Geranium species from different habitats show contrasting molecular patterns and physiological responses to phytochome-mediated detection of neighbor cues.
Abstract
Plants growing at high densities elongate their shoots to reach for light, a response known as the shade avoidance syndrome (SAS). Phytochrome-mediated detection of far-red light reflection from neighboring plants activates growth-promoting molecular pathways leading to SAS. However, it is unknown how plants that complete their life cycle in the forest understory and are shade tolerant prevent SAS when exposed to shade. Here, we show how two wild Geranium species from different native light environments regulate contrasting responses to light quality cues. A comparative RNA sequencing approach unveiled the molecular underpinnings of their contrasting growth responses to far-red light enrichment. It also identified differential phytochrome control of plant immunity genes and confirmed that far-red enrichment indeed contrastingly affects resistance against Botrytis cinerea between the two species. Furthermore, we identify a number of candidate regulators of differential shade avoidance. Three of these, the receptor-like kinases FERONIA and THESEUS1 and the non-DNA binding bHLH protein KIDARI, are functionally validated in Arabidopsis thaliana through gene knockout and/or overexpression studies. We propose that these components may be associated with either showing or not showing shade avoidance responses.
INTRODUCTION
Plants have evolved a variety of strategies to deal with environmental stresses. Oftentimes, the underlying regulatory circuits are very well understood in model plants, while alternative strategies in species with a different evolutionary history are not. One example of this is the way in which deetiolated plants adjust to light quality changes.
Plants absorb blue (400–500 nm) and red (600–700 nm) light, but not far-red (700–800 nm) light for photosynthesis. Due to this preferential absorption by leaves the red:far-red light ratio (R:FR) declines in dense vegetation, which is a signal for neighbor proximity. In response, shade intolerant plants prioritize enhanced elongation of their leaf-bearing organs (stems and petioles) over branching, lift their leaves to a more vertical position (hyponasty) to bring their leaves to the top of the canopy, and flower early, a phenomenon known as the shade avoidance syndrome (SAS; reviewed in Pierik and Testerink, 2014; Fraser et al., 2016). Low R:FR-induced shade avoidance is absent in shade-tolerant plants that thrive on the forest floor and cannot outcompete the tall trees surrounding them (Gommers et al., 2013; Valladares and Niinemets, 2008).
Changes in R:FR light are sensed by the phytochrome family of photoreceptors in all higher plants. These photoconvert from the active, FR-absorbing form (Pfr) to the inactive, R-absorbing form (Pr) and vice versa. In direct sunlight (R:FR = 1.1), Pfr localizes to the nucleus, where it binds to a set of basic helix-loop-helix (bHLH) transcription factors, the PHYTOCHROME INTERACTING FACTORs (PIFs). In canopy shade (R:FR < 0.4), PIFs are released when Pfr is photoconverted to Pr and regulate cell elongation by enhancing transcription of growth-promoting genes (Quail, 2002). PIFs directly regulate the expression of genes encoding cell wall modifying proteins, such as XYLOGLUCAN ENDOTRANSGLUCOSYLASE/HYDROLASE15 (XTH15) (Hornitschek et al., 2009). PIFs also directly target genes encoding proteins involved in controlling auxin homeostasis and signaling, which is a major hormone controlling hypocotyl elongation of Arabidopsis thaliana seedlings during shade avoidance (Tao et al., 2008; Keuskamp et al., 2010; Li et al., 2012; Hornitschek et al., 2012; Nozue et al., 2015). PIF4 interacts with growth repressing DELLA proteins (Djakovic-Petrovic et al., 2007; de Lucas et al., 2008) and BRASSINAZOLE RESISTANT1 (BZR1; Oh et al., 2012), which are targets of two other growth-regulating hormones, gibberellin and brassinosteroids, respectively.
SAS is a widespread strategy that is accompanied by controlled suppression of jasmonic acid (JA)- and salicylic acid-mediated defenses against pathogens and herbivores (de Wit et al., 2013; Moreno et al., 2009), thus prioritizing consolidation of light capture over other stresses. Arabidopsis plants exposed to low R:FR light are more susceptible to the necrotrophic pathogen Botrytis cinerea, as a consequence of stabilization of jasmonate ZIM-domain (JAZ) transcriptional repressors, when DELLA proteins are degraded (Chico et al., 2014; Cerrudo et al., 2012; de Wit et al., 2013).
The molecular pathways that stimulate elongation growth would be maladaptive in forest understory plants. Indeed, variation in low R:FR-induced stem elongation rates has been documented between (Morgan and Smith, 1978; Gilbert et al., 2001) and within (Sasidharan et al., 2009; Coluccio et al., 2011; Jiménez-Gómez et al., 2010; Filiault and Maloof, 2012) species. Species from forest understories, in addition to suppressing low R:FR-induced stem elongation, have evolved shade tolerance strategies, such as optimal leaf morphology for low-light photosynthesis (Morgan and Smith, 1978) and tolerance to biotic and abiotic stresses (reviewed in Valladares and Niinemets, 2008). Currently, it remains unknown how shade tolerant species inhibit SAS while being exposed to potent SAS-inducing light signals (Gommers et al., 2013). Identifying the molecular mechanisms underpinning suppression of SAS has great potential to (1) show how different strategies of adaptive plasticity can be regulated and (2) inform crop-breeding programs targeted at minimizing yield losses, which are caused by wasteful carbon investments in nonharvestable stems (Carriedo et al., 2016) and suppressed immunity.
Here, we study responses to changing R:FR light ratios in two Geranium species from contrasting habitats: G. robertianum (herb Robert), which grows in a wide amplitude of conditions including forest understories, and G. pyrenaicum (hedge cranesbill), which occurs in open habitats. In response to low R:FR light conditions, these two species showed highly contrasting growth, transcriptome, and pathogen defense responses. We identified novel putative SAS regulators, which were subsequently confirmed in gene functional studies in Arabidopsis.
RESULTS
G. pyrenaicum and G. robertianum Express Opposite Growth Responses to Low R:FR
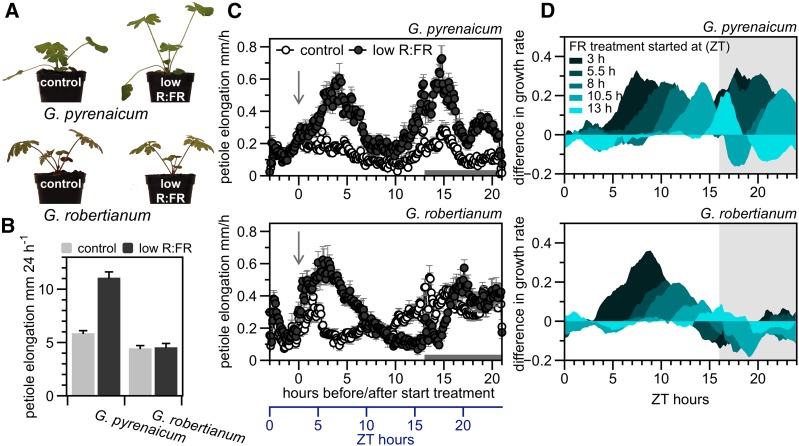
To study the regulatory pathways that suppress shade avoidance, we exposed two wild Geranium species from contrasting habitats to FR-enriched (low R:FR = 0.2) versus control (R:FR = 1.8) white light conditions. G. pyrenaicum expressed the classic shade avoidance response by elongating its petioles, whereas this was not apparent in G. robertianum (Figures 1A and 1B). Detailed petiole growth kinetics over 24 h (Figure 1C) and 48 h (Supplemental Figure 1A) show that both species initially enhance petiole elongation upon low R:FR treatment. Nevertheless, G. robertianum does not show enhanced petiole elongation in low R:FR toward the end of the day and during the night (Figure 1C), regardless of the start time of the treatment (Figure 1D; Supplemental Figure 2). This results in no net difference between the treatments after 24 h in G. robertianum. By contrast, G. pyrenaicum is always able to rapidly induce petiole elongation upon exposure to low R:FR, independent of the time of day, although it suppresses petiole elongation in the night period when treatments started at 17:30 or 20:00 (Figure 1D; Supplemental Figure 2).
Figure 1.
Characterization of G. pyrenaicum and G. robertianum as Model Species with Opposite Petiole Elongation Responses in Low R:FR Light.
(A) G. pyrenaicum and G. robertianum plants exposed to control white light (R:FR = 1.8) or a low R:FR (R:FR = 0.2) treatment for 5 d.
(B) Elongation (mm 24 h−1) of the second petiole of G. pyrenaicum and G. robertianum leaves, exposed to control white light or low R:FR conditions. Data represent means + se, n = 7.
(C) Growth rates of G. pyrenaicum and G. robertianum petioles in mm h−1 over 24 h, data for every 6 min. Plants grown in either control white light (open circles) or a low R:FR treatment (dark circles), starting at time point 0 (ZT = 3 h, indicated by an arrow). Data represent means ± se, n = 6. The gray bar at the x axis indicates the night period.
(D) The difference between petiole growth rates (mm h−1) in control and low R:FR treatments with a different starting point during the photoperiod, for a 24-h period (differences for the ZT = 3 h starting point are taken from data presented in [C]), in G. pyrenaicum (upper graph) and G. robertianum (lower graph). Differences between the means of the two treatments are smoothed using exponential smoothing, n = 6. The gray area represents the night period.
In both species, petioles elongated mainly at the apical part, just under the leaf lamina (Supplemental Figures 1B and 1C). Based on these results, we harvested the most responsive apical part of the petiole after 2 and 11.5 h of low R:FR treatment and corresponding controls grown in white light for a transcriptome analysis to investigate the gene expression profiles associated with these growth differences.
De Novo Assembly of Geranium Transcriptomes and Comparison to Plant Model Species
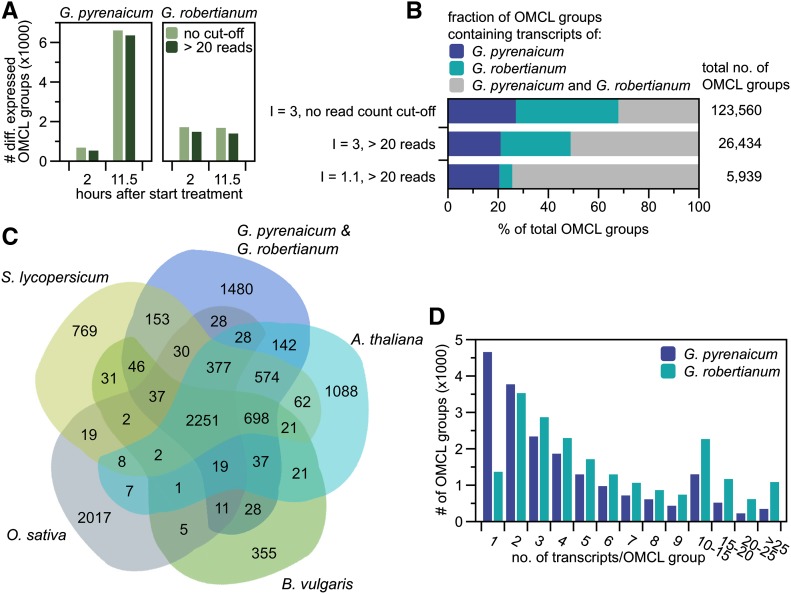
Two Geranium reference transcriptomes were assembled de novo from combined Illumina-sequenced normalized and non-normalized libraries to establish maximum transcript coverage. Using the Markov Clustering Algorithm (OMCL; Enright et al., 2002) with an inflation factor (I) of 3.0, transcripts were clustered into putatively orthologous transcript groups. All low-abundant OMCL groups (<20 reads) were removed without affecting the number of differentially expressed OMCL groups during further analysis (Figures 2A and 2B). Of the 26,434 OMCL groups, 51% was shared by the two species (Figure 2B), and lowering the I increased this to 74%, at the cost of solely G. robertianum OMCL groups (∼28% with I = 3.0 to ∼5% with I = 1.1; Figure 2B), indicating that this species has many transcript variants, while being considered a diploid, as is G. pyrenaicum (Warburg, 1938; Tofts, 2004). Accordingly, most OMCL groups contained only one G. pyrenaicum, but more than one G. robertianum transcript (Figure 2D; I = 3.0 and <20 reads).
Figure 2.
Geranium Transcriptome Assembly and Clustering of Putatively Orthologous Transcripts.
(A) Effect of the read count cutoff of 20 on the number of differentially expressed OMCL groups (I = 3.0 and > 20 reads, low R:FR versus control, P < 0.01) for both species at the two time points.
(B) Fractions (in %) of ortho Markov-clustered groups containing transcripts of G. pyrenaicum, G. robertianum, or both, with two different inflation factors (3.0 versus 1.1) and with or without an applied read count cutoff of 20 for at least one of the treatments/time points. The total number of OMCL groups for the three different settings is given at the right side of the graph.
(C) Ortholog transcript distribution over six species (G. pyrenaicum, G. robertianum, Arabidopsis, B. vulgaris, O. sativa, and S. lycopersicum) in a plant kingdom-wide clustering, using OMCL (I = 1.1, Geranium read counts > 20). G. pyrenaicum and G. robertianum OMCL groups are pooled for the figure.
(D) OMCL group sizes (no. of transcripts) for both species separate (I = 3.0 and > 20 counts).
Geranium transcriptomes were compared with model species Arabidopsis, tomato (Solanum lycopersicum), beet (Beta vulgaris) and rice (Oryza sativa), using OMCL clustering with I = 1.1 (and >20 reads for Geranium transcripts). As shown in Figure 2C, 2251 OMCL groups were shared among all six species, 1480 covered only Geranium transcripts, and only two OMCL groups were present in all species but not identified in Geranium. Our two species showed strong overlap with beet (∼88%), tomato (∼82%), and Arabidopsis (∼77%).
Transcriptome Analysis of G. pyrenaicum and G. robertianum Exposed to Low R:FR Light
To identify transcriptional changes upon 2 or 11.5 h of low R:FR in the two species, we mapped Illumina sequenced reads of the non-normalized libraries (constructed from the apical half of petioles in control or low R:FR light at the two time points) to the newly constructed transcriptomes. Read counts of all contigs in an OMCL group were summed before further statistical analysis.
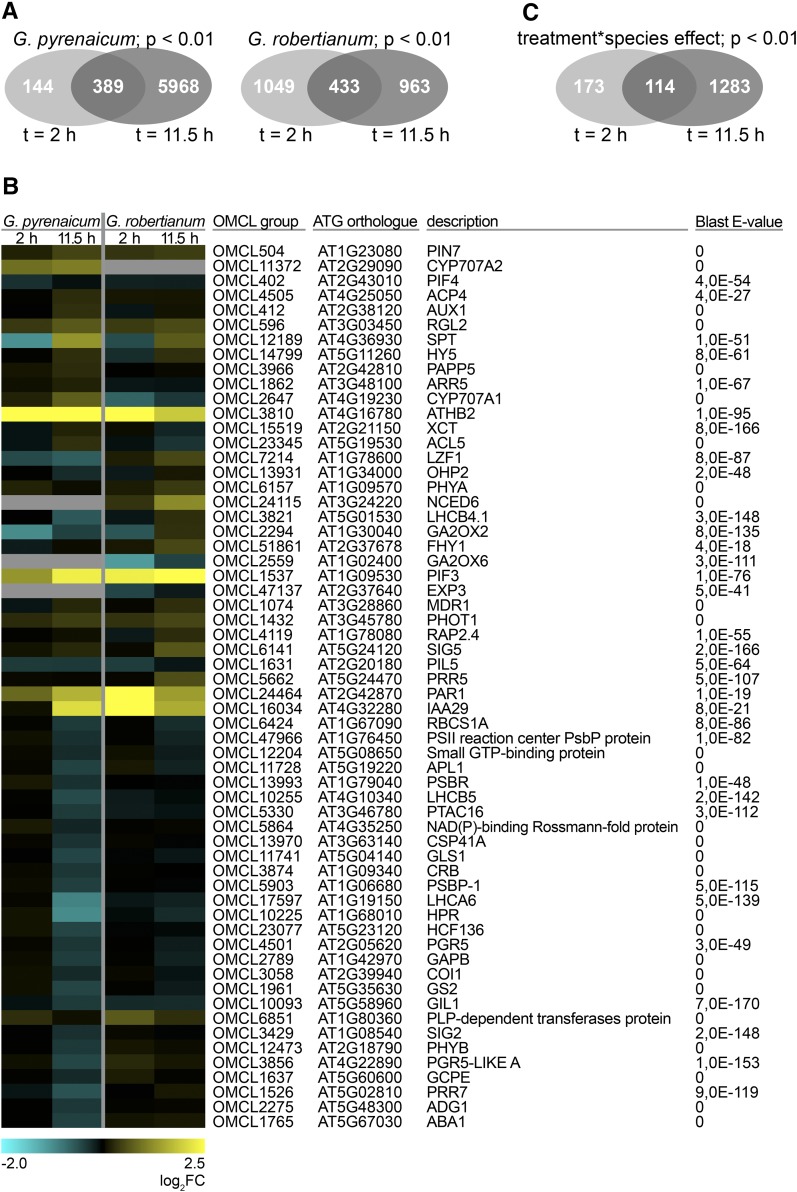
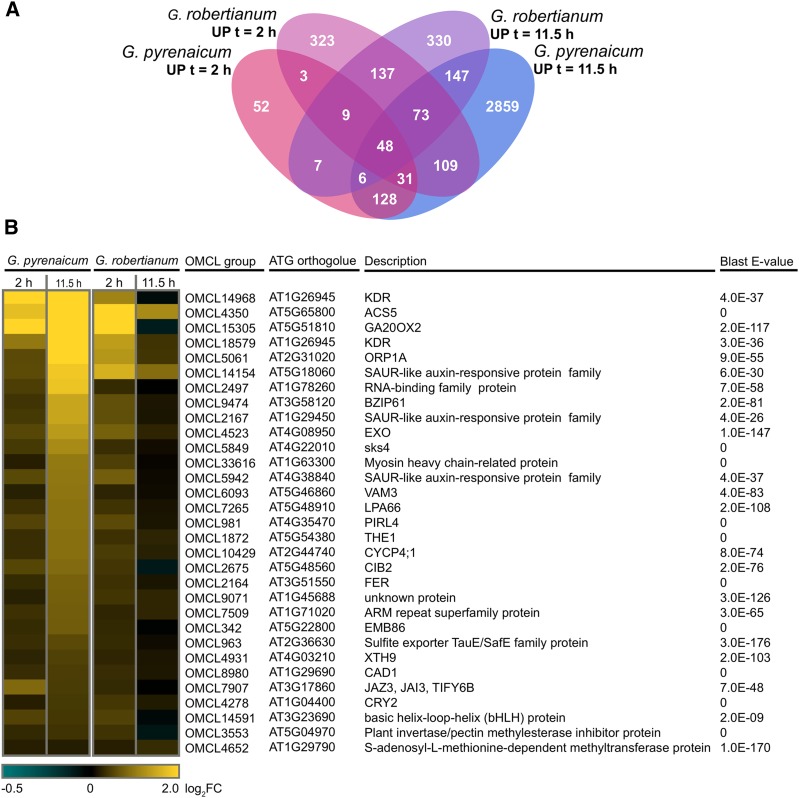
In G. pyrenaicum, 533 OMCL groups were differentially regulated after 2 h of low R:FR light, and this increased after 11.5 h to 6.357 (Figure 3A). In G. robertianum, the numbers of differentially expressed OMCL groups were comparable at the two time points (1.482 at t = 2 h; 1.396 at t = 11.5 h; Figure 3A). Matching the contrasting petiole elongation phenotypes of these species in low R:FR light, expression of Geranium orthologs of several SAS-associated genes (binned to the Gene Ontology [GO] terms “R or FR light signaling pathway,” “response to R or FR light,” “shade avoidance,” “response to R light,” “response to FR light,” “R light signaling pathway,” and/or “FR light signaling pathway” in Arabidopsis) was different between the two species, especially at time point t = 11.5 h (Figure 3B). GO clustering of all up- and downregulated OMCL groups (Supplemental Figure 3) shows that both species upregulate “shade avoidance”-related, as well as several hormone- and cell wall-related genes. Photosynthesis-associated ontologies were enriched among downregulated OMCL groups, which would be in accordance with previous studies that show how phytochrome inactivation can repress photosynthetic capacity (Toledo-Ortiz et al., 2010; Yang et al., 2016).
Figure 3.
Low R:FR Light Differentially Affects G. pyrenaicum and G. robertianum Gene Expression.
(A) Venn diagrams representing overlap of the differentially expressed OMCL groups upon the low R:FR treatment (up- and downregulated, P < 0.01, qCML method, followed by an exact test) between the two different time points for G. pyrenaicum (upper) and G. robertianum (lower) separately.
(B) Heat map representing expression values of Geranium orthologs of Arabidopsis genes appointed to the GO terms “R or FR light signaling pathway,” “response to R or FR light,” “shade avoidance,” “response to R light,” “response to FR light,” “R light signaling pathway,” and/or “FR light signaling pathway,” which are significantly (P < 0.01, qCML method, followed by an exact test) induced or repressed by a low R:FR treatment at at least one time point (2 or 11.5 h) in at least one species (G. pyrenaicum or G. robertianum). Colors represent the log2 fold change upon the low R:FR treatment. The ATG code and description of the closest Arabidopsis ortholog is given for each Geranium OMCL group, with matching E-value of the BLAST hit. Gray areas represent the absence of this OMCL group in this species.
(C) Venn diagram showing the number of OMCL groups with a significant treatment*species interaction (P < 0.01, glmLRT) and overlap between the two time points.
As shown by the large number of OMCL groups with a significant treatment*species interaction, most differences in low R:FR-induced expression between the species occurred in the later time point (1.397 OMCL groups, compared with 287 at t = 2 h; Figure 3C), matching the growth patterns shown in Figure 1.
A GO analysis on the OMCL groups with a significant treatment*species interaction (Supplemental Figure 4) revealed that after 2 h of low R:FR, the OMCL groups with a species*treatment interaction larger than zero (which represents significantly higher log2 fold changes for G. robertianum than G. pyrenaicum) were overrepresented in the GO category for shade avoidance. At the second time point, the OMCL groups with a negative interaction were overrepresented in SAS-related GO categories, such as brassinosteroid metabolic process, cell wall compounds, and response to light stimulus.
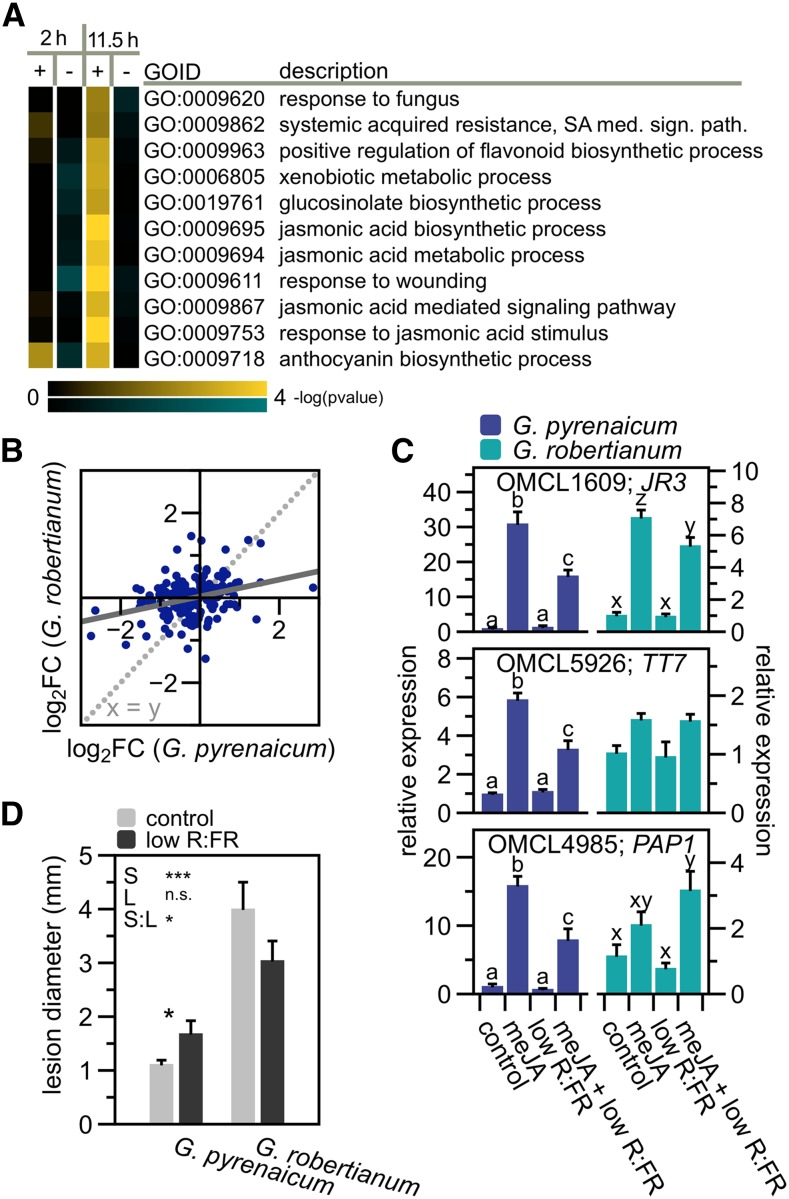
JA-Mediated Defenses Are Repressed by Low R:FR in G. pyrenaicum but Enhanced in G. robertianum
Among the GO categories with a strong positive interaction effect in the evening, we identified several defense-associated ontologies (Supplemental Figure 4; Figure 4A). Many OMCL groups with possible functions in the JA-mediated defense against herbivores or necrotrophic pathogens appeared to be downregulated after 11.5 h of low R:FR treatment in G. pyrenaicum, but not, or to a lesser extent, in G. robertianum (Figure 4B; Supplemental Figure 5A). Downregulation of defense pathways by low R:FR has also been described in Arabidopsis (Cargnel et al., 2014; Cerrudo et al., 2012; de Wit et al., 2013) and tomato (Cortés et al., 2016). Geranium orthologs of JA-inducible genes JASMONIC ACID RESPONSIVE3 (JR3), TRANSPARENT TESTA7 (TT7), and PRODUCTION OF ANTHOCYANIN PIGMENT1 (PAP1) were transcriptionally induced by a methyl-JA treatment, but this induction was suppressed by simultaneous exposure to low R:FR light in G. pyrenaicum (Figure 4C), similar to what was previously shown in Arabidopsis (de Wit et al., 2013; Supplemental Figure 5B). Interestingly, G. robertianum lacks the reduction in defenses observed in G. pyrenaicum. These data hint at differential control of immunity by R:FR between the two species. To examine the biological consequences of these differences, cotyledons of both species were infected with spores of the necrotrophic pathogen B. cinerea under control and low R:FR light. Consistent with the gene expression patterns, lesion diameters caused by the pathogen increased in the low R:FR treatment in G. pyrenaicum, but decreased in G. robertianum (Figure 4D). These data indicate that the divergence of R:FR responses between these species extends to pathogen resistance. G. robertianum shows higher background susceptibility to B. cinerea than G. pyrenaicum, and this may be associated with differences in leaf structure between the two species. Low R:FR-mediated petiole elongation in the seedlings (cotyledon petioles), however, remained unaffected by the B. cinerea treatment and was pronounced in G. pyrenaicum and absent in G. robertianum (Supplemental Figure 5C).
Figure 4.
Low R:FR Light Differentially Affects G. pyrenaicum and G. robertianum Immunity.
(A) Heat map of the gene ontology clusters overrepresenting transcripts with a positive species*treatment interaction (log2 fold changes are significantly higher in G. robertianum compared with G. pyrenaicum) at t = 11.5 h, and a role in plant defenses against pests. Colors represent the -log of the adjusted P value, where blue is a negative interaction (G. pyrenaicum fold change > G. robertianum fold change) and yellow is a positive interaction (G. pyrenaicum < G. robertianum).
(B) Scatterplot of the log fold changes (low R:FR versus control) in G. pyrenaicum (x axis) and G. robertianum (y axis) of transcripts attributed to the gene ontology clusters presented in (A).
(C) Expression data of the Geranium orthologs of JR3, TT7, and PAP1 upon 100 μM methyl-JA, low R:FR (R:FR = 0.2, 2 h), or combined treatment, relative to the control (R:FR = 1.8, mock) and the reference gene (ortholog of PDF1), in G. pyrenaicum (light blue) and G. robertianum (dark blue) leaves (petiole + lamina). Chemicals (250 μL) were sprayed on the leaf at the start of the light treatment. Data represent means + se, n = 5 (biological replicates are a pool of the second leaf of two individual plants) Different letters represent significant differences (two-way ANOVA and post-hoc Tukey test, P < 0.05).
(D) Lesion diameters (mm) of a B. cinerea infection (2 μL of 2.5 × 105 spores mL−1) on G. pyrenaicum and G. robertianum cotyledons, in a control or low R:FR light environment, measured 3 d after inoculation. Data represent means ±se, n = 28. Asterisks represent significant differences (two-way ANOVA and post-hoc Tukey test, P < 0.05).
Candidate OMCL Group Selection for Differential Low R:FR-Induced Growth Patterns
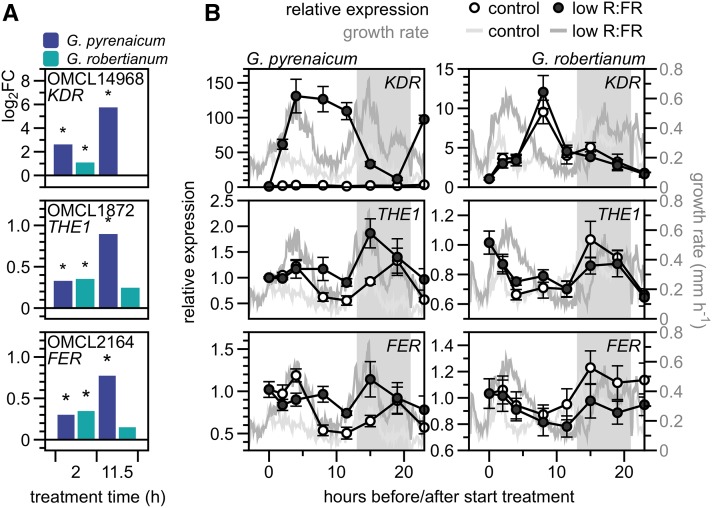
To find OMCL groups that might be functionally associated with the low R:FR-induced growth patterns of the Geranium species, we selected the groups upregulated in both time points in G. pyrenaicum, but only in the early time point in G. robertianum (Figure 5A). Among this selection of 31 OMCL groups (Figure 5B) were orthologs of some known downstream SAS components, such as GIBBERELLIC ACID 20 OXIDASE2 (GA20OX2), XYLOGLUCAN ENDOTRANSGLUCOSYLASE/HYDROLASE9 (XTH9), and several SMALL AUXIN REGULATED RNAs (SAURs). In a search for potential upstream differential components, we selected bHLH transcription factor KIDARI (KDR) and two members of the family of Catharanthus roseus receptor-like kinases (CrRLKs): THESEUS1 (THE1) and FERONIA (FER) (Figure 6A). Transcription of these OMCL groups was analyzed using RT-qPCR in more detailed kinetics and plotted together with petiole elongation rates (Figure 6B). KDR expression was rapidly and strongly upregulated by low R:FR in G. pyrenaicum and dropped during the night. Low R:FR-induced transcription of THE1 and FER was slower and weaker than KDR, peaking at the beginning of the night. All three OMCL groups are hardly upregulated in the low R:FR-treated G. robertianum petioles, and expression is even slightly inhibited in the night. Expression of a number of other OMCL groups from the cluster of 31, including GA20OX2 and transcription factors BZIP61 and CIB2, was also confirmed with RT-qPCR to match the RNA seq patterns (Supplemental Figures 6A and 6B).
Figure 5.
Selection of Candidate Genes from the Transcriptome Analysis to Explain Low R:FR-Induced Growth Patterns in Geraniums.
(A) Venn diagram presenting OMCL groups significantly upregulated in low R:FR (R:FR = 0.2) versus control (R:FR = 1.8) (adjusted P < 0.01, read counts > 20) in both species after 2 (red shades) or 11.5 h (blue shades).
(B) The selection of 31 OMCL groups, upregulated in G. pyrenaicum after 2 and 11.5 h, and in G. robertianum after 2 h (from 5A), with the closest Arabidopsis ortholog, description, and BLAST E-value. Colors represent the log2 fold change.
Figure 6.
G. pyrenaicum and G. robertianum Differentially Express KDR, THE1, and FER in Low R:FR Light over Time.
(A) Log2 fold changes of OMCL14968 (KDR ortholog), OMCL1872 (THE1), and OMCL2164 (FER) in G. pyrenaicum (dark-blue bars) and G. robertianum (turquoise bars). Asterisks represent significant transcriptional regulation (low R:FR versus control, adjusted P < 0.01, qCML method, followed by an exact test).
(B) Relative OMCL group expression over time in G. pyrenaicum (left panels) and G. robertianum (right panels) petioles in control (open circles) and low R:FR (dark circles) on the left y axis. Data are relative to the reference gene (ortholog of PDF1) and to t = 0 (10:00 am) and represent means ± se, n = 5 (biological replicates are a pool of the second petiole of three individual plants). Right y axis and gray lines represent growth rates in mm h−1 over the same time period in control (light gray) and low R:FR (darker gray) conditions (same data as Figure 1C). Gray area represents the night period.
Heterologous Studies in Arabidopsis Confirm a Role for KDR, THE1, and FER in SAS
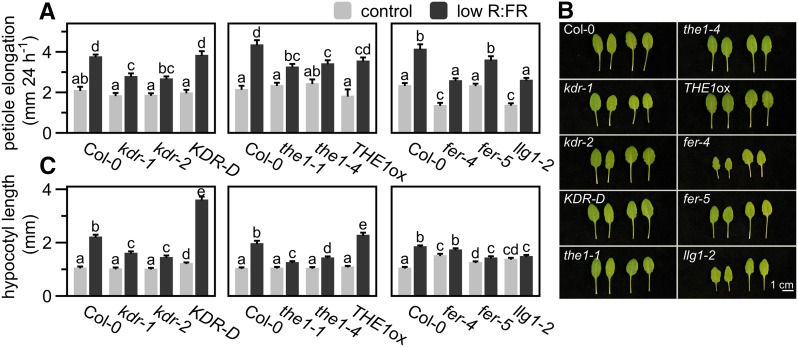
To functionally test the importance of KDR, THE1, and FER in the regulation of SAS, we continued in the model species Arabidopsis because transgenic methods are currently unavailable in Geranium species. Petioles of a KDR knockout (kdr-1; Supplemental Figure 7A) and knockdown (kdr-2; Supplemental Figure 7A) line had reduced responses to low R:FR treatment, whereas the activation tagged line KDR-D (Supplemental Figure 7B) was similar to Col-0 wild type (Figures 7A and 7B). In young seedlings, these differences were more extreme. Hypocotyls of kdr-1 and kdr-2 were strongly inhibited in low R:FR-induced elongation and KDR-D was hypersensitive to low R:FR (Figure 7C).
Figure 7.
Functional Analysis of KDR, THE1, FER, and LLG1 in Low R:FR-Exposed Arabidopsis Rosettes and Seedlings Reveals Functions in Regulating Elongation of Petioles and Hypocotyls.
(A) and (B) Petiole elongation (mm) of several KDR, THE1, FER, and LLG1 deficient and overexpressing lines compared with the wild-type control (Col-0), grown in control white light (R:FR = 1.8, light gray bars) or low R:FR (R:FR = 0.2, dark gray bars) conditions for 24 h (data represent means + se, n = 10. Different letters indicate significant differences; P < 0.05, two-way ANOVA with post-hoc Tukey test) (A), with representative photographs of leaves grown in control (left pairs) or low R:FR (right pairs) for 5 d (B).
(C) Hypocotyl length (mm) of Arabidopsis lines shown in (A) after 4 d of control white light or low R:FR treatment. Data represent means + se, n = 40. Different letters indicate significant differences (P < 0.05, two-way ANOVA with post-hoc Tukey test).
The THE1 knockout the1-4, loss-of-function missense mutant the1-1, and the HERKULES1 (HERK1; a related RLK that was previously shown to act redundantly to THE1 in regulating hypocotyl, petiole and lamina cell growth; Guo et al., 2009) THE1 double mutant herk1 the1 showed significantly reduced low R:FR-induced petiole elongation (Figures 7A and 7B; Supplemental Figures 8A and 8B). On the other hand, THE1ox responded similarly to the wild type (Figures 7A and 7B). Hypocotyls of the1-1, the1-4, and herk1 the1 seedlings showed severe low R:FR-induced elongation defects, whereas THE1ox showed an exaggerated response compared with Col-0 (Figure 7C; Supplemental Figure 8C). Consistent with the known THE1-HERK1 redundancy, the herk1 single mutant responded similar to Col-0 (Supplemental Figures 8A to 8C).
Adult FER knockdown (fer-5) and knockout (fer-4) plants, as well as the mutant of the FER coreceptor LORELEI-LIKE GPI-AP1 (LLG1; Li et al., 2015) llg1-2, all displayed petiole elongation responses to low R:FR that were similar to the wild type (Figures 7A and 7B). However, these mutations dramatically reduced low R:FR-induced hypocotyl elongation: fer-4, fer-5, and llg1-2 seedlings lacked this response completely (Figure 7C). Interestingly, although all our candidate genes were found in petiole tissue in rosette-stage Geranium, their functional role is particularly striking in Arabidopsis seedlings.
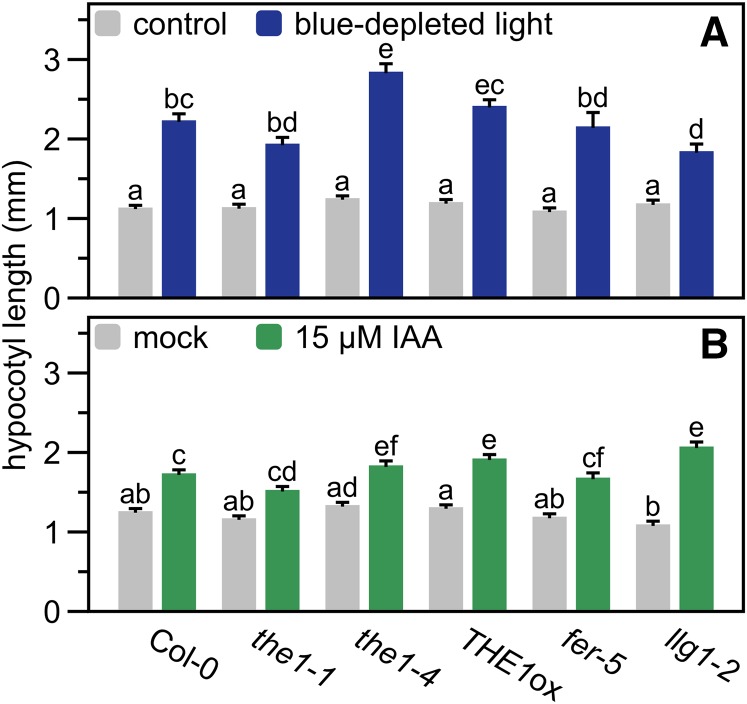
We exposed the1-1, the1-4, THE1ox, fer-5, and llg1-2 seedlings to blue-depleted light (Figure 8A), another shade cue that induces strong elongation (Keuskamp et al., 2011), or 15 μM indole-3-acetic acid (IAA, auxin; Figure 8B). Even though variation in the responses existed, all genotypes would still strongly elongate hypocotyls in these treatments with some variation between different mutants for the same gene, but without a consistent reduction as seen in the low R:FR treatment. This indicates that these RLKs are particularly important for low R:FR-induced hypocotyl elongation, rather than being generic hypocotyl elongation regulators.
Figure 8.
RLKs Have No Function in Blue Light-Depleted- and Auxin-Mediated Hypocotyl Elongation.
Hypocotyl length (mm) of Arabidopsis THE1-, FER-, and LLG1-deficient and -overexpressing lines and the wild-type control (Col-0) after 4 d of control white light (±60 μmol m−2 s−1 blue) and blue light depleted (±4 μmol m−2 s−1 blue light) (A), or mock (0.1% DMSO) and 15 μM IAA treatment (in 0.1% DMSO) (B). Data represent means + se, n = 40. Different letters indicate significant differences (P < 0.05, two-way ANOVA with post-hoc Tukey test).
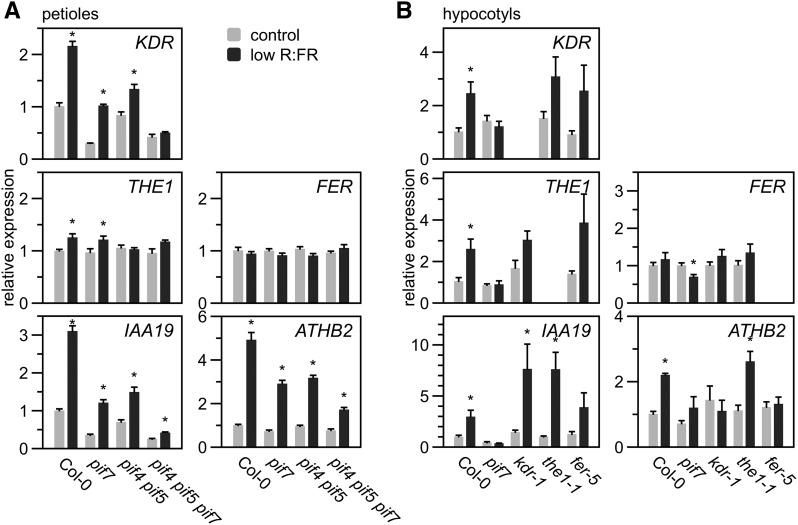
Finally, we established whether these novel SAS regulators are transcriptionally regulated in response to low R:FR in Arabidopsis as observed in G. pyrenaicum. We analyzed the expression of the KDR, THE1, FER, and SAS marker genes IAA19 and ATHB2 in petioles of wild-type Arabidopsis, as well as pif7, pif4 pif5, and pif4 pif5 pif7 mutants, using RT-qPCR. KDR expression was induced in petioles and was abolished solely in the pif4 pif5 pif7 triple mutant, while IAA19 and ATHB2 expression in low R:FR was partly, but not completely, dependent on PIF4, PIF5, and PIF7 (Figure 9A). THE1 transcription increased only marginally in low R:FR light in Col-0 and pif7, while FER, HERK1, and HERK2 expression remained completely unchanged by the light treatment (Figure 9A; Supplemental Figure 8D) in Arabidopsis.
Figure 9.
Transcriptional Regulation of KDR, THE1, and FER in Low R:FR Light-Exposed Arabidopsis.
Relative expression of KDR, THE1, FER, IAA19, and ATHB2 in petioles of Col-0, pif7, pif4 pif5, or pif4 pif5 pif7 exposed to control white (R:FR = 1.8) or low R:FR light (R:FR = 0.2) for 4 h (values are relative to reference genes and Col-0 in control treatment, data represent means + se, n = 6 biological replicates) (A) and in hypocotyls of Col-0, pif7, kdr-1, the1-1, and fer-5 exposed to the same treatments for 24 h (values are relative to reference genes and Col-0 in control conditions; data represent means + se, n = 4 biological replicates) (B). Asterisks indicate significant differences between the treatments (Student’s t test, P < 0.05).
Since kdr, the1, and fer mutations resulted in clear hypocotyl elongation defects in low R:FR-exposed seedlings, we next analyzed the expression of these candidate genes, as well as of IAA19 and ATHB2 in hypocotyls of the wild type, pif7 (with severely suppressed SAS in seedlings; Li et al., 2012), kdr-1, the1-1, and fer-5 (Figure 9B). KDR and THE1 expression were, similar to IAA19, induced by low R:FR in all genotypes but pif7, which lacked this response completely. FER expression remained unchanged in all genotypes but was slightly repressed in low R:FR-exposed pif7 seedlings. Surprisingly, ATHB2 induction by low R:FR light was absent in pif7, kdr-1, and fer-5, but not in the1-1.
Consistent with these expression data, results summarized from previously published transcriptome studies also indicated that low R:FR light, auxin treatments, and pif mutations have marginal effects on expression of FER, THE1, and related RLKs (Supplemental Figure 9). These relatively weak inductions found in published data sets may be related to their use of whole shoots or seedlings: Two recent studies on organ-specific gene expression also find more clear induction of THE1 and FER expression in Arabidopsis hypocotyls (Das et al., 2016; Kohnen et al., 2016).
DISCUSSION
Our transcriptomics approach in wild species with opposite ecologies and consequently contrasting light quality responses has identified molecular patterns associated with contrasting ecological strategies and novel regulators of shade avoidance responses. G. pyrenaicum and G. robertianum appear to be a promising comparative model system to study SAS and its suppression, both at a growth-physiological and a transcriptomic level.
The two species respond differently to low R:FR not only at the level of plant architecture, but also in immunity (summarized in Figure 10). It has been shown in Arabidopsis, and other shade avoiding species, that low R:FR light conditions reduce defense responses against herbivorous insects and both necrotrophic and biotrophic pathogens that involve JA- or salicylic acid-mediated signaling (de Wit et al., 2013; Cargnel et al., 2014; Cerrudo et al., 2012; Moreno et al., 2009; Izaguirre et al., 2006). We show here that G. robertianum, which occurs in forest understories, enhances its immunity against the fungal pathogen B. cinerea upon shade detection, rather than the reduced immunity observed in shade avoiding Arabidopsis and G. pyrenaicum. The mechanism through which low R:FR reduces pathogen resistance in shade-intolerant plants is not yet fully established but involves upregulation of the JAZ proteins, which are negative regulators of the JA pathway (Moreno et al., 2009; Cerrudo et al., 2012; Campos et al., 2016). Indeed, the Geranium ortholog of JAZ3 (OMCL7907) is induced by low R:FR in the evening in G. pyrenaicum, but not in G. robertianum (Figure 5B). We speculate that differential JAZ transcript accumulation between the two Geranium species in response to phytochrome inactivation under low R:FR could lead to the observed differential control of immunity.
Figure 10.
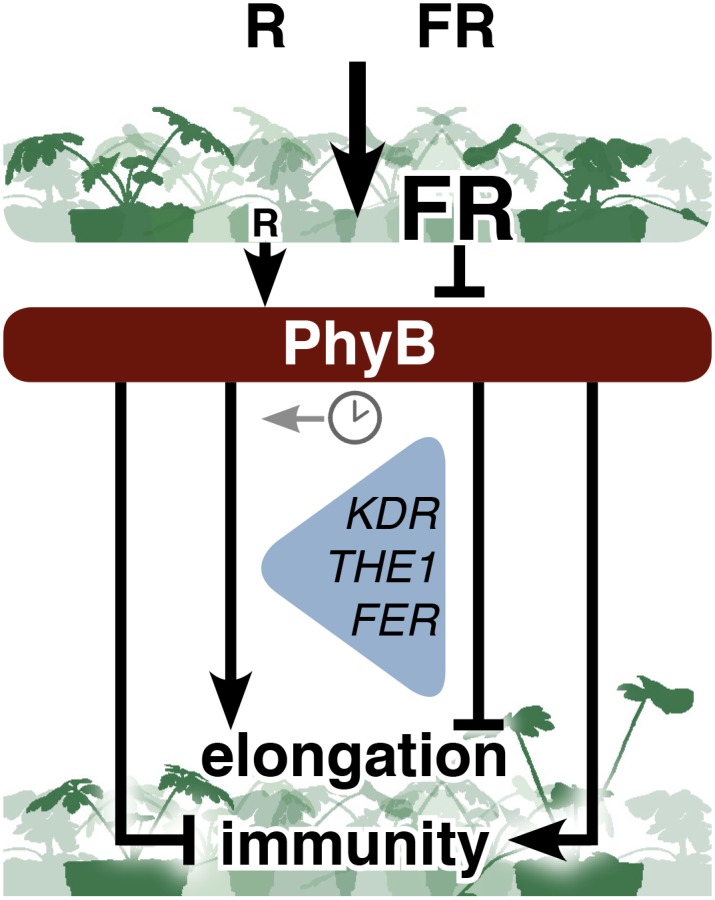
Schematic Overview of How R:FR Light in Plant Canopies Differentially Affects Elongation and Immunity in G. robertianum (Left) and G. pyrenaicum (Right) upon Phytochrome Deactivation.
KDR, THE1, and FER act downstream of PhyB to regulate elongation and are differentially regulated between the two species. In G. robertianum, petiole growth and the transcriptional induction of these genes are suppressed at a specific time of the day (indicated with the clock symbol). In Arabidopsis, these three proteins are positive regulators of low R:FR-induced growth, and KDR and THE1 are transcriptionally induced in a PIF-dependent manner.
From the Geranium transcriptome, we identified KDR, FER, and THE1 as novel candidate genes regulating shade avoidance in response to low R:FR and their involvement was confirmed in heterologous gene functional studies in Arabidopsis (Figure 7). The atypical non-DNA binding bHLH transcription factor KDR has previously been shown to bind, and functionally inhibit, another atypical bHLH protein, LONG HYPOCOTYL IN FAR-RED LIGHT1 (HFR1) (Hyun and Lee, 2006; Hong et al., 2013), a well-studied suppressor of PIF4 and PIF5 function (Hornitschek et al., 2009). Furthermore, KDR transcription is directly regulated by the PIF4-BZR1 complex (Oh et al., 2012; Bai et al., 2012). Our expression data in Arabidopsis pif mutants showed that KDR expression in petioles depends on functional PIF4, PIF5, and PIF7 in both white light and low R:FR (Figure 9A), while in hypocotyls of young seedlings it solely depends on PIF7. Nevertheless, KDR had not previously been identified as a low R:FR-inducible functional regulator of SAS. The strong differences in expression levels between the Geranium species and its presumed interaction with a modulator of PIF activity, HFR1, make KDR an interesting candidate regulating the two different strategies in the shade. However, since we could not identify HFR1 orthologs in the Geranium transcriptomes, it is possible that KDR in these species interacts with other regulators, potentially other bHLH proteins, to suppress PIF-induced elongation in low R:FR. HFR1 was also not detected in the transcriptome of another wild species, Rumex palustris, which shows pronounced elongation when flooded, accompanied by transcriptional KDR induction (van Veen et al., 2013).
A second novel node of regulation identified here for SAS is that of RLKs. The THE1 and FER proteins regulate cell elongation (Lindner et al., 2012; Wolf and Höfte, 2014) but were not previously associated with photoreceptor responses. THE1 acts during cell expansion (Guo et al., 2009; Hématy et al., 2007) and is considered a cell wall integrity sensor, enhancing ROS production when cell wall integrity is lost (Denness et al., 2011). THE1 is redundant to HERK1 in regulating general vegetative growth (Guo et al., 2009). However, we show here that HERK1 plays no apparent role in SAS, whereas THE1 is a potent regulator independent of HERK1, as indicated by the single and double mutant data (Supplemental Figures 8A to 8C). In Arabidopsis, low R:FR light affected THE1 expression in hypocotyls, possibly due to the presence of a PBE-box, a putative binding region for PIF1, PIF3, PIF4, and PIF5, in the THE1 promoter (Oh et al., 2012; Pfeiffer et al., 2014; Martín et al., 2016). We show that this induction depends on PIF7, positioning THE1 expression downstream of PIF7. Mutation of the1 had no effect on the expression of the shade marker genes, suggesting that THE1 might act in an independent branch of the PIF7-controlled pathways, possibly acting at the cell wall during low R:FR-induced cell expansion that drives hypocotyl elongation. Low R:FR-induced elongation requires cell wall modification (Sasidharan et al., 2014, 2011) and we speculate that released polysaccharides from the reorganizing cell wall could act as ligands to activate THE1 (Lindner et al., 2012).
FER regulates female fertility through rupture of the growing pollen tube (Escobar-Restrepo et al., 2007; Duan et al., 2014) but has also been associated with vegetative growth (Guo et al., 2009; Li et al., 2015). Although FER expression is low R:FR-inducible in the two Geranium species, its expression in Arabidopsis is unaffected by low R:FR. The fer and llg1 mutant data, nevertheless, indicate an important role of this receptor complex in shade avoidance of Arabidopsis hypocotyls, and the loss of ATHB2 induction in fer-5 shows that FER is somehow connected to part of the canonical SAS-associated gene expression network in Arabidopsis.
Our data suggest a specific role for these RLKs in low R:FR light-mediated elongation. Auxin levels are typically enhanced in low R:FR conditions (de Wit et al., 2015; Zheng et al., 2016; Tao et al., 2008; Keuskamp et al., 2010), and this is a key route to shoot elongation. However, IAA-induced hypocotyl elongation was not disturbed in any of the studied RLK mutants nor was IAA19 expression affected in the1-1 and fer-5, suggesting that the identified RLKs are not in the established shade avoidance control module of PIF-mediated auxin synthesis and response. How the phytochrome pathway regulates these RLKs, which putative ligands are controlling their activity in shade avoidance and which factors might be the kinase targets are all topics that future studies should elucidate.
Interestingly, FER has been directly associated with plant immunity (Kessler et al., 2010; Masachis et al., 2016) and THE1 can regulate pathogen defense-related genes (Hématy et al., 2007). It remains to be determined if these factors also mediate shade avoidance–defense crosstalk.
In conclusion, our study using wild plants with different evolutionary histories has identified novel regulators of shade avoidance that had not been identified in Arabidopsis. The studies in Arabidopsis support the importance of these genes in SAS expression, but also hint that different organs and/or life cycle stages might express different modes of regulation. This study also shows that different species regulate the same components in different ways, consistent with differential control of shade avoidance.
METHODS
Plant Material and Growth Conditions
Geranium pyrenaicum (Cruydt-hoeck, Nijeberkoop, The Netherlands) and Geranium robertianum (Ecoflora, Halle, Belgium) seeds were germinated on polyethylene beads and water for 6 and 11 d, respectively, at saturated relative humidity, 20°C, under long-day conditions (16 h day, 8 h night; 180 μmol m−2 s−1 PAR; R:FR = 1.8). Seedlings were transferred to 70-mL pots with potting soil (mix Z2254; Primasta) and grown for two more weeks in long-day conditions (20°C; 70% relative humidity).
Different Arabidopsis thaliana genotypes (Col-0 and pif7 [Leivar et al., 2008], pif4 pif5 [Lorrain et al., 2008], pif4 pif5 pif7 [de Wit et al., 2015], the1-1 and THE1ox [Hématy et al., 2007], the1-4, herk1, and herk1 the1 [Guo et al., 2009], kdr-1 [SALK_048383C, Supplemental Figure 7A], kdr-2 [SALK_033495C, Supplemental Figure 7A], KDR-D [Hyun and Lee, 2006; Supplemental Figure 7B], fer-5 [SALK_029056C], fer-4, and llg1-2 [Li et al., 2015]) were grown as previously described (de Wit et al., 2012), with a 9-h-light (180 μmol m−2 s−1 PAR)/15-h-dark photoperiod until ready for experiments at 4 weeks. For seedling experiments, seeds of the same Arabidopsis genotypes were surface-sterilized with chlorine gas, stratified (4 d dark, 4°C), and germinated on 0.5 Murashige and Skoog agar (0.8% v/w) plates with MES buffer (1 g/L). Germination was started with a 2-h light pulse, followed by 24 h darkness, and plates were then transferred to a long-day photoperiod as above. Treatments started 48 h after the light pulse.
Light Treatments
Low R:FR light conditions were obtained by supplementing standard growth chamber light (R:FR = 1.8; Philips HPI) with far-red LEDs (730-nm peak; Philips Greenpower Research modules), to obtain a R:FR of 0.2 without changing PAR. Blue light-depleted (±4 μmol m−2 s−1) conditions were obtained by filtering higher intensity white light using a Lee Medium Yellow 010 filter, to similar PAR as in controls (180 μmol m−2 s−1 for rosette plants, 110 μmol m−2 s−1 for seedlings). Light spectra are presented in Supplemental Figure 10. Treatments started at 10:00 am.
Growth Measurements
Geranium and Arabidopsis petiole elongation was measured over a set period with a digital caliper (tx - t0). In Geranium, petiole growth rate kinetics were monitored using linear variable displacement transducers (type ST 200; Schlumberger Industries; Pierik et al., 2011; Voesenek et al., 2003). The linear variable displacement transducer was attached to the lamina-leaf junction of the second leaf 18 h prior to the start of the light treatment.
In Arabidopsis, hypocotyl length was measured at the end of a 4-d treatment using digital (600 dpi) images and ImageJ.
Statistical Analysis
Growth and bioassay data were analyzed by two-way ANOVA preceded by Levene’s test to verify equal variances (P > 0.05), followed by a post-hoc Tukey test. Arabidopsis gene expression data were analyzed by a Student’s t test preceded by an f-test to verify equal variances. If needed, data were ln transformed. All analyses were conducted in R and Microsoft Excel.
RNA Isolation and Gene Expression
For RT-qPCR, the apical 1 cm of the petiole of the second Geranium leaf of three individual plants (Figure 6; petiole elongation response) or the whole second leaf (petiole and lamina) of two individual plants (Figure 4C; pathogen defense of the leaf) were pooled as biological replicates. RNA was extracted using a Qiagen RNeasy kit with on-column DNaseI treatment. cDNA was synthesized using the Superscript III reverse transcriptase kit (Invitrogen) with RNase inhibitors and random primers. Quantitative RT-PCR was performed with SyberGreen Supermix (Bio-Rad) in a Viia7 PCR. The Geranium homolog of the PP2A subunit PDF1 was used as a reference gene. Relative expression was calculated using the 2−ΔΔCt method. Primers are listed in Supplemental Table 1. For Arabidopsis experiments in petioles, RNA was extracted from six pooled petioles (∼5 mm) of three plants per biological replicate, after 4 h of light treatment. For Arabidopsis experiments in seedlings, 40 to 45 hypocotyls were harvested and pooled per biological replicate, after 24 h of light treatment. Extraction and analysis were similar as for the geraniums; the average Ct of TUBULIN, APT1, and AT1G13320 (petioles), or the CT of At1G13320 solely (hypocotyls), was used as a reference. Expression data of RLK-encoding genes from previously published experiments on Arabidopsis rosettes (heat map in Supplemental Figure 9) was obtained using the Genevestigator program and contains data from Leivar et al. (2012), Hornitschek et al. (2012), Tao et al. (2008), de Wit et al. (2013), Sessa et al. (2005), Ciolfi et al. (2013), Delker et al. (2010), Goda et al. (2008), Armstrong et al. (2004), and Okushima et al. (2005).
Illumina Sequencing and Bioinformatics
RNA was derived from the Geraniums as described. The replicates were a pool of 12 individual plants harvested in three separate experiments. The harvest time points were 2 and 11.5 h after the start of the far-red treatment (12:00 and 21:30, respectively). Normalized libraries for de novo assembly were constructed by Vertis Biotechnologie from pooled material of control, low R:FR, blue light-depleted, and green filter treated petioles and laminas, after 2, 11.5, and 24 h, for both species separately, using the kinetic denaturation-reassociation technique.
Illumina sequencing (paired-end, 101-bp reads) was performed by Macrogen, followed by de novo assembly (Trinity method; Grabherr et al., 2011) of the reference transcriptomes and alignment of the experimental transcripts (RSEM-based abundance estimation).
Putatively orthologous transcript groups within the two species were constructed using OrthoMCL clustering (as described in Li et al., 2003; Enright et al., 2002), with inflation factor 1.1 (for kingdom-wide OMCL, to increase the ability to detect putatively orthologous groups) or 3.0 (for expression analysis) and the similarity matrix of an all-versus-all discontiguous megaBLASTn of the two (or six) transcriptomes as input. The best BLAST hit with the Arabidopsis transcriptome was used to name Geranium OMCL groups.
For statistical analysis, read counts of all transcripts in an OMCL group were summed and a cutoff of minimally 20 reads in at least one treatment was applied to filter out low abundant families. To test for differentially expressed genes in single-factorial comparisons, a negative binomial model of OMCL group read numbers was made using the quantile-adjusted conditional maximum likelihood (qCML) method, followed by an exact test, including Benjamini and Hochberg’s algorithm to control false discovery rates, of the EdgeR package (Bioconductor) (Robinson et al., 2010). The Cox-Reid profile-adjusted likelihood method in combination with a generalized linear model (likelihood ratio test [glmLRT]) of the EdgeR package was used in multifactor comparisons (McCarthy et al., 2012). A P value cutoff for differentially expressed genes was set at 0.01.
For GO analysis, differentially expressed Geranium OMCL groups, with a BLAST E-value < 10−10 with Arabidopsis genes, were tested for GO term enrichment using the R package GOseq (Young et al., 2010), with correction for the total length of all transcripts in the Geranium OMCL group.
Pathogen Assays
Necrotrophic fungus Botrytis cinerea (strain B0510) was grown on half-strength potato dextrose agar (BD Difco) for 2 weeks before conidia were harvested in potato dextrose broth (BD Difco). G. pyrenaicum and G. robertianum seedlings (6 and 11 d after germination, respectively) were transferred to 19-mL pots with potting soil, and on the following day, a 2-μL droplet of 2.5 × 105 spores/mL of B. cinerea was applied to each cotyledon. Plants were kept at saturated relative humidity in a control white or low R:FR treatment. Lesion diameters were measured 3 d after inoculation.
Pharmacological Treatments
For the methyl-JA treatment, 30 min prior to the start of the light treatments, a solution of 100 μM methyl-JA or mock (0.1% ethanol) with 0.1% Tween 20 was sprayed on the plants. The IAA treatments of seedlings were performed as described previously (Keuskamp et al., 2011), with slight adjustments. Briefly, 200 μL of a concentrated, sterile IAA solution (600 μM) was applied to the plates to create a film on top of the agar, with a final concentration of 15 μM (0.1% DMSO).
Accession Numbers
Raw sequencing files and transcript shotgun assemblies (accession numbers HAGG01000001 to HAGG01168191 for G. pyrenaicum, and HAGH01000001 to HAGH01300137 for G. robertianum) are stored at the European Nucleotide Archive and combined under study accession number PRJEB18552 (http://www.ebi.ac.uk/ena/data/view/PRJEB18552). OMCL group assignment of the transcripts and RNA-seq expression data are available in the ArrayExpress database (www.ebi.ac.uk/arrayexpress) under accession number E-MTAB-5371.
Supplemental Data
Supplemental Figure 1. A detailed analysis of low R:FR-induced growth over the time of day and in Geranium petioles.
Supplemental Figure 2. Low R:FR light suppresses petiole growth in G. robertianum, but not G. pyrenaicum, at a fixed time of the day.
Supplemental Figure 3. Gene Ontology analysis on low R:FR induced and repressed Geranium OMCL groups.
Supplemental Figure 4. Plant processes differentially regulated in low R:FR by the two Geranium species
Supplemental Figure 5. Low R:FR light affects MeJA-induced transcript abundance in Geranium and Arabidopsis, and pathogen infection does not affect Geranium growth in the shade.
Supplemental Figure 6. RT-qPCR on Geranium transcripts and validation of RNA sequencing.
Supplemental Figure 7. Confirmation of kdr mutants.
Supplemental Figure 8. HERK1 has no function in the Arabidopsis shade avoidance syndrome.
Supplemental Figure 9. RLK gene expression is hardly affected by low R:FR, pif mutations and IAA in previous transcriptome studies.
Supplemental Figure 10. Light spectra of the treatments used in this study.
Supplemental Table 1. Primer list.
Supplementary Material
Acknowledgments
We thank Herman Höfte, Alice Cheung, Yanhai Yin, and Christian Fankhauser for kindly providing Arabidopsis seeds. We thank Saskia van Wees, Irene Vos, and Merel Steenbergen for providing B. cinerea strains and helping with the pathogen assays. Funding was provided by the Netherlands Organisation for Scientific Research to C.M.M.G. (no. 022.001.018) and R.P. (VIDI Grant 864.12.003).
AUTHOR CONTRIBUTIONS
C.M.M.G., R.P., and L.A.C.J.V. conceived the project. C.M.M.G., R.P., and D.H.K. designed the experiments. C.M.M.G., D.H.K., S.B., E.R., and I.T.K. performed the experiments. C.M.M.G. and H.v.V. analyzed the RNA-seq data. C.M.M.G. and R.P. wrote the manuscript.
Glossary
- R:FR
red:far-red light ratio
- SAS
shade avoidance syndrome
- bHLH
basic helix-loop-helix
- JA
jasmonic acid
- OMCL
Markov Clustering Algorithm
- GO
Gene Ontology
- qCML
quantile-adjusted conditional maximum likelihood
Footnotes
Articles can be viewed without a subscription.
References
- Armstrong J.I., Yuan S., Dale J.M., Tanner V.N., Theologis A. (2004). Identification of inhibitors of auxin transcriptional activation by means of chemical genetics in Arabidopsis. Proc. Natl. Acad. Sci. USA 101: 14978–14983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bai M.-Y., Shang J.-X., Oh E., Fan M., Bai Y., Zentella R., Sun T.-P., Wang Z.-Y. (2012). Brassinosteroid, gibberellin and phytochrome impinge on a common transcription module in Arabidopsis. Nat. Cell Biol. 14: 810–817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campos M.L., Yoshida Y., Major I.T., de Oliveira Ferreira D., Weraduwage S.M., Froehlich J.E., Johnson B.F., Kramer D.M., Jander G., Sharkey T.D., Howe G.A. (2016). Rewiring of jasmonate and phytochrome B signalling uncouples plant growth-defense tradeoffs. Nat. Commun. 7: 12570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cargnel M.D., Demkura P.V., Ballaré C.L. (2014). Linking phytochrome to plant immunity: low red : far-red ratios increase Arabidopsis susceptibility to Botrytis cinerea by reducing the biosynthesis of indolic glucosinolates and camalexin. New Phytol. 204: 342–354. [DOI] [PubMed] [Google Scholar]
- Carriedo L.G., Maloof J.N., Brady S.M. (2016). Molecular control of crop shade avoidance. Curr. Opin. Plant Biol. 30: 151–158. [DOI] [PubMed] [Google Scholar]
- Cerrudo I., Keller M.M., Cargnel M.D., Demkura P.V., de Wit M., Patitucci M.S., Pierik R., Pieterse C.M.J., Ballaré C.L. (2012). Low red/far-red ratios reduce Arabidopsis resistance to Botrytis cinerea and jasmonate responses via a COI1-JAZ10-dependent, salicylic acid-independent mechanism. Plant Physiol. 158: 2042–2052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chico J.-M., Fernández-Barbero G., Chini A., Fernández-Calvo P., Díez-Díaz M., Solano R. (2014). Repression of jasmonate-dependent defenses by shade involves differential regulation of protein stability of MYC transcription factors and their JAZ repressors in Arabidopsis. Plant Cell 26: 1967–1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ciolfi A., Sessa G., Sassi M., Possenti M., Salvucci S., Carabelli M., Morelli G., Ruberti I. (2013). Dynamics of the shade-avoidance response in Arabidopsis. Plant Physiol. 163: 331–353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coluccio M.P., Sanchez S.E., Kasulin L., Yanovsky M.J., Botto J.F. (2011). Genetic mapping of natural variation in a shade avoidance response: ELF3 is the candidate gene for a QTL in hypocotyl growth regulation. J. Exp. Bot. 62: 167–176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cortés L.E., Weldegergis B.T., Boccalandro H.E., Dicke M., Ballar C.L. (2016). Trading direct for indirect defense ? Phytochrome B inactivation in tomato attenuates direct anti-herbivore defenses whilst enhancing volatile-mediated attraction of predators. New Phytol. 212: 1057–1071. [DOI] [PubMed] [Google Scholar]
- Das D., St Onge K.R., Voesenek L.A.C.J., Pierik R., Sasidharan R. (2016). Ethylene- and shade-induced hypocotyl elongation share transcriptome patterns and functional regulators. Plant Physiol. 172: 718–733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delker C., Pöschl Y., Raschke A., Ullrich K., Ettingshausen S., Hauptmann V., Grosse I., Quint M. (2010). Natural variation of transcriptional auxin response networks in Arabidopsis thaliana. Plant Cell 22: 2184–2200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Lucas M., Davière J.-M., Rodríguez-Falcón M., Pontin M., Iglesias-Pedraz J.M., Lorrain S., Fankhauser C., Blázquez M.A., Titarenko E., Prat S. (2008). A molecular framework for light and gibberellin control of cell elongation. Nature 451: 480–484. [DOI] [PubMed] [Google Scholar]
- Denness L., McKenna J.F., Segonzac C., Wormit A., Madhou P., Bennett M., Mansfield J., Zipfel C., Hamann T. (2011). Cell wall damage-induced lignin biosynthesis is regulated by a reactive oxygen species- and jasmonic acid-dependent process in Arabidopsis. Plant Physiol. 156: 1364–1374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Wit M., Kegge W., Evers J.B., Vergeer-van Eijk M.H., Gankema P., Voesenek L.A.C.J., Pierik R. (2012). Plant neighbor detection through touching leaf tips precedes phytochrome signals. Proc. Natl. Acad. Sci. USA 109: 14705–14710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Wit M., Ljung K., Fankhauser C. (2015). Contrasting growth responses in lamina and petiole during neighbor detection depend on differential auxin responsiveness rather than different auxin levels. New Phytol. 208: 198–209. [DOI] [PubMed] [Google Scholar]
- de Wit M., Spoel S.H., Sanchez-Perez G.F., Gommers C.M.M., Pieterse C.M.J., Voesenek L.A.C.J., Pierik R. (2013). Perception of low red:far-red ratio compromises both salicylic acid- and jasmonic acid-dependent pathogen defences in Arabidopsis. Plant J. 75: 90–103. [DOI] [PubMed] [Google Scholar]
- Djakovic-Petrovic T., de Wit M., Voesenek L.A.C.J., Pierik R. (2007). DELLA protein function in growth responses to canopy signals. Plant J. 51: 117–126. [DOI] [PubMed] [Google Scholar]
- Duan Q., Kita D., Johnson E.A., Aggarwal M., Gates L., Wu H.-M., Cheung A.Y. (2014). Reactive oxygen species mediate pollen tube rupture to release sperm for fertilization in Arabidopsis. Nat. Commun. 5: 3129. [DOI] [PubMed] [Google Scholar]
- Enright A.J., Van Dongen S., Ouzounis C.A. (2002). An efficient algorithm for large-scale detection of protein families. Nucleic Acids Res. 30: 1575–1584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Escobar-Restrepo J.-M., Huck N., Kessler S., Gagliardini V., Gheyselinck J., Yang W.-C., Grossniklaus U. (2007). The FERONIA receptor-like kinase mediates male-female interactions during pollen tube reception. Science 317: 656–660. [DOI] [PubMed] [Google Scholar]
- Filiault D.L., Maloof J.N. (2012). A genome-wide association study identifies variants underlying the Arabidopsis thaliana shade avoidance response. PLoS Genet. 8: e1002589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fraser D.P., Hayes S., Franklin K.A. (2016). Photoreceptor crosstalk in shade avoidance. Curr. Opin. Plant Biol. 33: 1–7. [DOI] [PubMed] [Google Scholar]
- Gilbert I.R., Jarvis P.G., Smith H. (2001). Proximity signal and shade avoidance differences between early and late successional trees. Nature 411: 792–795. [DOI] [PubMed] [Google Scholar]
- Goda H., et al. (2008). The AtGenExpress hormone and chemical treatment data set: experimental design, data evaluation, model data analysis and data access. Plant J. 55: 526–542. [DOI] [PubMed] [Google Scholar]
- Gommers C.M.M., Visser E.J.W., St Onge K.R., Voesenek L.A.C.J., Pierik R. (2013). Shade tolerance: when growing tall is not an option. Trends Plant Sci. 18: 65–71. [DOI] [PubMed] [Google Scholar]
- Grabherr M.G., et al. (2011). Full-length transcriptome assembly from RNA-Seq data without a reference genome. Nat. Biotechnol. 29: 644–652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo H., Li L., Ye H., Yu X., Algreen A., Yin Y. (2009). Three related receptor-like kinases are required for optimal cell elongation in Arabidopsis thaliana. Proc. Natl. Acad. Sci. USA 106: 7648–7653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hématy K., Sado P.-E., Van Tuinen A., Rochange S., Desnos T., Balzergue S., Pelletier S., Renou J.-P., Höfte H. (2007). A receptor-like kinase mediates the response of Arabidopsis cells to the inhibition of cellulose synthesis. Curr. Biol. 17: 922–931. [DOI] [PubMed] [Google Scholar]
- Hong S.-Y., Seo P.J., Ryu J.Y., Cho S.-H., Woo J.-C., Park C.-M. (2013). A competitive peptide inhibitor KIDARI negatively regulates HFR1 by forming nonfunctional heterodimers in Arabidopsis photomorphogenesis. Mol. Cells 35: 25–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hornitschek P., Kohnen M.V., Lorrain S., Rougemont J., Ljung K., López-Vidriero I., Franco-Zorrilla J.M., Solano R., Trevisan M., Pradervand S., Xenarios I., Fankhauser C. (2012). Phytochrome interacting factors 4 and 5 control seedling growth in changing light conditions by directly controlling auxin signaling. Plant J. 71: 699–711. [DOI] [PubMed] [Google Scholar]
- Hornitschek P., Lorrain S., Zoete V., Michielin O., Fankhauser C. (2009). Inhibition of the shade avoidance response by formation of non-DNA binding bHLH heterodimers. EMBO J. 28: 3893–3902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hyun Y., Lee I. (2006). KIDARI, encoding a non-DNA Binding bHLH protein, represses light signal transduction in Arabidopsis thaliana. Plant Mol. Biol. 61: 283–296. [DOI] [PubMed] [Google Scholar]
- Izaguirre M.M., Mazza C.A., Biondini M., Baldwin I.T., Ballaré C.L. (2006). Remote sensing of future competitors: impacts on plant defenses. Proc. Natl. Acad. Sci. USA 103: 7170–7174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiménez-Gómez J.M., Wallace A.D., Maloof J.N. (2010). Network analysis identifies ELF3 as a QTL for the shade avoidance response in Arabidopsis. PLoS Genet. 6: e1001100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kessler S.A., Shimosato-Asano H., Keinath N.F., Wuest S.E., Ingram G., Panstruga R., Grossniklaus U. (2010). Conserved molecular components for pollen tube reception and fungal invasion. Science 330: 968–971. [DOI] [PubMed] [Google Scholar]
- Keuskamp D.H., Pollmann S., Voesenek L.A.C.J., Peeters A.J.M., Pierik R. (2010). Auxin transport through PIN-FORMED 3 (PIN3) controls shade avoidance and fitness during competition. Proc. Natl. Acad. Sci. USA 107: 22740–22744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keuskamp D.H., Sasidharan R., Vos I., Peeters A.J.M., Voesenek L.A.C.J., Pierik R. (2011). Blue-light-mediated shade avoidance requires combined auxin and brassinosteroid action in Arabidopsis seedlings. Plant J. 67: 208–217. [DOI] [PubMed] [Google Scholar]
- Kohnen M.V., Schmid-Siegert E., Trevisan M., Petrolati L.A., Sénéchal F., Müller-Moulé P., Maloof J., Xenarios I., Fankhauser C. (2016). Neighbor detection induces organ-specific transcriptomes, revealing patterns underlying hypocotyl-specific frowth. Plant Cell 28: 2889–2904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leivar P., Monte E., Al-Sady B., Carle C., Storer A., Alonso J.M., Ecker J.R., Quail P.H. (2008). The Arabidopsis phytochrome-interacting factor PIF7, together with PIF3 and PIF4, regulates responses to prolonged red light by modulating phyB levels. Plant Cell 20: 337–352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leivar P., Tepperman J.M., Cohn M.M., Monte E., Al-Sady B., Erickson E., Quail P.H. (2012). Dynamic antagonism between phytochromes and PIF family basic helix-loop-helix factors induces selective reciprocal responses to light and shade in a rapidly responsive transcriptional network in Arabidopsis. Plant Cell 24: 1398–1419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li C., et al. (2015). Glycosylphosphatidylinositol-anchored proteins as chaperones and co-receptors for FERONIA receptor kinase signaling in Arabidopsis. eLife 4: 1–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li L., et al. (2012). Linking photoreceptor excitation to changes in plant architecture. Genes Dev. 26: 785–790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li L., Stoeckert C.J. Jr., Roos D.S. (2003). OrthoMCL: identification of ortholog groups for eukaryotic genomes. Genome Res. 13: 2178–2189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindner H., Müller L.M., Boisson-Dernier A., Grossniklaus U. (2012). CrRLK1L receptor-like kinases: not just another brick in the wall. Curr. Opin. Plant Biol. 15: 659–669. [DOI] [PubMed] [Google Scholar]
- Lorrain S., Allen T., Duek P.D., Whitelam G.C., Fankhauser C. (2008). Phytochrome-mediated inhibition of shade avoidance involves degradation of growth-promoting bHLH transcription factors. Plant J. 53: 312–323. [DOI] [PubMed] [Google Scholar]
- Martín G., Leivar P., Ludevid D., Tepperman J.M., Quail P.H., Monte E. (2016). Phytochrome and retrograde signalling pathways converge to antagonistically regulate a light-induced transcriptional network. Nat. Commun. 7: 11431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masachis S., Segorbe D., Turrà D., Leon-Ruiz M., Fürst U., El Ghalid M., Leonard G., López-Berges M.S., Richards T.A., Felix G., Di Pietro A. (2016). A fungal pathogen secretes plant alkalinizing peptides to increase infection. Nat. Microbiol. 1: 16043. [DOI] [PubMed] [Google Scholar]
- McCarthy D.J., Chen Y., Smyth G.K. (2012). Differential expression analysis of multifactor RNA-Seq experiments with respect to biological variation. Nucleic Acids Res. 40: 4288–4297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moreno J.E., Tao Y., Chory J., Ballaré C.L. (2009). Ecological modulation of plant defense via phytochrome control of jasmonate sensitivity. Proc. Natl. Acad. Sci. USA 106: 4935–4940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morgan D.C., Smith H. (1978). The relationship between phytochrome-photoequilibrium and development in light grown Chenopodium album L. Planta 142: 187–193. [DOI] [PubMed] [Google Scholar]
- Nozue K., Tat A.V., Kumar Devisetty U., Robinson M., Mumbach M.R., Ichihashi Y., Lekkala S., Maloof J.N. (2015). Shade avoidance components and pathways in adult plants revealed by phenotypic profiling. PLoS Genet. 11: e1004953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oh E., Zhu J.-Y., Wang Z.-Y. (2012). Interaction between BZR1 and PIF4 integrates brassinosteroid and environmental responses. Nat. Cell Biol. 14: 802–809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okushima Y., et al. (2005). Functional genomic analysis of the AUXIN RESPONSE FACTOR gene family members in Arabidopsis thaliana: unique and overlapping functions of ARF7 and ARF19. Plant Cell 17: 444–463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfeiffer A., Shi H., Tepperman J.M., Zhang Y., Quail P.H. (2014). Combinatorial complexity in a transcriptionally centered signaling hub in Arabidopsis. Mol. Plant 7: 1598–1618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pierik R., Testerink C. (2014). The art of being flexible: how to escape from shade, salt, and drought. Plant Physiol. 166: 5–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pierik R., De Wit M., Voesenek L.A. (2011). Growth-mediated stress escape: convergence of signal transduction pathways activated upon exposure to two different environmental stresses. New Phytol. 189: 122–134. [DOI] [PubMed] [Google Scholar]
- Quail P.H. (2002). Phytochrome photosensory signalling networks. Nat. Rev. Mol. Cell Biol. 3: 85–93. [DOI] [PubMed] [Google Scholar]
- Robinson M.D., McCarthy D.J., Smyth G.K. (2010). edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 26: 139–140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sasidharan R., Chinnappa C.C., Voesenek L.A.C.J., Pierik R. (2009). A molecular basis for the physiological variation in shade avoidance responses: a tale of two ecotypes. Plant Signal. Behav. 4: 528–529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sasidharan R., Keuskamp D.H., Kooke R., Voesenek L.A.C.J., Pierik R. (2014). Interactions between auxin, microtubules and XTHs mediate green shade- induced petiole elongation in Arabidopsis. PLoS One 9: e90587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sasidharan R., Voesenek L.A., Pierik R. (2011). Cell wall modifying proteins mediate plant acclimatization to biotic and abiotic stresses. Crit. Rev. Plant Sci. 30: 548–562. [Google Scholar]
- Sessa G., Carabelli M., Sassi M., Ciolfi A., Possenti M., Mittempergher F., Becker J., Morelli G., Ruberti I. (2005). A dynamic balance between gene activation and repression regulates the shade avoidance response in Arabidopsis. Genes Dev. 19: 2811–2815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tao Y., et al. (2008). Rapid synthesis of auxin via a new tryptophan-dependent pathway is required for shade avoidance in plants. Cell 133: 164–176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tofts R.J. (2004). Geranium robertianum L. J. Ecol. 92: 537–555. [Google Scholar]
- Toledo-Ortiz G., Huq E., Rodríguez-Concepción M. (2010). Direct regulation of phytoene synthase gene expression and carotenoid biosynthesis by phytochrome-interacting factors. Proc. Natl. Acad. Sci. USA 107: 11626–11631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valladares F., Niinemets Ü. (2008). Shade tolerance, a key plant feature of complex nature and consequences. Annu. Rev. Ecol. Evol. Syst. 39: 237–257. [Google Scholar]
- van Veen H., Mustroph A., Barding G.A., Vergeer-van Eijk M., Welschen-Evertman R.A.M., Pedersen O., Visser E.J.W., Larive C.K., Pierik R., Bailey-Serres J., Voesenek L.A.C.J., Sasidharan R. (2013). Two Rumex species from contrasting hydrological niches regulate flooding tolerance through distinct mechanisms. Plant Cell 25: 4691–4707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voesenek L.A.C.J., Jackson M.B., Toebes A.H.W., Huibers W., Vriezen W.H., Colmer T.D. (2003). De-submergence-induced ethylene production in Rumex palustris: regulation and ecophysiological significance. Plant J. 33: 341–352. [DOI] [PubMed] [Google Scholar]
- Warburg A.E.F. (1938). Taxonomy and relationship in the Geraniales in the light of their cytology. New Phytol. 37: 130–159. [Google Scholar]
- Wolf S., Höfte H. (2014). Growth control: a saga of cell walls, ROS, and peptide receptors. Plant Cell 26: 1848–1856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang D., Seaton D.D., Krahmer J., Halliday K.J. (2016). Photoreceptor effects on plant biomass, resource allocation, and metabolic state. Proc. Natl. Acad. Sci. USA 113: 7667–7672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young M.D., Wakefield M.J., Smyth G.K., Oshlack A. (2010). Gene ontology analysis for RNA-seq: accounting for selection bias. Genome Biol. 11: R14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng Z., Guo Y., Chen W., Ljung K., Noel J.P., Chory J. (2016). Local auxin metabolism regulates environment-induced hypocotyl elongation. Nat. Plants 2: 1–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.