OsCUL3a interacts with and degrades OsNPR1 to regulate innate immunity in rice.
Abstract
Cullin3-based RING E3 ubiquitin ligases (CRL3), composed of Cullin3 (CUL3), RBX1, and BTB proteins, are involved in plant immunity, but the function of CUL3 in the process is largely unknown. Here, we show that rice (Oryza sativa) OsCUL3a is important for the regulation of cell death and immunity. The rice lesion mimic mutant oscul3a displays a significant increase in the accumulation of flg22- and chitin-induced reactive oxygen species, and in pathogenesis-related gene expression as well as resistance to Magnaporthe oryzae and Xanthomonas oryzae pv oryzae. We cloned the OsCUL3a gene via a map-based strategy and found that the lesion mimic phenotype of oscul3a is associated with the early termination of OsCUL3a protein. Interaction assays showed that OsCUL3a interacts with both OsRBX1a and OsRBX1b to form a multisubunit CRL in rice. Strikingly, OsCUL3a interacts with and degrades OsNPR1, which acts as a positive regulator of cell death in rice. Accumulation of OsNPR1 protein is greater in the oscul3a mutant than in the wild type. Furthermore, the oscul3a osnpr1 double mutant does not exhibit the lesion mimic phenotype of the oscul3a mutant. Our data demonstrate that OsCUL3a negatively regulates cell death and immunity by degrading OsNPR1 in rice.
INTRODUCTION
Plants have evolved a two-layered immune system to protect themselves from pathogen attack: PAMP-triggered immunity and effector-triggered immunity (ETI) (Schwessinger and Zipfel, 2008; Faulkner and Robatzek, 2012). One hallmark of ETI is the hypersensitive response, in which the infection site rapidly undergoes programmed cell death (PCD) that arrests the growth and spread of the pathogen (Wu et al., 2014; Zebell and Dong, 2015). Given its severe consequences for cell fate and its effectiveness in halting pathogen ingress, PCD must be tightly regulated in the absence of pathogen attack (Mukhtar et al., 2016).
The ubiquitin/proteasome pathway is a major system for the selective degradation of proteins in eukaryotes. Previous studies in plants have demonstrated that the ubiquitin/proteasome pathway is an essential component of the regulatory networks controlling many important cellular processes, including growth and development (Figueroa et al., 2005; Hu et al., 2014), as well as defense responses (Zeng et al., 2004). Among the three major enzymes in this pathway, the E3 ubiquitin ligases are responsible for controlling the specificity of target protein recognition and degradation via the 26S proteasome. A prominent subset of the E3 ligases are the Cullin-RING E3 ligases (CRLs), in which the Cullin3 (CUL3) protein acts as a scaffold for the binding of both a Ring-finger domain protein, RING-BOX1 (RBX1), and a substrate adaptor BTB/POZ domain protein. These proteins together constitute a CRL3 complex (Hua and Vierstra, 2011). CUL3 is known to function in the regulation of PCD in animals. The death-associated protein kinase (DAPK) enzyme is an apoptosis mediator involved in interferon-induced PCD in mammalian cells. When triggered by interferon, the BTB domain protein KLHL20, which acts as the adaptor between CUL3 and DAPK, is sequestered in promyelocytic leukemia nuclear bodies, thus preventing its interaction with DAPK and stabilizing the kinase (Lee et al., 2010). In Drosophila melanogaster, CUL3-BTBSPOP mediates degradation of the Jun kinase phosphatase Puckered, thereby inducing tumor necrosis factor-dependent apoptosis (Liu et al., 2009). In Arabidopsis thaliana, simultaneous disruption of both CUL3a and CUL3b causes an embryo-lethal phenotype that affects both embryo patterning and endosperm development (Figueroa et al., 2005; Thomann et al., 2005). Whether CUL3a and CUL3b are involved in rice (Oryza sativa) PCD is not known.
In plants, the transcription coactivator NPR1 (NONEXPRESSOR OF PATHOGENESIS-RELATED GENES1) is a master regulator of both basal and systemic acquired resistance (SAR) and was first identified from an Arabidopsis mutant impaired in SAR (Cao et al., 1994, 1997). Overexpression of AtNPR1 in Arabidopsis leads to enhanced disease resistance to both bacterial and oomycete pathogens in a dose-dependent manner (Cao et al., 1998). Although AtNPR1 acts as a positive regulator of immunity, its 26S proteasome-mediated turnover is essential for its full coactivator activity in SAR (Spoel et al., 2009; Fu et al., 2012; Saleh et al., 2015). Similarly, OsNPR1 or NH1 overexpression rice plants have enhanced resistance to the bacterial pathogen Xanthomonas oryzae pv oryzae (Xoo), elevated expression levels of pathogenesis-related (PR) genes, and a benzothiadiazole-mediated cell death phenotype (Chern et al., 2005; Yuan et al., 2007; Bai et al., 2011). Although OsNPR1 has been identified as a positive regulator of resistance against bacterial pathogens, its protein turnover and molecular mechanism remain largely unknown.
In this study, we identified a rice lesion mimic mutant, oscul3a, in a mutant collection. Using a map-based cloning strategy, we isolated the gene that encodes OsCUL3a. Unlike Arabidopsis, in which CUL3a and CUL3b are largely functionally redundant, the lesion mimic mutant oscul3a showed a severe cell death phenotype at the tillering stage and had enhanced resistance to both the rice blast pathogen Magnaporthe oryzae and the bacterial blight pathogen Xoo. We establish that OsCUL3a interacts with OsRBX1a and OsRBX1b to form a CUL-Ring-like E3 ubiquitin ligase complex. We found that this complex promotes the degradation of OsNPR1 via the 26S proteasome. The cell death phenotype was blocked by the knockout of OsNPR1 in the oscul3a background. Our results demonstrate that OsCUL3a negatively regulates cell death and plant innate immunity by interacting with and degrading OsNPR1 in rice.
RESULTS
Phenotypic Characterization of the oscul3a Mutant
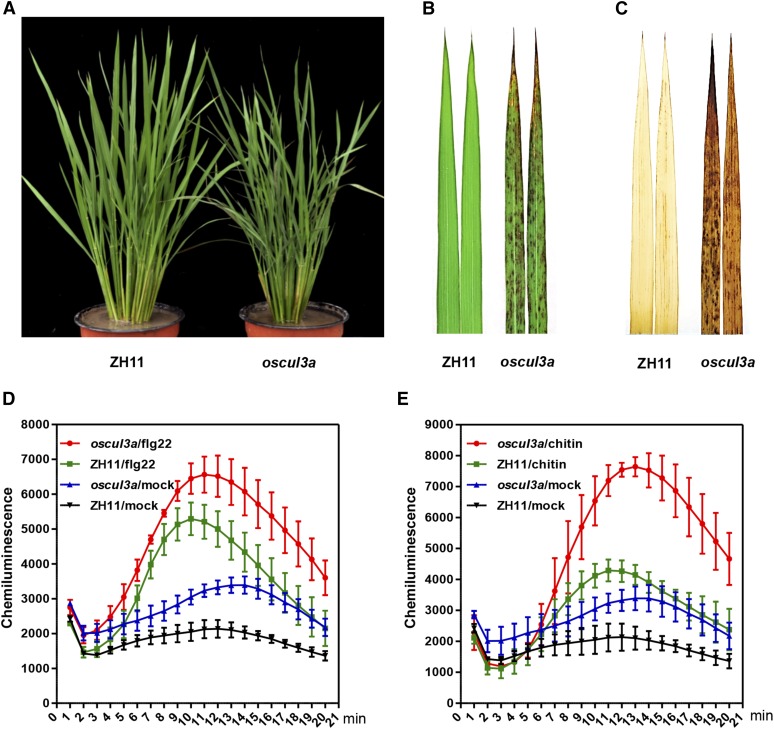
The rice oscul3a mutant was identified in a population of M2 mutant lines of the rice cultivar Zhonghua11 (ZH11) mutagenized by EMS treatment. oscul3a has reddish brown lesions scattered over the entire surface of its leaves (Figures 1A and 1B). Under field conditions in summer, the lesions appeared on the lower (older) leaf tips ∼45 d post-sowing (dps) (Supplemental Figure 1). These lesions spread gradually, eventually extending to whole leaves, leaf sheaths, and glumes. When oscul3a was grown in a greenhouse, the lesions were visible by ∼60 dps. Moreover, the number of the lesions was much less for oscul3a plants in the greenhouse than in the field. These variations in both the timing of lesion appearance and in lesion number suggest that cell death in oscul3a is influenced by growth conditions such as light intensity and temperature. To determine whether the development of lesions in oscul3a involves altered H2O2 accumulation, we performed 3,3 C-diaminobenzidine (DAB) staining, which reflects the formation of reddish-brown formazan precipitates indicative of H2O2 accumulation. DAB staining was much more abundant on oscul3a leaves than on ZH11 leaves (Figure 1C).
Figure 1.
Identification of the Lesion Mimic Mutant oscul3a.
(A) oscul3a mutant and ZH11 plants were grown in the field and photographed at 60 dps.
(B) Leaves of oscul3a mutant and ZH11 plants at 60 dps.
(C) DAB staining of the oscul3a mutant and ZH11 leaves shown in (B).
(D) ROS accumulation dynamics in oscul3a mutant and ZH11 plants after flg22 and water (mock) treatments. Error bars represent the se; n = 3.
(E) ROS accumulation dynamics in oscul3a mutant and ZH11 plants after chitin and water (mock) treatments. Error bars represent the se; n = 3.
To further investigate whether the reactive oxygen species (ROS) signaling pathway is impaired in oscul3a, we compared the dynamics of flg22- or chitin-induced ROS generation in ZH11 and oscul3a using a chemical luminescence assay. The ROS production rate was faster in oscul3a than in ZH11 in response to both flg22 and chitin treatments (Figures 1D and 1E). ROS levels peaked ∼11 min after both treatments, and these levels were nearly 1.5 times higher for the flg22 treatment and 2.0 times higher for the chitin treatment in oscul3a than in ZH11. In the control (which was treated with water), the basal ROS level was ∼1.3 times higher in oscul3a than in ZH11 (Figures 1D and 1E). These results indicate that the ROS signaling pathway is enhanced in oscul3a.
oscul3a Confers Enhanced Resistance to Both M. oryzae and Xoo
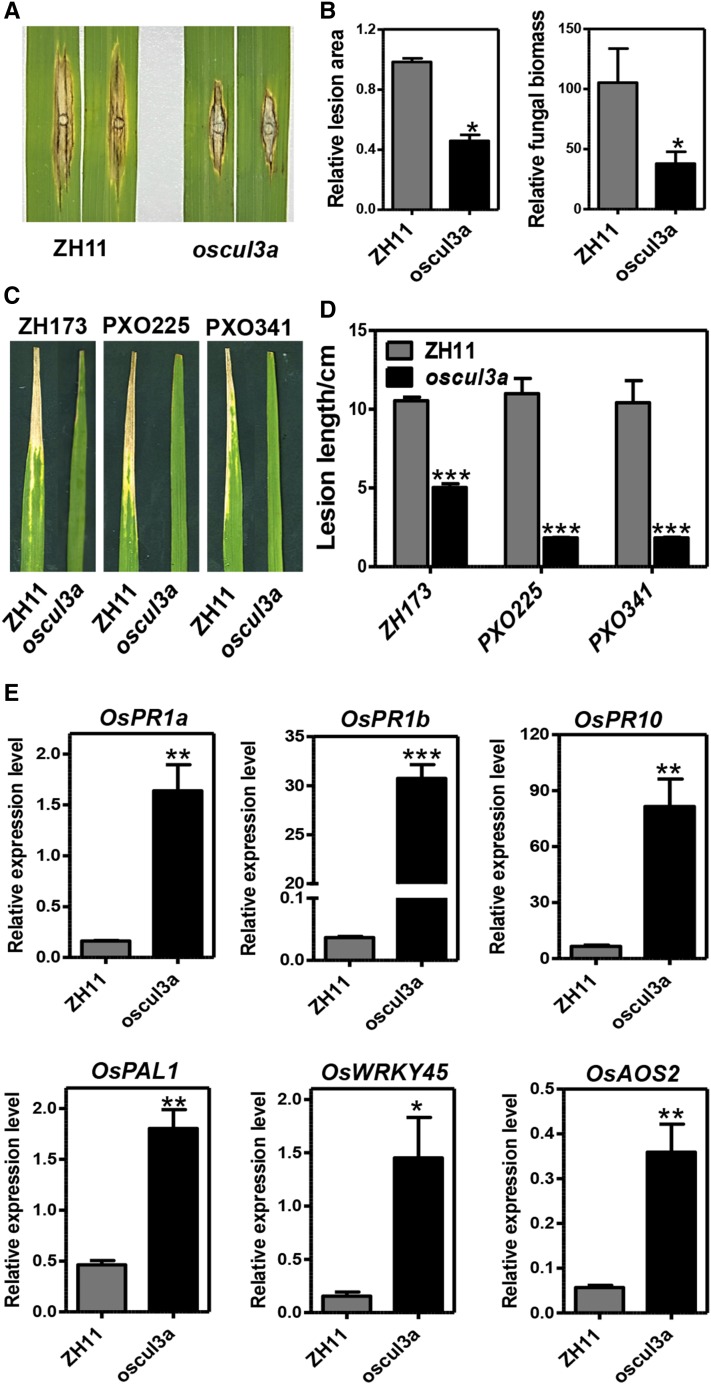
Given that oscul3a had hypersensitive response-like lesions and accumulated elevated levels of ROS following treatment with PAMPs, we speculated that oscul3a may exhibit enhanced resistance to rice pathogens. To test this possibility, oscul3a and ZH11 were challenged with the virulent M. oryzae isolate RB22. Punch inoculation assays revealed that both lesion area and fungal biomass in the lesions were significantly lower in oscul3a than in ZH11 (Figures 2A and 2B). In another test, fully expanded flag leaves of ZH11 and oscul3a were inoculated with the Xoo isolates ZH173, PXO225, and PXO341. At 15 d postinoculation (dpi), the lesion lengths were much shorter on oscul3a plants than on ZH11 plants (Figures 2C and 2D).
Figure 2.
The oscul3a Mutant Displays Enhanced Resistance to Both M. oryzae and Xoo.
(A) and (B) Punch inoculation of oscul3a mutant and ZH11 plants with the compatible M. Oryzae isolate RB22.
(A) Leaves were photographed at 12 dpi.
(B) Lesion area (left) and fungal biomass (right) of the inoculated leaves of oscul3a mutant and ZH11 plants as shown in (A) at 12 dpi. Error bars represent the se; n = 3 (t test between mutant and ZH11 values; *P < 0.05).
(C) and (D) Leaves of oscul3a mutant and ZH11 plants were inoculated with three compatible Xoo isolates.
(C) Leaves at 15 dpi.
(D) Lesion lengths on the leaves at 15 dpi. Error bars represent the se; n = 3 (t test between mutant and ZH11 values; ***P < 0.001).
(E) Total RNA was extracted from the leaves (second from the top) of oscul3a mutant and ZH11 plants at 60 dps. qRT-PCR was used to analyze the expression of OsPR1a, OsPR1b, OsPR10, OsPAL1, OsWRKY45, and OsAOS2. Data were normalized to the expression level of the constitutively expressed OsACTIN gene. Error bars represent the se; n = 3 (t test between mutant and ZH11 values; *P < 0.05, **P < 0.01, and ***P < 0.001).
To investigate the mechanism of enhanced resistance in oscul3a, we studied the transcription level of PR genes using qRT-PCR. OsPR1a, OsPR1b, OsPR10, OsPAL1, and OsWRKY45, which are known to be involved in the salicylic acid (SA) signaling pathway (Park et al., 2012), were all significantly upregulated in oscul3a (Figure 2E). The expression of OsAOS2, which encodes allene oxide synthase (a key enzyme in the jasmonic acid biosynthetic pathway), was ∼7-fold higher in oscul3a than in ZH11. These results suggest that oscul3a confers enhanced disease resistance by enhancing flg22- and chitin-triggered ROS accumulation and possibly by activating the SA and jasmonic acid signaling pathways in rice.
Map-Based Cloning of OsCUL3a
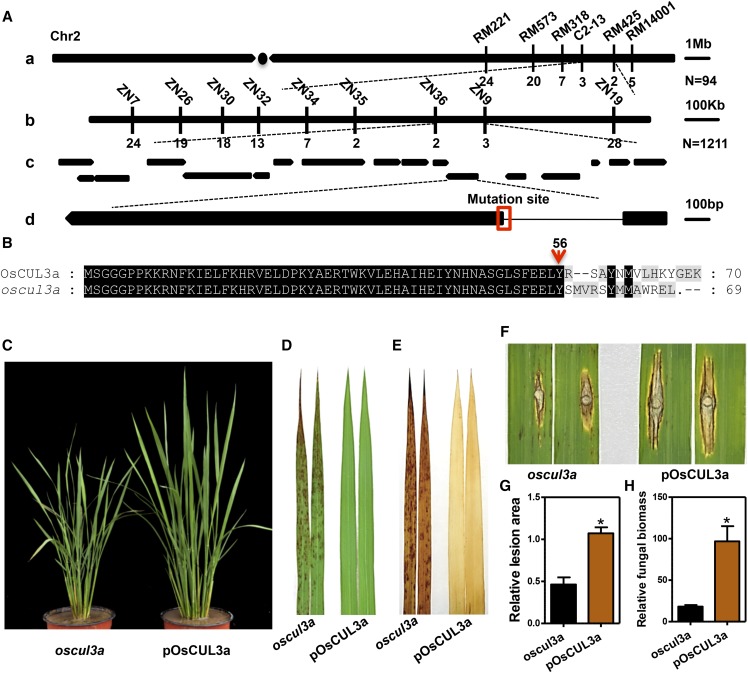
Genetic analysis revealed that the lesion mimic phenotype of oscul3a was controlled by a single recessive nuclear gene (normal:lesion mimic = 1566:434, χ2 = 1.53). To isolate the gene responsible for the oscul3a mutant phenotypes, an F2 mapping population (∼12,000 plants) was constructed by crossing oscul3a with cultivar Nanjing11. Bulked segregant analysis (BSA) revealed that six markers on the long arm of chromosome 2 cosegregated with the phenotype of oscul3a. Those markers were then used for genotyping 94 recessive individuals from the F2 mapping population. Linkage analysis localized the candidate mutation within an interval of 1.7 Mb between markers C2-13 and RM425 (Figure 3A, a). Nine additional polymorphic InDel markers of oscul3a were developed to enable higher resolution genotyping of the 1211 individual recessive F2 plants. The oscul3a locus was finally narrowed to a 123-kb region flanked by markers ZN36 and ZN9 (Figure 3A, b). According to the rice genome annotation project database (http://rice.plantbiology.msu.edu/), the 123-kb region contains 18 open reading frames (ORFs) (Figure 3A, c).
Figure 3.
Map-Based Cloning of oscul3a.
(A) Delimitation of the candidate genomic region of oscul3a. (a) Preliminary mapping of the oscul3a gene using 94 recessive F2 plants with simple sequence repeat markers. The numbers under the linkage map represent the number of recombinants. (b) Fine mapping of the oscul3a locus with the InDel markers. The numbers under the linkage map represent the number of recombinants. (c) The arrows denote the candidates (ORFs) within the genomic region between the ZN36 and ZN9 markers. (d) Structure of the OsCUL3a gene and the mutation site. The line represents the intron; black boxes represent the exons.
(B) Protein alignment between wild-type OsCUL3a and the deduced 69 amino acid product in oscul3a. The red arrow indicates the frameshift mutation site.
(C) Phenotype of 2-month-old complemented T1 plants grown in the field. pOsCUL3a complementation plants were developed by transforming the whole genomic fragment of OsCUL3a into the oscul3a mutant background. pOsCUL3a plants were photographed at 60 dps.
(D) Leaves (second from the top) of oscul3a mutant and pOsCUL3a plants at 60 dps.
(E) DAB staining of the oscul3a mutant and pOsCUL3a leaves shown in (D).
(F) to (H) Punch inoculation of oscul3a mutant and pOsCUL3a plants with the compatible M. oryzae isolate RB22.
(F) Leaves were photographed at 12 dpi.
(G) Lesion area of the inoculated leaves at 12 dpi. Error bar represent the se; n = 3 (t test; *P < 0.05).
(H) Fungal biomass of the inoculated leaves at 12 dpi. Error bars represent the se; n = 3 (t test; *P < 0.05).
To identify the oscul3a mutation, genomic DNA of the 18 ORFs were PCR amplified from both oscul3a and ZH11. Sequencing of these ORFs from both genotypes enabled the identification of an 11-bp substitution and an 8-bp deletion in LOC_Os02g51180 of oscul3a; both polymorphisms were in the junction region between the first intron and the second exon of this ORF. We then sequenced cDNA of LOC_Os02g51180 from both genotypes. This analysis revealed that the mutations disrupted the recognition site (AG) for intron splicing, causing incorrect splicing of 28 bp in the second exon of LOC_Os02g51180 in oscul3a (Supplemental Figure 2A). This splicing error was predicted to introduce a premature stop codon at the 69th amino acid, resulting from a frame shift (Figure 3B). LOC_Os02g51180 was predicted to encode a protein with a Cullin domain (32–637) and a Nedd8 domain (664–736) (Supplemental Figure 2B) and it was previously named OsCUL3a (Gingerich et al., 2005). This protein, the putative OsCUL3a, was highly conserved in dicots and monocots and shared 89.71% similarity at the amino acid sequence level with CUL3a in Arabidopsis. The rice genome contains 13 Cullin-like genes. Phylogenetic analysis of the Cullin proteins in rice and Arabidopsis revealed that OsCUL3a shows a high level of homology with AtCUL3a (79%) and AtCUL3b (77.9%) (Supplemental Figure 3).
The rice genome encodes three highly homologous Cullin3-like proteins (Supplemental Figure 4). All three genes were constitutively expressed, with the greatest abundance in leaf and sheath tissues (Supplemental Figure 5). To determine whether the mutation identified in OsCUL3a is responsible for the formation of the necrotic lesions in oscul3a plants, we performed a complementation test by transforming oscul3a calli with the full genomic DNA sequence of OsCUL3a. The transgene included the entire coding sequence and was driven by its native promoter. The resulting complemented plants are referred to as pOsCUL3a. In total, 21 independently transformed transgenic lines were generated. The presence of both the transgene and the original oscul3a allele was confirmed by DNA sequencing (Supplemental Figure 6). No lesions appeared on leaves of pOsCUL3a plants at any point in the life cycle, and the growth of pOsCUL3a was same as that of ZH11 (Figures 3C and 3D; Supplemental Figure 7).
To confirm that the elevated accumulation of H2O2 and disease resistance phenotypes of oscul3a result specifically from the oscul3a mutation, we subsequently performed DAB staining and rice blast inoculation with the oscul3a and pOsCUL3a plants. The increased H2O2 accumulation phenotype was abolished in pOsCUL3a (Figure 3E), and pOsCUL3a plants were more susceptible than oscul3a plants to the compatible blast isolate RB22 (Figure 3F). The lesion area and the relative fungal biomass in the lesions were significantly larger in pOsCUL3a than in oscul3a (Figures 3G and 3H). These results indicate that the lesion mimic phenotype of oscul3a is caused by the early termination of OsCUL3a protein and that OsCUL3a negatively regulates lesion formation (cell death) and disease resistance in rice.
OsCUL3a Interacts with Both OsRBX1a and OsRBX1b in Vivo
Cullin proteins can assemble with RBX and various adaptors to form CRLs in eukaryotes (Vierstra, 2009). Rice has two genes that encode RBX-like proteins, OsRBX1a and OsRBX1b, which share 92.7% amino acid sequence identity. Like Cullin proteins, RBX proteins are highly conserved in dicots and monocots. OsRBX1a and OsRBX1b exhibit 72.3 and 74.7% amino acid sequence similarity with AtRBX1 in Arabidopsis, respectively. Comparison of these three proteins indicated that their N-terminal regions are variable and that their C-terminal regions are highly conserved. AtRBX1 has 24 amino acids in its middle region that are absent in OsRBX1a/b (Supplemental Figure 8).
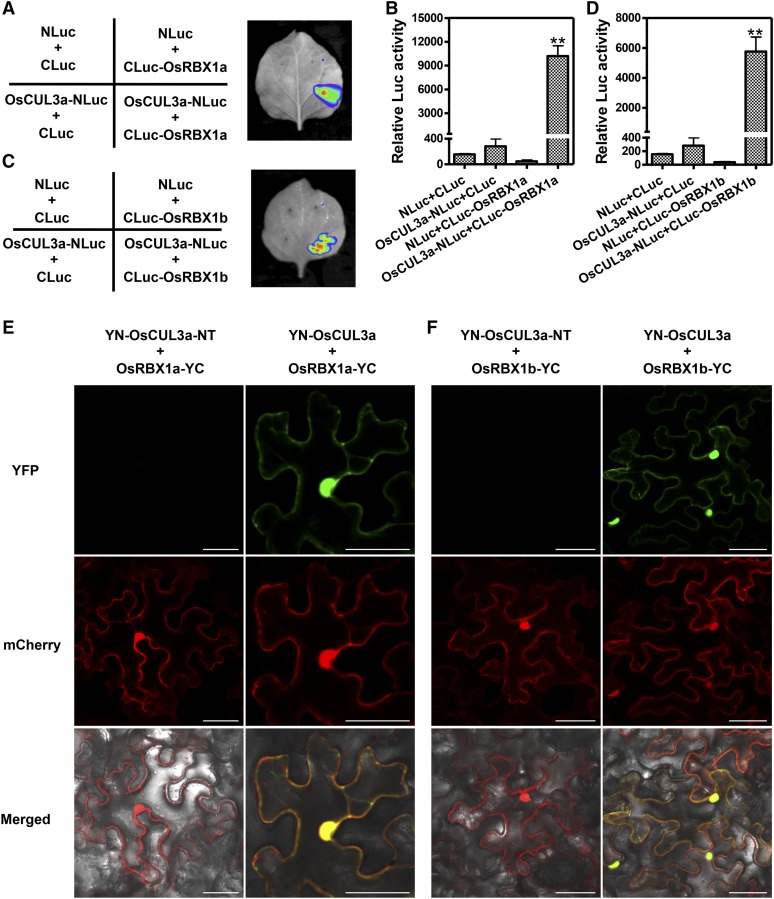
To evaluate the biochemical function of OsCUL3a, we first used the firefly luciferase complementation imaging (LCI) system to determine whether it can interact with OsRBX1a/b (Chen et al., 2008). Strong fluorescence was detected with a low-light imaging system in Nicotiana benthamiana leaves in which OsCUL3a-NLuc and CLuc-OsRBX1a were coexpressed (Figure 4A). The relative luciferase (LUC) activity in these cells was nearly 10,000. By contrast, coexpression of the control combinations (NLuc+CLuc, OsCUL3a-NLuc+CLuc, and NLuc+CLuc-OsRBX1a) showed only background levels of LUC activity (Figure 4B). OsCUL3a could also interact with OsRBX1b (Figures 4C and 4D).
Figure 4.
OsCUL3a Assembles with OsRBX1a or OsRBX1b to Form a CULLIN-RING-Like E3 Ligase Complex.
(A) OsCUL3a interacts with OsRBX1a as indicated by LCI assay. OsCUL3a-NLuc and CLuc-OsRBX1a were transiently expressed in N. benthamiana by coinfiltration; NLuc and CLuc were the negative controls. Luminescence was monitored with a low-light, cooled, CCD imaging apparatus at 3 dai.
(B) Quantification of LUC activity in the leaves shown in (A). Error bars represent the se; n = 3 (t test between OsCUL3a-NLuc+CLuc-OsRBX1a and control group value; **P < 0.01).
(C) OsCUL3a interacts with OsRBX1b as indicated by LCI assay. OsCUL3a-NLuc and CLuc-OsRBX1b were transiently expressed in N. benthamiana by coinfiltration; NLuc and CLuc were the negative controls. Luminescence was monitored with a low-light, cooled, CCD imaging apparatus at 3 dai.
(D) Quantification of LUC activity in the leaves shown in (C). Error bars represent the se; n = 3 (t test between OsCUL3a-NLuc+CLuc-OsRBX1b and control group value; **P < 0.01).
(E) BiFC assays showing the interaction between OsCUL3a and OsRBX1a in N. benthamiana. The OsCUL3a and OsRBX1a proteins were fused to the N- or C-terminal fragment of eYFP and transiently expressed in N. benthamiana. The YFP signal was evaluated via confocal microscopy at 3 dai. The N terminus of OsCUL3a was used as the negative control. Bar = 50 µm.
(F) BiFC assays for the interaction between OsCUL3a and OsRBX1b in N. benthamiana. The OsCUL3a and OsRBX1b proteins were fused to the N- or C-terminal fragment of eYFP and transiently expressed in N. benthamiana. The YFP signal was evaluated via confocal microscopy at 3 dai. The N terminus of OsCUL3a was used as the negative control. Bar = 50 µm.
To further confirm the interaction of OsCUL3a and OsRBX1a/b, we performed a yeast two-hybrid (Y2H) assay with full-length versions of OsRBX1a/b, full-length OsCUL3a, and six truncated versions of OsCUL3a containing different domain segments (Supplemental Figure 9A). Interestingly, OsCUL3a interacted only with OsRBX1a in yeast; no interaction between the full-length OsCUL3a and OsRBX1b was detected. Still, we found that the truncated version of OsCUL3a that retained its Cullin N-terminal domain was able to interact strongly with both OsRBX1a and OsRBX1b. By contrast, the C-terminal Cullin homology domain of OsCUL3a interacted only weakly with OsRBX1b, and the N-terminal domain of OsCUL3a did not interact with OsRBX1a or OsRBX1b (Supplemental Figure 9B). We next performed BiFC assays in N. benthamiana. Cells coexpressing YN-OsCUL3a and OsRBX1a-YC had strong yellow fluorescence throughout the whole cell; this fluorescence perfectly merged with the empty mCherry signal (Figure 4E, second row). However, no signal was detected in cells coexpressing YN-OsCUL3a-NT (N-terminal: 1–564) and OsRBX1a-YC under the same conditions (Figure 4E, first row). As shown in Figure 4F, the BiFC assay showed that OsCUL3a could also interact with OsRBX1b. The YN-OsCUL3a and OsRBX1a/b-YC BiFC signal pattern was consistent with the whole-cell distribution patterns of OsCUL3a and OsRBX1a/b in rice protoplasts and N. benthamiana leaves (Supplemental Figure 10). Taken together, these results indicate that OsCUL3a associates with OsRBX1a/b in vivo and is distributed throughout rice cells.
OsCUL3a Interacts with OsNPR1 and Promotes its Degradation via the 26S Proteasome
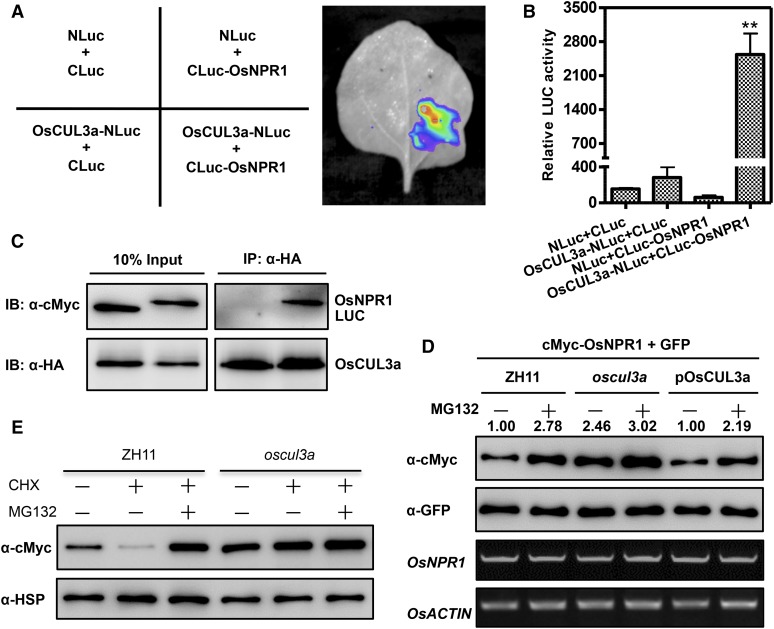
OsCUL3a exhibits high amino acid sequence identity to AtCUL3a. In Arabidopsis, AtCUL3a-mediated degradation of AtNPR1 monomers in the nucleus has dual roles in regulating immunity (Spoel et al., 2009). The rice genome contains five homologs of AtNPR1, which are named NH1-5 in rice. Overexpression of NH1 in rice results in resistance to Xoo, induction of the expression of PR genes such as OsPR1, OsPR10, and OsPAL1, and enhancement of BTH-mediated cell death (Chern et al., 2005). We therefore speculated that NH1 (OsNPR1) may be a substrate of OsCUL3a in rice. To test this hypothesis, we evaluated the interaction between OsCUL3a and OsNPR1 using the LCI system. As shown in Figure 5A, OsCUL3a interacted strongly with OsNPR1 in N. benthamiana. Cells coexpressing OsCUL3a-NLuc and CLuc-OsNPR1 exhibited significantly higher relative LUC activity than the controls (Figure 5B). We next conducted a coimmunoprecipitation (co-IP) assay in rice protoplasts. Myc-OsNPR1, but not Myc-CLuc, was coimmunoprecipitated with HA-OsCUL3a (Figure 5C). These results demonstrate that OsCUL3a interacts with OsNPR1 in vivo.
Figure 5.
OsCUL3a Interacts with OsNPR1 and Promotes Its Degradation in Vivo.
(A) OsCUL3a interacts with OsNPR1 as indicated by LCI assay. OsCUL3a-NLuc and CLuc-OsNPR1 were transiently expressed in N. benthamiana by coinfiltration; NLuc and CLuc were the negative controls. Luminescence was monitored with a low-light, cooled, CCD imaging apparatus at 3 dai.
(B) Quantification of the LUC activity in the leaves shown in (A). Error bars represent the se; n = 3 (t test between OsCUL3a-NLuc+CLuc-OsNPR1 and control group value; **P < 0.01).
(C) OsCUL3a interacts with OsNPR1 in a co-IP assay. Myc-tagged OsNPR1 or LUC was transiently expressed with HA-tagged OsCUL3a in rice protoplasts by cotransfection. Following total protein extraction, samples were immunoprecipitated with anti-HA antibody. Crude and immunoprecipitated proteins were analyzed with anti-HA or anti-Myc antibodies.
(D) OsCUL3a is required for OsNPR1 degradation in rice protoplasts. OsNPR1 was fused with the Myc tag and coexpressed with GFP in rice protoplasts isolated from ZH11, oscul3a, and pOsCUL3a plants. The transfected protoplasts were treated with or without MG132 for 20 h. Total protein was detected with anti-Myc and anti-GFP antibodies. The expression of OsNPR1 and OsACTIN was analyzed by RT-PCR. The relative abundance of OsNPR1 was calculated by comparing to GFP using ImageJ software. The OsNPR1/GFP relative quantity in ZH11 without MG132 was defined as 1.0 (lane 1).
(E) OsCUL3a-mediated degradation of OsNPR1 via the 26S proteasome. Myc-OsNPR1 was transiently expressed in rice protoplasts isolated from ZH11 and oscul3a plants with or without cycloheximide and MG132 treatments. Total protein was extracted at 48 h after transfection for immunoblotting analysis with anti-cMyc and anti-HSP antibodies.
We next determined whether OsNPR1 is a substrate for OsCUL3a-mediated ubiquitination despite the transcript accumulation of OsNPR1 is similar in ZH11, the oscul3a mutant, and pOsCUL3a plants (Supplemental Figure 11). Myc-OsNPR1 and GFP were transiently coexpressed in rice protoplasts isolated from ZH11, oscul3a, and pOsCUL3a plants with or without MG132 treatment. GFP was used to normalize the efficiency of both transformation and protein expression. Immunoblotting analysis indicated that the OsNPR1 protein level was lower in ZH11 and pOsCUL3a protoplasts than in oscul3a protoplasts and the relative band intensity in oscul3a over the control ZH11 was 2.46 (Figure 5D, lanes 1 and 3 in the top gel). Pretreatment with MG132 stabilized OsNPR1 not only in ZH11 and pOsCUL3a but also in oscul3a mutant (Figure 5D, lanes 2, 4, and 6 in the first panel), suggesting that OsNPR1 might be also regulated by another degradation pathway in rice. To further confirm OsNPR1 is a substrate of OsCUL3a, we performed a cycloheximide chase assay. In the absence of new protein biosynthesis, the amount of Myc-OsNPR1 decreased rapidly in ZH11, and this degradation could be suppressed by the MG132 treatment (Figure 5E, lanes 2 and 3 in the first panel). By contrast, the Myc-OsNPR1 protein level remained the same in oscul3a (Figure 5E, lanes 4 to 6 in the first panel). These results demonstrate that OsCUL3a can promote OsNPR1 degradation via the 26S proteasome system in rice.
Knockout of OsNPR1 in the oscul3a Background Abolishes the Cell Death Phenotype
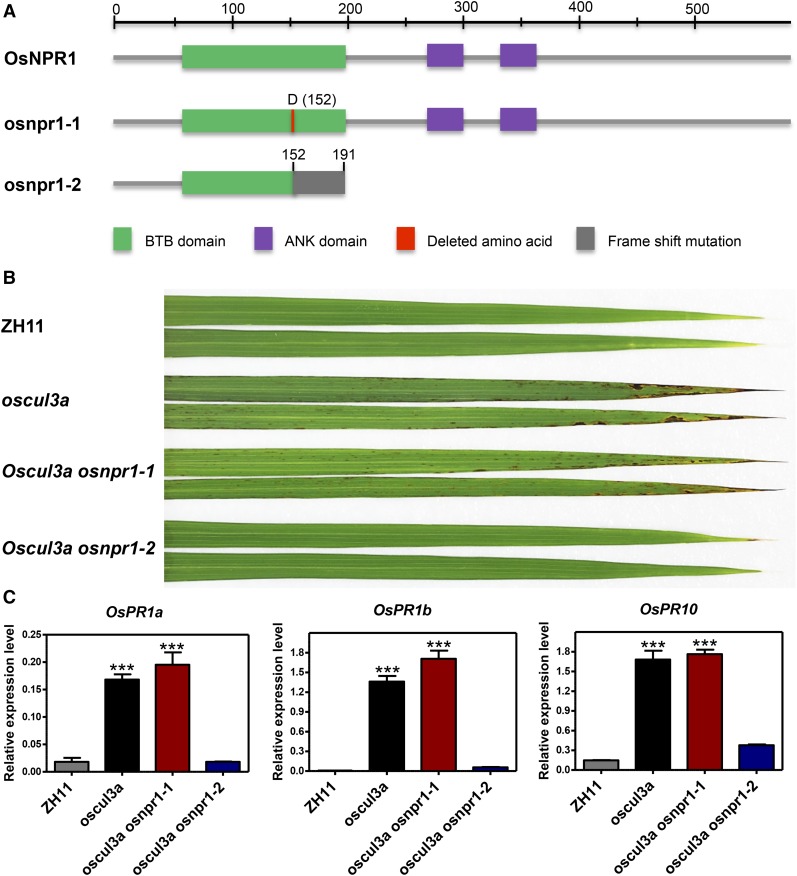
After determining that OsNPR1 is a substrate for OsCUL3a-mediated ubiquitination and is degraded by the 26S proteasome in rice, we speculated that the cell death phenotype (lesion mimic phenotype) and the elevated expression levels of PR genes in oscul3a may result from an increased accumulation of OsNPR1. To evaluate this possibility, we knocked out OsNPR1 in the oscul3a background using CRISPR/Cas9 methods (Wang et al., 2014) and obtained a series of mutant plants. After sequencing analysis, four distinct types of oscul3a osnpr1 double mutants were identified (Supplemental Figure 12A). Whereas oscul3a osnpr1-1 had a single amino acid deletion at position 152, the other three double mutants, oscul3a osnpr1-2, oscul3a osnpr1-3, and oscul3a osnpr1-4, contained a single base insertion that caused an early termination of OsNPR1 translation at the same position (Supplemental Figure 12A). To determine whether these mutant alleles were functional, we amplified the full-length cDNA of OsNPR1 from all of the mutants and fused them with the Myc tag. The derived constructs containing the OsNPR1 cDNAs from these mutants were transiently expressed in rice protoplasts. Immunoblot analysis indicated that osnpr1-1 was the same size as the wild-type OsNPR1 (∼85 kD); however, osnpr1-2, -3, and -4 were much smaller (∼35 kD) compared with the wild type (Supplemental Figure 12B). Phenotype evaluations in greenhouses revealed that the single base insertion in OsNPR1 in the three double mutants significantly reduced the cell death of oscul3a. However, the amino acid deletion in oscul3a osnpr1-1 did not affect OsNPR1 function in rice; the double mutant still showed cell death phenotypes similar to those of oscul3a (Figure 6B). In addition, the induction of OsPR1a, OsPR1b, and OsPR10 expression in oscul3a was abolished in oscul3a osnpr1-2 double mutants (Figure 6C). Together with the in vivo degradation data, these results indicate that OsCUL3a negatively regulates cell death in an OsNPR1-dependent manner.
Figure 6.
Characterization of the oscul3a osnpr1 Double Mutant.
(A) Domain structures of wild-type OsNPR1 and OsNPR1 from each of the two types of CRISPR/Cas9-derived (osnpr1) mutants.
(B) ZH11, oscul3a, and oscul3a osnpr1 double mutants were grown in a greenhouse, and leaves (second from the top) were collected and photographed at 60 dps. The cell death phenotype of the oscul3a mutant was abolished in the oscul3a osnpr1-2 double mutants.
(C) The expression levels of OsPR1a, OsPR1b, and OsPR10 in leaves (as in [B]) were analyzed using qRT-PCR and were normalized against OsACTIN. Error bars represent the se; n = 3 (t test between ZH11 and oscul3a value or ZH11 and oscul3a osnpr1-1 value; ***P < 0.001).
DISCUSSION
OsCUL3a Negatively Regulates Cell Death and Immunity in Rice
As the most major multisubunit E3 ligase, CRLs have been extensively studied in eukaryotes. CUL3, which assembles with the BTB domain-containing protein to recognize substrates, is known to be involved in regulating cell death in animals and Drosophila (Liu et al., 2009; Lee et al., 2010). The model plant Arabidopsis has two CUL3 proteins, AtCUL3a and AtCUL3b. In 2005, two research groups reported that AtCUL3a and AtCUL3b are functionally redundant in embryo development (Figueroa et al., 2005; Gingerich et al., 2005). Under normal growth conditions, the single homozygous atcul3a and atcul3b mutants are virtually indistinguishable from their wild type. However, knockout of both AtCUL3a and AtCUL3b results in an arrest in embryogenesis at much earlier stages (Figueroa et al., 2005; Gingerich et al., 2005). This embryo-lethal phenotype of the cul3a cul3b mutant was also confirmed by another group, who used different alleles of AtCUL3a and AtCUL3b mutants (Thomann et al., 2005). Spoel et al. (2009) demonstrated that both AtCUL3a and AtCUL3b are involved in the turnover of AtNPR1 for immune responses and SAR establishment.
Like in Arabidopsis, the rice genome encodes three CUL3 proteins. However, their function in rice immunity has not been fully understood. In this study, we found that the lesion mimic and enhanced immunity phenotypes of the oscul3a mutant are caused by the early termination of the OsCUL3a protein in rice (Figure 3). Loss of OsCUL3a function leads to the accumulation of OsNPR1, which causes PCD in the mutant plants (Figures 5D and 6). In addition, we also found that the seed setting rate was dramatically lower for oscul3a than for ZH11 in the field. It is unclear whether the reduction of seed setting rate in the mutant is due to embryonic development failure or to the autoactivation of immune responses in the rice plants. Because the rice genome contains three genes that encode CUL3-like proteins (Supplemental Figure 3), it is possible that OsCUL3a has unique functions in cell death control and immunity in comparison with the two orthologs in Arabidopsis.
OsCUL3a Associates with OsRBX1 and BTB Proteins to Form E3 Ubiquitin Ligase Complexes in Rice
Five types of CRLs have been found in eukaryotes. Among these, Cullin proteins function as the scaffold that assembles with RBX and different adaptors to recognize substrates. The human genome codes for two RBX proteins that can interact with CUL1, CUL2, CUL3, CUL4, and CUL5 (Ohta et al., 1999; Chen et al., 2000). The Saccharomyces cerevisiae genome contains only one gene encoding an RBX-like protein, which also interacts with all Cullin proteins in vivo (Ohta et al., 1999). In Arabidopsis, AtCUL3a and AtCUL3b interact with RBX1 and dozens of BTB proteins to form CRL complexes (Figueroa et al., 2005; Gingerich et al., 2005). Although many F-box and BTB proteins are known to participate in various developmental processes, there is little direct evidence for the existence of CRLs in rice (Sasaki et al., 2003; Jiang et al., 2013; Zhou et al., 2016). Based on Y2H, LCI, and BiFC analysis, we showed that OsCUL3a associates with both OsRBX1a and OsRBX1b in vivo. LCI and co-IP assays further demonstrated that OsCUL3a interacts with OsNPR1 in rice, suggesting that rice has conserved CUL3-type CRLs (Figures 4 and 5; Supplemental Figure 9).
Our subcellular localization results show that OsRBX1a, OsRBX1b, and OsCUL3a are distributed throughout the cell (Supplemental Figure 10), indicating that they could work together to participate in many biological processes. The human RBX protein ROC2 is critical in regulating apoptosis; when overexpressed in human cells, ROC2 can protect the cells from apoptosis induced by redox agents. At the same time, ROC2 appears to inhibit or delay metal ion-induced cytochrome c release and caspase activation (Duan et al., 1999). ROC2 also has a specific function in protecting brain cells against ROS-induced damage in mice (Yang et al., 2001). In this study, we found that both OsRBX1a and OsRBX1b can interact with OsCUL3a (Figure 4) but that they exhibit different interaction patterns with OsCUL3a in Y2H assays (Supplemental Figure 9). OsRBX1a, but not OsRBX1b, can interact with the full-length OsCUL3a. By contrast, the Cullin N-terminal domain segment of OsCUL3a, which lacks the Nedd8 domain, can strongly interact with both OsRBX1a and OsRBX1b (Supplemental Figure 9). Whether OsRBX1a and OsRBX1b can help regulate cell death and immunity in rice warrants further study.
OsCUL3a Negatively Regulates Cell Death and Immunity in an OsNPR1-Dependent Manner
In Arabidopsis, the AtNPR1-mediated immunity pathway has been extensively studied and many important components have been identified in the last two decades (Furniss and Spoel, 2015). AtNPR1 accumulation in the nucleus is required for basal defense gene expression and resistance, whereas its subsequent turnover is essential for establishing SAR. In an elegant study, researchers demonstrated that AtNPR1 paralogs AtNPR3 and AtNPR4 act as SA receptors and play a role in basal resistance, ETI, and SAR (Fu et al., 2012). They showed that in an SA-deficient background, AtNPR1 is degraded by AtCUL3AtNPR4, leading to enhanced disease susceptibility. When challenged with pathogens, the increase of SA inside the lesion facilitates AtCUL3AtNPR3-mediated degradation of AtNPR1, causing effector-triggered PCD. In the neighboring cells, AtNPR1 is not degraded due to the lower level of SA, which inhibits AtNPR1-AtNPR3 interaction and the accumulation of AtNPR1 to suppress PCD and establish SAR (Fu et al., 2012). These results clearly demonstrate the function of AtCUL3 and AtNPR1, AtNPR3, and AtNPR4 proteins in SA perception and innate immunity in Arabidopsis.
In rice, the function of OsCUL3 and OsNPR1 protein is still elusive. It was reported that overexpression of its ortholog protein OsNPR1 in rice plants also results in enhanced resistance to Xoo, the induction of PR gene expression, and the development of lesion-mimic lesions on leaves under greenhouse conditions (Chern et al., 2005; Yuan et al., 2007; Bai et al., 2011). Chern et al. (2016) recently reported that OsNPR1 positively regulates resistance by activating the expression of cysteine-rich receptor-like kinases in rice. In this study, we found that OsNPR1 protein overaccumulates in the oscul3a mutant and that the overaccumulation may cause the cell death and enhanced disease resistance phenotypes in the mutant (Figure 5). This conclusion is also supported by the genetic evidence that mutation of OsNPR1 in the oscul3a background abolishes the cell death phenotypes and the induction of PR gene expression (Figures 6B and 6C). Taken together, these results demonstrate that OsCUL3a negatively regulates cell death and immunity in an OsNPR1-dependent manner. This is consistent with the findings in Arabidopsis that AtCUL3 is important for the function of AtNPR3 and AtNPR4 and loss of both AtNPR3 and AtNPR4 genes leads to accumulation of higher levels of NPR1 (Fu et al., 2012).
It is noteworthy that there are differences in the NPR1-mediated PCD and resistance in Arabidopsis and rice. The functions of SA and SAR in rice resistance are still controversial due to the fact that rice has a high level of SA (Yang et al., 2004). In Arabidopsis, AtNPR1 is a negative regulator of PCD; the protein is overaccumulated in the atnpr3 atnpr4 double mutant, which cannot undergo PCD in response to pathogen infection. By contrast, our data show that OsNPR1 positively regulates PCD in rice: The cell death phenotype disappears in the oscul3a osnpr1 double mutant (Figure 6). Moreover, the expression of PR genes including OsPR1a, OsPR1b, and OsPR10 were activated in oscul3a mutant plants (Figure 2E), whereas the expression of PR genes in AtNPR1 overexpression Arabidopsis plants is not observed until the plants are infected with pathogens (Cao et al., 1998). Nevertheless, further investigation is warranted to determine the function of OsCULs and OsNPRs and their relationship in rice immunity and SA perception.
Working Model for OsCUL3a Mediation of Cell Death and Immunity in Rice
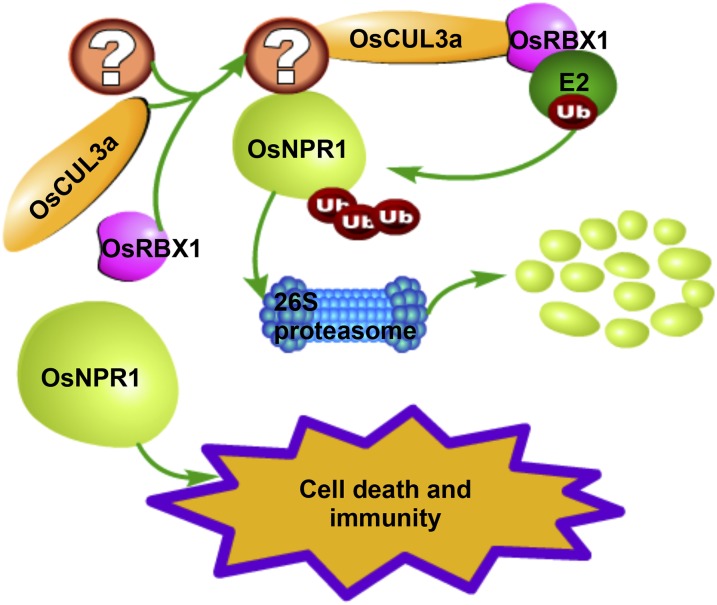
Based on our findings and those of previous reports, we present a working model for OsCUL3a-mediated cell death and immunity (Figure 7). In our model, OsCUL3a assembles with OsRBX1 and an unknown BTB domain-containing protein to form an E3 ligase complex that then mediates the ubiquitination of OsNPR1. OsNPR1 is subsequently degraded by the ubiquitin/26S proteasome system. In the oscul3a mutant plants, OsNPR1 overaccumulates, which leads to spontaneous cell death and activation of immunity that inhibits pathogen infection. Our study provides new functions of OsCUL3a in the regulation of PCD and immunity via OsNPR1 in rice.
Figure 7.
A Proposed Model of OsCUL3a Action.
OsCUL3a assembles with OsRBX1 and an unknown BTB domain-containing protein to form an E3 ligase complex that promotes the degradation of OsNPR1 via the 26S proteasome system. OsNPR1 highly accumulates in oscul3a mutant plants, resulting in cell death and immunity.
METHODS
Plant Materials and Growth Conditions
The oscul3a mutant was obtained from a mutant population generated by EMS treatment of the japonica rice (Oryza sativa) cultivar ZH11. For genetic tests, the BC1F2 population derived from the backcross between oscul3a and ZH11 were planted in a summer field. About 2000 individual plants were selected randomly for phenotype analysis at 60 dps. For map-based cloning, an F2 population was constructed by crossing oscul3a with the indica rice cultivar Nanjing 11. F2 populations and parents were seeded on a seedbed in a greenhouse and 25-d-old seedlings were then transplanted into a paddy field. For disease and other phenotype characterizations, the sterilized rice seeds were germinated on 0.5× Murashige and Skoog medium. Plants were grown in a greenhouse with a photoperiod of 14-h white light (452 μmol/m2/s) and 10 h dark at 28°C.
DAB Staining
For H2O2 measurement, leaves (second from the top) were collected at 60 dps and immediately submerged in a 1 mg/mL solution of DAB and incubated for 8 h in the dark at room temperature. Samples were then decolored in 95% boiling ethanol for 10 min and soaked for 48 h in 95% ethanol until all of the chlorophyll had been removed. The cleared leaves were then photographed.
Measurement of ROS
ROS was measured following PAMP treatment (flg22 and chitin) as previously described (Schwacke and Hager, 1992; Park et al., 2012). Briefly, leaves (second from the top) at 60 dps were punched into disks (0.25 cm2), which were submerged in distilled water overnight. Three disks per sample were then placed in a 1.5-mL microcentrifuge tube containing 100 μL luminol (Bio-Rad Immun-Star horseradish peroxidase substrate 170-5040), 1 μL horseradish peroxidase (Jackson ImmunoResearch), and 100 nM flg22 or 8 nM hexa-N-acetyl-chitohexaose; distilled water was used as a control. The luminescence was immediately measured at 10-s intervals over a period of 20 min in a Glomax 20/20 luminometer (Promega). Three biological replications (three disks for each replication) were performed for each sample of the three treatments.
Pathogen Infection
Rice plants were inoculated with Xoo by the leaf-clipping method. The ZH173, PXO225, and PXO341 strains were separately suspended in distilled water and adjusted to 109 viable cells/mL (OD600 = 1). Scissors were dipped into the bacterial suspensions and then used to remove the distal tips (5 cm) of flag leaves. At least three individual plants and three tillers of each plant were inoculated with one strain. The infected plants were grown in a growth chamber with a photoperiod of 14-h white light (452 μmol/m2/s) and 10 h dark at 28°C. Disease was scored by measuring the lesion length at 15 dpi.
To evaluate rice blast disease resistance, leaves (second from the top) at 60 dps were inoculated with Magnaporthe oryzae strain RB22 by a punch method (Ono et al., 2001; Park et al., 2012). In brief, the leaves were wounded with a mouse ear punch, and 10 μL of an M. oryzae spore solution (5 × 105 spores mL−1) was applied to the injured area, which was then sealed with cellophane tape. The inoculated plants were transferred to a growth chamber with a photoperiod of 12 h light and 12 h dark, 28°C, and 80% relative humidity. Lesion area and fungal biomass were measured at 12 dpi.
Map-Based Cloning of oscul3a and Complementation
Initially, a BSA method was used for linkage analysis. After BSA, additional InDel markers surrounding the preliminary linkage locus were designed based on the sequence polymorphism between japonica cultivar Nipponbare and indica cultivar 9311. The newly developed InDel markers were then used to narrow down the oscul3a region by genotyping 1211 recessive individual F2 plants. Genomic DNA fragments in the candidate region were amplified and sequenced. For complementation of the oscul3a mutant, a 6916-bp genomic fragment containing the entire ORF, 2555-bp upstream of the start codon, and 1656-bp downstream of the termination codon for OsCUL3a was obtained by PCR using the primers listed in Supplemental Table 1. The PCR product was then recombined in the binary vector pCAMBIA1300 using an In-Fusion Advantage Cloning kit (catalog no. PT4065; Clontech). The resultant expression construct was transformed into oscul3a by Agrobacterium tumefaciens-mediated transformation.
Y2H Assays
Various combinations of bait (pGBKT7) and prey (pGADT7) constructs were transformed into yeast strain AH109 (Supplemental Figure 9). Protein-protein interactions were evaluated according to the manufacturer’s recommendations (Clontech Yeast Protocols Handbook). Primers used in the generation of the Y2H constructs are listed in Supplemental Table 1.
BiFC Assay
The full-length cDNA sequence of OsCUL3a was amplified via PCR and cloned into the pSPYNE (R) 173 vector. OsRBX1a and OsRBX1b were similarly amplified and cloned into the pSPYCE (M) vector (Waadt et al., 2008). The constructs were transiently expressed in Nicotiana benthamiana by an agroinfiltration method. Three days after infiltration (dai), YFP fluorescence was observed using a Carl Zeiss confocal microscope. Primers used in the generation of the BiFC constructs are listed in Supplemental Table 1.
Co-IP Assay
The full-length cDNA sequences of OsCUL3a and OsNPR1 were amplified via PCR and fused with sequences encoding HA tag and Myc tag driven by 35S promoter, respectively. The constructs were transiently expressed in rice protoplasts as previously described (Chen et al., 2006). Total protein was extracted with 0.6 mL of native buffer supplemented with 50 µM MG132 (50 mM Tris-MES, pH 8.0, 0.5 M sucrose, 1 mM MgCl2, 10 mM EDTA, and plant protease inhibitor cocktail from Roche) and then immunoprecipitated with anti-HA magnetic beads (Thermo Scientific) according to the manufacturer’s instructions. The immunoprecipitated protein was separated via SDS-PAGE (10% gel) and analyzed by immunoblotting analysis with anti-HA (Roche; 11666606001) or anti-Myc antibodies (Roche; 11667203001). After incubating with a secondary antibody (ThermoFisher; 31430) for 1 h, the immunoblot signal was visualized with the Immobilon Western HRP substrate (Merck Millipore; WBKLS0100).
In Vivo OsNPR1 Degradation Assay
Plasmids containing 4×Myc-OsNPR1 and GFP were cotransformed into rice protoplasts isolated from ZH11, oscul3a, and pOsCUL3a. GFP was used as an internal control to normalize the efficiency of both transformation and protein expression. Samples were collected for protein and RNA extraction at 48 h after transfection. Protein levels were analyzed via immunoblotting with anti-Myc (Roche; 11667203001) and anti-GFP antibodies (Roche; 11814460001). Immunoblot signal visualization was performed as described above. For RNA expression analysis, RT-PCR was performed. For the suppression of 26S proteasome activity, the inhibitor MG132 was added to the buffer to a final concentration of 50 µM at 20 h before sampling. DMSO was used as the control.
qRT-PCR Expression Analysis of PR Genes
Total RNA was isolated from leaves (second from the top) with TRIzol reagent (Invitrogen). cDNA was synthesized with a ReverTra Ace kit (Toyobo) and was then used as a template for qRT-PCR, which was performed with a SYBR Premix ExTaq kit (Takara) on a Bio-Rad iQ2 system. Three biological replicates (three plants for each replicate) and two technical replicates were performed for each sample. The expression level of the target genes was normalized to that of the ACTIN gene. All the experiments were performed in triplicate. See Supplemental Table 1 for all primers used here.
LCI Assay
The LCI assay was performed as described previously (Chen et al., 2008). Primers used in the generation of the LCI constructs are listed in Supplemental Table 1.
oscul3a osnpr1 Double Mutant Construction Using the CRISPR/Cas9 System
oscul3a osnpr1 double mutants were constructed using previously described methods (Shan et al., 2014). In brief, the target site-containing sequence primers C-OsNPR1 were combined by annealing and then were cloned into the sgRNA expression vector pOsU3-sgRNA at the AarI site. The recombinant plasmid pOsU3-sgRNA-OsNPR1 was cotransformed into rice protoplasts isolated from oscul3a with the Cas9 expression plasmid pJIT163-2NLSCas9. The genomic DNA of transformed protoplasts was PCR amplified using primer CS-OsNPR1 (Supplemental Table 1). The PCR products were cloned into the pEASY-Blunt vector (TransGen Biotech) and then a total of 24 randomly selected colonies were further analyzed by sequencing to validate the activity of sgRNA. For stable rice transformation, plasmids of pOsU3-sgRNA-OsNPR1, pJIT163-2NLSCas9, and the hygromycin-containing pAct1-HPT were cotransformed into rice calli using the biolistic method. Embryogenic calli (6 to 9 weeks old) of oscul3a were bombarded using a PDS1000/He particle bombardment system. Mutant transgenic plants were identified as described above.
Protein Subcellular Localization Assay
The full-length CDS of OsCUL3a, OsRBX1a, and OsRBX1b were PCR amplified and fused with the coding sequence of EGFP. The recombinant proteins were transiently coexpressed with empty mCherry in rice protoplast or N. benthamiana leaves. The EGFP signal was detected with laser scanning confocal microscope (ZEISS 750) 36 h after transfection in rice protoplasts or 72 h after infiltration in N. benthamiana. Primers used in the generation of the constructs are listed in Supplemental Table 1.
Phylogenetic Analyses
The amino acid sequences of the Cullin proteins were downloaded from the TIGR website (http://rice.plantbiology.msu.edu/) and the TAIR website (http://www.arabidopsis.org/). Sequence alignment was performed with ClustalW (Supplemental Data Set 1). A neighbor-joining method implemented in MEGA5 was used to generate the phylogenetic tree; the bootstrap values indicated at the nodes in the phylogenetic tree are based on 1000 replications.
Accession Numbers
Sequence data from this article can be found in the GenBank/EMBL databases under the following accession numbers: OsCUL3a, XP_015626686.1; AtCUL1, NP_567243.1; AtCUL2, NP_171797.2; AtCUL3a, NP_174005.1; AtCUL3b, NP_177125.3; AtCUL4, NP_568658.1; OsRBX1a, XP_015625781.1; OsRBX1b, XP_015618780.1; AtRBX1, NP_001154725.1; OsNPR1, XP_015622114.1; LOC_Os01g27150.1, XP_015619069.1; LOC_Os01g27160.1, XP_015622492.1; LOC_Os01g50980.1, EEE55267.1; LOC_Os01g70920.1, XP_015625736.1; LOC_Os03g57290.1, XP_015632421.1; LOC_Os04g40830.1, XP_015635799.1; LOC_Os04g55000.1, XP_015634924.1; LOC_Os04g55030.1, XP_015636933.1; LOC_Os05g05700.1, XP_015639865.1; LOC_Os05g10580.1, EEE62617.1; LOC_Os08g07400.1, XP_015650664.1; and LOC_Os12g13360.1, ABA96903.1.
Supplemental Data
Supplemental Figure 1. Cell death phenotypes of the oscul3a mutant under field conditions.
Supplemental Figure 2. Sequence alignments between ZH11 and the oscul3a mutant.
Supplemental Figure 3. Phylogenetic analysis of CUL proteins in Arabidopsis and rice.
Supplemental Figure 4. Protein alignment of OsCUL3a, OsCUL3b, and OsCUL3c.
Supplemental Figure 5. OsCUL3a, OsCUL3b, and OsCUL3c are constitutively expressed in rice.
Supplemental Figure 6. Confirmation of the pOsCUL3a plants by sequencing.
Supplemental Figure 7. Cell death phenotype of oscul3a is rescued by OsCUL3a.
Supplemental Figure 8. Protein alignment between AtRBX1, OsRBX1a, and OsRBX1b.
Supplemental Figure 9. OsCUL3a interacts with OsRBX1a and OsRBX1b in yeast.
Supplemental Figure 10. Subcellular localization of OsCUL3a, OsRBX1a, and OsRBX1b.
Supplemental Figure 11. OsNPR1 transcriptional level in ZH11, oscul3a mutant, and pOsCUL3a plants.
Supplemental Figure 12. Characterization of different types of the osnpr1 mutants created by CRISPR/Cas9.
Supplemental Table 1. Primers used in this study.
Supplemental Data Set 1. Text file of the alignment used in the phylogenetic analysis shown in Supplemental Figure 3.
Supplementary Material
Acknowledgments
We thank Jianli Wu for providing Xoo strains. This project was supported by grants from the National Key Transgenic Program (2016ZX08001002), the Agricultural Science and Technology Innovation Program of Chinese Academy of Agricultural Sciences (CAAS-ASTIP-2013-CNRRI), the National Natural Science Foundation of China (#31571944), and the China Association for Science and Technology.
AUTHOR CONTRIBUTIONS
Q.L., Y.N., Y.Z., N.Y., X.Z., W.W., D.C., and X.W. performed the experiments and analyzed the data. Q.L., Y.N., Y.Z., G.-L.W., S.C., and L.C. designed the project and wrote the manuscript.
Glossary
- ETI
effector-triggered immunity
- PCD
programmed cell death
- DAPK
death-associated protein kinase
- SAR
systemic acquired resistance
- ZH11
Zhonghua11
- dps
days post-sowing
- DAB
3,3 C-diaminobenzidine
- ROS
reactive oxygen species
- dpi
days postinoculation
- SA
salicylic acid
- BSA
bulked segregant analysis
- ORF
open reading frame
- LCI
luciferase complementation imaging
- Y2H
yeast two-hybrid
- co-IP
coimmunoprecipitation
- dai
days after infiltration
Footnotes
Articles can be viewed without a subscription.
References
- Bai W., Chern M., Ruan D., Canlas P.E., Sze-To W.H., Ronald P.C. (2011). Enhanced disease resistance and hypersensitivity to BTH by introduction of an NH1/OsNPR1 paralog. Plant Biotechnol. J. 9: 205–215. [DOI] [PubMed] [Google Scholar]
- Cao H., Bowling S.A., Gordon A.S., Dong X. (1994). Characterization of an Arabidopsis mutant that is nonresponsive to inducers of systemic acquired resistance. Plant Cell 6: 1583–1592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao H., Glazebrook J., Clarke J.D., Volko S., Dong X. (1997). The Arabidopsis NPR1 gene that controls systemic acquired resistance encodes a novel protein containing ankyrin repeats. Cell 88: 57–63. [DOI] [PubMed] [Google Scholar]
- Cao H., Li X., Dong X. (1998). Generation of broad-spectrum disease resistance by overexpression of an essential regulatory gene in systemic acquired resistance. Proc. Natl. Acad. Sci. USA 95: 6531–6536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen A., Wu K., Fuchs S.Y., Tan P., Gomez C., Pan Z.Q. (2000). The conserved RING-H2 finger of ROC1 is required for ubiquitin ligation. J. Biol. Chem. 275: 15432–15439. [DOI] [PubMed] [Google Scholar]
- Chen H., Zou Y., Shang Y., Lin H., Wang Y., Cai R., Tang X., Zhou J.M. (2008). Firefly luciferase complementation imaging assay for protein-protein interactions in plants. Plant Physiol. 146: 368–376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen S., Tao L., Zeng L., Vega-Sanchez M.E., Umemura K., Wang G.L. (2006). A highly efficient transient protoplast system for analyzing defence gene expression and protein-protein interactions in rice. Mol. Plant Pathol. 7: 417–427. [DOI] [PubMed] [Google Scholar]
- Chern M., Fitzgerald H.A., Canlas P.E., Navarre D.A., Ronald P.C. (2005). Overexpression of a rice NPR1 homolog leads to constitutive activation of defense response and hypersensitivity to light. Mol. Plant Microbe Interact. 18: 511–520. [DOI] [PubMed] [Google Scholar]
- Chern M., Xu Q., Bart R.S., Bai W., Ruan D., Sze-To W.H., Canlas P.E., Jain R., Chen X., Ronald P.C. (2016). A genetic screen identifies a requirement for cysteine-rich-receptor-like kinases in rice NH1 (OsNPR1)-mediated immunity. PLoS Genet. 12: e1006049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duan H., Wang Y., Aviram M., Swaroop M., Loo J.A., Bian J., Tian Y., Mueller T., Bisgaier C.L., Sun Y. (1999). SAG, a novel zinc RING finger protein that protects cells from apoptosis induced by redox agents. Mol. Cell. Biol. 19: 3145–3155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faulkner C., Robatzek S. (2012). Plants and pathogens: putting infection strategies and defence mechanisms on the map. Curr. Opin. Plant Biol. 15: 699–707. [DOI] [PubMed] [Google Scholar]
- Figueroa P., Gusmaroli G., Serino G., Habashi J., Ma L., Shen Y., Feng S., Bostick M., Callis J., Hellmann H., Deng X.W. (2005). Arabidopsis has two redundant Cullin3 proteins that are essential for embryo development and that interact with RBX1 and BTB proteins to form multisubunit E3 ubiquitin ligase complexes in vivo. Plant Cell 17: 1180–1195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu Z.Q., Yan S., Saleh A., Wang W., Ruble J., Oka N., Mohan R., Spoel S.H., Tada Y., Zheng N., Dong X. (2012). NPR3 and NPR4 are receptors for the immune signal salicylic acid in plants. Nature 486: 228–232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furniss J.J., Spoel S.H. (2015). Cullin-RING ubiquitin ligases in salicylic acid-mediated plant immune signaling. Front. Plant Sci. 6: 154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gingerich D.J., Gagne J.M., Salter D.W., Hellmann H., Estelle M., Ma L., Vierstra R.D. (2005). Cullins 3a and 3b assemble with members of the broad complex/tramtrack/bric-a-brac (BTB) protein family to form essential ubiquitin-protein ligases (E3s) in Arabidopsis. J. Biol. Chem. 280: 18810–18821. [DOI] [PubMed] [Google Scholar]
- Hu X., Kong X., Wang C., Ma L., Zhao J., Wei J., Zhang X., Loake G.J., Zhang T., Huang J., Yang Y. (2014). Proteasome-mediated degradation of FRIGIDA modulates flowering time in Arabidopsis during vernalization. Plant Cell 26: 4763–4781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hua Z., Vierstra R.D. (2011). The cullin-RING ubiquitin-protein ligases. Annu. Rev. Plant Biol. 62: 299–334. [DOI] [PubMed] [Google Scholar]
- Jiang L., et al. (2013). DWARF 53 acts as a repressor of strigolactone signalling in rice. Nature 504: 401–405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee Y.R., Yuan W.C., Ho H.C., Chen C.H., Shih H.M., Chen R.H. (2010). The Cullin 3 substrate adaptor KLHL20 mediates DAPK ubiquitination to control interferon responses. EMBO J. 29: 1748–1761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J., et al. (2009). Analysis of Drosophila segmentation network identifies a JNK pathway factor overexpressed in kidney cancer. Science 323: 1218–1222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mukhtar M.S., McCormack M.E., Argueso C.T., Pajerowska-Mukhtar K.M. (2016). Pathogen tactics to manipulate plant cell death. Curr. Biol. 26: R608–R619. [DOI] [PubMed] [Google Scholar]
- Ohta T., Michel J.J., Schottelius A.J., Xiong Y. (1999). ROC1, a homolog of APC11, represents a family of cullin partners with an associated ubiquitin ligase activity. Mol. Cell 3: 535–541. [DOI] [PubMed] [Google Scholar]
- Ono E., Wong H.L., Kawasaki T., Hasegawa M., Kodama O., Shimamoto K. (2001). Essential role of the small GTPase Rac in disease resistance of rice. Proc. Natl. Acad. Sci. USA 98: 759–764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park C.H., Chen S., Shirsekar G., Zhou B., Khang C.H., Songkumarn P., Afzal A.J., Ning Y., Wang R., Bellizzi M., Valent B., Wang G.L. (2012). The Magnaporthe oryzae effector AvrPiz-t targets the RING E3 ubiquitin ligase APIP6 to suppress pathogen-associated molecular pattern-triggered immunity in rice. Plant Cell 24: 4748–4762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saleh A., Withers J., Mohan R., Marqués J., Gu Y., Yan S., Zavaliev R., Nomoto M., Tada Y., Dong X. (2015). Posttranslational modifications of the master transcriptional regulator NPR1 enable dynamic but tight control of plant immune responses. Cell Host Microbe 18: 169–182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sasaki A., Itoh H., Gomi K., Ueguchi-Tanaka M., Ishiyama K., Kobayashi M., Jeong D.H., An G., Kitano H., Ashikari M., Matsuoka M. (2003). Accumulation of phosphorylated repressor for gibberellin signaling in an F-box mutant. Science 299: 1896–1898. [DOI] [PubMed] [Google Scholar]
- Schwacke R., Hager A. (1992). Fungal elicitors induce a transient release of active oxygen species from cultured spruce cells that is dependent on Ca(2+) and protein-kinase activity. Planta 187: 136–141. [DOI] [PubMed] [Google Scholar]
- Schwessinger B., Zipfel C. (2008). News from the frontline: recent insights into PAMP-triggered immunity in plants. Curr. Opin. Plant Biol. 11: 389–395. [DOI] [PubMed] [Google Scholar]
- Shan Q., Wang Y., Li J., Gao C. (2014). Genome editing in rice and wheat using the CRISPR/Cas system. Nat. Protoc. 9: 2395–2410. [DOI] [PubMed] [Google Scholar]
- Spoel S.H., Mou Z., Tada Y., Spivey N.W., Genschik P., Dong X. (2009). Proteasome-mediated turnover of the transcription coactivator NPR1 plays dual roles in regulating plant immunity. Cell 137: 860–872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomann A., Brukhin V., Dieterle M., Gheyeselinck J., Vantard M., Grossniklaus U., Genschik P. (2005). Arabidopsis CUL3A and CUL3B genes are essential for normal embryogenesis. Plant J. 43: 437–448. [DOI] [PubMed] [Google Scholar]
- Vierstra R.D. (2009). The ubiquitin-26S proteasome system at the nexus of plant biology. Nat. Rev. Mol. Cell Biol. 10: 385–397. [DOI] [PubMed] [Google Scholar]
- Waadt R., Schmidt L.K., Lohse M., Hashimoto K., Bock R., Kudla J. (2008). Multicolor bimolecular fluorescence complementation reveals simultaneous formation of alternative CBL/CIPK complexes in planta. Plant J. 56: 505–516. [DOI] [PubMed] [Google Scholar]
- Wang Y., Cheng X., Shan Q., Zhang Y., Liu J., Gao C., Qiu J.L. (2014). Simultaneous editing of three homoeoalleles in hexaploid bread wheat confers heritable resistance to powdery mildew. Nat. Biotechnol. 32: 947–951. [DOI] [PubMed] [Google Scholar]
- Wu L., Chen H., Curtis C., Fu Z.Q. (2014). Go in for the kill: How plants deploy effector-triggered immunity to combat pathogens. Virulence 5: 710–721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang G.Y., Pang L., Ge H.L., Tan M., Ye W., Liu X.H., Huang F.P., Wu D.C., Che X.M., Song Y., Wen R., Sun Y. (2001). Attenuation of ischemia-induced mouse brain injury by SAG, a redox-inducible antioxidant protein. J. Cereb. Blood Flow Metab. 21: 722–733. [DOI] [PubMed] [Google Scholar]
- Yang Y., Qi M., Mei C. (2004). Endogenous salicylic acid protects rice plants from oxidative damage caused by aging as well as biotic and abiotic stress. Plant J. 40: 909–919. [DOI] [PubMed] [Google Scholar]
- Yuan Y., Zhong S., Li Q., Zhu Z., Lou Y., Wang L., Wang J., Wang M., Li Q., Yang D., He Z. (2007). Functional analysis of rice NPR1-like genes reveals that OsNPR1/NH1 is the rice orthologue conferring disease resistance with enhanced herbivore susceptibility. Plant Biotechnol. J. 5: 313–324. [DOI] [PubMed] [Google Scholar]
- Zebell S.G., Dong X. (2015). Cell-cycle regulators and cell death in immunity. Cell Host Microbe 18: 402–407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeng L.R., Qu S., Bordeos A., Yang C., Baraoidan M., Yan H., Xie Q., Nahm B.H., Leung H., Wang G.L. (2004). Spotted leaf11, a negative regulator of plant cell death and defense, encodes a U-box/armadillo repeat protein endowed with E3 ubiquitin ligase activity. Plant Cell 16: 2795–2808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou F., et al. (2016). Corrigendum: D14-SCF(D3)-dependent degradation of D53 regulates strigolactone signalling. Nature 532: 402. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.