Abstract
Background: Mutations in the epidermal growth factor receptor (EGFR) tyrosine kinase domain (TKD) are associated with response and resistance to targeted therapy. The EGFR mutation status in patients with advanced oral and oropharyngeal squamous cell carcinoma (OOSCC) was evaluated. A systematic literature review was undertaken to summarize current evidence and estimate the overall prevalence of EGFR TKD mutations in patients with head and neck squamous cell carcinoma (HNSCC). Materials and Methods: Genomic DNA was extracted from formalin-fixed, paraffin-embedded tumor samples of 113 patients with OOSCC. Pyrosequencing was performed to investigate mutations in EGFR exons 18 to 21. Medline databases were searched for relevant studies. Studies reporting mutations in the EGFR TKD in HNSCC were eligible for inclusion in the systematic review. Results: No mutations in the EGFR TKD were observed in 113 samples of OOSCC. A total of 53 eligible studies were included in the systematic review. In total, from the review, 117 patients harboring a total of 159 EGFR TKD mutations were reported among 4122 patients with HNSCC. The overall prevalence of EGFR TKD mutations in HNSCC was 2.8%. Conclusion: Large-scale studies are warranted to provide further evidence regarding the mutation status of EGFR in patients with HNSCC.
Keywords: Oral and oropharyngeal cancer, EGFR kinase domain, mutations, pyrosequencing, review
Head and neck squamous cell carcinoma (HNSCC) remains a challenging disease despite intensive clinical and translational research (1-3). A subset of head and neck cancer is caused by human papillomavirus (HPV) and represents a biologically distinct entity (4). In the past decades, several treatment strategies have been applied to treat HNSCC, however, survival outcomes have not substantially changed, emphasizing the need for more personalized medicine (5-8). Many efforts have, therefore, been made to identify predictive biomarkers and tailor treatment to the individual patient based on their own genetic and molecular profile.
The epidermal growth factor receptor (EGFR) is a transmembrane cell surface receptor belonging to the human epidermal growth factor receptor (HER) family of receptor tyrosine kinases. EGFR overexpression occurs in more than 90% of HNSCCs and has been correlated with poor outcome (9). Robust preclinical evidence underlines the role of EGFR in the development of HNSCC, showing that EGFR activation triggers several downstream signaling pathways that play a crucial role in cancer pathogenesis (3,10,11). In this context, strategies for inhibition of EGFR signaling using monoclonal antibodies and tyrosine kinase inhibitors (TKIs) have been investigated intensively in clinical trials. De novo or acquired resistance to EGFR-targeted therapy, however, has led to a modest survival benefit for patients with HNSCC, while up-to-date predictive biomarkers of treatment response remain elusive (8,12,13).
In non-small cell lung carcinoma (NSCLC), patients with activating mutations in the EGFR tyrosine kinase domain (TKD) are sensitive to small-molecule EGFR TKIs such as gefitinib, erlotinib, and afatinib (14-17). Given that mutations in the EGFR TKD may help in the selection of patients for EGFR TKIs or other targeted therapies, the EGFR mutation status in treatment-naïve patients with locally advanced oral and oropharyngeal squamous cell carcinoma (OOSCC) was retrospectively evaluated. In addition, a systematic literature review was undertaken to summarize current evidence regarding the EGFR mutation status in HNSCC. The present study aimed to estimate the overall prevalence of EGFR TKD mutations in patients with HNSCC and whether differences in the prevalence of EGFR mutations exist between patients with HNSCC across different countries and geographic regions.
Materials and Methods
Population of cohort study. This study included 113 patients diagnosed with primary locally advanced OOSCC who underwent neoadjuvant chemoradiation followed by tumor resection at the Department of Radiotherapy and Department of Cranio-Maxillofacial and Oral Surgery at the Medical University of Vienna between 2000 and 2009. All of them had: biopsy-proven OOSCC, available pretreatment biopsy tumor tissues, clinical TNM stage III or IV disease, no distant metastasis, no previous history of head and neck cancer, performance status and laboratory parameters permitting chemoradiotherapy and surgery, and had undergone complete resection (R0) of the primary tumor. The multimodal treatment comprised neoadjuvant chemotherapy with mitomycin C (15-20 mg/m2, i.v. bolus injection on day 1) and 5-fluorouracil (750 mg/m2/day, continuous infusion on days 1-5), concurrent with radiation therapy delivered over 5 weeks up to a total dose of 50 Gy (25 fractions of 2 Gy per day) followed by post-treatment radical surgery. Surgery was performed 4-8 weeks after the end of radiotherapy. Patients were followed-up regularly for further 5 years.
Patient data were obtained from the Vienna General Hospital Patient Information System (AKIM). Clinical and pathological TNM staging was based on the seventh edition of the classification Union for International Cancer Control (UICC) (18). The surgical specimens were histopathologically evaluated by means of an institutional standard protocol. Pathological complete response (pCR) was defined by the absence of residual cancer within both the primary tumor site and regional lymph nodes. A study-specific patient number was given to patients to protect their identity. The Ethics Committee of the Medical University of Vienna approved this research (approval number: 774/2008).
HPV testing. E6/E7-specific quantitative reverse transcriptase-polymerase chain reaction (qRT-PCR) was used to determine the HPV16/18 status. Results were corroborated by p16 (cyclin-dependent kinase inhibitor 2A) immunohistochemical staining.
EGFR mutation testing. EGFR mutations were studied in DNA extracted from formalin-fixed, paraffin-embedded tissue blocks routinely archived at the Department of Pathology at the Medical University of Vienna. For each patient, one section of an appropriate tumor block was stained with hematoxylin and eosin to confirm the presence of viable carcinoma cells. DNA was extracted from tumor samples confirmed to have >50% cancer cells. From each block, 5-μm-thick sections were cut for DNA extraction performed using the QIAamp DNA FFPE Tissue Kit (Qiagen, Hilden, Germany) according to the manufacturer’s protocol. EGFR mutation analysis was carried out with Therascreen® Pyro Kit (Qiagen, Hilden, Germany). The EGFR kit enables detection and quantitation of the most common mutations in codon 719 (exon 18), exon 19 deletions, codon 768 and 790 (exon 20) and codon 858-861 (exon 21) of the EGFR gene.
After the extraction of genomic DNA, EGFR was amplified by PCR with HotStarTaq DNA Polymerase (Qiagen) after the DNA concentration of each sample was adjusted to 2 ng/μl. The PCR conditions were 95˚C for 15 min for the initial activation of HotStarTaq DNA Poymerase, followed by a 3-step-cycling: denaturation at 95˚C for 20 s, annealing at 53˚C for 30 s and extension at 72˚C for 20 s for 42 cycles. Finally, incubation at 72˚C for 5 min was accomplished for the final extension. After amplification, the immobilization of PCR products on Streptavidin Sepharose High-Performance (GE Healthcare, Uppsala, Sweden) beads was performed using a volume of 10 μl PCR product which was added to a 24-well PCR plate containing 70 μl master mix [2 μl Streptavidin Sepharose High-Performance beads, 40 μl of PyroMark Binding Buffer (Qiagen) and 28 μl water]. The next step was the preparation of single-stranded DNA with a PyroMark Q24 Vacuum Workstation (Qiagen) and the annealing of the sequencing primer (included in the Therascreen® Pyro Kit; Qiagen) to the template. Pyrosequencing of the samples was then carried out on a PyroMark Q24 MDx system (Qiagen). The results were analyzed with PyroMark Q24 software (version 2.0.6; Qiagen). Pyrosequencing results in the initial round of sequencing were confirmed by subsequent runs of independent PCRs and pyrosequencing, as well as by Sanger sequencing.
Systematic literature review: Data sources, search strategy, selection of studies, and data extraction. This study followed the Preferred Reporting Items for Systematic Reviews and Meta-Analyses (PRISMA) guidelines (19). Medline databases (hosts: PubMed and OVID) from inception up to October 20, 2016 were searched for relevant studies using the key words “head and neck cancer”, “EGFR”, and “mutation”. No search restriction was applied. The complete search strategy can be found in Appendix A. In addition, manual searches were conducted on the web and by reviewing the reference lists of the retrieved articles.
Studies reporting the mutation status of the EGFR TKD in tumor tissues of patients with HNSCC were eligible for inclusion in the systematic review. For quantitative synthesis, only studies reporting the prevalence of EGFR TKD mutations in HNSCC were considered. Letters and unpublished research were not included in the present review. Case reports were considered as qualitative evidence. Two reviewers (CP and RP) independently carried out study selection and data extraction. Any disagreements between reviewers were resolved by consensus involving a third reviewer (JE). The reviewers independently screened all records that were identified by the search strategy. Duplicate publications were excluded both electronically and manually. The selected records were pooled, retrieved as full-text publications, and assessed for eligibility. The two reviewers independently extracted data from each eligible study using a predefined data-abstraction sheet. The following data were collected: name of the first author, year of publication, study location, characteristics of study cohorts (sample size, tumor stage, tumor site), source of tumor profiled, exon location and type of EGFR mutations, detection methods, prognostic effect of EGFR mutations, and the prevalence of EGFR mutations. The PRISMA flow diagram was used to describe the study selection processes.
Statistical analysis. For the cohort study, patient characteristics were summarized using descriptive statistics. Categorical variables are described with frequencies and percentages. Patient demographic, clinical, and tumor characteristics were tested for association with pathological resonse using the chi-square test for categorical variables. Overall survival was defined as the time from surgery to death from any cause or to date of last follow-up. The Kaplan–Meier method was used for overall survival assessment and the log-rank test to compare differences in survival between groups. A two-sided p-value of less than 0.05 was considered statistically significant.
The systematic review was quantitatively analyzed to pool the overall prevalence of EGFR mutations in HNSCC. Subgroup analysis was performed to assess the prevalence of EGFR mutations according to geographic region. Prevalence of EGFR mutations was defined as the proportion of patients with EGFR-mutated tumors among patients who underwent the a mutation testing and was assessed as percentage with the 95% confidence interval (CI) (20). Subgroups of geographic regions (Europe, North America, Southeast Asia, and South Asia) were generated if two or more studies on a specific geographic region were present. Study location was defined based on the country where the patients were recruited in the study. Statistical analysis was performed using the Statistical Package for the Social Sciences (SPSS®, version 21.0; IBM Corp., Armonk, NY, USA).
Results
Description of patient cohort and survival analysis. The clinical and histopathological characteristics of 113 study patients are presented in Table I. The median age of patients was 58 years (range=24-79 years) and most of the patients were male (73%) and current smokers (83%). The primary tumor was predominantly located in the oral cavity (86%). Pathological complete response to neoadjuvant chemoradio-therapy was observed in 46 patients (41%). Nine patients (8%) were HPV-positive. Among HPV-positive patients, two (22%) achieved a complete pathological response. Of 16 oropharyngeal tumours, two samples (13%) were positive for HPV, compared to seven HPV-positive samples (7%) out of 97 oral cavity tumors. At 2 years, the overall survival rate of the cohort was 66% and at 5 years 46%. The median follow-up time was 4.6 years by which time 56 (50%) patients had died. The overall survival of the 113 patients with OOSCC was assessed according to the pathological response status using the Kaplan–Meier method (Figure 1). The median overall survival was significantly higher in patients with tumors showing pathological complete response compared with those having non-complete tumor regression (7.9 versus 2.8 years respectively, log-rank p=0.001).
Table I. Patient clinical and histopathological characteristics.

pCR, Pathological complete response; HPV, human papillomavirus.
Figure 1. Kaplan–Meier estimates of the probability of overall survival in 113 patients with oral and oropharyngeal cancer according to pathological response to neoadjuvant treatment.

Mutation status of EGFR. The kinase domain of EGFR (exons 18-21) was analyzed in tumor samples from 113 treatment-naïve patients with primary locally advanced OOSCC. Using pyrosequencing technology, no EGFR mutations were detected in any of the 113 tumor tissues (Figure 2).
Figure 2. Representative pyromarks of wild-type EGFR in exons 18-21.

Characteristics of studies included in the systematic review. Overall, 53 eligible studies were included in the present systematic review. The literature search of databases and reference lists yielded 1203 records. After removing duplicates, 886 records remained to be screened. After screening of titles and abstracts, 824 studies that reported on unrelated topics were excluded. The full-text papers of the remaining 62 studies were reviewed in depth. A total of nine studies did not provide sufficient information regarding the EGFR TKD mutation status and were discarded, thus 53 studies were included in the qualitative review and the quantitative synthesis. The PRISMA flow diagram was used to illustrate the study selection process (Figure 3).
Figure 3. Flow diagram of the study selection process.

Table II shows the main characteristics of the studies included in the systematic review. All studies were published between 2005 and 2013 in peer-reviewed journals. Most of the studies were conducted in the USA (n=19), followed by the Republic of Korea (n=5), and India (n=4). The source for tumor DNA mutation profiling was fresh frozen tumor samples (n=26), formalin-fixed paraffin-embedded tumor tissue (n=23), or both (n=4). Three studies investigated the prognostic impact of EGFR mutation status on HNSCC survival using the Kaplan–Meier method/Cox proportional hazards regression model (21-23).
Table II. Main characteristics of 53 eligible studies included in the systematic review.

NA, Not available; NS, non-significant; EGFR, epidermal growth factor receptor; TKD, tyrosine kinase domain; OC, oral cavity; OP, oropharynx; H&N, head and neck; UADT, upper aerodigestive tract (included oral cavity, pharynx, larynx and oesophagus); FFPE, formalin-fixed paraffinembedded; OS, overall survival; DFS, disease-free survival; TR, treatment response; TCGA, The Cancer Genome Atlas; ICGC, International Cancer Genome Consortium; MALDI-TOF MS, matrix-assisted laser desorption ionization time-of-flight mass spectrometry; MFI, median fluorescence intensity; HRMA, high resolution melting analysis; SSCP, single-strand conformational polymorphism; NGS, next-generation sequencing.
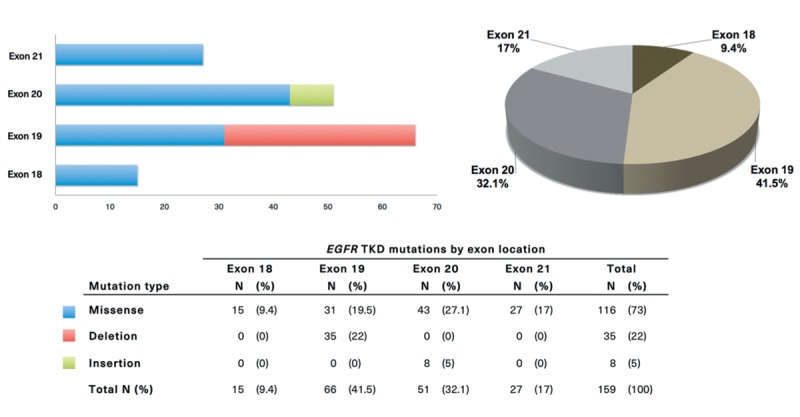
Overall prevalence of EGFR TKD mutations in HNSCC and distribution of mutations by exon location and type. In total, 117 patients harboring a total of 159 EGFR TKD mutations were reported among 4122 patients with HNSCC. The overall pooled prevalence of EGFR TKD mutations in HNSCC was 2.8% (95% CI=2.4-3.4%). The distribution of mutations in the EGFR TKD categorized by exon location and mutation type is shown in Figure 4. Among EGFR-mutated HNSCCs, the majority of TKD mutations were located in exon 19 (41.5%, 95% CI=33.8-49.6%), followed by exon 20 (32.1%, 95% CI=25.0-40.0%), exon 21 (17.0%, 95% CI=11.7-23.9%), and exon 18 (9.4%, 95% CI=5.6-15.3%). Of all EGFR mutations, missense mutations in exons 18-21 occurred in 73% (95% CI=65.2-79.6%), followed by deletions in exon 19 (22%, 95% CI=16.0-29.4%) and insertion mutations in exon 20 (5%, 95% CI=2.4-10.0%). The missense mutations T790M in exon 20 and L858R in exon 21 well-known in NSCLC occurred in 7.5% (95% CI=4.1-13.1%) and 2.5% (95% CI=0.8-6.7%) of all EGFR mutations, respectively.
Figure 4. Distribution of mutations in the EGFR kinase domain categorized by exon location and mutation type.
Prevalence of EGFR TKD mutations in HNSCC by study location. The EGFR mutation prevalence classified by study location (countries and geographic regions) is shown in Table III. The prevalence of EGFR-mutated HNSCCs was estimated in four geographic regions: the highest prevalence was shown in Southeast Asia (4.9%, 95% CI=3.7-6.4%), followed by North America (2.7%, 95% CI=2.1-3.6%; all 19 studies were conducted in the USA), and Europe (1.3%, 95% CI=0.7-2.6%), whereas in South Asia (all four studies were conducted in India), no EGFR TKD mutations were detected. Additionally, one study from South America stated the finding of two EGFR mutations in 45 tumor samples (prevalence 4.4%, 95% CI=0.8-16.4%) and one study from Australia reported one EGFR mutation in 60 tumor samples (prevalence 1.7%, 95% CI=0.1-10.1%). When considering EGFR TKD mutations in individual countries, the Republic of Korea had the highest prevalence with 15.1% (95% CI=11.2-20.1%), followed by the Czech Republic (6.9%, 95% CI=1.2-24.2%), and Greece (3.3%, 95% CI=0.8-9.9%).
Table III. Prevalence of epidermal growth factor receptor (EGFR) kinase domain mutations in patients with head and neck cancer by country and geographic region.

*The study of Temam et al. (72) included patients from two distinct study locations: 41 patients from France and 93 from the United States, who were categorized into European and North American populations, respectively. The study of Bahassi et al. (55) was not considered for prevalence estimation (case report).
Discussion
In this study, pyrosequencing technology was used to identify the EGFR mutation status in pretreatment tumor samples of patients with locally advanced OOSCC. No EGFR TKD mutations were observed among 113 cases of OOSCC. The results of this cohort study showed, however, a strong association of pathological complete response to neoadjuvant treatment with improved overall survival of patients with OOSCC, thus indicating the need for discovery of predictive biomarkers.
The identification of activating mutations in the EGFR TKD in a subset of NSCLC and their association with substantial sensitivity to gefitinib, erlotinib or afatinib represents an important milestone in the therapy of this malignancy (14,17,24). Driven by the paradigm in NSCLC, several studies in HNSCC attempted to define the mutational spectrum of the EGFR TKD. To date, genomic data from whole exome sequencing and targeted next-generation sequencing studies have provided a comprehensive landscape of genomic alterations in HNSCC (25-27). In a recent study, Ock et al. using targeted next-generation sequencing identified EGFR TKD mutations in 19 out of 71 (26.7%) HNSCCs (28). The Cancer Genome Atlas data from whole exome sequencing of HNSCCs demonstrated, however, that only one out of 279 (0.4%) tumor samples from HNSCCs harbored a missense mutation in the EGFR TKD (25). Given this background, the present systematic review aimed to summarize current evidence regarding the EGFR mutation status in HNSCC. Based on the quantitative data analysis, this study demonstrated that the overall prevalence of EGFR TKD mutations in HNSCC is 2.8%. This study revealed that the EGFR mutation prevalence in patients with HNSCC varies modestly across geographic regions, with the highest prevalence shown (4.9%) in Southeast Asia and the lowest in South Asia (0%). The EGFR mutation prevalence within the population of Southeast Asia varies by country, from approximately 1% in Taiwan to 15% in the Republic of Korea. In addition, this study showed that the EGFR mutation status in HNSCC has been insufficiently assessed worldwide as evident from the limited number of studies conducted in Australia (n=1) and South America (n=1), and the lack of data from several large geographic regions, particularly Africa, Central America, the Middle East, and Central Asia. Therefore, it is apparent that large-scale and multicenter studies are necessary to provide more definitive answers regarding the prevalence of EGFR mutations across geographic regions and countries and to assess their potential clinical value in patients with HNSCC.
In this systematic review, the overall EGFR mutation status in HNSCC according to exon location and mutation type was explored. The data showed that the most prevalent EGFR kinase domain mutations, accounting for 73% of all EGFR mutations in HNSCC, are missense mutations in exons 18-21. The L858R substitution, well-known in NSCLC, which comprises about 40% of all EGFR mutations in NSCLC and is associated with sensitivity to EGFR TKIs, was found in only 2.5% of all EGFR-mutated HNSCCs (29). The missense mutation T790M in exon 20, which is associated with acquired resistance to EGFR TKIs in about half of all patients with NSCLC, was found in 7.5% of all EGFR mutations in HNSCC (17). In-frame deletions in exon 19, which account for about 45% of all EGFR mutations in NSCLC and are linked to responsiveness to EGFR TKIs, were observed in 22% of all EGFR-mutated HNSCCs (24). Insertion mutations in exon 20, which occur in about 3% of all EGFR mutations in NSCLC and are frequently associated with resistance to EGFR TKIs, were observed in 5% of all EGFR mutations in HNSCC (30). Taken together, it is clear that substantial differences exist between HNSCC and NSCLC regarding the distribution of mutations within exons 18-21 of the EGFR TKD. Unlike NSCLC, EGFR mutations in HNSCC do not involve specific hotspots but are rather scattered throughout exons 18 to 21. Thus, mutation screening in HNSCC should not be limited to the NSCLC hotspot regions in exons 19 and 21 of EGFR. Moreover, given that the overall prevalence of EGFR TKD mutations in HNSCC is 2.8%, it is challenging to identify specific EGFR mutations related to response or resistance to anti-EGFR therapy or other targeted therapies (31).
The present cohort study has some weaknesses, including its retrospective nature and the relatively small sample size. Additionally, next-generation sequencing methods to compare and validate the results of the EGFR mutation testing by pyrosequencing were not used. However, recent studies have shown that pyrosequencing has the ability to detect EGFR mutations at a low ratio of mutant to wild-type alleles and thus provides high analytical sensitivity for identifying EGFR mutations (32,33). The systematic review is limited in several ways. Firstly, high heterogeneity has to be assumed across the study populations given the differences in study location, tumor site, stage and interventions. Secondly, various mutation testing methods with different sensitivities in detecting EGFR mutations were used across studies. Thirdly, a number of studies limited their EGFR mutation testing to hotspot regions in exons 19 and 21, thus the true prevalence of EGFR mutations might in fact be under-reported. Taken together, the results of the quantitative synthesis should be interpreted cautiously.
In the emerging era of personalized medicine, the identification of clinically useful prognostic and, most importantly, predictive biomarkers to guide treatment decision in patients with cancer is urgently needed. In this study, no mutations were detected using pyrosequencing when analyzing the EGFR TKD mutation status in a cohort of 113 patients with advanced OOSCC. In addition, the systematic review demonstrated that EGFR TKD mutations are rare in HNSCC, with an overall prevalence of 2.8% and modest variation in the prevalence across countries and geographic regions. Large-scale studies are warranted to provide further up-to-date evidence regarding the mutation status of EGFR in patients with HNSCC and to investigate whether the EGFR mutation profile of individual tumors is associated with sensitivity or resistance to targeted therapy.
Conflicts of Interest
None declared.
Financial Disclosure
None.
Grant Support
This project was supported by fund of the Oesterreichische Nationalbank, Anniversary Fund, Project Number 13469.
Acknowledgements
The Author thanks Martin Filipits, Anita Brandstetter, and Ágnes Bilecz for their excellent technical support. The Author thanks Robert Pillerstorf and Johannes Engelmann for their contribution to the systematic review with regard to study selection and data extraction.
References
- 1.Barak V, Meirovitz A, Leibovici V, Rachmut J, Peretz T, Eliashar R, Gross M. The diagnostic and prognostic value of tumor markers (CEA, SCC, CYFRA 21-1, TPS) in head and neck cancer patients. Anticancer Res. 2015;35:5519–5524. [PubMed] [Google Scholar]
- 2.Muramatsu H, Akimoto T, Maebayashi K, Kita M, Mitsuhashi N. Prognostic significance of dysadherin and E-cadherin expression in patients with head and neck cancer treated by radiation therapy. Anticancer Res. 2008;28:3859–3864. [PubMed] [Google Scholar]
- 3.Cohen EE. Role of epidermal growth factor receptor pathway-targeted therapy in patients with recurrent and/or metastatic squamous cell carcinoma of the head and neck. J Clin Oncol. 2006;24:2659–2665. doi: 10.1200/JCO.2005.05.4577. [DOI] [PubMed] [Google Scholar]
- 4.Tahtali A, Hey C, Geissler C, Filman N, Diensthuber M, Leinung M, Stover T, Wagenblast J. HPV status and overall survival of patients with oropharyngeal squamous cell carcinoma–a retrospective study of a German head and neck cancer center. Anticancer Res. 2013;33:3481–3485. [PubMed] [Google Scholar]
- 5.Warnakulasuriya S. Global epidemiology of oral and oropharyngeal cancer. Oral Oncol. 2009;45:309–316. doi: 10.1016/j.oraloncology.2008.06.002. [DOI] [PubMed] [Google Scholar]
- 6.Dang RP, Le VH, Miles BA, Teng MS, Genden EM, Bakst RL, Gupta V, Zhang DY, Demicco EG, Posner MR, Misiukiewicz KJ. Clinical outcomes in patients with recurrent or metastatic human papilloma virus-positive head and neck cancer. Anticancer Res. 2016;36:1703–1709. [PubMed] [Google Scholar]
- 7.Matsumoto F, Valdecanas DN, Mason KA, Milas L, Ang KK, Raju U. The impact of timing of EGFR and IGF-1R inhibition for sensitizing head and neck cancer to radiation. Anticancer Res. 2012;32:3029–3035. [PubMed] [Google Scholar]
- 8.Cohen EE, Kane MA, List MA, Brockstein BE, Mehrotra B, Huo D, Mauer AM, Pierce C, Dekker A, Vokes EE. Phase II trial of gefitinib 250 mg daily in patients with recurrent and/or metastatic squamous cell carcinoma of the head and neck. Clin Cancer Res. 2005;11:8418–8424. doi: 10.1158/1078-0432.CCR-05-1247. [DOI] [PubMed] [Google Scholar]
- 9.Rubin Grandis J, Melhem MF, Gooding WE, Day R, Holst VA, Wagener MM, Drenning SD, Tweardy DJ. Levels of TGF-alpha and EGFR protein in head and neck squamous cell carcinoma and patient survival. J Natl Cancer Inst. 1998;90:824–832. doi: 10.1093/jnci/90.11.824. [DOI] [PubMed] [Google Scholar]
- 10.Chen LF, Cohen EE, Grandis JR. New strategies in head and neck cancer: understanding resistance to epidermal growth factor receptor inhibitors. Clin Cancer Res. 2010;16:2489–2495. doi: 10.1158/1078-0432.CCR-09-2318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Stoehlmacher-Williams J, Obermann L, Ehninger G, Goekkurt E. Polymorphisms of the epidermal growth factor receptor (EGFR) and survival in patients with advanced cancer of the head and neck (HNSCC) Anticancer Res. 2012;32:421–425. [PubMed] [Google Scholar]
- 12.Cohen EE, Rosen F, Stadler WM, Recant W, Stenson K, Huo D, Vokes EE. Phase II trial of ZD1839 in recurrent or metastatic squamous cell carcinoma of the head and neck. J Clin Oncol. 2003;21:1980–1987. doi: 10.1200/JCO.2003.10.051. [DOI] [PubMed] [Google Scholar]
- 13.Soulieres D, Senzer NN, Vokes EE, Hidalgo M, Agarwala SS, Siu LL. Multicenter phase II study of erlotinib, an oral epidermal growth factor receptor tyrosine kinase inhibitor, in patients with recurrent or metastatic squamous cell cancer of the head and neck. J Clin Oncol. 2004;22:77–85. doi: 10.1200/JCO.2004.06.075. [DOI] [PubMed] [Google Scholar]
- 14.Lynch TJ, Bell DW, Sordella R, Gurubhagavatula S, Okimoto RA, Brannigan BW, Harris PL, Haserlat SM, Supko JG, Haluska FG, Louis DN, Christiani DC, Settleman J, Haber DA. Activating mutations in the epidermal growth factor receptor underlying responsiveness of non-small-cell lung cancer to gefitinib. N Engl J Med. 2004;350:2129–2139. doi: 10.1056/NEJMoa040938. [DOI] [PubMed] [Google Scholar]
- 15.Paez JG, Janne PA, Lee JC, Tracy S, Greulich H, Gabriel S, Herman P, Kaye FJ, Lindeman N, Boggon TJ, Naoki K, Sasaki H, Fujii Y, Eck MJ, Sellers WR, Johnson BE, Meyerson M. EGFR mutations in lung cancer: correlation with clinical response to gefitinib therapy. Science. 2004;304:1497–1500. doi: 10.1126/science.1099314. [DOI] [PubMed] [Google Scholar]
- 16.Pao W, Miller V, Zakowski M, Doherty J, Politi K, Sarkaria I, Singh B, Heelan R, Rusch V, Fulton L, Mardis E, Kupfer D, Wilson R, Kris M, Varmus H. EGF receptor gene mutations are common in lung cancers from "never smokers" and are associated with sensitivity of tumors to gefitinib and erlotinib. Proc Natl Acad Sci USA. 2004;101:13306–13311. doi: 10.1073/pnas.0405220101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hirsch FR, Suda K, Wiens J, Bunn PA Jr. New and emerging targeted treatments in advanced non-small-cell lung cancer. Lancet. 2016;388:1012–1024. doi: 10.1016/S0140-6736(16)31473-8. [DOI] [PubMed] [Google Scholar]
- 18.Sobin LH, Wittekind C (eds.) International Union Against Cancer (UICC) TNM Classification of Malignant Tumours, Sixth Edition. New. 2002;York:Wiley. [Google Scholar]
- 19.Moher D, Liberati A, Tetzlaff J, Altman DG, Group P. Preferred reporting items for systematic reviews and meta-analyses: the PRISMA statement. PLoS Med. 2009;6:e1000097. doi: 10.1371/journal.pmed.1000097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Newcombe RG. Two-sided confidence intervals for the single proportion: comparison of seven methods. Stat Med. 1998;17:857–872. doi: 10.1002/(sici)1097-0258(19980430)17:8<857::aid-sim777>3.0.co;2-e. [DOI] [PubMed] [Google Scholar]
- 21.Hama T, Yuza Y, Saito Y, J Ou, Kondo S, Okabe M, Yamada H, Kato T, Moriyama H, Kurihara S, Urashima M. Prognostic significance of epidermal growth factor receptor phosphorylation and mutation in head and neck squamous cell carcinoma. Oncologist. 2009;14:900–908. doi: 10.1634/theoncologist.2009-0058. [DOI] [PubMed] [Google Scholar]
- 22.Murray S, Bobos M, Angouridakis N, Nikolaou A, Linardou H, Razis E, Fountzilas G. Screening for EGFR mutations in patients with head and neck cancer treated with gefitinib on a compassionate-use program: A Hellenic Cooperative Oncology Group Study. J Oncol. 2010;2010:709678. doi: 10.1155/2010/709678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Na II, Kang HJ, Cho SY, Koh JS, Lee JK, Lee BC, Lee GH, Lee YS, Yoo HJ, Ryoo BY, Yang SH, Shim YS. EGFR mutations and human papillomavirus in squamous cell carcinoma of tongue and tonsil. Eur J Cancer. 2007;43:520–526. doi: 10.1016/j.ejca.2006.09.025. [DOI] [PubMed] [Google Scholar]
- 24.Sharma SV, Bell DW, Settleman J, Haber DA. Epidermal growth factor receptor mutations in lung cancer. Nat Rev Cancer. 2007;7:169–181. doi: 10.1038/nrc2088. [DOI] [PubMed] [Google Scholar]
- 25.Cancer Genome Atlas N. Comprehensive genomic characterization of head and neck squamous cell carcinomas. Nature. 2015;517:576–582. doi: 10.1038/nature14129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Seiwert TY, Zuo Z, Keck MK, Khattri A, Pedamallu CS, Stricker T, Brown C, Pugh TJ, Stojanov P, Cho J, Lawrence MS, Getz G, Bragelmann J, DeBoer R, Weichselbaum RR, Langerman A, Portugal L, Blair E, Stenson K, Lingen MW, Cohen EE, Vokes EE, White KP, Hammerman PS. Integrative and comparative genomic analysis of HPV-positive and HPV-negative head and neck squamous cell carcinomas. Clin Cancer Res. 2015;21:632–641. doi: 10.1158/1078-0432.CCR-13-3310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Stransky N, Egloff AM, Tward AD, Kostic AD, Cibulskis K, Sivachenko A, Kryukov GV, Lawrence MS, Sougnez C, McKenna A, Shefler E, Ramos AH, Stojanov P, Carter SL, Voet D, Cortes ML, Auclair D, Berger MF, Saksena G, Guiducci C, Onofrio RC, Parkin M, Romkes M, Weissfeld JL, Seethala RR, Wang L, Rangel-Escareno C, Fernandez-Lopez JC, Hidalgo-Miranda A, Melendez-Zajgla J, Winckler W, Ardlie K, Gabriel SB, Meyerson M, Lander ES, Getz G, Golub TR, Garraway LA, Grandis JR. The mutational landscape of head and neck squamous cell carcinoma. Science. 2011;333:1157–1160. doi: 10.1126/science.1208130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ock CY, Son B, Keam B, Lee SY, Moon J, Kwak H, Kim S, Kim TM, Jeon YK, Kwon SK, Hah JH, Lee SH, Kwon TK, Kim DW, Wu HG, Sung MW, Heo DS. Identification of genomic mutations associated with clinical outcomes of induction chemotherapy in patients with head and neck squamous cell carcinoma. J Cancer Res Clin Oncol. 2016;142:873–883. doi: 10.1007/s00432-015-2083-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Tsiambas E, Lefas AY, Georgiannos SN, Ragos V, Fotiades PP, Grapsa D, Stamatelopoulos A, Kavantzas N, Patsouris E, Syrigos K. EGFR gene deregulation mechanisms in lung adenocarcinoma: A molecular review. Pathol Res Pract. 2016;212:672–677. doi: 10.1016/j.prp.2016.06.005. [DOI] [PubMed] [Google Scholar]
- 30.Cheng L, Alexander RE, Maclennan GT, Cummings OW, Montironi R, Lopez-Beltran A, Cramer HM, Davidson DD, Zhang S. Molecular pathology of lung cancer: key to personalized medicine. Mod Pathol. 2012;25:347–369. doi: 10.1038/modpathol.2011.215. [DOI] [PubMed] [Google Scholar]
- 31.Cassell A, Grandis JR. Investigational EGFR-targeted therapy in head and neck squamous cell carcinoma. Expert Opin Investig Drugs. 2010;19:709–722. doi: 10.1517/13543781003769844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Querings S, Altmuller J, Ansen S, Zander T, Seidel D, Gabler F, Peifer M, Markert E, Stemshorn K, Timmermann B, Saal B, Klose S, Ernestus K, Scheffler M, Engel-Riedel W, Stoelben E, Brambilla E, Wolf J, Nurnberg P, Thomas RK. Benchmarking of mutation diagnostics in clinical lung cancer specimens. PLoS One. 2011;6:e19601. doi: 10.1371/journal.pone.0019601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Dufort S, Richard MJ, Lantuejoul S, de Fraipont F. Pyrosequencing, a method approved to detect the two major EGFR mutations for anti-EGFR therapy in NSCLC. J Exp Clin Cancer Res. 2011;30:57. doi: 10.1186/1756-9966-30-57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Feldman R, Gatalica Z, Knezetic J, Reddy S, Nathan CA, Javadi N, Teknos T. Molecular profiling of head and neck squamous cell carcinoma. Head Neck. 2016;38(Suppl 1):E1625–1638. doi: 10.1002/hed.24290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Chau NG, Li YY, Jo VY, Rabinowits G, Lorch JH, Tishler RB, Margalit DN, Schoenfeld JD, Annino DJ, Goguen LA, Thomas T, Becker H, Haddad T, Krane JF, Lindeman NI, Shapiro GI, Haddad RI, Hammerman PS. Incorporation of next-generation sequencing into routine clinical care to direct treatment of head and neck squamous cell carcinoma. Clin Cancer Res. 2016;22:2939–2949. doi: 10.1158/1078-0432.CCR-15-2314. [DOI] [PubMed] [Google Scholar]
- 36.Wu ES, Park JY, Zeitouni JA, Gomez CR, Reis IM, Zhao W, Kwon D, Lee E, Nelson OL, Lin HY, Franzmann EJ, Savell J, McCaffrey TV, Goodwin WJ, Hu JJ. Effect of actionable somatic mutations on racial/ethnic disparities in head and neck cancer prognosis. Head Neck. 2016;38:1234–1241. doi: 10.1002/hed.24420. [DOI] [PubMed] [Google Scholar]
- 37.Huang KK, Jang KW, Kim S, Kim HS, Kim SM, Kwon HJ, Kim HR, Yun HJ, Ahn MJ, Park KU, Ramnarayanan K, McPherson JR, Zhang S, Rhee JK, Vettore AL, Das K, Ishimoto T, Kim JH, Koh YW, Kim SH, Choi EC, Teh BT, Rozen SG, Kim TM, Tan P, Cho BC. Exome sequencing reveals recurrent REV3L mutations in cisplatin-resistant squamous cell carcinoma of head and neck. Sci Rep. 2016;6:19552. doi: 10.1038/srep19552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Vettore AL, Ramnarayanan K, Poore G, Lim K, Ong CK, Huang KK, Leong HS, Chong FT, Lim TK, Lim WK, Cutcutache I, McPherson JR, Suzuki Y, Zhang S, Skanthakumar T, Wang W, Tan DS, Cho BC, Teh BT, Rozen S, Tan P, Iyer NG. Mutational landscapes of tongue carcinoma reveal recurrent mutations in genes of therapeutic and prognostic relevance. Genome Med. 2015;7:98. doi: 10.1186/s13073-015-0219-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kim HS, Kwon HJ, Jung I, Yun MR, Ahn MJ, Kang BW, Sun JM, Kim SB, Yoon DH, Park KU, Lee SH, Koh YW, Kim SH, Choi EC, Koo DH, Sohn JH, Kim B, Kwon NJ, Yun HJ, Lee MG, Lee JH, Kim TM, Kim HR, Kim JH, Paik S, Cho BC. Phase II clinical and exploratory biomarker study of dacomitinib in patients with recurrent and/or metastatic squamous cell carcinoma of head and neck. Clin Cancer Res. 2015;21:544–552. doi: 10.1158/1078-0432.CCR-14-1756. [DOI] [PubMed] [Google Scholar]
- 40.Pickering CR, Zhang J, Neskey DM, Zhao M, Jasser SA, Wang J, Ward A, Tsai CJ, Ortega Alves MV, Zhou JH, Drummond J, El-Naggar AK, Gibbs R, Weinstein JN, Wheeler DA, Wang J, Frederick MJ, Myers JN. Squamous cell carcinoma of the oral tongue in young non-smokers is genomically similar to tumors in older smokers. Clin Cancer Res. 2014;20:3842–3848. doi: 10.1158/1078-0432.CCR-14-0565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wang DS, Lai HC, Huang JM. Epidermal growth factor receptor mutations in Chinese patients with laryngeal squamous cell carcinoma. Acta Otolaryngol. 2014;134:631–635. doi: 10.3109/00016489.2013.879741. [DOI] [PubMed] [Google Scholar]
- 42.Tan DS, Wang W, Leong HS, Sew PH, Lau DP, Chong FT, Krisna SS, Lim TK, Iyer NG. Tongue carcinoma infrequently harbor common actionable genetic alterations. BMC Cancer. 2014;14:679. doi: 10.1186/1471-2407-14-679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.McBride SM, Rothenberg SM, Faquin WC, Chan AW, Clark JR, Ellisen LW, Wirth LJ. Mutation frequency in 15 common cancer genes in high-risk head and neck squamous cell carcinoma. Head Neck. 2014;36:1181–1188. doi: 10.1002/hed.23430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Mehta DT, Annamalai T, Ramanathan A. Lack of mutations in protein tyrosine kinase domain coding exons 19 and 21 of the EGFR gene in oral squamous cell carcinomas. Asian Pac J Cancer Prev. 2014;15:4623–4627. doi: 10.7314/apjcp.2014.15.11.4623. [DOI] [PubMed] [Google Scholar]
- 45.Boeckx C, Weyn C, Vanden Bempt I, Deschoolmeester V, Wouters A, Specenier P, Van Laer C, Van den Weyngaert D, Kockx M, Vermorken JB, Peeters M, Pauwels P, Lardon F, Baay M. Mutation analysis of genes in the EGFR pathway in head and neck cancer patients: implications for anti-EGFR treatment response. BMC Res Notes. 2014;7:337. doi: 10.1186/1756-0500-7-337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Nagalakshmi K, Jamil K, Pingali U, Reddy MV, Attili SS. Epidermal growth factor receptor (EGFR) mutations as biomarker for head and neck squamous cell carcinomas (HNSCC) Biomarkers. 2014;19:198–206. doi: 10.3109/1354750X.2014.895852. [DOI] [PubMed] [Google Scholar]
- 47.Gaykalova DA, Mambo E, Choudhary A, Houghton J, Buddavarapu K, Sanford T, Darden W, Adai A, Hadd A, Latham G, Danilova LV, Bishop J, Li RJ, Westra WH, Hennessey P, Koch WM, Ochs MF, Califano JA, Sun W. Novel insight into mutational landscape of head and neck squamous cell carcinoma. PLoS One. 2014;9:e93102. doi: 10.1371/journal.pone.0093102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Argiris A, Ghebremichael M, Gilbert J, Lee JW, Sachidanandam K, Kolesar JM, Burtness B, Forastiere AA. Phase III randomized, placebo-controlled trial of docetaxel with or without gefitinib in recurrent or metastatic head and neck cancer: an Eastern Cooperative Oncology Group trial. J Clin Oncol. 2013;31:1405–1414. doi: 10.1200/JCO.2012.45.4272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Maiti GP, Mondal P, Mukherjee N, Ghosh A, Ghosh S, Dey S, Chakrabarty J, Roy A, Biswas J, Roychoudhury S, Panda CK. Overexpression of EGFR in head and neck squamous cell carcinoma is associated with inactivation of SH3GL2 and CDC25A genes. PLoS One. 2013;8:e63440. doi: 10.1371/journal.pone.0063440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Lechner M, Frampton GM, Fenton T, Feber A, Palmer G, Jay A, Pillay N, Forster M, Cronin MT, Lipson D, Miller VA, Brennan TA, Henderson S, Vaz F, O’Flynn P, Kalavrezos N, Yelensky R, Beck S, Stephens PJ, Boshoff C. Targeted next-generation sequencing of head and neck squamous cell carcinoma identifies novel genetic alterations in HPV+ and HPV– tumors. Genome Med. 2013;5:49. doi: 10.1186/gm453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.India Project Team of the International Cancer Genome C. Mutational landscape of gingivo-buccal oral squamous cell carcinoma reveals new recurrently-mutated genes and molecular subgroups. Nat Commun. 2013;4:2873. doi: 10.1038/ncomms3873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Pickering CR, Zhang J, Yoo SY, Bengtsson L, Moorthy S, Neskey DM, Zhao M, Ortega Alves MV, Chang K, Drummond J, Cortez E, Xie TX, Zhang D, Chung W, Issa JP, Zweidler-McKay PA, Wu X, El-Naggar AK, Weinstein JN, Wang J, Muzny DM, Gibbs RA, Wheeler DA, Myers JN, Frederick MJ. Integrative genomic characterization of oral squamous cell carcinoma identifies frequent somatic drivers. Cancer Discov. 2013;3:770–781. doi: 10.1158/2159-8290.CD-12-0537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Fanjul-Fernandez M, Quesada V, Cabanillas R, Cadinanos J, Fontanil T, Obaya A, Ramsay AJ, Llorente JL, Astudillo A, Cal S, Lopez-Otin C. Cell–cell adhesion genes CTNNA2 and CTNNA3 are tumour suppressors frequently mutated in laryngeal carcinomas. Nat Commun. 2013;4:2531. doi: 10.1038/ncomms3531. [DOI] [PubMed] [Google Scholar]
- 54.Bontognali S, Pless M, Brutsche MH, Fischer C, Rochlitz C, Buess M. Analysis of the EGFR mutation status in head and neck squamous cell carcinoma before treatment with gefitinib. Onkologie. 2013;36:161–166. doi: 10.1159/000349941. [DOI] [PubMed] [Google Scholar]
- 55.Bahassi el M, Li YQ, Wise-Draper TM, Deng L, Wang J, Darnell CN, Wilson KM, Wells SI, Stambrook PJ, Rixe O. A patient-derived somatic mutation in the epidermal growth factor receptor ligand-binding domain confers increased sensitivity to cetuximab in head and neck cancer. Eur J Cancer. 2013;49:2345–2355. doi: 10.1016/j.ejca.2013.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Smilek P, Neuwirthova J, Jarkovsky J, Dusek L, Rottenberg J, Kostrica R, Srovnal J, Hajduch M, Drabek J, Klozar J. Epidermal growth factor receptor (EGFR) expression and mutations in the EGFR signaling pathway in correlation with anti-EGFR therapy in head and neck squamous cell carcinomas. Neoplasma. 2012;59:508–515. doi: 10.4149/neo_2012_065. [DOI] [PubMed] [Google Scholar]
- 57.Tan EH, Goh C, Lim WT, Soo KC, Khoo ML, Tan T, Tan DS, Ang MK, Ng QS, Tan PH, Lim A, Hwang J, Teng YH, Lim TH, Tan SH, Baskaran N, Hui KM. Gefitinib, cisplatin, and concurrent radiotherapy for locally advanced head and neck cancer: EGFR FISH, protein expression, and mutational status are not predictive biomarkers. Ann Oncol. 2012;23:1010–1016. doi: 10.1093/annonc/mdr327. [DOI] [PubMed] [Google Scholar]
- 58.Friedland P, Thomas A, Naran A, Amanuel B, Grieu-Iacopetta F, Carrello A, Harnett G, Meyer C, Phillips M. Human papillomavirus and gene mutations in head and neck squamous carcinomas. ANZ J Surg. 2012;82:362–366. doi: 10.1111/j.1445-2197.2011.05791.x. [DOI] [PubMed] [Google Scholar]
- 59.Szabo B, Nelhubel GA, Karpati A, Kenessey I, Jori B, Szekely C, Petak I, Lotz G, Hegedus Z, Hegedus B, Fule T, Dome B, Timar J, Tovari J. Clinical significance of genetic alterations and expression of epidermal growth factor receptor (EGFR) in head and neck squamous cell carcinomas. Oral Oncol. 2011;47:487–496. doi: 10.1016/j.oraloncology.2011.03.020. [DOI] [PubMed] [Google Scholar]
- 60.Hsieh CH, Chang JW, Hsieh JJ, Hsu T, Huang SF, Liao CT, Wang HM. Epidermal growth factor receptor mutations in patients with oral cavity cancer in a betel nut chewing-prevalent area. Head Neck. 2011;33:1758–1764. doi: 10.1002/hed.21665. [DOI] [PubMed] [Google Scholar]
- 61.Morris LG, Taylor BS, Bivona TG, Gong Y, Eng S, Brennan CW, Kaufman A, Kastenhuber ER, Banuchi VE, Singh B, Heguy A, Viale A, Mellinghoff IK, Huse J, Ganly I, Chan TA. Genomic dissection of the epidermal growth factor receptor (EGFR)/PI3K pathway reveals frequent deletion of the EGFR phosphatase PTPRS in head and neck cancers. Proc Natl Acad Sci USA. 2011;108:19024–19029. doi: 10.1073/pnas.1111963108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Agrawal N, Frederick MJ, Pickering CR, Bettegowda C, Chang K, Li RJ, Fakhry C, Xie TX, Zhang J, Wang J, Zhang N, El-Naggar AK, Jasser SA, Weinstein JN, Trevino L, Drummond JA, Muzny DM, Wu Y, Wood LD, Hruban RH, Westra WH, Koch WM, Califano JA, Gibbs RA, Sidransky D, Vogelstein B, Velculescu VE, Papadopoulos N, Wheeler DA, Kinzler KW, Myers JN. Exome sequencing of head and neck squamous cell carcinoma reveals inactivating mutations in NOTCH1. Science. 2011;333:1154–1157. doi: 10.1126/science.1206923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Szymanska K, Levi JE, Menezes A, Wunsch-Filho V, Eluf-Neto J, Koifman S, Matos E, Daudt AW, Curado MP, Villar S, Pawlita M, Waterboer T, Boffetta P, Hainaut P, Brennan P. TP53 and EGFR mutations in combination with lifestyle risk factors in tumours of the upper aerodigestive tract from South America. Carcinogenesis. 2010;31:1054–1059. doi: 10.1093/carcin/bgp212. [DOI] [PubMed] [Google Scholar]
- 64.Van Damme N, Deron P, Van Roy N, Demetter P, Bols A, Van Dorpe J, Baert F, Van Laethem JL, Speleman F, Pauwels P, Peeters M. Epidermal growth factor receptor and K-RAS status in two cohorts of squamous cell carcinomas. BMC Cancer. 2010;10:189. doi: 10.1186/1471-2407-10-189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Keller J, Shroyer KR, Batajoo SK, Zhao HL, Dong LM, Hayman MJ, Chan EL. Combination of phosphorylated and truncated EGFR correlates with higher tumor and nodal stage in head and neck cancer. Cancer Invest. 2010;28:1054–1062. doi: 10.3109/07357907.2010.512602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Huang SF, Chuang WY, Chen IH, Liao CT, Wang HM, Hsieh LL. EGFR protein overexpression and mutation in areca quid-associated oral cavity squamous cell carcinoma in Taiwan. Head Neck. 2009;31:1068–1077. doi: 10.1002/hed.21067. [DOI] [PubMed] [Google Scholar]
- 67.Carlson M, Wuertz B, Lin J, Taylor R, Ondrey F. Exons 19 and 21 of epidermal growth factor receptor are highly conserved in squamous cell cancer of the head and neck. Int J Otolaryngol. 2009;2009:649615. doi: 10.1155/2009/649615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Jin T, Yu CX, Jin T, Yu CX, Lei DP, Liu DY, Xu FL, Lu YT, Pan XL. Identification of epidermal growth factor receptor (EGFR) exon 20 single nucleotide polymorphism in Chinese squamous cell carcinoma of head and neck (SCCHN) Acta Otolaryngol. 2009;129:1306–1312. doi: 10.3109/00016480802620662. [DOI] [PubMed] [Google Scholar]
- 69.Schwentner I, Witsch-Baumgartner M, Sprinzl GM, Krugmann J, Tzankov A, Jank S, Zwierzina H, Loeffler-Ragg J. Identification of the rare EGFR mutation p.G796S as somatic and germline mutation in white patients with squamous cell carcinoma of the head and neck. Head Neck. 2008;30:1040–1044. doi: 10.1002/hed.20831. [DOI] [PubMed] [Google Scholar]
- 70.Sheikh Ali MA, Gunduz M, Nagatsuka H, Gunduz E, Cengiz B, Fukushima K, Beder LB, Demircan K, Fujii M, Yamanaka N, Shimizu K, Grenman R, Nagai N. Expression and mutation analysis of epidermal growth factor receptor in head and neck squamous cell carcinoma. Cancer Sci. 2008;99:1589–1594. doi: 10.1111/j.1349-7006.2008.00861.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Chiang WF, Liu SY, Yen CY, Lin CN, Chen YC, Lin SC, Chang KW. Association of epidermal growth factor receptor (EGFR) gene copy number amplification with neck lymph node metastasis in areca-associated oral carcinomas. Oral Oncol. 2008;44:270–276. doi: 10.1016/j.oraloncology.2007.02.008. [DOI] [PubMed] [Google Scholar]
- 72.Temam S, Kawaguchi H, El-Naggar AK, Jelinek J, Tang H, Liu DD, Lang W, Issa JP, Lee JJ, Mao L. Epidermal growth factor receptor copy number alterations correlate with poor clinical outcome in patients with head and neck squamous cancer. J Clin Oncol. 2007;25:2164–2170. doi: 10.1200/JCO.2006.06.6605. [DOI] [PubMed] [Google Scholar]
- 73.Lemos-Gonzalez Y, Paez de la Cadena M, Rodriguez-Berrocal FJ, Rodriguez-Pineiro AM, Pallas E, Valverde D. Absence of activating mutations in the EGFR kinase domain in Spanish head and neck cancer patients. Tumour Biol. 2007;28:273–279. doi: 10.1159/000110425. [DOI] [PubMed] [Google Scholar]
- 74.Perrone F, Suardi S, Pastore E, Casieri P, Orsenigo M, Caramuta S, Dagrada G, Losa M, Licitra L, Bossi P, Staurengo S, Oggionni M, Locati L, Cantu G, Squadrelli M, Carbone A, Pierotti MA, Pilotti S. Molecular and cytogenetic subgroups of oropharyngeal squamous cell carcinoma. Clin Cancer Res. 2006;12:6643–6651. doi: 10.1158/1078-0432.CCR-06-1759. [DOI] [PubMed] [Google Scholar]
- 75.Chung CH, Ely K, McGavran L, Varella-Garcia M, Parker J, Parker N, Jarrett C, Carter J, Murphy BA, Netterville J, Burkey BB, Sinard R, Cmelak A, Levy S, Yarbrough WG, Slebos RJ, Hirsch FR. Increased epidermal growth factor receptor gene copy number is associated with poor prognosis in head and neck squamous cell carcinomas. J Clin Oncol. 2006;24:4170–4176. doi: 10.1200/JCO.2006.07.2587. [DOI] [PubMed] [Google Scholar]
- 76.Willmore-Payne C, Holden JA, Layfield LJ. Detection of EGFR- and HER2-activating mutations in squamous cell carcinoma involving the head and neck. Mod Pathol. 2006;19:634–640. doi: 10.1038/modpathol.3800552. [DOI] [PubMed] [Google Scholar]
- 77.Cohen EE, Lingen MW, Martin LE, Harris PL, Brannigan BW, Haserlat SM, Okimoto RA, Sgroi DC, Dahiya S, Muir B, Clark JR, Rocco JW, Vokes EE, Haber DA, Bell DW. Response of some head and neck cancers to epidermal growth factor receptor tyrosine kinase inhibitors may be linked to mutation of ERBB2 rather than EGFR. Clin Cancer Res. 2005;11:8105–8108. doi: 10.1158/1078-0432.CCR-05-0926. [DOI] [PubMed] [Google Scholar]
- 78.Lee JW, Soung YH, Kim SY, Nam HK, Park WS, Nam SW, Kim MS, Sun DI, Lee YS, Jang JJ, Lee JY, Yoo NJ, Lee SH. Somatic mutations of EGFR gene in squamous cell carcinoma of the head and neck. Clin Cancer Res. 2005;11:2879–2882. doi: 10.1158/1078-0432.CCR-04-2029. [DOI] [PubMed] [Google Scholar]


