Interaction with the gene-specific transcription factor WER recruits the histone chaperone NRP1 to the GL2 promoter, where it coactivates GL2 transcription in Arabidopsis root hairs.
Abstract
NUCLEOSOME ASSEMBLY PROTEIN1 (NAP1) defines an evolutionarily conserved family of histone chaperones and loss of function of the Arabidopsis thaliana NAP1 family genes NAP1-RELATED PROTEIN1 (NRP1) and NRP2 causes abnormal root hair formation. Yet, the underlying molecular mechanisms remain unclear. Here, we show that NRP1 interacts with the transcription factor WEREWOLF (WER) in vitro and in vivo and enriches at the GLABRA2 (GL2) promoter in a WER-dependent manner. Crystallographic analysis indicates that NRP1 forms a dimer via its N-terminal α-helix. Mutants of NRP1 that either disrupt the α-helix dimerization or remove the C-terminal acidic tail, impair its binding to histones and WER and concomitantly lead to failure to activate GL2 transcription and to rescue the nrp1-1 nrp2-1 mutant phenotype. Our results further demonstrate that WER-dependent enrichment of NRP1 at the GL2 promoter is involved in local histone eviction and nucleosome loss in vivo. Biochemical competition assays imply that the association between NRP1 and histones may counteract the inhibitory effect of histones on the WER-DNA interaction. Collectively, our study provides important insight into the molecular mechanisms by which histone chaperones are recruited to target chromatin via interaction with a gene-specific transcription factor to moderate chromatin structure for proper root hair development.
INTRODUCTION
Eukaryotic chromatin is a hierarchically packaged superstructure that is highly dynamic in response to varying cellular requirements. The basic structural and functional unit of chromatin is the nucleosome, which consists of 145 to 147 bp of DNA wrapped around a globular histone octamer in roughly 1.65 turns. The octamer is composed of the central (H3-H4)2 tetramer and two flanking H2A/H2B dimers, each with distinct sites of interaction with the wrapping DNA (Luger et al., 2012). Nucleosome assembly establishes the chromatin structure and ensures DNA stability, while nucleosome disassembly releases the DNA template from histones, allowing for diverse metabolic processes such as replication, transcription, and repair. During nucleosome assembly and disassembly, histone chaperones, a large family of proteins with histone binding activity, function to prevent spontaneous aggregation between oppositely charged histones and DNA under physiological conditions. Based on their affinities for different histones, members of this large family are classified as either H2A/H2B or H3/H4 histone chaperones (De Koning et al., 2007; Avvakumov et al., 2011; Zhu et al., 2013; Zhou et al., 2015).
NUCLEOSOME ASSEMBLY PROTEIN1 (NAP1) was originally isolated from eggs of Xenopus laevis. This important H2A/H2B histone chaperone was shown to facilitate nucleosome assembly in vitro (Laskey et al., 1978). Later, NAP1 was found to associate with H2A/H2B and facilitate nucleosome disassembly in coordination with other chromatin factors (Levchenko and Jackson, 2004; Lorch et al., 2006). The Arabidopsis thaliana genome encodes four NAP1 homologs, NAP1;1 to NAP1;4 (Liu et al., 2009), and two NAP1-RELATED PROTEIN (NRP) members, NRP1 and NRP2 (Zhu et al., 2006). Arabidopsis NAP1 and NRP share conserved protein domains (Zhou et al., 2015) and are both required for somatic homologous recombination, the predominant pathway for repair of DNA double-strand breaks (Gao et al., 2012; Zhou et al., 2016). However, loss-of-function mutants of NAP1s and NRPs displayed different phenotypes during plant development (Zhu et al., 2006; Liu et al., 2009). For example, the double mutant defective in both NRP1 and NRP2 (nrp1-1 nrp2-1) showed abnormal phenotypes including short roots and ectopic root hairs (Zhu et al., 2006). The nrp1-1 nrp2-1 double mutant also showed decreased expression levels of GLABRA2 (GL2), which encodes a homeodomain-leucine zipper transcription factor that plays key roles in root hair patterning (Zhu et al., 2006). In contrast, the mutant with the deletion of three NAP1 homologs showed normal root hairs and a GL2 expression level similar to that in the wild type (Liu et al., 2009). Thus, further research is required to determine the specific role of NRPs in root hair patterning, as well as to characterize their underlying mechanisms.
Root hairs, which develop from epidermal cells, are important for plant anchorage, microbial interactions, and nutrient acquisition (Hofer, 1991; Grierson et al., 2014). In Arabidopsis, the epidermal cells are longitudinally arranged along the root and differentiate into hair or non-hair cells in a position-dependent manner. The cells located outside an anticlinal cortical cell wall (H-position) develop into hair cells (H-cells), while those located outside a periclinal cortical cell wall (N-position) develop into non-hair cells (N-cells). Two membrane-localized receptor-like protein kinases, SCRAMBLED (SCM) and BRASSINOSTEROID INSENSITIVE1, function in the signal transduction of position information from different cortex cells to ensure the acquisition of proper epidermal cell fates (Kwak et al., 2005; Kwak and Schiefelbein, 2007; Kuppusamy et al., 2009).
In addition, transcription factor networks and phytohormone signaling pathways play important roles in the specification and formation of cells in the root epidermis (reviewed in Grierson et al., 2014). Specifically, GL2 is the central regulator of epidermal cell fate determination and inhibits hair formation in N-cells. GL2 expression is regulated by a transcription factor complex that consists of an R2R3-type MYB-domain transcription factor WEREWOLF (WER), one of two redundant basic helix-loop-helix (bHLH) transcription factors (GL3 and ENHANCER OF GL3 [EGL3]), and the WD40-repeat transcription factor TRANSPARENT TESTA GLABRA1 (TTG1) (reviewed in Grierson et al., 2014). WER is specifically expressed in N-position cells, and the WER-containing transcription factor complex directly binds to and activates GL2, another R2R3-type MYB gene, MYB23, and the R3-type MYB gene CAPRICE (CPC). The MYB23 protein also binds to its own promoter, forming a positive feedback loop (Kang et al., 2009). Intriguingly, although the CPC protein is expressed in N-position cells, it moves to neighboring H-position cells to repress GL2 expression, thus allowing the corresponding cells to acquire the H-cell fate (Kurata et al., 2005). Notably, the CPC protein negatively regulates the expression of the CPC gene, thus forming a negative feedback loop (Lee and Schiefelbein, 2002). Conversely, although GL3 and EGL3 proteins are expressed in H-position cells, they move to adjacent N-position cells to determine their N-cell fate (Bernhardt et al., 2005; Cheng et al., 2014). The activity of the MYB-bHLH-WD40-repeat transcription factor complex can be modulated by other factors via protein-protein interactions. For example, BRASSINOSTEROID INSENSITIVE2, a brassinosteroid-relevant GSK3-like kinase, can phosphorylate EGL3 and TTG1 to suppress the activity of the WER-GL3/EGL3-TTG1 complex in N-position cells (Cheng et al., 2014). GL2-EXPRESSION MODULATOR (GEM) is another factor that interacts with TTG1 and negatively modulates GL2 expression (Caro et al., 2007). Recently, the GL2 protein was shown to directly bind to and suppress the expression of five bHLH transcription factor genes, including ROOT HAIR DEFECTIVE6 (RHD6) (Lin et al., 2015), a key factor involved in root hair formation mediated by the phytohormones auxin and ethylene (Masucci and Schiefelbein, 1994).
Root hair patterning and formation also show plasticity in their responses to environmental signals. The fates of root epidermis cells can be regulated by salt stress (Wang et al., 2008), phosphorus or iron availability (Zhang et al., 2003; Müller and Schmidt, 2004), and nanoparticle exposure (García-Sánchez et al., 2015). In many cases, a change in root hair fate can be reversible, which represents a coordinated strategy for environmental adaptation and implicates the involvement of epigenetic regulation. Notably, GEM was shown to play a role in regulating histone H3 acetylation and H3K9 methylation in chromatin at the GL2 and CPC loci (Caro et al., 2007). Two members of the histone deacetylase family, HISTONE DEACETYLASE18 (HDA18) and HDA6, affect epidermal cell fate by modulating the transcript levels of GL2-centered network genes through changing their local histone acetylation status (Liu et al., 2013; Li et al., 2015). Three-dimensional fluorescence in situ hybridization analyses revealed that the chromatin at the GL2 locus is in an open state in N-position cells but in a closed state in H-position cells (Costa and Shaw, 2006). Intriguingly, the chromatin state of the GL2 gene was found to reset at mitosis and respecify during the following G1 phase (Costa and Shaw, 2006). Understanding the molecular mechanisms underlying changes in chromatin state at the GL2 locus is of great importance.
In this study, we report that the histone chaperone NRP1 directly interacts with the WER transcription factor to fully activate the expression of GL2 through mediating histone eviction and nucleosome disassembly. Our results represent an important link between a cell type-specific transcription factor and a histone chaperone for epigenetic regulation of cell fate determination in the root epidermis of Arabidopsis.
RESULTS
NRPs Are Required for Full GL2 Expression
In our previous study, we reported the downregulation of GL2 and the ectopic root hair phenotype in the nrp1-1 nrp2-1 double mutant (Zhu et al., 2006). Here, using quantitative RT-PCR, we confirmed that GL2 was downregulated by ∼2-fold in the roots of nrp1-1 nrp2-1 at 12 d after germination (DAG). Meanwhile, the transcript levels of GL2 upstream regulators such as WER, CPC, TTG1, GL3, EGL3, and MYB23, which encode the members of the MYB-bHLH-WD40 complex, and those of other regulators such as GEM and SCM, were unchanged (Figure 1A). To further determine the GL2 expression at the cellular level, we introduced the GUS expression reporter pGL2:GUS (Szymanski et al., 1998) into the nrp1-1 nrp2-1 double mutant plants. Consistent with the quantitative RT-PCR analysis, GUS staining was obviously weaker in N-position cells of nrp1-1 nrp2-1 than in those of the wild type (Figures 1B and 1C), but the spatial expression patterns of GL2 were largely similar in both types of plants (Figures 1B and 1C).
Figure 1.
NRPs Are Required for Full Expression of GL2.
(A) Relative expression levels of root hair-related genes in nrp1-1 nrp2-1 double mutant. Roots of the wild type and nrp1-1 nrp2-1 were collected at 12 DAG for RNA isolation and quantitative RT-PCR. Values are normalized to ACTIN2. Error bars show sd from three biological replicates. Mean values of relative gene expression levels in nrp1-1 nrp2-1 compared with that in the wild type (set as 1) are shown with error bars. Asterisk indicates statistically significant difference (P < 0.01).
(B) Expression patterns of GL2:GUS reporter in root tips of the wild type (WT) and nrp1-1 nrp2-1 double mutant. Bar = 50 μm.
(C) Transverse sections were prepared with the root tips of the wild type and nrp1-1 nrp2-1 harboring reporter GL2:GUS. Bars = 20 μm.
(D) Time course of NRP1 induction. Gene expression in wild-type and transgenic plants harboring ES:YFP-NRP1 in nrp1-1 nrp2-1 background at 12 DAG was induced by 4 μM estradiol. Roots were collected at the indicated times for RNA isolation and quantitative RT-PCR examination. Values are normalized to ACTIN2. Error bars show sd from three biological replicates. Mean values of relative NRP1 expression levels in transgenic plants compared with that in the wild type (set to 1) are shown with error bars.
(E) Time course of relative changes in GL2 and TTG1 transcript levels, determined using the same RNA as that used in (D). Mean values of relative GL2 and TTG1 expression levels in induced transgenic plants compared with those in uninduced ones (set to 1) are shown with error bars.
Next, we examined the recovery of GL2 transcript levels after short-term expression of exogenous estradiol-induced YFP-NRP1 (ES:YFP-NRP1), which can completely rescue the phenotypes of nrp1-1 nrp2-1 mutant plants (Zhu et al., 2006). Upon estradiol induction, the transcript level of NRP1 rose quickly and reached a plateau after 6 h (Figure 1D). Meanwhile, the transcript level of GL2 quickly increased and eventually stabilized at about twice the preinduction level after 4 h, similar to the expression level of GL2 in the wild type (Figure 1E). The induction time observed here was far shorter than the duration of the cell cycle and DNA replication in Arabidopsis root cells (17 h in the meristematic zone and 30 h in the elongation zone) (Hayashi et al., 2013), suggesting that NRP1-mediated GL2 activation precedes cellular division/differentiation in root hair determinacy. As a control, TTG1, whose expression was unaffected in nrp1-1 nrp2-1, showed little change in its expression level during the estradiol induction (Figure 1E). Examination of NRP1, TTG1, and GL2 expression in wild-type plants revealed that estradiol has no detectable side effect (Supplemental Figure 1), which is in agreement with the original report establishing the ES-inducible system in plants (Zuo et al., 2000).
Collectively, our results indicate that GL2 transcription can rapidly respond to NRP1 expression. Although NRP1 was dramatically overexpressed under the estradiol-inducing system, its overload did not lead to GL2 overexpression. This latter observation suggests that NRPs are necessary but not rate-limiting to activate GL2 expression.
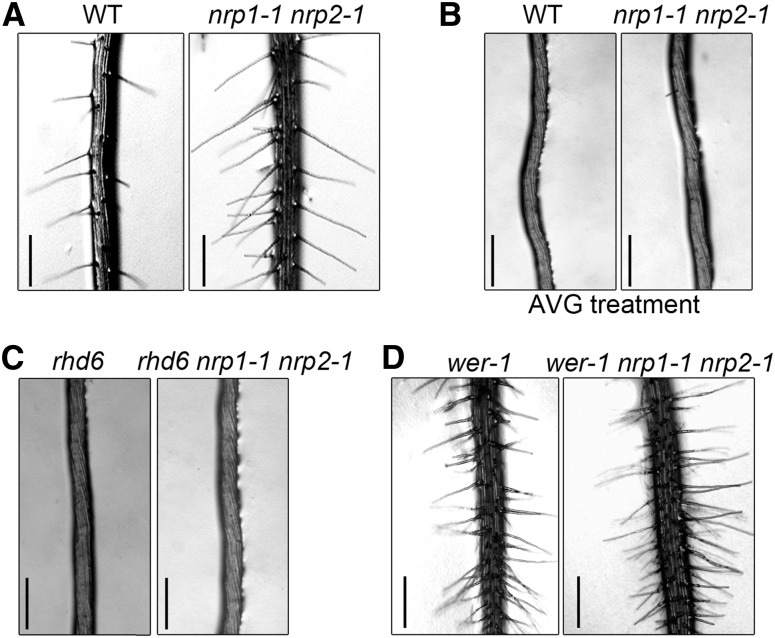
NRPs Act Upstream of Ethylene/RHD6 and in Conjunction with WER-Containing Complex to Regulate Root Hair Development
In order to better understand NRPs function in root hair development, we tested whether NRPs affect the ethylene and auxin pathways, both of which are important for root hair formation. Treatment with an ethylene synthesis inhibitor, aminoethoxyvinylglycine (AVG), inhibited nearly all the root hairs of nrp1-1 nrp2-1, similar to those of the wild type (Figures 2A and 2B), indicating that the ethylene pathway acts downstream of NRP activity. The GFP reporter driven by the auxin response promoter DR5 (DR5:GFP), which monitors auxin distribution (Ottenschläger et al., 2003), showed that the auxin distribution patterns were similar between nrp1-1 nrp2-1 and the wild type (Supplemental Figure 2). A mutation in RHD6, which is associated with the ethylene and auxin pathways, converted the ectopic root hair phenotype of nrp1-1 nrp2-1 into the hairless state (Figure 2C). Transcripts of ethylene/auxin-related genes (Masucci and Schiefelbein, 1996) and RHD6 remained at nearly wild-type levels in nrp1-1 nrp2-1 (Supplemental Figure 3). These molecular and genetic analyses indicated that ethylene/RHD6 pathways act downstream of NRPs in root hair formation.
Figure 2.
NRPs Act Upstream of Ethylene/RHD6 Pathways and in Conjunction with WER-Containing Transcription Factor Complex.
The wild type (WT) and nrp1-1 nrp2-1 were vertically cultured in normal culture medium (A) and medium containing 25 μM AVG (B), respectively. Mutants of rhd6 (C) and wer-1 (D) were introgressed into nrp1-1 nrp2-1 to obtain rhd6 nrp1-1 nrp2-1 and wer-1 nrp1-1 nrp2-1 triple mutants, respectively. Plants were grown vertically in normal culture medium. Images were taken at 12 DAG. Bars = 0.5 mm.
The WER-GL3/EGL3-TTG1 transcription factor complex is known to directly activate GL2 expression in N-cells, while replacement of WER by CPC in the complex inhibits GL2 expression in H-cells (Grierson et al., 2014). Because NRP1 also functions as an activator of GL2, we wondered whether NRP1 acts in conjunction with the WER-containing complex. We then introgressed the wer-1 mutant into nrp1-1 nrp2-1. The wer-1 nrp1-1 nrp2-1 triple mutant plants displayed a hairy root phenotype, which was overall similar to the wer-1 single mutant (Lee and Schiefelbein, 1999) but more severe than the nrp1-1 nrp2-1 double mutant (Figure 2D), suggesting that NRPs may act to regulate root hair in a WER-dependent genetic pathway. The nrp1-1 nrp2-1 double mutant has fewer root hairs than the wer-1 nrp1-1 nrp2-1 triple mutant, suggesting other redundant chromatin factors could be involved in the regulation of root hair development.
To verify the roles of NRPs in cell specification of the root epidermis, hair cells (H-cells) and non-hair cells (N-cells) were quantified as previously reported (Simon et al., 2007) for different genotypes and treatment conditions tested in our study (Table 1). We found that there were more H-cells in the untreated nrp1-1 nrp2-1, wer-1, and wer-1 nrp1-1 nrp2-1 mutants, which is consistent with the hairy phenotype of these mutants. Together, our results suggest that NRPs act in conjunction with WER, a key activator of GL2, in cell fate determination of hair cells and non-hair cells.
Table 1. Cell-Type Specification in the Root Epidermis.
| H-Position |
N-Position |
|||
|---|---|---|---|---|
| H-Cells (%) | N-Cells (%) | H-Cells (%) | N-Cells (%) | |
| Wild type | 97.5 ± 1.5 | 2.5 ± 1.5 | 1.8 ± 1.1 | 98.2 ± 1.1 |
| nrp1-1 nrp2-1 | 96.9 ± 1.8 | 3.1 ± 1.8 | 42.0 ± 9.8 | 58.0 ± 9.8 |
| nrp1-1 nrp2-1 (ES:YFP-NRP1) | 96.8 ± 1.2 | 3.2 ± 1.2 | 2.5 ± 1.4 | 97.5 ± 1.4 |
| nrp1-1 nrp2-1 (ES:YFP-NRP1-mN) | 97.7 ± 2.2 | 2.3 ± 2.2 | 44.3 ± 6.2 | 55.7 ± 6.2 |
| nrp1-1 nrp2-1 (ES:YFP-NRP1-ΔC) | 95.8 ± 2.8 | 4.2 ± 2.8 | 43.2 ± 7.7 | 56.8 ± 7.7 |
| Wild type (AVG treated) | 1.4 ± 1.2 | 98.6 ± 1.2 | 0.9 ± 0.7 | 99.1 ± 0.7 |
| nrp1-1 nrp2-1 (AVG treated) | 1.9 ± 1.5 | 98.1 ± 1.5 | 1.3 ± 1.1 | 98.7 ± 1.1 |
| rhd6 | 2.2 ± 2.2 | 97.8 ± 2.2 | 1.9 ± 0.9 | 98.1 ± 0.9 |
| rhd6 nrp1-1 nrp2-1 | 2.7 ± 1.3 | 97.3 ± 1.3 | 2.4 ± 1.0 | 97.6 ± 1.0 |
| wer-1 | 95.6 ± 3.3 | 4.4 ± 3.3 | 94.4 ± 2.5 | 5.6 ± 2.5 |
| wer-1 nrp1-1 nrp2-1 | 97.2 ± 2.9 | 2.8 ± 2.9 | 95.6 ± 1.8 | 4.4 ± 1.8 |
Values indicate the mean ± sd of at least three independent experiments. In each experiment, the percentage of H- or N-cells is determined from at least 10 roots for each sample.
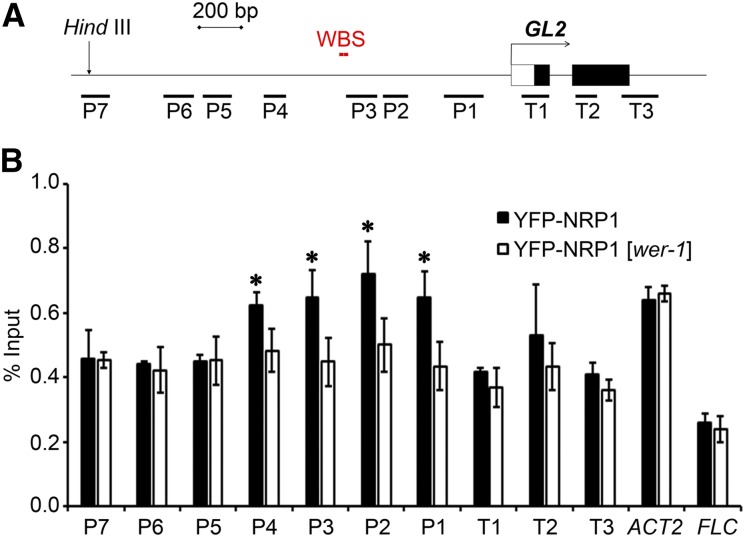
NRP1 Is Recruited to the GL2 Promoter in a WER-Dependent Manner
Previously, we reported that NRP1 directly binds to the chromatin of the GL2 locus (Zhu et al., 2006). Another study showed that a 2.1-kb DNA fragment of the GL2 promoter was able to functionally control GL2 transcription (Szymanski et al., 1998). To investigate in detail the occupancy of NRP1 at GL2, we designed seven primer pairs within the 2.1-kb promoter segment (P1 to P7) and three primer pairs downstream from the transcription initiation site (T1 to T3) for chromatin immunoprecipitation (ChIP)-PCR analyses (Figure 3A). Transgenic plants harboring ES:YFP-NRP1 in the nrp1-1 nrp2-1 background (Zhu et al., 2006) were used in the ChIP assay, and we found that YFP-NRP1 was clearly enriched at the regions from P1 to P4, which are close to the WER binding site (WBS; Song et al., 2011) of the GL2 promoter (Figures 3A and 3B). To investigate whether the enrichment of NRP1 at the GL2 promoter was dependent on WER, we analyzed YFP-NRP1 binding activity in the wer-1 mutant background. Interestingly, the enrichment of YFP-NRP1 in the P1-to-P4 regions of GL2 significantly decreased in the wer-1 background (Figure 3B), indicating that NRP1 binds to the GL2 promoter in a WER-dependent manner. The other examined regions of GL2 as well as the ACT2 and FLC genes, used as controls, did not show any significant difference of YFP-NRP1 binding between the wild type and wer-1 (Figure 3). To further evaluate the specific enrichment of NRP1 at the GL2 promoter, we also examined NAP1;3, a major isoform of the NAP1 family proteins in Arabidopsis (Liu et al., 2009). In contrast to YFP-NRP1, no obvious enrichment of YFP-NAP1;3 was observed at the examined regions of GL2 (Supplemental Figure 4), which is consistent with the normal root hair pattern and GL2 transcription level in the loss-of-function nap1 mutant (Liu et al., 2009).
Figure 3.
Enrichment of NRP1 at GL2 Promoter Is Dependent on WER.
(A) Schematic representation of structures of GL2 promoter and first two exon/introns. Black boxes represent exons, white box represents the untranscribed region, lines represent promoter and introns. Number-labeled bars (P1–P7 and T1–T3) represent regions amplified by primer pairs corresponding to numbers on x axis of graph below. The HindIII site represents start of GL2 functional promoter and WBS indicates the location of the WER binding sites.
(B) Enrichment of YFP-NRP1 at the GL2 promoter. Roots of plants vertically grown and collected at 12 DAG were fixed with formaldehyde for ChIP analysis using polyclonal anti-GFP antibody. Error bars show sd from three biological replicates. Asterisks indicate statistically significant difference of YFP-NRP1 enrichment at GL2, FLC, and ACTIN2 (ACT2) between the wild type and wer-1 (P < 0.05).
NRP1 Physically Interacts with WER
Because both WER and NRP1 act as activators of GL2 and they share overlapping binding regions in the GL2 promoter, we wondered whether NRP1 could physically interact with WER. To test this, we purified GST-WER recombinant protein and used it as the bait in a pull-down assay. We found that YFP-NRP1 expressed in the transgenic plants was specifically retained by GST-WER beads, but not by GST beads (Figure 4A). We further verified the interaction between WER and NRP1 via a bimolecular fluorescence complementation (BiFC) assay, in which Yn and Yc, the complementary moieties of YFP, were fused to NRP1 and WER, respectively. The YFP signal was detected only in tobacco leaf cells coexpressing NRP1 and WER fusion proteins but not in the negative controls (Figure 4B).Next, we expressed and purified the full-length recombinant NRP1 and WER without tags from Escherichia coli (Supplemental Figure 5) and analyzed their interaction by size exclusion chromatography (SEC). The elution volume (Ve) of the WER protein was 15.6 mL (Figure 4C), which corresponds to the estimated size of WER. The Ve of NRP1 was 11.5 mL (Figure 4C), which is far away from the estimated size of a monomer but closer to the estimated size of a dimer of the NRP1 protein. The difference between the effective Ve value (11.5 mL) and the estimated Ve of 15 mL for a dimer of NRP1 suggests that the NRP1 dimer is not folded in a globular conformation. Interestingly, when NRP1 and WER were mixed at a molar ratio of 1:1, the Ve shifted to a single peak at 10.6 mL (Figure 4C), which indicates that NRP1 can form a stable complex with WER at a 1:1 molar ratio. When NRP1 and WER were mixed at a molar ratio of 1:2, the complex also eluted at a constant Ve at 10.6 mL and an additional peak corresponding to excessive WER was observed at 15.6 mL (Supplemental Figure 6). These SEC analyses revealed that NRP1 and WER can directly interact in vitro and form a stable protein complex at 1:1 molar ratio. Collectively, our results demonstrate that the histone chaperone NRP1 directly interacts with WER in vitro and in vivo, providing molecular evidence for the WER-dependent recruitment of NRP1 to the GL2 promoter.
Figure 4.
NRP1 Directly Interacts with Transcription Factor WER in Vitro and in Vivo.
(A) Pull-down assay. Protein extracts from transgenic plants expressing YFP-NRP1 were incubated with beads coated with GST and GST-WER. Quantity/purity of GST and GST-WER beads were analyzed by SDS-PAGE gel stained by Coomassie blue (CBB) (left panel). Two percent of the input and pull-down fractions were analyzed by immunoblot using polyclonal anti-GFP antibody (indicated by the arrowhead, right panel).
(B) BiFC analysis of interaction between NRP1 and WER in tobacco leaf cells. Bars = 50 μm.
(C) SEC profiles (Superdex 200 10/300 GL) of NRP1 (blue) and WER (red), as well as their mixture at a molar ratio of 1:1 (black).
Crystal Structure of NRP1 Reveals That It Dimerizes via Its N-Terminal α-Helix
To give more insight into molecular basis of NRP1 dimer formation, we determined the crystal structure of NRP1. Using the multiwavelength anomalous diffraction method, we determined the crystal structure of NRP1 using a truncated version of the protein (amino acids 19–225) lacking both N and C termini and with two L-to-M mutations (L20M and L220M) for selenium-methionine (Se-Met) labeling (Supplemental Table 1). We found that two NRP1 molecules form a dimer via hydrophobic interactions between the two antiparallel N-terminal long α-helices (19–77 amino acids) and that an earmuff domain is attached to each end of the α-helix (Figures 5A and 5B; PDB code 5DAY). The earmuff domain contains six α-helices, four-stranded antiparallel β-sheets, and disordered fragments between β4/α5 (163–184 amino acids) and α5/α6 (192–203 amino acids) (dashed lines in Figure 5C). Protein sequence alignment revealed that NRP1 and NRP2 are more similar to human TEMPLATE-ACTIVATING FACTOR-Iβ (HsTAF-Iβ) (41% identity) than to yeast VACUOLAR PROTEIN SORTING75 (ScVPS75) (22% identity) and yeast NAP1 (ScNAP1) (16% identity). Different from NRP1 and NRP2, Arabidopsis NAP1;1 (21% identity) and yeast NAP1 (Park and Luger, 2006) contain an extra accessory domain between the dimerization domain and the earmuff domain (Supplemental Figure 7). Consistent with this, the overall structure of NRP1 showed higher similarity to that of HsTAF-Iβ (PDB code 2E50; Muto et al., 2007) with root-mean-square derivation (r.m.s.d.) of 2.40 Å over 285 amino acids than to those of ScNAP1 (PDB code 2AYU; Park and Luger, 2006) with r.m.s.d. of 7.34 Å over 254 amino acids and ScVPS75 (PDB code 3DM7; Tang et al., 2008) with r.m.s.d. of 6.76 Å over 268 amino acids (Supplemental Figure 8). Notably, when superimposing earmuff domains, the N-terminal long α-helix backbones of these four proteins showed distinct directions (Supplemental Figure 8). Furthermore, the ScNAP1 backbone helix showed a sigmoid shape in the top view (Park and Luger, 2006), whereas that of NRP1 was nearly straight (Supplemental Figure 8).
Figure 5.
Crystal Structure Shows That NRP1 Forms a Dimer through Its N-Terminal α-Helices.
(A) Overall structure of NRP1 (19–225 amino acids) dimer shown as ribbon diagram. Dimerization domain (19–78 amino acids) is shown in blue. Structures of α2-α4 (79–106 amino acids), β1–β4 (107–156 amino acids), and α5–α7 (157–225 amino acids) in earmuff domain are shown in green, yellow, and red, respectively.
(B) Structure of dimerization domain of NRP1. Hydrophobic amino acids within two antiparallel N-terminal long α-helices are labeled in black (α1) and red (α1’).
(C) Labeled structure of earmuff domain. Dashed lines show peptides (163–184 amino acids) and (192–203 amino acids) between β4 and α6.
The structure also provides insight about the potential histone binding sites. A previous study showed that mutations of N (S162A/K164A/D165A), O (T191A/T194A/D195A), or P (D202A/E203A/E206A) on the surface of HsTAF-Iβ decreased its histone binding activity (Muto et al., 2007). Based on the comparison of sequence and structure with HsTAF-Iβ, we found that the amino acids within the N, O, and P mutations are mostly conserved in NRP1, with seven of them clearly visualized on the NRP1 crystal structure (Supplemental Figures 7 and 9). We marked these seven amino acids (E161, T188, T191, D192, D205, E206, and D209) on the NRP1 structure, corresponding, respectively, to S162, T191, T194, D195, D202, E203, and E206 of HsTAF-Iβ, to show the putative interaction surface for histones binding (Supplemental Figure 9). Together, the structure analysis indicates that NRP1 forms a dimer and that the conserved negatively charged surface is consistent with its histone binding activity.
Disruption of Dimerization or Acidic C Terminus of NRP1 Diminishes Its Interaction with Histones and with WER
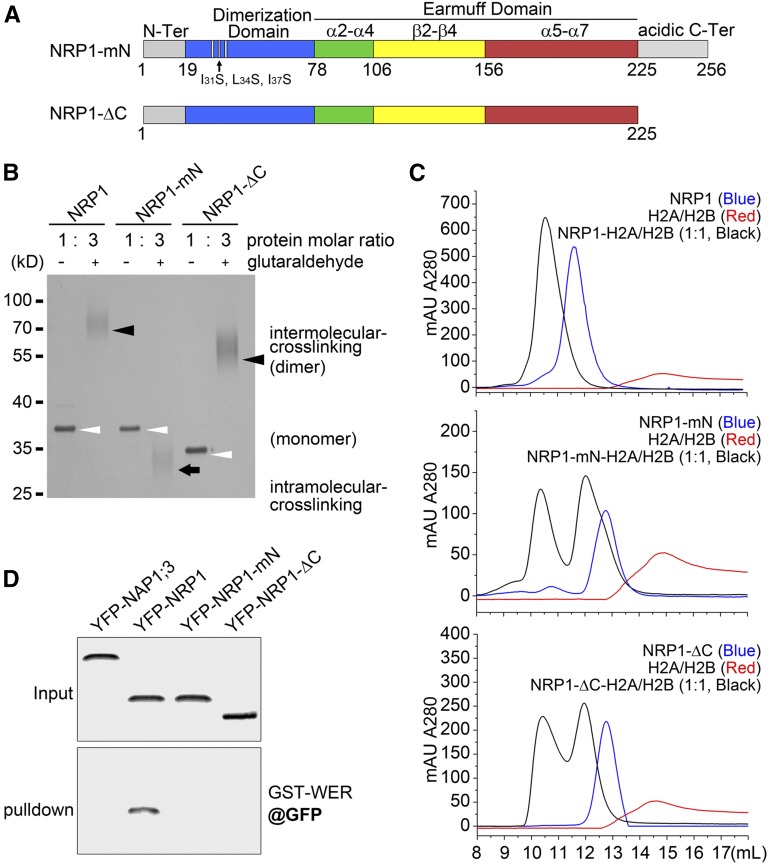
The crystal structure of NRP1 revealed an essential role of the N-terminal long α-helix for NRP1 protein dimerization. For further verification, we mutated the first three hydrophobic amino acids (Ile-31, Leu-34, and Ile-37) within the N-terminal α-helix of NRP1 to the hydrophilic residue Ser. This mutant was designated NRP1-mutated N-terminus (NRP1-mN) (Figure 6A). The effect of the mutation was first examined in a glutaraldehyde-mediated coupling assay because glutaraldehyde can cause intermolecular cross-linking. Treatment with glutaraldehyde resulted in dimerization of NRP1, and the dimer was detected in the denaturing gel electrophoresis analysis (Figure 6B). In contrast, NRP1-mN totally lost the ability to dimerize and displayed a distinct pattern in the electrophoresis analysis due to intramolecular cross-linking (Figure 6B), indicating that the long α-helix is critical for NRP1 dimerization.
Figure 6.
Disruption of Dimerization or Acidic C Terminus of NRP1 Diminishes Its Interaction with Histones and with WER.
(A) Diagrammatic representations of NRP1-mN and NRP1-ΔC. In NRP1-mN, three hydrophobic amino acids (Ile-31, Leu-34, and Ile-37) in dimerization domain were replaced by hydrophilic Ser residue. In NRP1-ΔC, acidic C terminus (226–256 amino acids) was deleted.
(B) Glutaraldehyde coupling of NRP1 and its mutants. Open arrowheads indicate monomer of each uncoupled protein. Black arrowheads indicate dimer of coupled proteins formed by intermolecular cross-linking. Black arrow indicates coupled form of NRP1-mN, likely formed by intramolecular cross-linking. Bands of cross-linked proteins broaden in the SDS-PAGE, so 3-fold glutaraldehyde-treated proteins were loaded in the gel compared with the controls to provide a clear image.
(C) SEC profiles (Superdex 200 10/300 GL) of NRP1, NRP1-mN, and NRP1-ΔC (each in blue) and H2A/H2B dimer (red), as well as their mixtures (molar ratio of 1:1 in black). Buffer was 20 mM Tris-HCl, pH 8.0, and 150 mM NaCl. Note that NRP1 and H2A/H2B dimer complex eluted as a single peak at 1:1 molar ratio, indicating that a NRP1 dimer could stably bind two molecules of H2A/H2B dimer. Neither NRP1-mN nor NRP1-ΔC mixed with H2A/H2B dimer displayed a single peak in SEC profiles.
(D) Pull-down assay. Protein extracts of transgenic plants overexpressing YFP-NAP1;3, YFP-NRP1, YFP-NRP1-mN, and YFP-NRP1-ΔC were incubated with beads coated with GST-WER. Input and pull-down fractions were analyzed by immunoblot using polyclonal anti-GFP antibody.
The C terminus of NRP1 (226–256 amino acids) is flexible and highly enriched with acidic residues (Asp and Glu), which explains why we were unable to obtain crystals of NRP1 with the acidic C terminus. To analyze its function, we constructed a truncated mutant lacking the acidic C terminus (amino acids 1–225, designated NRP1-ΔC). The dimeric form of NRP1-ΔC was observed in the presence of glutaraldehyde (Figure 6B), indicating that the C-terminal acidic tail is not required for the dimerization of NRP1.
Although the recombinant NRP1, NRP1-mN, and NRP1-ΔC proteins were stable (Supplemental Figure 5), we further checked their secondary structure by circular dichroism spectroscopy. We found that NRP1, NRP1-mN, and NRP1-ΔC display similar circular dichroism spectra, indicating that the mutation/deletion probably did not affect the secondary structure of NRP1 (Supplemental Figure 10).
In yeast, one NAP1 dimer can bind one (H2A/H2B)2 unconventional tetramer, so the stoichiometric ratio of the NAP1 monomer to H2A/H2B dimer is 1:1 (D’Arcy et al. 2013). Our SEC assay showed that the mixture of NRP1 and the H2A/H2B dimer at 1:1 molar ratio was eluted in a single symmetric peak of Ve at 10.5 mL (Figure 6C, upper panel). Although NRP1 and H2A/H2B at a 1:2 molar ratio showed an asymmetric peak and those at 2:1 failed to form a single peak (Supplemental Figure 11), both mixtures showed a peak at 10.5 mL similar to that at 1:1 molar ratio. These results indicated that one NRP1 homodimer could associate with two H2A/H2B heterodimers to form a stable complex. In contrast, although NRP1-mN and NRP1-ΔC formed a similar complex with H2A/H2B dimer as NRP1 did at 10.5 mL, a single symmetric SEC peak was not obtained in the assays with either NRP1-mN and H2A/H2B or NRP1-ΔC and H2A/H2B mixtures at 1:1 (Figure 6C, middle and lower panels), 1:2, or 2:1 molar ratios (Supplemental Figure 11), indicating that neither NRP1-mN nor NRP1-ΔC could form a complex with the H2A/H2B dimer as strongly as did NRP1.
Next, we examined whether NRP1-mN and NRP1-ΔC would affect the NRP1-WER interaction. The SEC analysis showed that both NRP1-mN and NRP1-ΔC formed complexes with WER, but excessive WER can be clearly observed for the mixture of WER and NRP1-ΔC, indicating a reduced ability of complex formation due to the C-terminal deletion of the NRP1 protein (Supplemental Figure 12). The equilibrium dissociation constant (Kd) values of WER-NRP1, WER-NRP1-mN, and WER-NRP1-ΔC were measured as 1.70, 2.18, and 2.99 μM, respectively, by electrophoretic mobility shift assay (EMSA; Supplemental Figure 13), indicating that not only C-terminal deletion but also the N-terminal mutation of NRP1 decreases its binding activity with WER.
Lastly, we investigated the effect of mutations by pull-down assays using transgenic plant lines expressing YFP-NRP1-mN or YFP-NRP1-ΔC. We selected those lines expressing fusion proteins at similar levels compared with that in ES:YFP-NRP1 transgenic plants. Fluorescence microscopy observation confirmed that similar to YFP-NRP1 both YFP-NRP1-mN and YFP-NRP1-ΔC were mainly localized in the nucleus (Supplemental Figure 14). In a pull-down assay, YFP-NRP1 was retained by GST-WER beads, whereas YFP-NRP1-mN, YFP-NRP1-ΔC, and YFP-NAP1;3 were not (Figure 6D). This is consistent with the reduced binding affinity of these mutant proteins to WER. Compared with the SEC and EMSA assays using purified recombinant proteins, the pull-down assay from total plant protein extracts is more restricted, which likely explains the total absence of YFP-NRP1-mN and YFP-NRP1-ΔC in the pull-down detection. Taken together, our results establish that either the disruption of dimerization or the deletion of the acidic C terminus of NRP1 impairs its interaction with histones as well as with WER.
Dimerization and the Acidic C Terminus Are Essential for NRP1 Function in Planta
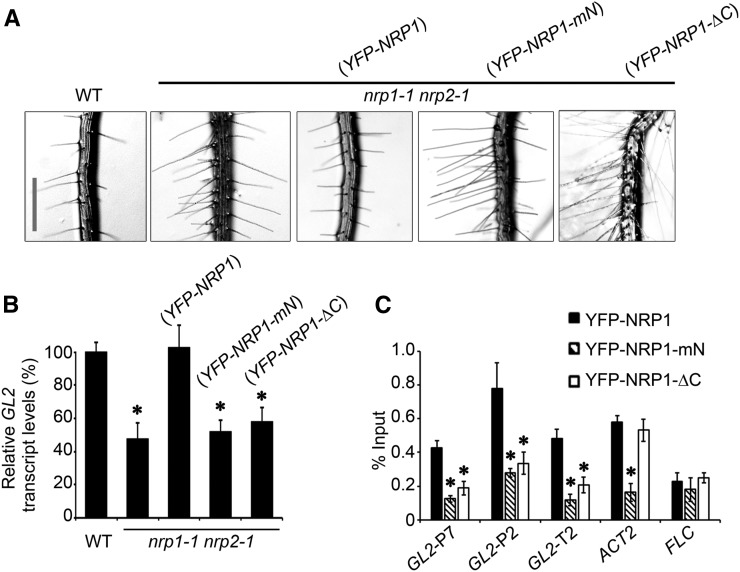
Next, we examined the function of the N-terminal α-helix and the acidic C-terminal tail of NRP1 in planta by transgenic plant rescue experiments. After estradiol induction, YFP-NRP1 fully rescued the phenotype of nrp1-1 nrp2-1, whereas YFP-NRP1-mN and YFP-NRP1-ΔC did not (Figure 7A, Table 1), although they were expressed at similar levels as YFP-NRP1 (Figure 6D). Consistent with this, the transgenic plants expressing NRP1-mN or NRP1-ΔC failed to fully activate GL2 transcription (Figure 7B).
Figure 7.
NRP1-mN and NRP1-ΔC Could Not Promote Full GL2 Expression, Leading to Loss of NRP1 Function in Planta.
(A) Root hair phenotypes of wild-type (WT), nrp1-1 nrp2-1, and transgenic plants harboring ES:YFP-NRP1, ES:YFP-NRP1-mN, and ES:YFP-NRP1-ΔC in nrp1-1 nrp2-1 background, respectively. Plants were grown vertically in culture medium supplemented with 4 μM estradiol. Images were taken at 12 DAG. Bar = 0.5 mm.
(B) Relative transcription of GL2 in roots of plants in (A). Values are normalized to ACTIN2. Error bars show sd from three biological replicates. Mean values of the relative GL2 levels compared with that in the wild type (set to 100%) are shown with error bars. Asterisks indicate statistically significant difference of relative GL2 transcription between the wild type and nrp1-1 nrp2-1 (P < 0.01).
(C) Enrichment of YFP-NRP1, YFP-NRP1-mN, or YFP-NRP1-ΔC at GL2 promoter. Roots from transgenic plants in (A) were fixed with formaldehyde for ChIP analysis using polyclonal anti-GFP antibody. Error bars show sd from three biological replicates. Asterisks indicate statistically significant difference of enrichment at GL2, FLC, and ACT2 between YFP-NRP1 and YFP-NRP1-mN/YFP-NRP1-ΔC (P < 0.01).
We then performed a ChIP analysis to investigate the recruitment of NRP1-mN and NRP1-ΔC to GL2. The relative binding efficiency of YFP-NRP1-ΔC to the examined GL2 regions was significantly decreased (Figure 7C). The relative binding efficiency of YFP-NRP1-mN was also decreased in all examined GL2 regions as well as at ACT2 but not FLC (Figure 7C). Together, these data indicate that both NRP1-mN and NRP1-ΔC failed to enrich at GL2, to activate GL2 transcription, and to rescue the phenotype of nrp1-1 nrp2-1, together providing strong evidence supporting that both NRP1 dimerization and its acidic tail are critical for the function of NRP1 in planta.
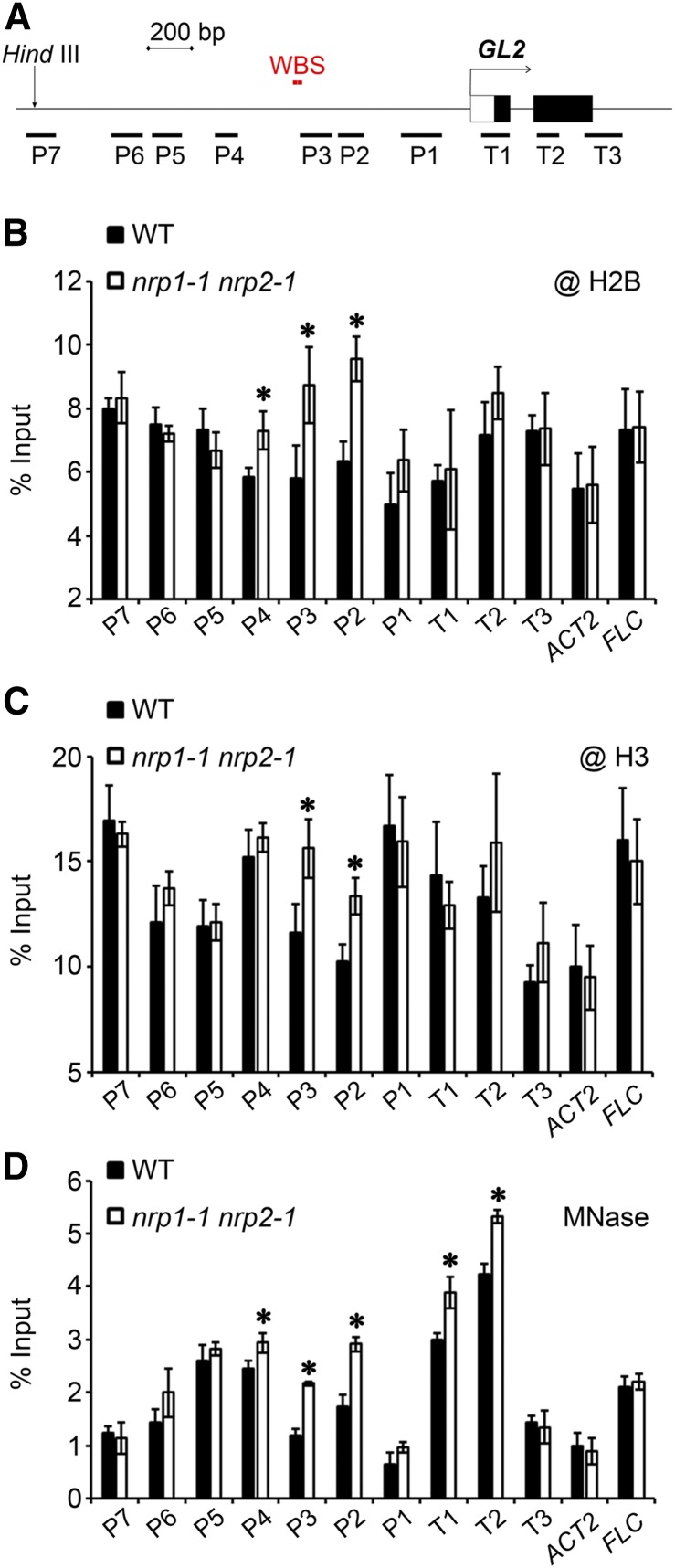
NRPs Promote Histone Eviction and Nucleosome Removal at GL2 Promoter
In order to get insight about mechanism of NRP function at chromatin level, we tested the histone occupancy and nucleosome density at GL2 (Figure 8A). Roots of wild-type and nrp1-1 nrp2-1 plants at 12 DAG were collected and antibodies against histone H2B or H3 were used in ChIP assays. Comparing the relative histone occupancy in nrp1-1 nrp2-1 with that in the wild type, we found an obvious enrichment of H2B at regions P2 to P4 (Figure 8B) and a clear enrichment of H3 at regions P2 and P3 of GL2 (Figure 8C). The other examined GL2 regions as well as ACT2 and FLC did not show significant differences between nrp1-1 nrp2-1 and the wild type (Figures 8B and 8C). These findings indicated that NRPs may promote the removal of histones at the GL2 promoter regions around the WBSs.
Figure 8.
NRPs Promote Histone Release and Decrease Nucleosome Density at GL2 Promoter.
(A) Schematic representation of structures of GL2 promoter and first two exons.
(B) and (C) Histone occupancy at promoter regions of GL2. Roots of vertically grown wild type and nrp1-1 nrp2-1 at 12 DAG were fixed with formaldehyde for ChIP analysis using commercial antibodies against H2B (B) and H3 (C). Error bars show sd from three biological replicates. Asterisks indicate statistically significant difference of histone enrichment at GL2, FLC, and ACT2 between the wild type and nrp1-1 nrp2-1 (P < 0.05).
(D) Nucleosome density at the GL2 promoter in nrp1-1 nrp2-1. Nuclei were isolated from roots of the wild type and nrp1-1 nrp2-1 at 12 DAG and were treated with MNase. The digested DNA fragments were purified for quantitative PCR analysis, and the intact genomic DNA without digestion were used as the input. Error bars show sd from three biological replicates. Asterisks indicate statistically significant difference of DNA level between the wild type and nrp1-1 nrp2-1 (P < 0.05).
We next investigated chromatin accessibility to the micrococcal nuclease (MNase) at GL2. MNase has a higher affinity for naked DNA than for nucleosomal DNA wrapped around histones. Consequently, the chromatin integrity after MNase attack is correlated with nucleosome density. The nuclei from wild-type and nrp1-1 nrp2-1 roots were purified and digested to mononucleosomes by MNase and then the DNA was purified and used in quantitative PCR analysis. The assay showed that the P2-P4 regions of GL2 are less accessible to MNase in nrp1-1 nrp2-1 than in the wild type (Figure 8D), which is in agreement with the impaired removal of histones around the WBS as observed in ChIP assay. Remarkably, the T1 and T2 regions of GL2 also showed a reduced accessibility to MNase in nrp1-1 nrp2-1 compared with that in the wild type (Figure 8D). It is possible that the impaired transcription has rendered the T1 and T2 regions, which are located just downstream from the transcription initiation site of GL2, more occupied and thus less accessible to MNase treatment. Similar to ChIP assay (Figures 8B and 2C), the other examined GL2 regions as well as ACT2 and FLC did not show significant difference between nrp1-1 nrp2-1 and the wild type in MNase assay (Figure 8D). Taken together, our results suggest that NRPs may facilitate histone eviction and nucleosome removal at GL2, particularly around the WBS within the promoter, in promoting GL2 transcription.
NRP1 Releases the Inhibitory Effect of Histones on WER-DNA Interaction
Next, we explored the molecular mechanism of WER-dependent NRP1 recruitment to the target DNA. First, we synthesized and used a 21-bp double-stranded DNA fragment (5′-GAAAATGCGGTTGGAGAATTA-3′), which contained one of the two WER binding sites within the GL2 promoter (Song et al., 2011), as the DNA substrate for WER in binding assays. In SEC analysis, when WER was added to DNA at a molar ratio of 1:1, a single peak containing both protein (measured at A280) and DNA (measured at A254) was observed before the elution of WER or DNA alone (Figure 9A), indicating that WER bound to DNA and formed a stable complex at 1:1 molar ratio.
Figure 9.
WER Forms a Stable Complex with Its Target DNA When NRP1 Associates with H2A/H2B Dimer.
(A) SEC profiles (Superdex 200 10/300 GL) of WER (A280 in cyan) and its DNA substrate (A254 in black), as well as their complex at 1:1 molar ratio (A254 in red; A280 in blue).
(B) EMSA of NRP1, H2A/H2B dimer, WER, and its substrate DNA. Lanes 1 to 14, 2 μM DNA (black arrow, set as relative molar ratio at 1). Lanes 2 to 4, WER titrated against DNA at molar ratios of 2:1, 4:1, and 6:1. Lanes 5 to 9, H2A/H2B dimer titrated against DNA at molar ratios of 1:1, 2:1, 4:1, 5:1, and 6:1, while amount of WER was held at 6:1 molar ratio. Lanes 10 to 14, NRP1 titrated against DNA at molar ratios of 1:1, 2:1, 4:1, 5:1, and 6:1, while WER and H2A/H2B dimer were both held at molar ratio of 6:1 against DNA. Lane 15, H2A/H2B dimer and NRP1 as in lane 14 without WER and its substrate DNA. All samples were separated on 6% native PAGE gel, stained with GelRed, and visualized by UV (upper panel). The GelRed-stained gel shows bands of DNA bound to H2A/H2B (black arrowhead) and DNA bound to WER (open arrowhead). Gel was further stained by CBB to show bands of NRP12-(H2A/H2B)4 (green arrowhead), NRP12-(H2A/H2B)2 (red arrowhead), and WER bound to DNA (purple arrowhead).
Next, we analyzed the roles of histones and NRP1 in the WER-DNA association by biochemical competition assays. WER formed a stable complex with its substrate DNA in EMSA (Figure 9B, lanes 1 to 4, upper panel [GelRed]). However, the WER-DNA complex was disrupted by adding incremental amounts of H2A/H2B, and histone-DNA complexes gradually formed (Figure 9B, lanes 5–9, upper panel), which is consistent with the lower Kd of H2A/H2B-DNA at 0.26 μM compared with that of WER-DNA at 0.55 μM (Supplemental Figure 13). Our observation implies that histones inhibit the WER-DNA interaction and that WER cannot break the histone-DNA association to access its binding site. When excess histones were added (H2A/H2B:DNA at a molar ratio of 6:1), the WER-DNA complex was barely detectable. In contrast, when increasing amounts of the histone chaperone NRP1 were added, then the WER-DNA band gradually recovered and increasing amount of the NRP1-H2A/H2B complex appeared (Figure 9B, lanes 10 to 14), indicating that NRP1 competitively removed out histones from the DNA binding. Based on the fact that stable NRP1-H2A/H2B complexes can form at both 1:1 and 1:2 molar ratios (Supplemental Figure 15) and that NRP1 can form a dimer, the protein complexes observed in Figure 9B were deduced as NRP12-(H2A/H2B)2 and NRP12-(H2A/H2B)4, which is in agreement with the previous study on the yeast NAP1 and H2A/H2B complex formation (D’Arcy et al. 2013). Lastly, we tested the effect of NRP1 on WER binding to DNA-H2A/H2B. We found that addition of WER alone to DNA-H2A/H2B caused aggregates, which remained in the gel slots (Supplemental Figure 16). In contrast, addition of NRP1-WER to DNA-H2A/H2B resulted in WER-DNA formation and dissociation of DNA-H2A/H2B complex (Supplemental Figure 16).
Together, our results indicate that the histone chaperone NRP1 plays a key role in WER binding to histone-associated DNA, likely through NRP1-histone binding and release of DNA accessibility.
DISCUSSION
Although histone chaperones are known to play important roles in nucleosome dynamics, little is known about the mechanism by which they are recruited to specific chromatin regions to exert their functions (Venkatesh and Workman, 2015; Zhou et al., 2015). In this study, we show that NRP1 directly induces GL2 activation and is enriched at the GL2 promoter in a WER-dependent manner. As revealed in our in vitro and in vivo experiments, WER directly interacts with the histone chaperone NRP1, indicating that NRP1 can be selectively recruited to target chromatin regions by the gene-specific transcription factor WER. In this way, histone chaperones are selectively recruited to target chromatin regions by gene-specific transcription factors, where they affect chromatin structure and gene expression during plant development.
Recruitment of NRP1 to the GL2 promoter
In this study, we showed that the histone chaperone NRP1 directly interacts with WER to activate GL2 expression in Arabidopsis roots. Nevertheless, in the nrp1-1 nrp2-1 double mutant, GL2 is still expressed but to a level roughly ∼50% of that in the wild type. The facts that the spatial pattern of pGL2:GUS expression remains largely similar in the nrp1-1 nrp2-1 double mutant as in the wild type and that the ES-induced NRP1 rapidly promotes GL2 expression strongly argue that the impaired GL2 expression is the cause rather than a consequence of the nrp1-1 nrp2-1 mutant root hair phenotype. It is known that GL2 functions in a dosage-dependent manner in regulation of root hair formation (Masucci et al., 1996). Our observed decrease of GL2 expression is consistent with ectopic root hair formation at N-position in the nrp1-1 nrp2-1 double mutant.
Meanwhile, the remaining expression of GL2 in nrp1-1 nrp2-1 also implies that WER can bind to and activate GL2 without NRPs. We found that in vitro WER alone cannot efficiently bind H2A/H2B-coated DNA, whereas NRP1-WER complex can (Supplemental Figure 16). Thus, the question arises: Without NRPs, how does WER bind to its target DNA within the chromatin context? In cells of eukaryotes, dynamic nucleosome assembly and disassembly occur along the genome chromatin; thus, the WBS on GL2 promoter could be transiently open and then WER would be able to bind directly to the accessible DNA.
In the nucleus of wild-type plants, WER and NRP1 form a complex, which may direct NRP1 to the GL2 promoter. This idea is supported by our observation that NRP1 enrichment around the WBS of GL2 is WER dependent. When WER binds to its target DNA, NRP1 could be released to associate with the nearby nucleosomal histones, resulting in histone eviction and nucleosome dissociation. Subsequently, the opened chromatin site may facilitate recruitment of other factors to form a stable WER-associated transcription complex (Grierson et al., 2014) in effective activation of GL2 transcription. Future studies will be required to investigate other factors involved as well as to explore timely activation process of GL2 transcription.
Regulation of Nucleosome Dynamics at GL2 Locus
Previously, the H3-H4 histone chaperone FASCIATA2 (FAS2), a subunit of the CHROMATIN ASSEMBLY FACTOR-1 complex in plants, was shown to be required for root hair patterning. In a fas2 mutant (in the Ler background), the chromatin was in an open state in most H-position cells and GL2 was ectopically expressed (Costa and Shaw, 2006). However, the requirement for FAS2 in root hair differentiation seems to be ecotype specific because the ectopic expression of GL2 and impaired root hair phenotype observed in fas2 (in the Ler background) in a previous study (Costa and Shaw, 2006) were not observed in fas2-4 (in the Col background) in our study (Supplemental Figure 17). This result suggested that FAS2 may play different roles in different Arabidopsis ecotypes. It is currently unknown how the FAS2 protein functions to regulate GL2 expression in the Ler background in Arabidopsis and whether or not FAS2 can bind to GL2 chromatin.
Our study showed that in the Col background NRP1 and NRP2 are involved in regulation of GL2 expression and root hair formation. Nevertheless, the root hair phenotype of the nrp1-1 nrp2-1 double mutant is obviously weaker than those of the wer-1 single and the wer-1 nrp1-1 nrp2-1 triple mutants, suggesting that redundant chromatin factors could be involved in GL2 activation. Other histone chaperones are likely candidates; for example, FACILITATES CHROMATIN TRANSCRIPTION, another conserved H2A/H2B histone chaperone, also functions in H2A/H2B eviction and nucleosome disassembly (Morillo-Huesca et al., 2010; Formosa, 2012). Chromatin remodeling factors are alternative candidates because they show ATP-dependent nucleosome assembly/disassembly activities. Also, there are several lines of evidence that chromatin-remodeling factors coordinate with histone chaperones (reviewed in Clapier and Cairns, 2009). For example, yeast NAP1 was found to facilitate transcription and nucleosome disassembly in the presence of the chromatin-remodeling complex RSC (Lorch et al., 2006). Our ChIP analyses revealed the accumulation of histone H2B and the retention of H3 at the GL2 promoter in nrp1-1 nrp2-1 compared with the wild type, suggesting that NRPs participate in the formation of histone-free chromatin regions during transcription. Whether or not NRPs cooperate with some chromatin-remodeling factors to regulate GL2 remains an interesting issue to be resolved in future research. Additionally, histone deacetylases were shown to modulate GL2 transcription by changing the local histone acetylation status (Liu et al., 2013; Li et al., 2015). Future genetic screening based on the ectopic root hair phenotype of the nrp1-1 nrp2-1 double mutant will help to elucidate the NRP1-dependent and -independent mechanisms for GL2 activation and to unravel the molecular interplay among different chromatin factors during transcriptional regulation and cell fate determination.
NRP1 May Regulate Different Target Genes through Different Mechanisms
As shown in the structural comparison (Supplemental Figure 8), the NRP1 protein shows the highest similarity to human TAF-Iβ (HsTAF-Iβ). Whereas WER recruits NRP1 to activate the expression of GL2, the interactions between HsTAF-Iβ and specific transcription factors repress gene expression (Suzuki et al., 2003; Miyamoto et al., 2003). HsTAF-Iβ masks the DNA binding activity of Sp1, a C2H2-type zinc finger transcription factor, and this inhibition of Sp1 eliminates its ability to activate its target genes (Suzuki et al., 2003). HsTAF-Iβ also interacts with the DNA binding domain of KLF5, a cardiovascular transcription factor, and thus negatively regulates the DNA binding, transactivation, and cell proliferative activities of KLF5 (Miyamoto et al., 2003). HsTAF-Iβ functions in gene repression by interacting with the DNA binding domain of the transcription factors Sp1 and KLF5, implying that the binding strength of HsTAF-Iβ to transcription factors is stronger than that of the transcription factors to the target DNA. In contrast, our results showed that the interaction between WER and NRP1 is weaker than that between WER and its target DNA. Moreover, when NRP1 is recruited to the GL2 promoter by WER, NRP1 is released from the WER-NRP1 complex and associates with histones to promote nucleosome removal and the WER-DNA interaction, thus activating GL2.
In addition to GL2, more genes were misregulated in the nrp1-1 nrp2-1 double mutant (Zhu et al., 2006). For example, PLETHORA2 (PLT2), which encodes an AP2 family transcription factor that is essential for root quiescent center specification and stem cell activity (Aida et al., 2004), was upregulated in nrp1-1 nrp2-1. NRP1 also binds to chromatin at the PLT2 locus (Zhu et al., 2006), suggesting that NRP1 represses the transcription of PLT2. It is possible that NRP1 functions via a mechanism similar to that of HsTAF-Iβ to repress transcription. Further identification of novel NRP1-interacting proteins will help to clarify whether interactions between histone chaperones and specific transcription factors are a conserved mechanism to recruit histone chaperones and will also uncover more molecular mechanisms by which NRPs regulate transcription.
Structural Basis of Arabidopsis NRP1 Function
In this study, we determined the crystal structure of Arabidopsis NRP1. The crystallography analysis showed that NRP1 forms a dimer through its long N-terminal α-helix, which is similar to other NAP1 family proteins, such as ScNAP1, ScVPS75, and HsTAF-Iβ. When its dimerization was disrupted, the mutant protein NRP1-mN failed to enrich at GL2 and to rescue the phenotypes of the nrp1-1 nrp2-1 double mutant (Figure 7). Similarly, the acidic C-terminal of NRP1 was also shown to be important for NRP1 function in vivo (Figure 7).
A structural comparison between Arabidopsis NRP1 (this study) and yeast NAP1 (Park and Luger, 2006) provided some insights into the molecular basis of the similarities and differences between NAP1 and NRPs (Supplemental Figure 8). The overall similarity in the structures of NRP1 and ScNAP1 suggested that they have homologous roles. For example, both can form dimers and show histone binding activity (Levchenko and Jackson, 2004; Lorch et al., 2006; Liu et al., 2009). However, their structural differences also imply that the specific function of NRP1 differs from that of ScNAP1. ScNAP1 contains a nuclear export sequence within the long helix backbone masked by an accessory domain and a nuclear localization sequence in the antiparallel β-sheet. This structure is consistent with the fact that ScNAP1 is a nucleo-cytoplasmic shuttling protein (Miyaji-Yamaguchi et al., 2003). Both the nuclear export sequence and the accessory domain are conserved in all Arabidopsis NAP1 homologs, but are not found in NRPs (Zhou et al., 2015). Although the role of the accessory domain remains unclear, its location at the side of helix backbone could explain why Arabidopsis NAP1 generally localizes in the cytoplasm (Liu et al., 2009). On the other hand, NRPs are mainly localized in the nuclei of Arabidopsis (Zhu et al., 2006), independently from whether or not the antiparallel interaction of two long α-helix backbones is disrupted (YFP-NRP1-mN; this study). The distinct cellular localization may explain why NRP1 specifically interacts with WER in vivo, but NAP1;3 does not (this study), even though the latter can also form a dimer and bind to histone H2A/H2B (Liu et al., 2009).
In conclusion, our results provide important insight into the molecular mechanisms by which histone chaperones are recruited to specific chromatin regions via interaction with a gene-specific transcription factor and how they act as coactivators to promote gene expression through histone eviction and nucleosome dissociation. The crystal structure and functional domain analysis of NRP1 provide useful information to understand the different specificities of NRP1 and NAP1. The NRP1-WER interaction identified in this study may constitute a significant step toward determining the structure of the NRP1-WER complex. Elucidating the structure of this complex will clarify the molecular functions of both the histone chaperone NRP1 and the cell type-specific transcription factor WER.
METHODS
Plant Materials and Growth Conditions
All Arabidopsis thaliana strains used in this study were in the Columbia (Col) background. The double mutant nrp1-1 nrp2-1 has been described previously (Zhu et al., 2006). The pGL2:GUS (N8851) and DR5:GFP (N9402) marker lines as well as the wer-1 (CS6349) and rhd6 (CS6347) mutant lines were obtained from the Arabidopsis Biological Resource Centre (http://www.arabidopsis.org). Plants were cultured in vitro on agar-solidified Murashige and Skoog medium M0255 (Duschefa) supplemented with 0.9% sucrose at 21°C under a 16-h-light/8-h-dark photoperiod in a Percival AR41L5 growth chamber in 60 to 90 μE⋅m−2⋅s−1 white light. Estradiol (Sigma-Aldrich; E2758) and the ethylene biosynthesis inhibitor AVG (Sigma-Aldrich; A6685) were included in the medium when needed.
RT-PCR Analysis
Three independent groups of plants were vertically grown on agar-solidified medium. For estradiol induction, 10 mL liquid Murashige and Skoog medium with 4 μM estradiol was added to plants at 12 DAG. From each group of plants, roots were collected as one biological replicate. RNA isolation and quantitative RT-PCR were performed as described previously (Zhang et al. 2015). Statistical analysis was performed using OriginPro 7.5 software (OriginLab) as previously reported (Gao et al., 2012). The primers used for RT-PCR are listed in Supplemental Table 2.
Histochemical GUS staining
GUS activity was assayed by incubating plant tissues in GUS staining buffer (Bu et al., 2014) for 6 h at 37°C. Transverse sections (5 μm) were cut from dehydrated roots as previously described (Masucci et al., 1996). The root tips or the transverse sections were observed directly under a Carl Zeiss Axio Imager A2 microscope.
Microscopy
The pattern of epidermal cell types was determined as previously reported (Simon et al., 2007). A light microscope (Axio Imager A2; Zeiss) was used to determine cell type and relative locations of epidermal cells. An epidermal cell was counted as an H-cell if any protrusion was visible, regardless of its length. The proportions of H- and N-cells in root epidermis were determined by examining at least 10 roots from each sample in three independent experiments.
ChIP Analysis
ChIP was performed as previously described (Johnson et al., 2002). Three independent groups of plants were vertically grown on agar-solidified medium. From each group of plants, ∼1 g of roots of 12-d-old seedlings were collected and fixed as one biological replicate. The antibodies used in ChIP were GFP (Invitrogen; A-11122), H3 (Abcam; ab1791), and H2B (Millipore; 07-371). Quantitative PCR was performed to determine the enrichment of DNA immunoprecipitated in the ChIP experiments, using primers listed in Supplemental Table 2. The efficiency values are the ratios determined by taking a fixed aliquot of the DNA extracted from the immunoprecipitated samples and the input. Error bars show sd from three independent experiments. Statistical analyses were performed using OriginPro 7.5 software (OriginLab) as previously reported (Gao et al., 2012). The primers used for RT-PCR are listed in Supplemental Table 2.
Protein Expression and Purification
NRP1-mN was generated from the open reading frame (ORF) of NRP1 (Zhu et al., 2006) using a Takara MutanBEST kit. NRP1-ΔC and NRP1 (19-225) were generated by PCR. The ORF of WER was generated by RT-PCR and subcloned into pGEXT-4T-1 (GE Healthcare). The purification of GST-WER from Escherichia coli was as described previously (Zhu et al., 2011). A DNA fragment encoding a 6×His-SUMO-tag was added to NRP1, NRP1 mutants, NAP1, and WER, and then the constructs were each subcloned into the pET28a vector (Novagen). The 6×His-SUMO-tagged proteins were expressed in the E. coli Rosetta (DE3) strain. Their expression was induced by adding 0.2 mM isopropyl β-d-thiogalactoside to E. coli cells at an OD600 of 0.6. After growth at 20°C for 18 h, cells were harvested, lysed by a high-pressure disruptor, and then purified using Ni-chelating sepharose. The 6×His-SUMO tag was cleaved by ulp1 protease, and the proteins without the tag were further purified by Q FF anion exchange (for NRP1, NRP1 mutants, and NAP1) or SP FF cation exchange (for WER), and finally by Superdex 200 16/60 (GE Healthcare) gel filtration chromatography. The purified NRP1, NRP1-mN, NRP1-ΔC, and NAP1 proteins were concentrated and stored at 20 mg/mL in 20 mM Tris-HCl, pH 8.0, and 150 mM NaCl. The full-length WER was concentrated and stored at 10 mg/mL in 20 mM Tris-HCl, pH 7.5, 500 mM NaCl, and 1 mM DTT. Se-Met-labeled NRP1 (amino acids 19–225) was produced by inhibiting the methionine biosynthesis pathway in the host cell.
The ORFs of HTA1 (AT5G54640) and HTB1 (AT1G07790), which encode Arabidopsis H2A and H2B, respectively, were generated by PCR. A DNA fragment (5′-AATAATTTTGTTTAACTTTAAGAAGGAGATATACATATG-3′) was added to the 5′ primer of HTB1 to create a TATA box and a ribosome binding site, and then HTA1 and HTB1 were connected and subcloned into the pET28a vector (Novagen). H2A and H2B were coexpressed in the E. coli BL21 (DE3) strain and purified by SP FF cation exchange and Superdex 75 16/60 (GE Healthcare) gel filtration chromatography. The purified H2A/H2B dimer was stored at 40 mg/mL in buffer (20 mM Tris-HCl, pH 8.0, and 1 M NaCl).
Constructs and Plant Transformation
The DNA fragments of NRP1-mN and NRP1-ΔC were fused in frame to the 3′-end of the ORF encoding YFP, resulting in YFP-NRP1-mN and YFP-NRP1-ΔC. The fusion constructs were cloned into the pER8 vector (Zuo et al., 2000) with an estradiol-inducible promoter, and the resulting ES:YFP-NRP1-mN and ES:YFP-NRP1-ΔC plasmids were then introduced into the double mutant nrp1-1 nrp2-1 via Agrobacterium tumefaciens GV1301 as previously described (Zhu et al., 2006).
Pull-Down Assay
The pull-down assay was performed as described previously (Zhang et al., 2015). Protein extracts from transgenic plants expressing YFP-NAP1;3, YFP-NRP1, YFP-NRP1-mN, and YFP-NRP1-ΔC were used in the pull-down assay. Briefly, matrix-bound GST-WER or GST recombinant proteins were mixed with protein extracts in PBS buffer containing 1× PBS, 0.2% Nonidet P-40, 1 mM DTT, and protease inhibitor cocktail (Roche) at 4°C for 4 h. After washing three times with wash buffer containing 20 mM Tris-HCl (pH 8.0), 0.5 M NaCl, 0.5% Nonidet P-40, 1 mM DTT, and protease inhibitor cocktail, the bead-bound fractions were separated on SDS-PAGE for immunoblotting using antibody against GFP (Invitrogen; A-11122).
BiFC Assay
The BiFC assays were performed as described previously (Bu et al., 2014). Leaves of 4- to 8-week-old Nicotiana benthamiana plants were coinfiltrated with Agrobacterium strain GV1301 carrying transgene constructs. Localization of BiFC fluorescence was observed using a confocal laser scanning microscope (LSM 710; Zeiss).
Crystallization and Data Collection
Crystals of native and Se-Met-substituted NRP1 (19–225 amino acids) were grown by hanging-drop vapor diffusion at 16°C. Briefly, 20 mg/mL NRP1 (19–225 amino acids) in 20 mM Tris-HCl, pH 8.0, and 150 mM NaCl was combined at a 1:1 ratio with reservoir solution containing 25% PEG400, 0.1 M HEPES, pH 7.6, and 0.2 M calcium chloride dihydrate. The crystals grew to a maximum size of 0.2 × 0.2 × 1 mm.
For data collection, crystals were flash frozen (100K) in the above solution supplemented with 25% glycerol. A total of 360 frames with 1° oscillation were collected for the native and Se-Met-labeled NRP1 (19-225 amino acids) crystals at the Shanghai Synchrotron Radiation Facility beamline 17U. The data were processed with HKL2000 (HKL Research). The space group of the crystals was P212121. The data collection and refinement statistics of native NRP1 (19–225 amino acids) are listed in Supplemental Table 1.
Structure Determination
The structure of Se-Met NRP1 (19–225 amino acids) was solved using multiwavelength anomalous diffraction. The positions of heavy atoms were determined and the model was built with the program Coot (Emsley and Cowtan, 2004). The structure model was refined against the native data set using REFMAC (CCP4 package) (Collaborative Computational Project, Number 4, 1994) and PHENIX/COOT. The structural images, electrostatic surface representation, and structure superpositions were created with PyMOL (http://pymol.sourceforge.net/). The structure of NRP1 (19–225 amino acids) has been deposited in wwPDB (http://deposit.wwpdb.org) under the PDB ID 5DAY.
Glutaraldehyde Cross-Linking
For the glutaraldehyde cross-linking analysis, 60 ng NRP1, NRP1-mN, or NRP1-ΔC protein was incubated at room temperature in buffer containing Tris-HCl, pH 8.0, 100 mN NaCl, 1 mM EDTA, and 10% glycerol, with or without 0.05% glutaraldehyde (Sigma-Aldrich; G5882). The loading buffer for SDS-PAGE was added to terminate the reaction, and the samples were heated at 98°C for 5 min before separation. Protein bands were detected by silver staining.
SEC Analysis
NRP1, NRP1-mN, NRP1-ΔC, and H2A/H2B dimers were mixed and incubated on ice for 30 min in a buffer containing 20 mM Tris-HCl, pH 8.0, and 150 mM NaCl. Mixtures were then loaded on Superdex 200 10/300 GL in the same buffer and the elution profiles were recorded. A 21-bp DNA fragment (5′-GAAAATGCGGTTGGAGAATTA-3′) was synthesized as the substrate DNA of WER. For gel filtration with DNA, the buffer was 20 mM Tris-HCl, pH 7.5, 150 mM NaCl, and 1 mM DTT.
MNase treatment
Three independent groups of plants were vertically grown on agar-solidified medium. From each group of 12-d-old seedlings, roots were collected as one biological replicate. Nuclei were extracted from roots and were treated with MNase as previously described (Li et al., 2014). Briefly, nuclei were digested with 0.02 units μL−1 MNase (Takara; D2910) for 15 min at room temperature. The digestion was stopped by the addition of EDTA to a final concentration of 4 mM and further treated with RNase A (Sigma-Aldrich; R6513) and Proteinase K (Roche; 03115836001). The digested DNA was purified with phenol/chloroform extraction and then precipitated with salts and ethanol in the presence of glycogen (Takara; D605A). Purified DNA was run on a 2% agarose gel, and fragments within 100 to 200 bp were collected and purified by a gel purification kit (Qiagen) and were used for quantitative PCR analysis using primers listed in Supplemental Table 2.
EMSA for Competition
The substrate DNA of WER was purified by PAGE gel. The EMSA buffer contains 20 mM Tris-HCl, pH 7.5, 150 mM NaCl, and 2 mM DTT. The DNA was dissolved to 2 mM in EMSA buffer and annealed to form double-stranded DNA by heating to 95°C for 10 min then to 5°C for 10 min. The DNA, WER, H2A/H2B, and NRP1 were diluted to 40, 40, 40, and 40 μM with EMSA buffer, respectively.
For Figure 9B, the final volume for each sample is 20 μL. DNA (1 μL) was titrated against WER at 2, 4, or 6 μL and incubated on ice for 30 min. The DNA (1 μL) and WER (6 μL) mixture was titrated against H2A/H2B at 1, 2, 4, 5, or 6 μL and incubated on ice for 30 min. The DNA (1 μL), WER (6 μL), and H2A/H2B (6 μL) mixture was titrated against NRP1 at 1, 2, 4, 5, or 6 μL and incubated on ice for 30 min.
For Supplemental Figure 16, the final volume for each sample is 20 μL. WER-NRP1 complex (20 μM) was prepared by adding equal volume of WER and NRP1 and incubated on ice for 30 min. DNA (1 μL) was titrated against H2A/H2B at 2, 3, and 4 μL and incubated on ice for 30min. WER or WER-NRP1 complex was added to DNA (1 μL) and H2A/H2B (4 μL) mixture, respectively, and incubated on ice for 30 min.
Samples were separated by 6% native PAGE gel in 0.5× TBE buffer at 4°C. The DNA and proteins in the native PAGE gel were visualized by GelRed and Coomassie Brilliant Blue staining, respectively.
EMSA Assay for Kd Measurement
The single-stranded DNA (5′-GAAAATGCGGTTGGAGAATTA-3′) was labeled FAM at 5′ end and annealed with single-stranded DNA (5′-TAATTCTCCAACCGCATTTTC-3′) to form double-stranded DNA. Double-stranded DNA (0.1 μM) labeled by FAM was mixed with H2A/H2B from 0.039 to 20 μM or WER from 0.00625 to 3.2 μM on ice for 30 min. Samples were separated by 6% native PAGE gel in 0.5× TBE buffer at 4°C. The DNA in the native PAGE gels were imaged by Typhoon FLA 9000 (GE Healthcare). The decreased quantity of DNA was calculated by ImageJ and the curves were fitted using GraphPad Prism5.
NRP1, NRP1-mN, or NRP1-ΔC (0.4 μM) was mixed with WER increased from 0.039 to 20 μM, and 0.4 μM NRP1 was mixed with H2A/H2B from 0.039 to 20 μM. The mixtures were incubated on ice for 30 min. Samples were separated by 6% native PAGE gel in 0.5× TBE buffer at 4°C. The proteins in the native PAGE gel were visualized by silver staining. The decreased quantity of protein was calculated by ImageJ and the curves were fitted using GraphPad Prism5.
Accession Numbers
Sequence data from this article can be found in the Arabidopsis Genome Initiative under the following accession numbers: WER (At5g14750), CPC (At2g46410), TTG1 (At5g24520), GL3 (At5g41315), EGL3 (At1g63650), MYB23 (AT5G40330), GEM1 (At2g22475), SCM (At1g11130), GL2 (At1g79840), NRP1 (At1g74560), NRP2 (At1g18800), RHD6 (At1g66470), and NAP1;3 (At5g56950).
Supplemental Data
Supplemental Figure 1. Transcript Levels of Examined Genes in Roots of the Wild Type.
Supplemental Figure 2. Auxin Reporter DR5:GFP Reveals Similar Auxin Distribution in Roots of the Wild Type and nrp1-1 nrp2-1.
Supplemental Figure 3. Similar Transcript Levels of Ethylene/Auxin-Related Genes and RHD6 in the Wild Type and nrp1-1 nrp2-1.
Supplemental Figure 4. YFP-NAP1;3 Is Not Enriched at the GL2 Promoter.
Supplemental Figure 5. Purified Recombinant Proteins used in This Study.
Supplemental Figure 6. Size Exclusion Chromatography of NRP1 and WER Mixture.
Supplemental Figure 7. Protein Sequence Alignment of NAP1 Family Members.
Supplemental Figure 8. Structural Comparison of AtNRP1 with ScNAP1, ScVps75, and HsTAF-Iβ.
Supplemental Figure 9. Charge Distribution of NRP1 (19–225 Amino Acids) Dimer.
Supplemental Figure 10. Circular Dichroism Spectroscopy of NRP1, NRP1-mN, and NRP1-ΔC.
Supplemental Figure 11. Size Exclusion Chromatography of the H2A/H2B Dimer with NRP1, NRP1-mN, or NRP1-ΔC.
Supplemental Figure 12. Size Exclusion Chromatography of WER-NRP1-mN and WER-NRP1ΔC.
Supplemental Figure 13. Binding Affinity by Electrophoretic Mobility Shift Assay.
Supplemental Figure 14. Both YFP-NRP1-mN and YFP-NRP1-ΔC Localize in the Nuclei, Like YFP-NRP1.
Supplemental Figure 15. Size Exclusion Chromatography of NRP1-H2A/H2B.
Supplemental Figure 16. The WER-NRP1 Complex More Efficiently Binds to H2A/H2B-Coated DNA Than WER Alone.
Supplemental Figure 17. Normal Root Hair Phenotype of fas2-4 Mutant in Col Background.
Supplemental Table 1. Crystallographic Statistics.
Supplemental Table 2. Primers Used in This Study.
Supplementary Material
Acknowledgments
This work was supported by National Basic Research Program of China (973 Program, Grant 2012CB910500) and by the National Natural Science Foundation of China (Grants NSFC 91519308, NSFC31570315, NSFC31370752, NSFC90919003, and NSFC31671341). The research was conducted within the context of the International Associated Laboratory Plant Epigenome Research. We thank Hong Ma, Hongquan Yang, and Hai Huang for critical reading of the manuscript.
AUTHOR CONTRIBUTIONS
A.D., Y.Z., and J.M. conceived and designed the research. Y.Z., L.R., Q.L., B.W., N.Z., Y.Y., C.Z., H.F., and L.Z. performed the experiments. Y.Z., L.R., and Q.L. analyzed the data. Y.Z., W.-H.S., J.M., and A.D. wrote the article. All authors read, revised, and approved the article.
Glossary
- DAG
days after germination
- AVG
aminoethoxyvinylglycine
- ChIP
chromatin immunoprecipitation
- WBS
WER binding site
- BiFC
bimolecular fluorescence complementation
- SEC
size exclusion chromatography
- Ve
elution volume
- Se-Met
selenium-methionine
- r.m.s.d.
root-mean-square derivation
- EMSA
electrophoretic mobility shift assay
- MNase
micrococcal nuclease
- ORF
open reading frame
References
- Aida M., Beis D., Heidstra R., Willemsen V., Blilou I., Galinha C., Nussaume L., Noh Y.S., Amasino R., Scheres B. (2004). The PLETHORA genes mediate patterning of the Arabidopsis root stem cell niche. Cell 119: 109–120. [DOI] [PubMed] [Google Scholar]
- Avvakumov N., Nourani A., Côté J. (2011). Histone chaperones: modulators of chromatin marks. Mol. Cell 41: 502–514. [DOI] [PubMed] [Google Scholar]
- Bernhardt C., Zhao M., Gonzalez A., Lloyd A., Schiefelbein J. (2005). The bHLH genes GL3 and EGL3 participate in an intercellular regulatory circuit that controls cell patterning in the Arabidopsis root epidermis. Development 132: 291–298. [DOI] [PubMed] [Google Scholar]
- Bu Z., Yu Y., Li Z., Liu Y., Jiang W., Huang Y., Dong A.W. (2014). Regulation of arabidopsis flowering by the histone mark readers MRG1/2 via interaction with CONSTANS to modulate FT expression. PLoS Genet. 10: e1004617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caro E., Castellano M.M., Gutierrez C. (2007). A chromatin link that couples cell division to root epidermis patterning in Arabidopsis. Nature 447: 213–217. [DOI] [PubMed] [Google Scholar]
- Cheng Y., Zhu W., Chen Y., Ito S., Asami T., Wang X. (2014). Brassinosteroids control root epidermal cell fate via direct regulation of a MYB-bHLH-WD40 complex by GSK3-like kinases. eLife, 10.7554/eLife.02525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clapier C.R., Cairns B.R. (2009). The biology of chromatin remodeling complexes. Annu. Rev. Biochem. 78: 273–304. [DOI] [PubMed] [Google Scholar]
- Collaborative Computational Project, Number 4 (1994). The CCP4 suite: programs for protein crystallography. Acta Crystallogr. D Biol. Crystallogr. 50: 760–763. [DOI] [PubMed] [Google Scholar]
- Costa S., Shaw P. (2006). Chromatin organization and cell fate switch respond to positional information in Arabidopsis. Nature 439: 493–496. [DOI] [PubMed] [Google Scholar]
- D’Arcy S., Martin K.W., Panchenko T., Chen X., Bergeron S., Stargell L.A., Black B.E., Luger K. (2013). Chaperone Nap1 shields histone surfaces used in a nucleosome and can put H2A-H2B in an unconventional tetrameric form. Mol. Cell 51: 662–677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Koning L., Corpet A., Haber J.E., Almouzni G. (2007). Histone chaperones: an escort network regulating histone traffic. Nat. Struct. Mol. Biol. 14: 997–1007. [DOI] [PubMed] [Google Scholar]
- Emsley P., Cowtan K. (2004). Coot: model-building tools for molecular graphics. Acta Crystallogr. D Biol. Crystallogr. 60: 2126–2132. [DOI] [PubMed] [Google Scholar]
- Formosa T. (2012). The role of FACT in making and breaking nucleosomes. Biochim. Biophys. Acta 1819: 247–255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- García-Sánchez S., Bernales I., Cristobal S. (2015). Early response to nanoparticles in the Arabidopsis transcriptome compromises plant defence and root-hair development through salicylic acid signalling. BMC Genomics 16: 341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao J., Zhu Y., Zhou W., Molinier J., Dong A., Shen W.H. (2012). NAP1 family histone chaperones are required for somatic homologous recombination in Arabidopsis. Plant Cell 24: 1437–1447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grierson C., Nielsen E., Ketelaarc T., Schiefelbein J. (2014). Root hairs. The Arabidopsis Book 12: e0172, doi/10.1199/tab.0172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayashi K., Hasegawa J., Matsunaga S. (2013). The boundary of the meristematic and elongation zones in roots: endoreduplication precedes rapid cell expansion. Sci. Rep. 3: 2723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hofer R.M. (1991). Root hairs. In Plant Roots: The Hidden Half, Waisel Y., Eshel A., Kafkafi U., eds (New York: Marcel Dekker), pp. 129–148. [Google Scholar]
- Johnson L., Cao X., Jacobsen S. (2002). Interplay between two epigenetic marks. DNA methylation and histone H3 lysine 9 methylation. Curr. Biol. 12: 1360–1367. [DOI] [PubMed] [Google Scholar]
- Kang Y.H., Kirik V., Hulskamp M., Nam K.H., Hagely K., Lee M.M., Schiefelbein J. (2009). The MYB23 gene provides a positive feedback loop for cell fate specification in the Arabidopsis root epidermis. Plant Cell 21: 1080–1094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuppusamy K.T., Chen A.Y., Nemhauser J.L. (2009). Steroids are required for epidermal cell fate establishment in Arabidopsis roots. Proc. Natl. Acad. Sci. USA 106: 8073–8076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurata T., et al. (2005). Cell-to-cell movement of the CAPRICE protein in Arabidopsis root epidermal cell differentiation. Development 132: 5387–5398. [DOI] [PubMed] [Google Scholar]
- Kwak S.H., Schiefelbein J. (2007). The role of the SCRAMBLED receptor-like kinase in patterning the Arabidopsis root epidermis. Dev. Biol. 302: 118–131. [DOI] [PubMed] [Google Scholar]
- Kwak S.H., Shen R., Schiefelbein J. (2005). Positional signaling mediated by a receptor-like kinase in Arabidopsis. Science 307: 1111–1113. [DOI] [PubMed] [Google Scholar]
- Laskey R.A., Honda B.M., Mills A.D., Finch J.T. (1978). Nucleosomes are assembled by an acidic protein which binds histones and transfers them to DNA. Nature 275: 416–420. [DOI] [PubMed] [Google Scholar]
- Lee M.M., Schiefelbein J. (1999). WEREWOLF, a MYB-related protein in Arabidopsis, is a position-dependent regulator of epidermal cell patterning. Cell 99: 473–483. [DOI] [PubMed] [Google Scholar]
- Lee M.M., Schiefelbein J. (2002). Cell pattern in the Arabidopsis root epidermis determined by lateral inhibition with feedback. Plant Cell 14: 611–618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levchenko V., Jackson V. (2004). Histone release during transcription: NAP1 forms a complex with H2A and H2B and facilitates a topologically dependent release of H3 and H4 from the nucleosome. Biochemistry 43: 2359–2372. [DOI] [PubMed] [Google Scholar]
- Li D.X., Chen W.Q., Xu Z.H., Bai S.N. (2015). HISTONE DEACETYLASE6-defective mutants show increased expression and acetylation of ETC1 and GL2 with small but significant effects on root epidermis cellular pattern. Plant Physiol. 168: 1448–1458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li G., Liu S., Wang J., He J., Huang H., Zhang Y., Xu L. (2014). ISWI proteins participate in the genome-wide nucleosome distribution in Arabidopsis. Plant J. 78: 706–714. [DOI] [PubMed] [Google Scholar]
- Lin Q., Ohashi Y., Kato M., Tsuge T., Gu H., Qu L.J., Aoyama T. (2015). GLABRA2 directly suppresses basic helix-loop-helix transcription factor genes with diverse functions in root hair development. Plant Cell 27: 2894–2906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu C., Li L.C., Chen W.Q., Chen X., Xu Z.H., Bai S.N. (2013). HDA18 affects cell fate in Arabidopsis root epidermis via histone acetylation at four kinase genes. Plant Cell 25: 257–269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Z., Zhu Y., Gao J., Yu F., Dong A., Shen W.H. (2009). Molecular and reverse genetic characterization of NUCLEOSOME ASSEMBLY PROTEIN1 (NAP1) genes unravels their function in transcription and nucleotide excision repair in Arabidopsis thaliana. Plant J. 59: 27–38. [DOI] [PubMed] [Google Scholar]
- Lorch Y., Maier-Davis B., Kornberg R.D. (2006). Chromatin remodeling by nucleosome disassembly in vitro. Proc. Natl. Acad. Sci. USA 103: 3090–3093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luger K., Dechassa M.L., Tremethick D.J. (2012). New insights into nucleosome and chromatin structure: an ordered state or a disordered affair? Nat. Rev. Mol. Cell Biol. 13: 436–447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masucci J.D., Rerie W.G., Foreman D.R., Zhang M., Galway M.E., Marks M.D., Schiefelbein J.W. (1996). The homeobox gene GLABRA2 is required for position-dependent cell differentiation in the root epidermis of Arabidopsis thaliana. Development 122: 1253–1260. [DOI] [PubMed] [Google Scholar]
- Masucci J.D., Schiefelbein J.W. (1994). The rhd6 mutation of Arabidopsis thaliana alters root-hair initiation through an auxin- and ethylene-associated process. Plant Physiol. 106: 1335–1346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masucci J.D., Schiefelbein J.W. (1996). Hormones act downstream of TTG and GL2 to promote root hair outgrowth during epidermis development in the Arabidopsis root. Plant Cell 8: 1505–1517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyaji-Yamaguchi M., Kato K., Nakano R., Akashi T., Kikuchi A., Nagata K. (2003). Involvement of nucleocytoplasmic shuttling of yeast Nap1 in mitotic progression. Mol. Cell. Biol. 23: 6672–6684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyamoto S., Suzuki T., Muto S., Aizawa K., Kimura A., Mizuno Y., Nagino T., Imai Y., Adachi N., Horikoshi M., Nagai R. (2003). Positive and negative regulation of the cardiovascular transcription factor KLF5 by p300 and the oncogenic regulator SET through interaction and acetylation on the DNA-binding domain. Mol. Cell. Biol. 23: 8528–8541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morillo-Huesca M., Maya D., Muñoz-Centeno M.C., Singh R.K., Oreal V., Reddy G.U., Liang D., Géli V., Gunjan A., Chávez S. (2010). FACT prevents the accumulation of free histones evicted from transcribed chromatin and a subsequent cell cycle delay in G1. PLoS Genet. 6: e1000964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Müller M., Schmidt W. (2004). Environmentally induced plasticity of root hair development in Arabidopsis. Plant Physiol. 134: 409–419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muto S., Senda M., Akai Y., Sato L., Suzuki T., Nagai R., Senda T., Horikoshi M. (2007). Relationship between the structure of SET/TAF-Ibeta/INHAT and its histone chaperone activity. Proc. Natl. Acad. Sci. USA 104: 4285–4290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ottenschläger I., Wolff P., Wolverton C., Bhalerao R.P., Sandberg G., Ishikawa H., Evans M., Palme K. (2003). Gravity-regulated differential auxin transport from columella to lateral root cap cells. Proc. Natl. Acad. Sci. USA 100: 2987–2991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park Y.J., Luger K. (2006). The structure of nucleosome assembly protein 1. Proc. Natl. Acad. Sci. USA 103: 1248–1253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simon M., Lee M.M., Lin Y., Gish L., Schiefelbein J. (2007). Distinct and overlapping roles of single-repeat MYB genes in root epidermal patterning. Dev. Biol. 311: 566–578. [DOI] [PubMed] [Google Scholar]
- Song S.K., Ryu K.H., Kang Y.H., Song J.H., Cho Y.H., Yoo S.D., Schiefelbein J., Lee M.M. (2011). Cell fate in the Arabidopsis root epidermis is determined by competition between WEREWOLF and CAPRICE. Plant Physiol. 157: 1196–1208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki T., Muto S., Miyamoto S., Aizawa K., Horikoshi M., Nagai R. (2003). Functional interaction of the DNA-binding transcription factor Sp1 through its DNA-binding domain with the histone chaperone TAF-I. J. Biol. Chem. 278: 28758–28764. [DOI] [PubMed] [Google Scholar]
- Szymanski D.B., Jilk R.A., Pollock S.M., Marks M.D. (1998). Control of GL2 expression in Arabidopsis leaves and trichomes. Development 125: 1161–1171. [DOI] [PubMed] [Google Scholar]
- Tang Y., Meeth K., Jiang E., Luo C., Marmorstein R. (2008). Structure of Vps75 and implications for histone chaperone function. Proc. Natl. Acad. Sci. USA 105: 12206–12211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Venkatesh S., Workman J.L. (2015). Histone exchange, chromatin structure and the regulation of transcription. Nat. Rev. Mol. Cell Biol. 16: 178–189. [DOI] [PubMed] [Google Scholar]
- Wang Y., Zhang W., Li K., Sun F., Han C., Wang Y., Li X. (2008). Salt-induced plasticity of root hair development is caused by ion disequilibrium in Arabidopsis thaliana. J. Plant Res. 121: 87–96. [DOI] [PubMed] [Google Scholar]
- Zhang C., Cao L., Rong L., An Z., Zhou W., Ma J., Shen W.H., Zhu Y., Dong A. (2015). The chromatin-remodeling factor AtINO80 plays crucial roles in genome stability maintenance and in plant development. Plant J. 82: 655–668. [DOI] [PubMed] [Google Scholar]
- Zhang Y.J., Lynch J.P., Brown K.M. (2003). Ethylene and phosphorus availability have interacting yet distinct effects on root hair development. J. Exp. Bot. 54: 2351–2361. [DOI] [PubMed] [Google Scholar]
- Zhou W., Gao J., Ma J., Cao L., Zhang C., Zhu Y., Dong A., Shen W.H. (2016). Distinct roles of the histone chaperones NAP1 and NRP and the chromatin-remodeling factor INO80 in somatic homologous recombination in Arabidopsis thaliana. Plant J. 88: 397–410. [DOI] [PubMed] [Google Scholar]
- Zhou W., Zhu Y., Dong A., Shen W.H. (2015). Histone H2A/H2B chaperones: from molecules to chromatin-based functions in plant growth and development. Plant J. 83: 78–95. [DOI] [PubMed] [Google Scholar]
- Zhu Y., Dong A., Meyer D., Pichon O., Renou J.P., Cao K., Shen W.H. (2006). Arabidopsis NRP1 and NRP2 encode histone chaperones and are required for maintaining postembryonic root growth. Plant Cell 18: 2879–2892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu Y., Dong A., Shen W.H. (2013). Histone variants and chromatin assembly in plant abiotic stress responses. Biochim. Biophys. Acta 1819: 343–348. [DOI] [PubMed] [Google Scholar]
- Zhu Y., Weng M., Yang Y., Zhang C., Li Z., Shen W.H., Dong A. (2011). Arabidopsis homologues of the histone chaperone ASF1 are crucial for chromatin replication and cell proliferation in plant development. Plant J. 66: 443–455. [DOI] [PubMed] [Google Scholar]
- Zuo J., Niu Q.W., Chua N.H. (2000). Technical advance: An estrogen receptor-based transactivator XVE mediates highly inducible gene expression in transgenic plants. Plant J. 24: 265–273. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.