TEAR1 is an active E3 ligase that controls leaf development by promoting CIN-like TCP activity by targeting the TCP repressor TIE1 for degradation via the ubiquitin-proteasome system.
Abstract
The developmental plasticity of leaf size and shape is important for leaf function and plant survival. However, the mechanisms by which plants form diverse leaves in response to environmental conditions are not well understood. Here, we identified TIE1-ASSOCIATED RING-TYPE E3 LIGASE1 (TEAR1) and found that it regulates leaf development by promoting the degradation of TCP INTERACTOR-CONTAINING EAR MOTIF PROTEIN1 (TIE1), an important repressor of CINCINNATA (CIN)-like TEOSINTE BRANCHED1/CYCLOIDEA/PCF (TCP) transcription factors, which are key for leaf development. TEAR1 contains a typical C3H2C3-type RING domain and has E3 ligase activity. We show that TEAR1 interacts with the TCP repressor TIE1, which is ubiquitinated in vivo and degraded by the 26S proteasome system. We demonstrate that TEAR1 is colocalized with TIE1 in nuclei and negatively regulates TIE1 protein levels. Overexpression of TEAR1 rescued leaf defects caused by TIE1 overexpression, whereas disruption of TEAR1 resulted in leaf phenotypes resembling those caused by TIE1 overexpression or TCP dysfunction. Deficiency in TEAR partially rescued the leaf defects of TCP4 overexpression line and enhanced the wavy leaf phenotypes of jaw-5D. We propose that TEAR1 positively regulates CIN-like TCP activity to promote leaf development by mediating the degradation of the TCP repressor TIE1.
INTRODUCTION
Compared with mobile animals, sessile plants evolved developmental plasticity as an important strategy to adapt to different environmental conditions (Gaillochet and Lohmann, 2015). Leaves are the main photosynthetic organs in plants and have high developmental plasticity (Szymanski, 2014; Ichihashi and Tsukaya, 2015). Although alteration of leaf size and shape is important for plant survival and global food security, the molecular mechanisms by which plants control the formation of leaves are not well understood.
A plant leaf develops from a group of cells called the leaf primordium, which is initiated at the periphery of the shoot apical meristem (SAM) (Ichihashi and Tsukaya, 2015; Sluis and Hake, 2015). Initially, all cells in the leaf primordium actively divide to convert the cylindrical peg-like primordium into a flat nascent leaf with proximal-distal, adaxial-abaxial, and medial-lateral polarities. Subsequently, cell division, cell differentiation, and cell expansion are temporally and spatially coordinated to convert the nascent leaf into a mature leaf with a specific leaf size and shape (Szymanski, 2014; Sluis and Hake, 2015). Several important transcription factors, such as class I KNOX family proteins, ASYMMETRIC LEAVES1 (AS1), AS2, the class III homeodomain-leucine zipper proteins, AUXIN RESPONSE FACTOR3 (ARF3), ARF4, KANADI, the YABBY family proteins, ANGUSTIFOLIA3, and CINCINNATA (CIN)-like TEOSINTE BRANCHED1/CYCLOIDEA/PCF (TCP) proteins, have been shown to regulate different stages of leaf development (Poethig and Sussex, 1985; Nath et al., 2003; Scanlon, 2003; Ha et al., 2010; Hay and Tsiantis, 2010; Piazza et al., 2010; Moon and Hake, 2011; Bolduc et al., 2012; Vercruyssen et al., 2014; Hur et al., 2015; Husbands et al., 2015; Sluis and Hake, 2015). Among these factors, CIN-like TCP genes play pivotal roles in leaf formation by controlling leaf cell division and differentiation (Nath et al., 2003; Koyama et al., 2007; Ori et al., 2007; Efroni et al., 2008; Koyama et al., 2010; Martín-Trillo and Cubas, 2010).
TCP proteins are plant-specific transcription factors and are conserved in the plant kingdom (Navaud et al., 2007). CIN was the first TCP transcription factor shown to control leaf cell differentiation and, thus, leaf flatness in Antirrhinum majus (Nath et al., 2003). In Arabidopsis thaliana, the TCP family consists of 24 members classified into 13 class I group TCPs and 11 class II group TCPs based on the conserved noncanonical basic helix-loop-helix domain. The class II TCPs are further grouped into eight CIN-like TCPs and three CYC/TB1-like TCPs (Martín-Trillo and Cubas, 2010). The CIN-like TCPs have high functional redundancy in Arabidopsis. Whereas the tcp single mutant shows no obvious leaf phenotypes, disruption of multiple CIN-like TCPs causes leaf curvature and wavy leaf margins in a dose-dependent manner (Schommer et al., 2008; Koyama et al., 2010), suggesting that TCP activity is central for the elaboration of leaf forms. The genetic alteration of TCP activity leads to different leaf sizes and shapes, further confirming that the tight control of TCP activity at both the temporal and spatial levels is fundamental in determining leaf form (Efroni et al., 2008; Shleizer-Burko et al., 2011).
Plants have evolved several strategies to tightly control the activity of CIN-like TCPs during leaf development. First, the activity of CIN-like TCPs is regulated at the transcriptional level. For example, we identified an activation-tagging mutant, iamt1-D, and found that indole-3-acetic acid (IAA) carboxyl methyltransferase (IAMT1) negatively regulates the transcripts of several CIN-like TCP genes to control leaf development by converting active IAA to inactive methyl IAA (Qin et al., 2005). Second, TCP activity is tightly regulated at the posttranscriptional level. Overexpression of the microRNA miR319 in the activation-tagging mutant jaw-D causes leaf curvature by targeting five CIN-like TCP genes (Palatnik et al., 2003). The regulation of TCP activity by miR319 is important for plant leaf development; the expression of TCPs with synonymous mutations that resulted in resistance to miR319-mediated cleavage caused severe leaf phenotypes or even seedling death in Arabidopsis and changed compound leaves into simple leaves in tomato (Palatnik et al., 2003, 2007; Ori et al., 2007). Third, the activity of CIN-like TCPs is modulated at the protein level. For example, ARMADILLO BTB ARABIDOPSIS PROTEIN1 may modify TCP24 activity to regulate leaf cell proliferation (Masuda et al., 2008). The SWI/SNF chromatin remodeling ATPase BRAHMA (BRM) mediates CIN-like TCP activity to reduce leaf sensitivity to cytokinin by increasing the expression of the negative cytokinin regulator ARABIDOPSIS RESPONSE REGULATOR16 (ARR16) (Efroni et al., 2013). We previously characterized a transcriptional repressor, TCP INTERACTOR-CONTAINING EAR MOTIF PROTEIN1 (TIE1) (Tao et al., 2013). The overexpression of TIE1 in the activation-tagging mutant tie1-D resulted in hyponastic leaves, whereas disruption of TIE1 caused epinastic leaves. TIE1 regulates leaf development by physically interacting with CIN-like TCPs. We further demonstrated that TIE1 is a key transcriptional repressor of TCP activity that acts as a bridge between TCPs and the corepressor TOPLESS (TPL) (Tao et al., 2013). However, the molecular mechanism by which TIE1 temporally and spatially regulates TCP activity is still unknown.
The ubiquitin (Ub)-proteasome system is a major protein degradation system that flexibly regulates the activity of transcription regulators and is important in hormone, stress, and other signaling pathways (Potuschak et al., 2003; Chen and Hellmann, 2013; Duplan and Rivas, 2014; Shabek and Zheng, 2014; Noir et al., 2015). Plants utilize the Ub-proteasome system for morphological plasticity and altering physiological states in response to internal and external stimuli via the rapidly changing proteome (Chen and Hellmann, 2013). The Ub-proteasome system comprises the 26S proteasome and three types of enzymes for substrate protein ubiquitination. These enzymes are the Ub-activating enzymes (E1), the Ub-conjugating enzymes (E2), and the Ub ligases (E3). Among them, E3 ligases are key for defining the specificity of substrates by transferring Ub from the E2-Ub intermediate to the target proteins. The four major types of E3 include RING (REALLY INTERESTING NEW GENE), CULLIN-RING, U-box, and HECT (HOMOLOGOUS TO E6-ASSOCIATED PROTEIN C TERMINUS) (Chen and Hellmann, 2013; Shabek and Zheng, 2014). The Arabidopsis genome is predicted to contain more than 460 RING-containing proteins (Stone et al., 2005). Although the function of several RING proteins has been studied extensively (Dong et al., 2006; Stone et al., 2006; Lau and Deng, 2012; Lazaro et al., 2012, 2015; Liu and Stone, 2013; Lee and Seo, 2015; Pauwels et al., 2015; Wang et al., 2015a), the E3 ligase activity and the targets of most RING proteins in Arabidopsis have yet to be elucidated.
Here, using a yeast two-hybrid screen with an Arabidopsis cDNA library, we identified a TIE1-interacting protein that we named TIE1-ASSOCIATED RING-TYPE E3 LIGASE1 (TEAR1). TEAR1 contains a C3H2C3-type RING domain, and we demonstrate that TEAR1 has E3 ligase activity and that TIE1 is an unstable protein. We further show that TIE1 is ubiquitinated in vivo and is degraded by the 26S proteasome. TEAR1 is colocalized with TIE1 in the nuclei and interacts with TIE1 in vivo and in vitro. Disruption of TEAR1 leads to defective leaves with wavy margins similar to those observed in tcp multiple knockout mutants and TIE1 overexpression lines. Our data suggest that TEAR1 plays important roles in the control of leaf development by enhancing TCP activity via degradation of the TCP repressor TIE1.
RESULTS
Identification and Cloning of TEAR1 and TEAR2
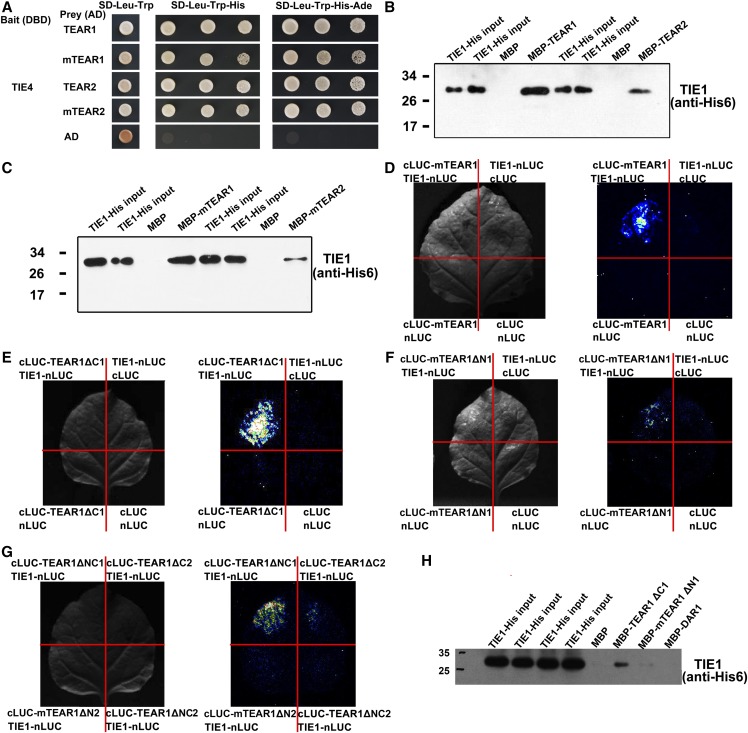
We previously identified TIE1 transcriptional repressors by characterizing an activation-tagging mutant, tie1-D, in Arabidopsis. TIE1 and its close homologs TIE3 and TIE4 play important roles in the leaf formation by controlling the activity of CIN-like TCP transcription factors. To further elucidate how TIE1 regulates TCP activity temporally and spatially, we searched for TIE1 interactors using yeast two-hybrid screening. Because the full-length TIE1 activated the reporter gene in yeast cells, whereas the full-length TIE4 did not (Tao et al., 2013), we used the full-length coding region of TIE4 as the bait to screen an Arabidopsis cDNA library. Sequencing analysis showed that the insert sequences of 13 positive clones encoded five different RING-type proteins. Among them, At1g53190 and At3g15070 were shown to be closely related in the phylogenetic tree (Supplemental Figures 1A and 1B and Supplemental Data Set 1). Therefore, we first focused our analysis on At1g53190 and At3g15070 and named them TEAR1 and TEAR2 for the reasons described below.
We cloned the full-length coding regions of both TEAR1 and TEAR2 to verify their interactions with TIE4. The results confirmed that both interacted with TIE4 in yeast cells (Figure 1A). TEAR1 and TEAR2 are predicted to contain a canonical RING domain with eight metal binding residues coordinating two zinc ions in a cross-brace arrangement (Supplemental Figure 1A; Stone et al., 2005). The RING domain in TEAR1 and TEAR2 is a C3H2C3-type or RING-H2 type containing two His residues at metal ligand positions 4 and 5 (Supplemental Figure 1A; Stone et al., 2005). To determine whether the RING domains of TEAR1 and TEAR2 are required for their interactions with TIE4, we mutated the two conserved His residues into Tyr residues in the RING domains to generate mTEAR1 and mTEAR2 (Supplemental Figure 1C). The yeast two-hybrid assays showed that both mTEAR1 and mTEAR2 were still able to interact with TIE4 (Figure 1A), suggesting that the RING domain was dispensable for the interactions of TIE4 with TEAR1 or TEAR2. The results are consistent with the previous reports that the RING domain forms a platform for interacting with ubiquitin conjugases (UBCs), while the other domains bind to substrate (Metzger et al., 2014).
Figure 1.
TIE1 Interacts with TEAR1 and TEAR2
(A) Testing of interactions between TIE4 and TEAR1 or TEAR2 using yeast two-hybrid screening. TIE4 was also tested with mTEAR1 or mTEAR2, in which two conserved His residues in the RING domain were mutated to Tyr residues. AD, activation domain; DBD, DNA binding domain. Transformed yeasts were spotted on control medium (SD-Leu-Trp) or selective medium (SD-Leu-Trp-His and SD-Leu-Trp-His-Ade) at dilutions of 10-, 100-, and 1000-fold. The empty vectors were used as controls.
(B) and (C) Pull-down assays to test TIE1 interaction with TEAR1, mTEAR1, TEAR2, or mTEAR2 in vitro. The TIE1-His proteins were detected with an anti-His monoclonal antibody, and MBP proteins were used as the negative control. The two lanes of TIE1-His input were the control for the following two lanes of MBP and MBP-TEAR1 or MBP and MBP-TEAR2.
(D) to (G) The firefly luciferase (LUC) complementation imaging assays of TIE1 interaction with mTEAR1 or different truncated TEAR1 proteins in vivo. The left pictures were taken under bright field, while the right LUC images were captured using a low-light cooled CCD imaging apparatus. LUC signals were detected in the combination of TIE1-nLUC and cLUC-mTEAR1. The other combinations, including TIE1-nLUC and cLUC, nLUC and cLUC-mTEAR1, and nLUC and cLUC, were used as negative controls. Interactions between TIE1 and different truncated TEAR1 proteins in firefly luciferase complementation imaging assays are shown in (E) to (G). The truncated TEAR1 constructs are schematically represented in Supplemental Figure 2D.
(H) Assessing TIE1 interactions with different truncated TEAR1 proteins by pull-down assays.
TEAR1 and TEAR2 Interact with TIE1 in Vitro and in Vivo
To determine whether TEAR1 and TEAR2 also interact with the TIE4 homolog TIE1, we first expressed a maltose binding protein (MBP) fusion with TEAR1 or TEAR2 and His-tagged TIE1 proteins. In vitro pull-down assays showed that both MBP-TEAR1 and MBP-TEAR2 could bind to TIE1, whereas the control MBP could not (Figure 1B). We then expressed MBP-mTEAR1 and MBP-mTEAR2 and assessed their interactions with TIE1, and the results showed that MBP-mTEAR1 and MBP-mTEAR2 could also pull down the TIE1 protein, further indicating that the RING domain was not required for the interaction of TIE1 with TEAR1 or TEAR2 (Figure 1C). The protein level of MBP-TEAR1, MBP-TEAR2, MBP-mTEAR1, and MBP-mTEAR2 was comparable to or lower than that of the control MBP, based on immunoblots using anti-MBP antibody (Supplemental Figures 2A and 2B). Considering that the RING domain is dispensable for TIE1 interactions with TEAR1 or TEAR2 (Figure 1C) and TEAR1 and TEAR2 may lead to TIE1 degradation in planta, we further investigated the interactions in vivo using mTEAR1 and mTEAR2 but not TEAR1 and TEAR2 in firefly luciferase complementation imaging assays. Clear fluorescence was observed in the leaf areas cotransformed with the C terminus of LUC fused to mTEAR1 or mTEAR2 and the N terminus of LUC fused to TIE1, whereas no obvious fluorescence was observed in the areas of the same leaf cotransformed with the control plasmids, further confirming that TIE1 interacts with TEAR1 and TEAR2 in vivo (Figure 1D; Supplemental Figure 2C).
Many RING-type proteins contain other domains in addition to RING domains that bind to E2s, while the other domains are possibly responsible for interaction with substrates (Stone et al., 2005; Metzger et al., 2014). TEAR1 and TEAR2 contain an unidentified domain associated with RING1 (DAR1), which is also present in At5g42940 (MBRL2; see below) (Stone et al., 2005). To identify which domain or region in TEAR1 could interact with TIE1, we first partitioned TEAR1 into the N terminus (TEAR1ΔC1) and the C terminus containing the RING domain with the two conserved His residues mutated to Tyr (mTEAR1ΔN1) (Supplemental Figure 2D). Firefly luciferase complementation imaging assays showed that the N terminus of TEAR1 played a major role in mediating the interaction with TIE1, while the C terminus containing the DAR1 domain (residues 374 to 411 in TEAR1) had a minor role in the interaction (Figures 1E and 1F). We then further partitioned TEAR1 into TEAR1ΔC2, TEAR1ΔNC1, TEAR1ΔNC2, and mTEAR1ΔN2 (Supplemental Figure 2D). The imaging assays showed that TEAR1ΔNC1, which included the amino acid residues from 141 to 260, was the major domain that interacted with TIE1 (Figure 1G), consistent with the above results indicating that the RING domain is not required for the interaction between TEAR1 and TIE1. To further confirm the results, we expressed MBP fusions with TEAR1ΔC1, mTEAR1ΔN1, or DAR1. The protein levels of MBP-TEAR1ΔC1, MBP-mTEAR1ΔN1, and MBP-DAR1 were comparable to that of the control MBP (Supplemental Figure 2E). In vitro pull-down assays showed that MBP-TEAR1ΔC1 could pull down TIE1, whereas MBP-mTEAR1ΔN1 could pull down only trace amounts of TIE1, and both MBP-DAR1 and the control MBP pulled down none (Figure 1H). These results were consistent with those obtained above in firefly luciferase complementation imaging assays. These findings demonstrate that TEAR1 utilizes the N terminus to recognize TIE1.
TEAR1 and TEAR2 Can Localize to Nuclei
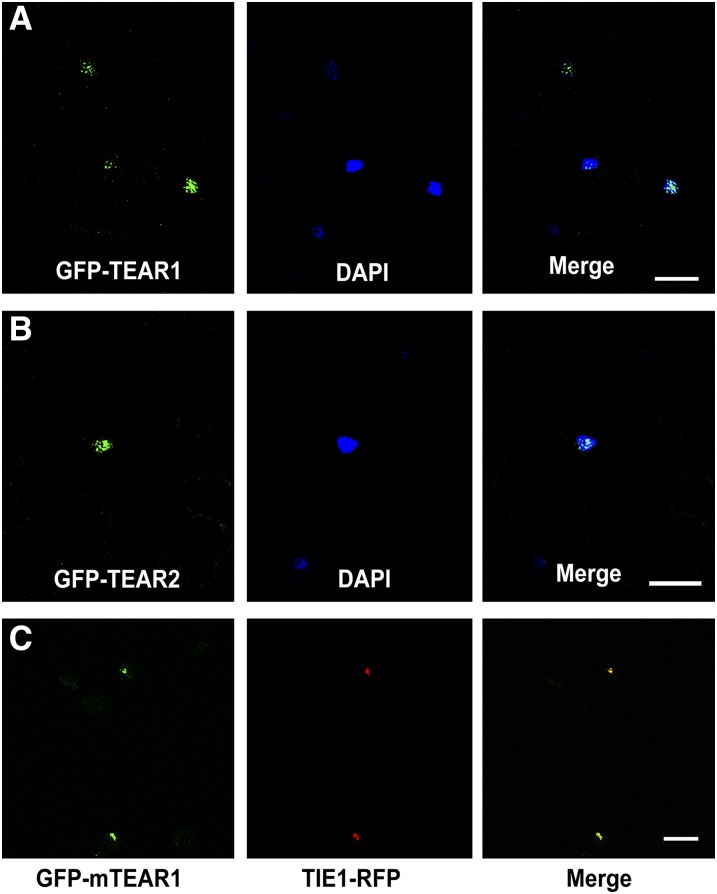
Both TEAR1 and TEAR2 are previously uncharacterized RING domain-containing proteins. To determine the subcellular localization of TEAR1 and TEAR2, we fused TEAR1 or TEAR2 to GFP. Transient expression of GFP-TEAR1 or GFP-TEAR2 in Nicotiana benthamiana leaves showed that both TEAR1 and TEAR2 could localize to nuclei (Figures 2A and 2B). When we coexpressed GFP-mTEAR1 and TIE1-RFP in Arabidopsis leaf protoplasts, mTEAR1 colocalized with TIE1 in nuclei (Figure 2C), further supporting the likelihood of interaction between TIE1 and TEAR1.
Figure 2.
TEAR1 and TEAR2 Localize to Nuclei.
(A) and (B) GFP-TEAR1 and GFP-TEAR2 fusion proteins were transiently expressed in N. benthamiana leaf cells. From left to right, the fluorescence of GFP, staining of nuclei by 4′,6-diamidino-2-phenylindole (DAPI), and merging of DAPI and GFP. Bars = 20 µm.
(C) GFP-mTEAR1 and TIE1-RFP were coexpressed in Arabidopsis wild-type protoplasts. From left to right, the fluorescence of GFP, fluorescence of RFP, and the merging of GFP and RFP. Bar = 50 µm.
The Expression of TEAR1 and TEAR2 Is Developmentally Regulated in the Leaves
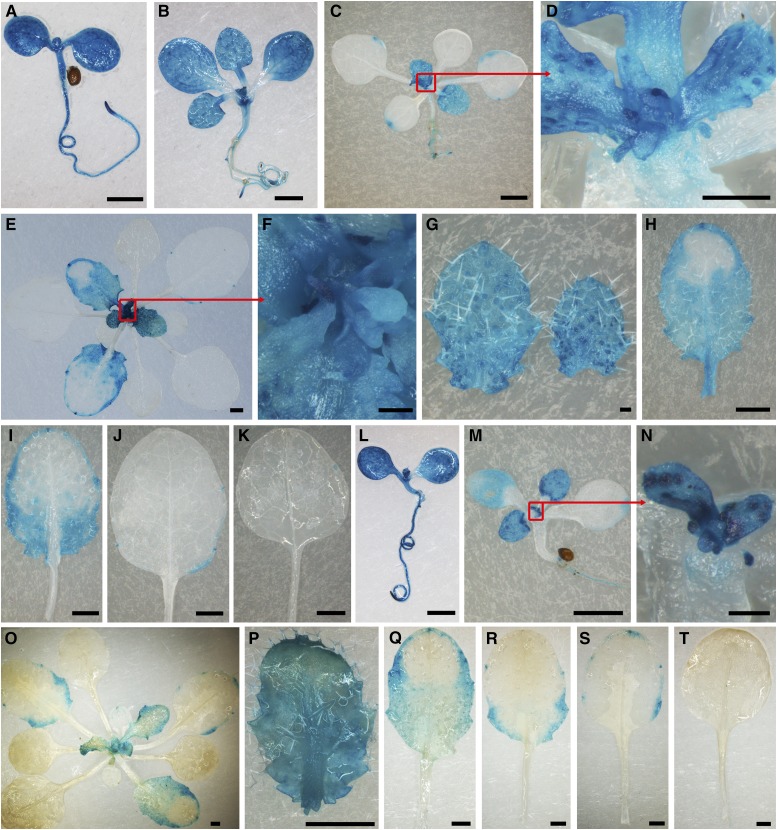
To determine the expression patterns of TEAR1 and TEAR2, we cloned the 2119-bp promoter of TEAR1 and the 1974-bp promoter of TEAR2. We generated the constructs TEAR1pro-GUS and TEAR2pro-GUS, in which the TEAR1 promoter or TEAR2 promoter was used to drive the GUS reporter gene. Eighteen TEAR1pro-GUS transgenic lines and 15 TEAR2pro-GUS lines displayed two types of staining patterns. GUS staining showed that both TEAR1 and TEAR2 were expressed predominantly in the SAM and in whole nascent and young leaves (Figures 3A to 3G and 3L to 3P). Interestingly, as the leaves aged, GUS staining gradually faded from the distal to proximal regions and the medial regions to margins in the leaves of TEAR1pro-GUS and TEAR2pro-GUS lines (Figures 3E to 3K and 3O to 3T). We observed the specific expression driven by the promoters of TEAR1 and TEAR2 in the proximal half (Figures 3I and 3R) and in the proximal leaf margins (Figures 3J and 3S) of moderately old leaves. Nearly no GUS staining was found in the old leaves (Figures 3K and 3T). The expression patterns of TEAR1 and TEAR2 overlapped with each other and with that of TIE1 (Tao et al., 2013), further providing temporal and spatial evidence for the interactions between TIE1 and TEAR1 or TEAR2. These data suggest that TEAR1 and TEAR2 may redundantly regulate leaf development by interacting with TIE1.
Figure 3.
Expression Patterns of TEAR1 and TEAR2.
(A) to (K) The expression pattern analysis of TEAR1 based on GUS staining in TEAR1pro-GUS-6.
(A) Six-day-old seedling.
(B) Ten-day-old seedling.
(C) Fifteen-day-old plant.
(D) Close-up view of SAM (boxed in [C]) from 15-d-old plant.
(E) Twenty-four-day-old plant.
(F) Close-up view of SAM (boxed in [E]) from 24-d-old plant.
(G) to (K) Close-up view of the young to old leaves from a 24-d-old plant.
(L) to (T) Expression pattern analysis of TEAR2 based on GUS staining in TEAR2pro-GUS-5.
(L) Six-day-old seedling.
(M) Ten-day-old seedling.
(N) Close-up view of SAM (boxed in [M]) from 10-d-old plant.
(O) Twenty-four-day-old seedling.
(P) to (T) Close-up view of the young to old leaves from a 24-d-old plant.
Bars = 1 mm in (A) to (C), (E), (H), (I) to (M), and (O) to (T) and 0.1 mm in (D), (F), (G), and (N).
TEAR1 and TEAR2 Are Functional E3 Ligases for Ubiquitination
RING domain-containing proteins were previously reported to independently function as E3 ligases (Stone et al., 2005). As TEAR1 and TEAR2 contain typical C3H2C3-type RING domains (Supplemental Figure 1A), we investigated whether TEAR1 and TEAR2 had E3 ligase activity. We expressed MBP-TEAR1, MBP-mTEAR1, MBP-TEAR2, and MBP-mTEAR2 in Escherichia coli cells and purified the fusion proteins. We first tested the protein level of MBP, MBP-TEAR1, MBP-mTEAR1, MBP-TEAR2, and MBP-mTEAR2 by immunoblots using anti-MBP before performing in vitro E3 ligase activity assays. The protein level of the control was comparable to or higher than that of MBP-TEAR1 or MBP-TEAR2 (Figure 4). Then we used the same amounts of the proteins to perform the assays. Clear Ub ladders were found in the lanes with MBP-TEAR1 or MBP-TEAR2 combined with E1, E2, and Ub, whereas no Ub ladders appeared in the lanes with combinations lacking E1, E2, or Ub and in the combination of MBP plus E1, E2, and Ub, indicating that TEAR1 and TEAR2 had E3 ubiquitination activity (Figure 4). However, Ub ladders were not present in the lanes with MBP-mTEAR1 or MBP-mTEAR2 plus E1, E2, and Ub (Figure 4). These results suggest that both TEAR1 and TEAR2 are active E3 ligases and that the RING domains are required for their E3 ligase activity.
Figure 4.
TEAR1 and TEAR2 Have E3 Ligase Activity.
E3 Ub ligase activity was tested using MBP-tagged TEAR1 (A) or MBP-TEAR2 (B) in the presence or absence of E1, E2, and Ub. An anti-Ub antibody was used to detect ubiquitinated proteins (top), and an anti-MBP was used to determine the amounts of MBP-TEAR1, MBP-TEAR2, MBP, MBP-mTEAR1, and MBP-mTEAR2 controls (bottom). Molecular mass markers in kilodaltons are indicated on the left.
TIE1 Is Ubiquitinated and Regulated by the Ub-Proteasome System in Planta
The E3 ubiquitination activity of TEAR1 and the interaction between TEAR1 and TIE1 demonstrated above suggested that TIE1 was likely ubiquitinated in planta and degraded by the 26S proteasome. To test this hypothesis, we first generated the construct 35Spro-TIE1-Flag, in which TIE1 was fused with 3×FLAG tags and driven by the CaMV 35S promoter. We transformed 35Spro-TIE1-Flag into wild-type plants. We obtained several transgenic plants showing more serrations, as observed in the TIE1 overexpression lines, indicating that FLAG-tagged TIE1 was functional (Supplemental Figure 3). We then used a tandem-repeated ubiquitin binding entities (TUBEs) method, which was previously developed to efficiently purify polyubiquitinated proteins from cell extracts in native conditions (Hjerpe et al., 2009). The polyubiquitinated proteins were isolated from the leaf cell extracts of 35Spro-TIE1-Flag transgenic lines or wild-type plants using TUBEs agarose resin. Immunoblot analysis using an anti-Ub antibody indicated that the polyubiquitinated proteins were successfully purified from either 35Spro-TIE1-Flag or wild type but not from the agarose control (Figure 5A). Moreover, the smeared ladders formed by the polyubiquitinated proteins were stronger after treatment with the proteasome inhibitor MG132 in both the 35Spro-TIE1-Flag and the wild type (Figure 5A). We then tested FLAG-tagged TIE1 using an anti-Flag antibody. Clear signals of polyubiquitinated TIE1 proteins were observed in the 35Spro-TIE1-Flag extracts, while no signals were found in the wild-type extracts (Figure 5B). Furthermore, the polyubiquitinated TIE1 proteins showed greater accumulation after MG132 treatment, indicating that TIE1 was regulated by the 26S proteasome (Figure 5B).
Figure 5.
TIE1 Was Ubiquitinated and Regulated by the Ub-Proteasome System.
(A) and (B) Ubiquitinated proteins were purified using the TUBEs resin. Agarose resin was used to detect nonspecific protein binding. Immunoblotting with anti-Ub (A) or anti-FLAG (B) antibodies was used to detect total ubiquitinated proteins or ubiquitinated Flag-tagged TIE1. Molecular mass markers in kilodaltons are indicated on the left.
(C) Flag-tagged TIE1 protein levels in 35Spro-TIE1-Flag transgenic lines after treatment with 50 µM CHX with or without 100 µM MG132 for the indicated durations. Proteins were detected by immunoblotting with an anti-FLAG monoclonal antibody. Relative amounts of proteins were determined by the ACTIN levels. Similar results were obtained in three repeated experiments.
To further confirm that TIE1 is degraded via the Ub-proteasome system, we detected the FLAG-tagged TIE1 levels in 35Spro-TIE1-Flag transgenic lines over time. After treatment with the protein biosynthesis inhibitor cycloheximide (CHX), the TIE1 bands were gradually attenuated over time, whereas the control ACTIN band was not (Figure 5C). After 24 h of treatment, few TIE1 proteins could be detected (Figure 5C). However, after treatment with both CHX and MG132, the TIE1 protein abundance was not changed, even 24 h after treatment, indicating that TIE1 degradation required the 26S proteasome (Figure 5C).
These results demonstrate that TIE1 is an unstable protein degraded by ubiquitination and the 26S proteasome.
TIE1 Levels Are Negatively Regulated by TEAR1
To investigate the biological roles of the interaction between TEAR1 and TIE1, we performed four experiments. First, we cotransformed TIE1-nLUC with cLUC-TEAR1/cLUC-TEAR2 or cLUC-mTEAR1/cLUC-mTEAR2 in two halves of the same N. benthamiana leaves. The fluorescence signals in the halves cotransformed with TIE1-nLUC and cLUC-mTEAR1 or cLUC-mTEAR2 were always stronger than those with TIE1-nLUC and cLUC-TEAR1 or cLUC-TEAR2 in six repeat experiments, suggesting that the cLUC-TEAR1 or cLUC-TEAR2 binding to TIE1-nLUC caused TIE1-nLUC degradation, resulting in the weak fluorescence signals, while cLUC-mTEAR1 or cLUC-mTEAR2 without E3 ligase activity did not (Figures 6A and 6B). Second, our previous study showed that TIE1 has transcriptional repression activity. Thus, we generated the reporter 35Spro-UAS-LUC, in which the luciferase gene LUC was driven by a CaMV 35S promoter fusion with six copies of the GAL4 binding sites (UAS), and the DBD-TIE1 construct, in which TIE1 fused to the GAL4 DNA binding domain (DBD) was driven by the CaMV 35S promoter. Compared with the control DBD, DBD-TIE1 indeed repressed the expression of the LUC reporter gene, consistent with the previous report (Tao et al., 2013). However, when we cotransformed DBD-TIE1 and 35Spro-UAS-LUC together with 35Spro-TEAR1, in which TEAR1 was driven by the CaMV 35S promoter, the repression of the reporter gene by TIE1 was significantly released, suggesting that TEAR1 could dampen the repression activity of TIE1 possibly by promoting TIE1 degradation (Figure 6C). Third, we transformed 35Spro-TEAR1 into wild-type Arabidopsis and obtained several TEAR1 overexpression lines. We crossed one line, 35Spro-TEAR1-4, with the TIE1 overexpression line 35Spro-TIE1-Flag-19, which produced small and serrated leaves. In the F2 generation, the 35Spro-TIE1-Flag-19 lines containing 35Spro-TEAR1-4 recovered a normal appearance, whereas the 35Spro-TIE1-Flag-19 siblings without 35Spro-TEAR1-4 still showed the small and serrated leaf phenotypes (Figures 6D and 6E). Although the level of TIE1 transcripts was similar, the TIE1 protein level in the 35Spro-TIE1-Flag-19 lines with 35Spro-TEAR1-4 was much lower than that in the 35Spro-TIE1-Flag-19 siblings without 35Spro-TEAR1-4 (Figure 6E). Finally, to further confirm that TEAR1 promotes TIE1 degradation, we performed TIE1 in vitro degradation assays using leaf extracts from 35Spro-TIE1-Flag or 35Spro-TEAR1 35Spro-TIE1-Flag plants. The TIE1 protein level was much lower in 35Spro-TEAR1 35Spro-TIE1-Flag than 35Spro-TIE1-Flag (Figures 6F and 6G), consistent with the result shown in Figure 6E. As time elapsed, the TIE1 degraded much faster in 35Spro-TEAR1 35Spro-TIE1-Flag than 35Spro-TIE1-Flag, while the proteasome inhibitor MG132 inhibited the TIE1 degradation (Figures 6F and 6G), suggesting that TEAR1 promotes TIE1 degradation through 26S proteasomes.
Figure 6.
TIE1 Protein Level Is Negatively Regulated by TEAR1.
(A) and (B) The fluorescence signals were monitored in leaf halves cotransformed with TIE1-nLUC and cLUC-mTEAR1 or mTEAR2 compared with TIE1-nLUC and cLUC-TEAR1 or TEAR2.
(C) Assay of transcriptional repression activity of TIE1. The ratio of LUC/REN was tested in N. benthamiana leaves using a GAL4/UAS-based system. In 35Spro-UAS-LUC, the luciferase gene LUC was driven by a CaMV 35S promoter without a TATA box fusion with six copies of the GAL4 binding sites (UAS); in 35Spro-G4DBD, the GAL4 DNA binding domain was driven by a CaMV 35S promoter; in 35Spro-G4DBD-TIE1, G4DBD fused with TIE1 was driven by a CaMV 35S promoter; in 35Spro-TEAR1, the coding sequence of TEAR1 was driven by a CaMV 35S promoter. The error bars represent the sd of three biological replicates; the asterisk means P value < 0.05.
(D) and (E) The phenotypes of 42-d-old wild type, 35Spro-TIE1-Flag×35Spro-TEAR1, and 35Spro-TIE1-Flag (D). In the F2 population of 35Spro-TIE1-Flag×35Spro-TEAR1, although the level of TIE1 transcripts was similar, the TIE1 protein level was much lower in the siblings of 35Spro-TIE1-Flag with 35Spro-TEAR1 than those without 35Spro-TEAR1 (E).
(F) and (G) Analysis of TIE1 protein degradation over time in 35Spro-TIE1-Flag line and 35Spro-TIE1-Flag with 35Spro-TEAR1 with or without MG132 treatment as indicated.
These results demonstrate that TEAR1 regulates TIE1 activity by negatively affecting TIE1 protein levels.
Disruption of TEAR1 Family Proteins Causes Leaf Defects
To further elucidate the function of TEAR1 and TEAR2 during leaf development, we generated a TEAR1 mutant, tear1, using the CRISPR/Cas9 system and identified a TEAR2 T-DNA insertion mutant named tear2 (Supplemental Figures 4A and 4B). The tear1 mutant contained a 1-bp insertion in the first exon of TEAR1 and caused premature translational termination (Supplemental Figure 4A), while the T-DNA insertion was located in the first exon of TEAR2 and led to TEAR2 loss of function (Supplemental Figure 4B). Neither mutant displayed obvious phenotypes. We then generated a tear1 tear2 double mutant by genetic crossing. No phenotypes were observed in tear1 tear2. Because TEAR1 and TEAR2 belong to a large RING domain-containing protein family (Supplemental Figures 1A and 1B), we hypothesized that other members including the previously identified MEDIATOR25 (MED25) BINDING RING-H2 PROTEIN1 (MBR1), MBR2, MBRL1, and MBRL2 (Iñigo et al., 2012) could be functionally redundant to TEAR1 and TEAR2. To test the hypothesis, we first cloned the promoters of MBR1 and MBRL1. The constructs of MBR1pro-GUS and MBRL1pro-GUS were generated by putting the GUS reporter gene under the control of the two promoters. GUS staining showed that the two genes were expressed in leaves (Supplemental Figure 5), in agreement with previous RT-PCR results (Iñigo et al., 2012). Furthermore, the GUS staining caused by the MBR1 and MBRL1 promoters was strong in young leaves and gradually diminished as the leaves aged, indicating that the expression of MBR1 and MBRL1 temporally and spatially overlapped with that of TEAR1 and TEAR2 (Supplemental Figure 5). We then investigated the interactions between six TEAR family proteins, i.e., TEAR1, TEAR2, MBR1, MBR2, MBRL1, and MBRL2, and three TIE family members i.e., TIE1, TIE3, and TIE4, which are reported to be functionally redundant in leaf development (Tao et al., 2013), to assess the specificity of the interactions between the two families. The firefly luciferase complementation imaging assays showed that TEAR1, TEAR2, MBR1, and MBR2 each interacted with all of the three TIEs (Supplemental Figure 6). However, MBRL1 had no interaction with TIE1, but interacted with TIE3 and TIE4 (Supplemental Figure 6). MBRL2 interacted only with TIE4 (Supplemental Figure 6). These results suggested that MBR1, MBR2, MBRL1, and MBRL2 could function redundantly with TEAR1 and TEAR2 and had some specificities for association with TIE family members.
To overcome the high functional redundancy of TEARs, we implemented a dominant-negative strategy by overexpression of mTEAR1 based on our above data indicating that mTEAR1 had no E3 ligase activity but interacted with TIE1. This system should allow mTEAR1 to compete for the binding sites with the endogenous TEAR family proteins. We generated the construct 35Spro-mTEAR1, in which mTEAR1 was driven by the CaMV 35S promoter. Interestingly, 15 35Spro-mTEAR1 transgenic lines produced defective leaves with excessive serrations and wavy margins similar to those observed in the TIE1 overexpression lines or tcp multiple mutants (Figures 7A to 7E) (Efroni et al., 2008; Koyama et al., 2010; Tao et al., 2013). These 35Spro-mTEAR1 lines also displayed other phenotypes, including semidwarfism, more branches, and low fertility, similar to those exhibited by the TIE1 overexpression lines at the adult stage (Supplemental Figure 7A). We then analyzed the leaf phenotypes of VC330 and AE284 transgenic lines, in which MBR1, MBR2, MBRL1, and MBRL2 were knocked down by two different tandem artificial microRNAs (Iñigo et al., 2012). Both VC330 and AE284 displayed severely wavy leaf margins like 35Spro-mTEAR1 lines (Supplemental Figure 7B), consistent with the previous observations (Iñigo et al., 2012). VC330 and AE284 also displayed semidwarfism and more branches, similar to 35Spro-mTEAR1 (Supplemental Figure 7C). However, the low fertility phenotype in 35Spro-mTEAR1 was not observed in VC330 or AE284 (Supplemental Figure 7C), suggesting that TEAR1 or TEAR2 may also play important roles at reproductive stage.
Figure 7.
Disruption of TEAR1 Family Proteins Leads to Defective Leaves and Genetic Interaction between TEARs and TCPs.
(A) to (D) Leaf phenotypes of 28-d-old wild type and three independent 35Spro-mTEAR1 transgenic lines. Bars = 1 mm
(E) Close-up views of the leaves from 28-d-old wild-type and two independent 35Spro-mTEAR1 transgenic plants. From left to right, the 5th and 6th leaves from the wild type and 35Spro-mTEAR1 transgenic lines. Bars = 1 mm.
(F) Phenotypes of 32-d-old wild type and two tear1 tear2 mbr1 mbr2 mbrl1 mbrl2 sextuple mutants. Bars = 1 mm.
(G) Close-up views of the 6th and 7th leaves from 32-d-old wild-type and two tear sextuple mutants. Bars = 1 mm.
(H) Phenotypes of 33-d-old wild type, VC330, jaw-5D, and VC330 jaw-5D double mutant. Bar = 1 cm.
(I) Close-up views of the 5th and 6th leaves from 33-d-old wild-type, VC330, jaw-5D, and VC330 jaw-5D double mutant. Bar = 1 mm.
(J) Phenotypes of 25-d-old wild type, VC330, 35Spro-mTCP4, and VC330 35Spro-mTCP4 double mutant. Bar = 1 cm.
(K) Close-up views of the 5th and 6th leaves from 25-d-old wild type, VC330, 35Spro-mTCP4, and VC330 35Spro-mTCP4 double mutant. Bar = 1 mm.
(L) Relative expression levels of the known TCP direct target genes in the leaves of wild type, VC330, and 35Spro-mTCP4. The expression levels of the genes in wild-type plants were set to 1.0. The error bars represent the sd of three biological replicates. Total RNA was extracted from the mixed leaf tissues of six wild-type, VC330, or 35Spro-mTCP4 plants. After reverse transcription, real-time PCR was performed three times to determine the relative expression levels of the TCP direct target genes.
To further confirm the functional significance of TEAR-related proteins during leaf development, we generated and identified mbr1, mbr2, mbrl1, and mbrl2 single mutants (Supplemental Figures 8A to 8D). No obvious leaf defects were observed in these single mutants. We then generated the multiple tear mutants by genetic crossing. Both the quadruple tear1 tear2 mbr2 mbrl1 mutant and the quintuple tear1 tear2 mbr2 mbrl1 mbrl2 mutant displayed no obvious defective-leaf phenotypes (Supplemental Figure 8E). However, the sextuple tear1 tear2 mbr1 mbr2 mbrl1 mbrl2 mutant did produce wavy and serrated leaves like those observed in 35Spro-mTEAR1 lines and VC330 (Figures 7F and 7G; Supplemental Figure 8E), further indicating that TEAR family proteins play pivotal roles during leaf development by targeting TIE proteins for degradation.
Genetic Interactions of TEAR and TCP Family Genes
To further explore the regulation of TCP activity by TEAR E3 ligases, we analyzed the genetic interactions of the mutants for TEAR and TCP family genes. We first crossed the TEAR family-deficient line VC330 and a jaw-5D mutant in which miR319b is overexpressed to cause the decrease of TCP2, TCP3, TCP4, TCP10, and TCP24 transcripts (Tao et al., 2013). Compared with the leaf curvature and wavy margins of VC330 or jaw-5D, the VC330 jaw-5D double mutants displayed much more severe phenotypes by producing highly unflattened leaves with severely wavy margins, suggesting that VC330 interacted with jaw-5D synergistically (Figures 7H and 7I). We then crossed VC330 to 35Spro-mTCP4 in which a miR319-resistant mTCP4 was overexpressed using CaMV 35S promoter (Tao et al., 2013) and small leaves were produced with smooth margins and very few trichomes as previously reported (Figures 7J and 7K) (Efroni et al., 2013). The VC330 35Spro-mTCP4 double mutant lines produced flattened leaves that had normal trichomes and were bigger than the leaves of 35Spro-mTCP4, indicating that overexpression of mTCP4 completely rescued the leaf curvature of VC330 and VC330 partially rescued the small size and few trichome defects in 35Spro-mTCP4 (Figures 7J and 7K). These results confirmed that TEAR family proteins control leaf development by positively regulating the activity of TCPs.
To further verify the regulation of TCP activity by TEAR family proteins, we investigated the expression of TCP target genes in TEAR-deficient line VC330 and 35Spro-mTCP4 using qRT-PCR. We selected LIPOXYGENESE2 (LOX2), ARR16, INDOLEACETIC ACID-INDUCED PROTEIN3 (IAA3), and SMALL AUXIN UP RNA gene At1g29460, all of which have been identified as TCP direct-target genes (Schommer et al., 2008; Koyama et al., 2010; Efroni et al., 2013). All four genes were downregulated significantly in VC330 but were upregulated in TCP4 overexpression line 35Spro-mTCP4 (Figure 7L). This is consistent with the previously reported results that the TCP target genes are repressed in TIE1 overexpression line tie1-D (Tao et al., 2013). The results again suggest that TEAR proteins regulate TCP activity by targeting the TCP-repressing TIE proteins for degradation.
DISCUSSION
In this study, we identified an E3 ligase, TEAR1, which contains a typical C3H2C3 RING-domain and controls leaf sizes and shapes by modulating CIN-like TCP activity. We showed that TEAR1 is a functional E3 ligase and interacts with the TCP repressor TIE1. We demonstrated that TIE1 can be modified by ubiquitination and degraded through the 26S proteasome. In addition, we propose a working model for TEAR1 function (Figure 8) in which TIE1 functions as a transcriptional repressor that recruits corepressor TPL/TPRs to CIN-like TCP transcription factors and thus suppresses TCP activity (Tao et al., 2013). TEAR1 functions as an active E3 ligase and targets the TIE1 protein for degradation via the Ub-proteasome system, releasing the TIE1 repression of CIN-like TCP activity to regulate leaf developmental plasticity (Figure 8).Our data indicated that TEAR1 is an important leaf developmental regulator that promotes CIN-like TCP activity by targeting the TCP repressor TIE1 for degradation.
Figure 8.
Working Model of TEAR1 Function.
TIE1 recruits corepressor TPL/TPRs to suppress CIN-like TCP activity as a transcriptional repressor (Tao et al., 2013). TEAR1 functions as an active E3 ligase and targets the TIE1 repressor for degradation through the Ub-26S proteasome system. The degradation of TIE1 by TEAR1 promotes leaf development by releasing the TIE1-mediated repression of CIN-like TCP activity. The cyan ellipse in the upper image indicates TIE1 protein that is not degraded by 26S proteasome, while the irregular cyan shape in the lower image represents TIE1 protein that is degraded by the 26S proteasome. The small red circles represent Ub. The question marks indicate the possible factor that could mediate the interaction between TPL/TPRs and HDA19.
RING-type E3 ligases play central roles in alteration of physiological and/or morphological properties of plants in response to different intrinsic and environmental signals (Zhang et al., 2005; Disch et al., 2006; Dong et al., 2006; Stone et al., 2006; Zhang et al., 2007; Liu et al., 2008; Qin et al., 2008; Lau and Deng, 2012; Sakai et al., 2012; Chen and Hellmann, 2013; Marino et al., 2013; Xia et al., 2013; Duplan and Rivas, 2014; Shabek and Zheng, 2014; Ding et al., 2015; Lazaro et al., 2015; Sarid-Krebs et al., 2015; Zhang et al., 2015). However, the Arabidopsis genome contains more than 460 RING-type proteins (Stone et al., 2005), most of which have yet to be characterized in detail. Here, we demonstrated that TEAR1 is a RING domain-containing protein and has E3 ligase activity. The expression of TEAR1 in leaves is developmentally regulated, with high levels in young leaves and low levels in old leaves. Disruption of TEAR1 causes obvious leaf developmental defects, indicating that TEAR1 plays pivotal roles in leaf development. As the tear1 tear2 double mutant displayed no obvious phenotypes, we hypothesize that TEAR1 functions redundantly not only with TEAR2 but also possibly with the other members of TEAR family proteins, including the previously identified MBR1, MBR2, MBRL1, and MBRL2 (Iñigo et al., 2012). MBR1 and MBR2 have been shown to be RING-type E3 ligases indirectly by associating with UBCs and promote flowering by targeting PHYTOCHROME AND FLOWERING TIME1 (PFT1)/MED25 for degradation (Iñigo et al., 2012). Interestingly, disruption of MBR1, MBR2, MBRL1, and MBRL2 by amiRNAs in VC330 and AE284 (Iñigo et al., 2012) resulted in leaves with wavy margins similar to those observed in the TEAR1 dominant-negative transgenic lines expressing mTEAR1. Our current results indicated that the sextuple tear mutant produced serrated leaves with wavy margins as well. These data suggest that MBR1, MBR2, MBRL1, and MBRL2 function in a redundant manner with TEAR1 and TEAR2 to regulate leaf development by mediating the proteasome degradation of the TCP repressor TIE1. Indeed, we found that MBR1, MBR2, MBRL1, and MBRL2 interact with TIE family proteins with different specificities. We also demonstrated that MBR1 and MBRL1 have overlapping expression patterns with TEAR1 and TEAR2. Together, our results confirm that MBR/TEAR family proteins have redundant roles in control of leaf development and uncover the molecular mechanism of the wavy leaf phenotypes in VC330 and AE284, which could not be explained by interactions of MBR1 or MBR2 with PFT1/MED25.
Transcriptional repressors containing ethylene-responsive element binding factor (ERF)-associated amphiphilic repression (EAR) motifs play essential roles in control of phytohormone signaling, stress responses, and other biological processes (Ohta et al., 2001; Hiratsu et al., 2002; Szemenyei et al., 2008; Pauwels et al., 2010; Causier et al., 2012; Wei et al., 2015). A central mechanism in signal transduction of several phytohormones is the E3 ligase-mediated degradation of transcriptional repressors. The removal of repressors releases the activity of key transcription factors in response to different hormonal signals (Szemenyei et al., 2008; Pauwels et al., 2010; Jiang et al., 2013; Zhou et al., 2013). For example, following detection of auxin, jasmonic acid (JA) or strigolactone, the EAR motif-containing transcriptional repressor AUXIN (AUX)/IAA, JAZ-NINJA complex, and SUPPRESSOR OF MAX2 1 (SMAX1)-LIKE (SMXL)/D53 are targeted to the proteasome for degradation and thus activate the signaling pathways of these phytohormones (Szemenyei et al., 2008; Pauwels et al., 2010; Jiang et al., 2013; Zhou et al., 2013; Soundappan et al., 2015; Wang et al., 2015b). Recently, auxin has been shown to trigger the degradation of AUX/IAA repressors that recruit corepressor TPL/TPRs to the MONOPTEROS (MP) transcription factor and thus alter the chromatin state by promotion of MP binding to SWI/SNF chromatin remodeling ATPases during flower development (Wu et al., 2015). CIN-like TCP transcription factors also recruit the SWI/SNF chromatin remodeling ATPase BRM to activate ARR16 during leaf development (Efroni et al., 2013). We previously identified TIE1 as a key repressor that recruits corepressor TPL/TPRs to CIN-like TCP transcription factors for the control of leaf development (Tao et al., 2013). In this study, we demonstrate that TIE1 is targeted for degradation by the E3 ligase TEAR1, suggesting that TEAR1 possibly causes a similar chromatin switch by promoting the release of CIN-like TCPs to recruit SWI/SNF chromatin remodeling ATPases during leaf development. The regulation of temporal and spatial TCP activity in the leaves is very important for formation of proper leaf sizes and morphologies in response to different environmental and internal signals (Ori et al., 2007; Efroni et al., 2008; Koyama et al., 2010; Shleizer-Burko et al., 2011). Our findings suggest that a chromatin state switch mediated by TEAR1, TIE1, BRM, and CIN-like TCPs could provide a simple and flexible way to regulate leaf cell division and differentiation and thus form proper leaf sizes and shapes in response to the ever-changing environmental conditions.
METHODS
Plant Materials and Growth Conditions
The ecotype Columbia-0 (Col-0) of Arabidopsis thaliana was used in this study. The seeds from wild-type, mutants including SALK_152632 (tear2), GABI_587C07 (mbr2), WiscDsLox461-464G17 (mbrl1), SAIL_756_C02 (mbrl2), the CRISPR/Cas9-generated tear1 and mbr1 and jaw-5D (Tao et al., 2013), the transgenic lines including 35Spro-mTCP4 (Tao et al., 2013), VC330, and AE284 (Iñigo et al., 2012), or different generations of crossed plants were placed on half-strength Murashige and Skoog medium with or without 50 µg/mL kanamycin or 10 µg/mL phosphinothricin. The plates with seeds were placed at 4°C for 2 d synchronization and were then incubated at 22°C ± 2°C under long-day conditions (16 h light and 8 h dark) with a light intensity of ∼170 μmol/m2/s using bulbs (Philips F17T8/TL841 17 W). After 7 to 10 d of growth, the green seedlings were transferred to soil and continually grown under the conditions described above. For the firefly luciferase complementation imaging and transient expression assays, Nicotiana benthamiana was grown in soil at 22°C ± 2°C under long-day conditions.
PCR Analysis and Gene Expression Assays
All the primers used in this study are listed in Supplemental Table 1.
For mutant analysis, TRIzol reagent (Invitrogen) was used to prepare total RNAs, and a SuperScript III kit (Invitrogen) was used for reverse transcription with the total RNA as a template according to the user’s manual. PCR or RT-PCR was performed using genomic DNA from mutants, plasmids, and the diluted cDNA as the templates. The cycling conditions of PCR or RT-PCR were 94°C for 30 s, 56°C to 58°C for 30 s, and 72°C for 60 to 120 s.
Generation of Binary Constructs and Transformation
The coding sequence of TEAR1 or TEAR2 was amplified from Arabidopsis leaf cDNA using the primer pair TEAR1-F/R or TEAR2-F/R. The amplified fragments were cloned into pENTR/D TOPO (Invitrogen) to generate pENTR-TEAR1 or pENTR-TEAR2, respectively. pENTR-mTEAR1 or pENTR-mTEAR2 was generated using PCR-based mutagenesis with the primer pair mTEAR1-F/R or mTEAR2-F/R from pENTR-TEAR1 or pENTR-TEAR2, respectively.
To generate the TIE1 overexpression line, the full-length coding region of TIE1 without a stop codon was amplified from Arabidopsis cDNA using primers TIE1-F and TIE1-Rnsc and was then cloned into pENTR/D-TOPO to generate pENTR-TIE1NS. The overexpression construct 35Spro-TIE1-FLAG was generated by an LR reaction between pENTR-TIE1NS and pK7FLAGWG2 (purchased from Ghent University). To establish the mTEAR1 overexpression lines, the 35Spro-mTEAR1 construct was generated by an LR reaction between pENTR-mTEAR1 and pK2GW7 (from Ghent University). For the overexpression of TEAR1, TEAR1 was first amplified from Arabidopsis genomic DNA using primers TEAR1 F-KpnI and TEAR1 R-SacI and then cloned into the KpnI-SacI-digested pQG110 (Qin et al., 2005; Guo et al., 2015).
To examine the TEAR1 or TEAR2 and MBR1 or MBRL1 expression pattern by GUS staining, 2119-bp TEAR1 or 1974-bp TEAR2 and 1914-bp MBR1 or 1424-bp MBRL1 promoter regions upstream of the ATG of TEAR1 or TEAR2 and MBR1 or MBRL1 were amplified from Arabidopsis genomic DNA with the primer pair TEAR1P-F/R or TEAR2P-F/R and MBR1P-F/R or MBRL1P-F/R, respectively. Then, the products were cloned into pENTR/D-TOPO to generate pENTR-TEAR1pro or pENTR-TEAR2pro and pENTR-MBR1pro or pENTR-MBRL1pro, respectively. TEAR1pro-GUS or TEAR2pro-GUS and MBR1pro-GUS or MBRL1pro-GUS were generated by an LR reaction between pKGWFS7 and pENTR-TEAR1pro or pENTR-TEAR2pro and pENTR-MBR1pro or pENTR-MBRL1pro, respectively.
Yeast Two-Hybrid Screening and Assays
The TIE4 coding sequence was amplified using the primer pair TIE4-F/R and then cloned into the vector pENTR/D TOPO to generate pENTR-TIE4. The bait construct DBD-TIE4 was generated by an LR reaction with pENTR-TIE4 and pDEST32 (Invitrogen). The cDNA-AD library was generated by TOPO cloning using the cDNA from a mixture of different organ tissues from Arabidopsis. Briefly, mRNA was prepared from Arabidopsis seedlings and flowers using the FastTrack MAG Maxi mRNA isolation kit (Invitrogen) and then reverse transcribed into cDNA. The cDNA products were cloned into pDONR222 (Invitrogen). The yeast strain AH109 (Clontech) was first transformed with the bait construct DBD-TIE4. Then, the cells containing the bait vector were transformed with 10 μg of the cDNA-AD library plasmid DNA mixture in the presence of 2 mg salmon sperm DNA. Cells were resuspended in 3 mL of 2×YPAD medium and allowed to recover at 30°C for 90 min. Next, the cells were plated on SD medium lacking leucine, tryptophan, histidine, and adenine (SD-Leu-Trp-His-Ade) supplemented with 3-amino-1,2,4-triazole for screening. Finally, the identity of the positive clones was determined by sequencing.
To further assess the interactions of TIE4 and TEARs or mTEARs, the prey constructs TEAR1-AD, TEAR2-AD, mTEAR1-AD, and mTEAR2-AD were generated by LR reactions with pDEST22. The bait DBD-TIE4 plasmids were cotransformed with each prey plasmid or the blank pDEST22 into the yeast strain AH109. Media supplemented with SD-Leu-Trp-His or SD-Leu-Trp-His-Ade and 5 mM 3-amino-1,2,4-triazole were used for selection.
Firefly Luciferase Complementation Imaging Assay
To generate the vectors pCAMBIA-nLUC-GW and pCAMBIA-cLUC-GW for firefly luciferase complementation imaging assays, attR1-ccdB-attR2 was amplified from pK2GW7 (Ghent University) using primers attR1 F-KpnI and attB2 R-XhoI and cloned into the KpnI and SalI site of pCAMBIA-nLUC or pCAMBIA-cLUC (Chen et al., 2008).
To test the interaction of TIE1 and mTEAR1 in vivo, TIE1-nLUC was generated by an LR reaction with pENTR-TIE1 and pCAMBIA-nLUC-GW. cLUC-mTEAR1 was generated by an LR reaction between pENTR-mTEAR1 and pCAMBIA-cLUC-GW. For the TEAR1 truncation assays, TEAR1△C1, mTEAR1△N1, TEAR1△C2, TEAR1△NC1, TEAR1△NC2, and mTEAR1△N2 were amplified using the primer pairs TEAR1-F/TEAR1△C1-R, mTEAR1△N1-F/TEAR1-R, TEAR1-F/TEAR1△C2-R, TEAR1△NC1-F/TEAR1△C1-R, TEAR1△NC2-F/R, and mTEAR1△N2-F/TEAR1-R, respectively. The fragments were first cloned into pENTR/D TOPO and then transferred into pCAMBIA-cLUC-GW by LR reactions to generate cLUC-TEAR1△C1, cLUC-mTEAR1△N1, cLUC-TEAR1△C2, cLUC-TEAR1△NC1, cLUC-TEAR1△NC2, and cLUC-mTEAR1△N2, respectively.
To detect the interaction between six TEAR family proteins, i.e., TEAR1, TEAR2, MBR1, MBR2, MBRL1, and MBRL2, and three TIE family members, i.e., TIE1, TIE3, and TIE4, mTEARs were amplified using corresponding primer pairs, and the genes were then used to generate pENTR vectors by LR reactions with pDONR P4-P1R, pENTR/D-TOPO, and pDONR P2R-P3 (Invitrogen). TIE1-nLUC, nLUC-TIE3, nLUC-TIE4, and cLUC-mTEARs were generated by LR reaction with the pENTR vectors and pCAMBIA-nLUC-GW, pCAMBIA-cLUC-GW, or pB7m34GW.
The above constructs were transformed into Agrobacterium tumefaciens GV3101. The different combinations shown in Figure 1 and Supplemental Figure 6 were coinfiltrated into N. benthamiana leaves. The plants were placed in the dark for 24 h followed by 48 h in a growth chamber under long-day conditions (16 h light and 8 h dark). Then, the infiltrated N. benthamiana leaves were sprayed with luciferin (100 mM) and kept in the dark for 10 min. The leaves were observed under a low-light cooled CCD imaging apparatus (Lumazone 1300B; Roper Bioscience).
Pull-Down Assay
The coding sequence of TIE1 was cloned into the EcoRI and SalI site of pET-28a(+) (Novagen) to generate pET28-TIE1-His. The coding sequences of TEAR1, TEAR2, mTEAR1, and mTEAR2 were cloned into the SalI site of pMAL-c2x (NEB). To generate the vector pMAL-GW, attR1-ccdB-attR2 was amplified from pK2GW7 (Ghent University) using primers attR1 F-XbaI and attB2 R-HindIII and cloned into the XbaI and HindIII site of pMAL-c2x (NEB). For TEAR1 truncation assay, DAR1 was amplified using the primer pair DAR1-F/DAR1-R. Then, the fragment was first cloned into pENTR/D TOPO to generate pENTR-DAR1. The pMAL-TEAR1△C1, pMAL-mTEAR1△N1, and pMAL-DAR1 constructs were generated by LR reactions with pMAL-GW and pENTR-TEAR1△C1, pENTR-mTEAR1△N1, and pENTR-DAR1, respectively.
The constructs were transformed into Escherichia coli BL21 (DE3) competent cells for protein expression. BL21 (DE3) cells were grown in LB medium at 37°C until the OD600 reached 0.5. The induction of protein expression was performed at 18°C for 12 h in the presence of 0.5 mM IPTG. Bacterial lysates contained ∼50 μg MBP-TEAR1, MBP-TEAR2, MBP-mTEAR1, MBP-mTEAR2, MBP-TEAR1△C1, MBP-mTEAR1△N1, or MBP-DAR1 fusion protein, and the control MBP proteins were mixed with lysates containing TIE1-His fusion protein. Amylose resin (30 μL; NEB) was added, and the mixtures were rotated at 4°C for 3 h. Beads were washed five times with the column buffer (20 mM Tris-HCl, pH 7.4, 200 mM NaCl, 1 mM EDTA, 1 mM PMSF, and 1× C-complete protease inhibitor [Roche]). The isolated proteins were then separated with 12% SDS-PAGE and detected by immunoblot analysis with an anti-His antibody (Sigma-Aldrich; catalog number H1029-2mL; 1:3000 dilution) or an anti-MBP antibody (NEB; catalog number E8032S, lot number 0101603; 1:10,000 dilution).
Subcellular Localization of TEAR1 and TEAR2
The 35Spro-GFP-TEAR1 and 35Spro-GFP-TEAR2 constructs were generated by LR reactions between pK7WGF2 and pENTR-TEAR1 or pENTR-TEAR2, respectively. Then, 35Spro-GFP-TEAR1 and 35Spro-GFP-TEAR2 were transformed into Agrobacterium GV3101, and the agrobacteria harboring the constructs were infiltrated into N. benthamiana leaves. The plants were then grown in the dark for 24 h followed by 48 h in a greenhouse under normal conditions. The transient GFP fluorescence in N. benthamiana leaf cells was observed under a Leica SPE confocal microscope. A 4′,6-diamidino-2-phenylindole (Sigma-Aldrich) solution was used to stain the nuclei.
For the colocalization of mTEAR1 and TIE1, TIE1-RFP was generated through a LR reaction between pENTR-TIE1 and pK7RWG2. The vectors TIE1-RFP and GFP-mTEAR1 were cotransformed into Arabidopsis protoplasts as described previously (Wu et al., 2009). Briefly, ∼2 × 105/mL protoplasts in 0.2 mL of MMG solution (0.4 M mannitol, 5 mM MgCl2, and 4 mM MES, pH 5.7) was transformed with 20 µg of a mixture of plasmid DNA using the polyethylene glycol method. The protoplasts were resuspended in 1 mL of W5 (1.54 mM NaCl, 1.25 mM CaCl2, 5 mM KCl, and 2 mM MES, pH 5.7) and incubated in a growth chamber for 16 h in the dark. GFP and RFP signals were observed under a Leica SPE confocal microscope.
Histochemical GUS Staining
Tissues from the TEAR1pro-GUS or TEAR2pro-GUS transgenic lines were soaked in 90% acetone solution for 20 min on ice. Then, the tissues were washed twice by phosphate buffer and incubated in GUS staining buffer containing 0.5 mg/mL 5-bromo-4-chloro-3-indolyl glucuronide. Samples were vacuum infiltrated for 10 min and were stained overnight in 37°C incubator. The staining buffer was then changed to 70% ethanol for decolorizing before microscopy analysis.
In Vitro Ubiquitination Assay
For the in vitro ubiquitination assay, ubiquitination reaction mixtures (30 µL) containing recombinant wheat (Triticum aestivum) E1 (GI: 136632), At-UBC10 (40 ng), recombinant plant ubiquitin (R&D Systems; 2 µg), and purified TEAR1 or TEAR2 (1 µg) fused with the MBP tag were mixed in a reaction buffer containing 50 mM Tris-HCl, pH 7.4, 5 mM MgCl2, 2 mM ATP, and 2 mM DTT (Xie et al., 2002). MBP, MBP-mTEAR1, and MBP-mTEAR2 were used as the negative controls for MBP-TEAR1 and MBP-TEAR2. The reactions were incubated at 30°C for 2 h and then stopped by the addition of 5× protein loading sample buffer (258 mM Tris-HCl, pH 6.8, 8% SDS, 40% glycerol, and 0.5% Coomassie Brilliant Blue). Then, the reaction product was further separated with 12% SDS-PAGE and was tested by immunoblot analysis with an anti-Ub antibody (Santa Cruz Biotechnology; catalog number sc-8017; lot number 1611; 1:2000 dilution), and input protein was detected with an anti-MBP antibody (NEB; catalog number E8032S, lot number 0101603; 1:10,000 dilution).
In Vivo and in Vitro Degradation Assays
For in vivo TIE1 degradation assay, 10-d-old 35Spro-TIE1-FLAG transgenic seedlings were treated by 50 µM CHX with or without 100 µM MG132 in 0.5× Murashige and Skoog liquid medium for a 24-h time course. Total proteins were extracted with extraction buffer (PBS and 1×Roche C-complete protease inhibitor) containing 1% Triton X-100, and the lysate was cleared by centrifugation at 20,000g for 10 min. Protein isolates were mixed with sample buffer and were separated with 12% SDS-PAGE. The separated proteins were then blotted onto nitrocellulose membranes, blocked with nonfat dry milk, and incubated with an anti-ACTIN antibody (Abmart; catalog number M20009M, lot number 224183; 1:2000 dilution) or anti-FLAG-HRP (M2; Sigma-Aldrich; catalog number A8592-2MG, lot number SLBH1183V; 1:10,000 dilution) to assess TIE1 degradation.
For in vitro degradation assays, total protein was extracted from 10-d-old seedling samples with native extraction buffer (50 mM Tris-MES, pH 8.0, 0.5 M sucrose, 1 mM MgCl2, 10 mM EDTA, 5 mM DTT, 1 mM PMSF, and 1× C-complete protease inhibitor [Roche]). Samples were incubated at 4°C temperature for various time periods. The extracted proteins were then blotted onto nitrocellulose membranes, blocked with nonfat dry milk, and incubated with an anti-ACTIN antibody (Abmart; catalog number M20009M, lot number 224183; 1:2000 dilution) or anti-FLAG-HRP (M2; Sigma-Aldrich; catalog number A8592-2MG, lot number SLBH1183V; 1:10,000 dilution). Protein gel blot pictures were scanned, and the intensity of the images was quantified by ImageJ (National Institutes of Health).
TUBEs Pull-Down Assay
The TUBEs pull-down assay was performed as described previously with modifications (Hjerpe et al., 2009). Briefly, total plant proteins were extracted using extraction buffer (50 mM Tris-HCl, pH 7.4, 150 mM NaCl, 10% glycerol, 1 mM PMSF, and 1× C-complete protease inhibitor [Roche]), and the mixtures were then centrifuged at 14,000g for 15 min at 4°C. For Ub immunoprecipitation assays, the deubiquitination inhibitor PR-619 (Tebu-bio) was added into the extraction buffer. Supernatants were precleared for 30 min at 4°C with agarose beads (Tebu-bio). Samples were separated into two fresh tubes either for TUBEs (tandem Ub binding entities; Tebu-bio) or for agarose beads (control) pull-down assays at 4°C. The pulled-down proteins were detected by immunoblot analysis with anti-Ub (Santa Cruz Biotechnology; catalog number sc-8017, lot number 1611; 1:2000 dilution) or anti-FLAG-HRP (M2; Sigma-Aldrich; catalog number A8592-2MG, lot number SLBH1183V; 1:10,000 dilution).
Transient Expression Assays in the Leaves of N. benthamiana
To generate the reporter vector 35Spro-UAS-LUC, 35Spro-UAS was amplified from the 35Spro-UAS-GUS reporter (Tao et al., 2013) and cloned into pDONR P4-P1R (Invitrogen) by a BP reaction. The coding region of the firefly luciferase gene LUC and the CaMV 35S terminator were cloned into pENTR/D-TOPO and pDONR P2R-P3 (Invitrogen), respectively. These three constructs were used to generate the reporter vector 35Spro-UAS-LUC by LR reactions with pK7m34GW. The effector vectors G4BD and G4BD-TIE1 were generated as described previously (Tao et al., 2013). The coding sequence of Renilla luciferase was cloned into pENTR/D-TOPO (Invitrogen) to generate pENTR-Rluc. Then, pENTR-TEAR1 and pENTR-Rluc were used in a LR reaction with pK2GW7 to generate 35Spro-TEAR1 and 35Spro-Rluc.
The reporter and effector constructs were transformed into Agrobacterium GV3101. Then, different combinations, e.g., 35Spro-UAS-LUC and G4BD, 35Spro-UAS-LUC and G4BD-TIE1, or 35Spro-UAS-LUC, G4BD-TIE1, and 35Spro-TEAR1, were coinfiltrated with 35Spro-Rluc and pCam-P19 into the same N. benthamiana leaves. The plants were incubated in the dark for 24 h followed by 36 h in a growth chamber under normal conditions. LUC and REN activity was measured using a Dual-Luciferase reporter kit (Promega).
Generation of the tear1 and mbr1 Mutants Using the CRISPR/Cas9 System
The sgRNA targets including GTTCCTAGCGCCTCCAAAAG in TEAR1 and GACGAACCCCAACCACCAAA in MBR1 were synthesized and cloned into the BbsI sites of AtU6-26SK vector (Feng et al., 2013). The chimeric RNA expression cassettes in AtU6-26SK between KpnI and SalI were subcloned into the KpnI and EcoRI sites of the pCambia1300 vector (Cambia) together with the SalI and EcoRI fragment of the Cas9 expression cassette (Feng et al., 2013). Then, the CRISPR constructs were transformed into wild-type Arabidopsis to obtain the tear1 and mbr1 mutants, respectively. The homozygous tear1 and mbr1 mutants were identified by sequencing.
Accession Numbers
Sequence data from this article can be found in the Arabidopsis Genome Initiative under the following accession numbers: TEAR1, At1g53190; TEAR2, At3g15070; MBR1, At2g15530; MBR2, At4g34040; MBRL1, At1g45180; MBRL2, At5g42940; TIE1, At4g28840; TIE3, At1g29010; TIE4, At2g34010; TCP4, At3g15030; LOX2, At3g45140; ARR16, AT2G40670; and IAA3, At1g04240.
Supplemental Data
Supplemental Figure 1. TEAR1 and TEAR2 belong to a RING domain-containing protein family.
Supplemental Figure 2. TIE1 interacts with TEAR1 and TEAR2 in vivo and in vitro.
Supplemental Figure 3. Leaf phenotypes of the 35Spro-TIE1-Flag transgenic lines.
Supplemental Figure 4. Identification of tear1 and tear2 mutants.
Supplemental Figure 5. Expression patterns of MBR1 and MBRL1.
Supplemental Figure 6. Interactions between six TEAR family proteins and three TIE family members.
Supplemental Figure 7. Disruption of TEAR family proteins leads to defective leaves.
Supplemental Figure 8. Generation and identification of mbr1, mbr2, mbrl1, and mbrl2 single mutants and the multiple tear mutants.
Supplemental Table 1. The list of primers used in this study.
Supplemental Data Set 1. Text file of the alignment used for the phylogenetic analysis shown in Supplemental Figure 1.
Supplementary Material
Acknowledgments
We thank Pablo Cerdán (Fundación Instituto Leloir, IIBBA-Consejo Nacional de Investigaciones Científicas y Técnicas, Argentina) for kindly providing the seeds of VC330 and AE284 and Qi Xie (Institute of Genetics and Developmental Biology, Chinese Academy of Sciences) for providing E1 and At-UBC10 for in vitro ubiquitination assay. We thank Li-Jia Qu and Hongya Gu (Peking University) for their valuable suggestions. This work was supported by the National Natural Science Foundation of China (Grant 31570284), the National Transformation Science and Technology Program (Grant 2016ZX08009003-003), and the National Key Basic Research Program of the People’s Republic of China (Grant 973-2012CB944801).
AUTHOR CONTRIBUTIONS
G.Q. conceived the project. G.Q. and J.Z. designed the experiments. G.Q., J.Z., B.W., R.Y., J.W., M.D., Z.C., and H.Y. performed the experiments. G.Q., J.Z., B.W., R.Y., J.W., M.D., Z.C., and H.Y. analyzed the data. G.Q. and J.Z. wrote the article.
Glossary
- SAM
shoot apical meristem
- IAA
indole-3-acetic acid
- TUBEs
tandem-repeated ubiquitin binding entities
- CHX
cycloheximide
Footnotes
Articles can be viewed without a subscription.
References
- Bolduc N., O’Connor D., Moon J., Lewis M., Hake S. (2012). How to pattern a leaf. Cold Spring Harb. Symp. Quant. Biol. 77: 47–51. [DOI] [PubMed] [Google Scholar]
- Causier B., Ashworth M., Guo W., Davies B. (2012). The TOPLESS interactome: a framework for gene repression in Arabidopsis. Plant Physiol. 158: 423–438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen H., Zou Y., Shang Y., Lin H., Wang Y., Cai R., Tang X., Zhou J.M. (2008). Firefly luciferase complementation imaging assay for protein-protein interactions in plants. Plant Physiol. 146: 368–376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen L., Hellmann H. (2013). Plant E3 ligases: flexible enzymes in a sessile world. Mol. Plant 6: 1388–1404. [DOI] [PubMed] [Google Scholar]
- Ding S., Zhang B., Qin F. (2015). Arabidopsis RZFP34/CHYR1, a ubiquitin E3 ligase, regulates stomatal movement and drought tolerance via SnRK2.6-mediated phosphorylation. Plant Cell 27: 3228–3244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Disch S., Anastasiou E., Sharma V.K., Laux T., Fletcher J.C., Lenhard M. (2006). The E3 ubiquitin ligase BIG BROTHER controls Arabidopsis organ size in a dosage-dependent manner. Curr. Biol. 16: 272–279. [DOI] [PubMed] [Google Scholar]
- Dong C.H., Agarwal M., Zhang Y., Xie Q., Zhu J.K. (2006). The negative regulator of plant cold responses, HOS1, is a RING E3 ligase that mediates the ubiquitination and degradation of ICE1. Proc. Natl. Acad. Sci. USA 103: 8281–8286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duplan V., Rivas S. (2014). E3 ubiquitin-ligases and their target proteins during the regulation of plant innate immunity. Front. Plant Sci. 5: 42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Efroni I., Blum E., Goldshmidt A., Eshed Y. (2008). A protracted and dynamic maturation schedule underlies Arabidopsis leaf development. Plant Cell 20: 2293–2306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Efroni I., Han S.K., Kim H.J., Wu M.F., Steiner E., Birnbaum K.D., Hong J.C., Eshed Y., Wagner D. (2013). Regulation of leaf maturation by chromatin-mediated modulation of cytokinin responses. Dev. Cell 24: 438–445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng Z., Zhang B., Ding W., Liu X., Yang D.L., Wei P., Cao F., Zhu S., Zhang F., Mao Y., Zhu J.K. (2013). Efficient genome editing in plants using a CRISPR/Cas system. Cell Res. 23: 1229–1232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaillochet C., Lohmann J.U. (2015). The never-ending story: from pluripotency to plant developmental plasticity. Development 142: 2237–2249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo D., Zhang J., Wang X., Han X., Wei B., Wang J., Li B., Yu H., Huang Q., Gu H., Qu L.J., Qin G. (2015). The WRKY transcription factor WRKY71/EXB1 controls shoot branching by transcriptionally regulating RAX genes in Arabidopsis. Plant Cell 27: 3112–3127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ha C.M., Jun J.H., Fletcher J.C. (2010). Control of Arabidopsis leaf morphogenesis through regulation of the YABBY and KNOX families of transcription factors. Genetics 186: 197–206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hay A., Tsiantis M. (2010). KNOX genes: versatile regulators of plant development and diversity. Development 137: 3153–3165. [DOI] [PubMed] [Google Scholar]
- Hiratsu K., Ohta M., Matsui K., Ohme-Takagi M. (2002). The SUPERMAN protein is an active repressor whose carboxy-terminal repression domain is required for the development of normal flowers. FEBS Lett. 514: 351–354. [DOI] [PubMed] [Google Scholar]
- Hjerpe R., Aillet F., Lopitz-Otsoa F., Lang V., England P., Rodriguez M.S. (2009). Efficient protection and isolation of ubiquitylated proteins using tandem ubiquitin-binding entities. EMBO Rep. 10: 1250–1258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hur Y.S., et al. (2015). Arabidopsis thaliana homeobox 12 (ATHB12), a homeodomain-leucine zipper protein, regulates leaf growth by promoting cell expansion and endoreduplication. New Phytol. 205: 316–328. [DOI] [PubMed] [Google Scholar]
- Husbands A.Y., Benkovics A.H., Nogueira F.T., Lodha M., Timmermans M.C. (2015). The ASYMMETRIC LEAVES complex employs multiple modes of regulation to affect adaxial-abaxial patterning and leaf complexity. Plant Cell 27: 3321–3335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ichihashi Y., Tsukaya H. (2015). Behavior of leaf meristems and their modification. Front. Plant Sci. 6: 1060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iñigo S., Giraldez A.N., Chory J., Cerdán P.D. (2012). Proteasome-mediated turnover of Arabidopsis MED25 is coupled to the activation of FLOWERING LOCUS T transcription. Plant Physiol. 160: 1662–1673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang L., et al. (2013). DWARF 53 acts as a repressor of strigolactone signalling in rice. Nature 504: 401–405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koyama T., Furutani M., Tasaka M., Ohme-Takagi M. (2007). TCP transcription factors control the morphology of shoot lateral organs via negative regulation of the expression of boundary-specific genes in Arabidopsis. Plant Cell 19: 473–484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koyama T., Mitsuda N., Seki M., Shinozaki K., Ohme-Takagi M. (2010). TCP transcription factors regulate the activities of ASYMMETRIC LEAVES1 and miR164, as well as the auxin response, during differentiation of leaves in Arabidopsis. Plant Cell 22: 3574–3588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lau O.S., Deng X.W. (2012). The photomorphogenic repressors COP1 and DET1: 20 years later. Trends Plant Sci. 17: 584–593. [DOI] [PubMed] [Google Scholar]
- Lazaro A., Valverde F., Piñeiro M., Jarillo J.A. (2012). The Arabidopsis E3 ubiquitin ligase HOS1 negatively regulates CONSTANS abundance in the photoperiodic control of flowering. Plant Cell 24: 982–999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lazaro A., Mouriz A., Piñeiro M., Jarillo J.A. (2015). Red light-mediated degradation of CONSTANS by the E3 ubiquitin ligase HOS1 regulates photoperiodic flowering in Arabidopsis. Plant Cell 27: 2437–2454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee K., Seo P.J. (2015). The E3 ubiquitin ligase HOS1 is involved in ethylene regulation of leaf expansion in Arabidopsis. Plant Signal. Behav. 10: e1003755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu H., Stone S.L. (2013). Cytoplasmic degradation of the Arabidopsis transcription factor abscisic acid insensitive 5 is mediated by the RING-type E3 ligase KEEP ON GOING. J. Biol. Chem. 288: 20267–20279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J., et al. (2008). Targeted degradation of the cyclin-dependent kinase inhibitor ICK4/KRP6 by RING-type E3 ligases is essential for mitotic cell cycle progression during Arabidopsis gametogenesis. Plant Cell 20: 1538–1554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marino D., Froidure S., Canonne J., Ben Khaled S., Khafif M., Pouzet C., Jauneau A., Roby D., Rivas S. (2013). Arabidopsis ubiquitin ligase MIEL1 mediates degradation of the transcription factor MYB30 weakening plant defence. Nat. Commun. 4: 1476. [DOI] [PubMed] [Google Scholar]
- Martín-Trillo M., Cubas P. (2010). TCP genes: a family snapshot ten years later. Trends Plant Sci. 15: 31–39. [DOI] [PubMed] [Google Scholar]
- Masuda H.P., Cabral L.M., De Veylder L., Tanurdzic M., de Almeida Engler J., Geelen D., Inzé D., Martienssen R.A., Ferreira P.C., Hemerly A.S. (2008). ABAP1 is a novel plant Armadillo BTB protein involved in DNA replication and transcription. EMBO J. 27: 2746–2756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Metzger M.B., Pruneda J.N., Klevit R.E., Weissman A.M. (2014). RING-type E3 ligases: master manipulators of E2 ubiquitin-conjugating enzymes and ubiquitination. Biochim. Biophys. Acta 1843: 47–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moon J., Hake S. (2011). How a leaf gets its shape. Curr. Opin. Plant Biol. 14: 24–30. [DOI] [PubMed] [Google Scholar]
- Nath U., Crawford B.C., Carpenter R., Coen E. (2003). Genetic control of surface curvature. Science 299: 1404–1407. [DOI] [PubMed] [Google Scholar]
- Navaud O., Dabos P., Carnus E., Tremousaygue D., Hervé C. (2007). TCP transcription factors predate the emergence of land plants. J. Mol. Evol. 65: 23–33. [DOI] [PubMed] [Google Scholar]
- Noir S., Marrocco K., Masoud K., Thomann A., Gusti A., Bitrian M., Schnittger A., Genschik P. (2015). The Control of Arabidopsis thaliana growth by cell proliferation and endoreplication requires the F-Box protein FBL17. Plant Cell 27: 1461–1476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohta M., Matsui K., Hiratsu K., Shinshi H., Ohme-Takagi M. (2001). Repression domains of class II ERF transcriptional repressors share an essential motif for active repression. Plant Cell 13: 1959–1968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ori N., et al. (2007). Regulation of LANCEOLATE by miR319 is required for compound-leaf development in tomato. Nat. Genet. 39: 787–791. [DOI] [PubMed] [Google Scholar]
- Palatnik J.F., Allen E., Wu X., Schommer C., Schwab R., Carrington J.C., Weigel D. (2003). Control of leaf morphogenesis by microRNAs. Nature 425: 257–263. [DOI] [PubMed] [Google Scholar]
- Palatnik J.F., Wollmann H., Schommer C., Schwab R., Boisbouvier J., Rodriguez R., Warthmann N., Allen E., Dezulian T., Huson D., Carrington J.C., Weigel D. (2007). Sequence and expression differences underlie functional specialization of Arabidopsis microRNAs miR159 and miR319. Dev. Cell 13: 115–125. [DOI] [PubMed] [Google Scholar]
- Pauwels L., et al. (2010). NINJA connects the co-repressor TOPLESS to jasmonate signalling. Nature 464: 788–791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pauwels L., et al. (2015). The RING E3 ligase KEEP ON GOING modulates JASMONATE ZIM-DOMAIN12 stability. Plant Physiol. 169: 1405–1417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piazza P., et al. (2010). Arabidopsis thaliana leaf form evolved via loss of KNOX expression in leaves in association with a selective sweep. Curr. Biol. 20: 2223–2228. [DOI] [PubMed] [Google Scholar]
- Poethig R.S., Sussex I.M. (1985). The cellular parameters of leaf development in tobacco: a clonal analysis. Planta 165: 170–184. [DOI] [PubMed] [Google Scholar]
- Potuschak T., Lechner E., Parmentier Y., Yanagisawa S., Grava S., Koncz C., Genschik P. (2003). EIN3-dependent regulation of plant ethylene hormone signaling by two arabidopsis F box proteins: EBF1 and EBF2. Cell 115: 679–689. [DOI] [PubMed] [Google Scholar]
- Qin F., et al. (2008). Arabidopsis DREB2A-interacting proteins function as RING E3 ligases and negatively regulate plant drought stress-responsive gene expression. Plant Cell 20: 1693–1707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qin G., Gu H., Zhao Y., Ma Z., Shi G., Yang Y., Pichersky E., Chen H., Liu M., Chen Z., Qu L.J. (2005). An indole-3-acetic acid carboxyl methyltransferase regulates Arabidopsis leaf development. Plant Cell 17: 2693–2704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakai T., Mochizuki S., Haga K., Uehara Y., Suzuki A., Harada A., Wada T., Ishiguro S., Okada K. (2012). The wavy growth 3 E3 ligase family controls the gravitropic response in Arabidopsis roots. Plant J. 70: 303–314. [DOI] [PubMed] [Google Scholar]
- Sarid-Krebs L., Panigrahi K.C., Fornara F., Takahashi Y., Hayama R., Jang S., Tilmes V., Valverde F., Coupland G. (2015). Phosphorylation of CONSTANS and its COP1-dependent degradation during photoperiodic flowering of Arabidopsis. Plant J. 84: 451–463. [DOI] [PubMed] [Google Scholar]
- Scanlon M.J. (2003). The polar auxin transport inhibitor N-1-naphthylphthalamic acid disrupts leaf initiation, KNOX protein regulation, and formation of leaf margins in maize. Plant Physiol. 133: 597–605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schommer C., Palatnik J.F., Aggarwal P., Chételat A., Cubas P., Farmer E.E., Nath U., Weigel D. (2008). Control of jasmonate biosynthesis and senescence by miR319 targets. PLoS Biol. 6: e230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shabek N., Zheng N. (2014). Plant ubiquitin ligases as signaling hubs. Nat. Struct. Mol. Biol. 21: 293–296. [DOI] [PubMed] [Google Scholar]
- Shleizer-Burko S., Burko Y., Ben-Herzel O., Ori N. (2011). Dynamic growth program regulated by LANCEOLATE enables flexible leaf patterning. Development 138: 695–704. [DOI] [PubMed] [Google Scholar]
- Sluis A., Hake S. (2015). Organogenesis in plants: initiation and elaboration of leaves. Trends Genet. 31: 300–306. [DOI] [PubMed] [Google Scholar]
- Soundappan I., Bennett T., Morffy N., Liang Y., Stanga J.P., Abbas A., Leyser O., Nelson D.C. (2015). SMAX1-LIKE/D53 family members enable distinct MAX2-dependent responses to strigolactones and karrikins in Arabidopsis. Plant Cell 27: 3143–3159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stone S.L., Hauksdóttir H., Troy A., Herschleb J., Kraft E., Callis J. (2005). Functional analysis of the RING-type ubiquitin ligase family of Arabidopsis. Plant Physiol. 137: 13–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stone S.L., Williams L.A., Farmer L.M., Vierstra R.D., Callis J. (2006). KEEP ON GOING, a RING E3 ligase essential for Arabidopsis growth and development, is involved in abscisic acid signaling. Plant Cell 18: 3415–3428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szemenyei H., Hannon M., Long J.A. (2008). TOPLESS mediates auxin-dependent transcriptional repression during Arabidopsis embryogenesis. Science 319: 1384–1386. [DOI] [PubMed] [Google Scholar]
- Szymanski D.B. (2014). The kinematics and mechanics of leaf expansion: new pieces to the Arabidopsis puzzle. Curr. Opin. Plant Biol. 22: 141–148. [DOI] [PubMed] [Google Scholar]
- Tao Q., Guo D., Wei B., Zhang F., Pang C., Jiang H., Zhang J., Wei T., Gu H., Qu L.J., Qin G. (2013). The TIE1 transcriptional repressor links TCP transcription factors with TOPLESS/TOPLESS-RELATED corepressors and modulates leaf development in Arabidopsis. Plant Cell 25: 421–437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vercruyssen L., et al. (2014). ANGUSTIFOLIA3 binds to SWI/SNF chromatin remodeling complexes to regulate transcription during Arabidopsis leaf development. Plant Cell 26: 210–229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang B., Duan C.G., Wang X., Hou Y.J., Yan J., Gao C., Kim J.H., Zhang H., Zhu J.K. (2015a). HOS1 regulates Argonaute1 by promoting transcription of the microRNA gene MIR168b in Arabidopsis. Plant J. 81: 861–870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L., Wang B., Jiang L., Liu X., Li X., Lu Z., Meng X., Wang Y., Smith S.M., Li J. (2015b). Strigolactone signaling in Arabidopsis regulates shoot development by targeting D53-like SMXL repressor proteins for ubiquitination and degradation. Plant Cell 27: 3128–3142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei B., Zhang J., Pang C., Yu H., Guo D., Jiang H., Ding M., Chen Z., Tao Q., Gu H., Qu L.J., Qin G. (2015). The molecular mechanism of sporocyteless/nozzle in controlling Arabidopsis ovule development. Cell Res. 25: 121–134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu F.H., Shen S.C., Lee L.Y., Lee S.H., Chan M.T., Lin C.S. (2009). Tape-Arabidopsis Sandwich - a simpler Arabidopsis protoplast isolation method. Plant Methods 5: 16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu M.F., Yamaguchi N., Xiao J., Bargmann B., Estelle M., Sang Y., Wagner D. (2015). Auxin-regulated chromatin switch directs acquisition of flower primordium founder fate. eLife 4: e09269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xia T., Li N., Dumenil J., Li J., Kamenski A., Bevan M.W., Gao F., Li Y. (2013). The ubiquitin receptor DA1 interacts with the E3 ubiquitin ligase DA2 to regulate seed and organ size in Arabidopsis. Plant Cell 25: 3347–3359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie Q., Guo H.S., Dallman G., Fang S., Weissman A.M., Chua N.H. (2002). SINAT5 promotes ubiquitin-related degradation of NAC1 to attenuate auxin signals. Nature 419: 167–170. [DOI] [PubMed] [Google Scholar]
- Zhang H., Cui F., Wu Y., Lou L., Liu L., Tian M., Ning Y., Shu K., Tang S., Xie Q. (2015). The RING finger ubiquitin E3 ligase SDIR1 targets SDIR1-INTERACTING PROTEIN1 for degradation to modulate the salt stress response and ABA signaling in Arabidopsis. Plant Cell 27: 214–227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X., Garreton V., Chua N.H. (2005). The AIP2 E3 ligase acts as a novel negative regulator of ABA signaling by promoting ABI3 degradation. Genes Dev. 19: 1532–1543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y., Yang C., Li Y., Zheng N., Chen H., Zhao Q., Gao T., Guo H., Xie Q. (2007). SDIR1 is a RING finger E3 ligase that positively regulates stress-responsive abscisic acid signaling in Arabidopsis. Plant Cell 19: 1912–1929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou F., et al. (2013). D14-SCF(D3)-dependent degradation of D53 regulates strigolactone signalling. Nature 504: 406–410. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.