Abstract
Staphylococcus aureus is an eminent human pathogen that can colonize the human host and cause severe life-threatening illnesses. This bacterium can reside in and infect a wide range of host tissues, ranging from superficial surfaces like the skin to deeper tissues such as in the gastrointestinal tract, heart and bones. Due to its multifaceted lifestyle, S. aureus uses complex regulatory networks to sense diverse signals that enable it to adapt to different environments and modulate virulence. In this minireview, we explore well-characterized environmental and host cues that S. aureus responds to and describe how this pathogen modulates virulence in response to these signals. Lastly, we highlight therapeutic approaches undertaken by several groups to inhibit both signaling and the cognate regulators that sense and transmit these signals downstream.
Keywords: Staphylococcus aureus, MRSA, pathogenesis, gene regulation
Staphylococcus aureus uses a complex regulatory network to adapt to different environments to support its different lifestyles.
INTRODUCTION
Staphylococcus aureus, aptly called a ‘Janus-faced’ bacterium (Broker, Holtfreter and Bekeredjian-Ding 2014), is a commensal organism and a debilitating pathogen. In the USA, ∼20% of the adult population carry S. aureus in their nares persistently, whereas ∼30% of the population is intermittently colonized by S. aureus (Wertheim et al.2005). Nasal carriage of S. aureus in children is substantially higher, ranging from 45% to 70% (Wertheim et al.2005). While colonization is typically not harmful to the host, S. aureus may breach innate host defenses and gain access to deeper tissues, causing a variety of superficial and invasive infections (Wertheim et al.2005; Tong et al.2015). For example, in healthy individuals in the community, S. aureus frequently causes minor skin and soft tissue infections such as impetigo, folliculitis and cutaneous abscesses. More rare but severe infections in the community include pyomyositis (Tong et al.2015), necrotizing fasciitis (Foster 1996; Tong et al.2015) and necrotizing pneumonia (Sader et al.2016; Kale and Dhawan 2016). In nosocomial settings, S. aureus can initiate infections at surgical sites or from implanted medical devices including artificial heart valves, catheters, prosthetic joints and orthopedic implants (Richards et al.1999; Brooks and Jefferson 2012; Hogan et al.2015; Tong et al.2015). During bacteremia, S. aureus circulates in blood and can seed vital organs (Archer et al.2011), resulting in disseminated infections such as endocarditis, osteomyelitis and descending urinary tract infections (Foster 1996; Wertheim et al.2005). The ability of this pathogen to persist in a wide variety of host niches ranging from skin (von Eiff et al.2001; Montgomery, David and Daum 2015) to abiotic devices (Scherr et al.2014) and deep-seated tissues makes it difficult to eradicate, resulting in recurrent infections.
Staphylococcus aureus has caused havoc in both the community and healthcare setting, resulting in a high socioeconomic burden in both developed and developing nations. For example, a large-scale study evaluating skin and soft tissue infections between 2001 and 2009 estimated treatment costs of hospitalized patients in the USA to vary between ∼$12 000 and $23 000 depending on the year and the patients’ age group (Suaya et al.2014). Management of S. aureus is complicated by the emergence of ‘super bugs’ that have become resistant to multiple antibiotics, as in the case of methicillin-resistant and vancomycin-resistant S. aureus (MRSA and VRSA, respectively). The average number of MRSA infections in the USA has been estimated to be ∼80 000 cases with a mortality rate of ∼11 000 individuals per year (Klevens et al.2007; Malani 2014). Studies suggest that total treatment costs for MRSA infections are on the order of double that of MSSA infections (Filice et al.2010). Thus, there is a critical need for new treatment strategies to manage S. aureus infections, especially infections with methicillin-resistant strains.
Importantly, S. aureus infections are most often derived from colonizing flora present on mucosal membranes or the skin of the infected host (Wertheim et al.2005). Inasmuch as the commensal and invasive lifestyles are radically different, it is likely that the bacterium undergoes extensive adaptation while transitioning between the two states. Thus, understanding how S. aureus regulates its virulence in response to host environments is crucial to devising effective treatment strategies.
Staphylococcal virulence regulation involves a complex web of global regulatory circuits that sense environmental signals and influence the activation of master regulators, which act alone and in concert to modulate gene expression. In addition to external stimuli, S. aureus responds to cell density by means of an autoinduced, quorum-sensing signal. In the following section, we provide a brief overview of staphylococcal autoinduced and environmental signaling systems. We will also introduce additional regulators that play into these networks, and discuss the specific host signals that they respond to.
First identified in 1986, the accessory gene regulatory (Agr) quorum-sensing, two-component system (TCS) is still the most characterized master regulator of virulence in S. aureus (Recsei et al.1986). Comprehensive reviews of this quorum-sensing system have been published (Lyon and Novick 2004; Novick and Geisinger 2008; Painter et al.2014; Singh and Ray 2014; Wang and Muir 2016). Briefly, S. aureus produces basal levels of a peptide signaling molecule called the auto-inducing peptide (AIP). Accumulation of AIP triggers a series of signal transduction events that in turn activate expression of the agr locus. The agr locus consists of two divergent promoters, P2 and P3, that encode AgrBDCA and the major regulatory RNA effector RNAIII, respectively. When bacterial cell density surpasses a certain threshold (quorum), accumulated AIP binds to the histidine kinase, AgrC, which in turn phosphorylates the response regulator AgrA. Activated AgrA can directly regulate virulence genes (Queck et al.2008), induce its own P2 promoter to increase the transcription of agrBDCA in a positive feedback loop and activate the adjacent P3 promoter to drive the transcription of RNAIII (Novick et al.1993). The remaining two genes in the agrP2 operon, agrD and agrB, respectively encode the AIP propeptide, and a transmembrane endopeptidase involved in the processing and export of the mature protein product.
RNAIII is the key effector molecule linking the Agr TCS and virulence. It is an RNA molecule that binds to the 5΄ region of target mRNAs and post-transcriptionally represses or activates virulence factors such as various toxins and immune modulatory proteins, either acting directly or by influencing their upstream regulators. One of the principal targets of RNAIII is another critical virulence regulator, the repressor of toxins (Rot). Rot positively and negatively modulates the activity of target promoters by directly binding to promoter elements (Said-Salim et al.2003; Geisinger et al.2006; Killikelly et al.2015). During the onset of infection, the agr locus is thought to be inactive due to the presence of few bacteria and low levels of AIP, resulting in high levels of Rot. Rot in turn upregulates the expression of immune evasion proteins and adhesins that help dodge first-line, innate immune defenses (Said-Salim et al.2003; Benson et al.2011, 2012; Xue et al.2012; Montgomery, David and Daum 2015; Mootz et al.2015). These virulence proteins are critical for the initial stages of the infection. Later, after infection is established and quorum is reached, RNAIII levels increase, Rot translation is inhibited, and toxins and exo-enzymes responsible for lysis of immune cells and tissue destruction are expressed (Said-Salim et al.2003; Mootz et al.2015).
The SarA protein family members are an additional set of global regulators with broad consequences on transcription of staphylococcal virulence genes (Cheung and Zhang 2002). SarA can directly bind the agr P2 and P3 promoters, albeit with different affinities, causing increased transcription of agrBDCA and higher abundance of RNAIII (Cheung et al.1992; Chien et al.1999). Evidence also exists that in binding to the agr P2 promoter, SarA bends DNA and enhances the ability of AgrA to activate the P2 and P3 promoters (Morfeldt, Tegmark and Arvidson 1996; Cheung, Eberhardt and Heinrichs 1997; Chien and Cheung 1998; Chien et al.1999). Additionally, SarA affects virulence independently of agr by binding directly to promoters of genes encoding for many virulence factors (Cheung and Ying 1994; Cheung, Eberhardt and Heinrichs 1997; Chan and Foster 1998; Sterba et al.2003).
Another critical regulator of S. aureus virulence is encoded by the saeRS locus (Giraudo et al.1994). Similar to agr, the sae locus encodes a TCS, SaeRS (Giraudo et al.1999). However, unlike Agr, which is a ‘self’-sensing system, SaeRS senses external stimuli and modulates virulence genes by binding to consensus sequences in promoter regions, directly influencing their transcription (Nygaard et al.2010; Sun et al.2010). SaeS serves as a sensor of environmental cues, and SaeR directly upregulates virulence in response to these signals (Montgomery, Boyle-Vavra and Daum 2010; Benson et al.2012; Olson et al.2013). While the sae locus is downstream of agr and is regulated by RNAIII via Rot (Li and Cheung 2008), it also has select functions that are epispastic to Agr (Novick and Jiang 2003). The transcription pattern of sae is complex; environmental signals such as changes in pH, high concentrations of sodium chloride and subinhibitory levels of certain antibiotics regulate its expression (Novick and Jiang 2003; Kuroda et al.2007). Additionally, sae promoter activity is affected by exposure to phagocytosis-related signals such as hydrogen peroxide and antimicrobial peptides produced by neutrophils such as alpha defensins (Geiger et al.2008; Flack et al.2014) and calprotectin (Cho et al.2015).
Despite our extensive knowledge of staphylococcal virulence factors and their regulation, subsequent treatments and vaccines based on this information have not been successful. The development of effective therapeutics is hampered by our limited understanding of in vivo signals that enhance or inhibit virulence. Results derived from in vitro studies or animal models of infection may not apply to the in vivo situation in humans. For instance, therapeutics under development that seek to inhibit in vitro expressed virulence effectors may not be effective for treatment of clinical infections in which they are not expressed or produced (Fowler and Proctor 2014). Thus, there is a critical need to understand signals in the human host that S. aureus encounters and adapts to, which results in its ability to modulate virulence. Below, we summarize several well-characterized host signals that are critical for S. aureus fitness, and address how the host modulates levels of such signals during an infection to inhibit S. aureus growth. We discuss how, in turn, S. aureus uses host signals as cues to modulate virulence and tolerate host stresses. Lastly, we highlight how knowledge of host signals and regulators critical for fitness of this pathogen has informed the development of therapeutics aimed at modifying and preventing S. aureus disease.
HOST SIGNALS AND STAPHYLOCOCCUS AUREUS RESPONSES
Molecular oxygen
Oxygen levels in host
Molecular oxygen (O2) is critical for S. aureus growth both in vitro and in host tissues. In vivo, O2 vary by tissue sites (Carreau et al.2011). For example, the arterial blood O2 content is 68–95 mmHg, while venous blood contains ∼40 mmHg O2 (Park, Myers and Marzella 1992). The skin possesses a wide range of O2 concentrations depending on the depth from the surface (8–35 mmHg). The intestinal lumen is completely anaerobic and contains <2 mmHg O2 (Zeitouni et al.2016), whereas critical organs such as the kidneys and liver contain relatively high levels of O2 (∼50–72 mmHg and ∼30—40 mmHg, respectively) (Brezis and Rosen 1995; Brooks et al.2004; Carreau et al.2011). Thus, tissues contain wide range of O2 levels, from being essentially anaerobic (intestines) to comparatively O2 replete (blood rich tissues). During an infection, rapid recruitment of energy-consuming immune cells such as activated neutrophils can increase O2 demands more than 50-fold (Gabig, Bearman and Babior 1979; Colgan and Taylor 2010), triggering oxygen deficiency (hypoxia) at sites of infection (Schaffer and Taylor 2015; Zeitouni et al.2016). Additionally, tissue-resident macrophages, dendritic cells and T cells induce inflammation, in turn altering vascular structures, leading to restricted blood flow to tissues and reducing O2 levels dramatically (Colgan and Taylor 2010).
Biofilms have also been shown to induce hypoxia (Lone et al.2015). Biofilms are complex microbial communities attached to surfaces or other cells that have a protective extracellular matrix, and can thus promote S. aureus colonization (Lister and Horswill 2014). Formation of biofilms by S. aureus on medical implants and host tissues makes this pathogen a leading cause of device-related infections, and results in dangerous, chronic and recurrent infections (Lister and Horswill 2014). In vitro experiments demonstrate that anaerobic conditions induce expression of ‘biofilm’ genes, as evidenced by induction of icaADBC (Cramton et al.2001), whose gene products lead to the production and transport of extracellular polysaccharide adhesins that help in attachment of bacterial cells to each other, to host cells and to surfaces (Vuong et al.2004; O’Gara 2007). Thus, depletion of O2 may either be a by-product of bacterial growth or a strategy employed by the bacterium to induce biofilm.
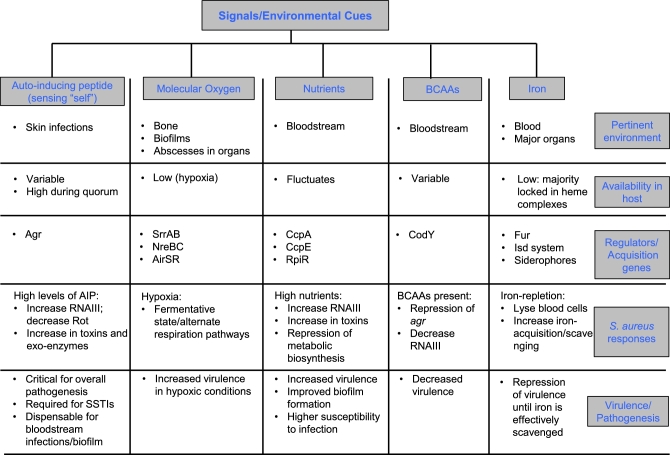
Osteomyelitis or infection of the bones is a low oxygen, biofilm-associated infection. Staphylococcus aureus is the major cause of osteomyelitis in adults and children, accounting for 70%–90% of infections in the latter (Bocchini et al.2006; Hatzenbuehler and Pulling 2011; Pendleton and Kocher 2015). Bone and bone marrow are considered hypoxic, due to low blood flow to these tissues (Mader et al.1980; Spencer et al.2014). Upon infection with S. aureus, O2 levels plummet further (Wilde et al.2015), similar to what happens with O2 levels in device-related S. aureus biofilms (described above). Importantly, in vitro studies indicate that hypoxic conditions increase S. aureus cytotoxin production, suggesting that reduced O2 states promote S. aureus pathogenesis (Wilde et al.2015). Moreover, S. aureus can induce hypoxia even in tissues that have relatively higher levels of O2, like the kidneys (Vitko, Spahich and Richardson 2015), leading to formation of O2-restricted microenvironments, such as abscesses. Staphylococcus aureus can then disseminate from these abscesses, become bacteremic and seed a variety of vital organs (Rubinstein 2008; Cheng et al.2009; Sheen et al.2010; Dahl, Hansen and Bruun 2013; Lister and Horswill 2014). Collectively, these observations suggest that S. aureus promotes hypoxia in tissues, which is a key signal for S. aureus biofilm formation and enhanced staphylococcal virulence (summarized in Fig. 1).
Figure 1.
Summary of host signals encountered by S. aureus and the response to these cues. Staphylococcus aureus senses ‘self’ or external environmental cues via various sensors and regulators that transmit these signals to alter metabolism and virulence.
Staphylococcus aureus responses to hypoxia
Under conditions of decreased oxygen, S. aureus readily uses nitrate and nitrite as its final oxygen acceptors. In the absence of these two terminal O2 acceptors, the bacterium will switch to fermentative metabolism (Burke and Lascelles 1975; Pagels et al.2010). Hypoxic or anaerobic conditions result in two major challenges: inability to replenish the NADH/NAD+ pools and inefficient ATP synthesis (Green and Paget 2004). Staphylococcus aureus is less versatile in comparison to other facultative aerobes such as Escherichia coli because it has less complex fermentative pathways and lacks cytochrome oxidases present in the latter (Burke and Lascelles 1975). However, it has several sensors by which it can quickly recognize hypoxia/anaerobiosis and turn on nitrate respiration and fermentation. Under anaerobic conditions, S. aureus upregulates genes in glycolysis, fermentation and anaerobic respiration and represses genes in the Krebs cycle—the main pathway responsible for NADH generation (Fuchs et al.2007). In addition, genes involved in nitrate and nitrite reduction pathways are upregulated (Fuchs et al.2007). Thus, concomitant with its ability to promote hypoxia, S. aureus has multiple regulatory pathways to respire in low oxygen conditions.
The staphylococcal respiratory response AB (SrrAB) TCS is critical for anaerobic growth of S. aureus in vitro (Throup et al.2001; Yarwood, McCormick and Schlievert 2001; Kinkel et al.2013). SrrAB was found bioinformatically due to its homology to the O2-responsive TCS in Bacillus subtilis called ResDE (Yarwood, McCormick and Schlievert 2001). The ligand responsible for SrrAB activation is currently unknown, although Kinkel et al. (2013) offer menaquinone as the most likely candidate based on various inducers of SrrAB. This hypothesis has been supported by Schlievert et al. (2013), who demonstrate that menaquinone analogs affect both S. aureus growth and alter toxin production in a SrrAB-dependent manner. SrrAB is induced under nitric oxide stress, detoxifies nitric oxide (Kinkel et al.2013; Grosser et al.2016) and is required for efficient biofilm formation (Ulrich et al.2007; Kinkel et al.2013). Several studies have demonstrated the contribution of SrrAB to S. aureus metabolism and pathogenesis. However, studies on the relationship between SrrAB and virulence produced seemingly conflicting results. Deletion of srrAB was shown to decrease bacterial recovery from infected kidneys in mice (Throup et al.2001), and an srrAB mutant was attenuated in osteomyelitis. These results suggest that SrrAB enhances virulence (Wilde et al.2015). In contrast, in vitro studies indicate that SrrAB represses virulence by negatively influencing agr P2/P3 and presumably virulence (Throup et al.2001; Pragman et al.2004). However, this apparent paradox was recently resolved by the demonstration that the in vivo attenuation of the mutant during osteomyelitis is independent of RNAIII (Wilde et al.2015). Both hypoxia and the srrA deletion resulted in enhanced expression of phenol-soluble modulins (PSMs) in an AgrA-dependent, but RNAIII-independent manner (Queck et al.2008). Thus, although SrrAB represses RNAIII in vitro, it is an activator of virulence during osteomyelitis.
NreBC is another TCS involved in O2 sensing and nitrogen regulation that was first identified in S. carnosus (Fedtke et al.2002). NreBC is encoded in an operon with NreA. The exact function of NreA is unclear although there are hints that it is a nitrate sensor (Hall and Ji 2013). The sensor histidine kinase NreB is a fumerate and nitrate reductase-type, cytoplasmic protein containing four conserved cysteine residues that together comprise an Fe-S cluster (Kamps et al.2004). The presence of O2 renders the NreBC TCS inactive due to oxidation of the Fe-S cluster, while the absence of O2 leads to reduction of the Fe-S cluster, causing dimerization and activation of the NreC response regulator, and ultimately induction of the nitrate reductase system (Kamps et al.2004; Hall and Ji 2013). Inactivation of NreBC abrogates the ability of S. aureus to reduce nitrate, forcing the bacterium to upregulate fermentative pathways for survival (Fedtke et al.2002; Schlag et al.2008; Yan et al.2011). Under anaerobic and nitrate respiration conditions, the NreABC locus was shown to induce nitrite and nitrate reductase genes (Schlag et al.2008). However, no phenotypes have been described for this system in vivo.
The third sensor of O2 in S. aureus is the AirSR/YhcSR TCS, a pleiotropic regulator that is essential for S. aureus survival (Sun et al.2005). It is involved in the positive regulation of the NreBC TCS during anaerobic growth of bacteria (Yan et al.2011). Expression of airSR increases by the addition of exogenous nitrate, but not nitrite, suggesting its exclusive role in nitrate respiration (Yan et al.2011). Consistent with this hypothesis, downregulation of airSR leads to poor growth of S. aureus under anaerobic growth in media containing nitrate (Yan et al.2011). Similar to NreBC, AirR binds to the promoters of the nitrate reductase gene, narG (Yan et al.2012). Likewise, the activity state of this TCS is determined by oxidation of an Fe-S cluster present in AirS (Sun et al. 2012b). Depletion of AirSR using antisense RNA interference results in decreased S. aureus survival in human blood, presumably owing to decreased S. aureus protease production. Additionally, proteases that are important for S. aureus pathogenesis are regulated by AirSR at the promoter level (Hall et al.2015).
In addition to directly sensing oxygen, S. aureus also produces Rex, a protein that senses NAD+/NADH pools, allowing the bacterium to monitor its metabolic state independently of O2 (Somerville and Proctor 2009). Rex activation leads to increased levels of enzymes involved in fermentative pathways, nitrate/nitrite reductases and srrAB (Pagels et al.2010). Lastly, the response regulator of the AgrAC TCS, AgrA, has been shown to modulate virulence factor production in response to oxygen levels in the cell. It is thought that under oxidative stress, intramolecular disulfide bonds between two cysteine residues within the AgrA active site impede AgrA DNA-binding activity, thus affecting transcription of various virulence factors (Sun et al. 2012a,b). Taken together, S. aureus has multiple regulatory proteins that interact in a complex manner to counteract low oxygen states in the host. These genetic elements perform dual functions by activating genes required to handle hypoxic stress and enable virulence by increasing expression of toxins and proteases.
Nutrients and metabolic signals
Availability in the host
Carbohydrates (carbon sources) are critical for cellular growth and metabolism. They serve as the precursors and metabolic intermediates in pathways such as glycolysis, the pentose phosphate pathway and the tricarboxylic acid (Krebs) cycle. Glucose is the preferred carbon source of most organisms (Monod 1942). In humans, glucose is produced and stored in the liver until it is transported into the bloodstream for distribution throughout the body. Glucose serves as the major energy source for many cell types and as a result, its homeostasis is carefully regulated (Nordlie, Foster and Lange 1999). This is not surprising given that glucose is the most abundant free carbohydrate in human serum (Psychogios et al.2011). In humans, blood glucose levels in the 80–130 mg/dL range are considered normal, while <70 and >200 mg/dL are indicative of hypoglycemia and hyperglycemia, respectively (Association AD 2016). To thrive under these diverse nutritional conditions, S. aureus tightly controls and modulates gene expression in a coordinated fashion based on particular environmental cues (Somerville and Proctor 2009). For example, under hypoxic states during an infection, S. aureus increases its glycolytic flux to balance the inefficient fermentation of carbohydrates. Likewise, to accommodate increased glucose consumption, S. aureus has adaptive mechanisms to increase its glucose uptake during infection (Vitko et al.2016).
Staphylococcus aureus pathogenesis seems to be closely linked to glucose availability in vitro and in humans. For instance, biofilm formation by S. aureus is enhanced by the addition of glucose to media (Waldrop et al.2014). In vivo studies have demonstrated that diabetic mice are more susceptible to S. aureus infections and are significantly deficient in clearing S. aureus compared to their non-diabetic counterparts (Rich and Lee 2005). Likewise, diabetic patients are at a higher risk for S. aureus pneumonia (Equils et al.2016) and are more susceptible to S. aureus-mediated foot infections (Dunyach-Remy et al.2016). Importantly, hospitalized patients who are hyperglycemic seem to be at a higher risk of S. aureus infection (Pomposelli et al.1998).
Staphylococcus aureus responses to carbohydrate availability and metabolism
In low glucose conditions, S. aureus assumes a low-energy ‘starvation’ state (Watson, Clements and Foster 1998). Watson et al. found that although over 99% of S. aureus cells lose viability in response to glucose starvation within the first few days of culture, the surviving population can remain viable for months. Cells in this long-term starvation state are smaller and denser than cells grown in the presence of glucose. Additionally, marked changes in RNA and protein synthesis profiles are observed during the early stages of nutrient starvation (Watson, Clements and Foster 1998). When starved cells are given complex medium containing glucose, they recover from their starvation state, rapidly increasing RNA synthesis and protein production to support growth (Clements and Foster 1998).
Staphylococcus aureus adapts to nutritionally diverse environments by prioritizing utility of primary versus secondary carbon sources. This process, best characterized in Bacillus subtilis, is known as carbon catabolite repression (CCR) (Titgemeyer and Hillen 2002; Warner and Lolkema 2003; Gorke and Stulke 2008). CcpA is a highly conserved transcription factor that plays important roles in CCR (Henkin et al.1991; Saier et al.1996). In response to the presence of rapidly metabolized carbon sources such as glucose or other glycolytic intermediates, HPr kinase phosphorylates the signaling intermediate HPr (Deutscher and Saier 1983). Phosphorylation allows HPr to complex with CcpA and together, this phospo-HPr-CcpA complex binds to catabolite responsive elements to modulate the expression of target genes (Deutscher et al.1995; Miwa et al.2000). Starvation-induced genes are among these target genes that have been shown to be regulated by CcpA in Gram-positive bacteria (Leboeuf et al.2000). Of note, serine phosphorylated Crh, an HPr homolog, has also been shown to complex with CcpA in CCR but this interaction is up to 10-fold weaker and results in a less robust phenotype (Galinier et al.1997; Martin-Verstraete, Deutscher and Galinier 1999). Notably, in S. aureus, the expression of RNAIII is significantly increased in the presence of glucose under constant pH, but not in a ΔccpA mutant, where the effect of glucose on RNAIII expression is markedly decreased (Seidl et al.2006). Collectively, these observations suggest that high glucose triggers a signal cascade through CcpA that upregulates RNAIII expression and ultimately modulates virulence gene expression.
Additionally, CcpA modulates the expression of genes involved in the glycolytic pathway through CCR. In response to high levels of glucose, CcpA represses the TCA cycle by downregulating the expression of critical TCA cycle enzymes (Strasters and Winkler 1963; Seidl et al.2008, 2009). Thus, as glucose is depleted from the media or is otherwise limited during nutrient starvation, the TCA cycle is progressively derepressed. This process is under the control of a second carbon catabolite protein, CcpE (Hartmann et al.2013). CcpE binds to citrate, the first intermediate of the TCA cycle, and adopts a predominantly tetrameric (active) state. Active CcpE binds to and regulates target promoters, including those of TCA cycle enzymes (Hartmann et al.2013; Ding et al.2014). Metabolomic, microarray and transcriptional analyses show that not only is CcpE involved in modulating the carbon flow through the TCA cycle, it is also a major regulator of virulence genes such as those involved in the synthesis of virulence factor capsular polysaccharides and superantigen-like proteins (Ding et al.2014). Whether this global regulation observed in the metabolomics analyses is due to its direct action on virulence gene promoters or indirectly due to its effects on metabolism is unknown but is likely influenced by both (Hartmann et al.2013; Ding et al.2014).
The pentose phosphate pathway (PPP) has also been implicated in linking metabolism to virulence, through the RpiR family of transcriptional repressors (Zhu et al.2011). The RpiR family was first identified as regulators of ribose metabolism in E. coli (Sorensen and Hove-Jensen 1996) but members of this family have since been linked to a number of other catabolism pathways, including the PPP, in both Gram-negative and Gram-positive bacteria (Jaeger and Mayer 2008; Daddaoua, Krell and Ramos 2009; Kohler, Choong and Rossbach 2011). Although RpiR family members have a C-terminal sugar isomerase-binding domain, the actual ligand is unknown. Of the three RpiR homologs present in S. aureus, only RpiRb and RpiRc appear to modulate PPP gene regulation. RpiRc is an important regulator of virulence (Zhu et al.2011; Balasubramanian et al.2016; Gaupp et al.2016). Recent work indicates that RpiRc senses metabolic shifts and represses virulence by modulating the expression of the agr locus. This results in the repression of RNAIII expression and thus increased translation of the repressor Rot (Balasubramanian et al.2016). Additional work suggests that the effect of RpiRc on agr and virulence gene expression occurs via repression of sarA, a positive regulator of agr and virulence (Gaupp et al.2016). Future work is required to elucidate the metabolic signal(s) responsible for activating RpiRc and to understand the molecular mechanism that governs the intersection between the PPP and RpiRc's contribution to pathogenesis.
In addition to central metabolism, amino acid availability plays a critical role in S. aureus pathogenesis. The branched chain amino acids (BCAAs) valine, leucine and isoleucine, along with GTP, initiate the repressive activity of CodY, a global metabolic regulator in S. aureus and many Gram-positive bacteria (Guedon et al.2001; Ratnayake-Lecamwasam et al.2001; Shivers and Sonenshein 2004; Tojo et al.2005; Sonenshein 2007). Upon sensing and binding intracellular GTP or BCAAs, the affinity of CodY toward consensus sequences (CodY binding boxes) increases. A dimerized CodY binds these cis-regulatory elements to control target gene expression (Shivers and Sonenshein 2004; den Hengst et al.2005; Levdikov et al.2006; Majerczyk et al.2008). As expected, CodY activity is at its highest in exponential growth phase where nutrients are in excess (Majerczyk et al.2008). As a result, metabolic pathways that are unnecessary in nutrient-replete environments are repressed. In S. aureus, the CodY regulon consists of over 200 genes, including biosynthesis genes of metabolic intermediates as well as those involved in virulence (Majerczyk et al.2008, 2010; Pohl et al.2009). Interestingly, CodY acts by direct binding to virulence gene promoters, and also indirectly through Agr. Although deletion of codY results in increased expression of agrBDCA and rnaIII, its low affinity to agr promoters suggests that direct transcriptional regulation is unlikely (Majerczyk et al.2008). Instead, it appears that CodY prevents premature activation of agr during exponential growth phase, despite the presence of phosphorylated AgrA (Roux et al.2014). Taken together, these data suggest that as GTP and/or BCAAs are depleted, CodY senses this change in nutritional state and progressively derepresses its target genes to increase the metabolic biosynthesis pathways and also regulate the expression virulence factors.
In summary, S. aureus must confront and adapt to diverse host environments, where levels of carbon-based nutrients naturally vary greatly. For example, when the nutrients are low, a subset of bacteria enters a low-energy long-term starvation state, which it encounters the nutrients that it needs. Factors such as CcpA, CcpE, RpiRc and CodY sense the changes in carbon state of the host and accordingly adjust the utilization of pathways involved in metabolism. Either in the process of or as a result of changes to the metabolic state, S. aureus differentially regulates virulence factor expression, thereby modifying its pathogenesis (Fig. 1).
Iron
Availability in the host
Iron is a vital nutrient across all domains of life. Although iron limitation inhibits cellular processes, iron abundance is toxic due to its highly reactive properties. As a result, iron metabolism in mammalian cells and in bacteria is tightly regulated to maintain homeostasis. Iron in vertebrates exists in four major forms: (i) as heme in hemoglobin, a tetrapyrrole molecule with high affinity for molecular oxygen; (ii) as iron–sulfur clusters in several critical enzymes; (iii) as extracellular storage molecules, such as transferrins found in serum and lactoferrins found in the lymphoid system (Hammer and Skaar 2011; Cassat and Skaar 2013); and (iv) intracellularly bound to ferritin (MacKenzie, Iwasaki and Tsuji 2008). Greater than 90% of iron in the host is intracellular, trapped in heme. As a result, free extracellular iron in human tissues is estimated to be around 10−18 M (Bullen, Rogers and Griffiths 1978), well below the concentration required for microbial life. Additionally, infection-induced inflammation leads to rapid decline in iron levels in blood serum (Cartwright et al.1946; Darton et al.2015). Finally, extracellular iron is often scavenged by host glycoproteins, further restricting iron availability for microbes during infection (Cassat and Skaar 2013). For example, NrampI, a phagosomal iron efflux pump that is important for bacterial clearance, is upregulated during certain infections (Loomis et al.2014). The process of depriving microbes of iron has been cleverly coined as ‘nutritional immunity’ (Hammer and Skaar 2011; Cassat and Skaar 2013).
Staphylococcus aureus responses to iron limitation
Staphylococcus aureus has evolved intricate mechanisms to counter iron deficiency. Here, we focus on two well-studied mechanisms of iron acquisition: siderophore-mediated acquisition and heme-iron acquisition. Similar to many other pathogens, S. aureus produces several low molecular weight scavenging proteins called siderophores, out of which staphyloferrins A and B are the best characterized (Konetschny-Rapp et al.1990; Hammer and Skaar 2011). These secreted factors capture extracellular iron bound to host glycoproteins by removing iron from loaded transferrins (Park et al.2005). Siderophores are essential for bacterial growth in media where transferrin is the sole source of iron (Park et al.2005). Once iron is removed from transferrins, the siderophore-bound iron is actively transported into the cell via ABC transporters (Skaar et al. 2004).
Although S. aureus culture filtrates have been long known to possess siderophore activity, Beasley et al. (2009) were the first to identify the genetic locus responsible for staphyloferrin A biosynthesis, called sfa. While this locus was important for S. aureus growth in iron-deplete media, it was dispensable for growth of the bacterium in serum, which is naturally iron deficient. This result was puzzling until the discovery that deletion of both sfa and a second poorly characterized siderophore operon (sbn) was required to abrogate S. aureus growth in serum. The sbn (siderophore biosynthesis gene cluster) operon contains nine genes encoding proteins required for biosynthesis of staphyloferrin B (Dale et al.2004). Inactivation of at least one of the genes in this operon, sbnE, abolishes siderophore activity in culture filtrates and leads to moderate reduction in S. aureus colonization of murine kidneys (Dale et al.2004).
While siderophores are adept at scavenging extracellular iron, the majority of iron in vertebrates is locked in complex with heme inside erythrocytes (Deiss 1983). Heme iron is obtained from lysis of erythrocytes by hemolysins and cytotoxins (Torres et al.2006, 2010; Spaan et al.2014, 2015). Following lysis, heme is captured and taken up by the iron-regulated surface determinant (Isd) system (Mazmanian et al.2003). This specialized system consists of the cell wall anchored surface proteins IsdABCH, the transporters IsdDEF and the cytoplasmic degradation enzymes IsdIG (Muryoi et al.2008). Briefly, the cell surface proteins IsdBH are important for binding hemoglobin to the surface of S. aureus (Torres et al.2006), and work together with IsdAC to extract heme. Extracted heme is transported across the membrane via two ABC transporter clusters: IsdDEF (Mazmanian et al.2003; Liu et al.2008) and HtsABC (Skaar et al. 2004). IsdIG then degrades heme, releasing iron Skaar, Gaspar and Schneewind (2004). In vivo, hts mutants are severely attenuated in their ability to colonize liver and kidneys of mice (Skaar et al. 2004). Likewise, isdB mutants demonstrate reduced ability to infect murine kidneys and spleen (Torres et al.2006).
The ferric uptake regulator (Fur) regulates iron metabolism in many Gram-negative and Gram-positive bacteria. By amplifying fur from S. aureus and expressing it recombinantly, Xiong et al. (2000) showed that Fur is involved in regulating genes in ferrichrome uptake and has an iron-binding site, similar to that of Fur found in other organisms. There are a series of excellent reviews summarizing decades of work on Fur-mediated regulation of iron metabolism (Hantke 2001; Troxell and Hassan 2013; Fillat 2014). Briefly, in the presence of iron, Fur directly binds Fe2+ and in its holoform, acts as a repressor of iron acquisition genes. In E. coli, Fur acts by repressing the small regulatory RNA, RyhB (Masse and Gottesman 2002). In iron-deplete conditions, RyhB is derepressed due to the inactivation of apo-Fur. Using an antisense base-pairing mechanism, RyhB rapidly upregulates expression of iron acquisition genes and shuts down production of non-essential proteins that use or store iron (Masse and Gottesman 2002).
While there are no reports of ryhB in S. aureus, iron homeostasis in S. aureus is clearly Fur dependent. Both the siderophore biosynthesis operons, sfa and sbn (Dale et al.2004; Cheung et al.2009), and the isd locus involved in heme acquisition are under Fur control (Torres et al.2010). Fur also connects iron metabolism and virulence gene expression in S. aureus: it positively impacts expression of immunomodulatory proteins such as coagulase, superantigen-like proteins and negatively regulates genes involved in virulence such as lipases and cytotoxins (Torres et al.2010). Importantly, fur mutants are attenuated for virulence in a murine pneumonia model of infection. Additionally, S. aureus lacking fur is more susceptible to neutrophil-mediated killing (Torres et al.2010).
In summary, S. aureus most likely encounters a gradient of iron concentrations when it traverses through different tissues. Under iron-starved conditions, S. aureus senses iron via Fur, upregulates siderophore and heme acquisition pathways, and represses virulence. When iron is abundant, either due to the natural reservoir of iron in the tissue or due to efficient acquisition of iron, S. aureus switches to a more pathogenic lifestyle characterized by enhanced virulence factor production (Fig. 1). While this review focuses solely on iron as a key element affecting S. aureus virulence, other metals such as manganese and zinc also alter S. aureus pathogenesis. Similar to S. aureus–iron interactions, specific regulatory proteins sense these metals and affect virulence, the host actively sequesters manganese and zinc, and S. aureus has evolved complicated transport mechanisms to acquire them (Cassat and Skaar 2012).
INHIBITION OF STAPHYLOCOCCUS AUREUS ENVIRONMENTAL SENSING: THERAPEUTIC POTENTIAL
Staphylococcus aureus relies on environmental cues derived from the host as it transitions between colonizing and invasive states. Accordingly, these cues are being targeted for the development of anti-S. aureus therapeutics involving inhibitory compounds, including natural and chemical inhibitors as well as antibodies that block environmental sensing. Both these regulatory mechanisms can be inhibited, albeit by different mechanisms. Here, we highlight the potential use of environmental sensing and signaling pathway inhibitors as novel anti-staphylococcal therapeutics.
By far, the largest category of therapeutics against S. aureus-sensing systems targets the Agr-mediated quorum-sensing system. As discussed above, Agr induces rapid and massive accumulation of harmful, tissue-degrading toxins and exo-enzymes that are critical for S. aureus pathogenesis (reviewed in Khan et al.2015). A small molecular inhibitor of Agr, called savarin (Staphylococcus aureus virulence inhibitor), was identified in a screen for compounds that attenuated agr P3 promoter activity. Extensive analysis of savarin revealed that it alters binding of AgrA to DNA, and attenuates skin ulcers and abscesses in murine models of infection. Resistance to savarin was not observed after in vitro and in vivo passage, enhancing its attractiveness as a therapeutic (Sully et al.2014). Others have undertaken an alternative approach by designing analogs or dominant negative AIP that competitively inhibit signal sensing. Tal-Gan et al. used an alanine-scanning approach to find mutations in AIP that disrupt binding and signaling of wild-type AIP via AgrC. These AIP mimetics were able to significantly reduce S. aureus hemolytic activity in vitro, suggesting efficient blockage of Agr-mediated signaling (Tal-Gan et al.2013). Likewise, monoclonal antibodies such as AP4-24H11 have been designed to ‘quench’ quorum sensing by binding and neutralizing AIP. AP4-24H11 has demonstrated protection in intradermal infections of mice and reduced S. aureus-mediated lethality in systemic infection models (Park et al.2007; Kirchdoerfer et al.2011). Lastly, an US Food and Drug Administration-approved, non-steroidal anti-inflammatory compound called diflunisal significantly attenuates S. aureus toxin production (Khodaverdian et al.2013), without altering bacterial growth (Hendrix et al.2016). This compound is thought to inhibit phosphorylation of AgrA by the sensor kinase AgrC, thus abrogating quorum-sensing and toxin levels (Khodaverdian et al.2013). Importantly, diflunisal has recently been shown to impede S. aureus cytotoxicity toward osteoblasts in vitro, due to reduced production of PSMs (Hendrix et al.2016). Moreover, this compound has promising efficacy in vivo, in that it can moderately attenuate S. aureus-mediated cortical bone destruction in a murine model of osteomyelitis (Hendrix et al.2016). Taken together, a variety of approaches exist to inhibit Agr that target different portions of the quorum-sensing cascade.
Agr inhibition is expected to be most effective as a therapeutic in clinical situations where this regulator is critical for pathogenesis (Fig. 1). CA-MRSA strains cause disease—primarily skin and soft tissue infections—in otherwise healthy community subjects. CA-MRSA strains have a ‘hyperactive’ agr locus and produce copious levels of toxins and proteases in vitro, in animal models of infection and in humans (Nastaly, Grinholc and Bielawski 2010; Date et al.2014). In contrast, S. aureus strains isolated from hospitalized patients frequently have mutations that inactivate or severely impair the activity of the Agr-TCS (Shopsin et al.2008; Traber et al.2008). Presumably, disruption of barrier functions by disease and clinical intervention in the hospital environment permit S. aureus strains that lack full virulence to cause infection. Furthermore, Agr dysfunction has been associated with persistent rather than resolving bacteremia, and mortality (Fowler et al.2004; Schweizer et al.2011), perhaps because killing by host and synthetic antimicrobials is reduced in agr-dysfunctional isolates (reviewed in Painter et al.2014). These observations suggest that there are situations in vivo where Agr activation is dispensable, or even deleterious for S. aureus. Thus, the clinical consequences of disabling Agr activity are not obvious; depending on the patient, efforts to use Agr and virulence as targets for new antimicrobials may be ill advised.
Recently, Arya et al. used a novel bioinformatics-based structural approach to design and synthesize a small molecule inhibitor of SarA (SarABI). As discussed above, SarA is a cytoplasmic transcription factor that activates genes critical for biofilm formation in an Agr-independent manner (Trotonda et al.2005). SarABI acts by binding to the DNA-binding domain of the transcription factor, forming a stable complex such that SarA's downstream regulatory events are blocked. Since SarA is a potent regulator of toxins and exo-enzymes and can act independently of Agr, SarABI may be useful to treat infections that are associated with low agr activity states (Arya et al.2015). One such scenario may be biofilm infections; activation of agr is thought to cause dispersal of the biofilm (Boles and Horswill 2008); and agr-defective cells are frequently recovered from biofilms on prosthetic devices in humans (Kiedrowski and Horswill 2011). Staphylococcus aureus biofilms are difficult to treat and are often the cause of recurring infections in humans (Parsek and Singh 2003; Harris and Richards 2006). Promisingly, SarABI is a potent inhibitor of biofilm development both in vitro (on abiotic surfaces) and in vivo (on rat vascular graft infections). Similar to agr inhibitors, SarABI does not restrict bacterial growth, suggesting that its use is likely to not elicit bacterial resistance in vivo (Arya et al.2015). Additional studies are urgently needed to determine the safety and efficacy of anti-SarA strategies such as SarABI.
Small molecules inhibitors have also been developed that antagonize critical metabolic pathways by targeting cytoplasmic rather than surface or secreted proteins. Tripathi et al. identified chemical inhibitors of the iron-scavenging siderophores called baulamycins. These compounds are natural antimicrobials that attenuate the function of the cytosolic synthetase involved in siderophore biosynthesis (Tripathi et al.2014). In addition, compound screening has identified a chemical inhibitor of SaeRS, apparently at the transcriptional level (Long et al.2013). Mechanisms by which these compounds inhibit their cognate receptors and downstream signaling are currently unknown.
In contrast to the numerous efforts undertaken to counteract S. aureus secreted proteins (reviewed in Missiakas and Schneewind 2016; Karauzum and Datta 2016; Giersing et al.2016; Lacey, Geoghegan and McLoughlin 2016), far fewer therapeutics target intracellular regulators of virulence or their signaling molecules. While regulators that control multiple virulence effector proteins make attractive target candidates, designing such counteractive therapeutics has been challenging. First, antibody-based neutralization approaches—the current golden standard for treating several infectious diseases—are ineffective against cytoplasmic regulatory proteins, as these are inaccessible to antibodies. Second, finding chemical inhibitors that traverse into the bacterial cytoplasm but leave host cells unharmed can be a challenging task. Third, S. aureus has an array of regulators that interact with each other in a complex manner and perform redundant functions (such as various TCSs responding to oxygen or the intricate network of proteins involved in toxin production). Lastly, the ligands of many regulators are unknown. As such, designing competitors or quenchers is an underdeveloped area of study that could hold great promise.
CONCLUSIONS
In this review, we highlight key features of Staphylococcus aureus adaptation to the host environment during infection (summarized in Fig. 1). We delved into tissue specific environments and metabolic stresses that the bacterium may encounter during infection. Understanding the signals and regulatory elements that alter S. aureus pathogenesis in response to environmental signals is crucial to developing novel therapeutics. Thus, basic and clinical research studies should account for differential production of S. aureus virulence factors under various environmental conditions and disease states. The results may inform the design of S. aureus vaccines and therapeutic trials. Additionally, a better understanding of factors specific to an individual's condition, such as site of infection, immune competency and the virulence potential of the infecting strain under these conditions, may pave the way for ‘personalized’ management of S. aureus infections.
Acknowledgments
We apologize to authors whose work relevant to this topic was not cited here due to space constraints.
FUNDING
The work on gene regulation in the Torres and Shopsin laboratories is supported in part by the National Institute of Allergy and Infectious Diseases (NIAID) of the National Institutes of Health (NIH) under award R01AI103268 to B.S. and V.J.T. D.B. was supported in part by a Jan Vilcek and David Goldfarb Endowed Fellowship (NYU School of Medicine). L.H. was supported in part by an UNCF-Merck Science Initiative Fellowship and a National Science Foundation Fellowship.
Conflict of interest. None declared.
REFERENCES
- American Database Association. Standards of medical care in diabetes. Diabetes Care 2016;39:S39–46.26696679 [Google Scholar]
- Archer NK, Mazaitis MJ, Costerton JW et al. . Staphylococcus aureus biofilms: properties, regulation, and roles in human disease. Virulence 2011;2:445–59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arya R, Ravikumar R, Santhosh RS et al. . SarA based novel therapeutic candidate against Staphylococcus aureus associated with vascular graft infections. Front Microbiol 2015;6:416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balasubramanian D, Ohneck EA, Chapman J et al. . Staphylococcus aureus coordinates leukocidin expression and pathogenesis by sensing metabolic fluxes via RpiRc. MBio 2016;7:e00818-00816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beasley FC, Vines ED, Grigg JC et al. . Characterization of staphyloferrin A biosynthetic and transport mutants in Staphylococcus aureus. Mol Microbiol 2009;72:947–63. [DOI] [PubMed] [Google Scholar]
- Benson MA, Lilo S, Nygaard T et al. . Rot and SaeRS cooperate to activate expression of the staphylococcal superantigen-like exoproteins. J Bacteriol 2012;194:4355–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benson MA, Lilo S, Wasserman GA et al. . Staphylococcus aureus regulates the expression and production of the staphylococcal superantigen-like secreted proteins in a Rot-dependent manner. Mol Microbiol 2011;81:659–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bocchini CE, Hulten KG, Mason EO Jr et al. . Panton-Valentine leukocidin genes are associated with enhanced inflammatory response and local disease in acute hematogenous Staphylococcus aureus osteomyelitis in children. Pediatrics 2006;117:433–40. [DOI] [PubMed] [Google Scholar]
- Boles BR, Horswill AR. Agr-mediated dispersal of Staphylococcus aureus biofilms. PLoS Pathog 2008;4:e1000052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brezis M, Rosen S. Hypoxia of the renal medulla–its implications for disease. N Engl J Med 1995;332:647–55. [DOI] [PubMed] [Google Scholar]
- Broker BM, Holtfreter S, Bekeredjian-Ding I. Immune control of Staphylococcus aureus - regulation and counter-regulation of the adaptive immune response. Int J Med Microbiol 2014;304:204–14. [DOI] [PubMed] [Google Scholar]
- Brooks AJ, Eastwood J, Beckingham IJ et al. . Liver tissue partial pressure of oxygen and carbon dioxide during partial hepatectomy. Brit J Anaesth 2004;92:735–7. [DOI] [PubMed] [Google Scholar]
- Brooks JL, Jefferson KK. Staphylococcal biofilms: quest for the magic bullet. Adv Appl Microbiol 2012;81:63–87. [DOI] [PubMed] [Google Scholar]
- Bullen JJ, Rogers HJ, Griffiths E. Role of iron in bacterial infection. Curr Top Microbiol Immunol 1978;80:1–35. [DOI] [PubMed] [Google Scholar]
- Burke KA, Lascelles J. Nitrate reductase system in Staphylococcus aureus wild type and mutants. J Bacteriol 1975;123:308–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carreau A, El Hafny-Rahbi B, Matejuk A et al. . Why is the partial oxygen pressure of human tissues a crucial parameter? Small molecules and hypoxia. J Cell Mol Med 2011;15:1239–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cartwright GE, Lauritsen MA, Humphreys S et al. . The anemia associated with chronic infection. Science 1946;103:72–3. [DOI] [PubMed] [Google Scholar]
- Cassat JE, Skaar EP. Metal ion acquisition in Staphylococcus aureus: overcoming nutritional immunity. Semin Immunopathol 2012;34:215–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cassat JE, Skaar EP. Iron in infection and immunity. Cell Host Microbe 2013;13:509–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan PF, Foster SJ. Role of SarA in virulence determinant production and environmental signal transduction in Staphylococcus aureus. J Bacteriol 1998;180:6232–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng AG, Kim HK, Burts ML et al. . Genetic requirements for Staphylococcus aureus abscess formation and persistence in host tissues. FASEB J 2009;23:3393–404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheung AL, Ying P. Regulation of alpha- and beta-hemolysins by the sar locus of Staphylococcus aureus. J Bacteriol 1994;176:580–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheung AL, Eberhardt K, Heinrichs JH. Regulation of protein A synthesis by the sar and agr loci of Staphylococcus aureus. Infect Immun 1997;65:2243–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheung AL, Koomey JM, Butler CA et al. . Regulation of exoprotein expression in Staphylococcus aureus by a locus (sar) distinct from agr. P Natl Acad Sci USA 1992;89:6462–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheung AL, Zhang G. Global regulation of virulence determinants in Staphylococcus aureus by the SarA protein family. Front Biosci 2002;7:d1825–42. [DOI] [PubMed] [Google Scholar]
- Cheung J, Beasley FC, Liu S et al. . Molecular characterization of staphyloferrin B biosynthesis in Staphylococcus aureus. Mol Microbiol 2009;74:594–608. [DOI] [PubMed] [Google Scholar]
- Chien Y, Manna AC, Projan SJ et al. . SarA, a global regulator of virulence determinants in Staphylococcus aureus, binds to a conserved motif essential for sar-dependent gene regulation. J Biol Chem 1999;274:37169–76. [DOI] [PubMed] [Google Scholar]
- Chien Y, Cheung AL. Molecular interactions between two global regulators, sar and agr, in Staphylococcus aureus. J Biol Chem 1998;273:2645–52. [DOI] [PubMed] [Google Scholar]
- Cho H, Jeong DW, Liu Q et al. . Calprotectin increases the activity of the SaeRS two component system and murine mortality during Staphylococcus aureus infections. PLoS Pathog 2015;11:e1005026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clements MO, Foster SJ. Starvation recovery of Staphylococcus aureus 8325-4. Microbiology 1998;144(Pt 7):1755–63. [DOI] [PubMed] [Google Scholar]
- Colgan SP, Taylor CT. Hypoxia: an alarm signal during intestinal inflammation. Nat Rev Gastroentero 2010;7:281–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cramton SE, Ulrich M, Gotz F et al. . Anaerobic conditions induce expression of polysaccharide intercellular adhesin in Staphylococcus aureus and Staphylococcus epidermidis. Infect Immun 2001;69:4079–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daddaoua A, Krell T, Ramos JL. Regulation of glucose metabolism in Pseudomonas: the phosphorylative branch and entner-doudoroff enzymes are regulated by a repressor containing a sugar isomerase domain. J Biol Chem 2009;284:21360–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dahl A, Hansen TF, Bruun NE. Staphylococcus aureus endocarditis with fast development of aortic root abscess despite relevant antibiotics. Heart Lung 2013;42:72–3. [DOI] [PubMed] [Google Scholar]
- Dale SE, Doherty-Kirby A, Lajoie G et al. . Role of siderophore biosynthesis in virulence of Staphylococcus aureus: identification and characterization of genes involved in production of a siderophore. Infect Immun 2004;72:29–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darton TC, Blohmke CJ, Giannoulatou E et al. . Rapidly escalating hepcidin and associated serum iron starvation are features of the acute response to typhoid infection in humans. PLoS Neglect Trop D 2015;9:e0004029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Date SV, Modrusan Z, Lawrence M et al. . Global gene expression of methicillin-resistant Staphylococcus aureus USA300 during human and mouse infection. J Infect Dis 2014;209:1542–50. [DOI] [PubMed] [Google Scholar]
- Deiss A. Iron metabolism in reticuloendothelial cells. Semin Hematol 1983;20:81–90. [PubMed] [Google Scholar]
- den Hengst CD, van Hijum SA, Geurts JM et al. . The Lactococcus lactis CodY regulon: identification of a conserved cis-regulatory element. J Biol Chem 2005;280:34332–42. [DOI] [PubMed] [Google Scholar]
- Deutscher J, Saier MH Jr. ATP-dependent protein kinase-catalyzed phosphorylation of a seryl residue in HPr, a phosphate carrier protein of the phosphotransferase system in Streptococcus pyogenes. P Natl Acad Sci USA 1983;80:6790–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deutscher J, Kuster E, Bergstedt U et al. . Protein kinase-dependent HPr/CcpA interaction links glycolytic activity to carbon catabolite repression in gram-positive bacteria. Mol Microbiol 1995;15:1049–53. [DOI] [PubMed] [Google Scholar]
- Ding Y, Liu X, Chen F et al. . Metabolic sensor governing bacterial virulence in Staphylococcus aureus. P Natl Acad Sci USA 2014;111:E4981–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunyach-Remy C, Ngba Essebe C, Sotto A et al. . Staphylococcus aureus toxins and diabetic foot ulcers: role in pathogenesis and interest in diagnosis. Toxins 2016;8:S1262–3636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Equils O, da Costa C, Wible M et al. . The effect of diabetes mellitus on outcomes of patients with nosocomial pneumonia caused by methicillin-resistant Staphylococcus aureus: data from a prospective double-blind clinical trial comparing treatment with linezolid versus vancomycin. BMC Infect Dis 2016;16:476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fedtke I, Kamps A, Krismer B et al. . The nitrate reductase and nitrite reductase operons and the narT gene of Staphylococcus carnosus are positively controlled by the novel two-component system NreBC. J Bacteriol 2002;184:6624–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Filice GA, Nyman JA, Lexau C et al. . Excess costs and utilization associated with methicillin resistance for patients with Staphylococcus aureus infection. Infect Cont Hosp Ep 2010;31:365–73. [DOI] [PubMed] [Google Scholar]
- Fillat MF. The FUR (ferric uptake regulator) superfamily: diversity and versatility of key transcriptional regulators. Arch Biochem Biophys 2014;546:41–52. [DOI] [PubMed] [Google Scholar]
- Flack CE, Zurek OW, Meishery DD et al. . Differential regulation of staphylococcal virulence by the sensor kinase SaeS in response to neutrophil-derived stimuli. P Natl Acad Sci USA 2014;111:E2037–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foster T. Staphylococcus. In Baron S. (ed.). Medical Microbiology. Galveston, TX: University of Texas Medical Branch at Galveston, 1996. [PubMed] [Google Scholar]
- Fowler VG Jr., Proctor RA. Where does a Staphylococcus aureus vaccine stand? Clin Microbiol Infect 2014;20Suppl 5:66–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fowler VG Jr., Sakoulas G, McIntyre LM et al. . Persistent bacteremia due to methicillin-resistant Staphylococcus aureus infection is associated with agr dysfunction and low-level in vitro resistance to thrombin-induced platelet microbicidal protein. J Infect Dis 2004;190:1140–9. [DOI] [PubMed] [Google Scholar]
- Fuchs S, Pane-Farre J, Kohler C et al. . Anaerobic gene expression in Staphylococcus aureus. J Bacteriol 2007;189:4275–89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gabig TG, Bearman SI, Babior BM. Effects of oxygen tension and pH on the respiratory burst of human neutrophils. Blood 1979;53:1133–9. [PubMed] [Google Scholar]
- Galinier A, Haiech J, Kilhoffer MC et al. . The Bacillus subtilis crh gene encodes a HPr-like protein involved in carbon catabolite repression. P Natl Acad Sci USA 1997;94:8439–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaupp R, Wirf J, Wonnenberg B et al. . RpiRc is a pleiotropic effector of virulence determinant synthesis and attenuates pathogenicity in Staphylococcus aureus. Infect Immun 2016;84:2031–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geiger T, Goerke C, Mainiero M et al. . The virulence regulator Sae of Staphylococcus aureus: promoter activities and response to phagocytosis-related signals. J Bacteriol 2008;190:3419–28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geisinger E, Adhikari RP, Jin R et al. . Inhibition of rot translation by RNAIII, a key feature of agr function. Mol Microbiol 2006;61:1038–48. [DOI] [PubMed] [Google Scholar]
- Giersing BK, Dastgheyb SS, Modjarrad K et al. . Status of vaccine research and development of vaccines for Staphylococcus aureus. Vaccine 2016;34:2962–6. [DOI] [PubMed] [Google Scholar]
- Giraudo AT, Raspanti CG, Calzolari A et al. . Characterization of a Tn551-mutant of Staphylococcus aureus defective in the production of several exoproteins. Can J Microbiol 1994;40:677–81. [DOI] [PubMed] [Google Scholar]
- Giraudo AT, Calzolari A, Cataldi AA et al. . The sae locus of Staphylococcus aureus encodes a two-component regulatory system. FEMS Microbiol Lett 1999;177:15–22. [DOI] [PubMed] [Google Scholar]
- Gorke B, Stulke J. Carbon catabolite repression in bacteria: many ways to make the most out of nutrients. Nat Rev Microbiol 2008;6:613–24. [DOI] [PubMed] [Google Scholar]
- Green J, Paget MS. Bacterial redox sensors. Nat Rev Microbiol 2004;2:954–66. [DOI] [PubMed] [Google Scholar]
- Grosser MR, Weiss A, Shaw LN et al. . Regulatory requirements for Staphylococcus aureus nitric oxide resistance. J Bacteriol 2016;198:2043–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guedon E, Serror P, Ehrlich SD et al. . Pleiotropic transcriptional repressor CodY senses the intracellular pool of branched-chain amino acids in Lactococcus lactis. Mol Microbiol 2001;40:1227–39. [DOI] [PubMed] [Google Scholar]
- Hall JW, Ji Y. Sensing and Adapting to Anaerobic Conditions by Staphylococcus aureus. Adv Appl Microbiol 2013;84:1–25. [DOI] [PubMed] [Google Scholar]
- Hall JW, Yang J, Guo H et al. . The AirSR two-component system contributes to Staphylococcus aureus survival in human blood and transcriptionally regulates sspABC operon. Front Microbiol 2015;6:682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hammer ND, Skaar EP. Molecular mechanisms of Staphylococcus aureus iron acquisition. Annu Rev Microbiol 2011;65:129–47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hantke K. Iron and metal regulation in bacteria. Curr Opin Microbiol 2001;4:172–7. [DOI] [PubMed] [Google Scholar]
- Harris LG, Richards RG. Staphylococci and implant surfaces: a review. Injury 2006;37Suppl 2:S3–14. [DOI] [PubMed] [Google Scholar]
- Hartmann T, Zhang B, Baronian G et al. . Catabolite control protein E (CcpE) is a LysR-type transcriptional regulator of tricarboxylic acid cycle activity in Staphylococcus aureus. J Biol Chem 2013;288:36116–28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hatzenbuehler J, Pulling TJ. Diagnosis and management of osteomyelitis. Am Fam Physician 2011;84:1027–33. [PubMed] [Google Scholar]
- Hendrix AS, Spoonmore TJ, Wilde AD et al. . Repurposing the nonsteroidal anti-inflammatory drug diflunisal as an osteoprotective, antivirulence therapy for Staphylococcus aureus osteomyelitis. Antimicrob Agents Ch 2016;60:5322–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henkin TM, Grundy FJ, Nicholson WL et al. . Catabolite repression of alpha-amylase gene expression in Bacillus subtilis involves a trans-acting gene product homologous to the Escherichia coli lacl and galR repressors. Mol Microbiol 1991;5:575–84. [DOI] [PubMed] [Google Scholar]
- Hogan S, Stevens NT, Humphreys H et al. . Current and future approaches to the prevention and treatment of staphylococcal medical device-related infections. Curr Pharm Des 2015;21:100–13. [DOI] [PubMed] [Google Scholar]
- Jaeger T, Mayer C. The transcriptional factors MurR and catabolite activator protein regulate N-acetylmuramic acid catabolism in Escherichia coli. J Bacteriol 2008;190:6598–608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kale P, Dhawan B. The changing face of community-acquired methicillin-resistant Staphylococcus aureus. Indian J Med Microbiol 2016;34:275–85. [DOI] [PubMed] [Google Scholar]
- Kamps A, Achebach S, Fedtke I et al. . Staphylococcal NreB: an O(2)-sensing histidine protein kinase with an O(2)-labile iron-sulphur cluster of the FNR type. Mol Microbiol 2004;52:713–23. [DOI] [PubMed] [Google Scholar]
- Karauzum H, Datta SK. Adaptive immunity against Staphylococcus aureus. Curr Top Microbiol Immunol 2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khan BA, Yeh AJ, Cheung GY et al. . Investigational therapies targeting quorum-sensing for the treatment of Staphylococcus aureus infections. Expert Opin Inv Drug 2015;24:689–704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khodaverdian V, Pesho M, Truitt B et al. . Discovery of antivirulence agents against methicillin-resistant Staphylococcus aureus. Antimicrob Agents Ch 2013;57:3645–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiedrowski MR, Horswill AR. New approaches for treating staphylococcal biofilm infections. Ann N Y Acad Sci 2011;1241:104–21. [DOI] [PubMed] [Google Scholar]
- Killikelly A, Benson MA, Ohneck EA et al. . Structure-based functional characterization of repressor of toxin (Rot), a central regulator of Staphylococcus aureus virulence. J Bacteriol 2015;197:188–200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kinkel TL, Roux CM, Dunman PM et al. . The Staphylococcus aureus SrrAB two-component system promotes resistance to nitrosative stress and hypoxia. MBio 2013;4:e00696–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirchdoerfer RN, Garner AL, Flack CE et al. . Structural basis for ligand recognition and discrimination of a quorum-quenching antibody. J Biol Chem 2011;286:17351–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klevens RM, Morrison MA, Nadle J et al. . Invasive methicillin-resistant Staphylococcus aureus infections in the United States. JAMA 2007;298:1763–71. [DOI] [PubMed] [Google Scholar]
- Kohler PR, Choong EL, Rossbach S. The RpiR-like repressor IolR regulates inositol catabolism in Sinorhizobium meliloti. J Bacteriol 2011;193:5155–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Konetschny-Rapp S, Jung G, Meiwes J et al. . Staphyloferrin A: a structurally new siderophore from staphylococci. Eur J Biochem 1990;191:65–74. [DOI] [PubMed] [Google Scholar]
- Kuroda H, Kuroda M, Cui L et al. . Subinhibitory concentrations of beta-lactam induce haemolytic activity in Staphylococcus aureus through the SaeRS two-component system. FEMS Microbiol Lett 2007;268:98–105. [DOI] [PubMed] [Google Scholar]
- Lacey KA, Geoghegan JA, McLoughlin RM. The role of Staphylococcus aureus virulence factors in skin infection and their potential as vaccine antigens. Pathogens 2016;5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leboeuf C, Leblanc L, Auffray Y et al. . Characterization of the ccpA gene of Enterococcus faecalis: identification of starvation-inducible proteins regulated by ccpA. J Bacteriol 2000;182:5799–806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levdikov VM, Blagova E, Joseph P et al. . The structure of CodY, a GTP- and isoleucine-responsive regulator of stationary phase and virulence in gram-positive bacteria. J Biol Chem 2006;281:11366–73. [DOI] [PubMed] [Google Scholar]
- Li D, Cheung A. Repression of hla by rot is dependent on sae in Staphylococcus aureus. Infect Immun 2008;76:1068–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lister JL, Horswill AR. Staphylococcus aureus biofilms: recent developments in biofilm dispersal. Front Cell Infect Microbiol 2014;4:178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu M, Tanaka WN, Zhu H et al. . Direct hemin transfer from IsdA to IsdC in the iron-regulated surface determinant (Isd) heme acquisition system of Staphylococcus aureus. J Biol Chem 2008;283:6668–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lone AG, Atci E, Renslow R et al. . Staphylococcus aureus induces hypoxia and cellular damage in porcine dermal explants. Infect Immun 2015;83:2531–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Long DR, Mead J, Hendricks JM et al. . 18beta-Glycyrrhetinic acid inhibits methicillin-resistant Staphylococcus aureus survival and attenuates virulence gene expression. Antimicrob Agents Ch 2013;57:241–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loomis WP, Johnson ML, Brasfield A et al. . Temporal and anatomical host resistance to chronic Salmonella infection is quantitatively dictated by Nramp1 and influenced by host genetic background. PLoS One 2014;9:e111763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lyon GJ, Novick RP. Peptide signaling in Staphylococcus aureus and other Gram-positive bacteria. Peptides 2004;25:1389–403. [DOI] [PubMed] [Google Scholar]
- MacKenzie EL, Iwasaki K, Tsuji Y. Intracellular iron transport and storage: from molecular mechanisms to health implications. Antioxid Redox Sign 2008;10:997–1030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mader JT, Brown GL, Guckian JC et al. . A mechanism for the amelioration by hyperbaric oxygen of experimental staphylococcal osteomyelitis in rabbits. J Infect Dis 1980;142:915–22. [DOI] [PubMed] [Google Scholar]
- Majerczyk CD, Sadykov MR, Luong TT et al. . Staphylococcus aureus CodY negatively regulates virulence gene expression. J Bacteriol 2008;190:2257–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Majerczyk CD, Dunman PM, Luong TT et al. . Direct targets of CodY in Staphylococcus aureus. J Bacteriol 2010;192:2861–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malani PN. National burden of invasive methicillin-resistant Staphylococcus aureus infection. JAMA 2014;311:1438–9. [DOI] [PubMed] [Google Scholar]
- Martin-Verstraete I, Deutscher J, Galinier A. Phosphorylation of HPr and Crh by HprK, early steps in the catabolite repression signalling pathway for the Bacillus subtilis levanase operon. J Bacteriol 1999;181:2966–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masse E, Gottesman S. A small RNA regulates the expression of genes involved in iron metabolism in Escherichia coli. P Natl Acad Sci USA 2002;99:4620–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mazmanian SK, Skaar EP, Gaspar AH et al. . Passage of heme-iron across the envelope of Staphylococcus aureus. Science 2003;299:906–9. [DOI] [PubMed] [Google Scholar]
- Missiakas D, Schneewind O. Staphylococcus aureus vaccines: deviating from the carol. J Exp Med 2016;213:1645–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miwa Y, Nakata A, Ogiwara A et al. . Evaluation and characterization of catabolite-responsive elements (cre) of Bacillus subtilis. Nucleic Acids Res 2000;28:1206–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monod J. Recherches sur la croissance des cultures bacteriennes (Research on the growth of bacterial cultures). Actua Sci Ind 1942;911:1–215. [Google Scholar]
- Montgomery CP, Boyle-Vavra S, Daum RS. Importance of the global regulators Agr and SaeRS in the pathogenesis of CA-MRSA USA300 infection. PLoS One 2010;5:e15177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montgomery CP, David MZ, Daum RS. Host factors that contribute to recurrent staphylococcal skin infection. Curr Opin Infect Dis 2015;28:253–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mootz JM, Benson MA, Heim CE et al. . Rot is a key regulator of Staphylococcus aureus biofilm formation. Mol Microbiol 2015;96:388–404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morfeldt E, Tegmark K, Arvidson S. Transcriptional control of the agr-dependent virulence gene regulator, RNAIII, in Staphylococcus aureus. Mol Microbiol 1996;21:1227–37. [DOI] [PubMed] [Google Scholar]
- Muryoi N, Tiedemann MT, Pluym M et al. . Demonstration of the iron-regulated surface determinant (Isd) heme transfer pathway in Staphylococcus aureus. J Biol Chem 2008;283:28125–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nastaly P, Grinholc M, Bielawski KP. Molecular characteristics of community-associated methicillin-resistant Staphylococcus aureus strains for clinical medicine. Arch Microbiol 2010;192:603–17. [DOI] [PubMed] [Google Scholar]
- Nordlie RC, Foster JD, Lange AJ. Regulation of glucose production by the liver. Annu Rev Nutr 1999;19:379–406. [DOI] [PubMed] [Google Scholar]
- Novick RP, Jiang D. The staphylococcal saeRS system coordinates environmental signals with agr quorum sensing. Microbiology 2003;149(Pt 10):2709–17. [DOI] [PubMed] [Google Scholar]
- Novick RP, Geisinger E. Quorum sensing in staphylococci. Annu Rev Genet 2008;42:541–64. [DOI] [PubMed] [Google Scholar]
- Novick RP, Ross HF, Projan SJ et al. . Synthesis of staphylococcal virulence factors is controlled by a regulatory RNA molecule. EMBO J 1993;12:3967–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nygaard TK, Pallister KB, Ruzevich P et al. . SaeR binds a consensus sequence within virulence gene promoters to advance USA300 pathogenesis. J Infect Dis 2010;201:241–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Gara JP. ica and beyond: biofilm mechanisms and regulation in Staphylococcus epidermidis and Staphylococcus aureus. FEMS Microbiol Lett 2007;270:179–88. [DOI] [PubMed] [Google Scholar]
- Olson ME, Nygaard TK, Ackermann L et al. . Staphylococcus aureus nuclease is an SaeRS-dependent virulence factor. Infect Immun 2013;81:1316–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pagels M, Fuchs S, Pane-Farre J et al. . Redox sensing by a Rex-family repressor is involved in the regulation of anaerobic gene expression in Staphylococcus aureus. Mol Microbiol 2010;76:1142–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Painter KL, Krishna A, Wigneshweraraj S et al. . What role does the quorum-sensing accessory gene regulator system play during Staphylococcus aureus bacteremia? Trends Microbiol 2014;22:676–85. [DOI] [PubMed] [Google Scholar]
- Park J, Jagasia R, Kaufmann GF et al. . Infection control by antibody disruption of bacterial quorum sensing signaling. Chem Biol 2007;14:1119–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park MK, Myers RA, Marzella L. Oxygen tensions and infections: modulation of microbial growth, activity of antimicrobial agents, and immunologic responses. Clin Infect Dis 1992;14:720–40. [DOI] [PubMed] [Google Scholar]
- Park RY, Sun HY, Choi MH et al. . Staphylococcus aureus siderophore-mediated iron-acquisition system plays a dominant and essential role in the utilization of transferrin-bound iron. J Microbiol 2005;43:183–90. [PubMed] [Google Scholar]
- Parsek MR, Singh PK. Bacterial biofilms: an emerging link to disease pathogenesis. Annu Rev Microbiol 2003;57:677–701. [DOI] [PubMed] [Google Scholar]
- Pendleton A, Kocher MS. Methicillin-resistant staphylococcus aureus bone and joint infections in children. J Am Acad Orthop Sur 2015;23:29–37. [DOI] [PubMed] [Google Scholar]
- Pohl K, Francois P, Stenz L et al. . CodY in Staphylococcus aureus: a regulatory link between metabolism and virulence gene expression. J Bacteriol 2009;191:2953–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pomposelli JJ, Baxter JK 3rd, Babineau TJ et al. . Early postoperative glucose control predicts nosocomial infection rate in diabetic patients. JPEN-Parenter Enter 1998;22:77–81. [DOI] [PubMed] [Google Scholar]
- Pragman AA, Yarwood JM, Tripp TJ et al. . Characterization of virulence factor regulation by SrrAB, a two-component system in Staphylococcus aureus. J Bacteriol 2004;186:2430–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Psychogios N, Hau DD, Peng J et al. . The human serum metabolome. PLoS One 2011;6:e16957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Queck SY, Jameson-Lee M, Villaruz AE et al. . RNAIII-independent target gene control by the agr quorum-sensing system: insight into the evolution of virulence regulation in Staphylococcus aureus. Mol Cell 2008;32:150–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ratnayake-Lecamwasam M, Serror P, Wong KW et al. . Bacillus subtilis CodY represses early-stationary-phase genes by sensing GTP levels. Genes Dev 2001;15:1093–103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Recsei P, Kreiswirth B, O’Reilly M et al. . Regulation of exoprotein gene expression in Staphylococcus aureus by agar. Mol Gen Genet 1986;202:58–61. [DOI] [PubMed] [Google Scholar]
- Rich J, Lee JC. The pathogenesis of Staphylococcus aureus infection in the diabetic NOD mouse. Diabetes 2005;54:2904–10. [DOI] [PubMed] [Google Scholar]
- Richards MJ, Edwards JR, Culver DH et al. . Nosocomial infections in medical intensive care units in the United States. National Nosocomial Infections Surveillance System. Crit Care Med 1999;27:887–92. [DOI] [PubMed] [Google Scholar]
- Roux A, Todd DA, Velazquez JV et al. . CodY-mediated regulation of the Staphylococcus aureus Agr system integrates nutritional and population density signals. J Bacteriol 2014;196:1184–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rubinstein E. Staphylococcus aureus bacteraemia with known sources. Int J Antimicrob Ag 2008;32(Suppl 1):S18–20. [DOI] [PubMed] [Google Scholar]
- Sader HS, Mendes RE, Jones RN et al. . Antimicrobial susceptibility patterns of community- and hospital-acquired methicillin-resistant Staphylococcus aureus from United States Hospitals: results from the AWARE Ceftaroline Surveillance Program (2012-2014). Diagn Microbiol Infect Dis 2016;86:76–9. [DOI] [PubMed] [Google Scholar]
- Said-Salim B, Dunman PM, McAleese FM et al. . Global regulation of Staphylococcus aureus genes by Rot. J Bacteriol 2003;185:610–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saier MH Jr., Chauvaux S, Cook GM et al. . Catabolite repression and inducer control in Gram-positive bacteria. Microbiology 1996;142(Pt2):217–30. [DOI] [PubMed] [Google Scholar]
- Schaffer K, Taylor CT. The impact of hypoxia on bacterial infection. FEBS J 2015;282:2260–6. [DOI] [PubMed] [Google Scholar]
- Scherr TD, Heim CE, Morrison JM et al. . Hiding in plain sight: interplay between staphylococcal biofilms and host immunity. Front Immunol 2014;5:37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schlag S, Fuchs S, Nerz C et al. . Characterization of the oxygen-responsive NreABC regulon of Staphylococcus aureus. J Bacteriol 2008;190:7847–58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schlievert PM, Merriman JA, Salgado-Pabon W et al. . Menaquinone analogs inhibit growth of bacterial pathogens. Antimicrob Agents Ch 2013;57:5432–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schweizer ML, Furuno JP, Sakoulas G et al. . Increased mortality with accessory gene regulator (agr) dysfunction in Staphylococcus aureus among bacteremic patients. Antimicrob Agents Ch 2011;55:1082–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seidl K, Goerke C, Wolz C et al. . Staphylococcus aureus CcpA affects biofilm formation. Infect Immun 2008;76:2044–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seidl K, Muller S, Francois P et al. . Effect of a glucose impulse on the CcpA regulon in Staphylococcus aureus. BMC Microbiol 2009;9:95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seidl K, Stucki M, Ruegg M et al. . Staphylococcus aureus CcpA affects virulence determinant production and antibiotic resistance. Antimicrob Agents Ch 2006;50:1183–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheen TR, Ebrahimi CM, Hiemstra IH et al. . Penetration of the blood-brain barrier by Staphylococcus aureus: contribution of membrane-anchored lipoteichoic acid. J Mol Med (Berl) 2010;88:633–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shivers RP, Sonenshein AL. Activation of the Bacillus subtilis global regulator CodY by direct interaction with branched-chain amino acids. Mol Microbiol 2004;53:599–611. [DOI] [PubMed] [Google Scholar]
- Shopsin B, Drlica-Wagner A, Mathema B et al. . Prevalence of agr dysfunction among colonizing Staphylococcus aureus strains. J Infect Dis 2008;198:1171–4. [DOI] [PubMed] [Google Scholar]
- Singh R, Ray P. Quorum sensing-mediated regulation of staphylococcal virulence and antibiotic resistance. Future Microbiol 2014;9:669–81. [DOI] [PubMed] [Google Scholar]
- Skaar EP, Humayun M, Bae T et al. . Iron-source preference of Staphylococcus aureus infections. Science 2004;305:1626–8. [DOI] [PubMed] [Google Scholar]
- Skaar EP, Gaspar AH, Schneewind O. IsdG and IsdI, heme-degrading enzymes in the cytoplasm of Staphylococcus aureus. J Biol Chem 2004;279:436–43. [DOI] [PubMed] [Google Scholar]
- Somerville GA, Proctor RA. At the crossroads of bacterial metabolism and virulence factor synthesis in Staphylococci. Microbiol Mol Biol R 2009;73:233–48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sonenshein AL. Control of key metabolic intersections in Bacillus subtilis. Nat Rev Microbiol 2007;5:917–27. [DOI] [PubMed] [Google Scholar]
- Sorensen KI, Hove-Jensen B. Ribose catabolism of Escherichia coli: characterization of the rpiB gene encoding ribose phosphate isomerase B and of the rpiR gene, which is involved in regulation of rpiB expression. J Bacteriol 1996;178:1003–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spaan AN, Vrieling M, Wallet P et al. . The staphylococcal toxins gamma-haemolysin AB and CB differentially target phagocytes by employing specific chemokine receptors. Nat Commun 2014;5:5438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spaan AN, Reyes-Robles T, Badiou C et al. . Staphylococcus aureus targets the duffy antigen receptor for chemokines (DARC) to lyse erythrocytes. Cell Host Microbe 2015;18:363–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spencer JA, Ferraro F, Roussakis E et al. . Direct measurement of local oxygen concentration in the bone marrow of live animals. Nature 2014;508:269–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sterba KM, Mackintosh SG, Blevins JS et al. . Characterization of Staphylococcus aureus SarA binding sites. J Bacteriol 2003;185:4410–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strasters KC, Winkler KC. Carbohydrate metabolism of Staphylococcus aureus. J Gen Microbiol 1963;33:213–29. [DOI] [PubMed] [Google Scholar]
- Suaya JA, Mera RM, Cassidy A et al. . Incidence and cost of hospitalizations associated with Staphylococcus aureus skin and soft tissue infections in the United States from 2001 through 2009. BMC Infect Dis 2014;14:296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sully EK, Malachowa N, Elmore BO et al. . Selective chemical inhibition of agr quorum sensing in Staphylococcus aureus promotes host defense with minimal impact on resistance. PLoS Pathog 2014;10:e1004174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun F, Li C, Jeong D et al. . In the Staphylococcus aureus two-component system sae, the response regulator SaeR binds to a direct repeat sequence and DNA binding requires phosphorylation by the sensor kinase SaeS. J Bacteriol 2010;192:2111–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun F, Liang H, Kong X et al. . Quorum-sensing agr mediates bacterial oxidation response via an intramolecular disulfide redox switch in the response regulator AgrA. P Natl Acad Sci USA 2012a;109:9095–100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun F, Ji Q, Jones MB et al. . AirSR, a [2Fe-2S] cluster-containing two-component system, mediates global oxygen sensing and redox signaling in Staphylococcus aureus. J Am Chem Soc 2012b;134:305–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun J, Zheng L, Landwehr C et al. . Identification of a novel essential two-component signal transduction system, YhcSR, in Staphylococcus aureus. J Bacteriol 2005;187:7876–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tal-Gan Y, Stacy DM, Foegen MK et al. . Highly potent inhibitors of quorum sensing in Staphylococcus aureus revealed through a systematic synthetic study of the group-III autoinducing peptide. J Am Chem Soc 2013;135:7869–82. [DOI] [PubMed] [Google Scholar]
- Throup JP, Zappacosta F, Lunsford RD et al. . The srhSR gene pair from Staphylococcus aureus: genomic and proteomic approaches to the identification and characterization of gene function. Biochemistry 2001;40:10392–401. [DOI] [PubMed] [Google Scholar]
- Titgemeyer F, Hillen W. Global control of sugar metabolism: a gram-positive solution. Anton Leeuw 2002;82:59–71. [PubMed] [Google Scholar]
- Tojo S, Satomura T, Morisaki K et al. . Elaborate transcription regulation of the Bacillus subtilis ilv-leu operon involved in the biosynthesis of branched-chain amino acids through global regulators of CcpA, CodY and TnrA. Mol Microbiol 2005;56:1560–73. [DOI] [PubMed] [Google Scholar]
- Tong SY, Davis JS, Eichenberger E et al. . Staphylococcus aureus infections: epidemiology, pathophysiology, clinical manifestations, and management. Clin Microbiol Rev 2015;28:603–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torres VJ, Pishchany G, Humayun M et al. . Staphylococcus aureus IsdB is a hemoglobin receptor required for heme iron utilization. J Bacteriol 2006;188:8421–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torres VJ, Attia AS, Mason WJ et al. . Staphylococcus aureus fur regulates the expression of virulence factors that contribute to the pathogenesis of pneumonia. Infect Immun 2010;78:1618–28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Traber KE, Lee E, Benson S et al. . agr function in clinical Staphylococcus aureus isolates. Microbiology 2008;154(Pt 8):2265–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tripathi A, Schofield MM, Chlipala GE et al. . Baulamycins A and B, broad-spectrum antibiotics identified as inhibitors of siderophore biosynthesis in Staphylococcus aureus and Bacillus anthracis. J Am Chem Soc 2014;136:1579–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trotonda MP, Manna AC, Cheung AL et al. . SarA positively controls bap-dependent biofilm formation in Staphylococcus aureus. J Bacteriol 2005;187:5790–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Troxell B, Hassan HM. Transcriptional regulation by Ferric Uptake Regulator (Fur) in pathogenic bacteria. Front Cell Infect Microbiol 2013;3:59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ulrich M, Bastian M, Cramton SE et al. . The staphylococcal respiratory response regulator SrrAB induces ica gene transcription and polysaccharide intercellular adhesin expression, protecting Staphylococcus aureus from neutrophil killing under anaerobic growth conditions. Mol Microbiol 2007;65:1276–87. [DOI] [PubMed] [Google Scholar]
- Vitko NP, Grosser MR, Khatri D et al. . Expanded glucose import capability affords Staphylococcus aureus optimized glycolytic flux during infection. MBio 2016;7:e00296-00216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vitko NP, Spahich NA, Richardson AR. Glycolytic dependency of high-level nitric oxide resistance and virulence in Staphylococcus aureus. MBio 2015;6:e00045-00015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- von Eiff C, Becker K, Metze D et al. . Intracellular persistence of Staphylococcus aureus small-colony variants within keratinocytes: a cause for antibiotic treatment failure in a patient with darier's disease. Clin Infect Dis 2001;32:1643–7. [DOI] [PubMed] [Google Scholar]
- Vuong C, Kocianova S, Voyich JM et al. . A crucial role for exopolysaccharide modification in bacterial biofilm formation, immune evasion, and virulence. J Biol Chem 2004;279:54881–6. [DOI] [PubMed] [Google Scholar]
- Waldrop R, McLaren A, Calara F et al. . Biofilm growth has a threshold response to glucose in vitro. Clin Orthop Relat R 2014;472:3305–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang B, Muir TW. Regulation of virulence in Staphylococcus aureus: molecular mechanisms and remaining puzzles. Cell Chem Biol 2016;23:214–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warner JB, Lolkema JS. CcpA-dependent carbon catabolite repression in bacteria. Microbiol Mol Biol R 2003;67:475–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watson SP, Clements MO, Foster SJ. Characterization of the starvation-survival response of Staphylococcus aureus. J Bacteriol 1998;180:1750–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wertheim HF, Melles DC, Vos MC et al. . The role of nasal carriage in Staphylococcus aureus infections. Lancet Infect Dis 2005;5:751–62. [DOI] [PubMed] [Google Scholar]
- Wilde AD, Snyder DJ, Putnam NE et al. . Bacterial hypoxic responses revealed as critical determinants of the host-pathogen outcome by TnSeq analysis of Staphylococcus aureus invasive infection. PLoS Pathog 2015;11:e1005341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiong A, Singh VK, Cabrera G et al. . Molecular characterization of the ferric-uptake regulator, fur, from Staphylococcus aureus. Microbiology 2000;146(Pt 3):659–68. [DOI] [PubMed] [Google Scholar]
- Xue T, You Y, Shang F et al. . Rot and Agr system modulate fibrinogen-binding ability mainly by regulating clfB expression in Staphylococcus aureus NCTC8325. Med Microbiol Immun 2012;201:81–92. [DOI] [PubMed] [Google Scholar]
- Yan M, Yu C, Yang J et al. . The essential two-component system YhcSR is involved in regulation of the nitrate respiratory pathway of Staphylococcus aureus. J Bacteriol 2011;193:1799–805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan M, Hall JW, Yang J et al. . The essential yhcSR two-component signal transduction system directly regulates the lac and opuCABCD operons of Staphylococcus aureus. PLoS One 2012;7:e50608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yarwood JM, McCormick JK, Schlievert PM. Identification of a novel two-component regulatory system that acts in global regulation of virulence factors of Staphylococcus aureus. J Bacteriol 2001;183:1113–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeitouni NE, Chotikatum S, von Kockritz-Blickwede M et al. . The impact of hypoxia on intestinal epithelial cell functions: consequences for invasion by bacterial pathogens. Mol Cell Pediatr 2016;3:14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu Y, Nandakumar R, Sadykov MR et al. . RpiR homologues may link Staphylococcus aureus RNAIII synthesis and pentose phosphate pathway regulation. J Bacteriol 2011;193:6187–96. [DOI] [PMC free article] [PubMed] [Google Scholar]