The RLA1/SMOS1 transcription factor acts as an integrator of the transcriptional complex directly downstream of the kinase GSK2 and plays an essential role in BR signaling and development in rice.
Abstract
Brassinosteroids (BRs) are plant-specific steroid hormones that control plant growth and development. Recent studies have identified key components of the BR signaling pathway in Arabidopsis thaliana and in rice (Oryza sativa); however, the mechanism of BR signaling in rice, especially downstream of GSK3/SHAGGY-like kinase (GSK2), remains unclear. Here, we identified a BR-insensitive rice mutant, reduced leaf angle1 (rla1), and cloned the corresponding gene. RLA1 was identical to the previously reported SMALL ORGAN SIZE1 (SMOS1), which was cloned from another allele. RLA1/SMOS1 encodes a transcription factor with an APETALA2 DNA binding domain. Genetic analysis indicated that RLA1/SMOS1 functions as a positive regulator in the BR signaling pathway and is required for the function of BRASSINAZOLE-RESISTANT1 (OsBZR1). In addition, RLA1/SMOS1 can interact with OsBZR1 to enhance its transcriptional activity. GSK2 can interact with and phosphorylate RLA1/SMOS1 to reduce its stability. These results demonstrate that RLA1/SMOS1 acts as an integrator of the transcriptional complex directly downstream of GSK2 and plays an essential role in BR signaling and plant development in rice.
INTRODUCTION
Brassinosteroids (BRs), a class of plant-specific steroid hormones, play diverse roles in growth and development, including hypocotyl elongation, shoot development, leaf development, root development, and male fertility, by regulating cell elongation, cell division, and cell differentiation (Clouse and Sasse, 1998; Yang et al., 2011). The BR signal transduction pathway has been intensively studied in Arabidopsis thaliana (Yang et al., 2011). In the absence of BRs, the BR receptor BRASSINOSTEROID INSENSITIVE1 (BRI1) is inhibited by a negative regulator BRI1 KINASE INHIBITOR1 (BKI1) binding to its C terminus (Li and Chory, 1997; Wang et al., 2005; Wang and Chory, 2006). The downstream negative regulator BRASSINOSTEROID INSENSITIVE2 (BIN2) phosphorylates BRI1-EMS SUPPRESSOR1 (BES1)/BRASSINAZOLE RESISTANT1 (BZR1) to inhibit their transcriptional activity (Li et al., 2001; Li and Nam, 2002; Wang et al., 2002; Yin et al., 2002). In the presence of BRs, the extracellular domain of BRI1 binds to BRs, leading to the dissociation of BKI1 from the plasma membrane to allow formation of an active receptor complex of BRI1 and BRI1-ASSOCIATED RECEPTOR KINASE1 (BAK1) through their trans-phosphorylation (Li et al., 2002; Wang and Chory, 2006; Wang et al., 2014). The phosphorylation activates BRI1-SUPPRESSOR1 through CONSTITUTIVE DIFFERENTIAL GROWTH1, resulting in dephosphorylation and inhibition of BIN2 (Mora-García et al., 2004; Tang et al., 2008; Kim et al., 2009; Yang et al., 2011). As a result, dephosphorylated BES1/BZR1 accumulates in the nucleus (Wang et al., 2002; Yin et al., 2002) and recruits other proteins such as BES1-INTERACTING MYC-LIKE1 (BIM1) and MYB30 to regulate BR-responsive gene expression (Yin et al., 2005; L. Li et al., 2009). Phosphorylated BKI1 can also act as a positive regulator by antagonizing the function of the 14-3-3 proteins on BES1/BZR1 to enhance their accumulation in the nucleus (Wang et al., 2011). Recently, a long isoform of BES1, BES1-L, was identified specifically in Arabidopsis; BES1-L promotes BR signaling more strongly than the canonical isoform BES1-S (Jiang et al., 2015).
In rice (Oryza sativa), some components of the BR signaling pathway have been identified, and the early BR signaling pathway in rice is relatively conserved with the BR signaling pathway in Arabidopsis. For example, OsBRI1 and OsBAK1 are orthologs of AtBRI1 and AtBAK1, respectively (Yamamuro et al., 2000; D. Li et al., 2009). In addition, Oryza sativa GLYCOGEN SYNTHASE KINASE3-LIKE GENE1 (OsGSK1) and GSK3/SHAGGY-like kinase (GSK2) are homologs of AtBIN2 and act as negative regulators of rice BR signaling (Koh et al., 2007; Tong et al., 2012). OsBZR1, a homolog of AtBES1/BZR1, plays a positive role in the rice BR signaling pathway (Bai et al., 2007).
Rice also has some unique transcription factors that play important roles specifically in rice BR signaling and plant architecture. The GRAS-family protein DWARF AND LOW-TILLERING (DLT) acts as a positive regulator downstream of GSK2 in the rice BR signaling pathway (Tong et al., 2009, 2012). LEAF AND TILLER ANGLE INCREASED CONTROLLER (OsLIC), a negative regulator in the rice BR signaling pathway, works antagonistically with OsBZR1 by binding to the promoter of OsBZR1 to repress its transcription. OsLIC also is a target of OsBZR1 and is repressed by OsBZR1 (Wang et al., 2008; Zhang et al., 2012). Other components found in rice may also be related to rice BR signaling. For example, GAI-RGA-SCR19 (OsGRAS19) and BRASSINOSTEROID UPREGULATED1 (OsBU1) play positive roles in BR signaling (Tanaka et al., 2009; Chen et al., 2013). However, how these transcription factors downstream of GSK2 work together in BR signaling is largely unknown.
In cereal crops, brassinosteroids play a key role in regulating plant architecture, which is an important agronomic trait that determines grain yield (Wang and Li, 2005; C. Zhang et al., 2014). For example, BR signaling tightly regulates leaf angle, an important aspect of plant architecture. The BR-deficient rice mutants, including Osdwarf4-1 and d61-7, have erect leaves; also, Osdwarf4-1 mutants have increased grain yield in high-density plantings (Morinaka et al., 2006; Sakamoto et al., 2006). Transgenic rice with slightly reduced expression of OsBRI1 had erect leaves and higher grain yield, but no change in grain size (Morinaka et al., 2006). A recent study discovered that BR signaling regulates leaf erectness in rice through the control of a specific U-Type cyclin and effects on abaxial sclerenchyma cell proliferation of the leaf joint regions (Sun et al., 2015). Therefore, uncovering the mechanisms of BR signaling in rice is important to control rice development and to increase grain yield.
In this study, we identified a dwarf BR-insensitive rice mutant, rla1 (reduced leaf angle1), with erect leaves. Through map-based cloning, we cloned RLA1, which encodes an APETALA2 (AP2)/ERF (ethylene-responsive element binding factor) family transcription factor. Genetic analysis indicated that RLA1 is a positive regulator in the BR signaling pathway and is required for OsBZR1 function, as indicated by the inability of Osbzr1-D to rescue the dwarf phenotype of rla1. Further biochemical study demonstrated that RLA1 interacts with OsBZR1 to enhance its transcription activity, and GSK2 can interact with and phosphorylate RLA1 to reduce its protein accumulation. We also found that RLA1 can interact with DLT. Therefore, our results suggest an intriguing mechanism that RLA1 is a novel and essential component that acts as an integrator of the transcriptional complex directly downstream of GSK2 in the rice BR-signaling pathway.
RESULTS
Characterization of the rla1 Mutant
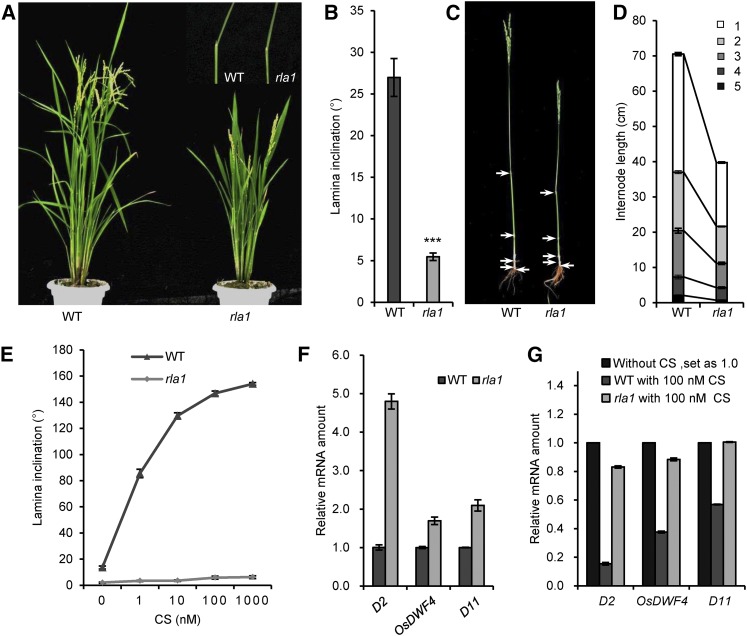
To obtain brassinosteroid-related mutants in rice, we screened a set of T-DNA insertion mutants and identified a dwarf mutant, rla1, with erect leaves. The rla1 mutant exhibited a typical BR loss-of-function phenotype with a semidwarf plant and erect leaves, compared with wild type (Nipponbare, japonica) (Figures 1A and 1B); this phenotype was similar to that of the BR-signaling mutants d61-1 and dlt (Yamamuro et al., 2000; Tong et al., 2009). The dwarf phenotype resulted from the reduced length of each internode (Figures 1C and 1D). In addition, we tested the sensitivity of rla1 to castasterone (CS), the most active BR in rice (Suzuki et al., 1995), and found that rla1 was less sensitive to CS than the wild type in lamina inclination assays (Figure 1E). Furthermore, we measured the expression levels of BR biosynthetic genes, including D2, OsDWF4, and D11, in the wild type and rla1 using RT-qPCR and found that these genes were upregulated in rla1 compared with the wild type (Figure 1F), indicating that the BR signaling is reduced in rla1. We also measured their expression after CS treatment both in the wild type and rla1 and found that the exogenous CS significantly reduced their transcription in the wild type, but not in rla1 (Figure 1G). These results demonstrated that BR signaling was significantly inhibited in the rla1 mutant.
Figure 1.
rla1 Is a BR-Insensitive Mutant.
(A) The morphological phenotypes of the wild type (WT) and rla1 mutants. The pictures at top right show the second lamina joints.
(B) The statistical data of the lamina angle of the second lamina joint. Data are means ± se (n = 25). The comparisons were determined by Student’s t test. ***P < 0.001.
(C) The internode phenotypes of the wild type and rla1 mutants. Arrows show the node positions.
(D) The statistical data of plant height and length of each internode of wild-type and rla1 plants. The numbers 1 to 5 indicate the first to fifth internodes, respectively. Data are means ± se (n = 20).
(E) The statistical data of lamina inclination responding to different CS concentrations in the wild type and rla1. The lamina joint of the first intact leaf were cut from the 1-week-old seedlings grown in the dark and treated with different CS concentrations for 48 h. Data are means ± se (n = 20).
(F) The relative transcript levels of D2, OsDWF4, and D11 in the 2-week-old wild-type and rla1 seedlings. The relative transcript level in wild type was defined as “1.” Data are means ± se (n = 3).
(G) The relative transcript levels of D2, OsDWF4, and D11 in the wild type and rla1 without or with 100 nM CS treatment for 3 h. Total RNAs were extracted from the lamina joints of the first intact leaf. Data are means ± se (n = 3).
Cloning of RLA1 and Complementation Analysis
We crossed the rla1 mutant with its wild type, Nipponbare, and found that the ratio of wild-type:mutant phenotypes in the F2 progeny was 359:109, close to 3:1 (χ2 = 0.729, P > 0.05), indicating that rla1 is a recessive mutation of a single locus. However, using thermal asymmetric interlaced PCR, we failed to isolate the flanking sequence of any potential T-DNA insertion. Since the T-DNA vector used for constructing the mutants contains a hygromycin-resistant gene, we next tested the mutants for hygromycin resistance. To that end, we planted the rla1 seedlings in a solution containing 50 mg/L hygromycin B, but these seedlings did not survive. This suggests that the rla1 mutant may not be caused by a T-DNA insertion, but by a somatic mutation during callus culture.
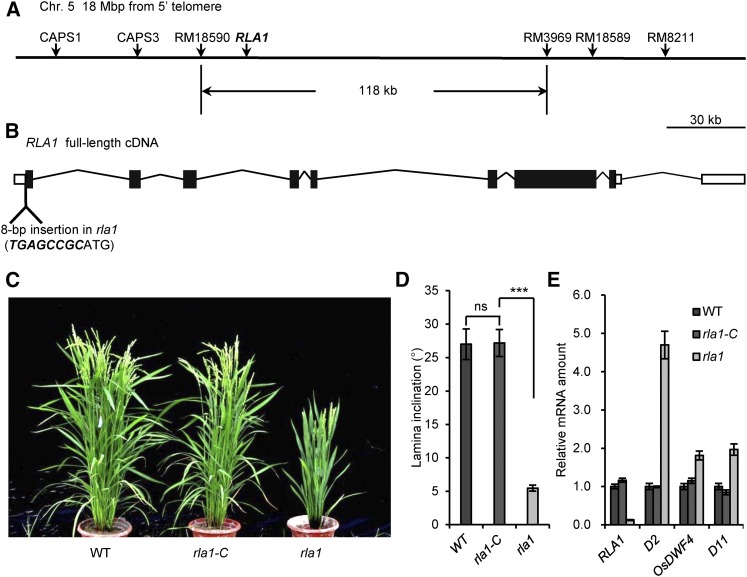
To clone the causal gene for rla1, we then conducted map-based cloning by creating an F2 population from a cross between rla1 and the indica variety 9311. Using 50 dwarf individuals from the F2 population, the rla1 locus was mapped to a 118-kb region on chromosome 5 between two molecular markers, RM18590 and RM3969 (Figure 2A). There are 18 predicted genes in this region, and we then performed RT-qPCR to test whether their transcript levels were affected in the rla1 mutant. We found that Os05g32270 was the most downregulated gene, with only 10% expression in rla1 compared with that in the wild type (Supplemental Figure 1). Genomic sequence analysis revealed an insertion of 8 bp before the first nucleotide of the Os05g32270 coding region (Figure 2B) in rla1. Therefore, we speculated that this small insertion might lead to the mutant phenotype of rla1.
Figure 2.
Map-Based Cloning of RLA1 and Complementation Analysis.
(A) Linkage map of the RLA1 locus. RLA1 is located 18 Mb from the 5′ telomere, between RM18590 and RM3969 on chromosome 5. The vertical arrows indicate the molecular marker positions.
(B) Schematic diagram of the RLA1 locus. The position of the rla1 mutation is shown. The inserted sequence is highlighted with italics and bold font.
(C) Phenotypic complementation by introduction of the full-length RLA1 genomic fragment into rla1 mutants. rla1-C, the transformant of RLA1 in the rla1 background. The rla1-C is a representative line for these transformants.
(D) The statistical data of the lamina angle of the second lamina joint of the wild type, rla1-C, and rla1. Data are means ± se (n = 20). The comparisons were determined by Student’s t test. ***P < 0.001, and “ns” means no significance.
(E) The transcript levels of RLA1, D2, OsDWF4, and D11 in the wild type, rla1-C, and rla1. The transcript level in the wild type was defined as “1.” Data are means ± se (n = 3).
To test this hypothesis, we created a construct containing a 7.5-kb wild-type genomic DNA fragment including the promoter and complete open reading frame regions of RLA1 and introduced it into the rla1 mutant to generate rla1-C plants. We found that most of the rla1-C transgenic plants (12 of 15 lines) were rescued to a wild type-like phenotype (Supplemental Figure 2), and the transcript levels of RLA1 and BR marker genes were reverted to that of the wild type (Figures 2C to 2E). Coincidentally, a previous study had reported the same gene locus (Aya et al., 2014), SMOS1 (SMALL ORGAN SIZE1), which was cloned from a mutant with a dwarf phenotype. They demonstrated that exogenous indole-3-acetic acid treatment could induce the expression of SMOS1 by OsARF1. Together, these findings demonstrated that RLA1/SMOS1 is the gene responsible for the mutant phenotype of rla1.
RLA1/SMOS1 Functions as a Positive Regulator in the BR Signaling Pathway
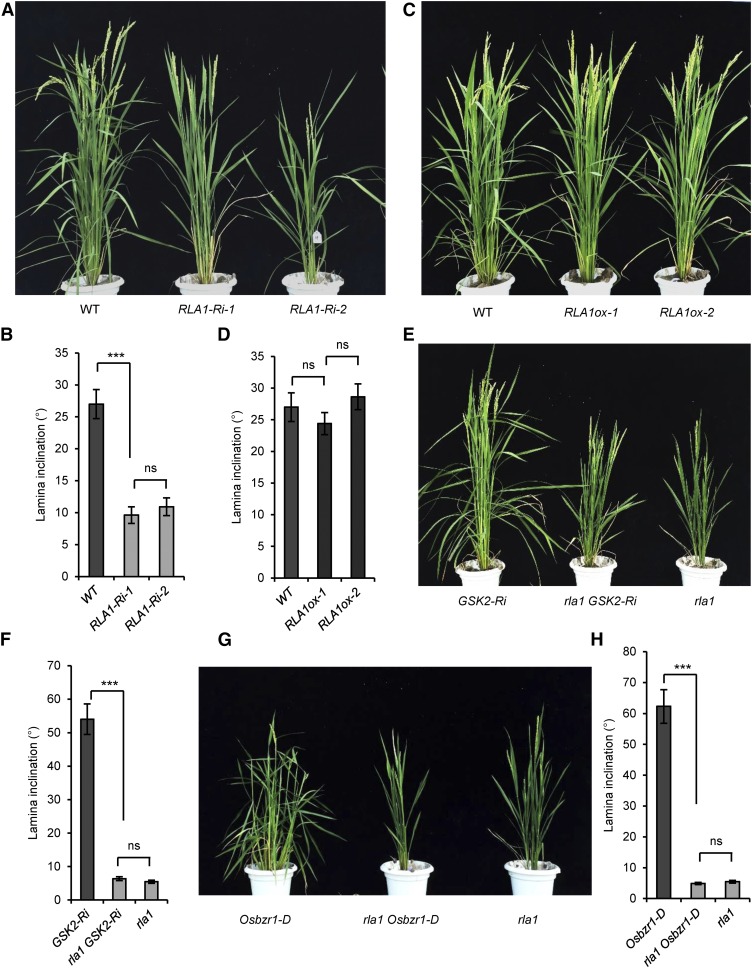
To study the function of RLA1, we generated RLA1 RNA interference lines (RLA1-Ri). The RLA1-Ri plants exhibit a typical BR loss-of-function phenotype similar to rla1 mutants (Figures 3A and 3B), and the RLA1 transcript level was reduced to 20% of the wild type (Supplemental Figure 3A). We also found that the BR biosynthetic genes were upregulated in the RLA1-Ri plants compared with the wild type (Supplemental Figure 3B), which is similar to rla1, indicating that RLA1 may play a positive role in the BR signaling pathway. To examine this, we generated RLA1-overexpressing rice (RLA1ox) with RLA1 driven by the cauliflower mosaic virus 35S promoter. We found that although the transcript levels of RLA1 reached 25-fold higher than that in the wild type (Supplemental Figure 3C), the RLA1ox plants did not display an enhanced leaf inclination phenotype (Figures 3C and 3D), in contrast to Osbzr1-D transgenic rice, which have a gain-of-function mutation of OsBZR1 (Figures 3G and 3H). We measured mRNA levels of the BR biosynthetic genes in the RLA1ox plants and found that the transcript levels of these genes were reduced slightly (Supplemental Figure 3D), suggesting that the enlarged BR signal may be not enough to obtain visible phenotypes. Overexpression of the wild-type RLA1 did not cause an obvious gain-of-function phenotype, such as an enlarged leaf inclination, which was similar to the overexpression of wild type AtBES1 and AtBZR1 in Arabidopsis (Wang et al., 2002; Yin et al., 2002).
Figure 3.
RLA1 Is Required for BR-Regulated Plant Growth and Acts as a Positive Regulator in BR Signaling.
(A) Phenotypes of the RLA1-Ri plants. Twenty-one independent RLA1-Ri lines displayed a similar phenotype.
(B) The statistical data of the lamina angle of the second lamina joint from (A). Data are means ± se (n = 20).
(C) Phenotypes of the RLA1ox lines. Fifteen independent transgenic lines displayed a similar phenotype.
(D) The statistic data of the lamina angle of the second lamina joint from (C). Data are means ± se (n = 20).
(E) Phenotypes of rla1, GSK2-Ri, and the double mutant rla1 GSK2-Ri plants.
(F) The statistic data of the lamina angle of the second lamina joint from (E). Data are means ± se (n = 20).
(G) Phenotypes of Osbzr1-D, rla1, and the double mutant rla1 Osbzr1-D plants.
(H) The statistical data of the lamina angle of the second lamina joint from (G). Data are means ± se (n = 20). For statistical data of (B), (D), (F), and (H), the comparisons were determined by Student’s t test. ***P < 0.001, and “ns” means no significance.
To explore the role of RLA1 in the BR signaling pathway, we crossed rla1 with GSK2-Ri and obtained the homozygous rla1 GSK2-Ri line. The rla1 GSK2-Ri plants displayed phenotypes similar to rla1 (Figures 3E and 3F), indicating that RLA1 acts downstream of GSK2. We also measured expression of BR biosynthetic genes of these three plants and found that the expression level in rla1 GSK2-Ri was similar to rla1, confirming our conclusion (Supplemental Figure 3E). In addition, we crossed rla1 with Osbzr1-D and obtained the rla1 Osbzr1-D homozygous lines. Surprisingly, Osbzr1-D could not rescue the mutant phenotype of rla1, as the rla1 Osbzr1-D plants showed erect leaves and a dwarf phenotype, which is similar to rla1 (Figures 3G and 3H), and the BR synthetic gene expression in rla1 Osbzr1-D is also similar to rla1 (Supplemental Figure 3F), suggesting that RLA1 is required for the OsBZR1-mediated regulation of rice development. These genetic studies demonstrated that RLA1 is a necessary positive regulator in the rice BR signaling pathway.
RLA1/SMOS1 Encodes a Transcriptional Regulator with an AP2 DNA Binding Domain
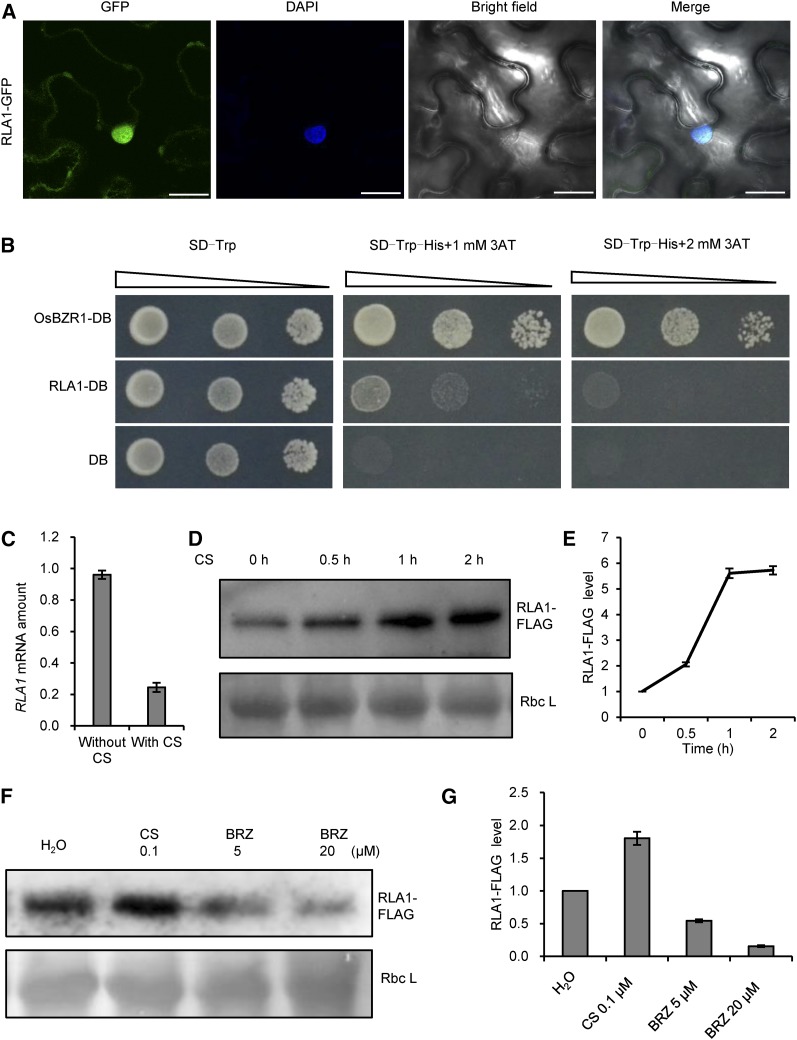
BLAST searches with the RLA1 amino acid sequence revealed that RLA1 contains an AP2/ERF domain and belongs to the DREB subfamily, suggesting that RLA1 might be a transcription factor and bind to the DRE (A/GCCGAC) (Sharoni et al., 2011). Furthermore, examination of RLA1 subcellular localization using a GFP fusion indicated that RLA1-GFP localized mainly in the nucleus and slightly in the cytoplasm in Nicotiana benthamiana pavement cells, which is similar to a previous study on SMOS1-GFP (Figure 4A) (Aya et al., 2014). To test whether RLA1 has transcription activity, we conducted transcription activation assays in yeast by fusing RLA1 with a DNA binding domain in the vector pGBKT7 and transforming it into the yeast strain AH109, which harbors a HIS3 reporter. The results indicated that RLA1 has transcription activity, but its activity is weaker than that of OsBZR1 (Figure 4B).
Figure 4.
Characterization of RLA1 Localization, Activity, and Levels, and Induction of RLA1.
(A) RLA1-GFP mainly localized in the nucleus. Bars = 25 μm.
(B) RLA1 has transcription-activation activity in yeast. The RLA1 coding sequence was cloned into the vector pGBKT7. OsBZR1 was used as a positive control. Transformed yeast were serially diluted and placed on SD (synthetic dropout medium) screening plates containing 3-aminotriazole (3AT).
(C) The transcript level of RLA1 in the wild-type plants treated with 1 μM CS (with CS) or control (without CS) for 2 h. Leaves of 2-week-old seedlings were used for the treatment. The transcript level of control was defined as “1.” Data are means ± se (n = 3).
(D) Time course of RLA1-FLAG protein levels in the RLA1ox line after 1 μM CS treatment. The Ponceau S-stained Rubisco large subunit (Rbc L) was used as a loading control.
(E) Quantification analysis for (D). The relative level of RLA1-FLAG at 0 h was defined as “1.” Data are means ± se (n = 3).
(F) RLA1-FLAG protein levels in the RLA1ox line grown on medium containing CS or BRZ for 8 d. Rbc L was used as a loading control.
(G) Quantification analysis for (F). The relative RLA1-FLAG protein level in plants without treatment was defined as “1.” Data are means ± se (n = 3).
To detect how RLA1 responds to BRs, we tested whether BRs regulate the expression or protein stability of RLA1. In wild-type plants, we found that CS treatment repressed RLA1 expression (Figure 4C). Using the RLA1ox transgenic lines with FLAG-tagged RLA1, we found that CS treatment promoted the accumulation of RLA1-FLAG protein (Figures 4D and 4E). By contrast, when the RLA1ox plants were grown on solution containing brassinozole (BRZ), a BR biosynthesis inhibitor, the RLA1-FLAG protein level decreased (Figures 4F and 4G). This indicated that BRs can enhance RLA1 protein accumulation, and the suppression of RLA1 expression by BR treatment may be caused by a negative feedback regulation, which apparently is a common regulatory mechanism in the BR signaling pathway. When BR signaling output is strong, as a negative feedback, the BR signaling could be suppressed by inhibiting the expression of genes encoding positive signaling components or BR biosynthetic enzymes. The inhibition of RLA1 expression by BR treatment may be due to RLA1 functioning as a positive component in BR signaling, which is similar to DLT and OsBRI1 (Yamamuro et al., 2000; Tong et al., 2009).
RLA1/SMOS1 Interacts with OsBZR1 and Increases Its Activity to Cooperatively Regulate Downstream Genes
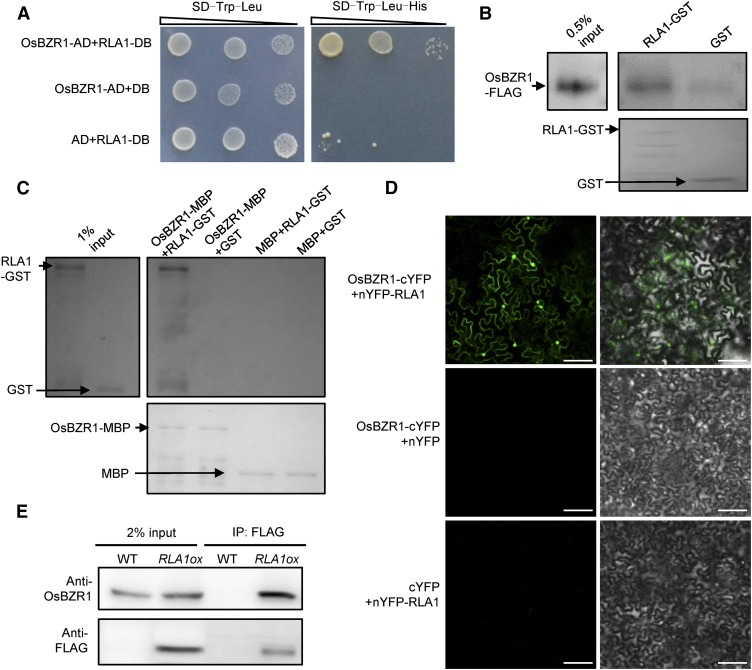
OsBZR1 is a transcription factor downstream of BR signaling and plays important roles in rice development (Bai et al., 2007). We generated OsBZR1 RNA interference lines (OsBZR1-Ri) and found that their phenotypes (reduced leaf angle but not dwarf) are similar to rla1 (Supplemental Figures 4A to 4C). Based on the above findings, we predicted that RLA1 might be a transcriptional regulator required for OsBZR1 function in the BR signaling pathway. Therefore, first, we used yeast two-hybrid assays to test their interaction and found that OsBZR1 and RLA1 can interact with each other (Figure 5A). Second, using semi-in vivo pull-down assays, we found that the RLA1 protein was able to pull down OsBZR1-FLAG protein (Figure 5B). OsBZR1-MBP proteins can also pull down RLA1-GST proteins in vitro (Figure 5C). Third, we conducted bimolecular fluorescence complementation (BiFC) assays with RLA1 fused to the N terminus of YFP (nYFP-RLA1) and OsBZR1 fused to the C terminus of YFP (OsBZR1-cYFP). When the Agrobacterium tumefaciens strains containing both the nYFP-RLA1 and the OsBZR1-cYFP constructs were injected into N. benthamiana leaves, a strong fluorescent signal was observed (Figure 5D). Then we conducted a coimmunoprecipitation (co-IP) assay using RLA1ox (fused with FLAG) plants and wild-type plants as control. The results showed that the OsBZR1 could be detected from the immunoprecipitated proteins of the RLA1ox plants (Figure 5E), indicating that RLA1 and OsBZR1 can interact with each other in vivo. These results indicate that RLA1 can interact with OsBZR1 in vitro and in vivo.
Figure 5.
RLA1 Interacts with OsBZR1 in Vitro and in Vivo.
(A) Interactions between RLA1 and OsBZR1 in yeast two-hybrid assays. The SD/-Trp-Leu-His medium contains 1 mM 3-aminotriazole.
(B) RLA1-GST proteins can pull down OsBZR1-D-FLAG from total protein extracts of Osbzr1-D-FLAG plants. RLA1-GST and GST were stained with Ponceau S as loading controls.
(C) OsBZR1-MBP can pull down RLA1-GST in vitro. OsBZR1-MBP and MBP were stained with Ponceau S as loading controls.
(D) Interaction between RLA1 and OsBZR1 in BiFC assays. Bars = 100 μm.
(E) Interaction between RLA1 and OsBZR1 in the co-IP assays. The proteins were extracted from wild-type or RLA1ox plants and immunoprecipitated by anti-FLAG M2 magnetic beads. Gel blots were probed with anti-FLAG or anti-OsBZR1 antibody.
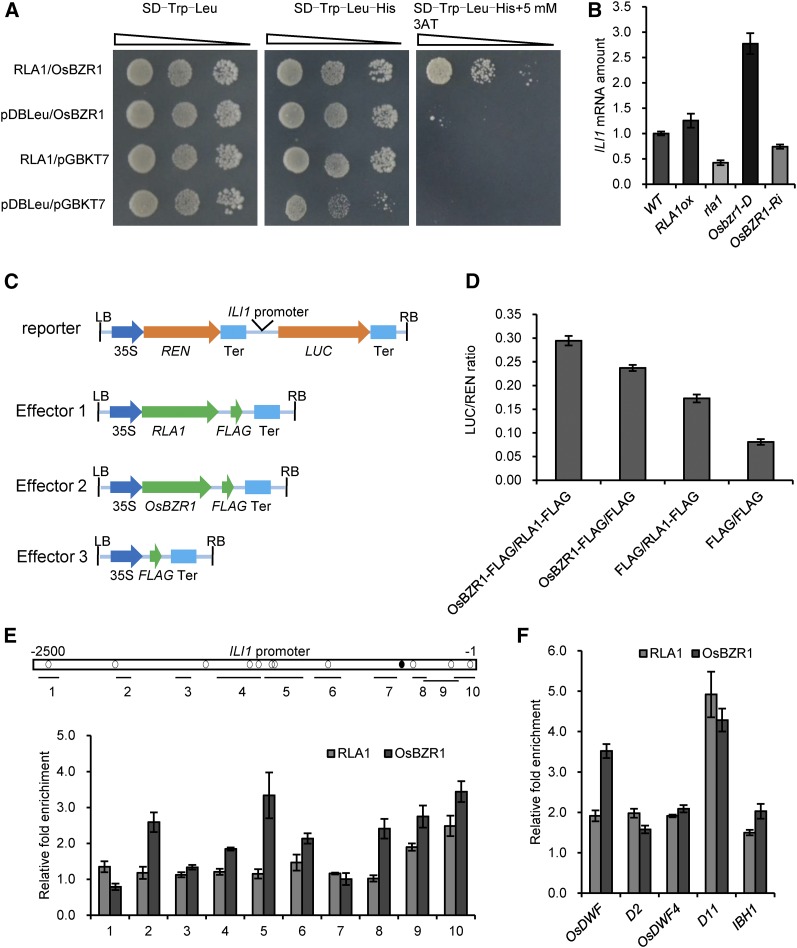
OsBZR1 can directly bind to the promoter regions of a number of genes to regulate their expression (Tong et al., 2009; Zhang et al., 2009; Zhang et al., 2012). To test whether RLA1 affects the transcriptional activity of OsBZR1, we performed transcriptional activation assay in yeasts. To test the activation ability of these proteins, we used the yeast strain AH109, harboring the reporter HIS3, and cotransformed RLA1 or OsBZR1 cloned into the DNA binding vectors pDBLeu and pGBKT7, which produce fusions of the respective proteins to a DNA binding domain for the HIS3 reporter. We cloned RLA1 into the pDBLeu vector and OsBZR1 into pGBKT7. The yeast transformed with both RLA1 and OsBZR1 grew better on medium lacking histidine than those transformed with pDBLeu and OsBZR1 (Figure 6A), suggesting that RLA1 can enhance the transcriptional activity of OsBZR1. Furthermore, we conducted dual-luciferase assays using N. benthamiana leaves as described (Cheng et al., 2014) to confirm that RLA1 can enhance the transcriptional activity of OsBZR1. ILI1 is a direct target of OsBZR1 in rice, and its expression is upregulated by BR treatment (Zhang et al., 2009). We measured the ILI1 expression level in the wild type, rla1, RLA1-ox, Osbzr1-D, and OsBZR1-Ri and found that the expression level of ILI1 is higher in RLA1ox and Osbzr1-D but is lower in rla1 and OsBZR1-Ri lines (Figure 6B). This indicated that ILI1 was regulated both by RLA1 and OsBZR1 in rice. We then used the promoter of ILI1 to drive the luciferase gene (LUC) as a reporter. RLA1-FLAG, OsBZR1-FLAG, and FLAG were used as the effectors (Figure 6C). When we transformed OsBZR1-FLAG into N. benthamiana leaves together with RLA1-FLAG, the LUC reporter expression was enhanced (Figure 6D). These findings suggested that RLA1 can enhance the transcriptional activity of OsBZR1.
Figure 6.
RLA1 Works with OsBZR1 and Enhances Its Transcriptional Activity.
(A) RLA1 enhances OsBZR1 transcriptional activity in yeast. OsBZR1-pGBKT7 and RLA1-pDBLeu were cotransformed in yeast.
(B) The transcript level of ILI1 in the wild type, RLA1ox, rla1, Osbzr1-D, and OsBZR1-Ri. The transcript level in the wild type was defined as “1.” Data are means ± se (n = 3).
(C) Schematic diagrams of the dual-luciferase reporter and effector constructs. The firefly luciferase (LUC) gene driven by the ILI1 promoter was used as the reporter. The Renilla luciferase (REN) reporter gene was controlled by the CaMV promoter (35S) and terminator (Ter). For the effectors, RLA1 and OsBZR1 were fused with FLAG.
(D) Transient gene expression assays in N. benthamiana mesophyll cells. The LUC reporter gene was cotransfected with OsBZR1-FLAG, RLA1-FLAG, or both. Data are means ± se (n = 3).
(E) ChIP assays on binding of RLA1 and OsBZR1 to the ILI1 promoter. Open box shows promoter region of ILI1, black circles show BR response element motifs, and white circles show putative E-box motifs. Regions analyzed by quantitative PCR are shown by short lines marked with numbers (1 to 10). The fold enrichment represents binding efficiency ratio of antibody/no antibody. Data are means ± se (n = 3).
(F) Binding of RLA1 and OsBZR1 to the promoters of OsDWF, D2, OsDWF4, D11, and IBH1. The fold enrichment represents binding efficiency ratio of antibody/no antibody. Data are means ± se (n = 3).
To confirm whether RLA1 is targeted to the promoter of ILI1, we performed chromatin immunoprecipitation (ChIP)-qPCR assays with the RLA1ox and Osbzr1-D transgenic plants using anti-FLAG antibody. Both RLA1 and OsBZR1 binding was enriched on positions 9 and 10 in the ILI1 promoter, which contain two OsBZR1 binding motif E-box (Figure 6E), suggesting that their binding elements may close to each other. Furthermore, we conducted ChIP assays to detect if RLA1 and OsBZR1 can bind the promoters of some BR-responsive genes together. The results indicated that RLA1 and OsBZR1 can bind the promoters of OsDWF, D2, OsDWF4, D11, and IBH1 (Figure 6F), indicating that RLA1 and OsBZR1 likely work together to regulate downstream gene expression. Taken together, RLA1 and OsBZR1 may cooperatively regulate downstream genes in the BR regulatory pathway through forming a transcriptional complex.
GSK2 Interacts with and Phosphorylates RLA1/SMOS1 to Regulate Its Stability
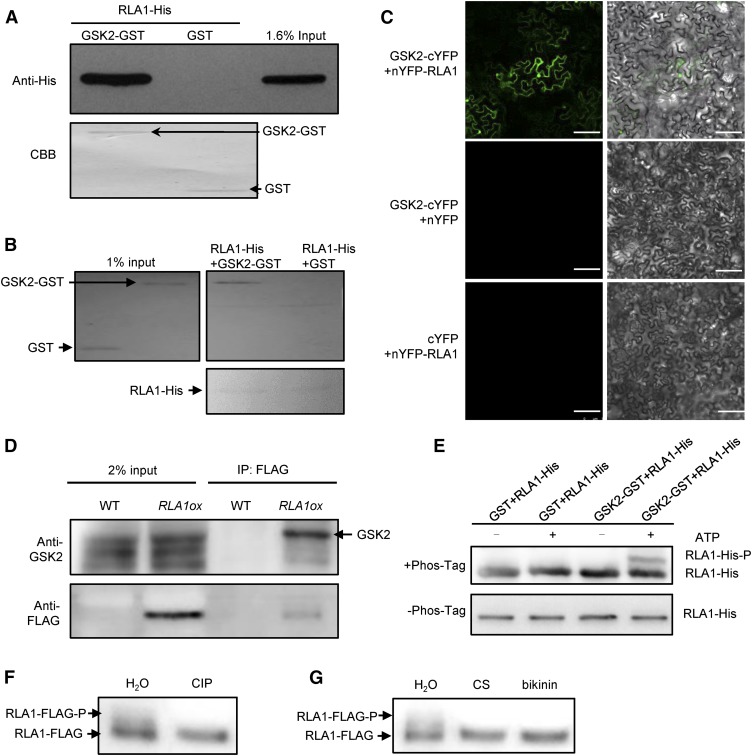
GSK2 is a negative regulator in rice BR signaling and it belongs to the GSK3-like kinase family, whose substrates containing a typical phosphorylation motif (Ser/Thr-X-X-X-Ser/Thr; X is any amino acid) (Youn and Kim, 2015). To understand how BR signaling regulates the accumulation of RLA1, we speculated that the negative regulator GSK2, which is a kinase, may modify RLA1 protein, as BIN2/GSK2 can phosphorylate and regulate a number of transcription factors in plants (D. Zhang et al., 2014). There are 10 potential phosphorylation motifs of GSK2 in RLA1 (Supplemental Figure 5) and these 10 motifs contain 18 potential phosphorylation sites. To test our hypothesis, we expressed and purified the RLA1-His and GSK2-GST fusion proteins in Escherichia coli and found that GSK2-GST can pull down RLA1-His (Figure 7A). Also, RLA1-His can pull down GSK2-GST (Figure 7B). We further conducted BiFC assays using N. benthamiana pavement cells and found that nYFP-RLA1 and GSK2-cYFP can interact with each other (Figure 7C). In addition, we conducted co-IP assays using RLA1ox (fused with FLAG) plants. GSK2 was detected in the immunoprecipitated proteins from the RLA1ox plants (Figure 7D), indicating that RLA1 and GSK2 can interact in vivo. We then performed in vitro kinase assays as described (Cai et al., 2014) to check whether GSK2 can phosphorylate RLA1. We used a Phos-tag approach, where phosphorylated proteins in the gel containing Phos-tag reagent are visualized as bands with slower migration compared with the corresponding dephosphorylated proteins. These assays showed that GSK2-GST can phosphorylate RLA1-His, as indicated by the shifted RLA1-His band detected with anti-His antibody (Figure 7E). To determine if RLA1 can be phosphorylated by GSK2 in vivo, first, we conducted IP assays using anti-FLAG beads from RLA1ox plants and treated the RLA1-FLAG protein with calf intestinal alkaline phosphatase (CIP). To avoid protein degradation, we added MG132 and a cocktail of proteinase inhibitors. To distinguish the phosphorylated and unphosphorylated RLA1-FLAG, we separated proteins by SDS-PAGE for a much longer time. We detected the phosphorylated RLA1-FLAG (RLA1-FLAG-P), and it disappeared when treated with CIP (Figure 7F). Second, we conducted immunoprecipitation assays using anti-FLAG beads from RLA1ox plants growing on medium containing CS and bikinin (GSK2 inhibitor). We found that CS and bikinin treatment can convert RLA1-FLAG-P to RLA1-FLAG (Figure 7G). These results indicated that BRs regulate RLA1 phosphorylation through GSK2 in vivo.
Figure 7.
GSK2 Interacts with and Phosphorylates RLA1 in Vitro and in Vivo.
(A) GSK2-GST proteins can pull down RLA1-His proteins by GST pull-down assays in vitro. GSK2-GST and GST were stained with Ponceau S as loading controls.
(B) RLA1-His proteins can pull down GSK2-GST proteins in vitro. RLA1-His proteins were stained with Ponceau S as loading controls.
(C) Interaction between RLA1 and GSK2 in BiFC assays. Bars = 100 μm.
(D) Interaction between RLA1 and GSK2 in co-IP assays. The proteins were extracted from wild-type or RLA1ox plants and immunoprecipitated by Anti-FLAG magnetic beads. Gel blots were probed with anti-FLAG or anti-GSK2 antibody.
(E) The in vitro kinase assay of RLA1 by GSK2 kinase using a Phos-tag gel. Proteins were detected by immunoblotting with anti-His antibodies. An equal amount of each recombinant protein was separated on the gel without the Phos-tag as a loading control.
(F) Immunoprecipitated RLA1-FLAG protein from RLA1ox plants was treated with CIP or water. The signal was detected by anti-FLAG.
(G) Immunoprecipitated RLA1-FLAG protein from the RLA1ox plants grown on medium containing 100 nM CS or 50 μM bikinin. The signal was detected by anti-FLAG.
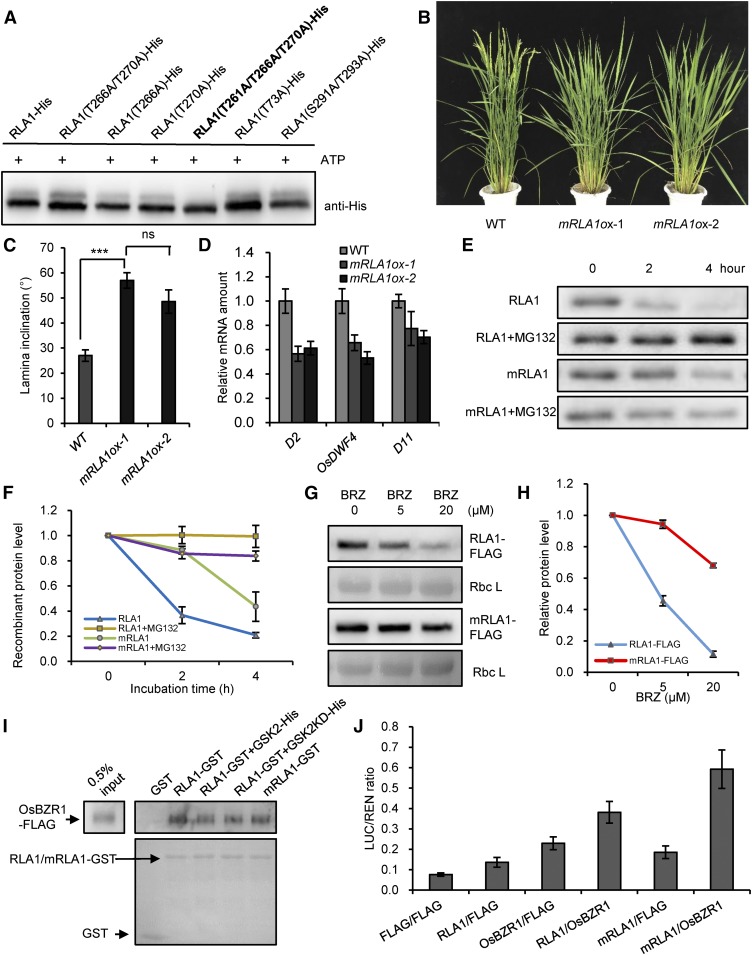
To identify the potential sites where GSK2 phosphorylates RLA1, we conducted mass spectroscopy analysis on RLA1 that had been phosphorylated by GSK2. This revealed several potential phosphorylation sites, including Thr-73, Thr-261, Thr-266, Thr-270, Ser-291, and Thr-293 (Supplemental Figures 6A to 6D). We then constructed mutated forms of RLA1 with alterations in these sites (Ser/Thr to Ala) and conducted kinase assays in vitro. We found that the RLA1T261A/T266A/T270A mutant protein could not be phosphorylated by GSK2 (Figure 8A), indicating that Thr-261, Thr-266, and Thr-270 are the major sites where GSK2 phosphorylates RLA1. To determine whether the phosphorylation of these sites is related to RLA1 function, we generated mRLA1ox (RLA1T261A/T266A/T270A) plants that expressed the mutant RLA1 driven by the CaMV 35S promoter. Interestingly, we found that the mRLA1ox line displayed BR-related gain-of-function phenotypes with enlarged leaf angles as compared with the wild type (Figures 8B and 8C), and the BR synthetic gene expression in the mRLA1ox lines was reduced greatly (Figure 8D), suggesting that BR signaling was enhanced in the mRLA1ox plants, which was different from the RLA1ox lines (Figure 3C). These results indicated that RLA1 was regulated by GSK2 through phosphorylation to transduce BR signals.
Figure 8.
BRs Regulate the Stability of RLA1 Protein through Phosphorylation by GSK2.
(A) GSK2 mainly phosphorylates RLA1 on Thr-261, Thr-266, and Thr-270. An equal amount of recombinant proteins were separated by SDS-PAGE and detected by anti-His antibody. The phosphorylated sites are highlighted with bold font.
(B) Phenotypes of the mRLA1ox (T261A/T266A/T270A) transgenic plants.
(C) The statistical data of the lamina angle of the second lamina joint of plants from (B). Data are means ± se (n = 20). The comparisons were determined by Student’s t test. ***P < 0.001, and “ns” means no significance.
(D) The transcript levels of D2, OsDWF4, and D11 in plants of (B). The transcript level in the wild type was defined as “1.” Data are means ± se (n = 3).
(E) Time course of degradation of RLA1-His and mRLA1-His (T261A/T266A/T270A) in the wild-type plants. In vitro cell-free degradation assays were conducted.
(F) Quantification analysis for (E). The relative levels of RLA1-His or mRLA1-His incubated with wild-type plant protein extracts at 0 h were defined as “1.” Data are means ± se (n = 3).
(G) RLA1-FLAG or mRLA1-FLAG protein levels in the RLA1ox or mRLA1ox lines grown on medium containing BRZ for 8 d. Rbc L was used as a loading control.
(H) Quantification analysis for (G). The relative levels of the RLA1-FLAG/mRLA1-FLAG grown on 0 μM BRZ were defined as “1.” Data are means ± se (n = 3).
(I) Different proteins (RLA1-GST with GSK2 or GSK2-KD proteins, mRLA1-GST) can pull down OsBZR1-D-FLAG from total protein extracts of the Osbzr1-D-FLAG plants. RLA1-GST/mRLA1-GST and GST were stained with Ponceau S as loading controls.
(J) Transient gene expression assays in rice protoplasts. The LUC reporter gene was cotransfected with OsBZR1-FLAG, RLA1-FLAG/mRLA1-FLAG, or both. Data are means ± se (n = 3).
Based on the observation that the RLA1 protein level was enhanced by BRs and inhibited by BRZ (Figures 4D to 4G), we speculated that BRs may regulate the stability of RLA1 through phosphorylation by GSK2. Using a cell-free protein degradation system as described (Wang et al., 2013), we incubated the recombinant proteins RLA1-His or mRLA1-His (RLA1T261A/T266A/T270A) with total protein extracts from the wild-type plants and monitored the amount of RLA1-His and mRLA1-His remaining in the reactions by immunoblotting after incubation at different time points. The results indicated that the degradation rate of the mRLA1-His was much slower than that of RLA1-His, similar to control, which contained 50 μM proteasome inhibitor MG132 (Figures 8E and 8F), indicating that the unphosphorylated form of RLA1 is more stable than the phosphorylated form. Furthermore, to determine the stability of mRLA1 protein in vivo, we detected the mRLA1-FLAG protein level in the mRLA1-ox transgenic plants growing on medium with different concentrations of BRZ (Figures 8G and 8H). The results showed that the degradation rate of the mRLA1-FLAG was much slower than the RLA1-FLAG when treated with BRZ, indicating the mRLA1 protein is more stable than the RLA1 in vivo.
To detect whether the phosphorylation of RLA1 by GSK2 can affect its interaction with OsBZR1, we conducted semi in-vivo pull-down assays to compare the interacting ability of the RLA1-OsBZR1 and the mRLA1–OsBZR1. We also checked the interaction of RLA1-OsBZR1 by adding GSK2 or GSK2-KD (a kinase-dead form of GSK2). The results indicated that the phosphorylation of RLA1 did not influence the interaction of RLA1 with OsBZR1 (Figure 8I). To confirm whether the mRLA1 transcription activity is increased, we conducted dual-luciferase assay using ILI1 promoter-driven LUC as a reporter in rice protoplasts. We found that mRLA1 can enhance the transcription activity of the complex with OsBZR1 more efficiently than RLA1 (Figure 8J). Together, these observations indicate that BRs regulate the stability of RLA1 through phosphorylation by GSK2 to control plant architecture.
DISCUSSION
RLA1/SMOS1 Is an Essential and Positive Component in the Rice BR Signaling Pathway
This study provided several lines of evidence to support the hypothesis that RLA1 is an essential component positively regulating BR signaling in rice. First, the rla1 mutant displayed BR-deficient phenotypes, including dwarfism and erect leaves, which are similar to the phenotypes of BR-insensitive mutants such as d61-1 and dlt (Yamamuro et al., 2000; Tong et al., 2009). Second, the rla1 mutant was insensitive to BR treatment both in the leaf inclination phenotype and in BR-responsive gene expression. Third, genetic analysis demonstrated that RLA1 is required for OsBZR1-mediated rice BR signaling, which differs from the pathway in Arabidopsis. More importantly, we demonstrated that BR signal transduction is dependent on the accumulation of RLA1. RLA1 can bind to the promoters of BR response genes together with OsBZR1. The accumulated RLA1 can interact with and work together with OsBZR1 to regulate BR-responsive gene expression, supporting the key role of RLA1 in mediating the BR signaling pathway in rice. Also, GSK2 can interact with and phosphorylate RLA1 to reduce its stability.
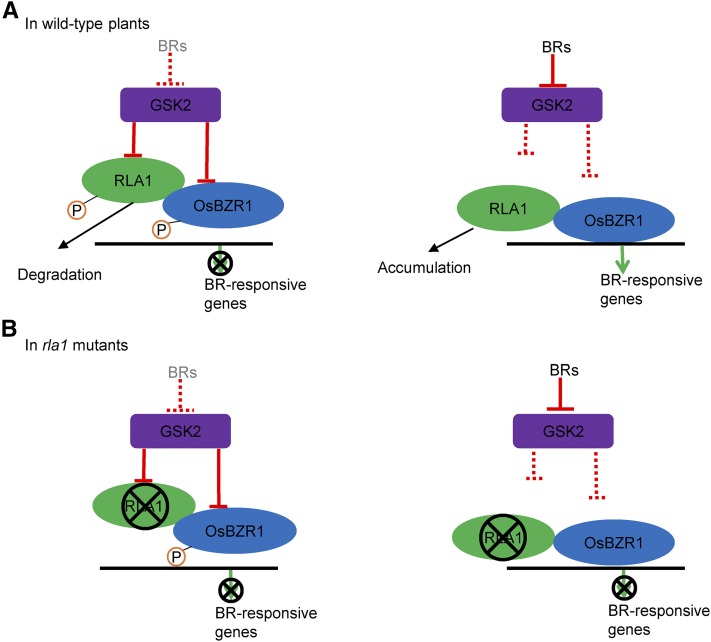
Based on our findings, we proposed a model to illustrate how RLA1 acts with OsBZR1 to regulate BR signaling and development in rice (Figure 9). In wild-type plants, with the low level of BRs, RLA1 and OsBZR1 are phosphorylated and inhibited by GSK2. The phosphorylated RLA1 is unstable, leading to decreased amounts of RLA1 protein (Figures 4F and 4G) and low transcriptional activity of the OsBZR1-RLA1 complex (Figure 9A, left). Following high level of BRs, the activity of GSK2 is inhibited, which leads to dephosphorylation of RLA1 and OsBZR1 and accumulation of RLA1. The accumulated RLA1 can promote the transcription activity of OsBZR1 to regulate BR-responsive gene expression and plant development (Figure 9A, right). In this model, RLA1 and OsBZR1 are direct targets of GSK2 and are required to regulate BR signaling and rice development. In rla1 plants, due to the lack of RLA1, the OsBZR1/RLA complex cannot form to regulate BR-responsive gene expression and rice development, either with or without BRs (Figure 9B). This model illustrates a mechanism for how transcription factors are coregulated by upstream kinases and work interdependently to transfer signals in plants. Although, in mammals, the Wnt/β-Catenin Signaling and Hippo signaling pathways all have the transcriptional complex and regulated by kinases, the mechanism of regulation is different from this model. For Wnt signaling, the TCF/β-Catenin complex controls transcriptional regulation and the protein stability of the transcriptional coactivator β-Catenin is regulated by the upstream kinases GSK3 through phosphorylation (Clevers and Nusse, 2012). For Hippo signaling, activity of the YAP (Yes-associated protein, Yki ortholog) and TAZ (transcriptional coactivator with PDZ binding motif) transcriptional coactivators is repressed through phosphorylation by upstream kinases LATS1/2 (large tumor suppressor kinase 1/2, Wts orthologs). Unphosphorylated YAP/TAZ can interact with TEAD family transcription factors to initiate gene expression and promote cell proliferation and organ growth (Zhao et al., 2008; Mo et al., 2014). However, whether the transcription factor TCF or TEAD is also regulated by upstream kinases in these signaling pathways remains unclear.
Figure 9.
A Proposed Model for RLA1 in Rice BR Signaling.
(A) In wild-type plants, low BR level leads to the phosphorylation of both RLA1 and OsBZR1 by GSK2. The phosphorylated RLA1 is targeted for degradation by an unknown mechanism, and BR-responsive gene expression is blocked (left). With high BR levels, GSK2 activity is inhibited, leading to the accumulation of unphosphorylated RLA1, which activates OsBZR1 transcriptional activity to regulate BR-responsive gene expression (right).
(B) In rla1 plants, because of the lack of RLA1, OsBZR1 cannot activate or inhibit the BR-responsive gene expression with (right) or without (left) BRs.
The Regulation of RLA Protein Stability Is Important for Its Function in BR Signaling
We demonstrated that GSK2 interacts with and phosphorylates RLA1 to regulate its stability. The overexpression of the wild-type RLA1 did not lead to a visible phenotype, which is likely caused by the strict regulation of protein stability by BR signaling. We created the rla1-C transgenic plants by introducing an RLA1 genomic fragment into rla1, and most of the mutant plants were rescued to a wild-type-like phenotype. In addition, the mRNA levels of RLA1 in some rla1-C plants were higher than in the wild type, but the leaf angles are similar to the wild type (Supplemental Figure 2), indicating that in the rla1-C plants, excessive RLA1 cannot accumulate due to the regulation of protein stability. Furthermore, we found that the mRLA1ox lines (a mutant form of RLA1, also fused with FLAG, which can’t be phosphorylated by GSK2 and is more stable than the wild type) displayed a BR-related gain-of-function phenotype with enlarged leaf angles and altered BR marker gene expression, compared with the wild type, indicating that a stable form of RLA1 can lead to a visible phenotype. This phenomenon is very similar to the overexpression of wild-type AtBES1 and AtBZR1 in Arabidopsis (Yin et al., 2002; Wang et al., 2002).
RLA1/SMOS1 May Be a Novel Connection Involved in Crosstalk between Auxin and BR Signaling
A previous study found that auxin can induce the transcription of RLA1/SMOS1 through direct promoter binding by OsARF1 (Aya et al., 2014), which indicated that the transcription of SMOS1/RLA1 is regulated by auxin. Previous studies showed that auxins and BRs have a synergistic effect on plant development. In rice, these studies reported that auxin can improve BR sensitivity in the rice lamina inclination assay, because auxin can induce the transcription of OsBRI1, which encodes a BR receptor, through direct promoter binding by OsARF11 and OsARF19 (Sakamoto et al., 2013; Zhang et al., 2015). In addition, a study of SMOS1 found that a rice homolog of tobacco phosphate-induced protein 1 (OsPHI-1) is a target gene of SMOS1/RLA1, and its knockdown lines exhibited a typical BR loss-of-function phenotype with a semidwarf plant and erect leaves. EXORDIUM, the homolog of OsPHI-1 in Arabidopsis, was induced by BRs (Coll-Garcia et al., 2004). These results all indicated that RLA1/SMOS1 is a component of the BR primary signaling pathway, but not a primary component of the auxin signaling pathway, which could regulate the expression of hundreds to thousands of genes. Therefore, the transcriptional regulation of RLA1/SMOS1 by auxin signaling is mediating their crosstalk, as claimed by Aya et al. (2014).
The Mechanism of BR Signaling Downstream of GSK3s Differs in Rice and Arabidopsis
The GSK3-like kinases in Arabidopsis and rice have several different targets in BR signaling and in diverse developmental processes (Youn and Kim, 2015), indicating significant differences between rice and Arabidopsis signaling pathways, especially in the transcriptional regulation downstream of the GSK3-like kinases. In Arabidopsis, BIM1 and MYB30 coactivate BES1, interacting with BES1 to cooperatively promote BR-induced genes (Yin et al., 2005; L. Li et al., 2009), but whether they are targets of BIN2 is unknown. Recent work identified the transcription factors MYBL2 (MYELOBLASTOSIS-LIKE2) and HAT1 (HOMEOBOX ARABIDOPSIS THALIANA1) as substrates of BIN2 in Arabidopsis (Ye et al., 2012; D. Zhang et al., 2014). Compared with the unphosphorylated form, MYBL2 phosphorylated by BIN2 is more stable and interacts with BES1 to suppress BES1-regulated gene expression (Ye et al., 2012). HAT1 works in a similar way to MYBL2 (D. Zhang et al., 2014), and BIN2-phosphorylated MYBL2 and HAT1 interact with BES1 to cooperatively inhibit BR-repressed gene expression (Ye et al., 2012; D. Zhang et al., 2014). In rice BR signaling, GSK2 directly phosphorylates three transcription factors, OsBZR1, DLT, and OsLIC. OsLIC negatively regulates BR signaling by inhibiting OsBZR1 transcription, and GSK1 regulates the subcellular localization of OsLIC by direct phosphorylation (Zhang et al., 2012). OsBZR1 has a similar function as its ortholog AtBES1/AtBZR1. Also, DLT positively regulates the rice BR-signaling pathway and can be directly phosphorylated by GSK2; BR treatment can induce DLT protein accumulation (Tong et al., 2009, 2012).
Besides BZR1 and DLT, RLA1 is another positive regulator required for rice BR signaling. Therefore, we speculated that these three positive regulators may form a transcriptional complex to regulate rice BR signaling. Because DLT and OsBZR1 cannot interact with each other (Tong et al., 2012), an adaptor may be needed to mediate their interaction. Using yeast two-hybrid assays and BiFC assays, we found that RLA1 can interact with DLT (Supplemental Figures 7A and 7B), suggesting that RLA1 may act as an adaptor to mediate the formation of the OsBZR1-RLA1-DLT complex, which can be phosphorylated by GSK2 respectively. However, further studies are needed to investigate the biochemical and genetic mechanism of how the BZR1, RLA1, and DLT proteins work together to mediate BR signaling and regulate rice development. Moreover, the genome-wide identification of binding sites of the RLA1-OsBZR1 complex and DLT will reveal the complicated transcriptional networks in BR-regulated development in rice, which deserves significant attention in the future.
METHODS
Plant Materials and Growth Conditions
The mutant rla1 was obtained from a T-DNA insertion population. The japonica (Oryza sativa) cultivar Nipponbare was the wild-type control. The plants were grown in the field. The young seedlings were grown in the incubator (Percival) at 28°C for 16 h (day, the light intensity was 120 μmol m−2 s−1) and 26°C for 8 h (night). The etiolated seedlings were grown in the incubator at 30°C under constant dark.
For leaf angle phenotype and the statistical analyses of the mature plants, we defined the flag leaf as the first leaf from the top, and we randomly selected the second lamina joints of the mature plants for analyses.
Vector Construction and Plant Transformation
For the complementation test, a 7.5-kb genomic region, including putative promoter and the complete open reading frame of RLA1, was obtained from the genomic DNA of Nipponbare through PCR, digested with EcoRI and SalI, and inserted into the vector pCAMBIA1300. The vector was introduced into Agrobacterium tumefaciens strain EHA105 and transformed into rla1. The primers used for cloning the RLA1 genomic sequence are listed in Supplemental Table 1.
For creating the RLA1ox and mRLA1ox transgenic plants, the coding sequence of RLA1 was amplified from cDNA of Nipponbare and inserted into the binary vector pCAMBIA1300. The mRLA1 was a mutated form of RLA1 created through PCR. The RLA1 and mRLA1 constructs were driven by the 35S promoter and fused with 3×FLAG tag at the C terminus. For making the RLA1-Ri transgenic plants, the region containing nucleotides 845 to 1044 of the RLA1 coding sequence (CDS) was amplified and inserted into the pTCK303 (Wang et al., 2004) driven by the 35S promoter. For making the OsBZR1-Ri transgenic plants, the region containing nucleotides 31 to 230 of the OsBZR1 CDS was amplified and inserted into the pTCK303 (Wang et al., 2004) driven by the 35S promoter. For making the GSK2-Ri transgenic plants, the region containing nucleotides 644-1053 of the GSK2 CDS was amplified and inserted into the pTCK303 driven by the 35S promoter. For creating the Osbzr1-D transgenic plants, the CDS of OsBZR1 was amplified from cDNA of Nipponbare and Pro-206 was mutated to leucine (Bai et al., 2007) and then inserted into the binary vector pCAMBIA1300. The Osbzr1-D constructs were driven by the native promoter and fused with 3×FLAG tag at the C terminus.
These vectors were introduced into the Agrobacterium strain EHA105 and transformed into Nipponbare.
RNA Extraction and RT-qPCR
Total RNA was extracted from plants using the Tiangen RNA pre plant kit (DP432), and the first-strand cDNA was synthesized using the Takara PrimeScript first-strand cDNA synthesis kit (2641A). For RT-qPCR, cDNAs were combined with SYBR master mix for PCR (S7563; Invitrogen). OsACTIN1 (Os03g50885) was used to normalize the data. The RT-qPCR was performed in triplicate with an Eppendorf cycler. Data were collected and analyzed with an Eppendorf real-time PCR detection system and software. The primers for RT-qPCR are listed in Supplemental Table 1.
CS and BRZ Treatment
For lamina inclination assays, the first intact leaf after germination (1.5 cm long with partial leaf blade and sheath) was excised from the 1-week-old etiolated seedlings of the wild type and rla1 and inserted vertically into a solution with different concentrations of CS or control solution and incubated in the dark for 72 h (Sun et al., 2015). At least 20 seedlings were used for each repeat. For determining marker gene expression, the laminar joints (0.25 cm long with partial leaf blade and sheath) were excised from the 1-week-old etiolated seedlings of the wild type and rla1 and immersed in solution with CS or control in the dark for 3 h. For protein level tests, the leaves of the 2-week-old RLA1ox seedlings were cut into 0.5-cm segments and immersed in 1 μM CS containing 0.1% Triton X-100 at different times.
For BRZ treatment, the 1-week-old plants were moved to medium with different concentrations of BRZ for 8 d.
Transcriptional Activation
The transcriptional activation assay was performed as described (Vert and Chory, 2006). The DNA binding domain vectors pDBLeu and pGBKT7 were used in this assay. The yeast strain AH109 harboring the HIS3 reporter gene was used to test the activation ability. For Figure 4B, OsBZR1 and RLA1 were cloned in the vector pGBKT7 and transformed into the AH109 strain to test transcriptional activation. The vector pGBKT7 was used as the negative control. For Figure 6A, OsBZR1 was cloned into pGBKT7 and RLA1 was cloned into pDBLeu. These two vectors or the corresponding empty vectors were cotransformed into AH109 and grown on medium lacking histidine and with 5 mM 3-aminotriazole.
Yeast Two-Hybrid Assays
The full-length coding sequence of OsBZR1 was cloned into the pAD502 vector, and the full-length coding sequence of RLA1 was cloned into the pDBLeu vector. These two constructs or the corresponding empty vectors were cotransformed into the yeast strain AH109 and grown on SD medium lacking Leu, Trp, and His.
In Vitro and Semi-in Vivo Pull-Down Assays
To test the interaction between RLA1 and GSK2, the full-length coding sequence of GSK2 was cloned into the pGEX-4T-1 vector and transformed into the Escherichia coli strain BL21 to get the GSK2-GST fusion proteins. The full-length coding sequence of RLA1 was cloned into the pET-28a (+) vector and transformed into BL21 to get the RLA1- His fusion proteins. A half microgram (μg) GST-GSK2 or GST was incubated with glutathione resin (L00206; Genscript) at 4°C for 1 h then 0.5 μg RLA1-His was added. The incubation continued for 2 h and the beads were washed several times. The beads were boiled in 1× SDS loading buffer and separated by 12% SDS-PAGE. The anti-His antibody (M20001; Abmart) was used to test the results. In the same way, 0.5 μg RLA1-His was incubated with TALON metal affinity resin (635502; Clontech) to pull down GST-GSK2 and detected by anti-GST (M20007; Abmart).
To test the interaction between RLA1 and OsBZR1, the full-length coding sequence of RLA1 was cloned into the pGEX-4T-1 vector and transformed into E. coli strain BL21 to get RLA1-GST fusion proteins. The OsBZR1-pETMALc-H vector was transformed into BL21 to get OsBZR1-MBP proteins. OsBZR1-MBP (0.5 μg) was incubated with amylose resin (E8021V; New England Biolabs) to pull down RLA1-GST. The 2-week-old Osbzr1-D transgenic seedlings were ground to powder in liquid nitrogen and solubilized with 2× protein extraction buffer (100 mM Tris·HCl, pH 7.5, 300 mM NaCl, 2 mM EDTA, pH 8.0, 1% Trion X-100, 10% [v/v] glycerol, and protease inhibitor mixtures [M307; AMRESCO]). The extracts were centrifuged at 12,000 rpm for 10 min and filtered with a syringe-driven filter unit (SLGP033RB; Millex). RLA1-GST or GST (0.5 μg) was incubated with glutathione resin (L00206; Genscript) for 1 h at 4°C then incubated with plant extracts from Osbzr1-D-FLAG for 2 h at 4°C. The beads were washed three times using 2× protein extraction buffer and boiled in 1× SDS loading buffer for analysis. The anti-FLAG (M20008; Abmart) antibody was used to test the results.
Transient Expression Assays in Nicotiana benthamiana Leaves and Rice Protoplasts
For BiFC assays, the nYFP-RLA1, GSK2-cYFP, or OsBZR1-cYFP constructs or corresponding empty vectors were transformed into Agrobacterium strain GV3101 and coinjected into young leaves of N. benthamiana. The fluorescence was observed by confocal microscopy (Leica) after growth for 1 d in darkness and 2 d under long-day conditions.
For subcellular localization, the RLA1-GFP construct was transformed in the same way and nuclei were stained with 2 μg/mL 4′,6-diamidino-2-phenylindole. GFP was also observed by confocal microscopy (Leica).
For dual-luciferase assay, the experiment was performed as described (Cheng et al., 2014). The ILI1 promoter (2.5 kb) was cloned in the pGreenII 0800-LUC vector as the reporter (Hellens et al., 2005). RLA1-FLAG, OsBZR1-FLAG, and the FLAG were cloned in the pCAMBIA1300 vector as effectors. The effectors and reporter were transformed into GV3101 and coinjected into N. benthamiana leaves. At 4 d after infiltration, the total proteins were extracted (E1910; Promega) for analysis.
The rice protoplast transformation was performed as described previously (Yoo et al., 2007).
In Vitro Kinase Assay
Each kinase assay used 0.5 μg RLA1-His or mutated fusion protein and 0.3 μg GSK2-GST or GST. Kinase reaction buffer was composed of 25 mM Tris (pH 7.4), 12 mM MgCl2, 1 mM DTT, and 1 mM ATP. The reactions were incubated at 37°C for 1 h and boiled with 5× SDS loading buffer then separated by SDS-PAGE with or without 50 μM Phos-tag (Kinoshita et al., 2006). The signals were detected with anti-His antibodies.
Determination of Phosphorylation Sites of RLA1 by GSK2 Kinase
RLA1-His was phosphorylated by GSK2-GST. The phosphorylated RLA1-His was recovered from the SDS-PAGE gel and subjected to in-solution alkylation/tryptic digestion followed by liquid chromatography/tandem mass spectrometry as described (Cai et al., 2014).
Cell-Free Protein Degradation Assay
Two-week-old wild-type seedlings were harvested and ground to a fine power in liquid nitrogen. Total protein was extracted in degradation buffer (25 mM Tris-HCl, pH 7.5, 10 mM NaCl, 10 mM MgCl2, 5 mM DTT, and 10 mM ATP). The same amount of extracts was added to the tubes containing equal amounts of recombinant proteins and incubated for different times.
Coimmunoprecipitation
The 2-week-old seedlings were ground to a fine powder in liquid nitrogen and solubilized with 2× extraction buffer (100 mM Tris-HCl, pH 7.5, 300 mM NaCl, 2 mM EDTA, pH 8.0, 1% Triton X-100, 10% glycerol, and a protease inhibitor cocktail). The extracts were centrifuged at 20,000g for 10 min, and the resultant supernatant was incubated with prewashed anti-FLAG M2 beads (Sigma-Aldrich) for 3 h at 4°C, and then the beads were washed four times with the 2× extraction buffer. The immunoprecipitates were eluted with 1× SDS sample buffer, separated on a 10% SDS-PAGE gel, transferred to nitrocellulose membrane (Amersham Biosciences), and detected with corresponding antibodies. Anti-GSK2 (AbP80050-A-SE) and anti-OsBZR1 (AbP80051-A-SE) were purchased from Beijing Protein Innovation.
ChIP Assays
The 2-week-old seedlings of RLA1ox and Osbzr1-D were cross-linked and used in ChIP assays. ChIP assays were performed as described previously (Zhu et al., 2012). Immunoprecipitation was performed with anti-FLAG antibody (M20008; Abmart). The RLA1-bound chromatin and OsBZR1-bound chromatin were isolated by incubation with Protein A Dynabeads (10002D; Invitrogen) and eluted with elution buffer. The ChIP products were purified and used for quantitative PCR with primers of the ILI1 promoter region and the other genes’ promoters. The primers are listed in Supplemental Table 1.
Accession Numbers
Sequence data from this article can be found in the GenBank/EMBL data libraries under the following accession numbers: RLA1, Os05g32270; OsBZR1, Os07g39220; GSK2 OS05g11730; ILI1, Os04g54900; DLT, OS06g03710; OsACTIN1, Os03g50885; D2, Os01g10040; OsDWF4, Os03g12660; D11, Os04g39430; OsDWF, Os03g40540; and IBH1, OS04g56500.
Supplemental Data
Supplemental Figure 1. Transcript Levels of Genes Located in the Region between the Two Markers RM18590 and RM3969 on Chromosome 5.
Supplemental Figure 2. Phenotypes and RLA1 Transcript Levels of the rla1-C Plants.
Supplemental Figure 3. Relative Gene Expression Analysis from Plants.
Supplemental Figure 4. Phenotypes of Ri-OsBZR1.
Supplemental Figure 5. There Are 10 Potential Phosphorylation Motifs of GSK2 in RLA1.
Supplemental Figure 6. Identification of RLA1 Phosphorylation Sites by GSK2 Kinase Using LC-MS/MS.
Supplemental Figure 7. RLA1 Interacts with DLT.
Supplemental Table 1. Primers Used in This Study.
Supplementary Material
Acknowledgments
We thank Yanhai Yin (Iowa State University) for providing the construct OsBZR1-pETMALc-H; Zhenying Cai (Fudan University) for helping to conduct in vitro kinase assays; and Yinwei Cheng and Yuan Wang (Fudan University) for helping with transient expression assays in N. benthamiana leaves. This work was supported by Grants 91535104 and 31430046 (to X.W.), and 2016YFD0100403, 31271684, and 31540080 (to S.S.) of the National Natural Science Foundation of China, Grant 2012CB114304 of the Ministry of Science and Technology of China (to X.W. and S.S.), and Grants 2662015PY020 and 2014RC002 of Huazhong Agricultural University (to X.W.).
AUTHOR CONTRIBUTIONS
S.Q. and S.S. conceived the project and design, acquired data, analyzed and interpreted data, and drafted and revised the article. L.W., C.L., Z.W., X.L., T.W., and L.L., acquired, analyzed, and interpreted data. W.T. and T.L. contributed unpublished essential data or reagents. X.W. conceived the project and design, analyzed and interpreted data, and drafted and revised the article.
Glossary
- BR
brassinosteroid
- CS
castasterone
- BRZ
brassinozole
- co-IP
coimmunoprecipitation
- BiFC
bimolecular fluorescence complementation
- ChIP
chromatin immunoprecipitation
- CIP
calf intestinal alkaline phosphatase
- CDS
coding sequence
References
- Aya K., Hobo T., Sato-Izawa K., Ueguchi-Tanaka M., Kitano H., Matsuoka M. (2014). A novel AP2-type transcription factor, SMALL ORGAN SIZE1, controls organ size downstream of an auxin signaling pathway. Plant Cell Physiol. 55: 897–912. [DOI] [PubMed] [Google Scholar]
- Bai M.Y., Zhang L.Y., Gampala S.S., Zhu S.W., Song W.Y., Chong K., Wang Z.Y. (2007). Functions of OsBZR1 and 14-3-3 proteins in brassinosteroid signaling in rice. Proc. Natl. Acad. Sci. USA 104: 13839–13844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai Z., Liu J., Wang H., Yang C., Chen Y., Li Y., Pan S., Dong R., Tang G., Barajas-Lopez Jde.D., Fujii H., Wang X. (2014). GSK3-like kinases positively modulate abscisic acid signaling through phosphorylating subgroup III SnRK2s in Arabidopsis. Proc. Natl. Acad. Sci. USA 111: 9651–9656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen L., Xiong G., Cui X., Yan M., Xu T., Qian Q., Xue Y., Li J., Wang Y. (2013). OsGRAS19 may be a novel component involved in the brassinosteroid signaling pathway in rice. Mol. Plant 6: 988–991. [DOI] [PubMed] [Google Scholar]
- Cheng Y., Zhu W., Chen Y., Ito S., Asami T., Wang X. (2014). Brassinosteroids control root epidermal cell fate via direct regulation of a MYB-bHLH-WD40 complex by GSK3-like kinases. eLife, 10.7554/eLife.02525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clevers H., Nusse R. (2012). Wnt/β-catenin signaling and disease. Cell 149: 1192–1205. [DOI] [PubMed] [Google Scholar]
- Clouse S.D., Sasse J.M. (1998). Brassinosteroids: essential regulators of plant growth and development. Annu. Rev. Plant Physiol. Plant Mol. Biol. 49: 427–451. [DOI] [PubMed] [Google Scholar]
- Coll-Garcia D., Mazuch J., Altmann T., Müssig C. (2004). EXORDIUM regulates brassinosteroid-responsive genes. FEBS Lett. 563: 82–86. [DOI] [PubMed] [Google Scholar]
- Hellens R.P., Allan A.C.A., Friel E.N., Bolitho K., Grafton K., Templeton M.D., Karunairetnam S., Gleave A.P., Laing W.A. (2005). Transient expression vectors for functional genomics, quantification of promoter activity and RNA silencing in plants. Plant Methods 1: 13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang J., Zhang C., Wang X. (2015). A recently evolved isoform of the transcription factor BES1 promotes brassinosteroid signaling and development in Arabidopsis thaliana. Plant Cell 27: 361–374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim T.W., Guan S., Sun Y., Deng Z., Tang W., Shang J.X., Sun Y., Burlingame A.L., Wang Z.Y. (2009). Brassinosteroid signal transduction from cell-surface receptor kinases to nuclear transcription factors. Nat. Cell Biol. 11: 1254–1260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kinoshita E., Kinoshita-Kikuta E., Takiyama K., Koike T. (2006). Phosphate-binding tag, a new tool to visualize phosphorylated proteins. Mol. Cell. Proteomics 5: 749–757. [DOI] [PubMed] [Google Scholar]
- Koh S., Lee S.C., Kim M.K., Koh J.H., Lee S., An G., Choe S., Kim S.R. (2007). T-DNA tagged knockout mutation of rice OsGSK1, an orthologue of Arabidopsis BIN2, with enhanced tolerance to various abiotic stresses. Plant Mol. Biol. 65: 453–466. [DOI] [PubMed] [Google Scholar]
- Li D., Wang L., Wang M., Xu Y.Y., Luo W., Liu Y.J., Xu Z.H., Li J., Chong K. (2009). Engineering OsBAK1 gene as a molecular tool to improve rice architecture for high yield. Plant Biotechnol. J. 7: 791–806. [DOI] [PubMed] [Google Scholar]
- Li J., Chory J. (1997). A putative leucine-rich repeat receptor kinase involved in brassinosteroid signal transduction. Cell 90: 929–938. [DOI] [PubMed] [Google Scholar]
- Li J., Nam K.H. (2002). Regulation of brassinosteroid signaling by a GSK3/SHAGGY-like kinase. Science 295: 1299–1301. [DOI] [PubMed] [Google Scholar]
- Li J., Nam K.H., Vafeados D., Chory J. (2001). BIN2, a new brassinosteroid-insensitive locus in Arabidopsis. Plant Physiol. 127: 14–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J., Wen J., Lease K.A., Doke J.T., Tax F.E., Walker J.C. (2002). BAK1, an Arabidopsis LRR receptor-like protein kinase, interacts with BRI1 and modulates brassinosteroid signaling. Cell 110: 213–222. [DOI] [PubMed] [Google Scholar]
- Li L., Yu X., Thompson A., Guo M., Yoshida S., Asami T., Chory J., Yin Y. (2009). Arabidopsis MYB30 is a direct target of BES1 and cooperates with BES1 to regulate brassinosteroid-induced gene expression. Plant J. 58: 275–286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mo J.S., Park H.W., Guan K.L. (2014). The Hippo signaling pathway in stem cell biology and cancer. EMBO Rep. 15: 642–656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mora-García S., Vert G., Yin Y., Caño-Delgado A., Cheong H., Chory J. (2004). Nuclear protein phosphatases with Kelch-repeat domains modulate the response to brassinosteroids in Arabidopsis. Genes Dev. 18: 448–460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morinaka Y., Sakamoto T., Inukai Y., Agetsuma M., Kitano H., Ashikari M., Matsuoka M. (2006). Morphological alteration caused by brassinosteroid insensitivity increases the biomass and grain production of rice. Plant Physiol. 141: 924–931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakamoto T., Morinaka Y., Inukai Y., Kitano H., Fujioka S. (2013). Auxin signal transcription factor regulates expression of the brassinosteroid receptor gene in rice. Plant J. 73: 676–688. [DOI] [PubMed] [Google Scholar]
- Sakamoto T., et al. (2006). Erect leaves caused by brassinosteroid deficiency increase biomass production and grain yield in rice. Nat. Biotechnol. 24: 105–109. [DOI] [PubMed] [Google Scholar]
- Sharoni A.M., Nuruzzaman M., Satoh K., Shimizu T., Kondoh H., Sasaya T., Choi I.R., Omura T., Kikuchi S. (2011). Gene structures, classification and expression models of the AP2/EREBP transcription factor family in rice. Plant Cell Physiol. 52: 344–360. [DOI] [PubMed] [Google Scholar]
- Sun S., Chen D., Li X., Qiao S., Shi C., Li C., Shen H., Wang X. (2015). Brassinosteroid signaling regulates leaf erectness in Oryza sativa via the control of a specific U-type cyclin and cell proliferation. Dev. Cell 34: 220–228. [DOI] [PubMed] [Google Scholar]
- Suzuki H., Fujioka S., Takatsuto S., Yokota T., Murofushi N., Sakurai A. (1995). Biosynthesis of brassinosteroids in seedlings of Catharanthus roseus, Nicotiana tabacum, and Oryza sativa. Biosci. Biotechnol. Biochem. 59: 168–172. [Google Scholar]
- Tanaka A., et al. (2009). BRASSINOSTEROID UPREGULATED1, encoding a helix-loop-helix protein, is a novel gene involved in brassinosteroid signaling and controls bending of the lamina joint in rice. Plant Physiol. 151: 669–680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang W., Kim T.W., Oses-Prieto J.A., Sun Y., Deng Z., Zhu S., Wang R., Burlingame A.L., Wang Z.Y. (2008). BSKs mediate signal transduction from the receptor kinase BRI1 in Arabidopsis. Science 321: 557–560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tong H., Liu L., Jin Y., Du L., Yin Y., Qian Q., Zhu L., Chu C. (2012). DWARF AND LOW-TILLERING acts as a direct downstream target of a GSK3/SHAGGY-like kinase to mediate brassinosteroid responses in rice. Plant Cell 24: 2562–2577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tong H., Jin Y., Liu W., Li F., Fang J., Yin Y., Qian Q., Zhu L., Chu C. (2009). DWARF AND LOW-TILLERING, a new member of the GRAS family, plays positive roles in brassinosteroid signaling in rice. Plant J. 58: 803–816. [DOI] [PubMed] [Google Scholar]
- Vert G., Chory J. (2006). Downstream nuclear events in brassinosteroid signalling. Nature 441: 96–100. [DOI] [PubMed] [Google Scholar]
- Wang H., Yang C., Zhang C., Wang N., Lu D., Wang J., Zhang S., Wang Z.X., Ma H., Wang X. (2011). Dual role of BKI1 and 14-3-3 s in brassinosteroid signaling to link receptor with transcription factors. Dev. Cell 21: 825–834. [DOI] [PubMed] [Google Scholar]
- Wang J., Jiang J., Wang J., Chen L., Fan S.L., Wu J.W., Wang X., Wang Z.X. (2014). Structural insights into the negative regulation of BRI1 signaling by BRI1-interacting protein BKI1. Cell Res. 24: 1328–1341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L., Xu Y., Zhang C., Ma Q., Joo S.H., Kim S.K., Xu Z., Chong K. (2008). OsLIC, a novel CCCH-type zinc finger protein with transcription activation, mediates rice architecture via brassinosteroids signaling. PLoS One 3: e3521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X., Chory J. (2006). Brassinosteroids regulate dissociation of BKI1, a negative regulator of BRI1 signaling, from the plasma membrane. Science 313: 1118–1122. [DOI] [PubMed] [Google Scholar]
- Wang X., Li X., Meisenhelder J., Hunter T., Yoshida S., Asami T., Chory J. (2005). Autoregulation and homodimerization are involved in the activation of the plant steroid receptor BRI1. Dev. Cell 8: 855–865. [DOI] [PubMed] [Google Scholar]
- Wang Y., Li J. (2005). The plant architecture of rice (Oryza sativa). Plant Mol. Biol. 59: 75–84. [DOI] [PubMed] [Google Scholar]
- Wang Y., Sun S., Zhu W., Jia K., Yang H., Wang X. (2013). Strigolactone/MAX2-induced degradation of brassinosteroid transcriptional effector BES1 regulates shoot branching. Dev. Cell 27: 681–688. [DOI] [PubMed] [Google Scholar]
- Wang Z., Chen C., Xu Y., Jiang R., Han Y., Xu Z., Chong K. (2004). A practical vector for efficient knockdown of gene expression in rice (Oryza sativa L.). PLANT. Mol. Biol. Rep. 22: 409–417. [Google Scholar]
- Wang Z.Y., Nakano T., Gendron J., He J., Chen M., Vafeados D., Yang Y., Fujioka S., Yoshida S., Asami T., Chory J. (2002). Nuclear-localized BZR1 mediates brassinosteroid-induced growth and feedback suppression of brassinosteroid biosynthesis. Dev. Cell 2: 505–513. [DOI] [PubMed] [Google Scholar]
- Yamamuro C., Ihara Y., Wu X., Noguchi T., Fujioka S., Takatsuto S., Ashikari M., Kitano H., Matsuoka M. (2000). Loss of function of a rice brassinosteroid insensitive1 homolog prevents internode elongation and bending of the lamina joint. Plant Cell 12: 1591–1606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang C.J., Zhang C., Lu Y.N., Jin J.Q., Wang X.L. (2011). The mechanisms of brassinosteroids’ action: from signal transduction to plant development. Mol. Plant 4: 588–600. [DOI] [PubMed] [Google Scholar]
- Ye H., Li L., Guo H., Yin Y. (2012). MYBL2 is a substrate of GSK3-like kinase BIN2 and acts as a corepressor of BES1 in brassinosteroid signaling pathway in Arabidopsis. Proc. Natl. Acad. Sci. USA 109: 20142–20147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin Y., Vafeados D., Tao Y., Yoshida S., Asami T., Chory J. (2005). A new class of transcription factors mediates brassinosteroid-regulated gene expression in Arabidopsis. Cell 120: 249–259. [DOI] [PubMed] [Google Scholar]
- Yin Y., Wang Z.Y., Mora-Garcia S., Li J., Yoshida S., Asami T., Chory J. (2002). BES1 accumulates in the nucleus in response to brassinosteroids to regulate gene expression and promote stem elongation. Cell 109: 181–191. [DOI] [PubMed] [Google Scholar]
- Yoo S.D., Cho Y.H., Sheen J. (2007). Arabidopsis mesophyll protoplasts: a versatile cell system for transient gene expression analysis. Nat. Protoc. 2: 1565–1572. [DOI] [PubMed] [Google Scholar]
- Youn J.H., Kim T.W. (2015). Functional insights of plant GSK3-like kinases: multi-taskers in diverse cellular signal transduction pathways. Mol. Plant 8: 552–565. [DOI] [PubMed] [Google Scholar]
- Zhang C., Bai M.Y., Chong K. (2014). Brassinosteroid-mediated regulation of agronomic traits in rice. Plant Cell Rep. 33: 683–696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang C., Xu Y., Guo S., Zhu J., Huan Q., Liu H., Wang L., Luo G., Wang X., Chong K. (2012). Dynamics of brassinosteroid response modulated by negative regulator LIC in rice. PLoS Genet. 8: e1002686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang D., Ye H., Guo H., Johnson A., Zhang M., Lin H., Yin Y. (2014). Transcription factor HAT1 is phosphorylated by BIN2 kinase and mediates brassinosteroid repressed gene expression in Arabidopsis. Plant J. 77: 59–70. [DOI] [PubMed] [Google Scholar]
- Zhang L.Y., et al. (2009). Antagonistic HLH/bHLH transcription factors mediate brassinosteroid regulation of cell elongation and plant development in rice and Arabidopsis. Plant Cell 21: 3767–3780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang S., Wang S., Xu Y., Yu C., Shen C., Qian Q., Geisler M., Jiang A., Qi Y. (2015). The auxin response factor, OsARF19, controls rice leaf angles through positively regulating OsGH3-5 and OsBRI1. Plant Cell Environ. 38: 638–654. [DOI] [PubMed] [Google Scholar]
- Zhao B., Ye X., Yu J., Li L., Li W., Li S., Yu J., Lin J.D., Wang C.Y., Chinnaiyan A.M., Lai Z.C., Guan K.L. (2008). TEAD mediates YAP-dependent gene induction and growth control. Genes Dev. 22: 1962–1971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu J.Y., Sun Y., Wang Z.Y. (2012). Genome-wide identification of transcription factor-binding sites in plants using chromatin immunoprecipitation followed by microarray (ChIP-chip) or sequencing (ChIP-seq). Methods Mol. Biol. 876: 173–188. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.