Elvira-Matelot, E., Bardou, F., Ariel, F., Jauvion, V., Bouteiller, N., Le Masson, I., Cao, J., Crespi, M.D., and Vaucheret, H. (2016). The nuclear ribonucleoprotein SmD1 interplays with splicing, RNA quality control, and posttranscriptional gene silencing in Arabidopsis. 28: 426–438.
A mistake was introduced when assembling the image of the top panel of Figure 4. The original image contained 10 lanes organized as shown in Figure 4 source data (see below); the three Col replicates for ATPase1 (top panel) were not loaded in three adjacent lanes due to a problem in the gel. To show triplicates adjacent to one another as in the bottom panel of Figure 4, lane 7, should have been cut and pasted in place of lane 3. Instead, lane 7 was deleted and lane 2 was duplicated and copied in place of lane 3. The corrected figure with all original data for each sample is shown here. The correction of this mistake does not alter the conclusions of the article. In addition, the legend of Figure 4 was amended to define the error bars and the values of the y axes of RNA quantification graphs.
Figure 4.
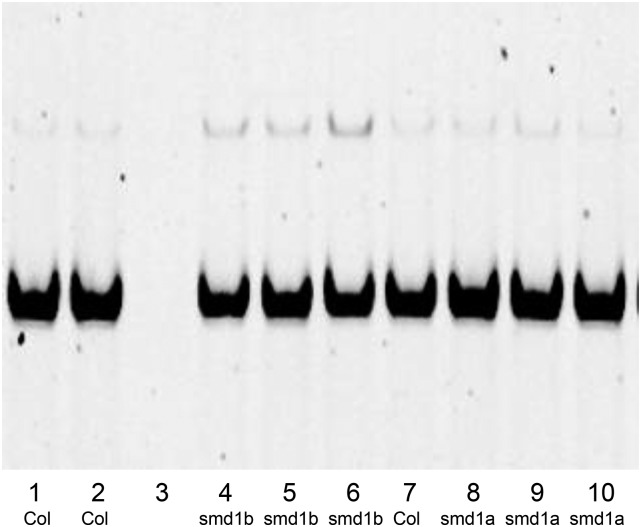
Source Data.
Lane 1, 1st Col replicate; lane 2, 2nd Col replicate; lane 3, empty lane (due to a problem in the gel); lane 4, 1st smd1b replicate; lane 5, 2nd smd1b replicate; lane 6, 3rd smd1b replicate; lane 7, 3rd Col replicate; lane 8, 1st smd1a replicate; lane 9, 2nd smd1a replicate; lane 10, 3rd smd1a replicate.
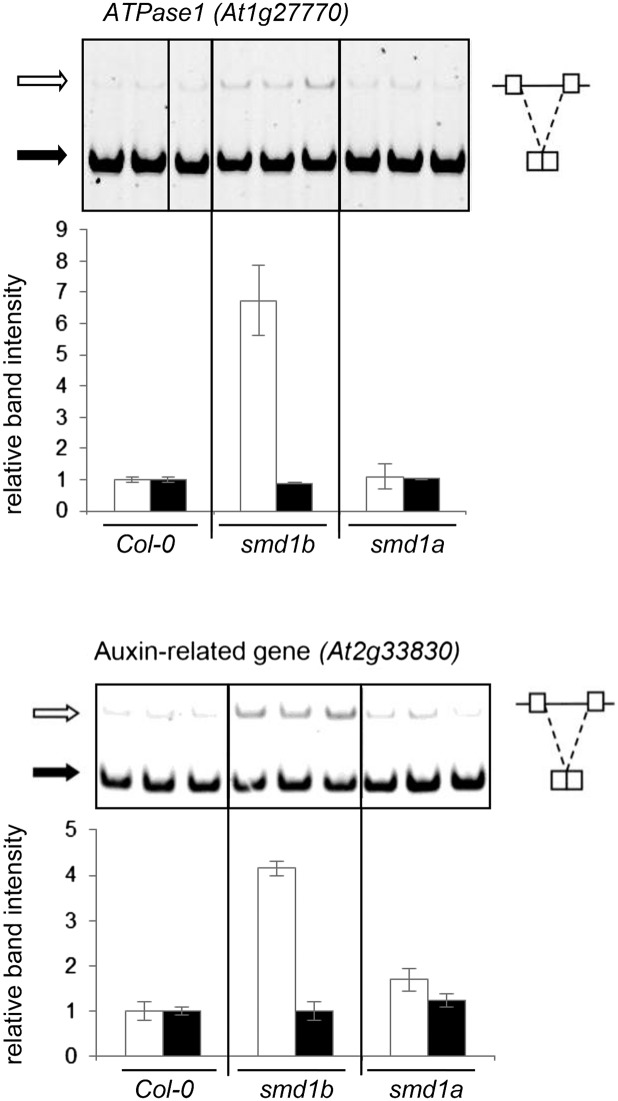
Figure 4.
Original: Endogenous RNA Accumulation in smd1 Mutants.
RT-PCR and quantification of RNA isoforms of the ATPase1 gene At1g27770 and auxin-related gene At2g33830 on PAGE gels. Black and white arrows indicate spliced and unspliced RNA, respectively. Increased intron retention is observed in smd1b but not smd1a.
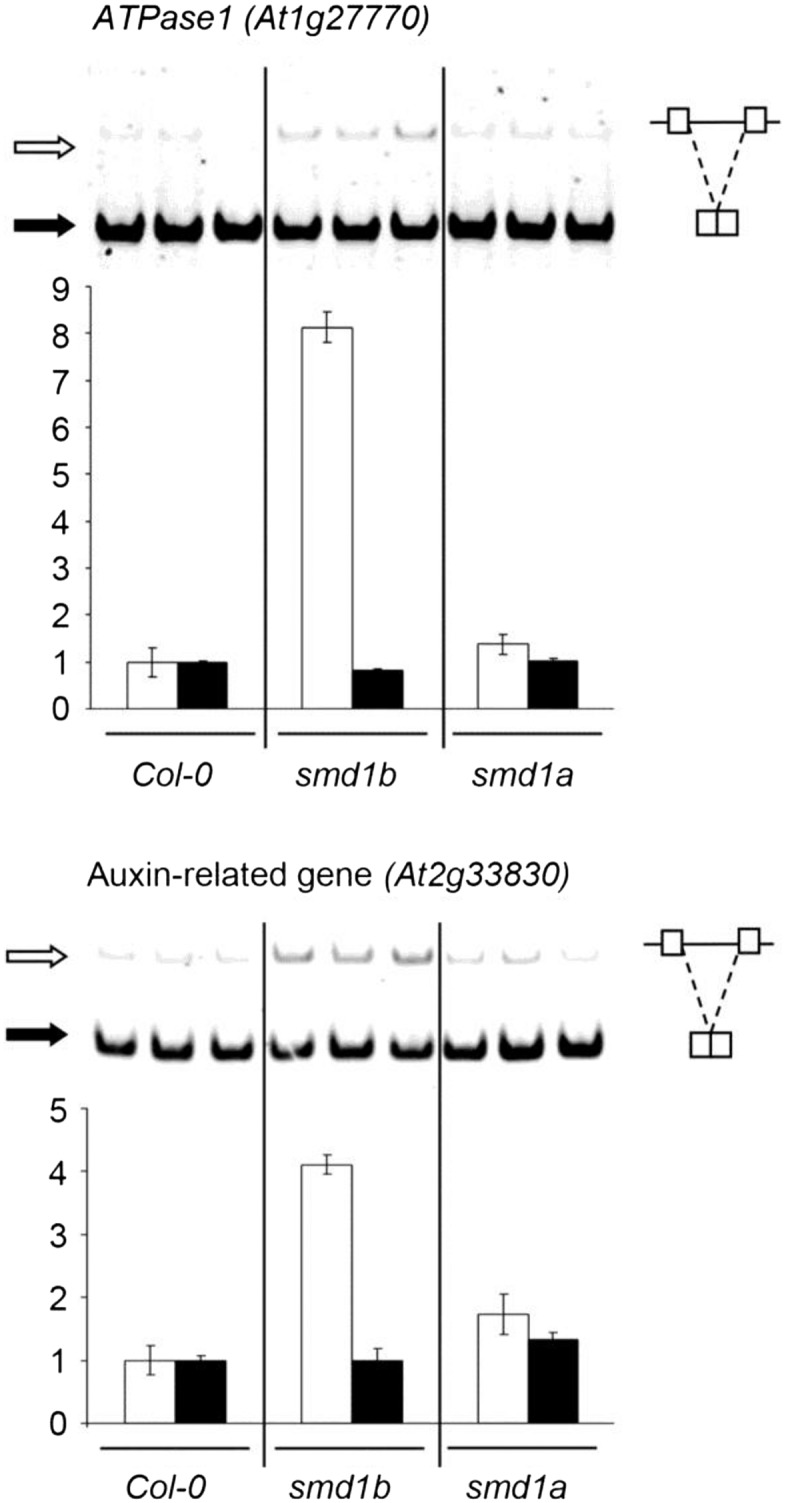
Figure 4.
Corrected: Endogenous RNA Accumulation in smd1 Mutants.
RT-PCR and quantification of RNA isoforms of the ATPase1 gene At1g27770 and auxin-related gene At2g33830 on PAGE gels. Black and white arrows indicate spliced and unspliced RNA, respectively. Values of both RNA species were arbitrarily set at 1 in Col-0. Error bars represent sd between triplicates. Increased intron retention in observed in smd1b but not smd1a.
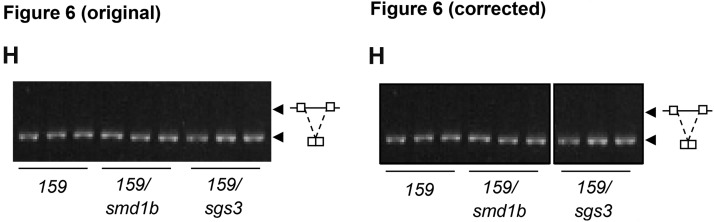
In addition, Figure 6H was mounted by assembling separate images from two different gels, and this was not indicated. Two boxes are now shown for clarification. This correction does not alter the conclusions of the article. The legend of Figure 6 remains unchanged.
Figure 6.
Transgene PTGS in smd1b Mutants.
(H) Analysis of GUS RNA splicing by RT-PCR using primers spanning the intron.
25S rRNA hybridization or ethidium bromide staining served as loading controls for high molecular weight RNA gel blots. U6 snRNA hybridization served as loading controls for low molecular weight RNA gel blots.
Co-first author Florian Bardou takes entire responsibility for making these mistakes and for not informing the other authors of the article about the manipulated images. Co-corresponding authors Martin Crespi and Hervé Vaucheret take responsibility for not reviewing the data completely before submitting the paper for publication. All authors approved this correction.
Footnotes
Articles can be viewed without a subscription.
Editor’s note: the corrected figures and accompanying text were reviewed by members of The Plant Cell editorial board.