ABSTRACT
Mitochondria are essential for cell growth and survival of most fungal pathogens. Energy (ATP) produced during oxidation/reduction reactions of the electron transport chain (ETC) Complexes I, III and IV (CI, CIII, CIV) fuel cell synthesis. The mitochondria of fungal pathogens are understudied even though more recent published data suggest critical functional assignments to fungal-specific proteins. Proteins of mammalian mitochondria are grouped into 16 functional categories. In this review, we focus upon 11 proteins from 5 of these categories in fungal pathogens, OXPHOS, protein import, stress response, carbon source metabolism, and fission/fusion morphology. As these proteins also are fungal-specific, we hypothesize that they may be exploited as targets in antifungal drug discovery. We also discuss published transcriptional profiling data of mitochondrial CI subunit protein mutants, in which we advance a novel concept those CI subunit proteins have both shared as well as specific responsibilities for providing ATP to cell processes.
KEYWORDS: antifungals, complex I subunits, discovery, mitochondria, targets
Mitochondrial drug targets: Functional relevance and specificity
There is ample evidence that fungi contribute to a huge burden of global infectious diseases and of these, invasive infections surpass deaths caused by drug-resistant tuberculosis or malaria.1 However, it is also clear that incidence and burden of all fungal diseases is even greater than referenced above. In Mexico alone, for example, the burden of the top 10 significant fungal infections totals well over 2 million, of which vaginal and recurrent vulvovaginal candidiasis (RVVC) account for ∼70%.2 RVVC infections are associated with high morbidity and lost work effort. There are several explanations for the high incidence of global mucosal, allergic, and invasive fungal infections (IFI). For the latter category, rapid, sensitive and specific diagnostic assays for Invasive Candidiasis (IC), Invasive Aspergillosis (IA), and mold infections such as fusariosis are still in development, although a new PCR assay may gain wide acceptance for the diagnosis of blood-borne candidiasis.3 As for therapeutic intervention, triazoles (ergosterol synthesis inhibitors) and echinocandins (β-1,3 glucan inhibitors) are the most popular antifungals in clinical use. However, the triazoles are fungistatic, select for resistant isolates, and cause drug-drug interactions. Newer versions are tailored older versions.4-6 While Cryptococcus neoformans and C. gattii have β-glucan and are inhibited by echinocandins, the concentrations are too high to be useful. Resistance is troublesome since that reduces the number of antifungals that can be used as an alternative therapy. Also, patient outcome is poorer with isolates that are resistant.6 To counter these problems, new antifungals should be developed against targets other than those of current antifungal therapeutics.
Amphotericin B was first identified in 1953, but it took about 15 y and a variety of expertise and expense before that initial observation was converted to a finished drug for distribution.7 The scientific disciplines that were needed for its clinical application included microbiology, organic chemistry, genetics (to develop the best producing strain), chemical engineering, pharmacology, biochemistry, and toxicology. Antifungal drug discovery even today requires the same expertise as well as time and money. Although many aspects of drug discovery have changed since amphotericin B was developed, challenges remain, particularly for antifungal drug development. For example, costs of new drug discovery have exploded. A 2014 report by the Tufts Center for the Study of Drug Development (CSDD) estimates that the cost of developing a new prescription drug is approximately $2.6 billion, representing a 145% increase since 2003, not including the additional $312 million for post-approval costs.8 Also, consider the costs of failure, which happen much more frequently than success. On the positive side, compound libraries are available for new drug discovery though more are needed. One of the most important developments in drug discovery with a high relevance to antifungal drug discovery has been the increase in chemical genetic and chemical genomic approaches to the identification of new chemical matter and the characterization of new drug targets.9 Genetic screens for targets (chemobiology) of mutant libraries offer read-outs of loss of tolerance (haploinsufficiency, HI) phenotypes. Or transcriptomics can be used to gain insights into compound mechanism of action. Genomics has allowed scientists to identify fungal-specific versus fungal-mammalian cell broadly conserved targets.
The mitochondrion is composed of about 1,000 proteins, most of which are nuclear-encoded and highly conserved among species.10 A recent characterization of the diversity of mitochondrial proteins has identified ∼16 functional categories, each of which correlate with their spatial organization within mitochondria.11 For example, proteins of the tricarboxylic cycle (TCA), β-oxidation, and the respiratory chain complexes (CI-CV) are spatially located at the inner membrane and mitochondrial matrix. Proteins associated with mitochondrial movement, import, and dynamics are located on the outer membrane. Mitochondrial functions such as respiration, ATP synthesis, movement, import, and morphology are highly conserved among diverse species. However, there is ample evidence herein that functional similarities (e.g., ATP formation) may include species-specific proteins associated with 5 of the 16 functional categories of proteins mentioned above.12 The task is to identify the species-specific mitochondrial proteins. With other fungi such as Neurospora crassa, Pichia pastoris and Yarrowia lipolytica, both conserved and fungal-specific proteins of mitochondria have been identified. These data have been rather easy to obtain, given the extensive data bases that allow comparisons across species including human mitochondria.13-17 If a search focuses upon new targets for antifungal drug discovery, specificity should be an initial descriptor followed by the verification of functions of the selected gene. The latter requires mutation/deletion of the encoding gene to profile dysfunction(s), and by inference, direct or indirect functions. In the case of fungal pathogens, an important finding may be that gene loss causes avirulence.
In the case of fungal-specific proteins, a reasonable question is what functional attributes are provided by this protein? Perhaps surprisingly in the case of mitochondrial proteins of fungi, proteins can be fungal-specific or identified from a single fungal genus, and in the latter case not all species of that genus.12 Among the most interesting of the mitochondrial proteins are those restricted to the CTG clade of the Ascomycetous Saccharomycotina which includes most Candida species but not C. glabrata.12,18 In the case of fungal or Candida spp, CTG clade-specific proteins, should drug discovery be toward targets of low or broad specificity? Which protein should be a reasonable target? Several CTG associated mitochondrial proteins are discussed below.
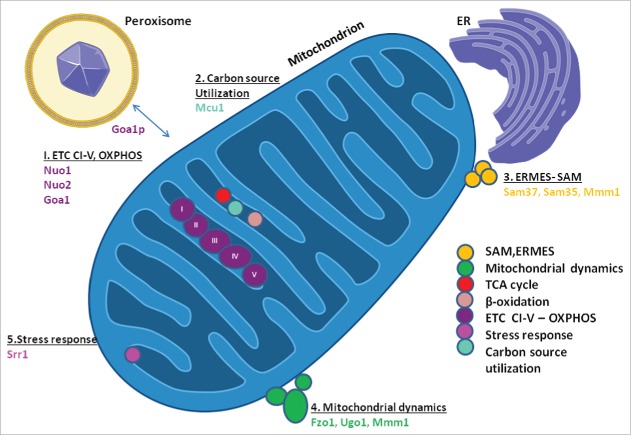
In this review, we focus upon those mitochondrial proteins that provide critical cell functions and target specificity. Our discussion will include fungal-specific proteins of the electron transport chain (ETC, complex I, CI), protein sorting/import, carbon source utilization, stress responses, and the mitochondrial dynamics of fusion/fission (Fig. 1).
Figure 1.
Fungal-specific proteins of mitochondria. Mitochondrial, fungal- specific proteins are distributed in 5 categories: 1) ETC, CI, OXPHOS, 2) Carbon source utilization, 3) ERMES and SAM, 4) Mitochondrial dynamics, and 5) Stress response signaling. The 11 fungal-specific genes are color-coded to reflect functional categories. In addition, the location of the TCA Cycle, β-oxidation components are placed spatially close to the ETC to emphasize relationships. Mutant responses are clustered for comparisons in several functional categories. Abbreviations: ETC, electron transport chain; CI, complex I; OXPHOS, oxidative phosphorylation; ERMES, ER-Mitochondrial Encounter Structure; SAM, Sorting and Assembly Machinery.
Complex I subunit proteins as drug targets
The electron transport chain is composed of 5 complexes, of which Complex I, III, and IV (CI, CIII, CIV) provide the proton gradients that result in ATP synthesis by CV. Electrons as a result of the oxidoreductase activity of CI combine with oxygen generating superoxides. If there is CI dysfunction, then a concomitant increase in superoxide occurs that is detrimental to cells. It is quite apparent that if cells are starved of mitochondrial ATP, cellular synthesis diminishes and events are triggered that result in cell death. CI deficient mutants accumulate mitochondrial ROS, lose chronological aging, and undergo apoptosis.18 Two examples are provided.19,20 The RAS-Cyr-PKA pathway is a major regulator of filamentation, biofilm formation, and virulence in C. albicans. Functioning of the Ras1-Cyr-PKA pathway requires a high cell energy status.19 The correlation of a functional Ras1 pathway and mitochondrial status was demonstrated using mitochondrial complex inhibitors. The Ras pathway was shown to interact with CI and CIV but not CII or the alternative oxidase pathway. The function of this pathway was assessed using filamentation as a phenotype.19 CII does not participate in the formation of a proton gradient as do the other complexes. As a second example, mitochondrial fusion/fission mutants of C. albicans display fragmented mitochondria and mitochondrial genome loss.20 A consequence of ATP decline in these mutants was an increase in susceptibility to azoles probably due to a reduced activity of energy-requiring efflux pumps. Our published data also correlate a loss of CI with increased susceptibility to fluconazole.21,22 In this case, we have shown that 10 of 12 CI mutants were hypersusceptible to fluconazole. The two CI subunits that were not hypersusceptible were not associated with a respiration function.21 Thus, loss of mitochondrial ATP may be a benefit of an inhibitor that could specifically block CI respiration activity and, correspondingly, would ensure a reduction in resistance to antifungals such as the azoles.
It would seem, therefore, that therapies to target mitochondria of fungal pathogens would be useful. The problem is choosing mitochondrial proteins to target therapeutically because of the extensive conservation among mammals and fungi. To address the issue of conservation among CI subunit proteins, we focused on 2 non-mammalian CI subunit proteins of C. albicans mitochondria that were first identified as NUXM and NUZM in Neurospora crassa.25 Both were only partially annotated functionally. NUXM is broadly conserved in C. albicans and other Candida spp, Aspergillus fumigatus, Neurospora crassa, Yarrowia lipolytica, Arabidopsis thaliana, Chlamydomonas reinhardtii, and other fungal pathogens such as Cryptococcus neoformans, C. gattii, and Fusarium spp. NUZM is fungal-specific.13-17,24,25 From the Candida Genome Database, we have identified NUXM as orf19.6607 (NUO1) and NUZM as orf19.287 (NUO2). We have shown that both proteins are NADH:Ubiquinone Oxidoreductases.26,27 These data identify CI fungal-specific subunit proteins, lending support to the development and exploitation of Nuo1p and Nuo2p as drug targets.
The electron transport chain (ETC) is composed of 5 complexes, 3 of which provide all mitochondrial ATP for cell activities. Of these, CI is the largest in molecular weight, generates about one-half of all mtATP. CI is comprised of a variable number of subunit proteins that are assembled by accessory proteins. The mitochondrial CI of C. albicans is composed of 39 subunit proteins, 37 of which are highly conserved across many species including mammals.18,26 As mentioned above, we have identified 2 subunits that are either solely fungal-specific or limited to fungi, green algae, and plants.27 Both are NADH:Ubiquinone Oxidoreductases, which we have named, Nuo1p and Nuo2p. As described below, their species-specificity and critical cell activities suggest that both proteins can be exploited as antifungal drug targets.
In addition to these CI subunit protein targets, Goa1p has been the subject of extensive study in our lab.12,27–31 Goa1p is not a CI subunit protein but does play a major role in CI NADH:ubiquinone oxidoreductase activity. However, its functional assignments in fact are also associated with peroxisomes. Thus, fatty acid β-oxidation, acetyl-CoA shuttle from peroxisomes to mitochondria, and peroxisome transporters indicate its dual organelle responsibilities.18,28 Goa1p is translocated to mitochondria during increases in cell reactive oxidant species (ROS).12 As mentioned above, Nuo1p and Nuo2p CI subunits are fungal-specific. Goa1p is one of 8 ETC other proteins (unpublished) that are only found in the CTG clade of the Saccharomycotina sub-phylum of Ascomycetes. Several of these proteins are required for optimum ATP production via mitochondrial respiration (unpublished). It is both convenient and interesting to categorize fungal mitochondrial ETC CI proteins in 4 groups that reflect their relationship to other fungi and organisms. The CI groups are: 1) broadly-specific, found in mammals (Ndh51p); 2) broadly specific in fungi, algae, plants but not in mammals (Nuo1p); 3) fungal-specific (Nuo2p); and 4) Candida-CTG-specific (Goa1p and others). Category 2-4 proteins perhaps suggest a role in host niche-specific growth, synthesis, and virulence, except that other members of the CTG clade are not usually pathogens.12 Below, we discuss CI subunit proteins and Goa1p.
The fungal specificity of these proteins to CI functions or as a CI regulatory protein (Goa1p) was mentioned above. One other CI subunit protein that we have included for functional comparisons in our studies is Ndh51p, which is broadly conserved among species including fungi and mammals.18,32 To determine their cellular functions, direct or indirect, we constructed knock-out strains of Nuo1, Nuo2, and Goa1. Common phenotypes included reduced mitochondrial respiration, ATP synthesis, and CI disassembly. These loss of function mutants also had reduced NADH:ubiquinone oxidoreductase activities. In addition, mutants of nuo1Δ and nuo2Δ and goa1Δ are avirulent in infected mice, have reduced interactions with human phagocytic cells, enhanced rates of apoptosis, and fail to maintain chronological aging.26,27
The CI subunits Nuo1 and Nuo2, as well as Goa1p share but have individualized functions. This observation was noted in transcriptional profiling of nuo1Δ, nuo2Δ, goa1Δ and ndh51Δ (Fig. 2)27 The transcriptional profiles of nuo1Δ and nuo2Δ showed that similar genes were upregulated (Fig. 2A). Those profiles included respiration, mitochondrial transporters, and TCA genes, while goa1Δ and ndh51Δ profiles were minimally affected (Fig. 2A). Differences in profiles were especially noticed among mutants in down regulated genes (Fig. 2B), including, glucanases, chitin synthases and chitinases, β-mannosyltransferases, protein mannosylation, and adhesin genes (nuo1Δ and nuo2Δ). The goa1Δ had a similar profile except that peroxisomal genes such as peroxisomal transporters and β-oxidation were significantly downregulated (Fig. 2B). For the ndh51Δ, downregulated genes of ergosterol synthesis were most prominent (Fig. 2B). These data strongly suggest a CI functional subunit specificity, which has not been reported in the fungal literature.
Figure 2.
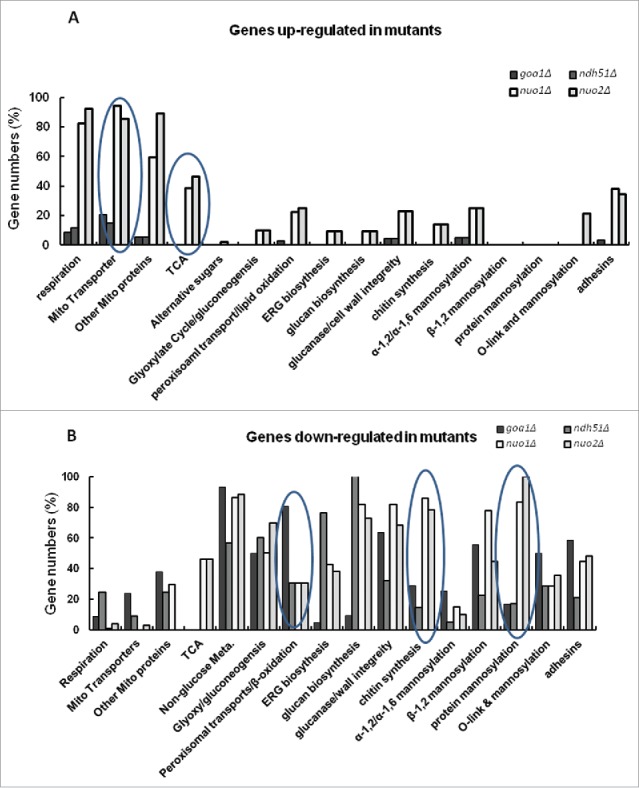
Transcriptional profiles of mitochondrial mutants. (A) Genes up-regulated in mitochondrial mutants, and (B) Genes down regulated in mitochondrial mutants genes. Shown are CI subunit mutants (nuo1Δ, nuo2Δ), CI regulatory protein (goa1Δ), and broadly conserved CI subunit (ndh51Δ). Several profiles in both A and B are circled to indicate differences among the 4 mutants.26
Biochemical studies of the cell wall in the goa1Δ have been recently published.31 A loss of β-1,2 linked mannosyltransferases and modifications in β-1,6 glucans was observed.31 The transcriptional changes of goa1Δ also correlate with the biochemical loss of bulk phosphomannan, as determined by GPC-MALLS analyses, as well as the loss of cell surface fibrils. The outer cell wall of the goa1Δ had fewer fibril extensions and the inner wall itself was more electron transparent (Fig. 3). However, synthesis of the inner polysaccharides β-1,3 and chitin were similar to parental cells, although the β-1,6 glucan had shorter side chains.31 We have initiated similar experiments with the nuo1Δ, nuo2Δ and ndh51Δ mutants. We hypothesize that the combination of fungal and novel subunit specificity, as well as their functional importance, point to Nuo1 and Nuo2 as attractive targets for antifungal drug discovery.
Figure 3.
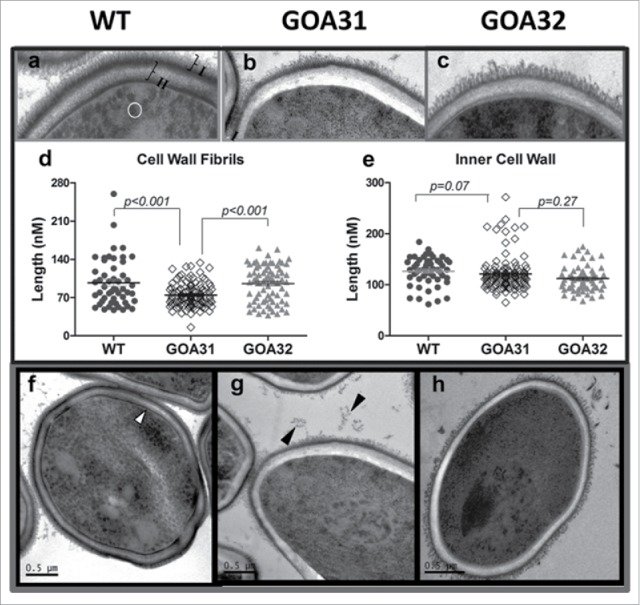
Goa1 mutants of C. albicans have altered cell wall architecture. C. albicans strains: a,f = WT; b,g = GOA31; c,h = GOA32. Differences are noted in the extent and length of the cell surface fibrils with GOA31 compared to the WT cells (“I,” Fig. 3a) and GOA32. Also, the inner cell wall (“II,” Fig. 3a) is more electron dense than that in GOA31, as well as the cell wall layer closest to plasma membrane (white arrow, Fig. 3f). Both are reduced only in GOA31. The average length of cell wall fibrils (d) and inner cell wall (e) are also determined. The fibrils are significantly shorter in GOA31 but the cell wall thickness is similar for all strains. In (d) each data point refers to an collection of 10 measurements of 10 cells for each strain using GIMP2.8 software. Vesicles (circle) in WT (Fig. 3a) are significantly reduced in GOA31. The black arrows may indicate sloughed cell wall material in Figure 3g.31 GOA31 = the goa1Δ; GOA32 = the GOA1 reconstituted strain in the GOA31 background.
Mitochondrial biogenesis and dynamics
These two functional categories have been studied in C. albicans and Aspergillus fumigatus laboratories, described below.33-36 Biogenesis usually includes both the SAM (Sorting and Assembly Machinery) and the ERMES (ER-Mitochondrial Encounter Structure) complexes of proteins that are vitally important in the insertion of proteins in mitochondria through the Transporters of the Outer Membrane (TOMs) (Fig. 1). Imported proteins are then transported to the matrix and inner membrane via the Transporters of the Inner Membrane (TIMs) (Fig. 1). The current belief is that the SAM and ERMES complexes form a “super” complex that is bridged by the Mdm10 protein.36 The SAM and ERMES super complex of proteins are often described as the “hub” for many mitochondrial activities, including lipid homeostasis, maintenance of a tubular mitochondrial morphology, inheritance, and regulating the process of fusion/fission of mitochondria, and trafficking (Fig. 1). For the SAM complex, the Sam57 subunit protein is required for optimum growth of C. albicans, virulence, protein insertion, and mitophagy, as well as interactions between mitochondria and the endoplasmic reticulum.33 Sam37 is also critical for cell wall integrity, as determined by the hypersensitivity of mutants to cell wall inhibitors and reduced cell wall β-1,3 and β-1,6 glucan. Thus, Sam37 is critically important for cells; importantly, there are significant structural differences in the fungal Sam37 as well as Sam35 compared to the metazoan homologues, suggesting exploitation of both fungal proteins as antifungal targets.36 Another protein is Sam51 which is however not required for virulence or viability.
A C. albicans ortholog of an ERMES complex protein (Mmm1p) is required for hyphal growth, mitochondrial genome maintenance, mitochondrial outer membrane translocase complex assembly, and mitochondrion-ER tethering/phospholipid transport. Importantly, and mmm1Δ is conditionally avirulent mutant (http://www.candidagenome.org/).37 Another of the ERMES complex protein subunits from C. albicans is Fzo1p a GTPase protein. The fzo1Δ mutant had fragmented mitochondria, loss of mitochondrial genome, phospholipid alterations, as well as heightened susceptibility to azoles and peroxide. Azole hypersusceptibility was associated with a down regulation of CDR efflux pumps. Mutant cells also had features of iron starvation. Virulence data are not reported for this mutant but are needed since the protein is fungal-specific.
Fusion/fission mitochondrial proteins in A. fumigatus
Fusion/fission of mitochondria is a requisite for mitochondrial dynamics and activities such as autophagy, maintenance of mitochondrial DNA, mitochondrial movement in cells, and morphology. Mitochondrial dynamics of fusion/fission have been recently reported in Aspergillus fumigatus.38 Fusion genes included Mgm1, Ugo1 and Fzo1 while fission genes studied were Drp1, Mdv1, Caf2, and Fis1. Of these 7 genes, the fusion genes Ugo1, Fzo1 and Mgm1 are partially conserved from S. cerevisiae to Aspergillus spp, and mutants are essential for viability of A. fumigatus, and avirulent in a Galleria mellonella infection model.38 Growth and sporulation of each fission mutant was substantially reduced compared to WT cells but unexplainably not essential for virulence. Also, the fission mutants demonstrated resistance to voriconazole and posaconizole but not caspofungin antifungals. Thus the retention of virulence and gain-of-function antifungal resistance might preclude them as targets, while the fusion proteins should be further studied as targets for antifungal drug discovery.
The mitochondrial Srr1p response regulator
Signal transduction pathways of 2-component proteins have been identified in bacteria, fungi and plants.39-41 One of the better known MAPK pathways that utilizes 2-component proteins is the Hog1p pathway. Environmental signals that trigger the HOG1 MAPK include high osmotic pressure and ROS. In the presence of either cue, phosphorylation of a histidine kinase, membrane sensor protein (Sln1p) occurs first. Phosphotransfer from Sln1p to a response regulator protein (Ypd1p) is in turn then relayed to a MAP Kinase (Hog1p), which locates to the nucleus where target proteins are activated to adapt cells to stress. The Ssk1p, along with a second response regulator (Skn7p), are cytoplasmic proteins. However, a mitochondrial response regulator (Srr1p) has been identified.39,40 Srr1p appears to be unique to the Candida clade, with the exception of C. krusei39 A knockout ssr1Δ is defective in hyphal development, adaptation to osmotic/oxidative stresses, and is avirulent. The participation of Srr1p in a signal pathway and its role in mitochondrial functions are unknown.
Alternative carbon source utilization
Glucose availability for pathogens like C. albicans is limiting during infections or during its lifestyle as a commensal on mucosal surfaces.36 In contrast, other metabolites such as N-acetylglucosamine (GlcNAc) are plentiful, in part most likely released by bacterial competitors at mucosal surfaces, such as in the human gut. There are a number of GlcNAc catabolic pathway genes that are required for a fully operational catabolism of this metabolite, including transporters and kinase/deacetylase/deaminase enzymes. Deletion of the MCU1 causes defects in the utilization of GlcNAc.42 MCU1 is actually part of a cluster of C. albicans genes that is required for the degradation of GlcNAc, non-fermentable carbon sources, and amino acids.42 A mcu1/mcu1Δ is defective in GlcNAc utilization, has impaired filamentation, and is avirulent. The subcellular localization of Mcu1p was determined using a TETp-regulated MCU1-GFP fusion. The signal co-localized with Mitotracker, indicating the protein resides in mitochondria. Of importance, Mcu1p is not found in higher eukaryotes and thus, would appear to fulfill the vital qualifications of an antifungal drug target. Immunprecipitation studies with Mcu1p-GFP and mass spectrometry identified 32 candidate (mostly metabolism) proteins as well as known mitochondrial proteins such as Tom70p. We await further developments in regard to this interesting protein complex.
Preclinical compounds: A mitochondrial inhibitor
This section of the review describes some of the new antifungals that are in the pipeline as well as those that are preclinical.43 A characteristic of many of these antifungals is that they inhibit targets not of the current vintage of antifungals. Of importance, one of the preclinical compounds is a mitochondrial inhibitor. Five categories of currently used or new antifungal drugs have been described.43 Each category of drugs is licensed for: 1) systemic treatment, 2) superficial treatment, 3) in clinical trials, 4) no longer in development, or no reports since 2012, and 5) in preclinical development.43 Category #1 and #2 compounds are those that we are most familiar (triazoles, echinocandins, and amphotericin B), and those remodeled from each category. A total of 26 new compounds are listed in categories #3-5, but 14 of those belong in category #4, which as stated above, have apparently not advanced in the clinical pipeline. The remaining compounds are in preclinical development or in clinical trials.43 Of those in preclinical development, the inhibitors of glycosylphosphatidylinositol GPI biosynthesis have been studied extensively because of their important functions and novel mode-of-action.44,45 GPI anchors, for instance, ensure binding of fungal cell wall glycoproteins to host receptors. These inhibitors cause endoplasmic reticulum [ER] stress and an unmasking of β-glucan which results in a heightened macrophage pro-inflammatory response.
Three of 4 lead compounds remain in preclinical development, a fourth has apparently been discontinued. Four of 5 compounds that target protein synthesis are apparently no longer in development.43 Those in clinical trial include, VT-1161 (ergosterol synthesis inhibitor), SCY-078 (β-glucan inhibitor), and nikkomycin Z (chitin synthesis inhibitor).46-48 The target of the fourth compound (F901318) is unknown, but it is in a phase 1 (biosafety) clinical trial that is underway in the US for the treatment of aspergillosis. Study results are not posted. Interestingly, SCY-078 binds differently to the β-1,3-glucan synthase target FKS and is active against strains with FKS-point mutants that are resistant to echinocandin. The Nikkomycin Z inhibitor may be useful against fungal pathogens like C. immitis which have higher cell wall chitin content. F90138 is in clinical trial also as mentioned. None of the compounds mentioned above target mitochondria except ilicicolin, discussed below.49-51
Ilicicolin is a CIII fungal-specific inhibitor
Ilicicolin-H is a polyketide natural product that has potent, broad-spectrum activity against species of Candida, Aspergillus, and Cryptococcus (Fig. 4). The in vitro mode-of-action of ilicicolin was nicely identified to the yeast cytochrome bc1 reductase of Complex III of the mitochondrial electron transport chain.49-51 Activity of the mammalian enzyme was unaffected, indicating that the compound is fungal-specific. Depending upon the fungal species, MICs ranged from 0.04-1.56 µg/mL. Those MIC values increased when assays for enzyme activity were done in the presence of plasma protein, meaning activity may be reduced also in vivo. Plasma protein binding is correlated to lipophilicity or acidity of similar compounds. Lead optimization was pursued to reduce binding while retaining in vivo antifungal activity. Seven lead optimized compounds were compared to the parental ilicicolin-H for activity and protein binding. Diacetate and C-19 cyclopropyl acetate derivatives had reduced binding to plasma proteins by 20-fold. We await further information on this compound.
Figure 4.
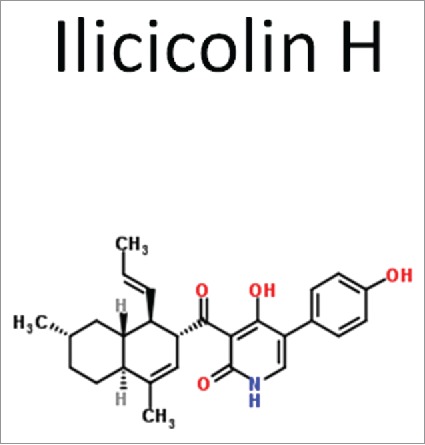
Ilicicolin H. A pentapeptide natural product produced by the fungus Gliocladium rosuem. The compound has good activity against Candida species, A. funigatus and Cryptococcus neoformans.
Resveratrol, a broad-spectrum inducer of apoptosis
Resveratrol is a natural polyphenolic (3,5,4′-trihydroxy-trans-stilbene) compound that has activity against a variety of pathogenic bacteria and fungi, including C. albicans, several Trichophyton species, and other dermatophytic species.52 An MIC of 20 μM was 2-fold higher than amphotericin B. Treatment in vitro caused the development of apoptosis through the caspase-dependent pathway which was associated with high levels of reactive oxygen species, and loss of membrane potential.
Conclusions and outlook
Two issues are quite clear. First, incidence of fungal infections is greater than previously believed. It is apparent that additions to the repertoire of antifungal drugs are critically needed to reduce the high incidence of fungal diseases and combat drug-resistant fungal pathogens. Patients with fungal diseases caused by drug resistant fungi have poorer outcomes than isolates caused by susceptible organisms. Second, there is a market for treating fungal diseases that should arouse the interests of Big Pharma and the biotech industry.53-56 The current world market for antifungal agents globally is in excess of US$6 billion. That is predicted to reach nearly $13.9 billion by 2018. Invasive fungal diseases can only be expected to increase in an aging population. The market just for treatment with current therapeutics is about ∼$1.3 billion (fluconazole and terbinafine (US$1bn), voriconazole (US$800M, itraconazole (US$900M), and caspofungin (US$600M), Ambisome (US∼$400M). We advocate for the discovery of new drug targets and, correspondingly, new compounds that inhibit these targets. Our intent herein has been to build an argument for targeting fungal-specific mitochondrial proteins since loss of these targets in gene deleted mutants is devastating to cell processes. We describe 11 such proteins and suggest that additional mitochondrial targets will be found using approaches briefly discussed. That these proteins are broadly distributed among pathogenic fungi is an advantage to their development as targets. Aside from the exploitation of mitochondrial as new drug targets, we suggest that functions of CI subunit proteins overlap but also differ substantially. That concept is quite new to our understanding of the underappreciated novelty of this organelle.
Disclosure of potential conflicts of interest
No potential conflicts of interest were disclosed.
Funding
Research in the Calderone Lab was supported by an NIH-NIAID grant (AI9029). DL is supported by bridge funding from the Biomedical Graduate Research Organization (BGRO).
References
- [1].Brown GD, Denning D, Gow N, Levitz S, Netea M, White T. Hidden killers: human fungal infections. Science Transl Med 2012; 4:Issue 165:165rv13. [DOI] [PubMed] [Google Scholar]
- [2].Corzo-Leon D, Armstrong-James D, Denning DW. Burden of serious fungal infections in Mexico. Mycoses 2015; 55(S5):34-44; http://dx.doi.org/ 10.1111/myc.12395 [DOI] [PubMed] [Google Scholar]
- [3].Clancy CJ, Nguyen MH. Diagnostic measures for blood-borne candidiasis. Methods Mil Biol 2016; 1356:215-38; http://dx.doi.org/ 10.1007/978-1-4939-3052-4_16 [DOI] [PubMed] [Google Scholar]
- [4].Ford C, Funt J, Abbey D, Issi L, Guiducci C, Martinez DA, Delorey T, Li BY, White TC, Cuomo C, et al.. The evolution of drug resistance in clinical isolates of Candida albicans. Elife 2015; 4:e00662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Pfaller MA, Diekema DJ. Epidemiology of invasive candidiasis: a persistent public health problem. Clin Microbiol Rev 2007; 20:133-63; PMID:17223626; http://dx.doi.org/ 10.1128/CMR.00029-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Pfaller MA. Antifungal drug resistance: mechanisms, epidemiology, and consequences for treatment. Amer J Med 2012; 125(1 Suppl):S3-13; PMID:22196207; http://dx.doi.org/ 10.1016/j.amjmed.2011.11.001 [DOI] [PubMed] [Google Scholar]
- [7].Dutcher J. The discovery and development of amphotericin B. Dis Chest 1968; 54, Suppl 1:40-42. [DOI] [PubMed] [Google Scholar]
- [8].(http://www.scientificamerican.com/article/cost-to-develop-new-pharmaceutical-drug-now-exceeds-2-5b : November 24, 2014. [Google Scholar]
- [9].Calderone R, Sun N, Gay-Andrieu F, Groutas W, Weerawarna P, Prasad S, Alex D, Li D. Antifungal drug discovery: the process and outcomes. Future Microbiol 2014; 9:791-05; PMID:25046525; http://dx.doi.org/ 10.2217/fmb.14.32 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Calderone R, Li D, Traven A. System level impact of mitochondria on virulence: to metabolism and beyond. FEMS Yeast Res 2015; 15:fovo27; http://dx.doi.org/ 10.1093/femsyr/fov027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Yang J-S, Kim J, Park S, Jeon J, Shin YE, Kim S. Spatial and functional organization of mitochondrial protein network. Sci Rep 2013; 3:1403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Bambach A, Fernandes MP, Ghosh A, Kruppa M, Alex D, Li D, Fonzi WA, Chauhan N, Sun N, Agrellos OA, et al.. Goa1p of Candida albicans localizes to the mitochondria during stress and is required for mitochondrial function and virulence. Eukaryot Cell 2009; 8:1706-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Videira A. Complex I from the fungus Neurospora crassa. Biochem Biophys Acta 1998; 1364:89-100; PMID:9593837 [DOI] [PubMed] [Google Scholar]
- [14].Gabaldon T, Rainey D, Huynen M. Tracing the evolution of a large protein complex in the eukaryotes, NADH:ubiquinone oxidoreductase. J Mol Biol 2005; 348:857-70; PMID:15843018; http://dx.doi.org/ 10.1016/j.jmb.2005.02.067 [DOI] [PubMed] [Google Scholar]
- [15].Abdrakhmanova A, Zickermann V, Bostina M, Radermacher M, Schagger H, Brandt U. Subunit composition of mitochondrial complex I from the yeast Yarrowia lipolytica. Biochim Piophys Acta 2004; 1658:148-56; http://dx.doi.org/ 10.1016/j.bbabio.2004.04.019 [DOI] [PubMed] [Google Scholar]
- [16].Bridges HR, Feamley I, Hirst J. The subunit composition of mitochondrial NADH:ubiquinoneoxidoreductase (Complex I) of Pichia pastoris. Mol Cell Proteomics 2010; 9:2318-26; PMID:20610779; http://dx.doi.org/ 10.1074/mcp.M110.001255 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Bridges HR, Grgic L, Harbour ME, Hirst J. The respiratory complexes I from the mitochondria of two Pichia species. Biochem J 2009; 422:151-59; PMID:19459785; http://dx.doi.org/ 10.1042/BJ20090492 [DOI] [PubMed] [Google Scholar]
- [18].Li D, Chen H, Florentino A, Alex D, Sikorski P, Fonzi WA, Calderone R. Enzymatic dysfunction of mitochondrial complex I of the Candida albicans goa1 mutant is associated with increased reactive oxidants and cell death. Eukaryot Cell 2011; 10:672-82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Grahl N, Demers EG, Lindsay AK, Harty CE, Willger SD, Piispanen AE, Hogan DA. Mitochondrial activity and Cyr1 are key regulators of Ras1 activation of Candida albicans virulence pathways. PLoS Pathog 2015; 11:e1005133; http://dx.doi.org/ 10.1371/journal.ppat.1005133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Thomas E, Roman E, Claypool S, Manzoor N, Pla J, Panwar SL. Mitochondria influence CDR1 efflux pump activity, Hog1-mediated oxidative stress pathway, iron homeostasis and ergosterol levels in Candida albicans. Antimicrob Agents Chemother 2013; 57:5580-99; PMID:23979757; http://dx.doi.org/ 10.1128/AAC.00889-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Sun N, Fonzi W, Chen H, Li X, Zhang L, Calderone R. Azole susceptibility and transcriptome profiling in Candida albicans mitochondrial electron transport chain complex I mutants. Antimicrob Agents Chemother 2013a; 57:532-42; http://dx.doi.org/ 10.1128/AAC.01520-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Sun N, Li D, Fonzi W, Li X, Zhang L, Calderone R. Multidrug-resistant transporter mdr1p-mediated uptake of a novel antifungal compound. Antimicrob Agents Chemother 2013b; 57:5931-39; http://dx.doi.org/ 10.1128/AAC.01504-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Pereira B, Videira A, Duarte M. Novel insights into the role of Neurospora crassa NDUFAF2, an evolutionarily conserved mitochondrial complex I assembly factor. Mol Cell Biol 2013; 33:2623-34; PMID:23648483; http://dx.doi.org/ 10.1128/MCB.01476-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Videra A, Duarte M. From NADH to ubiquione in Neurospora mitochondria. Biochim Biophys Acta 2002; 1555:187-91; PMID:12206913; http://dx.doi.org/ 10.1016/S0005-2728(02)00276-1 [DOI] [PubMed] [Google Scholar]
- [25].Videira A, Tropschug M, Werner S. Primary structure, in vitro expression and import into mitochondria of a 29-/21 kDa subunit of complex I from Neuropsora crassa. Biochem Biophys Res Commun 1990; 166:280-85; PMID:2137337; http://dx.doi.org/ 10.1016/0006-291X(90)91942-L [DOI] [PubMed] [Google Scholar]
- [26].Li D, Calderone R. Functional diversity of complex I subunits in Candida albicans mitochondria. Curr Genet 2016; 62:87-95; http://dx.doi.org/ 10.1007/s00294-015-0518-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].She X, Khamooshi K, Gao Y, Shen Y, Lv Y, Calderone R, Fonzi W, Liu W, Li D. Fungal-specific subunits of the Candida albicans mitochondrial complex I drive diverse cell functions including cell wall synthesis. Cell Microbiol 2015; 17(9):1350-64; PMID:25801605; http://dx.doi.org/ 10.1111/cmi.12438 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Chen H, Calderone R, Sun N, Wang Y, Li D. Caloric restriction restores the chronological life span of the Goa1 null mutant of Candida albicans in spite of high cell levels of ROS. Fungal Genet Biol 2012; 49:1023-31; PMID:23063955; http://dx.doi.org/ 10.1016/j.fgb.2012.09.007 [DOI] [PubMed] [Google Scholar]
- [29].She X, Zhang L, Chen H, Calderone R, Li D. Cell surface changes in the Candida albicans mitochondrial mutant goa1Δ are associated with reduced recognition by innate immune cells. Cell Microbiol 2013; 15:1572-84; PMID:23490206; http://dx.doi.org/ 10.1111/cmi.12135 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Khamooshi K, Sikorski P, Sun N, Calderone R, Li D. 2014. The Rbf1, Hfl1 and Dbp4 of Candida albicans regulate common as well as transcription factor-specific mitochondrial and other cell activities. BMC Genomics 2014; 15:56; PMID:24450762; http://dx.doi.org/ 10.1186/1471-2164-15-56 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].She X, Calderone R, Kruppa M, Lowman D, Williams D, Zhang L, Gao Y, Khamooshi K, Liu W, Li D. Cell wall β-mannosylation and mannan are reduced in the cell wall of the GOA1 mutant of the Candida albicans mitochondrial Complex 1. PLoS One 2016; 11(1):e0147175; http://dx.doi.org/ 10.1371/journal.pone.0147175 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].McDonough JA, Bhattacherjee V, Sadlon T, Hostetter MK. 2002. Involvement of Candida albicans NADH dehydrogenase complex I in filamentation. Fungal Biol Genet 2002; 36:117-27; http://dx.doi.org/ 10.1016/S1087-1845(02)00007-5 [DOI] [PubMed] [Google Scholar]
- [33].Qu Y, Jelicic , Pettolino F, Perry A, Lo TL, Hewitt VL, Bantun F, Beilharz TH, Peleg AY, Lithgow T, et al.. Mitochondria sorting and assembly machinery subunit Sam37 in Candida albicans: insight into the roles of mitochondria in fitness, cell wall integrity, and virulence. Eukary Cell 2012; 11:532-44; http://dx.doi.org/ 10.1128/EC.05292-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Shingu-Vazquez Traven A. Mitochondria and fungal pathogenesis: drug tolerance, virulence, and potential for antifungal therapy. Eukary Cell 2011; 10:1376-82; http://dx.doi.org/ 10.1128/EC.05184-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Dagley M, Gentle I, Bellharz T, Pettolino FA, Djordjevic JT, Lo TL, Uwamahoro N, Rupasinghe T, Tull DL, McConville M, et al.. Cell wall integrity is linked to mitochondria and phospholipid homeostasis in Candida albicans through the activity of the post-transcriptional regulator Ccr-Pop2. Mol Microbiol 2011; 79:968-89; PMID:21299651; http://dx.doi.org/ 10.1111/j.1365-2958.2010.07503.x [DOI] [PubMed] [Google Scholar]
- [36].Calderone R, Li D, Traven A. System-level impact of mitochondria on fungal virulence: to metabolism and beyond. FEMS Yeast Res 2015; 15:fov027; PMID:26002841; http://dx.doi.org/ 10.1093/femsyr/fov027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Becker J, Kauffman S, Hauser M, Huang L, Lin M, Sillaots S, Jiang B, Xu D, Roemer T. Pathway analysis of Candida albicans survival and virulence determinants in a murine infection model. PNAS USA 2010; 107:22044-49; PMID:21135205; http://dx.doi.org/ 10.1073/pnas.1009845107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Neubauer M, Zhu Z, Helmschroff C, Wagener N, Wagener J. Mitochondrial dynamics in the pathogenic mold Aspergillus fumigatus: therapeutic and evolutionary implications. Mol Microbiol 2015; 88:930-45; http://dx.doi.org/ 10.1111/mmi.13167 [DOI] [PubMed] [Google Scholar]
- [39].Chauhan N. Two-component phosphorelays in fungal mitochondria and beyond. Mitochondrion 2015: 22:60-65; PMID:25858273; http://dx.doi.org/ 10.1016/j.mito.2015.03.003 [DOI] [PubMed] [Google Scholar]
- [40].Desai C, Mavrianos J, Chauhan N. Candida albicans SRR1, a putative two-component response regulator gene, is required for stress adaptation, morphogenesis and virulence. Eukary Cell 2011; 10:1370-74; http://dx.doi.org/ 10.1128/EC.05188-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41].Defosse T, Sharma A, Mondal AK, Dugé de Bernonville T, Latgé JP, Calderone R, Giglioli-Guivarc'h N, Courdavault V, Clastre M, Papon N. Hybrid histidine kinases in pathogenic fungi. Mol Microbiol 2015; 95:914-24; PMID:25560420; http://dx.doi.org/ 10.1111/mmi.12911 [DOI] [PubMed] [Google Scholar]
- [42].Guan G, Wang H, Liang W, Cao C, Tao L, Naseem S, Konopka JB, Wang Y, Huang G. The mitochondrial protein Mcu1 plays important roles in carbon source utilization, filamentation, and virulence in Candida albicans. Fungal Genet Biol 2015; 81:150-59; PMID:25626172; http://dx.doi.org/ 10.1016/j.fgb.2015.01.006 [DOI] [PubMed] [Google Scholar]
- [43].Denning D, Bromley M. How to bolster the antifungal pipeline. Science 2015; 347:1414-16; PMID:25814567; http://dx.doi.org/ 10.1126/science.aaa6097 [DOI] [PubMed] [Google Scholar]
- [44].McLellan CA, Whitesell L, King OD, Lancaster AK, Mazitschek R, Lindquist S. Inhibiting GPI anchor biosynthesis in fungi stresses the endoplasmic reticulum and enhances immunogenicity. ACS Chem Biol 2012; 7:1520-8; PMID:22724584; http://dx.doi.org/ 10.1021/cb300235m [DOI] [PubMed] [Google Scholar]
- [45].Watanabe NA, Miyazaki M, Horii T, Sagane K, Tsukahara K, Hata K. E1210, a new broad-spectrum antifungal, suppresses Candida albicans hyphal growth through inhibition of glycosylphosphatidylinositol biosynthesis. Antimicrob Agents Chemother 2012; 56:960-71; PMID:22143530; http://dx.doi.org/ 10.1128/AAC.00731-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [46].Castanheira M, Messer SA, Rhomberg PR, Jones RN, Pfaller M, Activity of a novel echinocandin (CD101) tested against the most common Candida and Aspergillus species, including echinocandin-resistant and azole resistant mutants. 2014; Abstract M1082, ICAAC. [Google Scholar]
- [47].Moriyama B, Gordon LA, McCarthy M, Henning SA, Walsh TJ, Penzak SR. Emerging drugs and vaccines for candidemia. Mycoses 2014; 57:718-33; PMID:25294098; http://dx.doi.org/ 10.1111/myc.12265 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [48].Lamoth F, Alexander BD. Antifungal activities of SCY-078 (MK-3118) and standard antifungal agents against clinical non-Aspergillus mold isolates. Antimicrob Agents Chemother 2015; 59:4308-11; PMID:25896696; http://dx.doi.org/ 10.1128/AAC.00234-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [49].Singh S, Liu W, Li X, Li X, Chen T, Shafiee A, Dreikorn S, Hornak V, Meinz M, Onishi JC. Structure-activity relationship of cytochrome bc1 reductase inhibitor broad spectrum antifungal ilicicolin. Biorgan Med Chem Lett 2013; 23:3018-22; http://dx.doi.org/ 10.1016/j.bmcl.2013.03.023 [DOI] [PubMed] [Google Scholar]
- [50].Gutierrez-Cirlos E, Merbitz-Zahradnik T, Trumpower B. Inhibition of the yeast cytochrome bc1 complex by ilicictin, a novel inhibitor that acts at the Qn site of the bc1 complex. J Biol Chem 2004; 279:8708-14; PMID:14670947; http://dx.doi.org/ 10.1074/jbc.M311805200 [DOI] [PubMed] [Google Scholar]
- [51].Covian R, Turmpower B. Ilicicolin inhibition and binding at Center N of the dimeric cytochrome bc1 complex reveal electron transfer and regulatory interactions between monomers. J Biol Chem. 2009; 284:8614-20; PMID:19176478; http://dx.doi.org/ 10.1074/jbc.M808914200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [52].Lee J, Lee DG. Novel antifungal mechanism of resveratrol: apoptosis inducer in Candida albicans. Current Microbiol 2015; 70:383-90; PMID:25413604; http://dx.doi.org/ 10.1007/s00284-014-0734-1 [DOI] [PubMed] [Google Scholar]
- [53]. http://www.f2g.com/antifungal-markethttp://www.transparencymarketresearch.com/systemic-fungal-infection-therapeutics-market.html
- [54]. http://www.bccresearch.com/pressroom/phm/global-market-for-human-antifungal-therapeutics-to-reach-nearly-$13.9-billion-in-2018
- [55]. http://www.transparencymarketresearch.com/systemic-fungal-infection-therapeutics-market.html
- [56]. http://www.bccresearch.com/pressroom/phm/global-market-for-human-antifungal-therapeutics-to-reach-nearly-$13.9-billion-in-2018