The phytochrome-phytochrome interacting factor signaling node, which relays light signals to target genes, has been largely conserved during evolution and exhibits lineage-specific diversification.
Abstract
Across the plant kingdom, phytochrome (PHY) photoreceptors play an important role during adaptive and developmental responses to light. In Arabidopsis thaliana, light-activated PHYs accumulate in the nucleus, where they regulate downstream signaling components, such as phytochrome interacting factors (PIFs). PIFs are transcription factors that act as repressors of photomorphogenesis; their inhibition by PHYs leads to substantial changes in gene expression. The nuclear function of PHYs, however, has so far been investigated in only a few non-seed plants. Here, we identified putative target genes of PHY signaling in the moss Physcomitrella patens and found light-regulated genes that are putative orthologs of PIF-controlled genes in Arabidopsis. Phylogenetic analyses revealed that an ancestral PIF-like gene was already present in streptophyte algae, i.e., before the water-to-land transition of plants. The PIF homologs in the genome of P. patens resemble Arabidopsis PIFs in their protein domain structure, molecular properties, and physiological effects, albeit with notable differences in the motif-dependent PHY interaction. Our results suggest that P. patens PIFs are involved in PHY signaling. The PHY-PIF signaling node that relays light signals to target genes has been largely conserved during land plant evolution, with evidence of lineage-specific diversification.
INTRODUCTION
As photoautotrophic organisms, plants depend on light as an energy source and therefore have to adapt their growth and development to changing light conditions. To detect different aspects of their light environment, e.g., spectral composition or light intensity, plants are equipped with different types of photoreceptors. This is true for angiosperms, such as Arabidopsis thaliana, as well as for earlier diverging plant lineages such as ferns and mosses that do not reproduce by seeds but by spores (referred to hereafter as non-seed plants). Among these photoreceptors, the members of the phytochrome (PHY) family function as red (R) and far-red (FR) light receptors (Mathews, 2006; Li et al., 2011). Phytochromes exist in two different states that reversibly convert into each other by the absorption of light: the inactive Pr form, which has an absorption peak in R light (666 nm), and the active Pfr form, with maximal absorption in FR light (730 nm). The external R/FR light conditions are thus translated into an equilibrium of a wavelength-specific phytochrome Pfr/Ptot ratio (Ptot = Pfr + Pr) (Mancinelli, 1994).
The Arabidopsis genome encodes five phytochromes, which can be grouped into type I and type II phytochromes, represented by PHYTOCHROME A (PHYA) and PHYB-E, respectively. PHYB is the most important phytochrome under light conditions that result in a high Pfr/Ptot ratio and regulates seed germination, seedling deetiolation, induction of flowering, and responses to canopy shade or competition by neighboring plants. PHYA, the only type I phytochrome in eudicots, is most abundant in dark-grown seedlings and mediates germination and deetiolation under light conditions that induce a low Pfr/Ptot ratio (Kami et al., 2010; Li et al., 2011). Non-seed plant phytochrome paralogs have evolved independently of angiosperm phytochromes and cannot be assigned to either type I or type II (Mathews, 2010). They have been described as photoreceptors of phototropic and polarotropic growth, but also regulate response modes that are similar to Arabidopsis PHYA- or PHYB-dependent responses, such as R/FR light-reversible spore or gemma germination, or FR light-induced protonemata growth (Mathews, 2006; Hughes, 2013; Possart et al., 2014; Inoue et al., 2016).
As a first step in phytochrome signaling, activated phytochromes translocate from the cytosol into the nucleus. In Arabidopsis, PHYB possibly enters the nucleus bound to transcription factors (TFs) or using its own nuclear localization signal (NLS), whereas PHYA is transported into the nucleus by the paralogs FAR-RED ELONGATED HYPOCOTYL1 (FHY1) and FHY1-LIKE (FHL) (Chen et al., 2005; Kami et al., 2010; Li et al., 2011; Pfeiffer et al., 2012). Phytochromes of the liverwort Marchantia polymorpha, the moss Physcomitrella patens, and the fern Adiantum capillus-veneris also accumulate in the nucleus in a light-dependent manner; at least Pp-PHY1 nuclear transport depends on Pp-FHY1 (Tsuboi et al., 2012; Possart and Hiltbrunner, 2013; Inoue et al., 2016). Recently published work moreover indicated a light-mediated nuclear accumulation of PHYs from the green alga Micromonas pusilla (Duanmu et al., 2014).
Phytochrome downstream signaling in the nucleus has been intensively studied in angiosperms, but only little data are available on the mechanisms of phytochrome signaling in the nucleus in non-seed plants. In Arabidopsis, one branch of phytochrome signal transduction involves the phytochrome-mediated inhibition of the E3 ubiquitin ligase CONSTITUTIVE PHOTOMORPHOGENIC1 (COP1). COP1, in conjunction with SUPPRESSOR OF PHYA-105 1 (SPA1) or SPA1-related proteins, targets light-signaling TFs for proteasome-mediated degradation in the dark, a process that is inhibited by phytochromes in light (Kami et al., 2010; Li et al., 2011). In a concomitant signaling pathway, light-activated nuclear phytochromes bind and regulate members of the subfamily 15 of Arabidopsis basic helix-loop-helix (bHLH) TFs, designated as phytochrome interacting factors (PIFs) (Toledo-Ortiz et al., 2003; Leivar and Quail, 2011; Jeong and Choi, 2013). All Arabidopsis PIFs contain a highly conserved active PHYB binding (APB) motif, which is necessary and sufficient for specific interaction with PHYB (Khanna et al., 2004; Leivar and Monte, 2014). This interaction can be suppressed by point mutations in the APB motif, as demonstrated for At-PIF1, At-PIF3, At-PIF4, and At-PIF5 (Khanna et al., 2004; Shen et al., 2008). Two members of the At-PIF family, At-PIF1 and At-PIF3, moreover interact with PHYA, independently of the APB motif (Al-Sady et al., 2006; Shen et al., 2008). At-PIF3 contains an active PHYA binding (APA) motif, which is necessary for binding to PHYA and can be functionally impaired by the introduction of point mutations (Al-Sady et al., 2006; Shen et al., 2008). At-PIF1 also contains an APA motif, which, however, is different from the At-PIF3 APA motif (Shen et al., 2008; Krzymuski et al., 2014). The interaction of PIFs with light-activated nuclear phytochromes initiates the rapid phosphorylation of PIFs, which except for At-PIF7 results in their rapid degradation via the ubiquitin-proteasome system (Al-Sady et al., 2006; Kami et al., 2010; Leivar and Monte, 2014; Xu et al., 2015). PIF degradation is associated with a rapid colocalization of PIFs and phytochromes and the formation of nuclear bodies (NBs) (Bauer et al., 2004; Chen, 2008). In the dark, At-PIF1 inhibits seed germination, and several PIFs together promote skotomorphogenesis and inhibit photomorphogenesis of etiolated seedlings (Leivar and Quail, 2011; Leivar and Monte, 2014). Moreover, At-PIF4, At-PIF5, and At-PIF7 have been reported to promote the shade avoidance syndrome in deetiolated seedlings, and At-PIF3 and At-PIF4 regulate flowering time (Leivar and Quail, 2011; Casal, 2013; Leivar and Monte, 2014). In line with this, Arabidopsis higher order pif mutants exhibit constitutive photomorphogenic and light-hypersensitive seedling phenotypes as well as reduced shade avoidance syndrome; light-grown PIF overexpressors, on the other hand, show constitutively long hypocotyls and petioles, pale-green leaves, and early flowering (Fujimori et al., 2004; Khanna et al., 2007; Lorrain et al., 2008; Leivar et al., 2008a, 2008b; Shin et al., 2009; Leivar and Quail, 2011; Kumar et al., 2012). PIFs have been shown to possess TF activity (Leivar and Quail, 2011; Leivar and Monte, 2014). Genome-wide expression profiling of individual pif mutants or of the pif1 pif3 pif4 pif5 quadruple (pifq) mutant revealed an important role of PIFs during light-dependent regulation of gene expression and identified potential direct PIF target genes (Leivar et al., 2009; Shin et al., 2009; Hornitschek et al., 2012; Zhang et al., 2013). Light-activated phytochromes reverse PIF activities by inducing the rapid degradation of PIF proteins as well as by inhibiting their binding to target promoters, altogether changing the expression of PIF-regulated genes (Park et al., 2012; Jeong and Choi, 2013; Leivar and Monte, 2014; Xu et al., 2015).
Arabidopsis phytochromes regulate the expression of numerous genes related to phytohormone signaling or photosynthetic and metabolic changes that occur during photomorphogenesis (Leivar and Quail, 2011; Leivar and Monte, 2014). There have been few reports on a similar function of phytochromes in non-seed plants. Phytochromes from fern, moss, liverwort, and green algae have been shown to regulate the transcript levels of individual genes (Winands and Wagner, 1996; Christensen et al., 1998; Suzuki et al., 2001; Possart and Hiltbrunner, 2013; Inoue et al., 2016). Moreover, a recent approach identified R light-regulated genes in the moss P. patens that were misregulated in mutants deficient in phytochrome chromophore biosynthesis (Chen et al., 2012).
Homologs of all classical photoreceptors of angiosperms (PHYs, cryptochromes, phototropins, and UVR8) are present in non-seed plants, with the exception of ZEUTLUPE (ZTL) family proteins (Imaizumi et al., 2002; Suetsugu and Wada, 2003; Mathews, 2006; Holm et al., 2010; Wolf et al., 2010), but few homologs of signaling components have been described: The genome of the moss P. patens contains homologous sequences of COP1 and SPA1, and the characterization of the corresponding proteins has suggested partial functional conservation, indicating that an ancestral form of the PHY-COP1 pathway was already present in early land plants (Richardt et al., 2007; Rensing et al., 2008; Yamawaki et al., 2011; Ranjan et al., 2014); in addition, homologs of the TFs Arabidopsis HY5 and CONSTANS (CO) have been implicated in light and growth responses in P. patens (Yamawaki et al., 2011; Zobell et al., 2005). Although potential PIF homologs are encoded in the genome of the moss P. patens, little is known about the functions of PIFs in mosses (Carretero-Paulet et al., 2010; Richardt et al., 2010; Rösler et al., 2010; Feller et al., 2011; Jeong and Choi, 2013; Wu et al., 2014). Besides a recent publication that characterized the function of the solitary PIF in the liverwort M. polymorpha (Inoue et al., 2016), we know little about phytochrome signaling pathways in non-seed plants.
Here, we present evidence for the evolutionary conservation of a PIF-dependent phytochrome signaling pathway among land plants. Microarray analysis in the moss P. patens revealed global effects of R light on gene expression. The comparison with expression data from Arabidopsis showed a significant overlap with homologs of PIF-dependent genes. We identified and characterized potential functional PIF orthologs in P. patens and revealed molecular properties typical for this class of transcription factors as well as potential differences to angiosperm PIFs. Our data strongly suggest an important role of PIFs during light signaling in P. patens and the evolutionary conservation of PIF-dependent PHY signaling pathways.
RESULTS
The R Light Response in P. patens Is Characterized by Pausing Biosynthesis and Subsequent Derepression of Biosynthesis and Photosynthesis
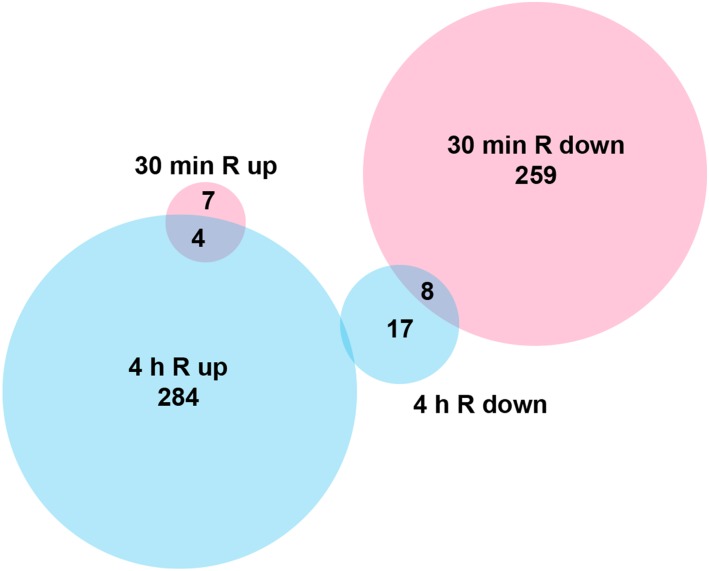
Phytochrome-mediated light signaling involves dramatic changes in gene expression in angiosperms, best studied in Arabidopsis. The dark-to-light transition also affects seedling morphogenesis in gymnosperms, suggesting that light regulates gene expression in all seed plants (Christensen et al., 2002; Mathews and Tremonte, 2012). To compare phytochrome signaling in Arabidopsis to that in a non-seed plant, we analyzed the effects of R light on the genome-wide gene expression in the moss P. patens. We performed microarray analysis on 6-week-old, 2-week dark-adapted P. patens plants that were either harvested directly in darkness or after subjection to R light treatment for 30 min and 4 h, respectively (Figure 1). Hierarchical clustering and principal component analyses of the array data (Supplemental Figures 1A and 1B) demonstrate that, as expected, the dark controls cluster, as well as the triplicates of the 30 min and 4 h R treatment. Compared with the dark control, the expression of 278 genes changed significantly after 30 min R light treatment, the majority being downregulated (96%). In contrast, upon 4 h in R light, 92% of the 313 differentially expressed genes (DEGs) were upregulated (Figure 1; Supplemental Data Set 1). Consequently, the majority of detected genes were differentially regulated between the 30 min and 4 h R light time points. Five DEGs were validated by quantitative PCR; they generally displayed good congruence with the microarray data (Supplemental Figure 1C).
Figure 1.
Early and Late P. patens R Light-Responsive Genes.
Venn diagram of DEGs and their directionality upon R light treatment. Microarray analysis was performed on dark-adapted P. patens gametophores subjected to R light treatment for 30 min (red) and 4 h (blue), respectively. Controls were harvested directly from plants grown in darkness.
By k-means clustering, we identified three major expression profiles: 295 genes were repressed in darkness as well as after 30 min R and became activated only after 4 h R (cluster 1, late activation); 65 genes were active during darkness and were repressed upon 30 min and 4 h R (cluster 2, dark active); 157 genes were active during darkness, repressed at 30 min R, and derepressed at 4 h R (cluster 3, temporarily repressed in R) (Supplemental Data Set 1).
The DEGs were grouped into functional categories according to Gene Ontology (GO) terms (Supplemental Figures 2 and 3). Cluster 2 (dark active) was enriched in genes related to amino acid metabolism and mitochondria, reflecting the provision of energy without light. Genes in cluster 3 (temporarily repressed) were mainly related to translation and other biosynthetic activity, which may be attributed to the transition from a dark-adapted state to growth in light. Cluster 1 genes (late active) could mainly be assigned to photosynthesis, namely, light harvesting and light reaction, carbon fixation, and plastid terms; at this point in time, the plants were reacting to available light on the transcriptional level, expressing genes involved in plastid-enabled carbon fixation (Supplemental Figure 3B). In line with a potential direct effect of R light signaling on transcription, we identified 30 TFs and transcriptional regulators (Supplemental Data Set 1, sheet “Summary DEGs,” column G). Among these, we assigned 11 to cluster 2 (dark active), 10 to cluster 3 (temporarily repressed), and 5 to cluster 1 (late active). Among the dark active TFs were two NACs, a family known to be involved in senescence and stress signaling. Among the late-activated genes, we identified a sigma-like factor, most probably instrumental in transcriptionally activating the plastid light response.
Putative PIFs Are Encoded in the Genome of P. patens
In Arabidopsis, phytochrome-mediated changes in gene expression depend on TFs such as PIFs. Previous studies comparing dark-grown Arabidopsis higher-order pif mutants with wild-type seedlings grown in R light identified genes that are regulated by PIFs during photomorphogenic development (Monte et al., 2004; Leivar et al., 2009; Shin et al., 2009). By comparing our microarray analysis with data from Arabidopsis, we identified potential P. patens homologs of genes that are regulated by PIFs in Arabidopsis. Among those R light-induced DEGs from P. patens for which we could determine potential Arabidopsis best-reciprocal-hit homologs (considered as potential orthologs, 98 in total), 46% (45) overlapped with DEGs described as R light-induced by Leivar et al. (2009) (Supplemental Data Set 2). The overlap was highly significant versus the background of all expressed genes (P < <0.001, Fisher’s exact test). Thirty-three DEGs were homologous to Arabidopsis genes that were regulated in a PIF-dependent manner (P < <0.001, Fisher’s exact test; Supplemental Data Set 2) (class 4 and 7 genes as defined by Leivar et al., 2009). Moreover, nine DEGs were homologous to Arabidopsis genes that had been described as direct PIF target genes (P < <0.001, Fisher’s exact test; Supplemental Data Set 2) (class 7 genes as defined by Leivar et al., 2009). We found very little overlap for R light-repressed genes. Altogether, the presence of putative PIF-dependent genes among R light-affected DEGs in P. patens pointed to a role of bHLH TFs during P. patens phytochrome signaling.
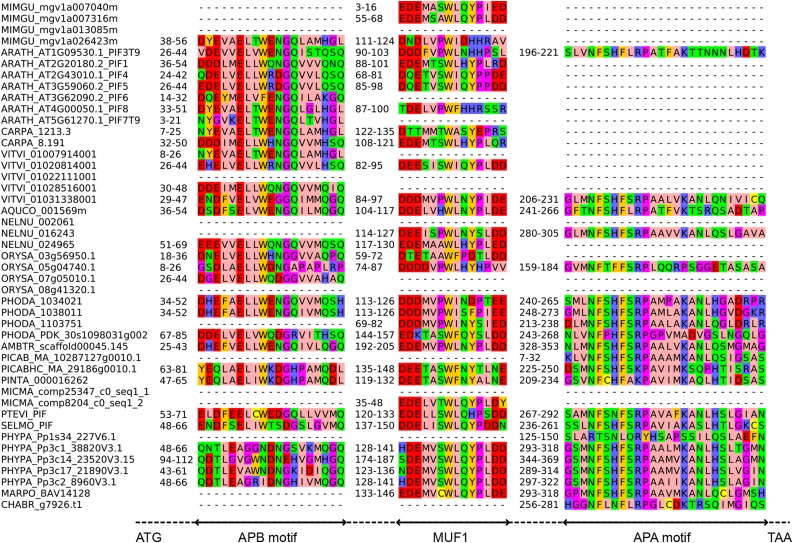
Based on the comparison of At-PIF3 and homologs from other seed plants, we defined the APA and APB consensus motifs and used these to search the P. patens genome and to analyze bHLH proteins that clustered together with At-PIF3 in a previously published phylogeny (Richardt et al., 2010). This revealed four potential PIF homologs, which we designated Pp-PIF1, Pp-PIF2, Pp-PIF3, and Pp-PIF4 (see Methods for more details). We compared the protein sequence of Pp-PIFs to each other and to bHLH proteins from selected algae, liverwort, lycophyte, fern, gymnosperm, and angiosperm (mono- and dicotyledonous) species (Supplemental Data Set 3). By de novo detection using MEME (Bailey et al., 2009), we derived APA and APB motifs. Using those, we tested all proteins for presence/absence of the APA and APB motifs. The APA motif defined by MEME was found to be conserved across the plant kingdom; it could be detected in At-PIF3 (but not At-PIF1) and, among others, the putative Pp-PIFs. In agreement with Inoue et al. (2016), we also found an APA in the Mp-PIF. Altogether, this indicated an early evolution of this protein motif. The APB motif could be unambiguously identified for a large fraction of the tested proteins, including At-PIF1, and At-PIF3 to 8. We also found a potential APB motif in all four putative Pp-PIFs, which, however, show weaker sequence similarity to APB motifs from Arabidopsis and other angiosperms than those show among each other (Figure 2). Accordingly, Inoue et al. (2016) reported that they did not find a canonical APB domain in P. patens PIF protein sequences. Furthermore, MEME analyses revealed three motifs of unknown function (MUF) (Figure 2; Supplemental Figures 4 and 5). One of these, MUF1, was recently described as playing a role in transcriptional activation in Arabidopsis PIFs, which is in line with its evolutionary conservation (Dalton et al., 2016). Proteins containing at least one motif (APA, APB, or MUF1, 2, or 3) could be detected in all land plants for which a sequenced genome was probed. Interestingly, we found an APA motif and MUF2 in one algal species, Chara braunii (CHABR) (Figure 2; Supplemental Figure 4). We also detected the MUFs in bHLH proteins across almost all analyzed land plant species. Notably, all four Pp-PIFs contained an APA, all three MUFs, and a putative APB motif, suggesting that these genes encode canonical PIF proteins.
Figure 2.
PIF Protein Motifs Are Highly Conserved across the Plant Kingdom.
Sequence alignment of APB, APA, and MUF1 detected in PIF bHLH proteins. The dashed line indicates intercalated regions. Species abbreviations are in a five letter code, where the first three letters represent the genus and the last two the species (e.g., ORYza SAtiva). For each motif, the respective positions in the protein sequence are indicated. See Supplemental Figure 4 for sequence alignment of MUF2 and 3. See Supplemental Data Set 3 for list of species and Supplemental File 1 for full-length alignment.
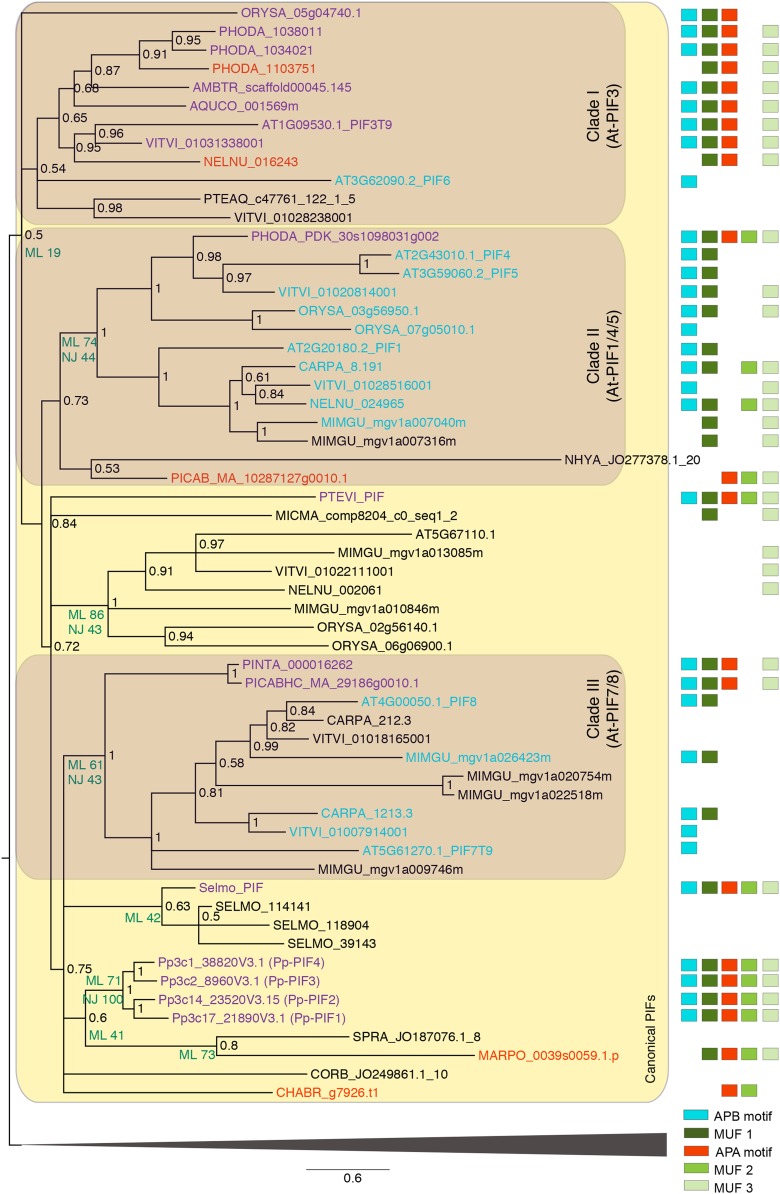
To further investigate the relationship of the protein sequences, we performed a phylogenetic analysis of plant bHLH transcription factors (Figure 3; Supplemental Figure 6, Supplemental Data Set 3, and Supplemental File 1). The clade harboring all sequences with APA/B motifs was not highly supported (Figure 3); however, it was consistently recovered regardless of alignment algorithm and phylogenetic inference method. Thus, there seems to be an overall structure of canonical PIF proteins, which is shared by the members of this clade and is probably derived from an ancestral PIF-like protein. In accordance with the presence of an APA motif, the sequence of the alga C. braunii was contained in the clade that harbored all known canonical PIF sequences (Figure 3) and might thus represent a PIF or PIF-like protein. Three more charophyte sequences derived from transcriptomes were also based in this clade, although they did not contain APA, APB, or MUF motifs (which, however, could be due to incomplete transcript representation). PIF proteins might thus have evolved before the land plant divergence.
Figure 3.
Excerpt of Phylogenetic Tree of Plant bHLH TFs: Clade Containing Canonical PIFs.
The phylogeny is based on Bayesian inference; support values (BI posterior probabilities) are shown at the nodes of the tree. Additional bootstrap support values from ML and NJ analyses are shown to the lower left of some nodes discussed in the text. Species abbreviations for all organisms except P. patens (Pp) and Arabidopsis (At) are in a five letter code, where the first three letters represent the genus and the last two the species (e.g., ORYza SAtiva) (see Supplemental Data Set 3 for list of species and Supplemental File 1 for full-length alignment). The sequence names contain the accession number, except for SELMO_PIF and PTEVI_PIF, for which the sequences have been assembled as described in Methods. Shaded areas highlight canonical PIFs (light shaded) and three major seed plant subclades (I–III; dark shaded) as described in the text. The presence of five motifs, APA, APB, and three motifs of unknown function (MUF), as inferred by MEME de novo motif detection, are depicted as boxes, shown on the right. Sequences containing the APB and APA motif are shown in purple. Sequences containing only the APB motif are shown in blue. Sequences containing only the APA motif are shown in red. The clade not containing any canonical PIFs (indicated by the triangular shape at the bottom) has been collapsed to enhance readability; for expansion of this clade, refer to Supplemental Figure 6.
Three major seed plant subclades were discernible in the phylogeny (Figure 3). Clade I contained At-PIF3; most sequences represented in this clade were APB/APA-containing proteins. With the exception of Phoenix dactylifera (date palm), all species were represented by a single protein; Mimulus guttatus (spotted monkey flower) and Carica papaya (papaya) were the only analyzed angiosperms not represented in this clade, which might be due to incomplete gene models.
Clade II contained At-PIF1/4/5 as well as other analyzed angiosperm sequences that harbored the APB (but not the APA) motif. The basal eudicot Aquilegia coerulea (Colorado blue columbine) and the basal angiosperm Amborella trichopoda were not represented in this clade, and the P. dactylifera sequence contained an APA motif. There was weak support for a conifer and charophyte sequence belonging to this clade; the latter might be a long-branch artifact. Clade III contained At-PIF7/8 and other angiosperm proteins containing the APB (but not the APA) motif, as in clade II. This clade contained no sequences from basal angiosperms, Liliopsida (monocots), or basal eudicots. However, it is sister to two conifer sequences containing the APB and APA motifs.
The relationship of non-seed plants (ferns, lycophyte, liverwort, moss, and charophyte algae), and to some extent conifers, with regard to those three clades could not be accurately inferred. In summary, most seed plant species harbored a canonical PIF3-like (APA/APB) and a canonical PIF1/4/5-like (APB-only) protein; the eudicots and conifers encoded additional APB-containing proteins. The non-seed plants analyzed encoded one to four potential PIFs, typically harboring the APA motif or the APA and APB motifs; there was no evidence for APB-only proteins found in these organisms. While our data and the recent report by Inoue et al. (2016) reveal that the M. polymorpha PIF contains only an APA motif, we found APA and putative APB motifs in P. patens PIFs.
Because they form a single clade, the Pp-PIF paralogs were acquired independently of seed plant PIF paralogs (Figure 3); most probably they were retained after whole-genome duplications (Rensing et al., 2007). We cloned the Pp-PIF1, 2, and 3 coding sequences by RT-PCR; Pp-PIF4 was cloned according to the gene model Pp1s147_126V6.1 (see Supplemental Methods for details). We amplified an additional splicing variant each for Pp-PIF2 (Pp-PIF2.2) and Pp-PIF4 (Pp-PIF4.2). Pp-PIF2.2 and Pp-PIF4.2 lacked the coding sequence of amino acids 51 to 90 and 1 to 108, respectively. Thus, both splicing variants do not contain the putative APB motif. In the following, these splicing variants are designated as Pp-PIF2ΔAPB and Pp-PIF4ΔAPB. The isoforms containing the putative APB motif are designated as Pp-PIF2 and Pp-PIF4, respectively. In contrast to PIFs from Arabidopsis, which consist of ∼450 amino acids, Pp-PIF genes encode for longer proteins of 702 (Pp-PIF1), 728 (Pp-PIF2), 688 (Pp-PIF2ΔAPB), 729 (Pp-PIF3), 772 (Pp-PIF4), and 664 (Pp-PIF4ΔAPB) amino acids, respectively.
Pp-PIF-Phytochrome Interaction Requires the APA but Not the APB Motif
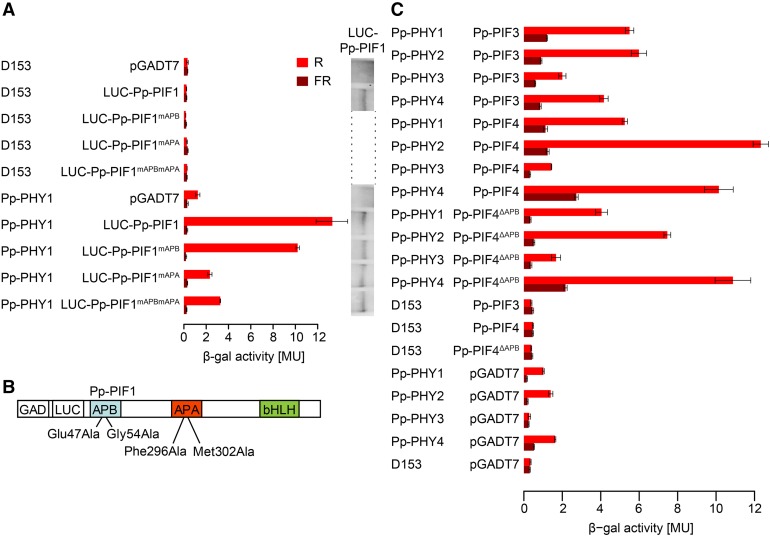
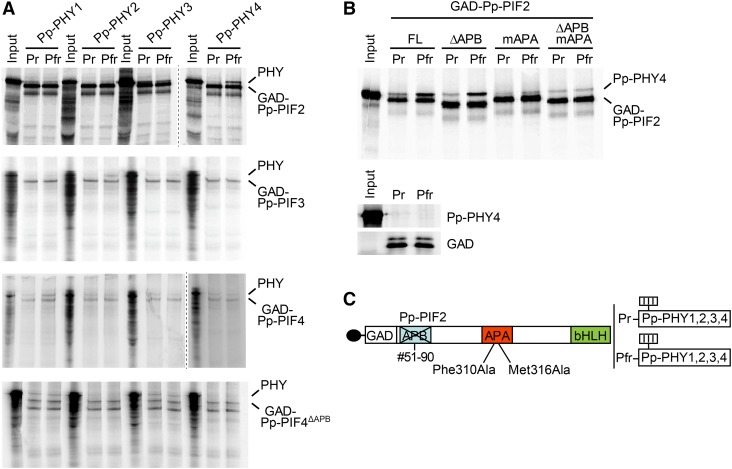
One important step in angiosperm phytochrome signaling is the interaction of light-activated phytochromes with PIFs. In a yeast-two-hybrid assay, we found that Pp-PIF1 and a truncated version that lacks the bHLH domain (Pp-PIF1ΔbHLH) interact with different phytochromes in a light-dependent manner. R light-activated PHY1 to 4 from P. patens and PHYA from Arabidopsis bound Pp-PIF1 and Pp-PIF1ΔbHLH. These interactions were strongly reduced when the phytochromes had been inactivated by FR light (Figure 4; Supplemental Figures 7 to 11). Also, Pp-PIF3, Pp-PIF4, and Pp-PIF4ΔAPB interacted with Pp-PHY1 to 4 in a light-dependent manner (Figure 4). Yeast transformed with Pp-PIF2 did not grow; thus, the interaction with phytochromes could not be investigated.
Figure 4.
Interaction of LUC-Pp-PIF1 with Light-Activated Pp-PHY1 in Yeast Requires the APA Motif.
(A) Full-length Pp-PIF1 fused to LUC interacts with P. patens PHY1 in a light-dependent manner. This interaction is abolished by mutations in the Pp-PIF1 APA motif. GAD plasmids (pGADT7) containing the coding sequence for Pp-PIF1 and mutated versions of Pp-PIF1 (Pp-PIF1mAPB, Pp-PIF1mAPA, and Pp-PIF1mAPBmAPA), respectively, fused to the GAL4 activation domain (GAD) and the coding sequence of luciferase (LUC) were used in yeast two-hybrid assays with GBD plasmids (D153) containing the coding sequence for Pp-PHY1 fused to the GAL4 DNA binding domain (GBD). Phytochromes were converted into the Pfr or Pr form by irradiating yeast cultures for 5 min with R (12 µmol m−2 s−1) or FR (12 µmol m−2 s−1) light. The β-galactosidase activity was measured after an additional incubation in the dark for 4 h. MU, Miller units. Bars indicate the mean of three biological replicates (i.e., three independent cultures were grown; each culture was measured in triplicate); error bars represent 95% confidence interval. The protein abundance of the wild type and mutated Pp-PIF1 in yeast was analyzed by immunoblot using an antibody specific to LUC. For complete immunoblot analyses of LUC-Pp-PIF1 and Pp-PHY protein abundance, refer to Supplemental Figure 8.
(B) Mutations inserted in Pp-PIF1 APB and APA motifs are shown schematically.
(C) Pp-PIF3, Pp-PIF4, and Pp-PIF4ΔAPB interact with Pp-PHY1, 2, 3, and 4 in a light-dependent manner. GAD-Pp-PIF3, 4, and 4ΔAPB versions and Pp-PHYs fused to GBD were used in yeast two-hybrid assays as described in (A).
To test the role of the APA motif in the Pp-PIF1-phytochrome interaction, we mutated Pp-PIF1 at positions Phe-296 and Met-302, corresponding to functionally important amino acids in the Arabidopsis PIF3-APA motif (Al-Sady et al., 2006), to alanine (Pp-PIF1mAPA). Although the putative APB motifs of Pp-PIFs are only weakly similar to the angiosperm consensus APB sequence, following the same rationale as for the APA motif, we mutated Glu-47 and Gly-54 (Al-Sady et al., 2006) to alanine (Pp-PIF1mAPB) to assess the motif’s potential role in PIF-PHY interaction. In addition, we generated a version containing all four mutations, designated Pp-PIF1mAPBmAPA (Figure 4; Supplemental Figure 7). Compared with wild-type Pp-PIF1, the interaction of Pp-PIF1mAPB with Pp-PHYs was only moderately affected. In contrast, mutation of the APA motif caused a significant drop in interaction of Pp-PIF1mAPA and Pp-PIF1mAPBmAPA with all Pp-PHYs (Figure 4; Supplemental Figures 7 to 9). A similar interaction pattern was observed for Pp-PIF1ΔbHLH, which lacks the bHLH domain, and its corresponding mutants (Supplemental Figure 10). Using fusion proteins of Pp-PIF1 and luciferase (Pp-PIF1-LUC), we confirmed the presence of similar protein levels for full-length Pp-PIF1 and its mutants by immunoblot analyses (Figure 4; Supplemental Figures 7 and 8).
We were able to extend our observations on Pp-PIF-phytochrome interactions by performing in vitro coimmunoprecipitation (co-IP) assays as previously described (Huq et al., 2004) (Figure 5). Pp-PIF2 strongly interacted with the Pfr form of Pp-PHY4 and weakly with the Pfr form of Pp-PHY2 (Figure 5A). Pp-PIF2 did not show a light-induced binding to Pp-PHY1 and Pp-PHY3. Despite their comparatively low expression levels, we also obtained a light-induced interaction for Pp-PIF3 with Pp-PHY2 and for Pp-PIF4 with Pp-PHY1 (Figure 5A). For Pp-PIF4ΔAPB, we obtained an interaction with Pp-PHY1 and Pp-PHY3, which was, however, independent of the light condition and therefore might be unspecific (Figure 5A). We further analyzed the role of the APA and the putative APB motif in the Pp-PIF2-Pp-PHY4 interaction. Pp-PIF2mAPA was generated according to studies on At-PIF3 by replacing Phe310 and Met316 with alanine (Al-Sady et al., 2006). To analyze the APB function, we used the Pp-PIF2 splicing variant (Pp-PIF2ΔAPB) described above. Using these mutants in in vitro co-IP assays, we found that the APA motif was essential for the light-dependent interaction between Pp-PIF2 and Pp-PHY4, as the mutation of APA completely abolished this interaction (Figure 5B). The deletion of the APB motif displayed minor, if any, effects on interaction between Pp-PIF2 and Pp-PHY4. Therefore, we conclude that Pp-PIF2 binds to the Pfr form of Pp-PHY4 through the APA motif, as observed for the interaction of Pp-PIF1 and all four Pp-PHYs in Yeast Two-Hybrid assays, altogether indicating a functional conservation of the APA motif in PIF-phytochrome interactions. We also attempted to test Pp-PIF1 for interaction with Pp-PHYs in in vitro co-IP assays, but could not show a light-regulated interaction of this protein. This may be due to a relatively low affinity of the protein that can be distinguished in yeast but is not detectable in in vitro co-IP assays.
Figure 5.
The APA Motif Mediates Interaction between Pp-PIF2 and Phytochromes from P. patens in Vitro.
(A) In vitro co-IP assays for Pp-PIFs and Pp-PHYs. Pp-PIFs and Pp-PHYs were transcribed and translated in vitro in the presence of 35S-methionine. Pp-PIFs fused to the GAD were used as bait. Pp-PHY1, 2, 3, or 4 were converted into the Pfr or Pr form by irradiating the expression mix with either R (17 µmol m−2 s−1) or FR (4.3 µmol m−2 s−1) light for 1 min and used as prey. Immunoprecipitated proteins were separated on a SDS-PAGE gel and detected by Typhoon phosphor imaging system.
(B) Pp-PIF2 interacts with light-activated Pp-PHY4 in vitro in an APA-dependent manner. GAD fusions of wild-type Pp-PIF2 and its mutated versions impaired in the APB (Pp-PIF2ΔAPB) or the APA (Pp-PIF2mAPA) motif, or both (Pp-PIF2ΔAPBmAPA), were used as bait in in vitro co-IP assays as described in (A). Pp-PHY4 was converted into the Pfr or Pr form by irradiation with either R (17 µmol m−2 s−1) or FR (4.3 µmol m−2 s−1) light for 1 min and used as prey. Small gel: empty GAD control.
(C) Schematic representation of mutated Pp-PIF2. For analysis of APB function, a Pp-PIF2 splicing variant lacking amino acids 51 to 90 (Pp-PIF2ΔAPB) was used. For analysis of APA function, amino acids corresponding to functionally relevant amino acids in At-PIF3 were mutated (Pp-PIF2mAPA).
In summary, based on their light-specific interaction with the photoreceptors, Pp-PIFs can be considered as potential factors of phytochrome downstream signaling in P. patens.
P. patens PIFs Localize to the Nucleus
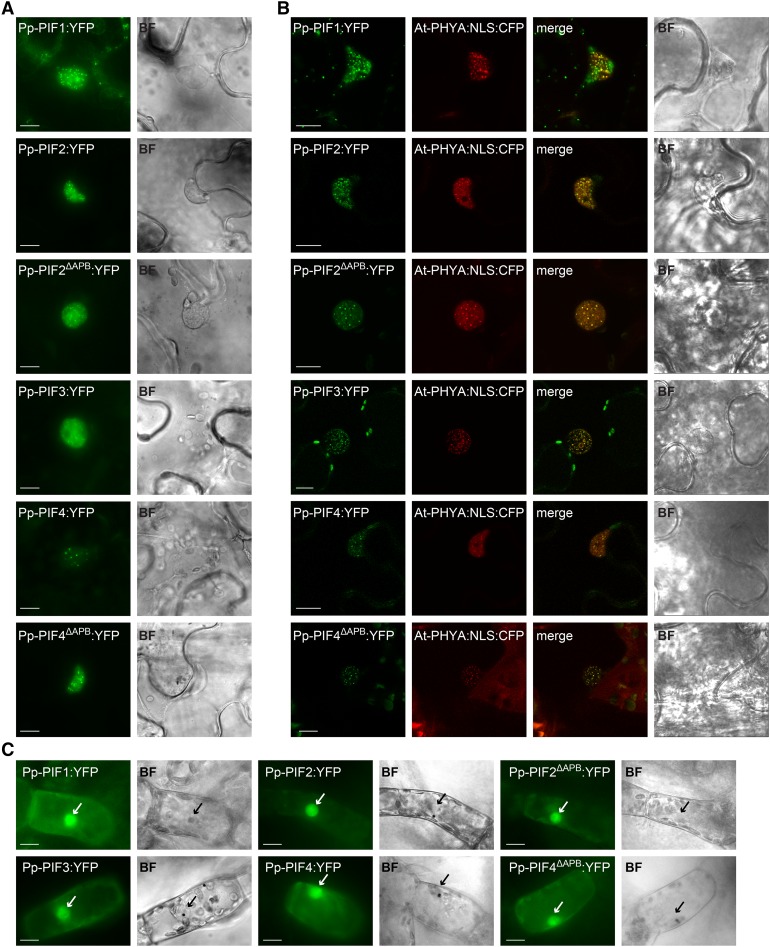
In Arabidopsis, PIFs localize to the nucleus, where they regulate gene expression. Their interaction with activated phytochromes is accompanied by the formation of nuclear bodies, a process that has been implicated in PIF degradation and signal transduction (Van Buskirk et al., 2012). In order to analyze the localization pattern of P. patens PIFs, we expressed YFP fusions of Pp-PIF1, Pp-PIF2, Pp-PIF2ΔAPB, Pp-PIF3, Pp-PIF4, and Pp-PIF4ΔAPB in Nicotiana benthamiana leaf cells. All six Pp-PIFs localized to the nucleus and formed nuclear bodies, resembling the typical localization of Arabidopsis PIFs (Figure 6A). In line with our observations on Pp-PIF-phytochrome interaction in yeast and in in vitro co-IP assays, we found that Pp-PIFs colocalized with Arabidopsis PHYA, which we had fused to an NLS in order to obtain visible amounts in the nucleus. All Pp-PIFs colocalized with At-PHYA-NLS in nuclear bodies (Figure 6B). The expression of full-length proteins was confirmed by immunoblot analysis (Supplemental Figure 12). Using particle bombardment, we transiently transformed P. patens protonema cells and observed a nuclear accumulation of Pp-PIF1, Pp-PIF2, Pp-PIF2ΔAPB, Pp-PIF3, Pp-PIF4, and Pp-PIF4ΔAPB similar to PIFs in Arabidopsis (Figure 6C). We conclude that PIFs from P. patens may act as nuclear phytochrome signaling components and, thus, similarly to Arabidopsis PIFs.
Figure 6.
PIFs from P. patens Localize to the Nucleus.
(A) Pp-PIF1, Pp-PIF2, Pp-PIF2ΔAPB, Pp-PIF3, Pp-PIF4, and Pp-PIF4ΔAPB localize in the nucleus. Leaves of N. benthamiana were transformed by infiltration with Agrobacterium tumefaciens containing a respective Pro35S:Pp-PIF:YFP construct. One day after transformation, the plants were transferred into darkness for another 1 to 3 d before epifluorescence microscopic analysis (n ≥ 20) of epidermal leaf cells. Bar = 10 µm; in all figure parts, BF = bright field.
(B) Pp-PIF1, Pp-PIF2, Pp-PIF2ΔAPB, Pp-PIF3, Pp-PIF4, and Pp-PIF4ΔAPB colocalize with At-PHYA:NLS in the nucleus. Leaves of N. benthamiana were cotransformed with the respective Pro35S:Pp-PIF:YFP and Pro35S:At-PHYA:NLS:CFP. One day after transformation, the plants were transferred into darkness for another 1 to 3 d and analyzed by epifluorescence /confocal microscopy (n ≥ 20; representative confocal images are shown). Bar = 10 µm.
(C) Pp-PIF1, Pp-PIF2, Pp-PIF2ΔAPB, Pp-PIF3, Pp-PIF4, and Pp-PIF4ΔAPB localize to the nucleus in P. patens. Protonema filaments were transiently transformed with the respective Pro35S:Pp-PIF:YFP using particle bombardment and incubated in darkness for 2 to 4 d before epifluorescence microscopy analysis (n ≥ 13). Arrows indicate nuclei. Bar = 10 µm.
P. patens PIFs Affect Phytochrome-Mediated Responses in Arabidopsis
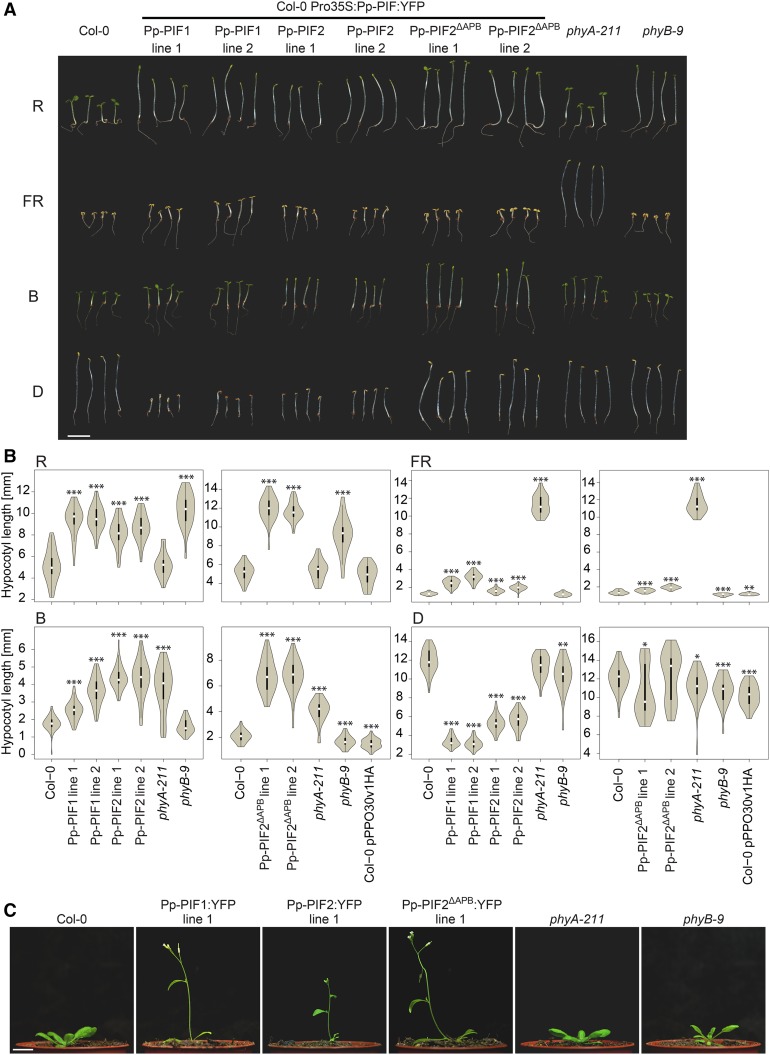
To further investigate potential functional conservation of P. patens and Arabidopsis PIFs, we expressed YFP fusions of Pp-PIF1, Pp-PIF2, and the Pp-PIF2 splicing variant, Pp-PIF2ΔAPB, under the control of the constitutive 35S promoter (Pro35S) in the Arabidopsis Columbia-0 background. In order to examine whether Pp-PIFs influence photomorphogenic responses, we analyzed seedling deetiolation in these overexpressor lines under different light conditions. The expression of all three Pp-PIFs led to a clear hyposensitive response in R light, with seedlings showing longer hypocotyls than the wild-type control (Figures 7A and 7B). We also observed elongated hypocotyl growth under blue light (B) in all three Pp-PIF overexpressor lines (Figures 7A and 7B). Moreover, Pp-PIF-OX seedlings partially failed to open their cotyledons in R and B light (Figure 7A). Pp-PIF-OX seedlings grown in FR light showed a similar, but much weaker, phenotype of elongated hypocotyls (Figures 6A and 6B). Dark-grown Pp-PIF-OX seedlings were etiolated, showing a typical apical hook and closed cotyledons, but Pp-PIF1-OX and Pp-PIF2-OX lines had shorter hypocotyls and partially exaggerated apical hooks in comparison to the wild-type control (Figures 7A and 7B). During our quantification of hypocotyl length phenotypes, we also tested lines transformed with the empty expression vector. These controls showed only minor (but in some replicates statistically significant) differences (Figure 7B). The effects of Pp-PIF overexpression resembled the phenotype of Arabidopsis PIF3- and PIF5-OX, suggesting the functionality of Pp-PIFs in Arabidopsis or an interference of Pp-PIFs with proper functioning of endogenous PIFs (Kim et al., 2003; Khanna et al., 2007). In adult plants, the expression of all three Pp-PIFs resulted in enhanced elongation growth and early flowering (Figure 7C). This phenotype was reminiscent of Arabidopsis PIF4- or PIF5-OX plants, again indicating an effect of Pp-PIFs on the endogenous signaling system (Fujimori et al., 2004; Khanna et al., 2007; Kumar et al., 2012). We detected a nuclear localization for all Pp-PIF YFP-fusions in etiolated seedlings (Supplemental Figure 13). The expression of full-length Pp-PIF proteins was confirmed by immunoblot analysis (Supplemental Figure 14A).
Figure 7.
Effects of PIFs from P. patens on Light Signaling in Arabidopsis.
(A) The response of Arabidopsis seedlings to light is impaired by Pp-PIF overexpression. Col-0 seedlings (control), Col-0 seedlings expressing Pro35S-driven Pp-PIF1:YFP, Pp-PIF2:YFP, or Pp-PIF2ΔAPB:YFP, as well as phyA-211 and phyB-9 seedlings, were grown for 4 d in R (22 µmol m−2 s−1), FR (3 µmol m−2 s−1), or B (8 µmol m−2 s−1) light or in darkness (D). PpPIF2ΔAPB lines 1 and 2 were grown in the same experiment, but on separate plates (for corresponding wild type, phyA-211, and phyB-9, see quantification in [B]). Bar = 5 mm.
(B) Pp-PIF overexpression affects hypocotyl length of Arabidopsis seedlings grown under different light conditions. Col-0, Pro35S-driven Pp-PIF1:YFP, Pp-PIF2:YFP or Pp-PIF2ΔAPB:YFP, phyA-211, and phyB-9 as well as Pro35S-YFP (pPPO30v1HA, empty vector control) expressing seedlings were grown as described in (A) before quantification of hypocotyl length. Data are shown as violin plots; white dots represent the median, and asterisks indicate P values (unpaired, two-tailed Student’s t test) of <0.05 (*), <0.01 (**), and <0.001 (***), respectively. The y axis scale was adjusted for maximal resolution in each plot.
(C) P. patens PIF overexpression results in early flowering in Arabidopsis. Col-0 plants expressing Pro35S-driven Pp-PIF1:YFP, Pp-PIF2:YFP, or Pp-PIF2ΔAPB:YFP, as well as phyA-211 and phyB-9, were grown for 21 d under standard greenhouse conditions. Bar = 1 cm.
Pp-PIFs Complement the Arabidopsis pif1 pif3 pif4 pif5 Quadruple Mutant Phenotype
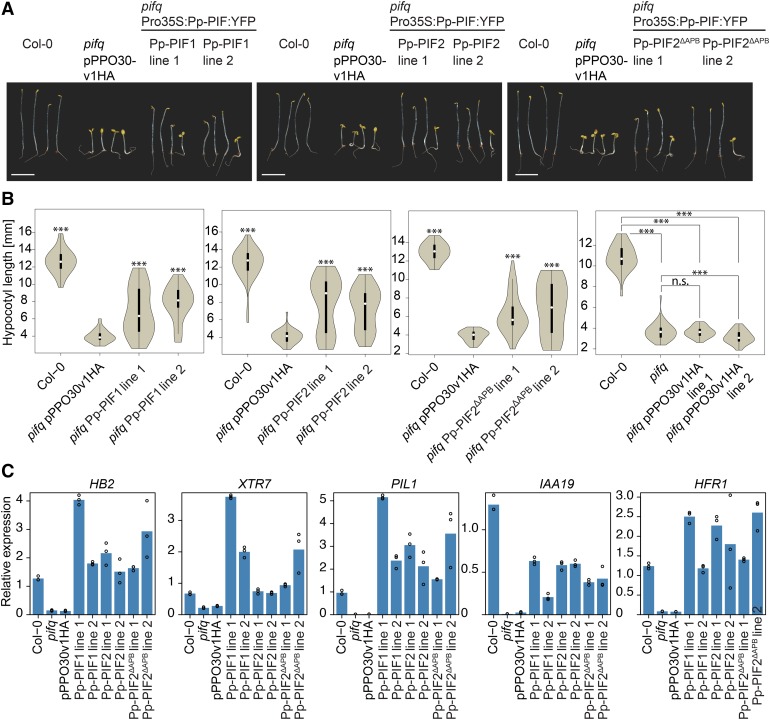
If Pp-PIFs acted as true PIFs, they should be able to substitute for endogenous PIFs in Arabidopsis. We therefore expressed YFP fusions of Pp-PIF1, Pp-PIF2, and Pp-PIF2ΔAPB under the control of the constitutive 35S promoter in the Arabidopsis pif1 pif3 pif4 pif5 quadruple (pifq) mutant. The expression of full-length Pp-PIF proteins was confirmed by immunoblot analysis (Supplemental Figure 14B). In darkness, seedlings of the Arabidopsis pifq mutant show a constitutive photomorphogenic (cop)-like phenotype of shortened hypocotyls and open apical hooks (Leivar et al., 2008a). Transforming the empty vector into the pifq background had only a minor effect on hypocotyl length (Figure 8B, right-most panel). All three PIFs from P. patens partially but significantly complemented this phenotype: Pp-PIF-overexpressing pifq mutants had an elongated hypocotyl and a partially closed apical hook (Figures 8A and 8B).
Figure 8.
PIFs from P. patens Rescue the cop-Like Phenotype of Arabidopsis pifq Mutant Seedlings.
(A) The cop-like phenotype of dark-grown Arabidopsis pifq mutant seedlings is partially complemented by Pp-PIF overexpression. Col-0 and pifq mutant seedlings expressing Pro35S-YFP (pPPO30v1HA, empty vector control) as well as pifq mutant seedlings expressing Pro35S-driven Pp-PIF1:YFP, Pp-PIF2:YFP, or Pp-PIF2ΔAPB:YFP were grown in darkness for 4 d. All transformed lines were analyzed in their segregating generation. Bar = 5 mm.
(B) Pp-PIF overexpression significantly complements the cop-like phenotype of Arabidopsis pifq mutant seedlings. Seedlings as described in (A) were used for quantification of hypocotyl length. Right panel shows comparison of empty vector control with pifq and Col-0 backgrounds. Data are shown as violin plots; white dots represent the median, and asterisks indicate P values (unpaired, two-tailed Student’s t test) of <0.05 (*), <0.01 (**), <0.001 (***), and >0.05 (n.s., not significant), respectively. The y axis scale was adjusted for maximal resolution in each plot.
(C) Pp-PIF overexpression restores the expression of PIF-dependent genes in Arabidopsis pifq mutants. The expression of the genes HB2, XTR7, PIL1, IAA19, and HFR1 was analyzed by quantitative PCR in 4 d dark-grown seedlings of Col-0, pifq, and pifq expressing Pro35S-YFP (pPPO30v1HA, empty vector control) or Pro35S-Pp-PIF1:YFP, Pro35S-Pp-PIF2:YFP, and Pro35S-Pp-PIF2ΔAPB:YFP, respectively. Data were normalized to PP2AA3 mRNA. All transformed lines were analyzed in their nonsegregating generation. Technical replicates (repeats within an experiment) are shown as circles; bars represent the mean. The y axis scale was adjusted for maximal resolution in each plot. Gene accession numbers are listed in the Supplemental Methods. For biological replicates, refer to Supplemental Figure 16.
A characteristic of most Arabidopsis PIFs in PIF-PHY signaling is their light-induced degradation (Bauer et al., 2004; Shen et al., 2007, 2008; Lorrain et al., 2008). We investigated the stability of Pp-PIFs upon R treatment in Arabidopsis pifq. Interestingly, Pp-PIFs appeared much more stable compared with At-PIF3 (Supplemental Figure 15), a behavior reminiscent of that of At-PIF7 (Leivar et al., 2008b).
Pp-PIF expression complemented the pifq phenotype not only at the morphological but also at the transcriptional level. By qPCR, we confirmed that the expression levels of five genes known to be downregulated in pifq (Zhang et al., 2013) were reconstituted or overcompensated when expressing Pp-PIF1, Pp-PIF2, or Pp-PIF2ΔAPB in the pifq background (Figure 8C; Supplemental Figure 16). Altogether, these results underline functionality of Pp-PIFs in Arabidopsis and corroborate functional conservation of PIFs in mosses and angiosperms.
DISCUSSION
R Light Broadly Affects Gene Expression in P. patens
Phytochromes from non-seed plants, e.g., from ferns and mosses, regulate light-dependent transient responses, such as phototropism, as well as developmental processes (Mathews, 2006; Hughes, 2013). However, in contrast to angiosperms and Arabidopsis in particular, little is known about the downstream components of phytochrome signaling in non-seed plants. We and others have previously demonstrated the evolutionary conservation of the first steps of phytochrome signaling since the earliest land plants by showing that phytochromes from ferns, mosses, and liverworts accumulate in the nucleus upon activation by light (Tsuboi et al., 2012; Possart and Hiltbrunner, 2013; Inoue et al., 2016). The data presented here suggest that the subsequent steps of phytochrome signaling in the moss P. patens also resemble those in the seed plant Arabidopsis. Genome-wide transcriptional profiling revealed conserved targets of phytochrome signaling in Arabidopsis and P. patens. Phylogenetic analysis and characterization of putative functional PIF orthologs from P. patens demonstrated that these proteins are similar to angiosperm PIFs in their molecular properties and physiological effects. While our manuscript was under review, Inoue and colleagues reported that the solitary PIF and PHY proteins in M. polymorpha interact and that Mp-PIF is involved in the regulation of Mp-PHY-dependent gene expression (Inoue et al., 2016). This is in line with our conclusion that PIF-PHY signaling nodes have been evolutionarily conserved since the earliest land plants.
R light-induced DEGs in P. patens clustered into dark-active, temporarily repressed, and late-active genes. This, together with the functional DEG classification, reveals the molecular processes that are induced by the transition from darkness to R light (Supplemental Figures 2 and 3). In dark-adapted plants, the high proportion of DEGs that were involved in amino acid metabolism indicates the use of nonphotosynthetic energy sources through the metabolization of amino acids, probably by the mitochondria. After 30 min R, this type of energy production is repressed. Photosynthesis is probably induced, which was, however, not yet detectable at the transcriptional level. After 4 h R light, biosynthesis is reactivated and photosynthetic functions are activated at the transcriptional level.
The comparison of our results to transcriptome analyses from Arabidopsis suggests similar effects of R light in P. patens. As described by Leivar et al. (2009), genes involved in cellular metabolism dominate among late (2 d) R light-repressed Arabidopsis genes, which is consistent with cluster 2 DEGs (dark active, i.e., genes repressed by R light) in this study. Moreover, photosynthesis- and chloroplast-related genes are the most abundant among R light-induced genes in Arabidopsis, resembling the function of cluster 1 DEGs (late active) (Leivar et al., 2009; Leivar and Monte, 2014). In contrast to Arabidopsis (Leivar et al., 2009), we found that R light affects the expression of only a few TFs and transcriptional regulators in P. patens. This might be partially attributed to the different proportions of TF genes in the genomes of non-seed plants and seed plants (e.g., 3% in P. patens versus 6% in Arabidopsis) (Lang et al., 2010). It may also indicate differences in the early light-induced gene network of Arabidopsis and P. patens. It is also worth mentioning that among the R-light regulated genes in P. patens, only the R-induced subset shows overlap with PIF-regulated genes during Arabidopsis deetiolation, even though R-repressed genes are much enriched in PIF-target genes in Arabidopsis (Leivar and Monte, 2014; Pfeiffer et al., 2014).
The Genome of P. patens Encodes Conserved PIF Proteins
Although the transcriptional regulation of TFs does not seem to be a prominent part of R light-regulated gene expression in P. patens, our results indicate an important role of PIF TFs: 46% of DEGs for which we could determine potential Arabidopsis homologs overlapped with Arabidopsis genes that had been described as indirect or direct targets of PIF-regulated seedling deetiolation (Class 4 or 7 genes as defined by Leivar et al., 2009). Thus, considering the evolutionary distance of these phytochrome systems, we have revealed a substantial overlap of effects on gene expression in P. patens and Arabidopsis.
In line with this, we identified four putative PIF functional orthologs in the genome of P. patens. The sequence conservation of these genes was highest in five regions, namely, the bHLH domain, the APA motif, as well in three MUFs, i.e., regions that are also highly conserved among PIF proteins from seed plants. In addition, we also identified a putative P. patens APB motif, in which, however, some of the amino acids known to be essential for PHY binding in Arabidopsis are not present. The strong conservation of these motifs is consistent with their importance for PIF function in Arabidopsis and suggests a functional conservation of PIFs across all land plants (Leivar and Quail, 2011; Leivar and Monte, 2014; Inoue et al., 2016).
PIF Evolution: From an Ancestral Gene in Charophytes to Complex Regulatory Nodes
Our phylogenetic analysis placed bHLH proteins from charophyte algae in the canonical PIF clade. In contrast to chlorophyte algae such as Chlamydomonas reinhardtii, some members of the paralogous charophytes share a common ancestor with land plants. While the C. braunii sequence harbors an APA motif and a MUF2, other available charophyte sequences were transcriptomic and therefore fragmentary in nature; hence, we cannot judge from our data whether other motifs might already have been present in these species. In any case, the Chara sequence in the PIF clade supports the notion that an ancestral PIF-like gene was already present in the last common ancestor of Charales and land plants. Interestingly, only Charales, Coleochaetales, and Zygnematophyceae, which are considered to be the algal groups most closely related to land plants (Wodniok et al., 2011; Timme et al., 2012), have representatives in this clade. An ancestral PIF-like gene, present before the water-to-land transition, would have been the basis for duplication and subfunctionalization during land plant evolution, eventually leading to the highly diversified situation present in many extant plants.
Non-seed plant genomes encode PIFs that contain APA and APB motifs, or APA only; they lack APB-only PIFs (Figure 3). From this, as well as from our interaction studies, we conclude that APA was the ancestral interaction motif. Interestingly, the solitaire M. polymorpha PIF contains an APA motif only, and the APB motifs of P. patens PIFs are not conserved in some amino acids essential for Arabidopsis PHY binding. Since mosses and liverworts are probably monophyletic (Wickett et al., 2014), we infer that either the APB motif has been lost in M. polymorpha or secondarily gained in P. patens.
Many seed plants encode at least two PIFs, one containing APA and APB, the other only APB. This could reflect the diversification of the phytochromes into PHYA and PHYB type that occurred in the common ancestor of gymnosperms and angiosperms (Mathews, 2010). The coevolution of phytochromes and phytochrome interacting factors enables a much more fine-grained regulation of red and far-red triggered signaling (Rensing et al., 2016). Interestingly, mosses also show evidence of phytochrome diversification (Li et al., 2015). Alternative splicing, as detected for Pp-PIF2 and Pp-PIF4, could potentially increase PIF diversity and entail more complex interaction patterns such as those that exist in seed plants.
Pp-PIFs Show Typical Interaction and Localization Properties as Well as Putative Neofunctionalization
Supporting the notion of functional conservation, P. patens PIFs resembled Arabidopsis PIFs in their molecular properties, showing light-dependent interaction with phytochromes. APA is the essential motif for the interaction of Pp-PIF1 and 2 with Pp-PHYs; while the binding of Pp-PIF1 and Pp-PIF2 to Pp-PHYs was completely abolished after mutation of APA, it was merely reduced when the APB motif was disabled. In Arabidopsis, the APA motif is essential for the interaction of PIFs with PHYA (Al-Sady et al., 2006; Leivar and Monte, 2014). At-PIF3 and At-PIF1, which contain different APA motifs, bind At-PHYA and act as PHYA downstream signaling components (Leivar and Monte, 2014). In motif detection and protein alignments (Figures 2 and 3), we identified the PIF3-type APA motif in PIF proteins in both non-seed plants and seed plants. One can speculate on an evolutionary conservation of the APA motif-dependent PIF-PHY interaction, which is a prerequisite for PIF3-modulated PHYA signaling in angiosperms. The binding of PIFs to PHYs via the APB motif, on the other hand, may not be relevant in P. patens phytochrome signaling and may have evolved only in seed plants. This is corroborated by the APA-dependent PHY-PIF interaction and the lack of an APB motif in M. polymorpha, as also shown by Inoue et al. (2016). The APB motifs detected in Pp-PIFs showed comparatively weak similarity to the angiosperm consensus APB sequence. The two amino acids that are essential for PIF-PHYB interaction in Arabidopsis are not conserved in Pp-PIF2 (Figure 2) (Khanna et al., 2004; Al-Sady et al., 2006). It is interesting that in Arabidopsis, the APB motif is also required for the interaction of several PIFs and DET1 (DE-ETIOLATED1) (Dong et al., 2014). The interaction of At-PIFs with At-DET1 is necessary to stabilize PIFs in darkness (Dong et al., 2014). The genome of P. patens contains three homologs of At-DET1 (Pp3c21_10400V3.1, Pp3c19_5540V3.1, and Pp3c11_15920V3.1); thus, a similar APB-dependent regulation of Pp-PIF abundance in P. patens could be envisioned. In Arabidopsis, the APB motif is also necessary for binding of PIF1 to COP1, which enhances recruitment of the COP1/SPA substrate HY5 (Xu et al., 2014). HY5 homologs are encoded in the P. patens genome; therefore, it can be speculated that a similar regulatory mechanism exists in P. patens (Xu et al., 2014; Yamawaki et al., 2011). Thus, the ancestral function of the APB motif may have been binding to DET1, COP1, and/or other factors. These interactions might also be important in P. patens; therefore, the overall sequence of the APB motif may have been preserved during evolution. Alternatively, the APB-like motif might have been independently gained in P. patens.
In the light of these scenarios, it is interesting that besides canonical PIFs containing all typical motifs, we identified splice variants of Pp-PIF2 and Pp-PIF4 that lack the complete APB motif (Pp-PIF2ΔAPB and Pp-PIF4ΔAPB). Notably, splicing variants that lack the APB motif have also been annotated for Arabidopsis PIF1 (AT2G20180.1) and PIF6 (AT3G62090.1 and AT3G62090.3). This potential splicing-mediated regulation of Pp-PIFs activity, together with the conservation of the APB motif, suggests a scenario in which the conditional removal of the APB motif might regulate the binding properties of Pp-PIFs and, thus, the quality of the response. The different combinations of APB and APA motifs in Pp-PIF splicing variants may be instrumental in a PIF regulatory network in P. patens. Such a network might be important not only during phytochrome signaling but also in other signaling cascades. Arabidopsis PIFs are involved in gibberellin (GA) signaling through their interaction with DELLA proteins. At-PIFs are thus important factors for GA downstream signaling and components of crosstalk between the light and GA signaling pathways (de Lucas et al., 2008; Feng et al., 2008; de Lucas and Prat, 2014). Although DELLA proteins were identified in P. patens, they do not seem to be functional in GA signaling (Sun, 2011). However, whether DELLAs from P. patens bind to Pp-PIFs and act in phytochrome downstream signaling remains elusive. Future work on the PIF-DELLA interplay in P. patens will help to assess the evolutionary conservation of signaling crosstalk.
A putative role of P. patens PIFs as regulators of phytochrome signaling was further supported by their localization to the nucleus. In N. benthamiana, they colocalized with At-PHYA-NLS and formed NBs, resembling the localization of Arabidopsis PIFs (Bauer et al., 2004; Chen, 2008). In a recent study, we have shown that light-activated phytochromes from P. patens also form NBs (Possart and Hiltbrunner, 2013). NBs have been associated with the PIF-phytochrome interaction and subsequent phytochrome-induced PIF degradation (Bauer et al., 2004; Chen, 2008). Despite numerous similarities between PIFs from Arabidopsis and from P. patens, the APB-independent interaction of Pp-PIF with Pp-PHY indicates that there might also be differences in PIF-PHY regulation between these two species. For example, in Arabidopsis, the APB-dependent interaction with PHYB is essential to inhibit binding of PIF3 to target promoters. It will be interesting to see whether the APA-mediated PIF-PHY interaction can lead to similar scenarios in P. patens or whether such regulatory mechanisms have evolved later in land plant evolution.
Moss PIFs Are Functional in Arabidopsis
Probably the most conclusive evidence that Pp-PIFs are bona fide PIFs came from expression in a heterologous system. First, overexpression of Pp-PIF1, Pp-PIF2, and Pp-PIF2ΔAPB in the Arabidopsis Col-0 wild-type background induced a phenotype reminiscent of the one observed for overexpression of Arabidopsis PIFs. The hyposensitive response of Pp-PIF-OX seedlings toward R light had been previously described for At-PIF3-OX, At-PIF4-OX, At-PIF5-OX, and At-PIF7-OX seedlings (Huq and Quail, 2002; Kim et al., 2003; Fujimori et al., 2004; Khanna et al., 2007; Leivar et al., 2008b). These Arabidopsis PIFs act as negative factors during PHYB-mediated photomorphogenesis (Kim et al., 2003; Fujimori et al., 2004; Khanna et al., 2007). Khanna et al. correlated the effect of At-PIF5 overexpression with reduced PHYB levels, indicating a regulation of PHYB abundance by endogenous At-PIF5 (Khanna et al., 2007). The phenotype of light-grown Arabidopsis Pp-PIF-OX seedlings suggested that PIFs from P. patens might interfere with endogenous phytochrome signaling in a similar way. The short-hypocotyl phenotype of dark-grown Pp-PIF-OX seedlings, counterintuitive at first, has also been described for At-PIF overexpressors: At-PIF5-OX seedlings show reduced hypocotyl length when grown in darkness, which has been attributed to elevated ethylene levels (Khanna et al., 2007). The behavior of Pp-PIF-OX plants at later developmental stages further supported a PIF-like effect of P. patens PIFs on light signaling in Arabidopsis. Very early flowering again phenocopied At-PIF5-OX and At-PIF4-OX plants (Fujimori et al., 2004; Kumar et al., 2012). In addition, this phenotype was reminiscent of the early flowering of Arabidopsis phyB and higher-order phy mutants (Reed et al., 1993; Strasser et al., 2010).
Second, Pp-PIFs were able to substitute for endogenous PIFs in Arabidopsis. Dark-grown seedlings of the Arabidopsis pifq mutant undergo a robust constitutive photomorphogenic development (Leivar et al., 2008a). Pp-PIFs, expressed in this mutant background, complemented the missing endogenous At-PIFs and largely rescued the cop-like phenotype. Moreover, Pp-PIFs also complemented At-PIFs at the molecular level by reconstituting the expression levels of PIF-dependent genes in the pifq mutant. In conclusion, PIFs from the moss P. patens can be considered functional in the angiosperm Arabidopsis.
It is interesting that Pp-PIFs are much more stable in light compared with At-PIF3 when expressed in the Arabidopsis pifq background. Pp-PIFs might thus be similar to At-PIF7, which is stable in red light (Leivar et al., 2008b). Alternatively, Pp-PIFs, despite being functional in Arabidopsis in darkness, might elude the typical PIF degradation processes in the heterologous species. It will be interesting to see if Pp-PIFs in P. patens are regulated through protein degradation and/or inhibition of binding to target promoters, similar to the situation in Arabidopsis, or through alternative mechanisms (Park et al., 2012; Leivar and Monte, 2014).
In summary, our data strongly suggest that Pp-PIFs play a role during light-induced adaptation and development in P. patens, similar to the function of PIFs in Arabidopsis. Thus, also in P. patens, nuclear phytochromes might regulate gene expression through the interaction with PIF proteins. An ancestral PIF node of phytochrome signaling apparently has been conserved during the course of evolution, further emphasizing its importance during light-dependent plant development. There is ever-increasing evidence that major network nodes of light signaling were already present in the earliest land plants. Since then, divergent evolution via gene duplication and subsequent sub- and neofunctionalization have led to modifications of the light signaling pathways. Such diversification apparently occurred in parallel in several land plant lineages.
METHODS
Cloning of Constructs
A description of DNA constructs can be found in the Supplemental Methods. Primers used for cloning are listed in Supplemental Table 1.
Microarray Analysis
Physcomitrella patens strain Gransden 2004 (Rensing et al., 2008) was cultivated on Knop agar plates under standard conditions (16/8-h light/dark photoperiod; bulb, Osram L 36 W/840-1, 70 µmol m−2 s−1 PAR) as previously described (Wolf et al., 2010). Four-week-old cultures were subjected to constant darkness (D) for 2 weeks before R light treatment (656 nm, 24-nm full width at half maximum [FWHM]; 12 µmol m−2 s−1). All samples, including the control (D), were harvested and immediately frozen in liquid nitrogen. An additional control experiment was set up using 3 weeks of darkness (Hiss et al., 2014). All experiments were conducted in triplicate. RNA was isolated using the Qiagen RNeasy plant mini kit. Amplification and labeling were performed using the Kreatech ampULSe kit, and quality control was done using a Nanodrop (Peqlab) and the Agilent Bioanalyzer 2100 with a plant RNA nano chip. Hybridization and data processing were performed as described previously (Wolf et al., 2010). The hierarchical clustering was generated with Genedata Analyst 7.5.7 [distance: positive correlation (1-r); linkage: average]. The principal component analysis was performed with Genedata Analyst 7.5.7 (use, covariance matrix; imputation, row mean). DEGs were identified based on unpaired cyber-t test with Benjamini-Hochberg false discovery rate correction (FDR; q < 0.05) (Benjamini and Hochberg, 1995; Long et al., 2001). One of the three 2-week control samples behaved aberrantly on the second component; the numbers of genes detected as differentially expressed therefore varied largely depending on the control group chosen. Since using all three 2-week control samples might have led to the detection of false positives, a control group consisting of five experiments (3× three weeks darkness, 2× two weeks darkness, devoid of the outlier) was used for the following analyses. The GO bias analyses used Fisher’s exact test to calculate P values. FDR-corrected q-values were calculated in R with the function p.adjust. Word cloud visualizations were created using the online tool wordle (http://www.wordle.net/). Word size was set proportional to the -log10(q-value), and overrepresented GO terms were colored dark green if q ≤ 0.0001 and light green if q > 0.0001. Underrepresented GO terms were colored dark red if q ≤ 0.0001 and light red if q > 0.0001.
Comparative Analysis of DEGs
For comparison between DEGs identified in our microarray study and genes identified as differentially expressed upon R light treatment in Arabidopsis (Leivar et al., 2009), we proceeded as follows. We used the P. patens cosmoss genome annotation v1.2 (https://www.cosmoss.org/physcome_project/wiki/Genome_Annotation/V1.2) and extracted the reciprocal best hit homolog from Arabidopsis thaliana (TAIR8) for every gene classified as expressed in the microarray analysis (Supplemental Data Sets 1 and 2). We then counted the number of Arabidopsis genes with the same locus identifier in the data of Leivar et al. (2009) for each of their seven classes of up- and downregulated genes, respectively. We did this analysis separately for the DEGs identified in the four groups (30 min R downregulated, 30 min R upregulated, 4 h downregulated, and 4 h upregulated) of our microarray study; we repeated this step for all expressed genes. Significant overlap was determined by a Fisher’s exact test comparing overlaps of DEGs to overlaps of all expressed genes.
Validation of Microarray Data by Quantitative PCR
P. patens cultivation, light treatments, and sampling were performed as described for the microarray analysis. Biological replicates (three per experiment) were generated by independent growth on individual plates. Three technical replicates were performed per biological replicate. RNA was isolated using the Qiagen RNeasy plant mini kit including an on-column DNase treatment. RNA was reverse-transcribed into first-strand cDNA using Superscript III (Invitrogen) and random hexamer primers (Thermo Fisher). Gene-specific oligonucleotides were designed utilizing Primer3 (Rozen and Skaletsky, 2000) with standardized melting temperature of 60°C and GC content of 50 to 60%; primer sequences can be found in the Supplemental Methods. Quantitative PCR was conducted using a SensiMix SYBR kit (Bioline) on a LightCycler480 (Roche). For each 25-μL reaction, 50 ng of RNA equivalent were used. Expression values (crossing point [Cp]) were normalized against the respective reference gene, employing the comparative Cp method. All values were converted into fold changes to time point zero. Thioredoxin (Pp1s545_10V6.1) (Hiss et al., 2014) showed Cp values suitable for all genes except for Lhc SR1 (Pp1s213_80V6.1), for which Pp1s215_36V6.1 (pectinesterase family protein) was chosen. One-tailed, heteroscedastic t tests were applied to test for significance. All analyses were performed with Analyst 7.5.7 (Genedata). Primers used for validation are listed in Supplemental Table 2.
Gene Identifiers of Pp-PIFs
For P. patens, cosmoss v1.6 (www.cosmoss.org) gene identifiers are as follows: Pp1s68_85V6.1 (Pp-PIF1), Pp1s69_37V6.1 (Pp-PIF2), Pp1s84_22V6.1 (Pp-PIF3), and Pp1s147_126V6.1 (Pp-PIF4). We used primers listed in the Supplemental Methods to clone coding sequences (CDS) from P. patens cDNA (see Supplemental Methods for details). For Pp-PIF4.1, we were not able to clone a CDS without a retained intron. Therefore, we cloned the 5′ part of the CDS by gene synthesis according to the annotated model (see Supplemental Methods for details). We verified that the cloned CDS sequences corresponded to the v1.6 reference annotation.
By sequence alignment, we identified the corresponding gene models in the most recent annotation (v3.3) of the P. patens genome. The CDS of Pp1s69_37V6.1 (Pp-PIF2) corresponds to that of Pp3c14_23520V3.5; the CDS of Pp1s84_22V6.1 (Pp-PIF3) corresponds to Pp3c2_8960V3.1. Pp1s68_85V6.1 (Pp-PIF1) corresponds to Pp3c17_21890V3.1; however, the protein sequence of the version 3.3 model is 20 amino acids longer than that of the version 1.6 model. For Pp1s147_126V6.1 (Pp-PIF4), the most similar annotated model is Pp3c1_38820V3.1. We used the v3.3 sequences for phylogenetic analysis.
Motif Detection and Alignment of PIFs from Selected Species
The bHLH domain and the APA and APB motifs were defined according to published information on Arabidopsis PIF3 (Toledo-Ortiz et al., 2003; Al-Sady et al., 2006). For sources and sequences of analyzed bHLH proteins, refer to Supplemental Table 1 and Supplemental File 1. Motif detection was performed using MEME (Bailey et al., 2009) on a subset of the sequences. Out of the five motifs detected N-terminally of the bHLH domain, two represented the known APB and APA domain. All five motifs were subsequently detected in the full sequence set with MAST and used as the basis for manual alignment. Sequence logos of all three motifs were created using WebLogo Version 2.8.2 (http://weblogo.berkeley.edu/) (Schneider and Stephens, 1990; Crooks et al., 2004).
Phylogenetic Analysis
Since the average size of the bHLH TF family in land plants is >100 members, we selected a few representative species for our analyses (Supplemental Data Set 3 and Supplemental File 1). Besides Arabidopsis and P. patens, we selected the genomic data sets for the eudicotyledons Carica papaya (belonging to the Brassicales like Arabidopsis, but not sharing the two most recent alpha and beta genome duplications of the Brassicaceae), Vitis vinifera (grapevine, Vitales; also no duplication since the gamma event), and Mimulus guttatus (to represent asterids), as well as Aquilegia coerula (Colorado blue columbine) and Nelumbo nucifera (sacred lotus) to represent the stem eudicotyledons. We also selected Oryza sativa (rice) and Phoenix dactylifera to represent Liliopsida (monocots) and Amborella trichopoda to represent basal angiosperms. For gymnosperms, we used Picea abies (Norway spruce) and Pinus taeda (loblolly pine). Selaginella moellendorffii (Lycopodiophyta) was included since it represents the only completed genome of the non-seed plants besides P. patens. To better represent non-seed plants, we also included transcriptomic data of ferns, mosses, and charophytes. The transcriptomes of the ferns Pteridium aquilinum (bracken fern) and Microlepia cf marginata, and genomic data of the liverwort Marchantia polymorpha and the charophyte alga Chara braunii (see Supplemental File 1 for sequence information) as well as from seven other charophyte algae (transcriptomic data) were used to search for bHLH proteins. In terms of individual sequences, the sequence entry for the PIF from S. moellendorffii, SELMO_PIF, was found in a BLAST search on the NCBI website using the APA consensus motif of seed plant PIFs as a query (http://www.ncbi.nlm.nih.gov/). SELMO_PIF was derived from ESTs (FE453350, FE430487, FE427154, FE430486, and FE453349) and the genome sequence (scaffold_54). It was retained for the analysis since it was different from the sequence present in the published filtered model 3. The assembled SELMO_PIF sequence is shown in Supplemental File 1. The Pteris vittata (chinese brake) sequence, PTEVI_PIF, was assembled from EST sequences (comp107614_c0_seq1 and comp107614_c0_seq2) that were found in a BLAST search on the P. vittata database on http://xselaginella.genomics.purdue.edu/cgi-bin/plant/blast_tmpl_soap.cgi using the APA consensus motif as a query. The assembled PTEVI_PIF sequence is shown in Supplemental File 1. As an outgroup, the unicellular alga Chlamydomonas reinhardtii was used. For further information on species and sequence resources, refer to Supplemental Data Set 3 and Supplemental File 1.
From this data set, all members of the bHLH TF family were selected as previously described (Lang et al., 2010) based on the presence of the PFAM HLH domain. In an iterative process, multiple sequence alignments were generated using MAFFT (Katoh et al., 2005) and neighbor-joining (NJ) phylogenies using Quicktree-SD (AWI-Bioinformatics). The alignments were manually curated using Jalview (Clamp et al., 2004) to remove short, fragmentary sequences. Furthermore, those subtrees were selected that contained all Arabidopsis PIFs (additional neighboring sequences were kept for reference and outgroup rooting). The final alignment was selected from several alignments generated with MAFFT, Dialign2 (Morgenstern, 1999), and MUSCLE (Edgar, 2004), using different options including profile alignment.
Alignments were manually evaluated based on the correct placement of the APA and APB motifs. The best alignment was generated using a two-step approach. First, all bHLH sequences containing APA and APB motifs were aligned using MUSCLE (Edgar, 2004). Using mafft in the profile mode, we then added the remaining bHLH sequences to the MUSCLE alignment (Supplemental File 1), retaining the alignment structure defined by MUSCLE, resulting in an alignment of 1879 positions.
The alignment was manually curated to remove positions of questionable quality or containing only a single sequence. Based on this alignment (964 positions), phylogenies were inferred using three different methods (NJ as mentioned above with 1000 bootstrap replicates). ProtTest (Abascal et al., 2005) was conducted to select the most suitable evolutionary model based on AIC/BIC, which turned out to be JTT (Jones et al., 1992), with data set frequencies and gamma distributed rates. Using this model, Bayesian inference (BI) and maximum likelihood (ML) inference were conducted. BI was performed using MrBayes (Ronquist and Huelsenbeck, 2003) with two hot and two cold chains for 3.7 million generations (sd of split frequencies < 0.01, 250 trees were discarded as burn-in). ML was conducted using RAxML (Stamatakis, 2014) starting from a random tree, generating 20 distinct trees to select the one with the best likelihood. Subsequently, 100 bootstrap inferences were performed and the support values drawn on the best tree. Trees were visualized using FigTree (http://tree.bio.ed.ac.uk/software/figtree/). The BI tree is shown as representative (Figure 3; Supplemental Figure 6), with support values from ML and NJ shown for nodes that are discussed in the text. It should be noted that the general structure of the tree, revealing a clade with all canonical PIFs, was recovered regardless of which alignment (we tested several more than the best alignment mentioned above) or inference method was used. The tree was rooted using the non-PIFs as an outgroup.
Yeast Two-Hybrid Analysis
Yeast two-hybrid analyses and ONPG assays were performed according to previously published protocols (Hiltbrunner et al., 2005). For all yeast two-hybrid assays, the yeast strain Y187 (Clontech) was used. The growth medium was supplemented with phycocyanobilin (PCB) purified from Spirulina (final concentration 10 µM) (Kunkel et al., 1993).
For evaluation of protein expression, transformed yeast cells were grown on plates supplemented with 10 µM PCB for 2 d in either constant R or FR light as previously described (Hiltbrunner et al., 2005). Total protein was extracted from yeast according to Printen and Sprague (Printen and Sprague, 1994). Protein extracts were used for SDS-PAGE and for electrotransfer to PVDF membranes. The membranes were stained with amido-black to confirm equal loading. For immunodetection, the membranes were blocked with 50 mM Tris-HCl, pH 7.5, 150 mM NaCl, 0.005% (v/v) Tween 20, and 5% (w/v) low fat milk powder and incubated with tag-specific antibodies (HA, Roche, diluted 1:1000; LUC, Sigma-Aldrich, diluted 1:2000) for 16 h at 4°C. After subsequent incubation with secondary antibodies (alkaline-phosphatase conjugate, Sigma-Aldrich, diluted 1:10,000), signal detection was performed by chemiluminescence reaction (ECL kit; Pierce) and x-ray film exposure.
In Vitro Co-IP
The in vitro co-IP assays were performed as previously described (Huq et al., 2004). Briefly, 250 ng of each plasmid was transcribed and translated in vitro in the presence of 35S-methionine using the TnT Quick Coupled transcription/translation system (catalog no. L1170; Promega). The GAD antibody (catalog no. sc-663; Santa Cruz Biotechnology) was first bound to Dynabeads (Life Technologies) (20 µL/µg antibody) for 30 min and washed twice with the binding buffer (1× PBS, BSA 1 μg/mL, 0.2% Nonidet P-40, and 1× protease inhibitor cocktail). To prepare the bait proteins, wild-type Pp-PIF2, Pp-PIF3, and Pp-PIF4 and various mutant Pp-PIF2 proteins were incubated with the Dynabeads bound to GAD antibody for 2 h, and the pellet was washed three times. To reconstruct phytochrome holoproteins as prey, the Pp-PHY1 to 4 expression mix was further incubated with 25 μM PCB (catalog no. P14137; Frontier Scientific) in the dark for 1 h. To prepare the active Pfr or inactive Pr forms of phytochromes, the Pp-PHY1 to 4 tubes were then illuminated with either R (17 µmol m−2 s−1) or FR (4.3 µmol m−2 s−1) light for 1 min, respectively, and incubated with GAD-Pp-PIF2, 3, or 4 in binding buffer for two additional hours. The beads were collected by magnet and washed thrice thoroughly with binding buffer and once with binding buffer without BSA. Immunoprecipitated proteins were heated at 65°C in a SDS sample buffer for 10 min, separated on a SDS-PAGE gel, and detected by Typhoon phosphor imaging system.
Transient Expression in Nicotiana benthamiana and P. patens
The transient transformation of N. benthamiana was performed as described previously (Grefen et al., 2008). Leaves from 4- to 6-week-old N. benthamiana plants were infiltrated at the adaxial sides with Agrobacterium tumefaciens strain C58 carrying plasmid coding for the respective fusion proteins. The p19 protein from tomato bushy stunt virus was used for suppression of transgene silencing (Grefen et al., 2008). After transformation with pPPO30:Pp-PIF1, pPPO30:Pp-PIF2, pPPO30:Pp-PIF2ΔAPB, pPPO30:Pp-PIF3, pPPO30:Pp-PIF4, or pPPO30:Pp-PIF4ΔAPB and cotransformation of these plasmids with pCHF40:At-PHYA-NLS, respectively, the wild tobacco plants were incubated in darkness for 1 to 3 d before epifluorescence or confocal microscopy analysis. For evaluation of protein expression, wild tobacco leaves of at least two independently transformed plants were harvested and combined in one protein extract. Leaves were immediately frozen in liquid nitrogen, and protein extraction and protein gel blotting were performed as described (Laemmli, 1970). Immunodetection of Pp-PIF proteins was performed using the monoclonal GFP antiserum (GFP, Abcam, diluted 1:1000) as primary antibody. Alkaline phosphatase-coupled (Sigma-Aldrich) anti-mouse antiserum was used as secondary antibody (diluted 1:10,000). Signal detection was performed by chemiluminescence reaction (ECL kit; Pierce) and x-ray film exposure.
Transient expression of Pp-PIFs in P. patens was done as follows. P. patens protonemata cultures (strain Gransden 2004) (Rensing et al., 2008) were placed on cellophane sheets covering Knop agar plates, covered with another sheet of cellophane to prevent upright growth, and incubated under standard growth conditions (16/8-h light/dark photoperiod; bulb, Philips TL70 F17T8/TL741, 50–70 µmol m−2 s−1 PAR) for 4 to 6 d. pUC1930:Pp-PIF1, pUC1930:Pp-PIF2, pUC1930:Pp-PIF2∆APB, pUC1930:Pp-PIF3, pUC1930:Pp-PIF4, or pUC1930:Pp-PIF4∆APB were transiently transformed into P. patens by particle bombardment with a biolistic particle delivery system. Gold particle diameter was 1 µm, helium pressure 700 kPa (7 bar), chamber vacuum pressure 80 kPa (0.8 bar), and target distance 5 cm. After bombardment, the samples were kept in darkness for 1 to 5 d before epifluorescence microscopic analysis.
Transformation of Arabidopsis
Arabidopsis lines expressing Pro35S:Pp-PIF1:YFP:TerRbcS, Pro35S:Pp-PIF2:YFP:TerRbcS, or Pro35S:Pp-PIF2ΔAPB:YFP:TerRbcS were obtained by Agrobacterium-mediated transformation (Clough and Bent, 1998; Davis et al., 2009). The selection for transgenic plants using the herbicide Butafenacil/Inspire (Syngenta Agro) was performed as previously described (Rausenberger et al., 2011). For evaluation of protein expression, Pp-PIF-OX seedlings were grown in darkness for 4 d before harvesting and preparation of protein extracts. For protein extraction, ∼100 mg of plant material was homogenized in liquid nitrogen using glass beads. Total protein was extracted from Arabidopsis seedlings according to Bauer et al. (2004). Protein blotting was performed as described above. Immunodetection was performed using an antibody specific to the GFP-tag or HA-tag (HA, Roche, diluted 1:1000; GFP, Abcam, diluted 1:1000). The detection of Actin protein using an Actin-specific antibody (Sigma-Aldrich, diluted 1:3000) is shown as loading control.
Phenotypic Analysis
Arabidopsis seeds were stratified for 2 to 10 d at 4°C and grown for 4 d on 0.5× Murashige and Skoog medium (Duchefa)/0.7% (w/v) agar at 22 to 24°C in darkness, continuous R (λmax, 671 nm; FWHM, 25 nm; light intensity [LI], 22 µmol m−2 s−1), FR (λmax, 742 nm; FWHM, 24 nm; LI, 3 µmol m−2 s−1), or B (λmax, 463 nm; FWHM, 22 nm; LI, 8 µmol m−2 s−1) light before phenotypic analysis of seedlings. LI was measured with SKYE SKR 1850 four-channel light sensor, channel 4 for R and B (∼410–710 nm), and channel 3 for FR (∼725–755 nm), respectively.
For the analysis of later developmental stages, Arabidopsis seeds were sown on soil. Plants were grown under standard conditions in the greenhouse. The analyses were performed partially with seed batches segregating or with seed batches homozygous for the insertion.
Gene Expression Analysis in Complemented Arabidopsis pifq Mutants by Quantitative PCR
mRNA abundance was analyzed using qPCR. Total RNA was isolated from dark-grown Arabidopsis seedlings 4 d after germination using the RNeasy mini kit (Qiagen), followed by first-strand cDNA synthesis. qPCR was performed with the ABsolute qPCR SYBR Green Mix (Thermo Fisher Scientific) using an ABI Prism7300 instrument (Applied Biosystems). Each qPCR reaction was repeated three times (technical replicates) for each of the three biological replicates. The standard curve method was used for the calculation of relative transcript quantities; expression was normalized against that of PP2AA3. Sequences of the respective primers are listed in Supplemental Table 2. Corresponding gene accession numbers are listed below.
Analysis of Pp-PIF Protein Levels in Arabidopsis pifq Mutants by Immunoblot
For analysis of PpPIF protein levels in Arabidopsis pifq lines, we performed immunoblots with total protein extracts from seedlings. Seedlings were grown 4 d in darkness and were harvested either directly or after irradiation with red light (22 µmol m−2 s−1) for 10 min, 30 min, 1 h, or 24 h. In addition, Arabidopsis Col-0 seedlings expressing Pro35S:At-PIF3:CFP were used to monitor light-dependent PIF degradation (Bauer et al., 2004). PIF proteins were detected as described above using an antibody specific to the GFP-tag (GFP, Abcam, diluted 1:1000); to check for equal loading, we used an anti-Actin antibody (Sigma-Aldrich, diluted 1:3000).
Microscopy Analysis and Image Processing
A Nikon ECLIPSE 90i (Nikon) equipped with YFP-, CFP-, and RFP-specific filter sets (Nikon) and a Photometrics Cool Snap ES camera was used for image acquisition with MetaMorph (version 6.2r5) software. A Leica TCS SP8 confocal microscope was used for image acquisition with LASAF software (Leica Microsystems); excitation was obtained with lasers of 405 nm (CFP) and 514 nm (YFP), respectively; emission was detected with sequential scan between 454 and 500 nm (CFP) and between 521 and 584 nm (YFP), respectively. ImageJ (version 1.44k; National Institute of Health), Photoshop CS5 (version 12.0.4.x32; Adobe), and LASAF Lite (Leica Microsystems) software were used for image processing.
Accession Numbers
Sequence data from this article can be found in the GenBank/EMBL, Phytozome (www.phytozome.net), or cosmoss (www.cosmoss.org) libraries under the following accession numbers: At-CO, At5g15840; At-COP1, At2g32950; At-DET1, At4g10180; At-FHL, At5g02200; At-FHY1, At2g37678; At-HB2, At4g16780; At-HFR1, At1g02340; At-HY5, At5g11260; At-IAA19, At3g15540; At-PHYA, At1g09570; At-PHYB, At2g18790; At-PIF1, At2g20180; At-PIF3, At1g09530; At-PIF4, At2g43010; At-PIF5, At3g59060; At-PIF6, At3g62090; At-PIF7, At5g61270; At-PIF8, At4g00050; At-PIL1, At2g46970; At-PP2AA3, At1g13320; At-SPA1, At2g46340; At-UVR8, At5g63860; At-XTR7, At4g14130; At-ZTL, At5g57360; Mp-PHY, LC093264; Mp-PIF, LC093265; Pp-COL1, AJ890106; Pp-COL2, AJ890107; Pp-COL3, AJ890108; Pp-COP1a, Pp1s135_17V6.1; Pp-FHY1, Pp3c17_1070; Pp-HY5a, Pp3c7_11360V1.1; Pp-HY5b, Pp1s80_72V6.3; Pp-PHY1, AB275304; Pp-PHY2, AB275305; Pp-PHY3, XM_001765983; Pp-PHY4, AB275307; Pp-PIF1, Pp1s68_85V6.1; Pp-PIF2, Pp1s69_37V6.1; Pp-PIF3, Pp1s84_22V6.1; Pp-PIF4, Pp1s147_126V6.1; Pp-SPAa, Pp1s59_66V6.1; and Pp-SPAb, Pp1s30_295V6.1. Microarray data from this article can be found in the ArrayExpress database under accession number E-MTAB-2227 (30 min and 4 h R treatment of P. patens).
Supplemental Data
Supplemental Figure 1. Global review and validation of microarray data.
Supplemental Figure 2. Word cloud visualization of DEGs after 30 min R light according to Gene Ontology terms.
Supplemental Figure 3. Word cloud visualization of DEGs after 4 h R light according to Gene Ontology terms.
Supplemental Figure 4. Sequence alignment of motifs of unknown function (MUF) 2 and 3 detected in PIF bHLH proteins.
Supplemental Figure 5. Sequence logos of the PIF APB motif, motifs of unknown function (MUF), and the APA motif.
Supplemental Figure 6. Excerpt of phylogenetic tree of plant bHLH TFs: clade not containing canonical PIFs.
Supplemental Figure 7. Interaction of LUC-Pp-PIF1 with light-activated Pp-PHY2-4 in yeast requires the APA motif.
Supplemental Figure 8. Immunoblot analyses of Pp-PIF1 and Pp-PHY protein abundance in yeast.
Supplemental Figure 9. Interaction of Pp-PIF1 with light-activated phytochromes.
Supplemental Figure 10. Interaction of Pp-PIF1ΔbHLH with light-activated phytochromes.
Supplemental Figure 11. Interaction of Pp-PIF1 and Pp-PIF1ΔbHLH with light-activated At-PHYA.
Supplemental Figure 12. Immunoblot analysis of Pp-PIF expression in N. benthamiana.
Supplemental Figure 13. Localization of PIFs from P. patens in Arabidopsis seedlings.
Supplemental Figure 14. Immunoblot analysis of Pp-PIF expression in Arabidopsis Columbia-0 and pifq mutant seedlings.
Supplemental Figure 15. Pp-PIF protein stability in Arabidopsis pifq mutants.
Supplemental Figure 16. Complementation of expression phenotypes in Arabidopsis pifq mutants through expression of Pp-PIFs.
Supplemental Methods. Cloning of constructs.
Supplemental Table 1. Primer list used for cloning.
Supplemental Table 2. Primer list used for qRT-PCR.
Supplemental File 1. Alignment of full-length bHLH protein sequences used for phylogenetic analysis.
Supplemental Data Set 1. DEGs from microarray analysis in P. patens after R light treatments.
Supplemental Data Set 2. Comparison of transcriptome analyses from P. patens and Arabidopsis (Leivar et al., 2009).
Supplemental Data Set 3. Overview of species and sequence resources used for phylogenetic analyses (for sequences of bHLH proteins shown in the phylogenetic trees, see also Supplemental File 1).
Supplementary Material
Acknowledgments
We thank Martina Krenz (University of Freiburg, Germany), Claudia König (University of Tübingen, Germany), and Laura Brodde (University of Tübingen, Germany) for supporting technical work. We thank Klaus Harter (Center for Plant Molecular Biology, University of Tübingen, Germany) for the provision of research facilities. This work was supported by grants from the German Research Foundation (DFG HI 1369/2-1 and HI 1369/4-1) to A.H., from the National Institute of Health (1R01 GM-114297) to E.H., and from the Human Frontier Science Program Organization (HFSP Research Grant RGP0025/2013) to A.H. and E.H. Work in the laboratory of A.H. was also supported by the Excellence Initiative of the German Federal and State Governments (EXC 294, Project C20). T.X. was supported by a fellowship from the China Scholarship Council. C.B. was supported by the Max Planck Society and DFG (SFB 1101).
AUTHOR CONTRIBUTIONS
A.P., E.H., S.A.R., and A.H. conceived the study. A.P., T.X., I.P., S.H., S.K., H.-M.H., L.W., and M.H. performed the experiments. A.P., T.X., I.P., S.H., M.H., C.B., S.A.R., and A.H. analyzed the data. A.P., S.A.R., and A.H. wrote the manuscript, with contributions from T.X., I.P., S.H., C.B., and E.H.
Glossary
- R
red
- FR
far-red
- B
blue
- TF
transcription factor
- bHLH
basic helix-loop-helix
- PIF
phytochrome interacting factor
- NB
nuclear body
- DEG
differentially expressed gene
- co-IP
coimmunoprecipitation
- GA
gibberellin
- GO
Gene Ontology
- Cp
crossing point
- CDS
coding sequence
- NJ
neighbor-joining
- BI
Bayesian inference
- ML
maximum likelihood
- PCB
phycocyanobilin
- LI
light intensity
- FWHM
full width at half maximum
References
- Abascal F., Zardoya R., Posada D. (2005). ProtTest: selection of best-fit models of protein evolution. Bioinformatics 21: 2104–2105. [DOI] [PubMed] [Google Scholar]
- Al-Sady B., Ni W., Kircher S., Schäfer E., Quail P.H. (2006). Photoactivated phytochrome induces rapid PIF3 phosphorylation prior to proteasome-mediated degradation. Mol. Cell 23: 439–446. [DOI] [PubMed] [Google Scholar]
- Bailey T.L., Boden M., Buske F.A., Frith M., Grant C.E., Clementi L., Ren J., Li W.W., Noble W.S. (2009). MEME SUITE: tools for motif discovery and searching. Nucleic Acids Res. 37: W202–W208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bauer D., Viczián A., Kircher S., Nobis T., Nitschke R., Kunkel T., Panigrahi K.C., Adám E., Fejes E., Schäfer E., Nagy F. (2004). Constitutive photomorphogenesis 1 and multiple photoreceptors control degradation of phytochrome interacting factor 3, a transcription factor required for light signaling in Arabidopsis. Plant Cell 16: 1433–1445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benjamini Y., Hochberg Y. (1995). Controlling the false discovery rate: a practical and powerful approach to multiple testing. J. R. Stat. Soc. B Met. 57: 289–300. [Google Scholar]
- Carretero-Paulet L., Galstyan A., Roig-Villanova I., Martínez-García J.F., Bilbao-Castro J.R., Robertson D.L. (2010). Genome-wide classification and evolutionary analysis of the bHLH family of transcription factors in Arabidopsis, poplar, rice, moss, and algae. Plant Physiol. 153: 1398–1412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casal J.J. (2013). Photoreceptor signaling networks in plant responses to shade. Annu. Rev. Plant Biol. 64: 403–427. [DOI] [PubMed] [Google Scholar]
- Chen M. (2008). Phytochrome nuclear body: an emerging model to study interphase nuclear dynamics and signaling. Curr. Opin. Plant Biol. 11: 503–508. [DOI] [PubMed] [Google Scholar]
- Chen M., Tao Y., Lim J., Shaw A., Chory J. (2005). Regulation of phytochrome B nuclear localization through light-dependent unmasking of nuclear-localization signals. Curr. Biol. 15: 637–642. [DOI] [PubMed] [Google Scholar]
- Chen Y.R., Su Y.S., Tu S.L. (2012). Distinct phytochrome actions in nonvascular plants revealed by targeted inactivation of phytobilin biosynthesis. Proc. Natl. Acad. Sci. USA 109: 8310–8315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christensen S., Tokuoka Y., Silverthorne J., Wada M. (1998). Phytochrome regulation of expression of mRNA encoding the major light-harvesting chlorophyll a/6-binding proteins of photosystem II in the haploid phase of Adiantum capillus-veneris. Plant Cell Physiol. 39: 647–654. [Google Scholar]
- Christensen S., LaVerne E., Boyd G., Silverthorne J. (2002). Ginkgo biloba retains functions of both type I and type II flowering plant phytochrome. Plant Cell Physiol. 43: 768–777. [DOI] [PubMed] [Google Scholar]
- Clamp M., Cuff J., Searle S.M., Barton G.J. (2004). The Jalview Java alignment editor. Bioinformatics 20: 426–427. [DOI] [PubMed] [Google Scholar]
- Clough S.J., Bent A.F. (1998). Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 16: 735–743. [DOI] [PubMed] [Google Scholar]
- Crooks G.E., Hon G., Chandonia J.M., Brenner S.E. (2004). WebLogo: a sequence logo generator. Genome Res. 14: 1188–1190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dalton J.C., Bätz U., Liu J., Curie G.L., Quail P.H. (2016). A modified reverse one-hybrid screen identifies transcriptional activation domains in PHYTOCHROME-INTERACTING FACTOR 3. Front. Plant Sci. 7: 881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis A.M., Hall A., Millar A.J., Darrah C., Davis S.J. (2009). Protocol: Streamlined sub-protocols for floral-dip transformation and selection of transformants in Arabidopsis thaliana. Plant Methods 5: 3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Lucas M., Prat S. (2014). PIFs get BRright: PHYTOCHROME INTERACTING FACTORs as integrators of light and hormonal signals. New Phytol. 202: 1126–1141. [DOI] [PubMed] [Google Scholar]
- de Lucas M., Davière J.M., Rodríguez-Falcón M., Pontin M., Iglesias-Pedraz J.M., Lorrain S., Fankhauser C., Blázquez M.A., Titarenko E., Prat S. (2008). A molecular framework for light and gibberellin control of cell elongation. Nature 451: 480–484. [DOI] [PubMed] [Google Scholar]
- Dong J., Tang D., Gao Z., Yu R., Li K., He H., Terzaghi W., Deng X.W., and Chen H.(2014). Arabidopsis DE-ETIOLATED1 represses photomorphogenesis by positively regulating phytochrome-interacting factors in the dark. Plant Cell 26: 3630–3645. [DOI] [PMC free article] [PubMed]
- Duanmu D., et al. (2014). Marine algae and land plants share conserved phytochrome signaling systems. Proc. Natl. Acad. Sci. USA 111: 15827–15832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edgar R.C. (2004). MUSCLE: a multiple sequence alignment method with reduced time and space complexity. BMC Bioinformatics 5: 113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feller A., Machemer K., Braun E.L., Grotewold E. (2011). Evolutionary and comparative analysis of MYB and bHLH plant transcription factors. Plant J. 66: 94–116. [DOI] [PubMed] [Google Scholar]
- Feng S., et al. (2008). Coordinated regulation of Arabidopsis thaliana development by light and gibberellins. Nature 451: 475–479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujimori T., Yamashino T., Kato T., Mizuno T. (2004). Circadian-controlled basic/helix-loop-helix factor, PIL6, implicated in light-signal transduction in Arabidopsis thaliana. Plant Cell Physiol. 45: 1078–1086. [DOI] [PubMed] [Google Scholar]
- Grefen C., Städele K., Růzicka K., Obrdlik P., Harter K., Horák J. (2008). Subcellular localization and in vivo interactions of the Arabidopsis thaliana ethylene receptor family members. Mol. Plant 1: 308–320. [DOI] [PubMed] [Google Scholar]
- Hiltbrunner A., Viczián A., Bury E., Tscheuschler A., Kircher S., Tóth R., Honsberger A., Nagy F., Fankhauser C., Schäfer E. (2005). Nuclear accumulation of the phytochrome A photoreceptor requires FHY1. Curr. Biol. 15: 2125–2130. [DOI] [PubMed] [Google Scholar]
- Hiss M., et al. (2014). Large-scale gene expression profiling data for the model moss Physcomitrella patens aid understanding of developmental progression, culture and stress conditions. Plant J. 79: 530–539. [DOI] [PubMed] [Google Scholar]
- Holm K., Källman T., Gyllenstrand N., Hedman H., Lagercrantz U. (2010). Does the core circadian clock in the moss Physcomitrella patens (Bryophyta) comprise a single loop? BMC Plant Biol. 10: 109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hornitschek P., Kohnen M.V., Lorrain S., Rougemont J., Ljung K., López-Vidriero I., Franco-Zorrilla J.M., Solano R., Trevisan M., Pradervand S., Xenarios I., Fankhauser C. (2012). Phytochrome interacting factors 4 and 5 control seedling growth in changing light conditions by directly controlling auxin signaling. Plant J. 71: 699–711. [DOI] [PubMed] [Google Scholar]
- Hughes J. (2013). Phytochrome cytoplasmic signaling. Annu. Rev. Plant Biol. 64: 377–402. [DOI] [PubMed] [Google Scholar]
- Huq E., Quail P.H. (2002). PIF4, a phytochrome-interacting bHLH factor, functions as a negative regulator of phytochrome B signaling in Arabidopsis. EMBO J. 21: 2441–2450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huq E., Al-Sady B., Hudson M., Kim C., Apel K., Quail P.H. (2004). Phytochrome-interacting factor 1 is a critical bHLH regulator of chlorophyll biosynthesis. Science 305: 1937–1941. [DOI] [PubMed] [Google Scholar]
- Imaizumi T., Kadota A., Hasebe M., Wada M. (2002). Cryptochrome light signals control development to suppress auxin sensitivity in the moss Physcomitrella patens. Plant Cell 14: 373–386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inoue K., Nishihama R., Kataoka H., Hosaka M., Manabe R., Nomoto M., Tada Y., Ishizaki K., Kohchi T. (2016). Phytochrome signaling is mediated by PHYTOCHROME INTERACTING FACTOR in the liverwort Marchantia polymorpha. Plant Cell 28: 1406–1421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeong J., Choi G. (2013). Phytochrome-interacting factors have both shared and distinct biological roles. Mol. Cells 35: 371–380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones D.T., Taylor W.R., Thornton J.M. (1992). The rapid generation of mutation data matrices from protein sequences. Comput. Appl. Biosci. 8: 275–282. [DOI] [PubMed] [Google Scholar]
- Kami C., Lorrain S., Hornitschek P., Fankhauser C. (2010). Light-regulated plant growth and development. Curr. Top. Dev. Biol. 91: 29–66. [DOI] [PubMed] [Google Scholar]
- Katoh K., Kuma K., Toh H., Miyata T. (2005). MAFFT version 5: improvement in accuracy of multiple sequence alignment. Nucleic Acids Res. 33: 511–518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khanna R., Huq E., Kikis E.A., Al-Sady B., Lanzatella C., Quail P.H. (2004). A novel molecular recognition motif necessary for targeting photoactivated phytochrome signaling to specific basic helix-loop-helix transcription factors. Plant Cell 16: 3033–3044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khanna R., Shen Y., Marion C.M., Tsuchisaka A., Theologis A., Schäfer E., Quail P.H. (2007). The basic helix-loop-helix transcription factor PIF5 acts on ethylene biosynthesis and phytochrome signaling by distinct mechanisms. Plant Cell 19: 3915–3929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J., Yi H., Choi G., Shin B., Song P.S., Choi G. (2003). Functional characterization of phytochrome interacting factor 3 in phytochrome-mediated light signal transduction. Plant Cell 15: 2399–2407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krzymuski M., Cerdán P.D., Zhu L., Vinh A., Chory J., Huq E., Casal J.J. (2014). Phytochrome A antagonizes PHYTOCHROME INTERACTING FACTOR 1 to prevent over-activation of photomorphogenesis. Mol. Plant 7: 1415–1428. [DOI] [PubMed] [Google Scholar]
- Kumar S.V., Lucyshyn D., Jaeger K.E., Alós E., Alvey E., Harberd N.P., Wigge P.A. (2012). Transcription factor PIF4 controls the thermosensory activation of flowering. Nature 484: 242–245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kunkel T., Tomizawa K., Kern R., Furuya M., Chua N.H., Schäfer E. (1993). In vitro formation of a photoreversible adduct of phycocyanobilin and tobacco apophytochrome B. Eur. J. Biochem. 215: 587–594. [DOI] [PubMed] [Google Scholar]
- Laemmli U.K. (1970). Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227: 680–685. [DOI] [PubMed] [Google Scholar]
- Lang D., Weiche B., Timmerhaus G., Richardt S., Riaño-Pachón D.M., Corrêa L.G.G., Reski R., Mueller-Roeber B., Rensing S.A. (2010). Genome-wide phylogenetic comparative analysis of plant transcriptional regulation: a timeline of loss, gain, expansion, and correlation with complexity. Genome Biol. Evol. 2: 488–503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leivar P., Quail P.H. (2011). PIFs: pivotal components in a cellular signaling hub. Trends Plant Sci. 16: 19–28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leivar P., Monte E. (2014). PIFs: systems integrators in plant development. Plant Cell 26: 56–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leivar P., Tepperman J.M., Monte E., Calderon R.H., Liu T.L., Quail P.H. (2009). Definition of early transcriptional circuitry involved in light-induced reversal of PIF-imposed repression of photomorphogenesis in young Arabidopsis seedlings. Plant Cell 21: 3535–3553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leivar P., Monte E., Oka Y., Liu T., Carle C., Castillon A., Huq E., Quail P.H. (2008a). Multiple phytochrome-interacting bHLH transcription factors repress premature seedling photomorphogenesis in darkness. Curr. Biol. 18: 1815–1823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leivar P., Monte E., Al-Sady B., Carle C., Storer A., Alonso J.M., Ecker J.R., Quail P.H. (2008b). The Arabidopsis phytochrome-interacting factor PIF7, together with PIF3 and PIF4, regulates responses to prolonged red light by modulating phyB levels. Plant Cell 20: 337–352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J., Li G., Wang H., Wang Deng X. (2011). Phytochrome signaling mechanisms. Arabidopsis Book 9: e0148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li F.W., Melkonian M., Rothfels C.J., Villarreal J.C., Stevenson D.W., Graham S.W., Wong G.K., Pryer K.M., Mathews S. (2015). Phytochrome diversity in green plants and the origin of canonical plant phytochromes. Nat. Commun. 6: 7852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Long A.D., Mangalam H.J., Chan B.Y.P., Tolleri L., Hatfield G.W., Baldi P. (2001). Improved statistical inference from DNA microarray data using analysis of variance and a Bayesian statistical framework. Analysis of global gene expression in Escherichia coli K12. J. Biol. Chem. 276: 19937–19944. [DOI] [PubMed] [Google Scholar]
- Lorrain S., Allen T., Duek P.D., Whitelam G.C., Fankhauser C. (2008). Phytochrome-mediated inhibition of shade avoidance involves degradation of growth-promoting bHLH transcription factors. Plant J. 53: 312–323. [DOI] [PubMed] [Google Scholar]
- Mancinelli A.L. (1994). The physiology of phytochrome action. In Photomorphogenesis in Plants, Kendrick R.E., Kronenberg G.M.H., eds (Dordrecht, The Netherlands: Kluwer Academic Publishers; ), pp. 211–269. [Google Scholar]
- Mathews S. (2006). Phytochrome-mediated development in land plants: red light sensing evolves to meet the challenges of changing light environments. Mol. Ecol. 15: 3483–3503. [DOI] [PubMed] [Google Scholar]
- Mathews S. (2010). Evolutionary studies illuminate the structural-functional model of plant phytochromes. Plant Cell 22: 4–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mathews S., Tremonte D. (2012). Tests of the link between functional innovation and positive selection at phytochrome A: the phylogenetic distribution of far-red high-irradiance responses in seedling development. Int. J. Plant Sci. 173: 662–672. [Google Scholar]
- Monte E., Tepperman J.M., Al-Sady B., Kaczorowski K.A., Alonso J.M., Ecker J.R., Li X., Zhang Y., Quail P.H. (2004). The phytochrome-interacting transcription factor, PIF3, acts early, selectively, and positively in light-induced chloroplast development. Proc. Natl. Acad. Sci. USA 101: 16091–16098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morgenstern B. (1999). DIALIGN 2: improvement of the segment-to-segment approach to multiple sequence alignment. Bioinformatics 15: 211–218. [DOI] [PubMed] [Google Scholar]
- Park E., Park J., Kim J., Nagatani A., Lagarias J.C., Choi G. (2012). Phytochrome B inhibits binding of phytochrome-interacting factors to their target promoters. Plant J. 72: 537–546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfeiffer A., Nagel M.K., Popp C., Wüst F., Bindics J., Viczián A., Hiltbrunner A., Nagy F., Kunkel T., Schäfer E. (2012). Interaction with plant transcription factors can mediate nuclear import of phytochrome B. Proc. Natl. Acad. Sci. USA 109: 5892–5897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfeiffer A., Shi H., Tepperman J.M., Zhang Y., Quail P.H. (2014). Combinatorial complexity in a transcriptionally centered signaling hub in Arabidopsis. Mol. Plant 7: 1598–1618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Possart A., Hiltbrunner A. (2013). An evolutionarily conserved signaling mechanism mediates far-red light responses in land plants. Plant Cell 25: 102–114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Possart A., Fleck C., Hiltbrunner A. (2014). Shedding (far-red) light on phytochrome mechanisms and responses in land plants. Plant Sci. 217–218: 36–46. [DOI] [PubMed] [Google Scholar]
- Printen J.A., Sprague G.F. Jr (1994). Protein-protein interactions in the yeast pheromone response pathway: Ste5p interacts with all members of the MAP kinase cascade. Genetics 138: 609–619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ranjan A., Dickopf S., Ullrich K.K., Rensing S.A., Hoecker U. (2014). Functional analysis of COP1 and SPA orthologs from Physcomitrella and rice during photomorphogenesis of transgenic Arabidopsis reveals distinct evolutionary conservation. BMC Plant Biol. 14: 178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rausenberger J., Tscheuschler A., Nordmeier W., Wüst F., Timmer J., Schäfer E., Fleck C., Hiltbrunner A. (2011). Photoconversion and nuclear trafficking cycles determine phytochrome A’s response profile to far-red light. Cell 146: 813–825. [DOI] [PubMed] [Google Scholar]
- Reed J.W., Nagpal P., Poole D.S., Furuya M., Chory J. (1993). Mutations in the gene for the red/far-red light receptor phytochrome B alter cell elongation and physiological responses throughout Arabidopsis development. Plant Cell 5: 147–157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rensing S.A., Ick J., Fawcett J.A., Lang D., Zimmer A., Van de Peer Y., Reski R. (2007). An ancient genome duplication contributed to the abundance of metabolic genes in the moss Physcomitrella patens. BMC Evol. Biol. 7: 130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rensing S.A., et al. (2008). The Physcomitrella genome reveals evolutionary insights into the conquest of land by plants. Science 319: 64–69. [DOI] [PubMed] [Google Scholar]
- Rensing S.A., Sheerin D.J., Hiltbrunner A. (2016). Phytochromes: more than meets the eye. Trends Plant Sci. 21: 543–546. [DOI] [PubMed] [Google Scholar]
- Richardt S., Lang D., Reski R., Frank W., Rensing S.A. (2007). PlanTAPDB, a phylogeny-based resource of plant transcription-associated proteins. Plant Physiol. 143: 1452–1466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richardt S., Timmerhaus G., Lang D., Qudeimat E., Corrêa L.G., Reski R., Rensing S.A., Frank W. (2010). Microarray analysis of the moss Physcomitrella patens reveals evolutionarily conserved transcriptional regulation of salt stress and abscisic acid signalling. Plant Mol. Biol. 72: 27–45. [DOI] [PubMed] [Google Scholar]
- Ronquist F., Huelsenbeck J.P. (2003). MrBayes 3: Bayesian phylogenetic inference under mixed models. Bioinformatics 19: 1572–1574. [DOI] [PubMed] [Google Scholar]
- Rösler J., Jaedicke K., Zeidler M. (2010). Cytoplasmic phytochrome action. Plant Cell Physiol. 51: 1248–1254. [DOI] [PubMed] [Google Scholar]
- Rozen S., Skaletsky H. (2000). Primer3 on the WWW for general users and for biologist programmers. Methods Mol. Biol. 132: 365–386. [DOI] [PubMed] [Google Scholar]
- Schneider T.D., Stephens R.M. (1990). Sequence logos: a new way to display consensus sequences. Nucleic Acids Res. 18: 6097–6100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen H., Zhu L., Castillon A., Majee M., Downie B., Huq E. (2008). Light-induced phosphorylation and degradation of the negative regulator PHYTOCHROME-INTERACTING FACTOR1 from Arabidopsis depend upon its direct physical interactions with photoactivated phytochromes. Plant Cell 20: 1586–1602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen Y., Khanna R., Carle C.M., Quail P.H. (2007). Phytochrome induces rapid PIF5 phosphorylation and degradation in response to red-light activation. Plant Physiol. 145: 1043–1051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shin J., Kim K., Kang H., Zulfugarov I.S., Bae G., Lee C.H., Lee D., Choi G. (2009). Phytochromes promote seedling light responses by inhibiting four negatively-acting phytochrome-interacting factors. Proc. Natl. Acad. Sci. USA 106: 7660–7665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stamatakis A. (2014). RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics 30: 1312–1313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strasser B., Sánchez-Lamas M., Yanovsky M.J., Casal J.J., Cerdán P.D. (2010). Arabidopsis thaliana life without phytochromes. Proc. Natl. Acad. Sci. USA 107: 4776–4781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suetsugu N., Wada M. (2003). Cryptogam blue-light photoreceptors. Curr. Opin. Plant Biol. 6: 91–96. [DOI] [PubMed] [Google Scholar]
- Sun T.P. (2011). The molecular mechanism and evolution of the GA-GID1-DELLA signaling module in plants. Curr. Biol. 21: R338–R345. [DOI] [PubMed] [Google Scholar]
- Suzuki T., Takio S., Yamamoto I., Satoh T. (2001). Characterization of cDNA of the liverwort phytochrome gene, and phytochrome involvement in the light-dependent and light-independent protochlorophyllide oxidoreductase gene expression in Marchantia paleacea var. diptera. Plant Cell Physiol. 42: 576–582. [DOI] [PubMed] [Google Scholar]
- Timme R.E., Bachvaroff T.R., Delwiche C.F. (2012). Broad phylogenomic sampling and the sister lineage of land plants. PLoS One 7: e29696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toledo-Ortiz G., Huq E., Quail P.H. (2003). The Arabidopsis basic/helix-loop-helix transcription factor family. Plant Cell 15: 1749–1770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsuboi H., Nakamura S., Schäfer E., Wada M. (2012). Red light-induced phytochrome relocation into the nucleus in Adiantum capillus-veneris. Mol. Plant 5: 611–618. [DOI] [PubMed] [Google Scholar]
- Van Buskirk E.K., Decker P.V., Chen M. (2012). Photobodies in light signaling. Plant Physiol. 158: 52–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wickett N.J., et al. (2014). Phylotranscriptomic analysis of the origin and early diversification of land plants. Proc. Natl. Acad. Sci. USA 111: E4859–E4868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winands A., Wagner G. (1996). Phytochrome of the green alga Mougeotia: cDNA sequence, autoregulation and phylogenetic position. Plant Mol. Biol. 32: 589–597. [DOI] [PubMed] [Google Scholar]
- Wodniok S., Brinkmann H., Glöckner G., Heidel A.J., Philippe H., Melkonian M., Becker B. (2011). Origin of land plants: do conjugating green algae hold the key? BMC Evol. Biol. 11: 104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolf L., Rizzini L., Stracke R., Ulm R., Rensing S.A. (2010). The molecular and physiological responses of Physcomitrella patens to ultraviolet-B radiation. Plant Physiol. 153: 1123–1134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu H.P., Su Y.S., Chen H.C., Chen Y.R., Wu C.C., Lin W.D., Tu S.L. (2014). Genome-wide analysis of light-regulated alternative splicing mediated by photoreceptors in Physcomitrella patens. Genome Biol. 15: R10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu X., Paik I., Zhu L., Huq E. (2015). Illuminating progress in phytochrome-mediated light signaling pathways. Trends Plant Sci. 20: 641–650. [DOI] [PubMed] [Google Scholar]
- Xu X., Paik I., Zhu L., Bu Q., Huang X., Deng X.W., Huq E. (2014). PHYTOCHROME INTERACTING FACTOR1 enhances the E3 ligase activity of CONSTITUTIVE PHOTOMORPHOGENIC1 to synergistically repress photomorphogenesis in Arabidopsis. Plant Cell 26: 1992–2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamawaki S., Yamashino T., Nakanishi H., Mizuno T. (2011). Functional characterization of HY5 homolog genes involved in early light-signaling in Physcomitrella patens. Biosci. Biotechnol. Biochem. 75: 1533–1539. [DOI] [PubMed] [Google Scholar]
- Zhang Y., Mayba O., Pfeiffer A., Shi H., Tepperman J.M., Speed T.P., Quail P.H. (2013). A quartet of PIF bHLH factors provides a transcriptionally centered signaling hub that regulates seedling morphogenesis through differential expression-patterning of shared target genes in Arabidopsis. PLoS Genet. 9: e1003244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zobell O., Coupland G., Reiss B. (2005). The family of CONSTANS-like genes in Physcomitrella patens. Plant Biol (Stuttg.) 7: 266–275. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.