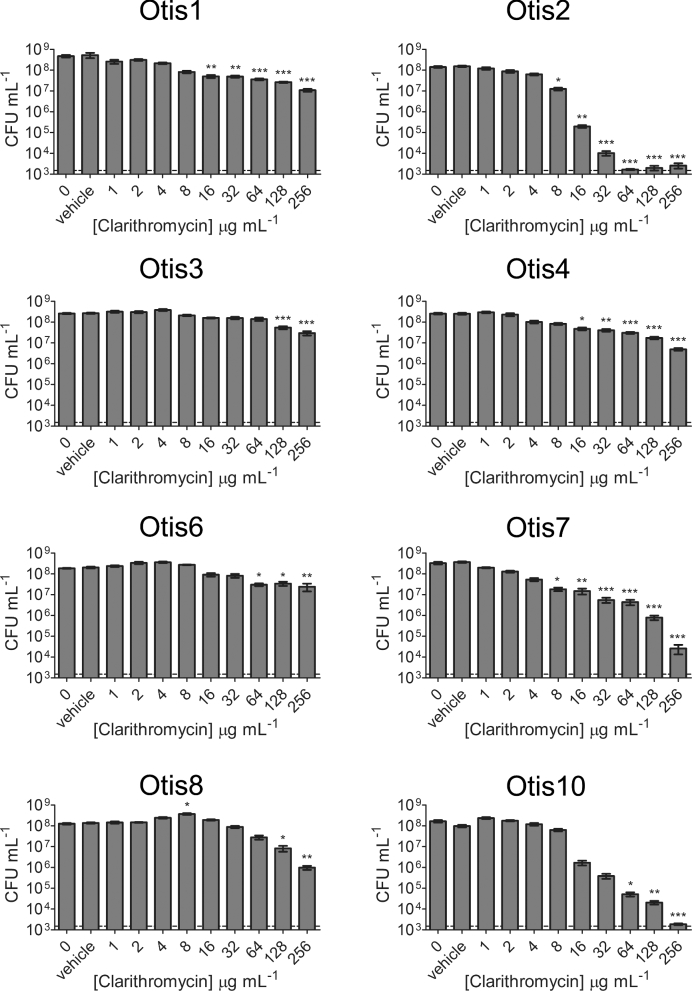
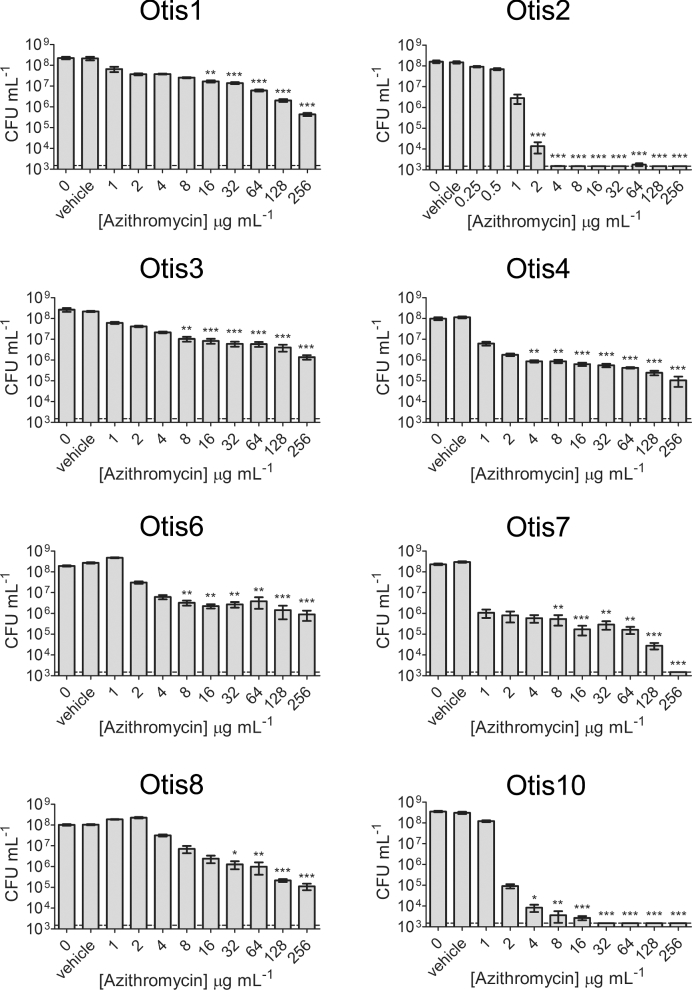
The macrolide antibiotics clarithromycin (1–256 μg mL−1) and azithromycin (0.25–256 μg mL−1) were also tested against the NTHi biofilms (Figs 5 and 6). Similar to the β-lactam antibiotic treatments, statistically significant killing generally occurred with higher clarithromycin and azithromycin concentrations; however, some strains were more sensitive to these antibiotics and some biofilms were eradicated at measured concentrations. Measureable MBECs of clarithromycin were determined for Otis2 (64 μg mL−1) and Otis10 (256 μg mL−1) (Fig. 5). MBECs of azithromycin were 4 μg mL−1 for Otis2, 256 μg mL−1 for Otis7 and 32 μg mL−1 for Otis10 (Fig. 6). These results suggest that macrolide antibiotics are slightly more effective against biofilms formed by some of these NTHi clinical isolates.
Figure 5.
Treatment of NTHi clinical strain biofilms with clarithromycin. A range of concentrations of clarithromycin were used to treat pre-established 24 h static biofilms. The results show the means with SEM of three experiments each done in triplicate. Statistical significance was determined by one-way ANOVA comparing the antibiotic-treated biofilms with the vehicle control (*P < 0.05; **P < 0.01; ***P < 0.001). The dashed line indicates the LOD.
Figure 6.
Azithromycin treatment of NTHi clinical strain biofilms. A range of concentrations of azithromycin were used to treat pre-established 24 h static biofilms. Biofilm bacteria were serially diluted and plated 24 h later in order to assess viability. Results pooled from three experiments done in triplicate are represented by the mean with SEM. Statistical significance was determined by one-way ANOVA comparing the antibiotic-treated biofilms with the vehicle control (*P < 0.05; **P < 0.01; ***P < 0.001). The LOD is indicated by the dashed line.
The MBEC results were summarized in Fig. 7. Unless specifically indicated on the graph, the MBEC was greater than the highest concentration tested for the antibiotic. The strains were ordered from highest to lowest biofilm biomass as determined by COMSTAT analyses of confocal images (Fig. 2b). Although all amoxicillin and ceftriaxone MBECs were beyond the highest concentrations measured for these experiments, some clarithromycin and azithromycin MBECs were measurable, within defined ranges, for biofilms with high biomass (Otis2) and low biomass (Otis10 and Otis7). Spearman correlations were calculated by changing the values calculated for biofilm thickness, CLSM biomass and CV biomass to percentages of the highest respective measurements, then comparing them to the MBEC values. MBECs higher than the greatest concentration tested were plotted as the next highest 2-fold dilution. No correlation coefficient had a P-value <0.39, suggesting that there was no correlation between the size of the biofilm and the ability to resist antibiotic treatment, regardless of the method for ranking biofilm size.
Figure 7.
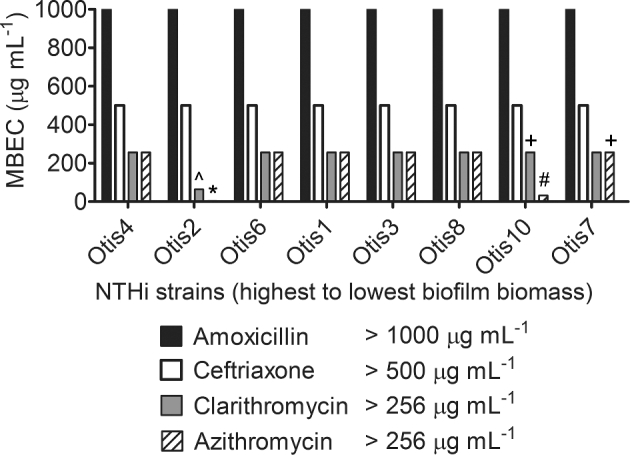
Antibiotic tolerance of NTHi clinical strains biofilms is not dependent on biofilm size. MBEC assays were performed on each strain with a range of concentrations of amoxicillin, ceftriaxone, clarithromycin and azithromycin. The MBECs, or antibiotic concentrations at which bacterial counts were reduced to or below the LOD (1.5 × 103 CFU mL−1), are shown and specifically indicated if less than or equal to the greatest concentration tested: ⁁ 64 μg mL−1, * 4 μg mL−1, + 256 μg mL−1, # 32 μg mL−1.
DISCUSSION
It has been well established that bacteria within biofilms are significantly more tolerant to antimicrobial treatment compared to planktonic state bacteria; proposed mechanisms for why this occurs include reduced metabolic activity of the bacteria within the biofilm, induction of biofilm-specific genes, the presence of a subpopulation of persister cells and a decreased ability of an antibiotic to penetrate the biofilm (Costerton, Stewart and Greenberg 1999; Anderl, Franklin and Stewart 2000; Mah and O'Toole 2001; Fux et al.2005; Slinger et al.2006; Lewis 2007). Reduced antimicrobial penetration could suggest that a larger biofilm would be better able to resist antibiotics than a smaller biofilm. In this study, we have demonstrated that for NTHi middle ear isolates, biofilm size does not play a role in the ability to resist eradication by clinically relevant antibiotics.
We isolated eight NTHi strains from the middle ear fluids of children undergoing tympanostomy tube placement. These strains were found to form biofilms that vary in biomass, thickness and structure (Fig. 2). Differences in biofilm formation by clinical NTHi strains have been previously observed by other groups as well (Murphy and Kirkham 2002; Garcia-Cobos et al.2014). Ranking the biofilms from highest to lowest biomass differed slightly between crystal violet and COMSTAT readouts. This was most notable for Otis10, which had the second highest biomass by crystal violet staining, yet the second lowest biomass by COMSTAT analysis of z-stack images. The staining for CLSM is not meant to label biofilm matrix components such as extracellular proteins; however, crystal violet stains all components contained in the biofilm. This suggested that the increased biomass in the crystal violet staining was due to a high amount of matrix staining. Other variations in biomass that we detected among the strains could likewise be due to differences in matrix components. Extracellular DNA, proteins (e.g. type IV pilin protein and adhesins) and various glycoforms of lipooligosaccharides from the bacterial outer membrane are important constituents of the NTHi biofilm extracellular matrix (Murphy and Kirkham 2002; Swords et al.2004; Jurcisek et al.2005; Gallaher et al.2006; Hong et al.2007; Jurcisek and Bakaletz 2007; Izano, Shah and Kaplan 2009). Further characterization of the clinical strain biofilm matrix and surface components is necessary in order to evaluate their antigenic heterogeneity and why the biofilms vary so greatly in their biomass and thickness.
We hypothesized that strains that formed biofilms with greater biomass, or that were thicker, would be more resistant to eradication by antibiotics than strains that formed thinner, or less dense, biofilms. However, the results of this study contradict this hypothesis. All eight strains were resistant to the β-lactam antibiotics amoxicillin and ceftriaxone at the maximum concentrations tested (Figs 3 and 4). A concentration of 1000 μg mL−1 (500×–2000× the MICs of the strains) was not sufficient to eradicate the biofilm bacteria. This is interesting because amoxicillin is generally the first antibiotic prescribed for OM, and yet it appears to have little to no effect on the in vitro biofilms formed by these clinical strains, despite their lack of β-lactamase production (Lieberthal et al.2013). Similarly, biofilms treated with 500 μg mL−1 ceftriaxone, a concentration over 15 000× greater than the MICs of the strains, experienced at most 1 to 2 logs of killing compared to the vehicle control. While statistically significant, these counts are still well above the LOD and cannot realistically be considered eradicated. Treatment with the macrolide antibiotic clarithromycin had little effect on all but two strains (Otis2 and Otis10, eradicated at 16× and 32× their MICs, respectively) (Fig. 5). Azithromycin was the most effective of the antibiotics tested with these strains. Interestingly, Otis2 was eliminated with only 4 μg mL−1 of azithromycin, which is only 8× its MIC (Fig. 6). This result is the most contradictory to our hypothesis since Otis2 forms a biofilm with size parameters on the higher end of the spectrum compared to the others. Therefore, from these results we concluded that there was not a correlation between biofilm size and the ability to resist antibiotic eradication, and that a reduced rate of biofilm penetration by the antibiotic is not likely a primary mechanism of tolerance, since smaller biofilms (e.g. Otis7) can survive antibiotic treatment as well as larger biofilms (e.g. Otis4).
Antibiotic treatment recommendations for acute OM (AOM) require the consideration of not only NTHi as a primary etiological agent, but also of the other most common bacterial causes, Streptococcus pneumoniae and Moraxella catarrhalis (Murphy et al.2009; Lieberthal et al.2013). The guidelines set by the American Academy of Pediatrics list amoxicillin (with or without clavulanate) and third-generation cephalosporins such as ceftriaxone and cefdinir as treatment options for AOM, and no longer recommend the use of macrolides (Lieberthal et al.2013). While the clinical NTHi isolates in this study demonstrate more sensitivity to macrolides than β-lactams, others have shown reduced clinical efficacy of macrolides (Dagan et al.2000; Tristram, Jacobs and Appelbaum 2007). Additionally, these drugs tend to accumulate in the host cells (e.g. macrophages); therefore, the higher concentrations required to have an impact on infections in the middle ear space may be more difficult to accomplish (Dagan et al.2000; Tristram, Jacobs and Appelbaum 2007). Orally administered antibiotics for AOM in general are not able to reach concentrations more than 10× the MIC in the middle ear (Belfield et al.2015). Our study and others have demonstrated that upwards of 100× the MIC is often required to kill bacteria in biofilms (Starner et al.2008; Qureishi et al.2014).
The question remains as to what mechanisms are involved that render these NTHi strains more tolerant to antibiotic treatment when in the biofilm state, and why some of the biofilms were more sensitive than others. Resistance to β-lactams is largely determined by the production of a β-lactamase for many species of bacteria. In Pseudomonas aeruginosa biofilms, β-lactamase production was found to be the only barrier to β-lactam diffusion through biofilms; as an uncharged antibiotic, β-lactams are unlikely to be impeded by interactions with extracellular matrix components (Walters et al.2003; Bagge et al.2004). Positively charged antibiotics, like aminoglycosides, have been observed to interact with negatively charged matrix components, obstructing the ability of the drug to reach the bacteria deep within the biofilm (Bagge et al.2004; Fux et al.2005). Azithromycin is a cationic antimicrobial agent and therefore may be subject to similar interactions within NTHi biofilms (Farmer, Li and Hancock 1992). Our biofilm characterization experiments indicate potential variability in biofilm extracellular matrix components. Proteins (e.g. type IV pilin protein and DNABII) and double-stranded DNA in particular have been found to be important matrix components for NTHi (Jurcisek and Bakaletz 2007; Izano, Shah and Kaplan 2009; Goodman et al.2011). Degradation of extracellular DNA and protein using DNase I and proteinase K, respectively, has been demonstrated to increase biofilm sensitivity to biocides and some antibiotics for certain NTHi strains (Izano, Shah and Kaplan 2009; Cavaliere et al.2014). While high biofilm density and/or thickness may not prevent the antibiotic from diffusing through the biofilm, the constituents of the extracellular matrix may provide a barrier to the bacteria or interact with the drugs in a way that hinders their action. The antigenic heterogeneity of NTHi as a species suggests that differences in matrix components or surface antigen expression among strains may contribute to differences in biofilm size and antimicrobial interactions (Garmendia et al.2012).
Bacteria within a biofilm are typically thought to have reduced metabolic activity (Walters et al.2003; Fux et al.2005; Lewis 2007). A proteomic analysis of NTHi strain 2019 in both the planktonic and biofilm states found that levels of proteins associated with metabolism, protein synthesis and aerobic respiration appeared to be downregulated in biofilm bacteria (Post et al.2014). This in particular would render the NTHi clinical strains more resistant to bactericidal antibiotics like amoxicillin and ceftriaxone, since the bacteria are less likely to be actively dividing.
In conclusion, we have observed that for clinical strains of NTHi, there is not a correlation between the size of the biofilm and the ability to withstand elimination by antibiotics. Further analysis of resistance mechanisms for these strains, as well as continued survey of the characteristics of other clinical isolate biofilms, will aid in elucidating what particular challenges we face in treating these biofilm-mediated infections.
Acknowledgments
The authors would like to thank Tina Swords for her assistance with middle ear fluid collection, and the members of the Swords lab for their support and advice.
FUNDING
This work was supported by the National Institute on Deafness and Other Communication Disorders at the National Institutes of Health (RO1 DC007444 and RO1 DC10051).
REFERENCES
- Anderl JN, Franklin MJ, Stewart PS. Role of antibiotic penetration limitation in Klebsiella pneumoniae biofilm resistance to ampicillin and ciprofloxacin. Antimicrob Agents Ch 2000;44:1818–24 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andrews JM. Determination of minimum inhibitory concentrations. J Antimicrob Chemoth 2001;48, Suppl 1, 5–16 [DOI] [PubMed] [Google Scholar]
- Bagge N, Hentzer M, Andersen JB et al. Dynamics and spatial distribution of beta-lactamase expression in Pseudomonas aeruginosa biofilms. Antimicrob Agents Ch 2004;48:1168–74 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belfield K, Bayston R, Birchall JP et al. Do orally administered antibiotics reach concentrations in the middle ear sufficient to eradicate planktonic and biofilm bacteria? A review. Int J Pediatr Otorhi 2015, 296–300 [DOI] [PubMed] [Google Scholar]
- Cavaliere R, Ball JL, Turnbull L et al. The biofilm matrix destabilizers, EDTA and DNaseI, enhance the susceptibility of nontypeable Haemophilus influenzae biofilms to treatment with ampicillin and ciprofloxacin. Microbiologyopen 2014;3:557–67 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costerton JW, Stewart PS, Greenberg EP. Bacterial biofilms: a common cause of persistent infections. Science 1999;284:1318–22 [DOI] [PubMed] [Google Scholar]
- Dagan R, Johnson CE, McLinn S et al. Bacteriologic and clinical efficacy of amoxicillin/clavulanate vs. azithromycin in acute otitis media. Pediatr Infect Dis J 2000;19:95–104 [DOI] [PubMed] [Google Scholar]
- Dickson G. Acute otitis media. Prim Care 2014;41:11–8 [DOI] [PubMed] [Google Scholar]
- Ehrlich GD, Veeh R, Wang X et al. Mucosal biofilm formation on middle-ear mucosa in the chinchilla model of otitis media. JAMA 2002;287:1710–5 [DOI] [PubMed] [Google Scholar]
- Erwin AL, Sandstedt SA, Bonthuis PJ et al. Analysis of genetic relatedness of Haemophilus influenzae isolates by multilocus sequence typing. J Bacteriol 2008;190:1473–83 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farmer S, Li ZS, Hancock RE. Influence of outer membrane mutations on susceptibility of Escherichia coli to the dibasic macrolide azithromycin. J Antimicrob Chemoth 1992;29:27–33 [DOI] [PubMed] [Google Scholar]
- Fux CA, Costerton JW, Stewart PS et al. Survival strategies of infectious biofilms. Trends Microbiol 2005;13:34–40 [DOI] [PubMed] [Google Scholar]
- Gallaher TK, Wu S, Webster P et al. Identification of biofilm proteins in non-typeable Haemophilus Influenzae. BMC Microbiol 2006;6:65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia-Cobos S, Moscoso M, Pumarola F et al. Frequent carriage of resistance mechanisms to beta-lactams and biofilm formation in Haemophilus influenzae causing treatment failure and recurrent otitis media in young children. J Antimicrob Chemoth 2014;69:2394–9 [DOI] [PubMed] [Google Scholar]
- Garmendia J, Marti-Lliteras P, Moleres J et al. Genotypic and phenotypic diversity of the noncapsulated Haemophilus influenzae: adaptation and pathogenesis in the human airways. Int Microbiol 2012;15:159–72 [DOI] [PubMed] [Google Scholar]
- Goodman SD, Obergfell KP, Jurcisek JA et al. Biofilms can be dispersed by focusing the immune system on a common family of bacterial nucleoid-associated proteins. Mucosal Immunol 2011;4:625–37 [DOI] [PubMed] [Google Scholar]
- Hall-Stoodley L, Hu FZ, Gieseke A et al. Direct detection of bacterial biofilms on the middle-ear mucosa of children with chronic otitis media. JAMA 2006;296:202–11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heydorn A, Nielsen AT, Hentzer M et al. Quantification of biofilm structures by the novel computer program COMSTAT. Microbiology 2000;146, (Pt 10), 2395–407 [DOI] [PubMed] [Google Scholar]
- Holder RC, Kirse DJ, Evans AK et al. One third of middle ear effusions from children undergoing tympanostomy tube placement had multiple bacterial pathogens. BMC Pediatr 2012;12:87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hong W, Pang B, West-Barnette S et al. Phosphorylcholine expression by nontypeable Haemophilus influenzae correlates with maturation of biofilm communities in vitro and in vivo. J Bacteriol 2007;189:8300–7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Izano EA, Shah SM, Kaplan JB. Intercellular adhesion and biocide resistance in nontypeable Haemophilus influenzae biofilms. Microb Pathog 2009;46:207–13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jurcisek J, Greiner L, Watanabe H et al. Role of sialic acid and complex carbohydrate biosynthesis in biofilm formation by nontypeable Haemophilus influenzae in the chinchilla middle ear. Infect Immun 2005;73:3210–8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jurcisek JA, Bakaletz LO. Biofilms formed by nontypeable Haemophilus influenzaein vivo contain both double-stranded DNA and type IV pilin protein. J Bacteriol 2007;189:3868–75 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaur R, Chang A, Xu Q et al. Phylogenetic relatedness and diversity of non-typable Haemophilus influenzae in the nasopharynx and middle ear fluid of children with acute otitis media. J Med Microbiol 2011;60:1841–8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klein JO. The burden of otitis media. Vaccine 2000;19, Suppl 1, S2–8 [DOI] [PubMed] [Google Scholar]
- Lacross NC, Marrs CF, Patel M et al. High genetic diversity of nontypeable Haemophilus influenzae isolates from two children attending a day care center. J Clin Microbiol 2008;46:3817–21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis K. Persister cells, dormancy and infectious disease. Nat Rev Microbiol 2007;5:48–56 [DOI] [PubMed] [Google Scholar]
- Lieberthal AS, Carroll AE, Chonmaitree T et al. The diagnosis and management of acute otitis media. Pediatrics 2013;131:e964–99 [DOI] [PubMed] [Google Scholar]
- Mah TF, O'Toole GA. Mechanisms of biofilm resistance to antimicrobial agents. Trends Microbiol 2001;9:34–9 [DOI] [PubMed] [Google Scholar]
- Murphy TF, Faden H, Bakaletz LO et al. Nontypeable Haemophilus influenzae as a pathogen in children. Pediatr Infect Dis J 2009;28:43–8 [DOI] [PubMed] [Google Scholar]
- Murphy TF, Kirkham C. Biofilm formation by nontypeable Haemophilus influenzae: strain variability, outer membrane antigen expression and role of pili. BMC Microbiol 2002;2:7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pichichero ME. Otitis media. Pediatr Clin North Am 2013;60:391–407 [DOI] [PubMed] [Google Scholar]
- Post D, Held JM, Ketterer MR et al. Comparative analyses of proteins from Haemophilus influenzae biofilm and planktonic populations using metabolic labeling and mass spectrometry. BMC Microbiol 2014;14:2322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qureishi A, Lee Y, Belfield K et al. Update on otitis media - prevention and treatment. Infect Drug Resist 2014;7:15–24 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slinger R, Chan F, Ferris W et al. Multiple combination antibiotic susceptibility testing of nontypeable Haemophilus influenzae biofilms. Diagn Microbiol Infect Dis 2006;56:247–53 [DOI] [PubMed] [Google Scholar]
- Sox CM, Finkelstein JA, Yin R et al. Trends in otitis media treatment failure and relapse. Pediatrics 2008;121:674–9 [DOI] [PubMed] [Google Scholar]
- Starner TD, Shrout JD, Parsek MR et al. Subinhibitory concentrations of azithromycin decrease nontypeable Haemophilus influenzae biofilm formation and Diminish established biofilms. Antimicrob Agents Ch 2008;52:137–45 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swords WE, Moore ML, Godzicki L et al. Sialylation of lipooligosaccharides promotes biofilm formation by nontypeable Haemophilus influenzae. Infect Immun 2004;72:106–13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tristram S, Jacobs MR, Appelbaum PC. Antimicrobial resistance in Haemophilus influenzae. Clin Microbiol Rev 2007;20:368–89 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walters MC, 3rd, Roe F, Bugnicourt A et al. Contributions of antibiotic penetration, oxygen limitation, and low metabolic activity to tolerance of Pseudomonas aeruginosa biofilms to ciprofloxacin and tobramycin. Antimicrob Agents Ch 2003;47:317–23 [DOI] [PMC free article] [PubMed] [Google Scholar]