Abstract
Cell-cycle entry is critical for homeostatic control in physiologic response of higher organisms but is not well understood. The antibody response begins with induction of naïve mature B cells, which are naturally arrested in G0/G1 phase of the cell cycle, to enter the cell cycle in response to antigen and cytokine. BLyS (BAFF), a cytokine essential for mature B cell development and survival, is thought to act mainly by attenuation of apoptosis. Here, we show that BLyS alone induces cell-cycle entry and early G1 cell-cycle progression, but not S-phase entry, in opposition to the cyclin-dependent kinase inhibitors p18INK4c. Independent of its survival function, BLyS enhances the synthesis of cyclin D2, in part through activation of NF-κB, as well as CDK4 and retinoblastoma protein phosphorylation. By convergent activation of the same cell-cycle regulators in opposition to p18INK4c, B cell receptor signaling induces cell-cycle entry and G1 progression in synergy with BLyS, but also DNA replication. The failure of BLyS to induce S-phase cell-cycle entry lies in its inability to increase cyclin E and reduce p27Kip1 expression. Antagonistic cell-cycle regulation by BLyS and p18INK4c is functionally linked to apoptotic control and conserved from B cell activation in vitro to antibody response in vivo, further indicating a physiologic role in homeostasis.
Keywords: BAFF, cyclin D2, cyclin E, cyclin-dependent kinase, B cell receptor signaling
Regulation of cell-cycle entry and exit critically controls homeostasis in physiologic responses of higher organisms. Our understanding of this process has been limited by a lack of experimental cellular systems that can be manipulated easily in vivo and ex vivo. Naïve mature B cells are naturally arrested in the G0/G1 phase of the cell cycle. They can be induced to enter the cell cycle in response to antigen and cytokine stimulation, and after clonal expansion they differentiate terminally to G1-arrested antibody-secreting plasma cells. Each successive step is controlled by the cell cycle in concert with apoptosis, and differentiation stage-specific B cells can be identified by cell surface markers and isolated for ex vivo analysis. The antibody response, therefore, is an exceptional mammalian system for elucidating cell-cycle control of the timing and magnitude of physiologic response.
In mammalian cells, cytokines and growth factors regulate cell-cycle entry and G1 to S phase cell-cycle progression mainly by modulating the balance between positive cell-cycle regulators [(cyclins and cyclin-dependent kinases (CDKs)] on the one hand and CDK inhibitors (CDKIs) on the other (1). One specific CDKI, p18INK4c (p18) (2, 3), is regulated by IL-6 (4) and is essential for the antibody response. p18 is required for G1 cell-cycle arrest and terminal differentiation of antibody-secreting plasma cells (5). It also may control cell-cycle entry at the beginning of an antibody response, because it attenuates B cell proliferation before and after immunization and in mitogenic stimulation in vitro (5, 7, 32). Moreover, p18-mediated cell-cycle control is functionally linked to homeostasis, as indicated by the acceleration of apoptosis of nonsecreting plasma-cytoid cells in the absence of p18 (5).
BLyS (BAFF) is a cytokine of the tumor necrosis factor family (8, 9), whose receptors (BR3, BCMA, and TACI) are expressed nearly exclusively on B cells (10-12). It is required for mature B cell development (12-15) and plasma cell survival (16), and it promotes the antibody response (17, 18) and Ig class switch recombination (19). A role for BLyS in the development of autoimmunity (20, 21) and the fatal plasma cell cancer, multiple myeloma (22, 23), also has been implicated. BLyS acts principally by attenuating apoptosis (18, 24) regardless of the cell-cycle status (18), presumably through activation of two NF-κB pathways (18, 25-27) and the downstream antiapoptotic Bcl-2 and Bcl-xL genes (18, 26, 28). Although it is generally assumed that attenuation of apoptosis underlies the diverse biological functions of BLyS, other possibilities have not been ruled out. BLyS alone does not induce S-phase cell-cycle entry (18). However, cyclin D2, the major D-type cyclin expressed in B cells and activated in B cell receptor (BCR) signaling (29, 30), is a target of NF-κB activation (31). This knowledge raises the possibility that BLyS may induce individual G1 cell-cycle regulators such as cyclin D2, although by a means that is insufficient to induce S phase entry. In this way, BLyS would cooperate with p18 in homeostatic control of B cell activation by regulating both the cell cycle and apoptosis.
To understand cell-cycle control of the antibody response better, we investigated the control of cell-cycle activation by p18 and BLyS in BCR signaling in vitro and in the T cell-independent antibody response in vivo. We present direct evidence that in freshly isolated mouse resting splenic B cells, BLyS alone induces cell-cycle entry and mid-G1 progression. Cell-cycle activation by BLyS is attenuated by CDKIs, p18 in early G1 and p27 in late G1. Both BLyS and p18 are required for optimal B cell survival during antigen stimulation in vitro and in the antibody response in vivo, further suggesting cooperative homeostatic cell-cycle control by BLyS and p18.
Materials and Methods
Mice and Isolation of Resting B Cells. p18+/+ and p18-/- mice (32) (7-11 weeks, age-matched) were immunized i.p. with 10 μg of 4-hydroxy-3-nitrophenylacetyl (NP) coupled to Ficoll (NP-Ficoll) (24:1; Biosearch) in 250 μl of PBS, alone or together with a daily i.p. injection of 10 μg of BLyS or PBS. p27-deficient (p27Δ51/Δ51) mice (33) were kindly provided by Andrew Koff (Memorial Sloan-Kettering Cancer Center, New York). Bcl-2 transgenic mice (Emμ-bcl-2-22) were purchased from The Jackson Laboratory. High-density (resting B) and low-density (activated B and plasma) cells were isolated from splenocytes from the 60-70% and 50-60% interfaces of a discontinuous Percoll gradient, respectively (18). The resting B cells were >96% pure based on the presence of B220, CD19, and IgM, and the absence of CD3.
B Cell Activation in Vitro. Resting B cells were cultured (4 × 105 cells/ml) in RPMI medium 1640 containing 10% heat-inactivated FCS (HyClone) in the absence or presence of goat anti-mouse IgM (5 μg/ml) (Jackson ImmunoResearch), alone or together with recombinant soluble human BLyS (50 ng/ml) as described in ref. 18. HIV-1 transactivator of transcription-cyclin E (34) was added to cultures together with a lipopolysaccharide inhibitor polymyxin B sulfate (200 units/ml) (Sigma), in the presence or absence of BLyS (30 ng/ml). Apoptotic cells were characterized with the annexin V FITC Apoptosis Detection Kit (Calbiochem). Flow cytometry was performed to determine the expression of CD21, CD23, and CD86 using FITC-anti-mouse CD21/CD35, phycoerythrin-anti-mouse CD23 (Fcε RII), and phycoerythrin-anti-mouse CD86 (B7-2) (Pharmingen), and changes in cell size by forward scattering analysis. Cell shape was visualized by phase contrast microscopy using a Zeiss Axioplan 2 microscope.
Analysis of BrdUrd Uptake and Carboxyfluorescein Diacetate Succinimidyl Ester (CFSE) Labeling. BrdUrd (5-bromo-2-deoxyuridine) uptake was determined by flow cytometry as described in ref. 18, in resting B cells after incubation with BrdUrd (5 μg/ml; Sigma) for 2 h or in low-density splenic B cells isolated from NP-Ficoll-immunized mice 2 h after i.p. injection of BrdUrd (50 mg per kg of body weight). Labeling resting B cells with CFSE (Molecular Probes) and analysis of cell division by the CFSE dilution by flow cytometry were performed as described in ref. 35. The CFSE-labeled cells were cultured in RPMI medium 1640 (4 × 105 cells per ml) in the absence or presence of anti-IgM (5 μg/ml) or daily addition of BLyS (50 ng/ml), or both.
Immunoblotting. Whole-cell lysates were prepared from viable resting B cells before and after in vitro incubation in a buffer containing 300 mM NaCl, 20 mM Hepes (pH 7.9), 0.2% Nonidet P-40, 1 mM MgCl2, 1 mM DTT, 20% glycerol, 1 μg/ml aprotinin, 1 μg/ml pepstatin, 1 μg/ml leupeptin, 2 mM sodium orthovana-date, 10 mM β-glycerol phosphate, and 1 mM PMSF. Proteins were resolved on a 4-12% NuPAGE gel (Invitrogen) and analyzed with one of the following antibodies: mouse monoclonal antibody to human retinoblastoma (Rb) (Pharmingen) or human CDK6 (Cell Signaling Technology, Beverly, MA); rabbit polyclonal antibodies to pSer807/811 of human Rb (Cell Signaling Technology), mouse CDK4, mouse cyclin D2, mouse p27, human CDK2, or rat cyclin E; or goat anti-human actin (all from Santa Cruz Biotechnology). Signals were developed with the enhanced chemiluminescence system (ECL, Amersham Pharmacia).
Real-Time RT-PCR. Total RNA was isolated from resting B cells before and after in vitro incubation by using the TRIzol reagent (Invitrogen). The first strand cDNA was synthesized by using SuperScript III (Invitrogen) and subjected to real-time RT-PCR by using the Assays-on-Demand gene expression mixes specific for cyclin D2 and 18S ribosomal RNA and the TaqMan Universal PCR Master Mix (Applied Biosystems). Reactions were carried out in the ABI PRISM 7900 HT Sequence Detection System (Applied Biosystems). The relative amount of products was determined by the comparative threshold cycle method according to the manufacturer's instructions.
Results
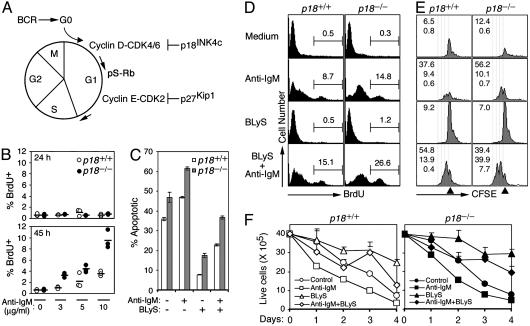
p18INK4c and BLyS Antagonistically Regulate the Cell Cycle and Cooperatively Control Homeostasis. To understand cell-cycle control of the antibody response, we investigated the control of cell-cycle entry by p18 and BLyS in BCR signaling in mouse resting splenic B cells. Stimulation of BCR by cross-linking with anti-IgM [3-10 μg/ml, or molar equivalents of the F(ab′)2 fragment] induced DNA replication in a dose-dependent manner by 45 h, as determined by pulse-labeling (2 h) with BrdUrd (Fig. 1 A and B). Removal of p18 greatly enhanced DNA replication (Fig. 1B), demonstrating that p18 is required to attenuate cell-cycle entry in BCR signaling.
Fig. 1.
Antagonistic cell-cycle control by p18 and BLyS. (A) Cell-cycle activation by BCR stimulation. p18 inhibits CDK4/6 in early G1, p27 inhibits cyclin E/CDK2 in late G1, and serine phosphorylation of Rb (pS-Rb) marks mid-G1 progression. (B) FACS analysis of BrdUrd uptake in p18+/+ and p18-/- resting B cells stimulated with indicated anti-IgM concentration for 24 (Upper) and 45 (Lower) h. The horizontal bars represent the average of three independent analyses. (C) The percentage of apoptotic (annexin V+/PI-) resting p18+/+ and p18-/- B cells stimulated with anti-IgM (5 μg/ml) or BLyS (50 ng/ml), or both, for 48 h. The error bars indicate three independent analyses. (D and E) BrdUrd uptake (D) and CFSE dilution (E) of cells incubated as in C for 4 days. The CFSE level in undivided cells (triangle), the reduction of CFSE by successive cell division (dotted line, right to left), and the percentage of cells in each cell division (numbers in each panel, top to bottom) are indicated. (F) The number of live cells was determined by trypan blue staining in triplicate cultures. The results in C-F are representative of four independent experiments.
However, p18-deficient (p18-/-) B cells were consistently more apoptotic than p18+/+ B cells, before and after stimulation with anti-IgM, even at a limiting concentration (5 μg/ml) (Fig. 1C). They also were less efficiently protected by BLyS (Fig. 1C). Because there are no appreciable differences in the proportions among resting B cell subsets between p18-/- and p18+/+ mice (X.H., M.D., and S.C.-K., unpublished data), p18 and BLyS may protect mature B cells from apoptosis through nonredundant mechanisms and cooperate in homeostatic control in BCR signaling.
To address this possibility, we confirmed that p18 attenuated the cell cycle in BCR signaling by BrdUrd pulse-labeling; twice as many p18-/- as p18+/+ B cells were in the S phase by day 4 of anti-IgM stimulation (Fig. 1D). Because p18 inhibits CDK4 and CDK6 in early G1 (2, 3), S-phase entry apparently was accelerated when the threshold for CDK4/6 activation was lowered by the removal of p18. As expected (18), BLyS alone did not induce DNA replication (Fig. 1D). However, it profoundly increased the proportion of replicating p18+/+ and p18-/- B cells in anti-IgM costimulation. This increase is not likely due to preferential protection of replicating B cells by BLyS, because anti-IgM activated B cells were less well protected by BLyS (Fig. 1C). Rather, these results suggest that BLyS enhances G1/S transition in BCR signaling in opposition to p18.
Cell division in BCR signaling was similarly accelerated by the absence of p18 because substantially more p18-/- than p18+/+ cells had divided once or twice in 4 days of anti-IgM stimulation as assessed by the dilution of CFSE (Fig. 1E). BLyS markedly increased the proportion of B cells that had completed at least two cell divisions in anti-IgM costimulation, and this increase was amplified in p18-/- B cells (Fig. 1E). However, accelerated division of p18-/- B cells seems to be counterbalanced by enhanced apoptosis, because live p18-/- and p18+/+ B cells accumulated comparably in anti-IgM stimulation and in costimulation with anti-IgM and BLyS, albeit to higher levels. (Fig. 1F). Considering the substantial increases in DNA replication and cell division in BLyS and anti-IgM costimulation (Fig. 1 D and E), these results cannot be explained solely by a survival function of BLyS. BLyS therefore may have additional functions acting in concert with p18 to control homeostasis of mature B cells in BCR signaling.
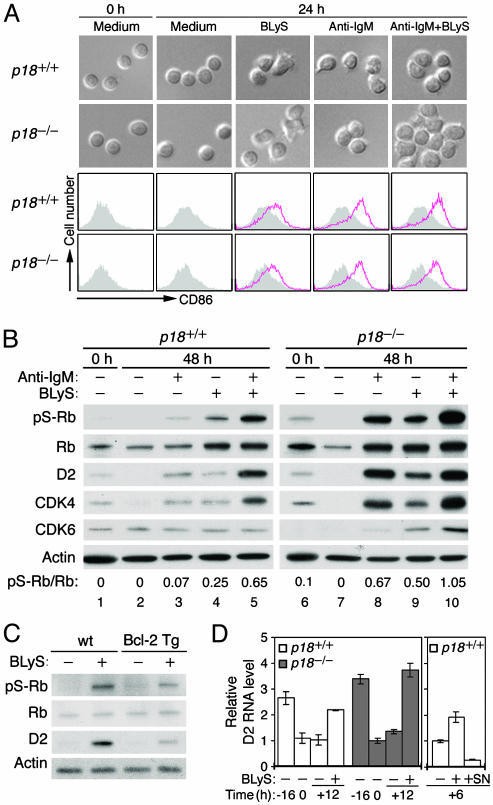
BLyS Induces Cell-Cycle Entry in Opposition to p18INK4c Through Activation of Cyclin D2 and CDK4. Activation of the cell cycle by BLyS without inducing G1/S transition is the most likely of these functions. Closer inspection of resting B cells within 24 h of BLyS stimulation revealed a change in cell shape, from spherical to oblong or even irregular (Fig. 2A). This change was accompanied by a modest increase in cell size based on forward-scattering analysis in flow cytometry (see Fig. 6, which is published as supporting information on the PNAS web site). Moreover, BLyS augmented CD23 expression as reported in ref. 36 (Fig. 6) and induced an early B cell activation marker, CD86, although also less compared with that in anti-IgM stimulation (Fig. 2 A). Neither the change in cell shape and size nor the induction of CD23 and CD86 by BLyS requires p18 (Fig. 2 A and data not shown). Thus, BLyS independently induces cell growth and cellular differentiation characteristic of early B cell activation by BCR stimulation. Together with the ability of BLyS to accelerate S-phase entry and cell division in BCR signaling (Fig. 1 D and E), these results suggest that in addition to attenuation of apoptosis, BLyS induces cell-cycle entry and limited G1 cell-cycle progression.
Fig. 2.
BLyS induces cell-cycle entry in opposition to p18. (A) Phase contrast microscopy and FACS analysis of CD86 expressed on resting p18+/+ and p18-/- B cells cultured in the absence (shaded area) or presence (red open area) of BLyS, anti-IgM, or both, for 0 or 24 h. (B and C) Immunoblot of pS-Rb, Rb, cyclin D2, CDK4, and CDK6, in cells cultured in A for 0 and 48 h (B) and in resting B cells isolated from Bcl-2 transgenic mice (Tg) and wild-type controls (wt) after stimulation with BLyS for 48 h (C). The ratio of pS-Rb to total Rb was determined by phosphoimaging (B). The actin level was used for a loading control. (D) Real-time RT-PCR analysis of cyclin D2 RNA levels in freshly isolated resting p18+/+ and p18-/- B cells (-16), after 16-h incubation in medium containing 0.5% serum (0), plus an additional 12 h in the absence (-) or presence (+) of BLyS (Left) or an additional 6 h in the absence (-) or presence (+) of BLyS plus SN50 (SN) (Right). Similar results were obtained without reducing the medium serum levels. The data are representative of four independent experiments.
To test this hypothesis, we investigated the regulation of cyclin D2 and its catalytic partners, CDK4 and CDK6, by BLyS and p18 near the onset of DNA replication on day 2. As a functional assay, we determined CDK4/6-specific phosphorylation of Ser807/811 on Rb (pS-Rb) (37, 38), which is indicative of early G1 cell-cycle progression. Cyclin D2 and CDK4 were modestly expressed in freshly isolated resting B cells and reduced to an undetectable level during incubation in the medium. They were increased coordinately by anti-IgM stimulation to modestly activate pS-Rb (Fig. 2B, lanes 1-3). Importantly, BLyS similarly elevated the levels of cyclin D2 and CDK4, as well as Rb. This increase led to greater phosphorylation of Rb than BCR signaling based on the ratios of pS-Rb to Rb (Fig. 2B, lane 4). Costimulation with BLyS and anti-IgM synergistically increased cyclin D2, CDK4, and pS-Rb (Fig. 2B, lane 5). This enhancement was not due to preservation or amplification of a specific subset of resting splenic B cell (T1, T2, marginal, and follicular), because the proportions among them were largely maintained in the presence of BLyS, while shifting in the absence of BLyS to favor T1 B cells that were less reliant on BLyS for survival (Fig. 6). The lack of CDK6 activation by BLyS and anti-IgM further indicates that CDK4 activation is specific and may be rate-limiting for serine phosphorylation of Rb. Collectively, these results demonstrate that BLyS alone induces pS-Rb by coordinated activation of cyclin D2 and CDK4 and that through convergent activation of these positive early G1 cell-cycle regulators BLyS and BCR signaling synergistically induces G1 cell-cycle progression.
The absence of p18 profoundly enhanced pS-Rb in BCR and BLyS signaling, alone or together (Fig. 2B, lanes 6-10). This enhancement was accompanied by a striking increase in the levels of cyclin D2, CDK4, Rb, and even CDK6 in BLyS signaling. Because DNA replication in p18-/- B cells on day 2 was modest in BCR signaling and barely detectable in BLyS signaling, these increases were restricted to G1 (Fig. 1B and data not shown). Given that the p18 function is dependent on CDK4 and not CDK6 (39), enhanced pS-Rb in the absence of p18 is mediated mainly by CDK4. Thus, p18 suppresses pS-Rb by inhibiting both the CDK4 catalytic activity and the synthesis of cyclin D2 and CDK4 proteins in either BCR or BLyS signaling. BLyS and p18 therefore antagonistically control cell-cycle entry through opposing regulation of cyclin D2, CDK4, and Rb phosphorylation.
BLyS Activates Cyclin D2 Independent of Its Survival Function Through NF-κB. To verify that BLyS induces cell-cycle entry independent of its survival function, the BLyS response was further characterized in resting splenic B cells isolated from mice transgenic for Bcl-2. Cyclin D2 protein and pS-Rb were enhanced by BLyS stimulation in these cells, although to a lesser extent than their wild-type littermate controls (Fig. 2C). The basis for the blunted BLyS response in B cell-overexpressing Bcl-2 is unknown, but BLyS clearly regulates cyclin D2 and pS-Rb independent of its survival function. BLyS activates NF-κB (18), which regulates both cyclin D2 and CD86 (31, 34). Real-time RT-PCR analysis revealed that the cyclin D2 RNA was expressed in resting B cells, reduced during overnight incubation in the medium regardless of serum levels (0.5-10%), and then increased by 12 h BLyS stimulation (2-fold in p18+/+ cells and 3- to 4-fold in p18-/- cells) to levels comparable with those before incubation (Fig. 2D). These results confirmed that BLyS activates cyclin D2 synthesis. The greater increase in the cyclin D2 protein by BLyS in the absence of p18 (Fig. 2B) is therefore due in part to enhanced RNA synthesis, and not solely to stabilization of the cyclin D2 protein in complex with CDK4. The time course parallels NF-κB activation in resting B cells and its diminution in vitro (18), consistent with NF-κB mediating the in vivo growth factor signals including BLyS for transcriptional activation of cyclin D2 in resting B cells. Indeed, inhibition of NF-κB activation with SN50, a cell-permeable peptide that blocks nuclear translocation of p50 (NF-κB1)-containing complexes, ablated the induction of cyclin D2 RNA by BLyS (Fig. 2D). BLyS therefore enhances cyclin D2 synthesis, at least in part, through NF-κB-mediated transcriptional activation independent of the BLyS survival function.
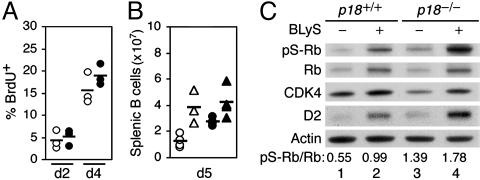
BLyS and p18INK4c Antagonistically Regulate Cyclin D2 and the Cell Cycle in the Antibody Response. The BLyS action is concentration-dependent (18). We therefore addressed the physiologic relevance of cell-cycle and homeostatic controls by p18 and BLyS in the antibody response to NP-Ficoll, a well characterized T cell-independent antigen. BrdUrd uptake and splenic B cell expansion after immunization was greater in p18-deficient mice than in their wild-type counterparts (Fig. 3 A and B). However, although administration of BLyS increased the number of splenic B cells as expected, the proportional increase in the p18-deficient mice was blunted, reaching a level similar to that of the BLyS-treated wild-type mice despite a higher baseline (Fig. 3B). These results suggest that p18 attenuates the cell cycle and cooperates with BLyS in homeostatic control of splenic B cell expansion in the NP antibody response, as in BCR signaling in vitro (Fig. 1 B and F).
Fig. 3.
BLyS and p18 antagonistically regulate the cell cycle in the antibody response. (A) FACS analysis of BrdUrd uptake in activated splenic B cells of p18+/+ and p18-/- mice on days (d) after NP-Ficoll immunization. (B) The number of total splenic B cells in p18+/+ and p18-/- mice on day 5 after immunization, with administration of PBS (circle) or BLyS (triangle). (C) Immunoblot of pS-Rb, Rb, CDK4, and cyclin D2 in activated p18+/+ and p18-/- splenic B cells isolated on day 4 after immunization, with or without BLyS administration. The results are representative of three independent experiments.
pS-Rb was detected along with a low level of cyclin D2 and strong CDK4 expression in splenic B cells at the height of replication on day 4 after immunization (Fig. 3C, lane 1). This finding indicates that CDK4 is activated by multiple in vivo signals, but induction of cyclin D2 is more specific and is rate-limiting for Rb phosphorylation in the antibody response. Consistent with this possibility, the cyclin D2 and Rb protein levels as well as pS-Rb were coordinately increased by BLyS (Fig. 3C, lane 2), more prominently in p18-/- splenic B cells than in their wild-type counterparts (Fig. 3C, lanes 3 and 4). Again, in keeping with regulation of CDK4 in vivo being more complex, the CDK4 level remained unchanged (Fig. 3C, lanes 1-4). BLyS and p18, therefore, antagonistically regulate cyclin D2 synthesis and Rb phosphorylation in the antibody response. The remarkable conservation of antagonistic cell-cycle regulation and cooperative homeostatic control by BLyS and p18, from cell-cycle entry in BCR signaling in vitro to asynchronously replicating splenic B cells in antigen-specific antibody response, indicate their physiologic role.
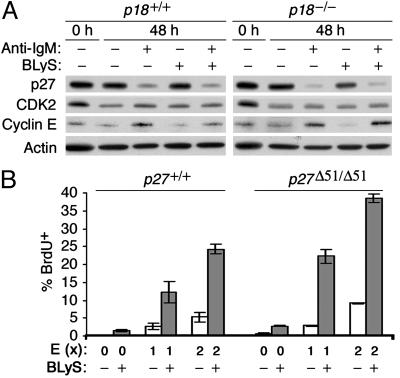
BLyS Fails to Induce G1/S Cell-Cycle Transition Due to Its Inability to Increase Cyclin E or Reduce p27. We then addressed the basis for the failure of BLyS to induce G1/S cell-cycle transition. Activation of resting B cells to enter the S phase by anti-IgM stimulation was accompanied by both an increase in cyclin E, which is necessary for S-phase entry, and a decrease in p27Kip1, which inhibits the catalytic activity of cyclin E/CDK2 (1) (Fig. 4A). This finding is consistent with observations in total splenic B cells (34, 40). In striking contrast, BLyS signaling modestly reduced cyclin E and sustained p27 expression, although maintaining CDK2 expression as BCR signaling (Fig. 4A). BLyS did not, however, interfere with the regulation of cyclin E and p27 by anti-IgM in costimulation.
Fig. 4.
BLyS fails to induce G1/S transition. (A) Immunoblot of p27, CDK2, and cyclin E in resting p18+/+ and p18-/- B cells cultured for 0 or 48 h as indicated. The level of actin was used for a loading control. (B) FACS analysis of BrdUrd uptake in resting p27+/+ and p27Δ51/Δ51 B cells cultured in the absence or presence of BLyS, 33 nM (1×) or 66 nM (2×) of HIV-1 transactivator of transcription cyclin E (E), or both, for 48 h in the presence of the lipopolysaccharide inhibitor. Similar results were obtained from five independent experiments regardless of the presence of the lipopolysaccharide inhibitor.
This set of findings poses two related questions. The first is whether p27 inhibits G1/S transition after cell-cycle entry is induced by BLyS. The second is whether cyclin E is required for the initial G1/S transition in response to BLyS, considering that in mouse embryonic fibroblasts cyclin E is required for cell-cycle reentry after serum starvation but not for cell-cycle progression (41). Transduction of a HIV-1 transactivator of transcription-cyclin E protein (34) into resting B cells led to a dose-dependent increase of S-phase cells (3% at 33 nM and 6% at 66 nM). Cyclin E therefore can bypass G1 activation and induce S-phase entry in resting B cells (Fig. 4B). Importantly, stimulation of HIV-1 transactivator of transcription-cyclin E transduced cells with BLyS profoundly increased the proportion of S-phase cells (25%) (Fig. 4B), demonstrating an extraordinary complementation between induction of cell-cycle entry by BLyS and expression of cyclin E. This complementation was amplified in p27-deficient B cells, as evidenced by the heightened proportion of S-phase cells (40%) (Fig. 4B). Thus, the failure of BLyS to drive cells past G1/S transition lies in its inability to increase cyclin E and reduce p27.
Discussion
This study reveals that BLyS, a key B cell survival factor, potently induces cell-cycle entry and early G1 progression and that p18, an early G1 CDKI required for plasma cell differentiation, also attenuates cell-cycle entry in BCR signaling. BLyS and p18 function antagonistically in G1 cell-cycle control and cooperatively in homeostatic control of B cell activation and the antibody response. On this basis, we propose a “homeostatic cell-cycle control” model (Fig. 5).
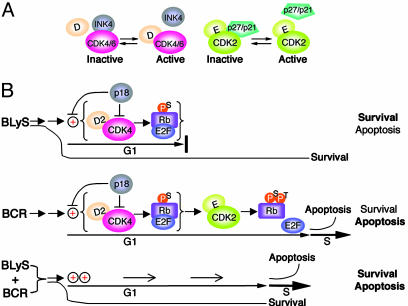
Fig. 5.
A model of homeostatic cell-cycle control. (A) Inactivation of CDK4/CDK6 by the INK4 CDKIs, and CDK2 by the Cip/Kip CDKIs. (B Top) BLyS induces cyclin D2 and CDK4 in opposition to p18, leading to Ser807/811 phosphorylation of Rb and early G1 progression, but not S-phase entry. The BLyS survival function is unaffected, and the outcome favors cell survival. By inhibiting both the CDK4 activity and the induction of cyclin D2 and CDK4, p18 attenuates pS-Rb and G1 progression as well as apoptosis. (Middle) BCR signaling induces serine phosphorylation of Rb as in BLyS signaling, which facilitates threonine phosphorylation of Rb by cyclin E/CDK2 and G1/S transition. Without intervention, the outcome favors apoptosis because of the release of an apoptosis factor. p18 attenuates the cell cycle and apoptosis as in BLyS signaling. (Bottom) BLyS and BCR costimulation leads to synergistic induction of cyclin D2 and CDK4, pS-Rb, accelerated G1 progression, and G1/S transition and increases the propensity of apoptosis. The balance between survival and apoptosis is subject to composite control by the strength of BCR and BLyS signaling and the presence of p18.
In this model, BLyS and BCR signaling cooperatively control cell-cycle entry and early G1 progression in opposition to p18 and diverge in the control of G1/S transition through differential regulation of cyclin E and p27. In addition, cell-cycle control by BLyS and p18 is linked functionally to apoptotic control. BLyS induces resting B cells to enter the cell cycle by activating the synthesis of cyclin D2, CDK4, and Rb, but not the S phase due to insufficient cyclin E and sustained p27 expression. Consequently, BLyS signaling does not lead to the release of a putative apoptotic factor, thereby preserving its full survival function. By contrast, BCR signaling induces sequential phosphorylation of Rb by cyclin D2-CDK4/6 and cyclin E-CDK2, G1/S transition and the release of the apoptotic factor. In BLyS and BCR costimulation, synergistic phosphorylation of Rb by cyclin D2-CDK4 accelerates early G1 progression and facilitates cyclin E CDK2 phosphorylation and the release of the putative apoptotic factor. This G1 acceleration, in turn, compromises the BLyS survival function, before and during G1/S transition. p18 effectively attenuates G1 progression by dual mechanisms, direct inhibition of the CDK4 activity and suppression of cyclin D2 and CDK4 synthesis, and attenuates the release of the putative antiapoptotic factor. In this way, the BLyS survival function is modulated by the control of cell-cycle entry and G1/S transition according to the strength of BCR and BLyS signaling and the expression of p18 and p27.
This model is supported by the following findings: (i) the p18 function is CDK4-dependent (39); (ii) cyclin D2 is the major D cyclin expressed in B cells (30) and is required for cell-cycle activation early in BCR signaling in vitro (29); (iii) cyclin D2 is transcriptionally activated by BLyS by means of NF-κB activation but independent of the BLyS survival function (Fig. 2); (iv) attenuation of apoptosis by BLyS is dose-dependent but cell-cycle-independent (18); and (v) cell-cycle and homeostatic controls of B cell expansion by BLyS and p18 is conserved from B cell activation in vitro (Figs. 1 and 2) to the antibody response (Fig. 3).
Incomplete cell-cycle activation by BLyS may in fact be a way both to prevent resting mature B cells from premature replication, thereby maintaining the resting B cell repertoire, and to facilitate their activation by antigen and cytokines. Precedent of complementary cell-cycle induction exists in cell-cycle control of T cell activation, where cell-cycle reentry and progression requires two sequential signals: T cell receptor stimulation, which induces cyclins and CDKs but not G1/S transition, and IL-2 signaling, which inactivates p27 (ref. 42 and A. Koff, personal communication). Reconstitution of S-phase entry by BLyS and cyclin E, particularly in p27-deficient B cells (Fig. 4), suggests that composite regulation of G1/S transition also may be a means of integrating cytokine signals for cell-cycle control in B cell activation. BLyS regulates nearly all functions of mature B cells (12-21), most likely according to signaling strength, considering that greater dependence on BLyS led to reduced competitiveness of autoantigen-engaged B cells (6). Cooperative cell-cycle and apoptotic control may underlie the diverse biological functions of BLyS. Additional experimental proof certainly will be required to validate this model, but the stage is set to explore further cell-cycle control of homeostasis in B cell immunity.
Supplementary Material
Acknowledgments
We thank David Hilbert and Human Genome Sciences for the recombinant human BLyS, Andrew Koff for the p27-deficient mice and insightful comments, and Lee Kiang for a critical reading of the manuscript. This work was supported by British Medical Research Council Program research grants (to I.C.M.M.) and National Institutes of Health Grants CA 80204 and AR 49436 and a Specialized Center of Research grant from the Leukemia and Lymphoma Society (to S.C.-K.).
Author contributions: X.H., M.D., and S.C.-K. designed research; X.H., M.D., L.K., S.E., and S.C.-K. performed research; X.H., M.D., and S.C.-K. analyzed data; S.C. and H.-c.L. contributed new reagents/analytic tools; and X.H., M.D., A.F.C., I.C.M.M., and S.C.-K. wrote the paper.
This paper was submitted directly (Track II) to the PNAS office.
Abbreviations: BCR, B cell receptor; CDK, cyclin-dependent kinase; CDKI, CDK inhibitor; CFSE, carboxyfluorescein diacetate succinimidyl ester; NP, 4-hydroxy-3-nitrophenylacetyl; NP-Ficoll, NP coupled to Ficoll; Rb, retinoblastoma protein; pS-Rb, phosphorylation of serine on Rb.
References
- 1.Sherr, C. J. & Roberts, J. M. (1999) Genes Dev. 13, 1501-1512. [DOI] [PubMed] [Google Scholar]
- 2.Guan, K. L., Jenkins, C. W., Li, Y., Nichols, M. A., Wu, X., O'Keefe, C. L., Matera, A. G. & Xiong, Y. (1994) Genes Dev. 8, 2939-2952. [DOI] [PubMed] [Google Scholar]
- 3.Hirai, H., Roussel, M. F., Kato, J. Y., Ashmun, R. A. & Sherr, C. J. (1995) Mol. Cell. Biol. 15, 2672-2681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Morse, L., Chen, D., Franklin, D., Xiong, Y. & Chen-Kiang, S. (1997) Immunity 6, 47-56. [DOI] [PubMed] [Google Scholar]
- 5.Tourigny, M. R., Ursini-Siegel, J., Lee, H., Toellner, K. M., Cunningham, A. F., Franklin, D. S., Ely, S., Chen, M., Qin, X. F., Xiong, Y., et al. (2002) Immunity 17, 179-189. [DOI] [PubMed] [Google Scholar]
- 6.Lesley, R., Xu, Y., Kalled, S. L., Hess, D. M., Schwab, S. R., Shu, H. B. & Cyster, J. G. (2004) Immunity 20, 441-453. [DOI] [PubMed] [Google Scholar]
- 7.Latres, E., Malumbres, M., Sotillo, R., Martin, J., Ortega, S., Martin-Caballero, J., Flores, J. M., Cordon-Cardo, C. & Barbacid, M. (2000) EMBO J. 19, 3496-3506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Moore, P. A., Belvedere, O., Orr, A., Pieri, K., LaFleur, D. W., Feng, P., Soppet, D., Charters, M., Gentz, R., Parmelee, D., et al. (1999) Science 285, 260-263. [DOI] [PubMed] [Google Scholar]
- 9.Schneider, P., MacKay, F., Steiner, V., Hofmann, K., Bodmer, J. L., Holler, N., Ambrose, C., Lawton, P., Bixler, S., Acha-Orbea, H., et al. (1999) J. Exp. Med. 189, 1747-1756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gross, J. A., Johnston, J., Mudri, S., Enselman, R., Dillon, S. R., Madden, K., Xu, W., Parrish-Novak, J., Foster, D., Lofton-Day, C., et al. (2000) Nature 404, 995-999. [DOI] [PubMed] [Google Scholar]
- 11.von Bulow, G. U. & Bram, R. J. (1997) Science 278, 138-141. [DOI] [PubMed] [Google Scholar]
- 12.Yan, M., Brady, J. R., Chan, B., Lee, W. P., Hsu, B., Harless, S., Cancro, M., Grewal, I. S. & Dixit, V. M. (2001) Curr. Biol. 11, 1547-1552. [DOI] [PubMed] [Google Scholar]
- 13.Gross, J. A., Dillon, S. R., Mudri, S., Johnston, J., Littau, A., Roque, R., Rixon, M., Schou, O., Foley, K. P., Haugen, H., et al. (2001) Immunity 15, 289-302. [DOI] [PubMed] [Google Scholar]
- 14.Harless, S. M., Lentz, V. M., Sah, A. P., Hsu, B. L., Clise-Dwyer, K., Hilbert, D. M., Hayes, C. E. & Cancro, M. P. (2001) Curr. Biol. 11, 1986-1989. [DOI] [PubMed] [Google Scholar]
- 15.Schiemann, B., Gommerman, J. L., Vora, K., Cachero, T. G., Shulga-Morskaya, S., Dobles, M., Frew, E. & Scott, M. L. (2001) Science 293, 2111-2114. [DOI] [PubMed] [Google Scholar]
- 16.O'Connor, B. P., Raman, V. S., Erickson, L. D., Cook, W. J., Weaver, L. K., Ahonen, C., Lin, L. L., Mantchev, G. T., Bram, R. J. & Noelle, R. J. (2004) J. Exp. Med. 199, 91-98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Balazs, M., Martin, F., Zhou, T. & Kearney, J. (2002) Immunity 17, 341-352. [DOI] [PubMed] [Google Scholar]
- 18.Do, R. K., Hatada, E., Lee, H., Tourigny, M. R., Hilbert, D. & Chen-Kiang, S. (2000) J. Exp. Med. 192, 953-964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Litinskiy, M. B., Nardelli, B., Hilbert, D. M., He, B., Schaffer, A., Casali, P. & Cerutti, A. (2002) Nat. Immunol. 3, 822-829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Khare, S. D., Sarosi, I., Xia, X. Z., McCabe, S., Miner, K., Solovyev, I., Hawkins, N., Kelley, M., Chang, D., Van, G., et al. (2000) Proc. Natl. Acad. Sci. USA 97, 3370-3374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mackay, F., Woodcock, S. A., Lawton, P., Ambrose, C., Baetscher, M., Schneider, P., Tschopp, J. & Browning, J. L. (1999) J. Exp. Med. 190, 1697-1710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Moreaux, J., Legouffe, E., Jourdan, E., Quittet, P., Reme, T., Lugagne, C., Moine, P., Rossi, J. F., Klein, B. & Tarte, K. (2004) Blood 103, 3148-3157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Novak, A. J., Darce, J. R., Arendt, B. K., Harder, B., Henderson, K., Kindsvogel, W., Gross, J. A., Greipp, P. R. & Jelinek, D. F. (2004) Blood 103, 689-694. [DOI] [PubMed] [Google Scholar]
- 24.Batten, M., Groom, J., Cachero, T. G., Qian, F., Schneider, P., Tschopp, J., Browning, J. L. & Mackay, F. (2000) J. Exp. Med. 192, 1453-1466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Claudio, E., Brown, K., Park, S., Wang, H. & Siebenlist, U. (2002) Nat. Immunol. 3, 958-965. [DOI] [PubMed] [Google Scholar]
- 26.Hatada, E. N., Do, R. K., Orlofsky, A., Liou, H. C., Prystowsky, M., MacLennan, I. C., Caamano, J. & Chen-Kiang, S. (2003) J. Immunol. 171, 761-768. [DOI] [PubMed] [Google Scholar]
- 27.Kayagaki, N., Yan, M., Seshasayee, D., Wang, H., Lee, W., French, D. M., Grewal, I. S., Cochran, A. G., Gordon, N. C., Yin, J., et al. (2002) Immunity 17, 515-524. [DOI] [PubMed] [Google Scholar]
- 28.Hsu, B. L., Harless, S. M., Lindsley, R. C., Hilbert, D. M. & Cancro, M. P. (2002) J. Immunol. 168, 5993-5996. [DOI] [PubMed] [Google Scholar]
- 29.Solvason, N., Wu, W. W., Parry, D., Mahony, D., Lam, E. W., Glassford, J., Klaus, G. G., Sicinski, P., Weinberg, R., Liu, Y. J., et al. (2000) Int. Immunol. 12, 631-638. [DOI] [PubMed] [Google Scholar]
- 30.Tanguay, D. A. & Chiles, T. C. (1996) J. Immunol. 156, 539-548. [PubMed] [Google Scholar]
- 31.Hinz, M., Krappmann, D., Eichten, A., Heder, A., Scheidereit, C. & Strauss, M. (1999) Mol. Cell. Biol. 19, 2690-2698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Franklin, D. S., Godfrey, V. L., Lee, H., Kovalev, G. I., Schoonhoven, R., Chen-Kiang, S., Su, L. & Xiong, Y. (1998) Genes Dev. 12, 2899-2911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kiyokawa, H., Kineman, R. D., Manova-Todorova, K. O., Soares, V. C., Hoffman, E. S., Ono, M., Khanam, D., Hayday, A. C., Frohman, L. A. & Koff, A. (1996) Cell 85, 721-732. [DOI] [PubMed] [Google Scholar]
- 34.Cheng, S., Hsia, C. Y., Leone, G. & Liou, H. C. (2003) Oncogene 22, 8472-8486. [DOI] [PubMed] [Google Scholar]
- 35.Lyons, A. B. & Parish, C. R. (1994) J. Immunol. Methods 171, 131-137. [DOI] [PubMed] [Google Scholar]
- 36.Gorelik, L., Cutler, A. H., Thill, G., Miklasz, S. D., Shea, D. E., Ambrose, C., Bixler, S. A., Su, L., Scott, M. L. & Kalled, S. L. (2004) J. Immunol. 172, 762-766. [DOI] [PubMed] [Google Scholar]
- 37.Knudsen, E. S. & Wang, J. Y. (1996) J. Biol. Chem. 271, 8313-8320. [DOI] [PubMed] [Google Scholar]
- 38.Zarkowska, T. & Mittnacht, S. (1997) J. Biol. Chem. 272, 12738-12746. [DOI] [PubMed] [Google Scholar]
- 39.Pei, X. H., Bai, F., Tsutsui, T., Kiyokawa, H. & Xiong, Y. (2004) Mol. Cell. Biol. 24, 6653-6664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Solvason, N., Wu, W. W., Kabra, N., Wu, X., Lees, E. & Howard, M. C. (1996) J. Exp. Med. 184, 407-417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Geng, Y., Yu, Q., Sicinska, E., Das, M., Schneider, J. E., Bhattacharya, S., Rideout, W. M., Bronson, R. T., Gardner, H. & Sicinski, P. (2003) Cell 114, 431-443. [DOI] [PubMed] [Google Scholar]
- 42.Firpo, E. J., Koff, A., Solomon, M. J. & Roberts, J. M. (1994) Mol. Cell. Biol. 14, 4889-4901. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.