Abstract
Mice lacking the transcription factor Nrl have no rod photoreceptors and an increased number of short-wavelength-sensitive cones. Missense mutations in NRL are associated with autosomal dominant retinitis pigmentosa; however, the phenotype associated with the loss of NRL function in humans has not been reported. We identified two siblings who carried two allelic mutations: a predicted null allele (L75fs) and a missense mutation (L160P) altering a highly conserved residue in the domain involved in DNA-binding-site recognition. In vitro luciferase reporter assays demonstrated that the NRL-L160P mutant had severely reduced transcriptional activity compared with the WT NRL protein, consistent with a severe loss of function. The affected patients had night blindness since early childhood, consistent with a severe reduction in rod function. Color vision was normal, suggesting the presence of all cone color types; nevertheless, a comparison of central visual fields evaluated with white-on-white and blue-on-yellow light stimuli was consistent with a relatively enhanced function of short-wavelength-sensitive cones in the macula. The fundi had signs of retinal degeneration (such as vascular attenuation) and clusters of large, clumped, pigment deposits in the peripheral fundus at the level of the retinal pigment epithelium (clumped pigmentary retinal degeneration). Our report presents an unusual clinical phenotype in humans with loss-of-function mutations in NRL.
Keywords: cone photoreceptor, retina, retinitis pigmentosa, transcription factor
The NRL gene encodes a basic motif-leucine zipper protein, which is a member of the Maf transcription factor family (1). In the adult retina, NRL is expressed in rod photoreceptors where it functions synergistically with the cone-rod homeobox transcription factor CRX to stimulate the expression of several proteins of the phototransduction cascade such as rhodopsin and the α and β subunits of rod-specific phosphodiesterase (2-7). NRL is essential for the normal development of photoreceptor cells as demonstrated by the analysis of Nrl knockout mice (Nrl-/-), which have a dramatically aberrant pattern of photoreceptor cell types (8). Rod photoreceptors are absent, and there is an increased number of short-wavelength-sensitive cones (S-cones; also referred to as blue cones).
The murine Nrl-/- phenotype resembles that of the rd7 mouse with a defect in the gene encoding another retina-specific transcription factor, Nr2e3 (9-11). Both Nrl-/- mice and rd7 mice have a greatly increased number of S-cones (8, 11). Nr2e3 is expressed in rod photoreceptors of developing and mature retina and functions as a transcriptional activator of rod-specific genes together with Nrl and Crx (12, 13). Humans with NR2E3 mutations have a greatly increased number of S-cones and a form of retinal degeneration that is variably named the enhanced S-cone syndrome, Goldmann-Favre syndrome, or clumped pigmentary retinal degeneration (14-17). These patients have enhanced S-cone function, diminished middle- and long-wavelength cone function, and absent rod function; their electroretinograms (ERGs) show similar amplitudes and delayed implicit times in response to white light stimuli when recorded under dark-adapted and light-adapted conditions, compatible with S-cone function (17-23).
Based on the increased number of S-cones and the reduced number of rods in both the Nrl-/- and rd7 mice, one would expect that the corresponding human NRL and NR2E3 phenotypes would also be similar to each other. However, no humans with an NRL-/- genotype have been reported (24). Rather, only dominant NRL mutations that are unlikely to be null alleles have been reported in humans to date (25-28). All of the published dominant NRL mutations are missense changes affecting one of three residues (S50, P51, and G122) (25-28). The patients with these mutations, unlike those with mutations in NR2E3, do not typically have extensive clumped pigment deposits or evidence of enhanced S-cone function (26-28). We conducted this study to search for loss-of-function NRL mutations among patients with various forms of presumably autosomal recessive retinal disease.
Methods
This study conformed to the tenets of the Declaration of Helsinki and was approved by internal review boards at the Massachusetts Eye and Ear Infirmary and Harvard Medical School. The patients were diagnosed after an ophthalmologic examination including ERGs. Full-field ERGs were recorded after maximal dilation of the pupil and at least 40 min of dark adaptation as reported in refs. 29-31. Patients with typical retinitis pigmentosa (RP) had elevated final dark-adaptation thresholds, abnormal rod and cone ERGs (responses to 0.5-Hz white-light flashes reduced in amplitude to <100 μV, and responses to 30-Hz white-light flashes reduced in amplitude to <50 μV and delayed in implicit time to >32 msec), retinal vessel attenuation, and bone spicule-pigment deposits in the periphery. Patients with atypical RP had normal or borderline normal final dark-adaptation thresholds, reduced but easily detectable ERG responses to 0.5-Hz white-light flashes (amplitudes 100-300 μV), minimal or no retinal vessel attenuation, and bone spicule-pigment deposits in most cases. Patients with Leber congenital amaurosis had severe visual dysfunction with severely reduced ERGs from early childhood. Patients with achromatopsia had a severe nonprogressive cone dysfunction from early childhood with 30-Hz cone ERG amplitudes that were not detectable without computer averaging (i.e., amplitudes <10 μV). Patients with cone-rod degeneration, when evaluated before the complete loss of their ERGs, had substantially less reduction in rod versus cone ERG amplitudes, such that the ratio of the amplitude of the 0.5-Hz rod-dominanted ERG versus the amplitude of the 30-Hz cone ERG was ≈30:1 or greater (the ratio in normal controls is ≈6:1). Patients with cone degeneration had progressively reduced acuity and cone ERGs that were substantially reduced with minimally reduced- or normal-amplitude rod ERGs. Patients with cone dysfunction typically had reduced visual acuity with no history of progression, normal fundi, and cone ERGs that indicated cone dysfunction with delayed implicit times and normal or reduced amplitudes. One patient with enhanced S-cone syndrome (patient 160-014) who was previously shown to carry the NR2E3 mutations R97H and IVS1-2A>C was included as a positive control (17).
Threshold static perimetry was performed with the 30-2 program of the Humphrey Field Analyzer by using the size III stimulus for conventional white-on-white (W/W) perimetry and the size V stimulus for blue-on-yellow (B/Y) perimetry (to isolate the short-wavelength-sensitive cone system). Sensitivity values were compared with age-matched norms provided by the instrument's database.
We used the Zeiss Stratus OCT3 instrument to obtain optical coherence tomography cross-sectional images of the retina through dilated pupils. Horizontal or vertical scans were centered on the fovea as viewed with the standard infrared background.
Blood samples were obtained from the patients and, in some cases, their relatives. Normal subjects were recruited as controls; many of the controls had eye examinations including ERGs that were within normal limits. Leukocyte DNA was purified by using standard procedures.
Exons containing the ORF of the NRL gene and their flanking intron splice sites were amplified by using the PCR with primer sequences published in ref. 27. The DNA sequences were obtained by direct sequencing with a dye-terminator cycle sequencing kit (Version 3.1) and an Applied Biosystems Model 3100 automated sequencer. The numbering of the bases of the cDNA and genomic sequences of NRL used in this paper is in accordance with GenBank entries NM_006177 and NT_026437, respectively.
Seven microsatellite markers (D14S548, D14S585, D14S990, D14S1430, D14S264, D14S1032, and D14S123) distributed between 2.2 Mb upstream and 2.5 Mb downstream of the NRL gene and six additional published highly polymorphic markers [HPRTB, FABP, CD4, MTH01, CYAR04, and LIPOL (32)] from different chromosomes were evaluated in DNA samples from selected patients to perform linkage, segregation, or parentage analysis. The primer sequences for these markers are reported in ref. 32 or are available at www.ncbi.nlm.nih.gov/genome/sts. The amplified DNA was radiolabeled with [32P] and diluted 1:1 with a solution of 95% formamide/20 mM EDTA/0.05% bromophenol blue/0.05% xylene cyanol. The fragments were separated through 6% denaturing polyacrylamide gels.
The human WT NRL and NRL-S50T (25) mutant coding sequences were cloned into the pcDNA4HisMax vector (Invitrogen). Site-directed mutagenesis was performed to create the NRL-P51S and NRL-L160P mutants, and the coding region of each construct was sequenced to establish that no additional changes had been introduced. Luciferase assays were performed by using a published method (25) with minor alterations. Briefly, human embryonic kidney 293 (HEK-293) cells (obtained from American Type Culture Collection, CRL-1573) were seeded at 4 × 104 cells per well in 24-well plates. Increasing amounts (0.0033, 0.01, 0.03, and 0.09 μg) of each plasmid containing the WT NRL insert or the NRL mutants S50T, P51S, or L160P were cotransfected with both a pGL2 plasmid containing the bovine rhodopsin promoter driving a luciferase cDNA sequence (p130-luc; 0.3 μg per well) and an expression plasmid containing the CRX cDNA sequence (pcDNA4-CRX; 0.1 μg per well). Empty pcDNA4 expression vector and cytomegalovirus-β-gal (0.1 μg per well) were included to control for the amount of transfected DNA and transfection efficiency, respectively. Transfections were performed by using FuGENE 6 (Roche Diagnostics). Luciferase assays (Luciferase Assay System, Promega) were repeated in triplicate at least twice and calculated as a fold change from the baseline (empty pcDNA4 expression vector). To estimate the level of protein produced by the WT and mutant NRL constructs, HEK-293 and COS-1 cells were seeded as above. Each construct was transfected into three wells (0.33 μg of DNA per well). Proteins from cells in each set of three wells were extracted and pooled, size-separated by SDS/PAGE, and transferred to nitrocellulose for immunoblot analysis by using polyclonal anti-NRL antibody (33).
Results
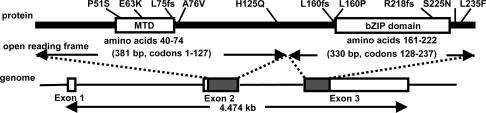
Identification of Sequence Variants. We evaluated the entire coding sequence and exon-flanking intron sequences of the NRL gene in 749 unrelated patients with various forms of retinal disease (Table 1) and 92 normal controls. Ten anisocoding changes (three frame-shift changes and seven missense changes) were discovered. Fig. 1 shows the locations of these changes in schematic diagrams of the gene and the protein. Table 2 summarizes the clinical findings of the patients identified with these changes.
Table 1. No. of unrelated patients screened.
| Diagnosis | No. |
|---|---|
| Autosomal recessive RP | 212 |
| Isolate RP | 301 |
| Atypical RP | 20 |
| Leber congenital amaurosis | 61 |
| Cone-rod degeneration | 96 |
| Achromatopsia | 12 |
| Cone degeneration | 10 |
| Cone dysfunction | 37 |
| Total | 749 |
Fig. 1.
Schematic diagram of the NRL protein, including the minimal transactivation domain (MTD) (39), the basic leucine zipper (bZIP) domain (40), and the intron/exon structure of the NRL gene. The locations of the 10 anisocoding changes identified in this study are indicated above the protein structure.
Table 2. Clinical findings of patients with DNA sequence changes found in the NRL gene.
| ERG amplitude, μV*
|
|||||||
|---|---|---|---|---|---|---|---|
| Patient | Protein | Nucleotide | Diagnosis | Age | Visual acuity in OD and OS | 0.5-Hz white OD and OS | 30-Hz white OD and OS |
| Likely pathogenic mutations | |||||||
| 048-096 | P51S/+† | 151 C>T | Autosomal dominant RP | 6 | 20/200, 20/70 | ND | ND |
| 003-001 | L75fs/ | 224_225insC | Clumped pigmentary RD | 43 | 20/40, 20/40 | 8, 6 | 0.9, 0.6 |
| 003-001 | L160P† | 479T>C | |||||
| Changes of uncertain significance | |||||||
| 078-059 | E63K/+ | 187G>A | Cone dysfunction syndrome | 19 | 20/100, 20/200 | 458, 396 | 37, 51 |
| 003-252 | A76V/+ | 227C>T | Autosomal recessive RP | 50 | 20/30, 20/30 | 4, 4 | 0.8, 0.9 |
| 115-031 | L160fs/+† | 459_477dup | Cone dysfunction syndrome | 24 | 20/100, 20/200 | 308, 264 | 4, 5 |
| 048-019 | R218fs/+† | 654delC | Leber congenital amaurosis | 23 | HM, HM | ND | 0.4, 0.5 |
| 115-039 | S225N/+ | 674G>A | Cone dysfunction syndrome | 44 | 20/200, 20/200 | 309, 212 | 39, 33 |
| Nonpathogenic changes | |||||||
| 182-003 | H125Q/+ | 375C>G | Cone dysfunction syndrome | 64 | 20/200, 20/200 | 306, 312 | 53, 53 |
| 246-007 | H125Q/+ | 375C>G | Atypical RP | 73 | 20/30, CF | 113, 127 | 17, 29 |
| 121-329 | L235F/+ | 703C>T | Isolate RP | 35 | 20/25, 20/30 | 11, 8 | 4, 3 |
OD, right eye OS, left eye; RD, retinal degeneration; HM, hand motions; CF, counting fingers at 4 ft (1 ft = 0.3 m); ND, nondetectable (i.e., < 10 μV); NA, not available.
Ranges of normal amplitudes are ≥350 μV for 0.5-Hz white ERGs and ≥50 μV for 30-Hz white ERGs.
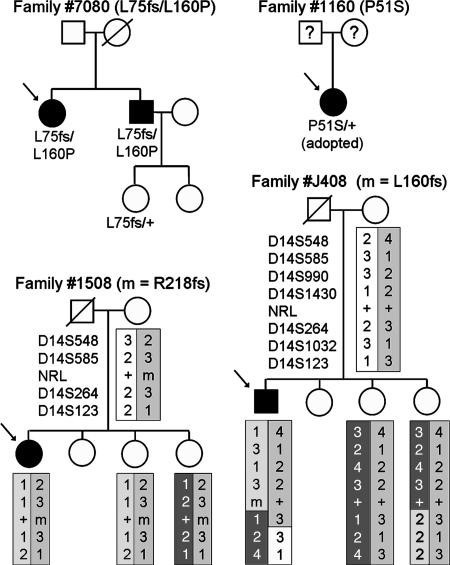
Schematic pedigrees are shown in Fig. 2.
Three of the sequence changes were interpreted as pathogenic mutations. One was the missense mutation P51S that was detected in a heterozygous female with severe retinal degeneration from early childhood. This patient was adopted and had no knowledge of her parents' ocular status. Because two previous missense mutations that altered the same residue (P51L and P51T) have been associated with autosomal dominant RP (26, 27), we interpreted the P51S allele as likely to be pathogenic. Another patient from a different family carried two mutations. One mutation was a 1-bp insertion in codon 75 (L75fs). This frameshift mutation resulted in a premature stop 19 codons downstream. It was interpreted as a null allele because this stop codon early in the reading frame would likely result in nonsense-mediated decay of the mutant RNA transcript, and, even if the RNA were translated, the resulting protein would have no basic leucine zipper domain. The second mutation was a missense change, L160P. Leucine at position 160 is a highly conserved residue among Maf proteins (data not shown). It is near the DNA-binding basic domain and is adjacent to an ancillary DNA-binding region (34), suggesting that the L160P mutation might affect DNA-binding-site recognition. In view of this location and especially because of the results of the in vitro functional assay of the encoded mutant protein (see below), we interpreted the L160P allele as likely to be pathogenic. The patient and her affected brother had recessive RP. Segregation analysis indicated that the L75fs and L160P mutations were allelic and that the two affected siblings were compound heterozygotes (Fig. 2); an unaffected daughter of the brother was an L75fs heterozygote.
Fig. 2.
Schematic pedigrees of four families with NRL sequence variants. Microsatellite marker alleles are presented for two families. Haplotypes were inferred and are displayed with differently shaded boxes. The affected patient in family 1160 was adopted, and information on all blood relatives was unavailable.
We did not have sufficient evidence, as described below, to classify five anisocoding changes as being either pathogenic or nonpathogenic. These changes (E63K, A76V, L160fs, R218fs, and S225N) were identified in heterozygotes with presumably recessive retinal disease (Fig. 1 and Table 2). A second mutation was not identified in any of these patients. Three of the changes (E63K, A76V, and S225N) altered residues that are not conserved among the members of the Maf protein family. They were found in one patient each with cone dysfunction syndrome or RP. These patients had no affected relatives, and segregation analysis was not performed. The two remaining changes were frameshift mutations (L160fs and R218fs). L160fs was a 19-bp duplication found in a patient with severe cone dysfunction. This mutation results in a premature stop codon; however, because the stop codon is in the last exon of NRL (Fig. 1), the transcribed mutant RNA is unlikely to be subject to nonsense-mediated decay. If the mutant RNA is translated, the corresponding mutant protein would likely have altered function because of 66 mutant residues at the carboxyl terminus and the disruption of a large part of the basic leucine zipper domain. Segregation analysis of the patient's family was complicated because the patient's father was deceased, and no affected relatives were available. Nevertheless, we evaluated the NRL gene and closely linked markers in three living family members (Fig. 2). The L160fs mutation was not found in any of them. Because the index patient's mother did not carry the L160fs mutation, it was either derived from the patient's deceased father or was a de novo mutation. Two paternally derived crossovers were detected among the patient and his siblings; in view of this, additional markers (HPRTB, FABP, CD4, MTH01, CYAR04, and LIPOL) from other chromosomes were evaluated, and the results were consistent with the designated parentage (data not shown). Whereas these results are consistent with L160fs being a dominant, pathogenic allele, the limited segregation analysis and the uncertainty about the phenotype of the deceased father precludes a definite interpretation of this allele.
The second frameshift mutation was a 1-bp deletion in codon 218 (R218fs) that creates a premature stop four codons downstream. This mutation would affect the carboxyl-terminal leucine-zipper region of NRL. The mutation was found in one heterozygote patient with Leber congenital amaurosis. The segregation analysis of NRL alleles in the family of this patient suggested that this mutation was not the cause of the patient's retinal disease (Fig. 2). Specifically, an unaffected sibling was found to share the same two NRL alleles as the patient.
Two missense changes, H125Q and L235F, were interpreted as nonpathogenic rare variants. H125Q was found in two unrelated heterozygotes with different, presumably autosomal, recessive phenotypes (atypical RP and cone dysfunction syndrome; Table 2). No second rare sequence variants were found in these patients. Residue H125 is not conserved among the members of the Maf protein family, and, in particular, a glutamine at the homologous position is present in at least one other member (Xenopus laevis L-Maf; GenBank accession no. AF202059). The L235F change was identified in one isolate RP case. The patient was a heterozygote, and no mutation was detected on the other allele. Residue L235 is only three residues upstream of the termination codon of NRL. It is also not conserved among the members of the Maf protein family, with other members having a phenylalanine at this position (e.g., Gallus gallus L-Maf; GenBank accession no. AF034570).
Three isocoding changes (L72L, c.G216A; S133S, c.399T; R147R, c.G441A) and five intronic changes (IVS1-37C>T, IVS1-25C>G, IVS2+84G>C, IVS2-37C>T, and IVS2-25C>G) were identified and were considered to be nonpathogenic. Four of them (S133S, R147R, IVS2+84G>C, and IVS2-25C>G) were found in multiple patients, although none was found at an allele frequency >1%. The remaining four variants were identified in one heterozygote patient each. None of these changes, and none of the other mutations or changes of uncertain significance described above, were predicted to affect RNA splicing, based on analysis of the DNA sequences with on-line computer software (www.fruitfly.org/seq_tools/splice.html) (35).
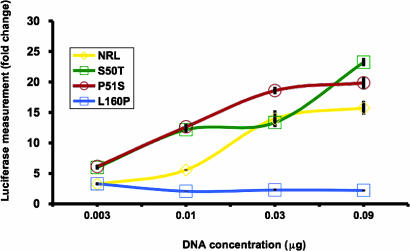
Rhodopsin Promoter Activity Assays of the NRL-P51S and NRL-L160P Mutants. To determine the effect of the P51S and L160P mutations, we performed transactivation assays to compare the abilities of the WT and mutant NRL proteins to stimulate the rhodopsin promoter driving a reporter gene (luciferase). Using an assay system similar to that used in ref. 25, we demonstrated that increasing amounts of DNA with the WT NRL sequence or the S50T and P51S mutants led to increases in rhodopsin promoter activity compared with an empty vector control (Fig. 3). The P51S and S50T mutants induced the rhodopsin promoter more intensively than WT NRL at the 0.003- and 0.01-μg concentrations (t test, P < 0.01). However, the L160P mutant only minimally activated the rhodopsin promoter at all concentrations tested. This level of promoter activation was statistically significantly lower than that induced by WT NRL at the 0.01-, 0.03-, and 0.09-μg amounts (t test, P < 0.001). To establish whether the low L160P activity was due to altered protein production, the constructs were individually transfected into HEK-293 and COS-1 cells and tested for their ability to express the encoded protein. The low amount of protein produced by HEK-293 cells could not be detected by immunoblotting; however, each plasmid was capable of producing its respective WT or mutant NRL protein in COS-1 cells (data not shown), suggesting that the L160P construct's poor activation of the rhodopsin promoter was not due to a defect in protein production.
Fig. 3.
Measure of the activation of the rhodopsin promoter by WT and mutant NRL proteins in the presence of CRX with transient transfection assays. DNA constructs encoding WT NRL, NRL-S50T, NRL-P51S, and NRL-L160P proteins were transfected into HEK-293 cells at increasing concentrations (0.0033, 0.01, 0.03, and 0.09 μg) in the presence of a constant amount of pcDNA4-CRX. Fold change is relative to the empty expression vector control. Error bars indicate the SE. SEs for the NRL, S50T, P51S, and L160P samples ranged from 0.17 to 2.75, 0.72 to 1.32, 0.52 to 2.06, and 0.17 to 0.44, respectively.
Clinical Assessment of the Patients. The patient with the P51S mutation had severe, autosomal dominant RP. She was an adopted female with no family history available. Visual acuity by history was 20/200 in each eye at 4 years of age, when nystagmus and a severe reduction of both rod and cone ERG responses were documented. At 36 years of age, she had nystagmus, a visual acuity of 20/80 OU, a tritan axis of confusion with the Farnsworth D-15 panel (consistent with more reduction of blue-cone function compared with red and green cone function), severely constricted visual fields, an elevation of the final dark-adaptation threshold by 2 log units, and markedly reduced rod and cone full-field ERGs (Table 2).
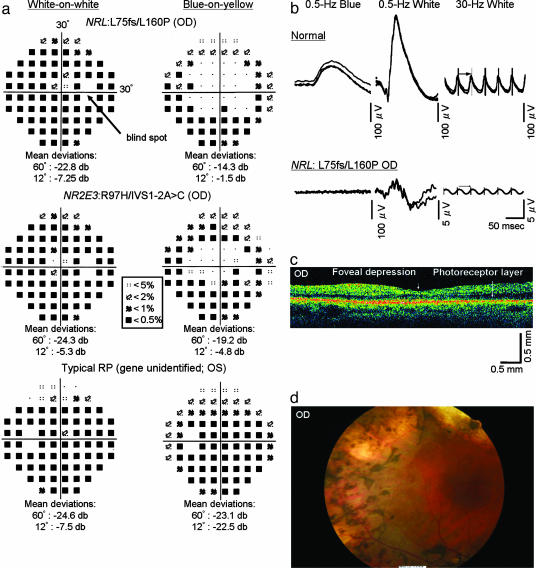
We were able to comprehensively evaluate only one of the two siblings with the L75fs and L160P mutations, the brother of patient 003-001 at 51 years of age. This patient was diagnosed as having clumped pigmentary retinal degeneration, a subtype of autosomal recessive RP. He had night blindness from early childhood. He noted a slowly progressive loss of his peripheral visual field. The patient was mildly myopic (refractive error of -2.0 diopters spherical equivalent). Best corrected visual acuities were 20/40 (right eye) and 20/200 (left eye); the left eye was considered to be amblyopic. Color vision evaluated with the Farnsworth D-15 panel and the Ishihara plates was normal. The Goldmann dynamic visual field testing showed constriction of the visual field, with the remaining central field having a diameter of ≈22° with the V-4e stimulus. Optical coherence tomography (Fig. 4b) showed that the overall retinal thickness was reduced, the photoreceptor layer was present, and the macula had a recognizable foveal depression. No macular cysts or schises were apparent. Funduscopy showed clumps of pigment in the periphery at the level of retinal pigment epithelium with shapes that are distinct from the bone spicule-shaped pigment deposits seen in patients with typical RP (Fig. 4). A more limited clinical evaluation of this patient's affected sibling showed similar visual acuities, Goldmann visual fields, and ERG amplitudes.
Fig. 4.
Clinical analyses of a patient with the recessive NRL mutations L75fs and L160P and two control patients. (a) Humphrey automated static perimetry performed on the right eye of a patient with recessive NRL mutations (Top), the right eye of a patient with enhanced S-cone syndrome with recessive NR2E3 mutations (Middle), and the left eye of a patient with typical RP and no identified mutations (Bottom). The fields were measured separately with W/W (Left) and B/Y (Right) protocols. Threshold deviations from normal controls are presented as different symbols (see code in central box). Below each field are the mean deviations in sensitivities in the central 60° and 12° compared with age-matched norms from the instrument's database. The location of the blind spot is shown in Top Left by an arrow. (b) Dark-adapted, full-field ERGs from a normal control individual (Upper) and the NRL patient (Lower). The columns show rod responses to 0.5-Hz blue flashes [1.2 log foot Lambert (ft.-L.), λmax = 440 nm] (Left), rod-plus-cone responses to 0.5-Hz white flashes (3.8 log ft.-L.) (Center), and cone responses to 30-Hz white flashes (3.8 log ft.-L.) (Right). Scale bars are at the lower-right corner of each ERG. For each test condition, two to four consecutive sweeps are shown to illustrate reproducibility, except for the 0.5-Hz white and 30-Hz white ERGs of the NRL patient, for which computer-averaged ERGs are shown because these ERGs were nondetectable otherwise. (c) Optical coherence tomography image of the right eye of the NRL patient. Scale bars are at the lower right corner. Arrowhead indicates foveal depression. Arrow indicates the preserved photoreceptor layer. (d) Fundus photo of the right eye of the NRL patient showing clumped pigment deposits in the periphery, attenuated retinal vessels, and regions of chorioretinal atrophy.
Full-field ERG recordings revealed a severe reduction of both rod and cone amplitudes (the amplitudes were only 4-5 μV in response to single white flashes, normal ≥350 μV; see Table 2 and Fig. 4). The considerable reduction in amplitudes precluded an evaluation of S-cone function by using ERG recordings with stimuli of different colors. However, the patient's relatively preserved visual acuity and central retinal function allowed us to evaluate S-cone function by using Humphrey static perimetry with differently colored test lights and backgrounds. We performed W/W and B/Y perimetry to compare blue cone (S-cone) mediated vision (B/Y perimetry) with that mediated by all three cone types (W/W perimetry). Similar W/W and B/Y perimetry measurements were also conducted in three additional control patients: a previously reported male with enhanced S-cone syndrome who had identified mutations in NR2E3 (patient 160-014; ref. 17) and two unrelated females with typical RP, no affected relatives, and no identified responsible gene defects. All four patients, including the patient with recessive NRL mutations, had similar visual acuities (20/30-20/60), normal color vision as determined with the Farnsworth D-15 panel, minimal or no evidence of cataract, and reduced ERG amplitudes to single white-light flashes (5, 25, 21, and 40 μV in the NRL, NR2E3, and two isolate RP patients, respectively; normal ≥350 μV). All patients had similar mean W/W sensitivity reductions in the central 12° (-4.5 to -7.5 dB). However, the NRL patient and the NR2E3 patient had mean B/Y sensitivity reductions of only -1.5 to -4.8 dB, whereas the typical RP patients had more severe B/Y reductions, -15.5 and -22.5 dB (the visual fields of only one of the two typical RP patients are shown in Fig. 4).
Discussion
We describe here patients identified with recessive, loss-of-function mutations in the NRL gene. This genotype and the corresponding phenotype would be expected to be homologous to that in previously reported transgenic knockout mice lacking Nrl (8). The Nrl-/- mice have no rod photoreceptors and a large excess of S-cones. The patients with the recessive NRL mutations have ERGs that were too severely reduced to evaluate the level of rod function versus S-cone function. However, based on our analysis of visual fields tested with light stimuli of different colors in one of the patients, we obtained evidence that S-cone function in the central retina was preserved in a manner similar to that found in a patient with enhanced S-cone syndrome who had comparably severe retinal degeneration caused by mutations in the NR2E3 gene, the only previously known cause of enhanced S-cone syndrome in humans. This pattern of cone function observed with recessive NR2E3 or NRL mutations is the reverse of preferential reduction of S-cone function that is found in patients with typical RP (36, 37). In addition to the preservation of S-cone function, patients with recessive NR2E3 or NRL mutations have a similar pattern of intraretinal pigmentation in their fundi. This pattern of abnormal pigment clumping in the periphery is the hallmark of a category of RP called clumped pigmentary retinal degeneration; it is found in ≈0.5% of RP cases (38). About half of all patients with clumped pigmentary retinal degeneration have mutations in the NR2E3 gene and are considered to have the enhanced S-cone syndrome (17). Based on our identification of only one family with recessive NRL mutations in our set of patients, it appears that mutations in NRL are a much less common cause of clumped pigmentary retinal degeneration than mutations in NR2E3.
We also report a NRL mutation, P51S, in a patient with dominant RP. This is the third reported mutation to affect residue P51 (26, 27). The affected patient since age 4 years has been symptomatic with severe loss of vision and severely reduced ERG amplitudes. In contrast to the recessive loss-of-function mutations in NRL, the dominant P51S mutation was not associated with clumped pigment deposits, and there was a preferential loss of blue cone function indicated by a tritan axis of confusion with the Farnsworth D-15 panel; these features are more typical of RP. The contrast in phenotypes produced by dominant versus recessive NRL mutations suggests that different mechanisms are responsible.
We identified seven additional anisocoding changes in NRL. This number is a considerable increase in the number of known anisocoding changes in this gene because alterations affecting only three different codons in NRL have been reported so far (25-28). Five of the newly identified changes (E63K, A76V, L160fs, R218fs, and S225N) were of uncertain significance. Our inability to determine the pathogenicity of these changes was due in part to the limited number of relatives of the index patients precluding an evaluation of whether the changes are strongly associated with specific diseases. Additional studies are required to delineate whether these sequences contribute to the development of retinal degeneration or dysfunction.
Acknowledgments
This work was supported by National Eye Institute Grants EY08683, EY00169, and EY11115 and the Foundation Fighting Blindness (FFB). J.S.F. is a recipient of a FFB-Canada postdoctoral fellowship. A.S. is a Harold F. Falls Collegiate Professor and a recipient of a Research to Prevent Blindness Senior Scientific Investigator award.
Author contributions: K.M.N., J.S.F., M.A.S., A.S., E.L.B., and T.P.D. designed research; K.M.N., J.S.F., and M.A.S. performed research; K.M.N., J.S.F., M.A.S., A.S., E.L.B., and T.P.D. analyzed data; and K.M.N., J.S.F., M.A.S., A.S., E.L.B., and T.P.D. wrote the paper.
Abbreviations: B/Y, blue-on-yellow; ERG, electroretinogram; HEK, human embryonic kidney; RP, retinitis pigmentosa; S-cones, short-wavelength-sensitive cones; W/W, white-on-white.
References
- 1.Swaroop, A., Xu, J. Z., Pawar, H., Jackson, A., Skolnick, C. & Agarwal, N. (1992) Proc. Natl. Acad. Sci. USA 89, 266-270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Rehemtulla, A., Warwar, R., Kumar, R., Ji, X., Zack, D. J. & Swaroop, A. (1996) Proc. Natl. Acad. Sci. USA 93, 191-195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kumar, R., Chen, S., Scheurer, D., Wang, Q. L., Duh, E., Sung, C. H., Rehemtulla, A., Swaroop, A., Adler, R. & Zack, D. J. (1996) J. Biol. Chem. 271, 29612-29618. [DOI] [PubMed] [Google Scholar]
- 4.Chen, S., Wang, Q. L., Nie, Z., Sun, H., Lennon, G., Copeland, N. G., Gilbert, D. J., Jenkins, N. A. & Zack, D. J. (1997) Neuron 19, 1017-1030. [DOI] [PubMed] [Google Scholar]
- 5.Mitton, K. P., Swain, P. K., Chen, S., Xu, S., Zack, D. J. & Swaroop, A. (2000) J. Biol. Chem. 275, 29794-29799. [DOI] [PubMed] [Google Scholar]
- 6.Lerner, L. E., Gribanova, Y. E., Ji, M., Knox, B. E. & Farber, D. B. (2001) J. Biol. Chem. 276, 34999-35007. [DOI] [PubMed] [Google Scholar]
- 7.Pittler, S. J., Zhang, Y., Chen, S., Mears, A. J., Zack, D. J., Ren, Z., Swain, P. K., Yao, S., Swaroop, A. & White, J. B. (2004) J. Biol. Chem. 279, 19800-19807. [DOI] [PubMed] [Google Scholar]
- 8.Mears, A. J., Kondo, M., Swain, P. K., Takada, Y., Bush, R. A., Saunders, T. L., Sieving, P. A. & Swaroop, A. (2001) Nat. Genet. 29, 447-452. [DOI] [PubMed] [Google Scholar]
- 9.Kobayashi, M., Takezawa, S., Hara, K., Yu, R. T., Umesono, Y., Agata, K., Taniwaki, M., Yasuda, K. & Umesono, K. (1999) Proc. Natl. Acad. Sci. USA 96, 4814-4819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Akhmedov, N. B., Piriev, N. I., Chang, B., Rapoport, A. L., Hawes, N. L., Nishina, P. M., Nusinowitz, S., Heckenlively, J. R., Roderick, T. H., Kozak, C. A., et al. (2000) Proc. Natl. Acad. Sci. USA 97, 5551-5556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Haider, N. B., Naggert, J. K. & Nishina, P. M. (2001) Hum. Mol. Genet. 10, 1619-1626. [DOI] [PubMed] [Google Scholar]
- 12.Cheng, H., Khanna, H., Oh, E. C., Hicks, D., Mitton, K. P. & Swaroop, A. (2004) Hum. Mol. Genet. 13, 1563-1575. [DOI] [PubMed] [Google Scholar]
- 13.Bumsted O'Brien, K. M., Cheng, H., Jiang, Y., Schulte, D., Swaroop, A. & Hendrickson, A. E. (2004) Invest. Ophthal. Vis. Sci. 45, 2807-2812. [DOI] [PubMed] [Google Scholar]
- 14.Haider, N. B., Jacobson, S. G., Cideciyan, A. V., Swiderski, R., Streb, L. M., Searby, C., Beck, G., Hockey, R., Hanna, D. B., Gorman, S., et al. (2000) Nat. Genet. 24, 127-131. [DOI] [PubMed] [Google Scholar]
- 15.Gerber, S., Rozet, J. M., Takezawa, S. I., dos Santos, L. C., Lopes, L., Gribouval, O., Penet, C., Perrault, I., Ducroq, D., Souied, E., et al. (2000) Hum. Genet. 107, 276-284. [DOI] [PubMed] [Google Scholar]
- 16.Milam, A. H., Rose, L., Cideciyan, A. V., Barakat, M. R., Tang, W. X., Gupta, N., Aleman, T. S., Wright, A. F., Stone, E. M., Sheffield, V. C. & Jacobson, S. G. (2002) Proc. Natl. Acad. Sci. USA 99, 473-478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sharon, D., Sandberg, M. A., Caruso, R. C., Berson, E. L. & Dryja, T. P. (2003) Arch. Ophthalmol. 121, 1316-1323. [DOI] [PubMed] [Google Scholar]
- 18.Marmor, M. F. (1989) Doc. Ophthalmol. 71, 265-269. [DOI] [PubMed] [Google Scholar]
- 19.Fishman, G. A. & Peachey, N. S. (1989) Ophthalmology 96, 913-918. [DOI] [PubMed] [Google Scholar]
- 20.Marmor, M. F., Jacobson, S. G., Foerster, M. H., Kellner, U. & Weleber, R. G. (1990) Am. J. Ophthalmol. 110, 124-134. [DOI] [PubMed] [Google Scholar]
- 21.Jacobson, S. G., Marmor, M. F., Kemp, C. M. & Knighton, R. W. (1990) Invest. Ophthalmol. Vis. Sci. 31, 827-838. [PubMed] [Google Scholar]
- 22.Jacobson, S. G., Roman, A. J., Roman, M. I., Gass, J. D. & Parker, J. A. (1991) Am. J. Ophthalmol. 111, 446-453. [DOI] [PubMed] [Google Scholar]
- 23.Hood, D. C., Cideciyan, A. V., Roman, A. J. & Jacobson, S. G. (1995) Vision Res. 35, 1473-1481. [DOI] [PubMed] [Google Scholar]
- 24.Acar, C., Mears, A. J., Yashar, B. M., Maheshwary, A. S., Andreasson, S., Baldi, A., Sieving, P. A., Iannaccone, A., Musarella, M. A., Jacobson, S. G., et al. (2003) Mol. Vis. 9, 14-17. [PubMed] [Google Scholar]
- 25.Bessant, D. A., Payne, A. M., Mitton, K. P., Wang, Q. L., Swain, P. K., Plant, C., Bird, A. C., Zack, D. J., Swaroop, A. & Bhattacharya, S. S. (1999) Nat. Genet. 21, 355-356. [DOI] [PubMed] [Google Scholar]
- 26.Martinez-Gimeno, M., Maseras, M., Baiget, M., Beneito, M., Antinolo, G., Ayuso, C. & Carballo, M. (2001) Hum. Mutat. 17, 520. [DOI] [PubMed] [Google Scholar]
- 27.DeAngelis, M. M., Grimsby, J. L., Sandberg, M. A., Berson, E. L. & Dryja, T. P. (2002) Arch. Ophthalmol. 120, 369-375. [DOI] [PubMed] [Google Scholar]
- 28.Bessant, D. A., Holder, G. E., Fitzke, F. W., Payne, A. M., Bhattacharya, S. S. & Bird, A. C. (2003) Arch. Ophthalmol. 121, 793-802. [DOI] [PubMed] [Google Scholar]
- 29.Berson, E. L., Gouras, P. & Gunkel, R. D. (1968) Arch. Ophthalmol. 80, 58-67. [DOI] [PubMed] [Google Scholar]
- 30.Reichel, E., Bruce, A. M., Sandberg, M. A. & Berson, E. L. (1989) Am. J. Ophthalmol. 108, 540-547. [DOI] [PubMed] [Google Scholar]
- 31.Andreasson, S. O., Sandberg, M. A. & Berson, E. L. (1988) Am. J. Ophthalmol. 105, 500-503. [DOI] [PubMed] [Google Scholar]
- 32.Alford, R. L., Hammond, H. A., Coto, I. & Caskey, C. T. (1994) Am. J. Hum. Genet. 55, 190-195. [PMC free article] [PubMed] [Google Scholar]
- 33.Swain, P. K., Hicks, D., Mears, A. J., Apel, I. J., Smith, J. E., John, S. K., Hendrickson, A., Milam, A. H. & Swaroop, A. (2001) J. Biol. Chem. 276, 36824-36830. [DOI] [PubMed] [Google Scholar]
- 34.Kerppola, T. K. & Curran, T. (1994) Oncogene 9, 3149-3158. [PubMed] [Google Scholar]
- 35.Reese, M. G., Eeckman, F. H., Kulp, D. & Haussler, D. (1997) J. Comput. Biol. 4, 311-323. [DOI] [PubMed] [Google Scholar]
- 36.Sandberg, M. A. & Berson, E. L. (1977) Invest. Ophthalmol. Vis. Sci. 16, 149-157. [PubMed] [Google Scholar]
- 37.Greenstein, V. C., Hood, D. C., Ritch, R., Steinberger, D. & Carr, R. E. (1989) Invest. Ophthalmol. Vis. Sci. 30, 1732-1737. [PubMed] [Google Scholar]
- 38.To, K. W., Adamian, M., Jakobiec, F. A. & Berson, E. L. (1996) Arch. Ophthalmol. 114, 950-955. [DOI] [PubMed] [Google Scholar]
- 39.Friedman, J. S., Khanna, H., Swain, P. K., DeNicola, R., Cheng, H., Mitton, K. P., Weber, C. H., Hicks, D. & Swaroop, A. (2004) J. Biol. Chem. 279, 47233-47241. [DOI] [PubMed] [Google Scholar]
- 40.Motohashi, H., O'Connor, T., Katsuoka, F., Engel, J. D. & Yamamoto, M. (2002) Gene 294, 1-12. [DOI] [PubMed] [Google Scholar]