Abstract
The biomechanical properties of populations of diseased cells are shown to have differences from healthy populations of cells, yet the overlap of these biomechanical properties can limit their use in disease cell enrichment and detection. We report a new microfluidic cell enrichment technology that continuously fractionates cells through differences in biomechanical properties, resulting in highly pure cellular subpopulations. Cell fractionation is achieved in a microfluidic channel with an array of diagonal ridges that are designed to segregate biomechanically distinct cells to different locations in the channel. Due to the imposition of elastic and viscous forces during cellular compression, which are a function of cell biomechanical properties including size and viscoelasticity, larger, stiffer and less viscos cells migrate parallel to the diagonal ridges and exhibit positive lateral displacement. On the other hand, smaller, softer and more viscous cells migrate perpendicular to the diagonal ridges due to circulatory flow induced by the ridges and result in negative lateral displacement. Multiple outlets are then utilized to collect cells with finer gradation of differences in cell biomechanical properties. The result is that cell fractionation dramatically improves cell separation efficiency compared to binary outputs and enables the measurement of subtle biomechanical differences within a single cell type. As a proof-of-concept demonstration, we mix two different leukemia cell lines (K562 and HL60) and utilize cell fractionation to achieve over 45-fold enhancement of cell populations, with high purity cellular enrichment (90% to 99%) of each cell line. In addition, we demonstrate cell fractionation of a single cell type (K562 cells) into subpopulations and characterize the variations of biomechanical properties of the separated cells with atomic force microscopy. These results will be beneficial to obtaining label-free separation of cellular mixtures, or to better investigate the origins of biomechanical differences in a single cell type.
1 Introduction
Cell separation is vital in several clinical diagnostic applications such as detection of cancer and infectious diseases (Chen et al. 2012; Hou et al. 2010). In hospitals and labs, cell separation is routinely carried out by centrifugation, size-exclusion and cell sorters based on fluorescent signals. The demand to obtain high purity cell population at low-cost spurred growing interest in exploring alternative cell separation methods based on microfluidics. For example electric field can be used to separate cells in microfluidic channels based on cell surface charge distribution (Vahey and Voldman 2008). Magnetic field is used to separate cells with different surface proteins that bind magnetic particles in microfluidic channels (Pamme and Wilhelm 2006). Also, acoustic field is used to separate cells of different sizes (Franke et al. 2010). In addition, research studies have shown that cell biomechanical properties including size (Vona et al. 2000), stiffness (Glenister et al. 2002; Suresh 2007; Xu et al. 2012) and viscoelasticity (Sawetzki et al. 2013) are biomarkers of diseases such as cancer and malaria. The changes in cell biomechanical properties are attributed to the transformation of cellular structures such as the cytoskeleton (Bongiorno et al. 2014; Brown et al. 2001; Wagner et al. 2006) and nucleus (Dahl et al. 2008). Therefore, cell biomechanical properties are phenotypes that could potentially be used for clinical diagnostics (Cross et al. 2007; Xu et al. 2012) and therapies (Sethu et al. 2006). Several recent studies have used hydrodynamic force (Gossett et al. 2012) and optical force (Otto et al. 2015) to distinguish cell phenotypes based on cell mechanical properties.
Utilizing differences in cell biomechanical properties for disease detection would be greatly aided by means to sort cells biomechanically. Biomechanical properties are intrinsic to the cell, and as such provide a label-free approach to enrichment without the need to discover and develop biomolecular reagents to aid detection (Bow et al. 2011; Guck et al. 2005; Wang et al. 2013). In addition, microfluidic platforms enable continuous sample processing, improved sensitivity, and utilize small sample volume. Recently, several microfluidic cell sorting approaches based on variations in cell biomechanical properties have been demonstrated (Hou et al. 2010; Hur et al. 2011; Zhang et al. 2012). However, the ultimate purity of the separated cells is limited by the intrinsic variability and overlap of the biomechanical properties of different cell types, even if the average properties are substantially different (Choi et al. 2007; Hur et al. 2011; Wang et al. 2013). As a result, biomechanical approaches to cell enrichment are in general inferior to the best results achieved by cell sorting methods based on antibody binding of magnetic (Kim and Soh 2009) or fluorescent signals (Cho et al. 2010; Wolff et al. 2003). Previously, we reported using diagonal ridges to separate cells based on cell size, stiffness (Wang et al. 2013), and viscoelasticity (Wang et al. 2015) in microfluidic devices with binary outputs. We optimized ridge geometries to improve cell separation. In this communication, we implement cell fractionation with similar ridge geometry by adding multiple outlets and aim to further boost the purity of the separated cells. Three outlets are designed to specifically collect the biomechanically overlapping cells at a central outlet and the biomechanically unique cells (corresponding to different cell types) at each of the two other outlets. We found the additional outlet significantly improved cell purity such that the cell enrichment factor is greater than 45-fold, approaching the purity achieved with single-color fluorescently activated cell sorting (Wolff et al. 2003). In addition, we utilized the method to subdivide a single cell type into subpopulations with distinct biomechanical phenotypes, which cannot be achieved using non-biomechanical methods.
In our current implementation of the sorting device, cells enter the microfluidic channel through a focused cell inlet. As cells flow through the microfluidic channel, the ridges compress the cells in rapid succession and alter their trajectory dependent on their biomechanical properties. The ridged channel design has been used in cell sorting based on stiffness (Wang et al. 2013), size (Choi et al. 2007; Mao and Alexeev 2011), adhesion (Choi et al. 2012), and viscosity (Wang et al. 2015); although in each case the specific sorting mechanisms are distinct. In cell sorting by stiffness, the cells are compressed by each ridge which causes translation of each cell laterally in a manner dependent on the intrinsic cell stiffness, the ridge parameters, such as angle and magnitude of compression, and the hydrodynamic drag force due to the fluid flow (Wang et al. 2013). In cell sorting by size, the cell trajectory entirely depends on the flow hydrodynamics since cells have no direct contact with the ridges. Cells of different sizes differentiate due to a combination of the cross-stream inertial migration and the secondary flow generated by the ridges (Mao and Alexeev 2011). In cell sorting by adhesion, the ridges are coated with adhesive molecules that interact with slow moving cells that express specific biomarkers to impart cell separation (Choi et al. 2012). More recently, cell sorting by differences in relaxation behavior has been reported, in which the cells are separated based on the relaxation time after high frequency compressions by the ridges (Wang et al. 2015). Viscous cells remain deformed and move perpendicular to the ridges while less viscous cells move along the ridges due to sustained cycles of relaxation and compression. In all these approaches, the devices utilized two outlets and can only obtain binary separation of cell mixtures.
2 Materials and Methods
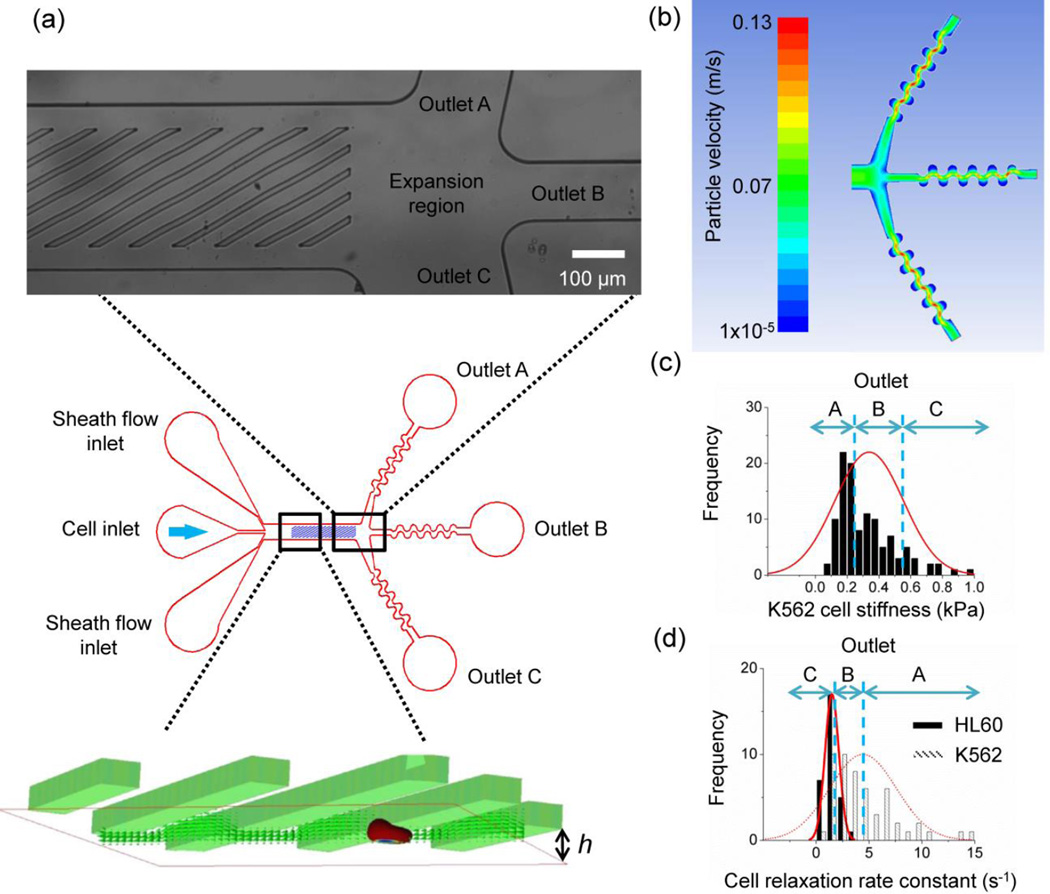
The microfluidic cell sorter utilizes a microchannel decorated with diagonal ridges on the top wall. The ridges are inclined at an angle to create hydrodynamic circulations underneath the ridge. Straight ridges along or across the channel will not generate effective flow circulation that results in cell lateral displacement. In addition, the ridges are designed to compress and translate cells normal to the flow direction. The ridges and the channel bottom surface form a gap, h, which is smaller than the cell diameters (Fig. 1a). The ridge angle was optimized for K562 and HL60 cells in a previous study. The ridge angle affects the trajectories of cells. For example, larger angle (45 degrees) reduces positive lateral displacement for larger and less viscous cells (K562) compared to smaller angle (30 degrees) (Wang et al. 2015). For cell separation in the current study, we employ 30 degree ridge angle. The overall channel length and width is 3,600 μm and 560 μm respectively. The ridge is 20 μm wide and distance between the ridges is 78 μm. The ridge width is chosen such that the cells are completed deformed under compression by the ridge. The distance between the ridges is designed to maximize lateral cell displacement (Wang et al. 2015).
Figure 1.
(a) A schematic that shows the cells entering the microfluidic channel are compressed by diagonal ridges in rapid succession and sorted into three outlets. The channel geometric parameters include gap size, h, ridge spacing, L, and channel height H. (b) Computational fluid dynamics simulations are used to engineer hydrodynamics near the outlets to ensure even splitting of the channel flow. The color gradient represents flow velocity vectors of particles. (c) A schematic describes cell fractionation of a single cell type (K562) based on the spread of cell stiffness. (d) A schematic describes cell fractionation of two cell types (K562 and HL60) based on the spread of cell relaxation.
We describe a microfluidic channel design that exploits cell stiffness and viscosity to sort cells into subpopulations of high purity. The microfluidic cell sorter is made using replica molding of polydimethylsiloxane (PDMS) on a permanent mold. The mold is created by two-step photolithography patterning of SU8 photoresist (Microchem) on a 4 inch diameter silicon wafer. After removal of PDMS from the mold, inlet and outlet holes are punched and the PDMS is subsequently bonded to a glass substrate to form the microfluidic channel. The substantial improvement in the purity of sorted cells is benefited from multiple outlets that are hydrodynamically balanced to fractionate the processed cells. The multiple outlets provide finer gradation of cells based on cell biomechanical properties compared to the binary outputs, akin to size exclusion chromatography fractions (Wen et al. 1996; Yamada and Seki 2005). As a proof-of-concept demonstration, we utilize three outlets within our microfluidic cell sorter (Fig. 1a). To improve the cell separation quality, we also incorporate two additional design elements: an expansion region after the ridged segment of the channel and outlets with serpentine architecture. The expansion region evenly divides channel flow among the outlets. The serpentine outlets are included to increase flow resistance and prevent flow biasing from uneven outlet sizes and external perturbations. We utilized computational fluid dynamics (Ansys Fluent) to design a balanced channel flow egress across all outlets (Fig. 1b, noting that the outlet c is substantially larger than outlets a and b in order to collect the long tail of cells exhibited in Fig 1c). The central outlet is designed to collect the cells that have moderate biomechanical properties (Fig. 1c). In the case of separation of two cell types, the central outlet can collect those cells that have overlapping biomechanical properties. As a result, the cells collected at the top and bottom outlets have pronouncedly different biomechanical properties (Fig. 1d).
Cell stiffness and relaxation are characterized with atomic force microscopy (Asylum Research) and cell size is recorded using optical microscopy (Nikon). The cell stiffness is represented by the average Young’s modulus and the cell viscosity is characterized by the relaxation rate constant. After cell indentation, the spherical tip indenter was held in place while the compression force was monitored for 5 seconds so that cell relaxation can be measured. The detailed description of AFM and cell sorting experimental methods is described in previous studies (Wang et al. 2015; Wang et al. 2013). Cell relaxation is used as a surrogate of apparent viscosity using the standard linear solid model including a spring and dashpot in parallel with another spring (Darling et al. 2007).
3 Results and discussion
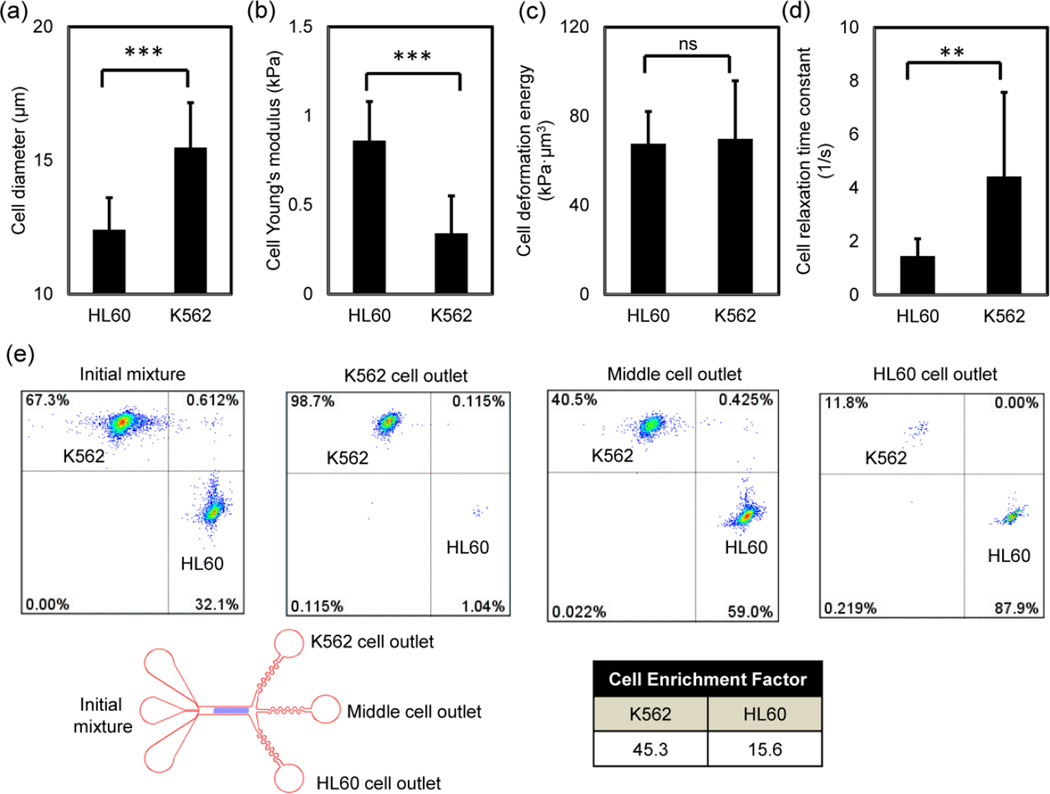
To demonstrate the cell sorting efficiency of the three-outlet channel, we tested cell sorting of two types of leukemia cell lines: K562 and HL60. The biomechanical properties of these two cell types and the results of cell separation using a binary output channel were previously reported (Wang et al. 2015). The cell size, Young’s modulus, size-adjusted elasticity (deformation energy) and the relaxation rate constants are shown in Fig. 2a, 2b, 2c and 2d respectively. The size-adjusted elasticity accounts for cell size and cell stiffness as a single parameter (Wang et al. 2015). We have demonstrated previously that K562 and HL60 cells are primarily sorted based on differences in cell relaxation due to a higher viscosity of HL60 cells. Cell mixtures are labelled fluorescently and mixed at 1 to 2 million cells per mL. The flow rate ranges from 0.0125 to 0.025 mL per minute. The detail of the cell separation setup and procedure is reported in a previous study (Wang et al. 2015). The cell sorting result with the cell fractionation is evaluated by flow cytometry and shown in Fig. 2e. To quantify cell sorting efficiency, we compute the cell enrichment factor, c.e.f. The c.e.f. accounts cell enrichment by normalizing ratios of cell proportions at outlets to the original cell proportion in the initial mixture. For example the enrichment for K562 cells is calculated as follows:
For the three outlet device, the c.e.f. for K562 and HL60 cells is 45.3 and 15.6 respectively. The fractionation improves the cell enrichment factor by an order of magnitude compared to the results utilizing binary outlets (Wang et al. 2015). The additional outlet in the middle collects those K562 and HL60 cells that have overlapping biomechanical properties. Therefore, the three-outlet channel enables a dramatic improvement in cell sorting purity. A video of the cell separation was obtained with high speed microscopy and shown in Supplemental Video 1.
Figure 2.
(a) The average cell diameter for K562 and HL60 cells are 15.5 ± 1.7 μm (n = 114) and 12.4 ± 1.2 μm (n = 36). (b) AFM measurements of cells’ Young’s modulus show that HL60 (E = 0.86 ± 0.22 kPa, n = 24) is stiffer than K562 (E = 0.34 ± 0.21 kPa, n = 114). c) The average values of Young’s modulus and cell diameter are used to calculate cell deformation energy. K562 and HL60 have similar deformation energy (69.8 ± 26.1 kPa·μm3, and 67.6 ± 14.5 kPa·μm3 respectively) when compressed with 9 μm channel gap height. d) AFM measurements of cell relaxation show HL60 (1.45 ± 0.65 s−1, n = 30) is slower in size recovery than K562 (4.42 ± 3.15 s−1, n = 52). t-test was used to analyze statistical significance: **: p < 0.001, ***: p < 0.0001, and ns: no significance. (e) Flow cytometric analysis of cell enrichment using a three-outlet channel. At the outlets, the cell enrichment factor for K562 and HL60 cells are 45.3 and 15.6 respectively. The purity of enriched K562 cells and HL60 cells are 98.7% and 87.9% respectively. The total number of cells initially is 22,000. The number of cells in the K562 outlet, the middle outlet and the HL60 outlet are 11,500, 4600, and 2100 respectively and rounded the nearest hundred.
In addition to enrichment of cell mixtures into individual cell types, the three-outlet channel can also be applied to fractionate a single cell type into distinct biomechanical phenotypes. Since biological cells are inherently heterogeneous in nature, their biomechanical properties may include large variations due to differences in cytoskeleton or nucleus (Dahl et al. 2008; Xu et al. 2012). For example, K562 cells have average Young’s modulus 0.34 kPa and a standard deviation 0.21 kPa (Fig. 2b). Utilizing the fractionation approach, we are able to obtain subpopulations of K562 cells with differing biomechanical properties.
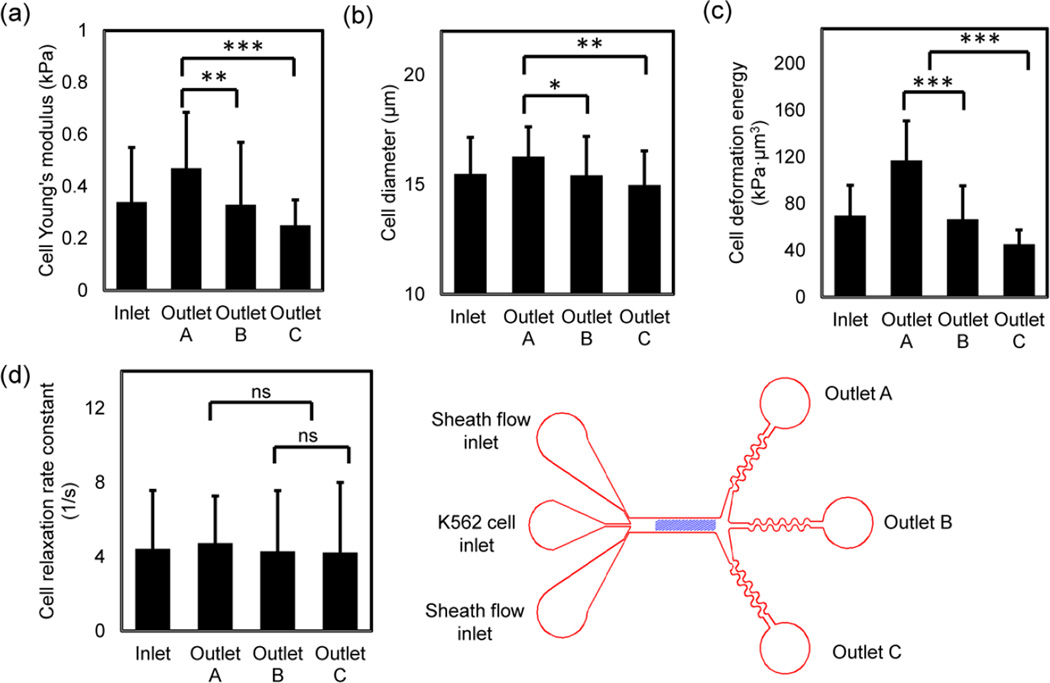
K562 cells were fractionated using the three-outlet channel and the separated cells were characterized with AFM and optical microscopy immediately after collection. The biomechanical properties of the K562 cells at inlet and three outlets are provided in Fig. 3. These data show that K562 cells can be successfully separated into three populations with different biomechanical properties, predominantly stiffness and size, but not significantly by cell relaxation. The size-adjusted elasticity (deformation energy) that combines the effects of cell size and stiffness is also significantly different among the cells collected at three outlets.
Figure 3.
(a) Atomic force microscopy measurements of cell Young’s modulus show that sorted K562 cells have different average Young’s modulus outlet A (E = 0.47 ± 0.21 kPa, n = 27), outlet B (E = 0.33 ± 0.24 kPa , n = 53) and outlet C (E = 0.25 ± 0.098 kPa, n = 38). The average Young’s modulus at inlet is 0.39 ± 0.21 kPa (n = 110). Spearman’s correlation analysis of the separated cells produces a p-value of 0 and a r-value of -0.42. (b) Cell sizes are different at three outlets with the largest average cell diameter in outlet A and the smallest average cell diameter in outlet C. The cell diameters in outlet A, B, C and inlet are 16.28 ± 1.4 μm (n = 27), 15.42 ± 1.78 μm (n = 53), 14.98 ± 1.56 μm (n = 38) and 15.2 ± 1.2 μm (n = 110) respectively. Spearman’s correlation analysis of the separated cells produces a p-value of 0.001 and a r-value of −0.29. (c) The deformation energy (size-adjusted elasticity) has the most pronounced differences at three outlets. The average deformation energy at outlet A, B, C and inlet are 117.14 ± 33.8 kPa·μm3, 66.7 ± 28.6 kPa·μm3, 45.4 ± 12.3 kPa·μm3, and 66.8 ± 21.6 kPa·μm3 respectively. Spearman’s correlation analysis of the separated cells produces a p-value of 0 and a r-value of −0.47. (d) Cell relaxation is slightly different at three outlets. The AFM measurement of cell relaxation rate constant at outlet A, B, C and inlet are 5.52 ± 4.2 s−1 (n = 20), 4.29 ± 3.3 s−1 (n = 19), 4.23 ± 3.8 s−1 (n = 17) and 4.46 ± 2.4 s−1 (n = 55) respectively. Spearman’s correlation analysis of the separated cells produces a p-value of 0.116 and a r-value of -0.22. t-test was used to analyze statistical significance, * p < 0.05, ** p < 0.01, and *** p < 0.001.
4 Conclusion
We show that cell enrichment through multiple-outlet biomechanica fractionation significantly improves the purity of sorted cells. The fractionation cell sorting circumvents the major challenge in biomechanical cell sorting caused by the inherent heterogeneity and significant overlap of cellular populations. The caveat is that the cells that show overlapping biomechanical properties and cells that have moderate biomechanical properties are collected in the same outlet and the separated cells are less than the initial number of cells in the mixture. Further development including implement additional outlet is likely to improve cell fractionation. Biomechanical fractionation may prove useful in preprocessing samples with low numbers of target cells for downstream analysis and detection, or to better understand the relationship of biomechanical phenotypes and molecular content and function.
Supplementary Material
Acknowledgments
This research was supported by NSF project number CBET-0932510, TI:GER program at Scheller College of Business at Georgia Tech, the Regenerative Engineering and Medicine Seed Grant, and the President’s Undergraduate Research Award (PURA) program at Georgia Tech. The authors would like to thank Dr. Wilbur Lam, Dr. Hang Lu and Dr. Peter Hesketh for helpful discussions.
References
- Bongiorno T, et al. Mechanical stiffness as an improved single-cell indicator of osteoblastic human mesenchymal stem cell differentiation. J Biomech. 2014;47:2197–2204. doi: 10.1016/j.jbiomech.2013.11.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bow H, et al. A microfabricated deformability-based flow cytometer with application to malaria. Lab Chip. 2011;11:1065–1073. doi: 10.1039/c0lc00472c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown MJ, Hallam JA, Colucci-Guyon E, Shaw S. Rigidity of circulating lymphocytes is primarily conferred by vimentin intermediate filaments. J Immunol. 2001;166:6640–6646. doi: 10.4049/jimmunol.166.11.6640. [DOI] [PubMed] [Google Scholar]
- Chen J, Li J, Sun Y. Microfluidic approaches for cancer cell detection, characterization, and separation. Lab Chip. 2012;12:1753–1767. doi: 10.1039/c2lc21273k. [DOI] [PubMed] [Google Scholar]
- Cho SH, Chen CH, Tsai FS, Godin JM, Lo YH. Human mammalian cell sorting using a highly integrated micro-fabricated fluorescence-activated cell sorter (mu FACS) Lab Chip. 2010;10:1567–1573. doi: 10.1039/c000136h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi S, Song S, Choi C, Park JK. Continuous blood cell separation by hydrophoretic filtration. Lab Chip. 2007;7:1532–1538. doi: 10.1039/b705203k. [DOI] [PubMed] [Google Scholar]
- Choi SY, Karp JM, Karnik R. Cell sorting by deterministic cell rolling. Lab Chip. 2012;12:1427–1430. doi: 10.1039/c2lc21225k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cross SE, Jin YS, Rao J, Gimzewski JK. Nanomechanical analysis of cells from cancer patients. Nat Nanotechnol. 2007;2:780–783. doi: 10.1038/nnano.2007.388. [DOI] [PubMed] [Google Scholar]
- Dahl KN, Ribeiro AJS, Lammerding J. Nuclear shape, mechanics, and mechanotransduction. CircRes. 2008;102:1307–1318. doi: 10.1161/CIRCRESAHA.108.173989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darling EM, Zauscher S, Block JA, Guilak F. A Thin-Layer Model for Viscoelastic, Stress-Relaxation Testing of Cells Using Atomic Force Microscopy: Do Cell Properties Reflect Metastatic Potential? Biophys J. 2007;92:1784–1791. doi: 10.1529/biophysj.106.083097. doi: http://dx.doi.org/10.1529/biophysj.106.083097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franke T, Braunmuller S, Schmid L, Wixforth A, Weitz DA. Surface acoustic wave actuated cell sorting (SAWACS) Lab Chip. 2010;10:789–794. doi: 10.1039/b915522h. [DOI] [PubMed] [Google Scholar]
- Glenister FK, Coppel RL, Cowman AF, Mohandas N, Cooke BM. Contribution of parasite proteins to altered mechanical properties of malaria-infected red blood cells. Blood. 2002;99:1060–1063. doi: 10.1182/blood.v99.3.1060. [DOI] [PubMed] [Google Scholar]
- Gossett DR, et al. Hydrodynamic stretching of single cells for large population mechanical phenotyping. Proc Natl Acad Sci U S A. 2012;109:7630–7635. doi: 10.1073/pnas.1200107109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guck J, et al. Optical deformability as an inherent cell marker for testing malignant transformation and metastatic competence. Biophys J. 2005;88:3689–3698. doi: 10.1529/biophysj.104.045476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hou HW, Bhagat AAS, Chong AGL, Mao P, Tan KSW, Han JY, Lim CT. Deformability based cell margination-A simple microfluidic design for malaria-infected erythrocyte separation. Lab Chip. 2010;10:2605–2613. doi: 10.1039/c003873c. [DOI] [PubMed] [Google Scholar]
- Hur SC, Henderson-MacLennan NK, McCabe ERB, Di Carlo D. Deformability-based cell classification and enrichment using inertial microfluidics. Lab Chip. 2011;11:912–920. doi: 10.1039/c0lc00595a. [DOI] [PubMed] [Google Scholar]
- Kim U, Soh HT. Simultaneous sorting of multiple bacterial targets using integrated Dielectrophoretic-Magnetic Activated Cell Sorter. Lab Chip. 2009;9:2313–2318. doi: 10.1039/b903950c. [DOI] [PubMed] [Google Scholar]
- Mao WB, Alexeev A. Hydrodynamic sorting of microparticles by size in ridged microchannels. Phys Fluids. 2011;23 [Google Scholar]
- Otto O, et al. Real-time deformability cytometry: on-the-fly cell mechanical phenotyping. Nat Meth. 2015;12:199–202. doi: 10.1038/nmeth.3281. [DOI] [PubMed] [Google Scholar]
- Pamme N, Wilhelm C. Continuous sorting of magnetic cells via on-chip free-flow magnetophoresis. Lab Chip. 2006;6:974–980. doi: 10.1039/b604542a. [DOI] [PubMed] [Google Scholar]
- Sawetzki T, Eggleton CD, Desai SA, Marr DWM. Viscoelasticity as a Biomarker for High-Throughput Flow Cytometry. Biophys J. 2013;105:2281–2288. doi: 10.1016/j.bpj.2013.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sethu P, Sin A, Toner M. Microfluidic diffusive filter for apheresis (leukapheresis) Lab Chip. 2006;6:83–89. doi: 10.1039/b512049g. [DOI] [PubMed] [Google Scholar]
- Suresh S. Biomechanics and biophysics of cancer cells. Acta Biomater. 2007;3:413–438. doi: 10.1016/j.actbio.2007.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vahey MD, Voldman J. An equilibrium method for continuous-flow cell sorting using dielectrophoresis. Anal Chem. 2008;80:3135–3143. doi: 10.1021/ac7020568. [DOI] [PubMed] [Google Scholar]
- Vona G, et al. Isolation by size of epithelial tumor cells - A new method for the immunomorphological and molecular characterization of circulating tumor cells. Am J Pathol. 2000;156:57–63. doi: 10.1016/S0002-9440(10)64706-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wagner B, Tharmann R, Haase I, Fischer M, Bausch AR. Cytoskeletal polymer networks: The molecular structure of cross-linkers determines macroscopic properties. Proc Natl Acad Sci U S A. 2006;103:13974–13978. doi: 10.1073/pnas.0510190103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang G, Crawford K, Turbyfield C, Lam W, Alexeev A, Sulchek T. Microfluidic cellular enrichment and separation through differences in viscoelastic deformation. Lab Chip. 2015;15:532–540. doi: 10.1039/c4lc01150c. [DOI] [PubMed] [Google Scholar]
- Wang GH, Mao WB, Byler R, Patel K, Henegar C, Alexeev A, Sulchek T. Stiffness Dependent Separation of Cells in a Microfluidic Device. PLoS One. 2013;8 doi: 10.1371/journal.pone.0075901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wen J, Arakawa T, Philo JS. Size-exclusion chromatography with on-line light-scattering, absorbance, and refractive index detectors for studying proteins and their interactions. Anal Biochem. 1996;240:155–166. doi: 10.1006/abio.1996.0345. [DOI] [PubMed] [Google Scholar]
- Wolff A, et al. Integrating advanced functionality in a microfabricated high-throughput fluorescent-activated cell sorter. Lab Chip. 2003;3:22–27. doi: 10.1039/b209333b. [DOI] [PubMed] [Google Scholar]
- Xu WW, Mezencev R, Kim B, Wang LJ, McDonald J, Sulchek T. Cell Stiffness Is a Biomarker of the Metastatic Potential of Ovarian Cancer Cells. PLoS One. 2012;7 doi: 10.1371/journal.pone.0046609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamada M, Seki M. Hydrodynamic filtration for on-chip particle concentration and classification utilizing microfluidics. Lab Chip. 2005;5:1233–1239. doi: 10.1039/b509386d. [DOI] [PubMed] [Google Scholar]
- Zhang WJ, et al. Microfluidics separation reveals the stem-cell-like deformability of tumor-initiating cells. Proc Natl Acad Sci U S A. 2012;109:18707–18712. doi: 10.1073/pnas.1209893109. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.