Abstract
The contractile actomyosin cytoskeleton and its connection to the plasma membrane are critical for control of cell shape and migration. We identify three STRIPAK complex components, FAM40A, FAM40B, and STRN3, as regulators of the actomyosin cortex. We show that FAM40A negatively regulates the MST3 and MST4 kinases, which promote the co-localisation of the contractile actomyosin machinery with the Ezrin/Radixin/Moesin family proteins by phosphorylating the inhibitors of PPP1CB, PPP1R14A-D. Using computational modelling, in vitro cell migration assays and in vivo breast cancer metastasis assays we demonstrate that co-localisation of contractile activity and actin-plasma membrane linkage reduces cell speed on planar surfaces, but favours migration in confined environments similar to those observed in vivo. We further show that FAM40B mutations found in human tumours uncouple it from PP2A and enable it to drive a contractile phenotype, which may underlie its role in human cancer.
Introduction
Cell migration is fundamental to multicellular organism development and pathology1, 2. The actin cytoskeleton controls cell migration by changing cell shape: actin polymerisation pushes the plasma membrane forward, whereas actomyosin contraction pulls the plasma membrane and generates hydrostatic pressure3, 4. Moreover, actin coupling to the surrounding matrix or neighbouring cells through cell adhesion molecules enables force transmission and cell migration5–8. Migrating cells have different morphologies depending on the balance of these factors9. Fibroblasts move with elongated morphologies and apply force to surrounding matrices through integrin-mediated adhesions. Leukocytes have more rounded and rapidly changing morphologies and move through interstitial collagen independently of integrins10, 11, but still depend on a contractile actomyosin cytoskeleton for migration. Invasive cancer cells can use both modes of migration10, 12. Despite knowing many of the factors that connect the actin cytoskeleton to integrins or the plasma membrane, the molecular ‘tuning’ mechanism that determines whether the actin network is predominantly coupled to the plasma membrane or integrins, and which is predicted to influence the mode of cancer cell migration, and thereby metastasis, remains unclear.
Actomyosin contractility is regulated by the phosphorylation of myosin light chain (MLC2/MYL9) on T18 and S196. MLC2 can be phosphorylated by many kinases, including MLCK, MRCKα&β, and ROCK1&26, and is dephosphorylated by protein phosphatase 1 (PP1) complexes13, 14. Targeting of the catalytic PPP1CB subunit to the actomyosin cytoskeleton requires PPP1R12A (MYPT1/MBS) and is negatively regulated by ROCK1/2-mediated phosphorylation of PPP1R12A6, 15. The contractile actomyosin network can transmit force to the surrounding matrix by being coupled to integrin complexes16. Alternatively, it can be coupled to the plasma membrane through linker proteins such as the Ezrin/Radixin/Moesin (ERM) family17, resulting cell morphology changes and generation of hydrostatic pressure. ERM protein linkage to the plasma membrane is regulated by phosphorylation of Ezrin (Thr567)/Radixin (Thr564)/Moesin (Thr558)17, which are also dephosphorylated by PP1 complexes18.
Protein phosphatase 2 (PP2) complexes contain multiple regulatory (B, B’, B’’, & B’’’/striatin) and scaffolding subunits19 and can also affect cell migration and actomyosin cytoskeleton20, 21. The striatin/cka regulatory subunit is required for the actomyosin-driven dorsal closure process in Drosophila and other subunits are implicated in cell morphology control22, 23. In mammals, core PP2A proteins such as striatin form part of the multi-subunit STRIPAK complex24–28, which also includes MST family kinases24. MST3, MST4 and SOK1 are involved in Golgi morphogenesis, cell polarity, ERM protein phosphorylation, and regulation of tyrosine phosphatases and can affect cell migration29–34.
Here we propose that the STRIPAK complex regulates the mode of cancer cell migration by controlling the activity of MST3/4, which locally co-ordinate the phosphorylation of ERM proteins and inhibit the dephosphorylation of myosin light chain leading to increased actin-membrane linkage.
Results
FAM40A and FAM40B regulate cell contractility
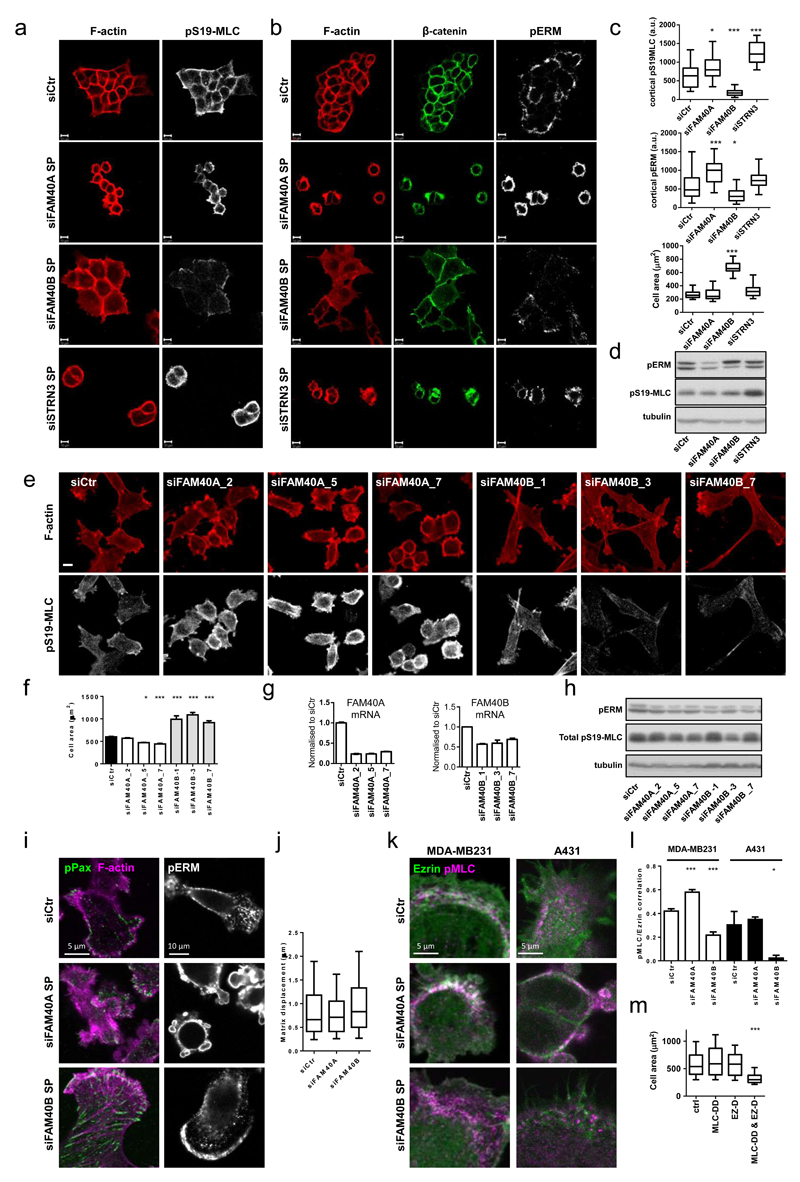
To identify regulators of actin-based cell migration, we performed a screen for genes required for border cell migration and actin organisation in follicle cells within the Drosophila egg chamber35. The results of this screen are presented in Supplementary Figure 1. Given that several of the genes involved in this well-studied cell migration system are also involved in cancer cell dissemination36–41, we hypothesized that the human homologues of the identified genes may play a role in the of cancer cell migration. We selected 85 human homologues of 43 Drosophila genes involved in border cell migration or follicle cell architecture for further analysis (Supplementary Table 1). Depletion of 32 of these genes in human A431 carcinoma cells led to altered F-actin organisation (Supplementary Figure 1c and Supplementary table 1). We focused on FAM40A, FAM40B, and STRN3 as they are known to form a physical complex24, 42. Depletion of Drosophila FAM40 and STRN (Cka/CG7392) delayed border cell migration (Supplementary Figure 1d&e), which was associated with perturbed localisation of pMoesin (Supplementary Figure 1f). FAM40A and STRN3 depletion in A431 cells led to increased cortical pS19-MLC, pT18pS19-MLC staining and phospho-ERM staining, frequent membrane blebs (Figure 1a-c, Supplementary Figure 2a&b and Supplementary Movie 1) and perturbed β-catenin and E-cadherin staining (Figure 1b and Supplementary Figure 2b). In contrast, FAM40B depleted cells had increased cell area, prominent lamellipodia, and less intense cortical pS19-MLC, pT18S19-MLC, and pERM staining (Figure 1a-c, Supplementary Figure 2a&b and Supplementary Movie 1). Multiple siRNAs against FAM40A, and FAM40B gave consistent phenotypes (Supplementary Figure 2c and d). As the total levels of pS19-MLC, pT18S19-MLC and pERM were not greatly changed in FAM40A or FAM40B depleted cells (Figure 1d), they likely regulate a pool of pMLC and pERM associated with the cell cortex.
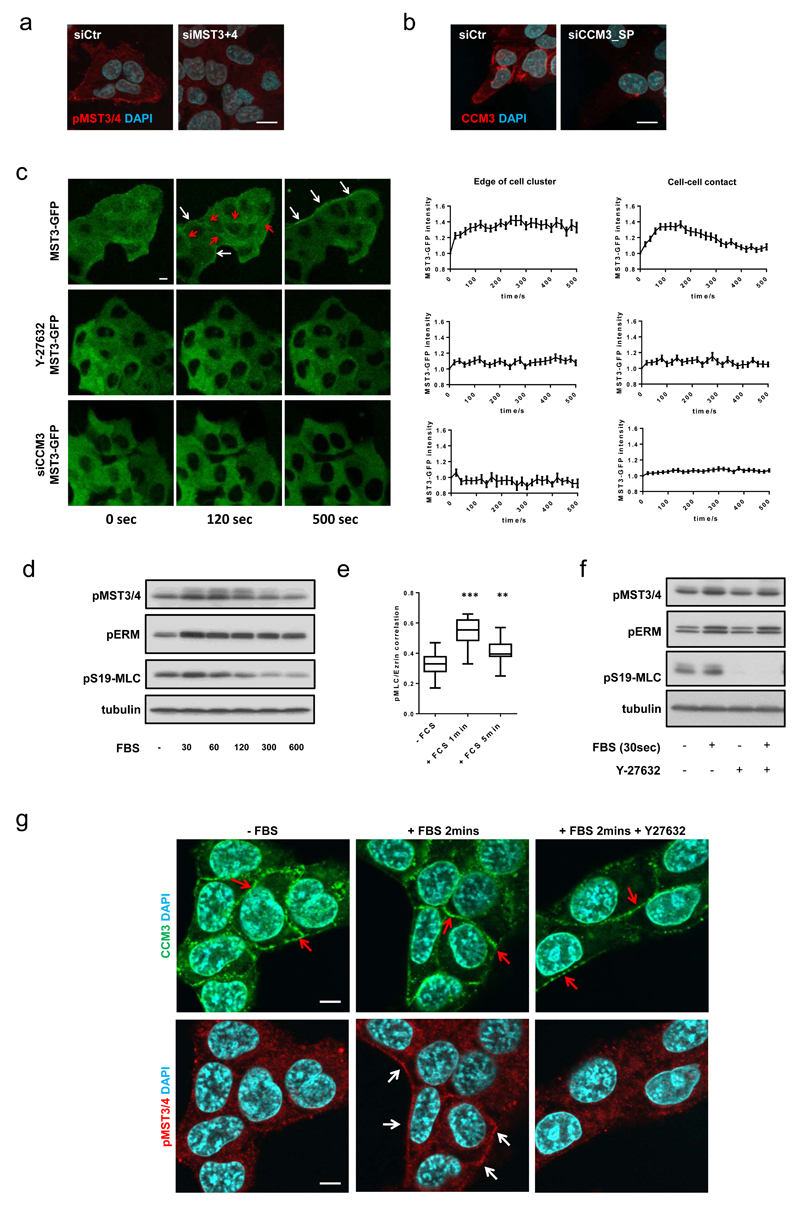
Figure 1. FAM40A, FAM40B and STRN3 regulate the cytoskeleton.
(a-b) Confocal sections of A431 cells depleted of FAM40A, FAM40B and STRN3 are shown plated on collagen-I/Matrigel 72h post-transfection and stained for pS19-MLC (white), β-catenin (green), pERM (white), and F-actin (red). (c) Quantification of A431 mean fluorescence of cortical pS19-MLC2 (upper panel) and cortical pERM (middle panel) intensities and cell area (lower panel). Box and whiskers graph: Line=Median, Box=distribution of 50% of values, Whiskers=distribution of min. and max. values. n=fields of cells; upper histogram (siCtr, 24; siFAM40A, 33; siFAM40B, 29; siSTRN3, 12; middle histogram (siCtr, 30; siFAM40A, 22; siFAM40B, 19; siSTRN3, 18); lower histogram (siCtr, 25; siFAM40A, 19; siFAM40B, 14; siSTRN3, 21). (d) Immunoblotting of pERM and pS19-MLC2 in depleted A431 cells. (e) MDA-MB231 cells are shown72h post-transfection - pS19-MLC2 (white) and F-actin (red) plated on collagen-I/matrigel. (f) MDA-MB231 cell area +/-s.e.m. (n=fields of cells; siCtr, 68; siFAM40A-2, 11; siFAM40A-5, 11; siFAM40A-7, 42; siFAM40B-1, 5; siFAM40B-3, 8; siFAM40B-7,30). (g) Knockdown efficiency +/-s.e.m of FAM40A and FAM40B mRNA using individual siRNA’s compared to control cells n=3 technical replicates. (h) Immunoblotting of pERM and pS19-MLC2 in depleted MDA-MB231 cells. (i) Immunofluorescence of pY118-paxillin (green), F-actin (magenta) and pERM (white) in depleted MDA-MB231 cells. (j) Analysis of matrix displacement (in microns) by MDA-MB231 cells transfected with the indicated siRNAs. Box and whiskers graph: Line=Median, Box=distribution of 50% of values, Whiskers=10-90 percentile. n=number of beads measured (siCtr, 3019; siFAM40A, 6657; siFAM40B, 6932). (k) Co-localisation analysis of pS19-MLC2 (magenta) and Ezrin-GFP (green) in depleted A431 and MDA-MB231 cells. (l) Quantification of pS19-MLC2/Ezrin-GFP co-localisation +/-s.e.m in MDA-MB231 cells (white bars, n=cells; siCtr,36; siFAM40A,24; siFAM40B,19) and A431 cells (black bars, n=fields of cells; siCtr,3; siFAM40A,2; siFAM40B,3). (m) MDA-MB231 cells were transfected with GFP, MLC2-T18D-S19D-GFP (MLC-DD), Ezrin-T567D (EZ-D) or double transfected with MLC2-T18D-S19D-GFP and Ezrin-T567D. GFP positive cells were analysed 72h after transfection for cell area. Box and whiskers graph: Line=Median, Box=distribution of 50% of values, Whiskers=10-90 percentile. (n=cells; Ctr,120; MLC,79; Ez,106; MLC&Ez,108). All statistical test were performed using 1way ANOVA, Sidak’s multiple comparison test, *P<0.05, **P<0.01, ***P<0.001. Scale bars, 10 μm, unless indicated otherwise.
To test if FAM40A&B influence cell morphology in cancer cells that have undergone epithelial-mesenchymal transition (EMT) we depleted FAM40A and FAM40B in MDA-MB231 cells, yielding similar phenotypes with the epitheliod A431 cell line (Figure 1e-I and Supplementary Figure 2d). FAM40A depletion increased cortical pS19-MLC and pERM staining, and reduced cell area (Figure 1F and Supplementary Fig. 2E). FAM40B depletion had the opposite effect, however pS19-MLC was still observed in stress fibres (Figure 1e) and pERM in a crescent in the lamellipod (Figure 1i). Although prominent focal adhesions were occasionally observed in FAM40B depleted cells (Figure 1i), quantitative analysis failed to reveal consistent changes in cell-ECM adhesion (Supplementary Figure 2f-h) and depletion of FAM40A or FAM40B did not affect the force-mediated matrix deformation by MDA-MB231 cells (Figure 1j).
FAM40A depletion promotes cortical localisation of both actomyosin and ERM proteins (Figure 1b,c,d,h,i and Supplementary Figure 2d,e). In contrast, when FAM40B is depleted pERM and pMLC localise in different parts of the cell. We confirmed this by measuring the spatial correlation co-efficient of pS19-MLC and Ezrin-GFP. MDA-MB231 cells have a spatial correlation co-efficient of ˜0.4, and this increases to ˜0.6 when FAM40A is depleted (Figure k&l – white bars). FAM40B depletion reduced the spatial correlation co-efficient to ˜0.2 in FAM40B depleted MDA-MB231 cells (Figure 1k&l). A more pronounced decrease in Ezrin-GFP/pS19-MLC co-localisation was observed in A431 cells (Figure 1k&l). As a control, the correlation of cytosolic GFP with pS19-MLC was ˜0.1 and did not vary when FAM40A and FAM40B were depleted (Supplementary Figure 2i). Structured Illumination Microscopy (SIM) demonstrated that the spatial correlation between non-muscle MyoIIa (MYH9) and Ezrin was regulated in the same manner as the pS19-MLC/Ezrin correlation (Supplementary Figure 2j). Gain of function analysis confirmed the relevance of co-ordinated MLC and ERM regulation to the contractile phenotype observed in FAM40A depleted cells. A reduction in cell area was only observed when expression of ‘activated’ phospho-mimetic mutants MLC-T18D,S19D (MLC-DD) and Ezrin-T567D (Ez-D) was combined (Figure 1m). These data indicate that co-ordinated regulation of pMLC and pERM is required to reduce cell area.
Interdependence of ROCK1&2 and FAM40A&B function
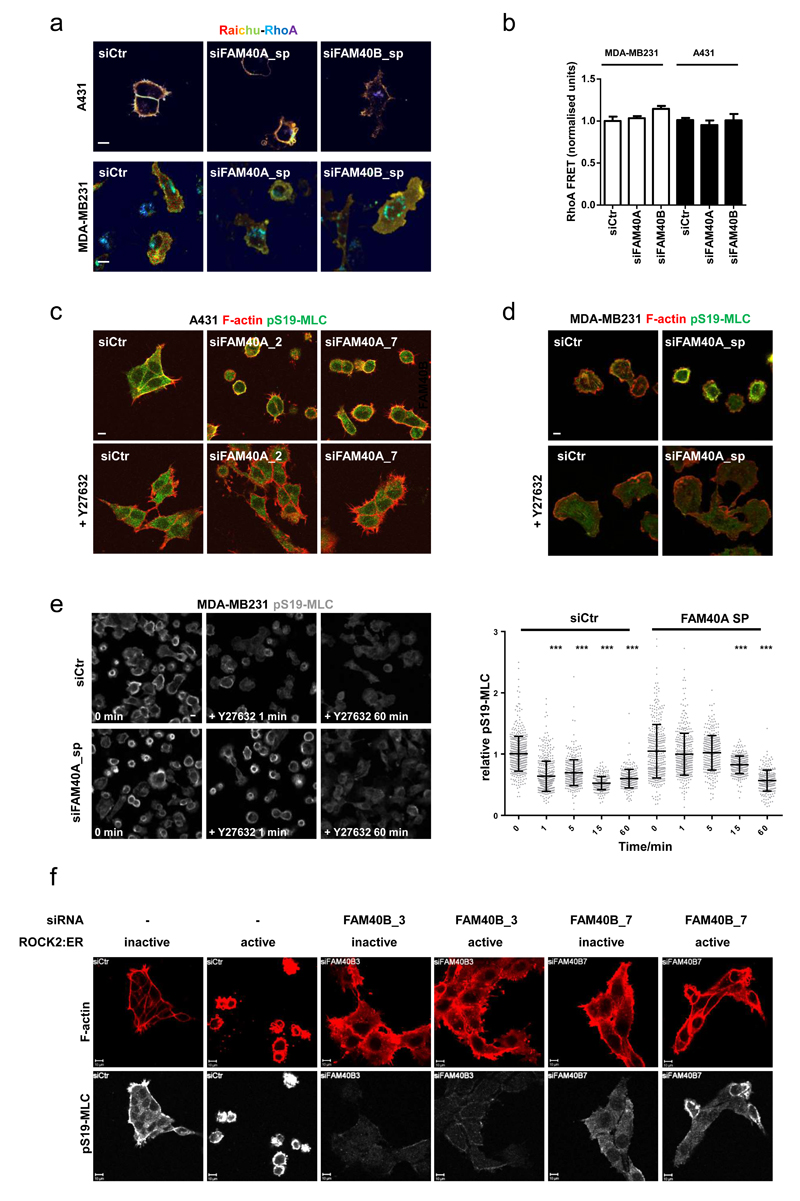
The data outlined above demonstrate that FAM40A and FAM40B are involved in the co-ordinated regulation of MLC and ERM proteins. As RhoA and its effectors, ROCK1&2, regulate both MLC and ERM protein phosphorylation43, we investigated the relationship between FAM40A&B and RhoA-ROCK signalling. Using a RhoA FRET biosensor we observed that RhoA activity at the cell cortex was not greatly changed by modulation of FAM40A or FAM40B levels (Figure 2a, 2b). We next investigated if there was any functional interaction between FAM40A, FAM40B, and ROCK1&2. The contractile phenotype (reduced cell area, increase cortical pS19-MLC, and membrane blebbing) of FAM40A-depleted cells was completely reversed by ROCK1&2 inhibition using Y27632 (Figures 2c, 2d). Temporal analysis revealed that the reduction in pS19-MLC levels following Y27632 treatment proceeded with slower kinetics in FAM40A depleted cells (Figure 2e)., indicating that the phosphatase for pS19-MLC could be less active in FAM40A depleted cells. Having established that ROCK function was required for the phenotype of FAM40A depleted cells, we tried the converse experiment by increasing ROCK2 function with a conditionally activated allele (ROCK2:ER – activated by 4-OH Tamoxifen)44. ROCK2:ER expressing cells became contracted when treated with 4-OHT, however this was prevented by FAM40B depletion (Figure 2f).
Figure 2. Inter-dependence of ROCK1&2 and FAM40A&B function.
(a) A431 and MDA-MB231 cells stably expressing RhoA-FRET biosensor were depleted for FAM40A and FAM40B and analysed for FRET signal. The images represent a ratiometric measurement, not FRET efficiency. (b) Ratiometric quantification of RhoA FRET signal +/-s.e.m in MDA-MB231 cells (white bars, n=fields of cells; siCtr, 15; siFAM40A, 10; siFAM40B, 14) and A431 cells (black bars, n=fields of cells; siCtr, 41; siFAM40A, 37; siFAM40B, 38). (c-d) A431 (c) and MDA-MB-231 (d) cells were transfected with siRNAs and replated for 48h. Cells were then treated with vehicle or 10μM ROCK inhibitor (Y-27632) for 18h before fixation. Staining of F-actin (red) and pS19-MLC2 (green) shows that ROCK1/2 inhibition rescues the contractile phenotypes observed for FAM40A. (e) Detailed temporal analysis revealed that the reduction in pS19-MLC levels following Y27632 treatment proceeded with slower kinetics in FAM40A depleted cells. A431 cells were depleted for FAM40A and plated on collagen-I/matrigels. Seventy-two hours post-transfection; the cells were treated with vehicle or 10μM Y-27632 and fixed at various time points. The cells were stained for pS19-MLC2 (images in left panel) and the mean fluorescence pS19-MLC2 intensities were quantified for individual cells (right panel). The intensities were normalised to time 0 (no Y-27632 treatment). n=cells; siCtr; (0, 504; 1, 498; 5, 399; 15, 291; 60, 243); siFAM40A (0, 380; 1, 402; 5, 332; 15, 312; 60, 254). (f) A431 cells expressing stable ROCK2:ER (active) or a kinase dead ROCK2:ER (inactive) were transfected with siRNA’s and then plated on collagen-I/matrigels. Cells were treated with 1μM tamoxifen for 18h before fixation. Staining shows F-actin (red) and pS19-MLC2 (white). All statistical test were performed using 1way ANOVA, Sidak’s multiple comparison test, *P<0.05, **P<0.01, ***P<0.001. All experiments were conducted at least 3 independent times. Scale bars, 10 μm.
MST3&4 act downstream of FAM40A
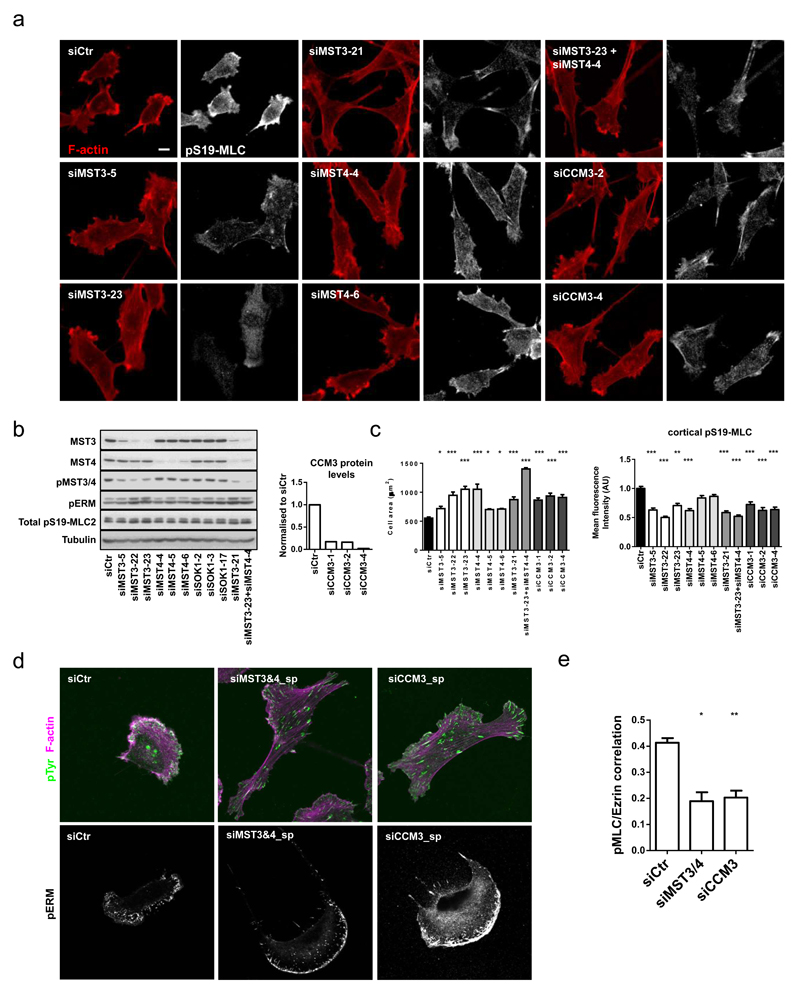
Having established that FAM40A&B are unlikely to be directly modulating the RhoA and ROCK1&2 activities, we sought other explanations for their cytoskeletal effects. FAM40A and FAM40B are part of the STRIPAK complex that contains both MST kinases and PP2A targeting subunits. Depletion of MST3 and MST4 and the CCM3 scaffold protein in MDA-MB231 cells, reduced cortical actomyosin, increased cell area, and generated an arc of pERM in the lamellipod (Figure 3a-d). Similar phenotypes were observed in A431 cells (Supplementary Figure 3a-d). MST4 depletion had a more modest effect than MST3 depletion (Figure 3a-d). Simultaneous depletion of MST3 and MST4 using either siRNAs targeting MST3 and MST4 independently or a single siRNA that targets both (MST3_21) resulted in a more pronounced phenotype (Figure 3a-c), which correlated with the reduction in active phosphorylated MST3&4 levels (compare Figure 3b and 3e - due to conservation of the activation loop a single phospho-antibody recognises both active MST3 and MST4). siRNA targeting the related kinases, MST1, MST2, and SOK1, did not affect cell morphology (Supplementary Figure 3e-h). Although cells lacking either MST3&4 or CCM3 had reduced cortical pS19-MLC and a more spread morphology, they still had pS19-MLC staining on stress fibres (Figure 3a), and reducing MST3&4 levels did not affect total pS19-MLC levels, but led to a slight increase in pERM (Figure 3b). Moreover, pS19-MLC and Ezrin-GFP where spatially segregated following depletion of MST3&4 and CCM3 (Figure 3e).
Figure 3. MST3, MST4 and CCM3 regulate the cytoskeleton in human cancer cells.
(a) Deconvolution of siRNA oligo’s in MDA-MB231 cells plated on collagen-I/matrigel. Cells were stained 72h post-transfection against pS19-MLC2 (white) and F-actin (red). (b) Immunoblotting analysis of MDA-MB231 cells after depletion of MST3, MST4 and SOK1. Knockdown efficiency of CCM3 protein using three individual siRNA oligo’s as compared to control MDA-MB-231 cells (right histogram). (c) Quantification of cell area +/-s.e.m (left histogram, n=5 fields of cells) and mean fluorescence cortical pS19-MLC2 intensities +/-s.e.m (right histogram, n=fields of cells; (siCtr, 37; siMST3-5, 5; siMST3-22, 5; siMST3-23, 5; siMST4-4, 5; siMST4-5, 5; siMST4-6, 5; siMST3-21, 5; siMST3-23+MST4-4, 9; siCCM3-1, 10; siCCM3-2, 13; siCCM3-4, 10) in depleted MDA-MB231 cells. (d) Immunofluorescence analysis of phospho-tyrosine (cl. 4G10) (green), F-actin (magenta) and pERM (white) in depleted MDA-MB231 cells. (e) Quantification of pS19-MLC2 and Ezrin-GFP co-localisation analysis +/-s.e.m in depleted MDA-MB231. n=cells; (Ctr, 57; siMST3&4, 25; siCCM3, 27). All statistical test were performed using 1way ANOVA, Sidak’s multiple comparison test, *P<0.05, **P<0.01, ***P<0.001. All experiments were conducted at least 3 independent times. Scale bars, 10 μm.
Epistasis experiments to test whether integrin function is required for the cell morphology changes following FAM40A or FAM40B depletion showed that depletion of ITGB1, TLN, and VCL neither prevented FAM40A depletion from promoting a more contractile phenotype, nor prevented FAM40B depletion from promoting a more spread phenotype (Supplementary Figure 3i). We conclude that FAM40A and FAM40B are not controlling cell morphology through direct regulation of cell-matrix adhesions (see also Figure 1 and Supplementary Figure 2f-h).
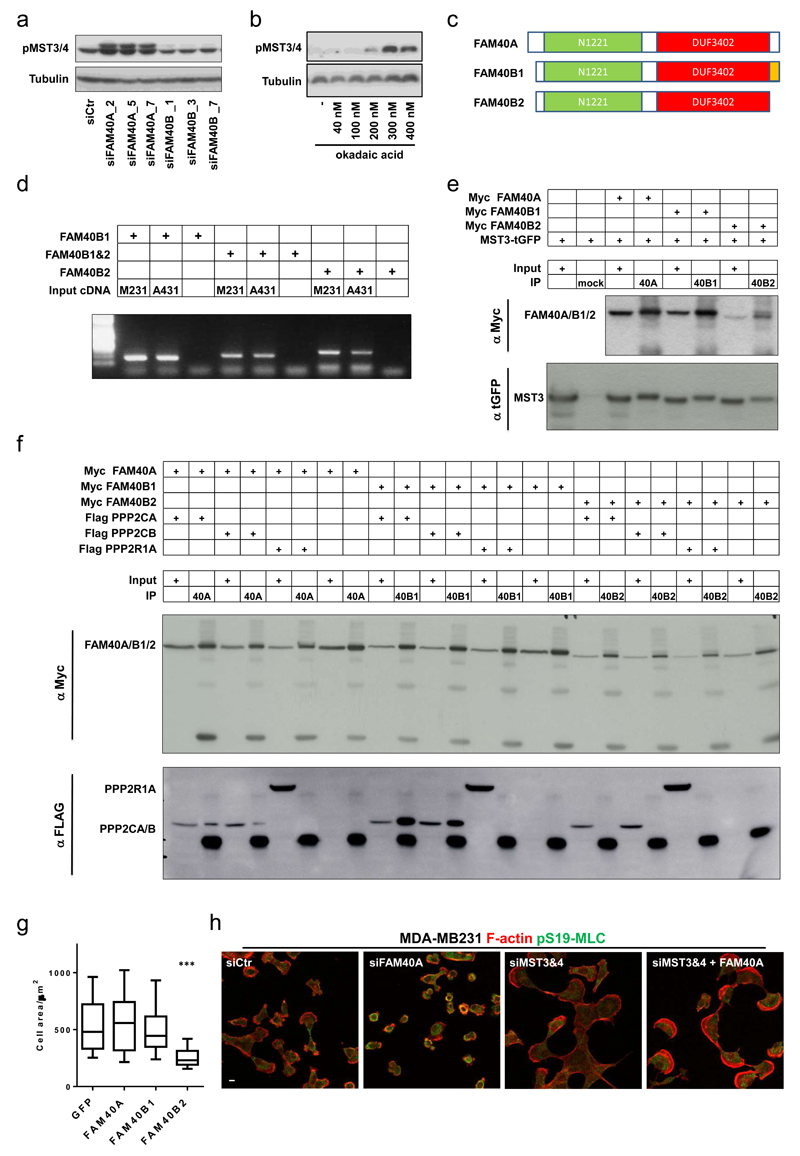
These data demonstrate that MST3&4 are involved in cytoskeletal regulation, either by controlling PP2A phosphatase activity which then regulates actomyosin, or because PP2A regulates the activity of MST3&4, which act ‘downstream’ of PP2A to regulate the cytoskeleton. To distinguish these possibilities we monitored the MST3&4 activities in response to perturbation of PP2A function and depletion of STRIPAK components. MST3&4 activities were markedly increased on FAM40A depletion and in response to the PP2A inhibitor okadaic acid (Figure 4a&b), indicating that PP2A and FAM40A negatively regulate the activity of MST3&4. To further investigate the underlying mechanism and reasons for the divergent phenotype of FAM40A and FAM40B depletion, we observed through sequence comparison of FAM40A and FAM40B that the N- and C-terminal regions were somewhat divergent and subject to alternative splicing (Figure 4c). We hypothesised that FAM40A might negatively regulate MST3/4 by recruiting PP2A catalytic subunits into a complex also containing the kinases. Further, we speculated that FAM40B1 and FAM40B2 isoforms might show differential binding to the PP2A catalytic subunits due to their divergent C-termini. After confirming that both FAM40B1 and FAM40B2 were expressed in MDA-MB231 and A431 cells (Figure 4d), we observed that FAM40A, FAM40B1, and FAM40B2 all interact with MST3 (Figure 4e). In contrast, the different FAM40 isoforms show differential binding to PP2A catalytic subunits. Although both FAM40A and FAM40B1 co-immunoprecipitate with PPP2CA and PPP2CB, FAM40B2 does not (Figure 4f). None of the FAM40 proteins interacted with PPP2R1A (Figure 4f). We propose that FAM40A and FAM40B1 negatively regulate MST3&4 by enabling the formation of a complex containing MST3&4 and PPP2CA&B, which dephosphorylate the activation loop of the kinases. FAM40B2 may act as a competitive inhibitor of FAM40A and FAM40B1 function. Indeed over-expression of FAM40B2, which cannot bind PP2A catalytic sub-units, induces contractility (Figure 4g). These data argue that FAM40A regulates cell morphology by controlling the PP2A-mediated inactivation of MST3&4. Consistent with this, the contractile phenotype of both FAM40A-depleted cells and okadaic acid-treated cells was not observed if MST3&4 were also depleted (Figure 4h and Supplementary Figure 3j).
Figure 4. The FAM40A/FAM40B1/PP2A complex negative regulates the activity of MST3&4.
(a) Immunoblotting analysis of active phosphorylated MST3&4 in depleted MDA-MB231 shows that FAM40A depletion induces activation of MST3&4 kinases. (b) Immunoblotting analysis shows that inhibition of the PP2A phosphatase using Okadaic acid induces activation of MST3&4 kinases. (c) Sequence comparison of FAM40A and FAM40B isoform 1 and 2 shows that the N- and C-terminal regions are slightly more divergent and subject to alternative splicing. Annotated domains; N1221 domain (green) and DUF3402 domain (red) as well as splice variant (orange) are depicted. (d) mRNA analysis shows that A431 and MD-MB231 cells express FAM40A and both isoforms of FAM40B (B1 and B2). (e) Co-immunoprecipitation analysis demonstrates that MST3 interact with FAM40A and both FAM40B1&2 isoforms. (f) Co-immunoprecipitation analysis demonstrates that FAM40A and FAM40B1 but not FAM40B2 interacts with both PPP2CA/B catalytic domains. The co-IPs also demonstrate that no binding to PPP1R1A takes place. (g) Overexpression of the FAM40B2 isoform is sufficient to induce a contractile phenotype with decrease in cell area. Box and Whiskers graph: Line=Median, Box=distribution of 50% of values, Whiskers=10-90% percentile. (n=cells; Ctr, 67; FAM40A, 82; FAM40B1, 65; FAM40B2, 57). Statistical test were performed using 1way ANOVA, Sidak’s multiple comparison test, ***P<0.001. (h) Epistasis experiment show that MST3&4 depletion rescues the contractile phenotype observed upon FAM40A depletion. MDA-MB231 cells were stained for F-actin (red) and pS19-MLC (green). All experiments were conducted at least 3 independent times. Scale bars, 10 μm.
CCM3 regulates the sub-cellular localisation of MST3&4
The data above indicate that MST3&4 are linked to the localised regulation of pMLC and pERM, but do not address the spatial localisation of MST3&4. Immuno-staining revealed enrichment of active phosphorylated MST3&4 around the outer edge of A431 cell clusters (Figure 5a). CCM3 was localised at the cell periphery, but also at cell-cell contacts (Figure 5b). Analysis of the dynamic localisation of MST3-GFP, MLC-GFP, and Ezrin-GFP in response to serum stimulation, which promotes ROCK-dependent actomyosin contractility45, revealed a transient MST3-GFP enrichment at cell-cell junctions around 100s and a more long-lived increase at the edges of cell clusters (Figure 5c, Supplementary Movie 2). MST3&4 activity was also increased (Figure 5d) and similar changes were seen in the localisation of both Ezrin-GFP and MLC-GFP (Supplementary Figure 4a, 4b). The correlation co-efficient of pS19-MLC and Ezrin and membrane blebbing were also increased by serum stimulation (Figure 5e and Supplementary Figure 4c). Disruption of actomyosin contractility using a ROCK inhibitor or CCM3 depletion prevented the MST3 translocation in response to serum stimulation (Figure 5c, Supplementary Movie 2). This was not linked to its biochemical activity as judged by pMST3&4 immunoblot analysis (Figure 5f) and CCM3 localisation was not dependent on actomyosin function (Figure 5g). We conclude that CCM3 and the actomyosin network combine to recruit MST3 to the edge of A431 cell clusters and cell-cell contacts.
Figure 5. CCM3 regulates the sub-cellular localisation of MST3&4.
(a-b) Immunofluorescence analysis reveals spatial localisation of phospho-MST3&4 (a) and CCM3 (b) at cell borders and cell-cell junctions. (c) Serum stimulation of serum starved A431-MST3-GFP cells induces an immediate translocation of MST3-GFP to the actomyosin network at the outer edge (white arrows) of A431 cell clusters and at cell-cell junctions (red arrows). Quantification of the serum-induced MST3-GFP translocation +/-s.e.m was performed by normalising the mean fluorescence intensity at cell border (left graph) and cell-cell junctions (right graph) to the mean fluorescence intensity of the adjacent cytosolic fraction. MST3-GFP translocation was inhibited by 10μM ROCK inhibitor (Y-27632) (middle panels) and by CCM3 depletion (lower panels). n = regions of individual measurements at edge of cluster (left panels); MST3-GFP, 79; Y27632, 48; siCCM3, 125. n = regions of individual measurements at cell-cell contacts (right panels); MST3-GFP, 70; Y27632, 71; siCCM3, 164. The values are pooled from three independent experiments. (d) Immunoblotting analysis demonstrating immediate activation of pMST3&4, pERM and pS19-MLC2 upon FBS stimulation. Serum starved A431 cells were stimulated with FBS and lysates were made at various time-points. (e) Quantification of pS19-MLC2 and Ezrin-GFP co-localisation analysis +/-s.e.m. Serum starved A431 cells were stimulated with FBS and fixed and at various time-points (n=20 cells). Box and whiskers graph: Line=Median, Box=distribution of 50% of values, Whiskers=min. to max. n=20 cells. Statistical test were performed using 1way ANOVA, Sidak’s multiple comparison test, **P<0.01, ***P<0.001. (f) Immunoblotting analysis demonstrating that activation of pMST3&4 and pERM is independent of ROCK activity. A431 cells were serum starved and treated overnight with vehicle or 10μM ROCK inhibitor (Y-27632). The cells were lysed 30 seconds after FBS stimulation. (g) Immunofluorescence analysis reveals spatial localisation of active pMST3&4 (red, white arrows) and CCM3 (green, red arrows) in serum starved, serum stimulated (2 minutes), and Y27632 pre-treated serum stimulated (2 minutes) A431 cells. All experiments were conducted at least 3 independent times. Scale bars, 10 μm.
FAM40B, MST3&4, and CCM3 antagonise 2D migration but are required for metastasis
Actomyosin contractility and ERM proteins are known to be involved in migration, however their role can be context dependent46. We utilised a computational model to investigate the effect of perturbing the co-localisation of actomyosin contraction and actin cortex to plasma membrane attachment in different matrix geometries45. The contractile machinery and connection to the plasma membrane were either co-located on the same side of the cell or on opposite sides (left and right side of boxes respectively). Three matrix geometries were considered: one corresponded to a 2D migration assay, the second to squeezing through a series of gaps, and the third mimicked an in vivo environment. We varied the relationship between actomyosin contractility and actin cortex to plasma membrane linkage. Co-localisation of contractile function and plasma membrane – actin cortex linkage was predicted to reduce the efficiency of cell migration on 2D surfaces (Figure 6a). This was because blebbing at the front of the cell disrupted the formation of lamellipodia. In contrast, this organisation was favourable for migration in matrix geometries that required cell squeezing through gaps (Figure 6a, middle and right panels). This suggested that a positive spatial correlation between actomyosin and cortex-membrane attachment, similar to FAM40A depletion, would be slightly detrimental to 2D migration but favourable for squeezing through gaps. In contrast, no correlation or an inverse correlation between these parameters favoured migration on a planar surface (Figure 6a, left panel). To test these predications, we performed migration assays on a rigid 2D substrate, through 8µm pores in a 6.5µm thick membrane, and tested the ability of cells to extravasate in vivo.
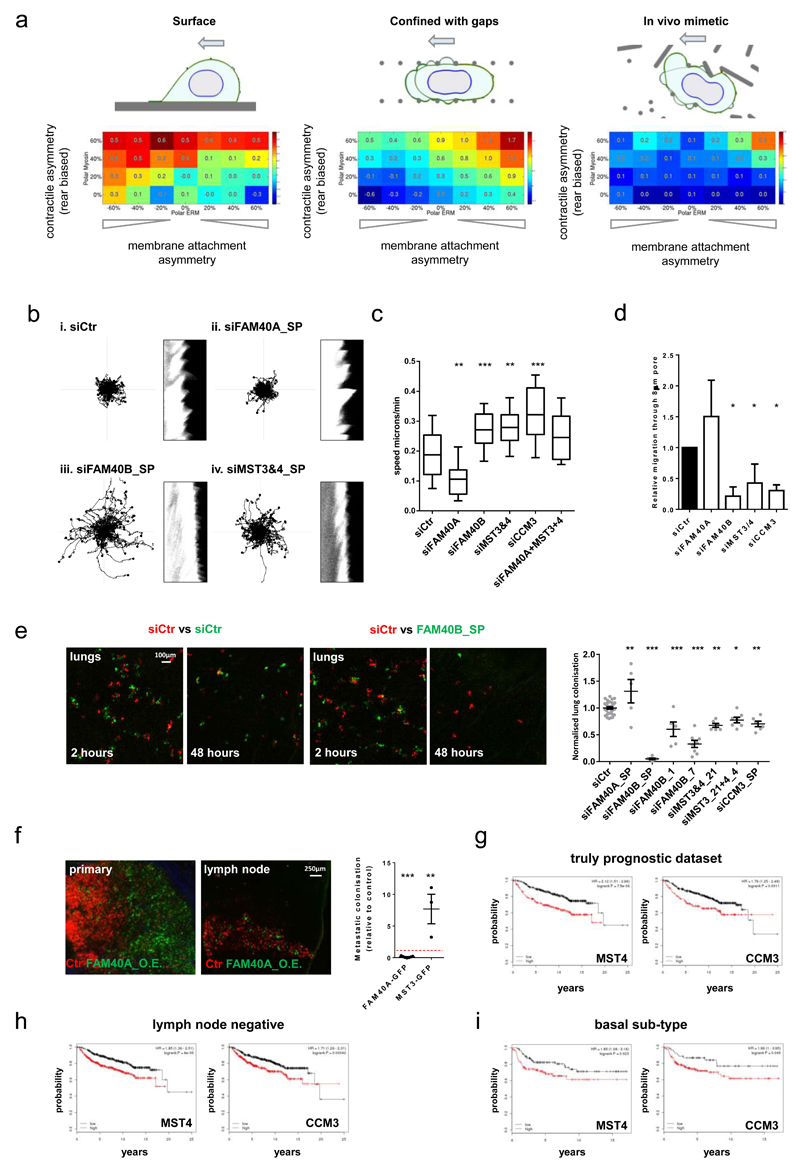
Figure 6. FAM40B, MST3&4, and CCM3 antagonise 2D migration but are required for metastasis.
(a) Matrix geometries investigated using computational model: flat 2D migration, squeezing through a series of gaps, in vivo mimetic environment. (b) 2D analysis of siRNA transfected MDA-MB231 cells (left panels; migration tracks, right panels; kymographs) Kymographs of the cell edge periodicity were generated from high resolution fluorescence images taken over 3minutes at 1second intervals. (c) Analyses of 2D migration speed: Box and whiskers graph: Line=Median, Box=distribution of 50% of values, Whiskers=20-80 percentile. (n=fields of cells (10x); Ctr,34; siFAM40A,37; siFAM40B,30; siMST3&4,35; siCCM3,32; siFAM40A & MST3&4,34). (d) Depletion of FAM40B, MST3&4 and CCM3 prevents MDA-MB231 cell migrating through 8μm pores 6.5µm deep. (n=4 experiments). (e) Mice were injected with mix of Che-expressing control cells and GFP-expressing siRNA-transfected cells. Images show tumour cells lodging the lung parenchyma. The metastatic burden was quantified 2h and 48h post injection by determining the ratio of GFP and Che positive tumour area and normalised to control-injected mice. n as “for Ctr, siFAM40A, siFAM40B-sp, siFAM40B-1, siFAM40B-7, siMST3_21, siMST3&4, siCCM3-sp, n= 31, 5, 5, 5, 8, 6, 7, 6 mice pooled from 7(Ctr), 2 (siFAM40A-sp, siFAM40B-1, siMST_21, siMST3&4, siCCM3-sp) and 3 (siFAM40B-sp, siFAM40B-7) experiments. (f) Mice were injected with a mix of control MDA-MB-231-Che and either MDA-MB-231-FAM40A-GFP or MST3-GFP cells in a ratio (1:1) into the mammary fat pad and the inguinal lymph nodes where imaged after 2-4 months. The spontaneous metastatic burden was quantified by determining the ratio of GFP and Che positive tumour area and normalised to the GFP/Che ratio of the primary tumour (right panel). Images of primary and lymph nodes are shown (right panels). Each mouse is represented in the chart. n=mice; (FAM40AGFP,7; MST3GFP,3). (g-i) Kaplan-Meier plots of distant metastasis free survival of breast cancer patient (n=1809 patients). The analysis includes (g) a truly prognostic dataset, (h) early stage lymph node negative patients (i) and aggressive basal subtype of breast cancer patients. The KM analyses for MST4 and CCM3 were performed using the publicly available datasets http://kmplot.com/analysis/. All statistical test were performed using 1way ANOVA, Sidak’s multiple comparison test, *P<0.05,**P<0.01,***P<0.001. All experiments were conducted at least 3 independent times. Scale bars as indicated.
Cells depleted of FAM40B, MST3&4 and CCM3 migrated faster and more persistently on 2D surfaces (Figure 6b-c and Supplementary Figure 5b) and formed stable lamellipodia (Supplementary Movie 3 and Figure 6b kymograph). In contrast, FAM40A depleted cells had lower cell speeds, were less persistent (Figure 6b&c and Supplementary Movie 3), and exhibited highly dynamic blebbing (Supplementary Movie 4 and kymograph in Figure 6b). These analyses are consistent with the computational prediction that weak co-localisation of actomyosin and actin cortex-membrane linkage favours 2D migration. Modelling also predicted that co-localisation of actomyosin contractility and actin-membrane attachment at the cell rear is favourable for squeezing through gaps (Figure 6a and modelling of the pore dimensions of transwell dishes in Supplementary Figure 5). FAM40B, MST3&4, and CCM3 depletion that disrupt the ability of cells to co-localise their actomyosin network with actin- membrane linkers (ERM proteins) reduce the ability of MDA-MB231 cells to migrate through 8µm pores (Figure 6d).
Our computational analysis showed that varying the co-localisation of the contractile machinery and actin-plasma membrane connection would have opposite effects on 2D surfaces and in vivo. FAM40B, MST3&4, and CCM3 depletion, which reduce pMLC/Ezrin co-location, would be expected to reduce migration in vivo. We tested this hypothesis by intravenously injecting mixed populations of control and target-gene-depleted MDA-MB-231 and evaluating their ability to extravasate and enter the lung parenchyma after 48h. When mCherry-expressing control cells were mixed with GFP-expressing control cells, an equal number of both cell types was detected after both 2 and 48hrs (Figure 6e). However, FAM40B depletion significantly reduced the number of cells in the lung after 48h. Similar results were achieved by targeting FAM40B, MST3&4 and CCM3 with additional siRNAs (Figure 6e). FAM40A depletion, which causes the opposite phenotype to FAM40B depletion, led to a small increase in the efficiency of lung colonisation (Figure 6e). We further examined the role of STRIPAK complexes in metastasis by generating cells that moderately over-expressed FAM40A or MST3 and testing their role to metastasize from a primary tumour in the mammary fat pad to the inguinal lymph nodes (over-expression levels shown in Supplementary Figure 6b). MST3 overexpression increased the ability of MDA-MB231 cells to metastasize (Figure 6f), whereas FAM40A expression had the opposite effect (Figure 6f).
Altered expression of STRIPAK components in cancer
Analysis of available human breast cancer datasets revealed elevated expression of MST3, MST4 and CCM3 in more aggressive breast cancer sub-types (Supplementary Figure 6c), with high levels of MST4 and CCM3 expression indicating a worse distant metastasis free survival within lymph node negative, grade 1, and grade 2 disease, and basal breast cancer (Figure 6h&i)47. The expression of other STRIPAK components, STRN3 and STRN4, decreased in aggressive breast cancer sub-types (Supplementary Figure 6c). Simultaneous depletion of all three STRNs in MDA-MB231 cells confirmed that they are negative regulators of MST3&4 (Supplementary Figure 6d).
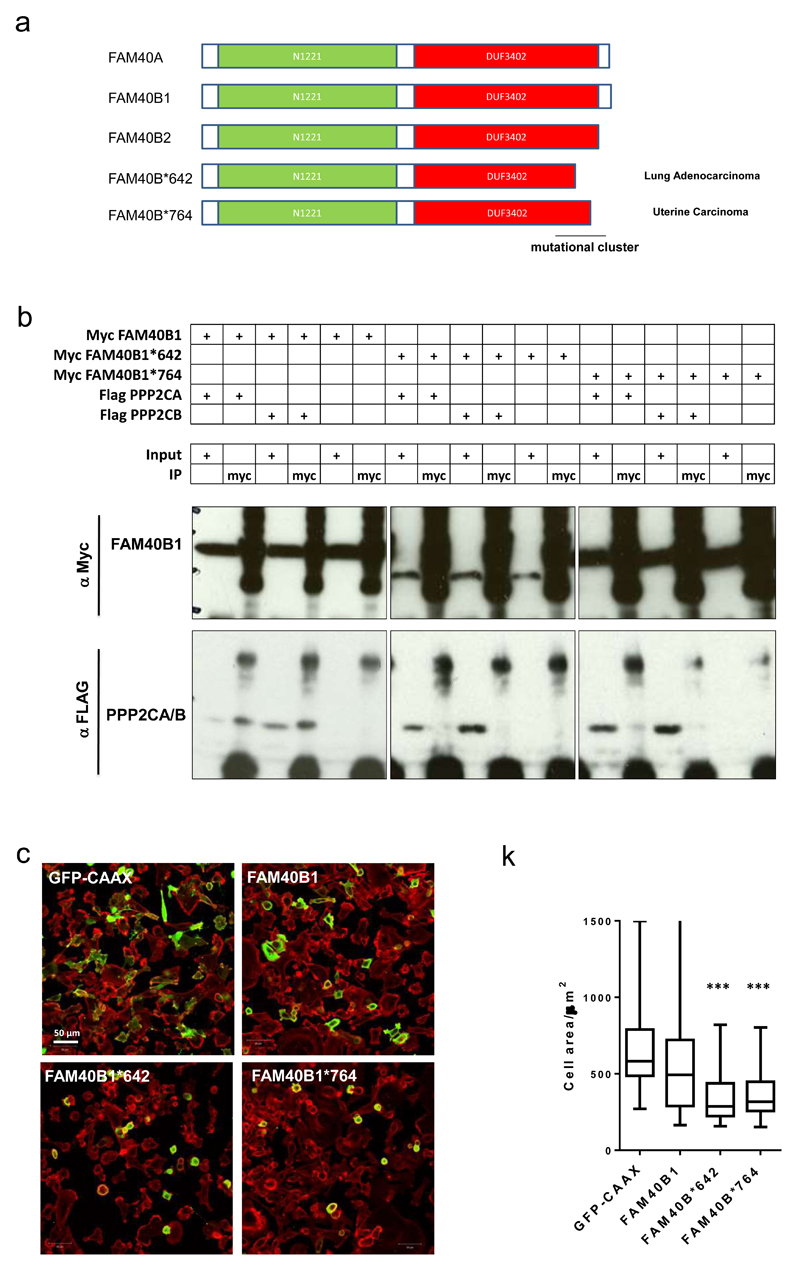
Analysis of data from The Cancer Genome Atlas resource; cBioPortal for Cancer Genomics revealed that FAM40B was more frequently amplified or mutated than FAM40A (links in Supplementary Figure 6e). Many FAM40B mutations cluster around the C-terminal region of the protein which binds to the catalytic sub-units of PP2A (Figure 7a) and we hypothesised that C-terminal truncation mutants would no longer bind PPP2CA and PPP2CB and would drive cell contraction when over-expressed. Indeed, the truncation mutants FAM40B-642stop and FAM40B-764stop are defective in binding to PP2A catalytic sub-units and drive cell contraction when over-expressed (Figures 7b&c).
Figure 7. Tumour-derived FAM40B truncation mutants drive cell contraction.
(a) Sequence comparison of FAM40A, FAM40B isoform 1 and 2, and two FAM40B point mutants 642stop and 764stop encoding two stop codons. The C-terminal regions harbour a mutational cluster. Annotated domains; N1221 domain (green) and DUF3402 domain (red) are depicted. (b) Co-immunoprecipitation analysis demonstrates that the two truncated FAM40B proteins FAM40B-642stop -and 764stop lose their ability to interact with PPP2CA/B catalytic domains. (c) Ectopically expression of FAM40B-642stop -and 764stop induce cell contraction. MDA-MB231 cells were transfected with GFP-CAAX (plasma membrane tag), Myc-FAM40B1, Myc-FAM40B-642stop and Myc-FAM40B-764stop. The cells were fixed and stained and images for F-actin (red) and GFP/Myc (green). Quantification of MDA-MB231 cell area +/-s.e.m upon overexpression of Myc-tagged constructs. n=cells; (GFP, 28; 40B1, 41; 40B*642, 52; 40B*764, 60). Statistical test were performed using 1way ANOVA, Sidak’s multiple comparison test, ***P<0.001. All experiments were conducted at least 3 independent times. Scale bars, 10 μm, unless indicated otherwise.
MST3&4 regulates PPP1CB to modulate the cytoskeleton
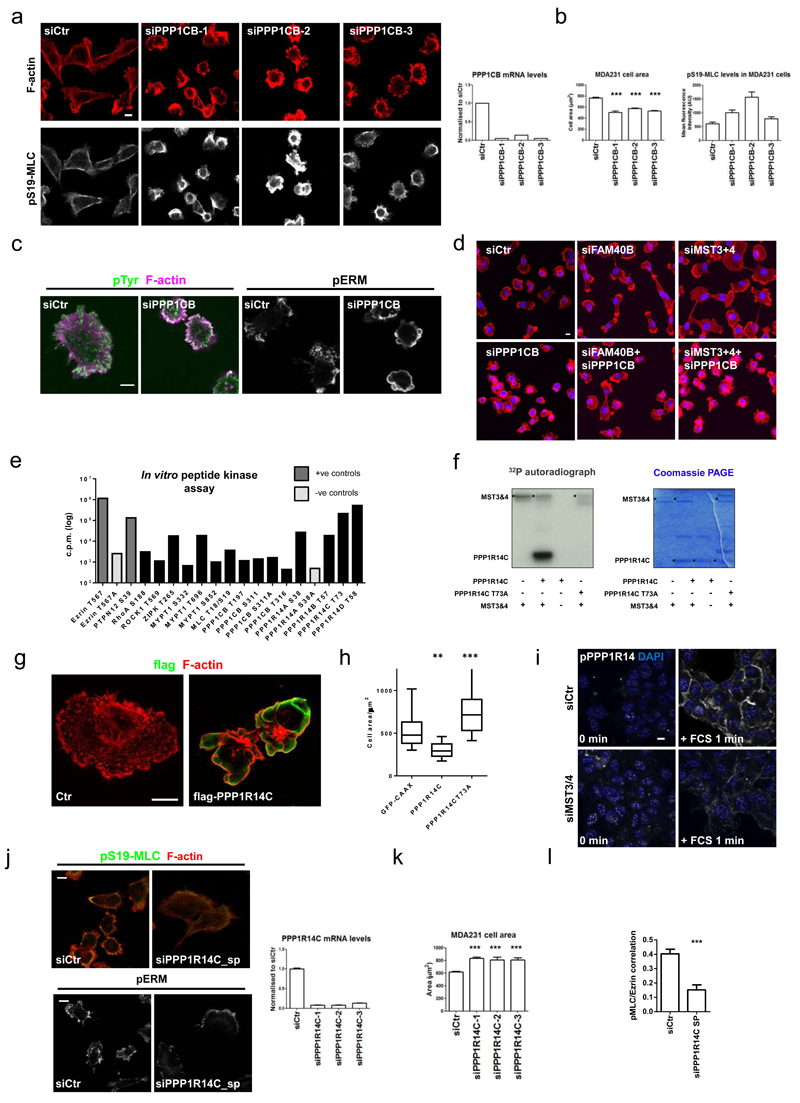
We next sought to identify MST3&4 substrates responsible for control of cell morphology. Although Ezrin is an MST4 substrate32, 48, overexpression of an active Ezrin T567D mutant did not phenocopy FAM40A depletion (Figure 1m), nor was it able to rescue FAM40B-, MST3&4-, or CCM3-depleted cell phenotypes (Supplementary Figure 7). We thus postulated that some of the 32 genes identified in Supplementary Figure 1 may be involved in control of the cytoskeleton by MST3&4. Depletion of PPP1CB in MDA-MB231 cells, but not other phosphatase components from the border cell migration screen, led to highly contracted blebbing cells (Figure 8a-d, Supplementary Figure 8a, and Supplementary Table 1). The effects of MST3&4 depletion could be reverted by depletion of PPP1CB (Figure 8d), suggesting that MST3&4 may regulate the cytoskeleton through modulation of PPP1CB function. PPP1CB could be regulated by phosphorylation in a similar manner to PPP1CA and PPP1CC, however recombinant MST3&4 were unable to phosphorylate the potential regulatory sites of PPP1CB (Ser311 and Thr316) (Figure 8e).
Figure 8. MST3&4 regulate PPP1CB to modulate the actomyosin cytoskeleton.
(a) PPP1CB siRNA in MDA-MB231 cells. Images of cells were stained 72h post-transfection against pS19-MLC2 (white) and F-actin (red). Knockdown efficiency of PPP1CB mRNA +/-s.e.m (right histogram, n = 3 technical replicates). (b) MDA-MB231 cell area +/-s.e.m (left histogram, n=4 fields of cells) and mean fluorescence intensity of cortical pS19-MLC2 +/-s.e.m (right histogram).(c) Immunofluorescence analysis of phospho-tyrosine (cl. 4G10) (green), F-actin (magenta) and pERM (white) in PPP1CB depleted MDA-MB231 cells. (d) Epistasis experiment combining PPP1CB depletion with FAM40B and MST3&4 depletion. F-actin (red) and DAPI (blue). (e) In vitro peptide phosphorylation by MST3&4 kinases are depicted by incorporated γ32P-ATP (cpm). The values are average of all the experiments conducted (see supplementary table 2). Ezrin-T567 and PTPN12-S39 are positive controls (dark grey), while alanine substitutions in Ezrin-T567A and PPP1R14A-S58A are negative controls (light grey). (f) MST3&4 in vitro kinase assay shows that MST3&4 only phosphorylates wild-type T73 and not the alanine mutated T73A version of full length PPP1R14C. Left panels show 32P autoradiograph. Right panel shows the coomassie staining of the gel. (g) Structured illumination microscopy (SIM) shows ectopically expressed flag-tagged PPP1R14C in MDA-MB231. Flag-PPP1R14C is stained using anti-flag antibody (in green). F-actin (red). (h) Box and whisker blot show cell area following GFP-CAAX, PPP1R14C, or PPP1R14C-T73Atransfection. Box and whiskers graph: Line=Median, Box=distribution of 50% of values, Whiskers=10-90 percentile. n=cells; (GFP, 38; R14C, 58; R14C-T73A, 67). (i) Phospho-PPP1R14 staining after 1 minutes serum stimulation of control A431 cells or MST3&4 depleted cells. (j) Immunofluorescence analysis (left panels) of pS19-MLC (green), F-actin (red) and pERM (white) in PPP1R14C depleted MDA-MB231 cells. Knockdown efficiency of PPP1R14C mRNA +/-s.e.m. as compared to control cells (right histogram, n=3 technical replicates). (k) Quantification of MDA-MB231 cell area +/-s.e.m. after depletion of PPP1R14C with multiple siRNA oligos (n=7 fields of cells (20x)). Unpaired student’s t-test (two-tailed), *P<0.05, **P<0.01, ***P<0.001. (l) Quantification of pS19-MLC2/Ezrin-GFP co-localisation analysis +/-s.e.m. (n=cells; siCtr, 21; 14Csp, 23). All statistical test were 1way ANOVA, Sidak’s multiple comparison test, if not mentioned differently, *P<0.05,**P<0.01,***P<0.001. All experiments were conducted at least 3 independent times. Scale bars, 10 μm.
A small-scale screen of peptides from PP1 regulators or cytoskeletal molecules was performed to identify MST3&4 substrates (Supplementary Table 2). The most highly phosphorylated peptides were from the PPP1R14 family of proteins (Figure 8e), which are negative regulators of PPP1CB. PPP1R14A can be positively regulated by phosphorylation 49 and PPP1R14C and PPP1R14D were phosphorylated by MST3&4 to similar levels as positive control peptides from Ezrin and PTPN12 (Figure 8e). We confirmed that MST3&4 phosphorylate PPP1R14C on S73 by kinase assays using wild-type and T73A full length recombinant protein (Figure 8f). PPP1R14C expression in MDA-MB231 cells was sufficient to drive membrane blebbing (Figure 8g), whereas mutation of the MST3&4 phosphorylation site to alanine (T73A) prevented regulation of membrane blebbing by PPP1R14C (Figure 8h). Expression of the wild-type and T73A mutant demonstrated that a phosho-PPP1R14A antibody cross-reacted with PPP1R14C (Supplementary Figure 8b) and could be used to monitor phosphorylation changes in PPP1R14 proteins. Activation of MST3&4 by serum stimulation for one minute led to increased pPPP1R14 staining at cell-cell contacts and around the edge of cell clusters (Figure 8i and Supplementary Figure 8c). This corresponds to the localisation of MST3 following serum stimulation (Figure 5), and was reduced by MST3&4 depletion (Figure 8i). PPP1R14C depletion reduced the cortical localisation of pS19-MLC and increased cell area in both A431 and MDA-MB231 cells (Figure 8j&k and Supplementary Figure 8d&e). PPP1R14C depleted cells also had broad pERM positive lamellipodia (Figure 8j) and reduced co-localisation of pS19-MLC and Ezrin (Figure 8l). These data establish PPP1R14C as an MST3&4 substrate that links phenotypes observed following manipulation of the STRIPAK/PP2A complex to PP1-mediated regulation of the cytoskeleton.
Discussion
Cancer cells can adopt different motility modes9–11: some use lamellipodia and an elongated morphology (mesenchymal), whereas others use either small F-actin rich protrusions or blebs to extend the plasma membrane forward (amoeboid). Cell motility mode can affect the ability of cancer cells to metastasize7, 8, 12, 50, 51 and we demonstrate that the STRIPAK complex regulates both how cancer cells move and their ability to metastasize through the function of MST3&4. FAM40A and STRN1,3,&4 antagonize the activity of MST3&4 by enabling the catalytic sub-units of the PP2A complex to de-phosphorylate the activation loop of MST3&4 (Supplementary Figure 8f). FAM40B2 cannot perform this function and acts as a competitive inhibitor of FAM40A. The effect of FAM40B depletion likely depends on the relative levels of FAM40B1 and FAM40B2. If significant levels of FAM40B2 are present, removing this from the system will favour negative regulation of MST3&4 by FAM40A. This could explain the divergent phenotypes reported when FAM40A and FAM40B are depleted22. CCM3 localises MST3 to the edge of cell clusters and cell-cell contacts. Active MST3&4 at the cell cortex can phosphorylate PPP1R14C and promote its ability to locally inhibit de-phosphorylation of MLC by PPP1CB. This elevates phosphorylated MLC in the cortical actin underlying the plasma membrane. This model is supported by the reduced rate of MLC dephosphorylation in cells with elevated MST3&4 activities (Figure 2e).
MST3&4 can also regulate ERM proteins at the cell cortex, likely through direct phosphorylation32. However, MST3&4 cannot be the only ERM kinases because the total levels of pERM do not change following MST3&4 depletion. Instead pERM is lost from the cell cortex but is prominent in the lamellipodia. SLK or PKC family members may be relevant ERM kinases in this context. Links between CCM3 and ERM proteins have been previously reported in endothelial cells52, 53. Although we do not find evidence of STRIPAK components acting upstream of RhoA in cancer cells of epithelial origin (Figure 2), we cannot completely exclude negative regulation of Rho signalling by the STRIPAK complex.
We propose that MST3&4 coordinate inhibition of MLC dephosphorylation and phosphorylation of ERM proteins at the cell cortex. Absence of MST3&4 prevents the colocalisation of pS19-MLC with ERM proteins. The spatial segregation of actomyosin machinery from ERM proteins prevents the contractile cytoskeleton from pulling on the plasma membrane efficiently. This facilitates cell spreading, reduces hydrostatic pressure, and leads to contractile force being coupled to integrins via focal adhesions, instead of being exerted on the plasma membrane. The slightly increased focal adhesion numbers that we observe upon MST3&4 and CCM3 depletion (Figure 3d) may reflect effects via ICAP154.
We observe significant correlations between activation of MST3&4 function and more aggressive breast cancer sub-types and poor prognosis. Cancer genome sequencing has identified frequent mutations in FAM40B, and based on mutation frequency and types, FAM40B was classified as an oncogene55. We demonstrate that FAM40B truncation mutants confer a dominant ability for FAM40B to promote a contractile cell phenotype. We propose that FAM40B mutants and high MST3&4 activity promote malignancy by coupling contractile cytoskeleton to the plasma membrane and generating significant intra-cellular hydrostatic pressure, which enables migration through confined spaces56, 57.
Methods
Cell culture and transfections
Human A431 and MDA-MB-231 cells were grown in DMEM supplemented with 10% FCS. All cell lines were frequently tested for mycoplasma and were negative. A431 cells were transfected using Dharmafect 2 (Dharmacon). MDA-MB-231 cells were transfected using Lipofectamine 2000 (Invitrogen). In brief, cells were plates at 60% confluence and subjected to transfection the following day using 25–50 nM siRNA. After 24 h of transfection the cells were re-plated onto plastic or a gel consisting of 4.25–4.75 mg ml−1 collagen I (BD Biosciences; catalogue no. 354249) and 2–2.5 mg ml−1 Matrigel (BD Biosciences; catalogue no. 354234). Cells were either fixed or lysed 48–72 h post-transfection. The phenotypic changes observed in the screen were blindly scored by a minimum of 3 people. The screen was performed three times in A431 cells. siRNA oligonucleotides were purchased from Dharmacon and Qiagen. A comprehensive list of all siRNA oligonucleotides used in this study can be found in Supplementary Table 3. Ectopic expression of fluorescently tagged proteins was generated by classical DNA transfection. In most cases cells were treated with antibiotics to generate stable pools of cells. Once stable, the cells were FACS sorted for further purification. All cloning was conducted using classical techniques and the Gibson Assembly cloning kit (NEB catalogue no. E5510S). A comprehensive list of all expression vectors used in this study can be found in Supplementary Table 3. The pEzrin–GFP and pEzrin-T567D-GFP vectors were gifts from R. Lamb, Cancer Research Centre, University of Liverpool, UK. The pEGFP-MLC, pROCK:ER and pROCK KD:ER vectors were gifts from M. F. Olson, Beatson Institute for Cancer Research, Glasgow, UK. The pEGFP-MST4 vector was a gift from F. Barr, University of Oxford, UK. The pEGFP-CCM3 vector was a gift from J. Zalvide, University of Santiago de Compostela, Spain. The Raichu–RhoA biosensor (1523x) was a gift from M. Matsuda, Kyoto University, Japan.
Western blotting, RNA extraction and quantitative real-time PCR
Western blotting was performed using standard techniques. A comprehensive list of all antibodies used in this study can be found in Supplementary Table 3. Total RNA was prepared using RNeasy (Qiagen), according to the manufacturer’s instructions. cDNA was generated from 2 μg total RNA using M-MLV H-point mutant reverse transcriptase (Promega) and random hexamer primers. qPCR was carried out on cDNA using EXPRESS SYBR GreenER SuperMix with Premixed ROX or Platinum SYBR Green qPCR SuperMix-UDG with ROX (Invitrogen), according to the manufacturer’s instructions. The housekeeping gene coding for glyceraldehyde-3-phosphate dehydrogenase (GAPDH) was used as a control. A comprehensive list of all qPCR primers used in this study can be found in Supplementary Table 3.
Immunoprecipitation assays
Myc-tagged FAM40A, FAM40B1 or FAM40B2 was transiently transfected into 293T cells plated on 10 cm plates together with tGFP–MST3, Flag–PPP2CA, Flag–PPP2CB or Flag–PPP2R1A. After 12 h of transfection the cells were lysed in TNN-HS buffer25 followed by centrifugation for 10 min at 16,000g at 4 °C. Lysates were incubated with 5 μg of anti-Myc antibody (9E10 clone) for 4 h tumbling at 4 °C in the presence of 50 μl of 50% slurry TNN-HS pre-washed protein-G Dyna beads (Invitrogen). Beads were collected by magnetic force and washed 4 times with 1 ml TNN-HS buffer. The immune-precipitates were analysed by SDS–PAGE and protein detection with anti-Flag–HRP-conjugated (Cell Signaling), anti-Myc (cl. 9E10) or Turbo-GFP (Evrogen) antibodies.
Recombinant protein and soluble peptide kinase assay
Assays were carried out in a final volume of 30 μl buffer containing 160 μM peptide or 400 ng recombinant protein substrate in 50 mM Hepes pH 7.5, 10 mM MgCl2, 1 mM EGTA, 0.01% Brij35. The phosphorylation reactions were initiated by the addition of 100 ng of kinase (MST3/4, Abcam) and ATP (10 μM cold plus 0.3 μl [32P]ATP 370 MBq per 60 μl). All phosphorylation reactions were carried out at 30 °C for 30 min. Analysis of recombinant protein substrate was performed by SDS–PAGE; 25 μl of the reaction loaded, the gel was fixed, stained and dried using standard methods. Analysis of peptide substrates was performed by transferring 25 μl to 2 cm square pieces of phosphor-cellulose p81 paper (Whatman). The papers were immediately immersed in 1%v/v phosphoric acid, washed 3 times for 5 min, washed once in acetone, dried and the radioactivity was measured by Cherenkov counting.
Fly strains and immunofluorescence of the egg chambers
UAS.IR transgenic RNAi lines (on the second or third chromosome) were obtained from the Vienna Drosophila RNAi Centre57 and driven in the border cells by the Gr1–Gal4 driver. Two independent RNAi lines for each gene were used to confirm the migration delay phenotype: Fam40/CG11526—106184 and 16211 (data not shown); Cka/CG7392—106971 and 35232 (data not shown). Twelve thousand siRNA lines crossed onto the hh.GAL4 driver were assumed to have no strong phenotype within the egg chamber (either border cell or follicular epithelium) if the flies were healthy and viable—these were not further analysed. The 1,393 lethal siRNA lines were further analysed for egg chamber phenotypes by crossing the Gr1.GAL4 driver; 594 of these lines were fertile and therefore not analysed further. The remaining 844 lines were analysed in more detail by crossing onto the actin>stop>Gal4 driver. Ovary dissection was performed in phosphate-buffered saline (PBS), followed by fixation in 4% paraformaldehyde (PFA) in PBS for 20 min. After fixation, ovaries were rinsed with PBS-T (1× PBS and 0.1% Triton X-100) and then incubated in blocking solution (1× PBS, 0.1% Triton X-100 and 5% goat serum) for at least 30 min. Ovaries were then incubated with the primary antibodies diluted in PBS-T and 5% goat serum overnight at 4 °C, washed with PBS-T, followed by secondary antibody staining. For phospho-MyoII staining, ovaries were fixed in 8% PFA for 10 min, blocked for 1 h in PBS-T with 5% BSA, and then incubated with the primary antibody at 4 °C overnight in PBS-T, 5% BSA. The following primary antibodies were used: rabbit anti-phospho-MLC2 (1:50; Cell Signaling), mouse anti-Dlg (4F3, 1:250; DSHB), rabbit anti-phospho-ERM (1:100; Cell Signaling). Goat secondary antibodies conjugated to Alexa Fluor 546 or 647 (Invitrogen) were used at a dilution of 1:500 in PBS-T and incubated for 2 h at room temperature. Ovaries were further stained with DAPI (Invitrogen) in PBS-T to visualize nuclei before mounting in Vectashield (Vector Labs). Images were acquired on a Leica SP5 confocal microscope using ×40 oil-immersion objectives, and were processed with Adobe Photoshop CS3.
Confocal microscopy
All fluorescent images were acquired using either Zeiss LSM510, LSM710 or LSM780 inverted confocal microscopes. For time-lapse analysis the cells were kept at 37 °C and 5 or 10% CO2. For immunofluorescence analysis the cells were fixed with 4% PFA in PBS and permeabilized using 0.2% Triton X-100 in PBS before blocking with 3% BSA in PBS. Cells were stained with primary antibodies overnight at 4 °C followed by DAPI, Phalloidin–TRITC (Sigma #P1951) and fluorescently conjugated secondary antibodies for 2–6 h at room temperature. When staining pERM proteins, the 4% PFA fixation was replaced by a 30 min, 10% TCA fixation on ice. A comprehensive list of all antibodies used in this study can be found in Supplementary Table 3. Cell phenotypes were determined manually by placing cells into categories depending on the organization of the actin cytoskeleton. FRET imaging was performed using a Zeiss LSM780 system using constructs and methods described previously58. Briefly, cells were excited using 458 nm light and the ratio of YFP to CFP fluorescence was determined. The quantification represents a ratiometric measurement, not FRET efficiency. Controls involving imaging after photo-bleaching of the acceptor and imaging CFP alone were performed to confirm that FRET was being measured (data not shown).
Structured illumination microscopy
Cells were seeded in 35 mm glass Mattek dishes and processed for immunostaining as described above. After staining they were then mounted in a 1:1 mixture of glycerol and MOWIOL. Imaging was performed using a Zeiss ELYRA PS1 system (Zeiss) using a Plan-Apochromat ×63/1.40 Oil DIC M27 objective. Data were processed using the structured illumination microscopy tools in the Zen 2011 software (Zeiss). Channel alignment was performed with 0.1 μm TetraSpeck fluorescent microspheres (Life Technologies).
Image analysis metrics
The cell area (±s.e.m.) of MDA-MB-231 cells was determined using Volocity software. Images were thresholded on the basis of F-actin staining and cells automatically identified. A431 grow in clumps and were not amenable to this method. The average cell area of A431 cells was quantified by dividing the total area of cells in a field of view (as determined using F-actin thresholding) by the number of nuclei in that field of view. The mean fluorescent intensity (±s.e.m.) of either pS19-MLC or pERM was determined using Volocity software. Images were thresholded on the basis of F-actin staining, cells automatically identified, and the mean intensity of staining in other fluorescent channels was extracted. Multiple fields of views were quantified for each independent experiment. The co-localization of Ezrin–GFP and pS19-MLC staining was determined following siRNA transfection of MDA-MB-231 or A431 cells stably expressing Ezrin–GFP. Confocal images were thresholded on the GFP channel to exclude any cells that were not expressing Ezrin–GFP; the correlation coefficient (r) of pixel intensity values for Ezrin–GFP and pS19-MLC fluorescence was extracted from ZEN software.
Matrix displacement analysis
For matrix displacement measurements, preparation of polyacrylamide gels of defined elastic moduli was performed as previously described59. Briefly, 3 kPa polyacrylamide gels were embedded with 0.2 μm fluorescent beads and coated with 0.1 mg ml−1 collagen I. Following siRNA transfection, MDA-MB-231 cells were then seeded on top of the 3 kPa matrices. One day after seeding the fluorescent beads underlying the cells were imaged before and after the addition of tryspin/EDTA and cytochalasinD/blebbistatin to disrupt all cell–matrix adhesions and actomyosin force generation. These images then enabled the matrix displacement to be tracked between the substrate with cells exerting force on it (before) and the relaxed state (after).
Quantification of GFP-tagged protein translocation
Fluorescently tagged A431 cells were plated on glass Mattek dishes and left overnight. The next day the medium was changed to serum-free medium and left overnight. FBS to a final concentration of 10% was added to the cells and image acquisition was initiated immediately. Image acquisition was performed at 37 °C and 10% CO2 on a Leica LSM780 inverted confocal microscope using a ×63 1.54 NA objective (Leica). Cells were imaged every 20 s for 12 min. Quantification of GFP-tagged protein at cell–cell junctions and cell borders was accomplished by measuring the mean fluorescence intensity within a 15 × 15 pixel area from each frame using ImageJ. The mean fluorescence intensity within that square was divided by the mean fluorescence intensity of a second square having a 15 × 15 pixel area placed adjacently to the first square, but within the cytosol of the cell. All values were then normalized to t = 0.
Phase-contrast time-lapse and migration analysis
Bright-field and epifluorescence time-lapse imaging of MDA-MB-231-H2b–mCherry cells was performed at 37 °C and 5% CO2 with an inverted microscope (Nikon ECLIPSE TE2000-E). Bright-field and epifluorescence images were taken every 5 min for 12 h through a ×10 PlanFluor, NA 0.3 Ph1, Nikon objective. The imaging system includes a Xenon PE300BF lamp, an Andor iXonEM+ DU-888 back-illuminated EMCCD scientific camera, and System Control Software MetaMorph Version 7.7.3.0 (Molecular Devices). Cell tracking was performed by tracking the H2b–mCherry-positive nuclei using MetaMorph software. Cell speed and persistence were quantified using Mathematica Software.
Transwell migration assays
MDA-MB-231 cells were transfected with siRNA as described above. The day after transfection the cells were trypsinized and seeded into Transwell inserts with 8 μm pores (Costar no. 3422) in serum-free media while the lower reservoir contained 10% FBS. After 48 h the cells were fixed and images were acquired of the cells that remained in the inserts and those that had migrated through the pores. The number of cells in each image was quantified using ImageJ, and the migrated cells/total cell number was calculated.
Morphometric analysis of focal adhesion
siRNA-transfected MDA-MB-231 cells were plated on glass, and fixed 72 h post-transfection with 4% PFA. Focal adhesions were stained using phospho-tyrosine antibody (clone 4G10). Image acquisition was performed on a Leica LSM510 inverted confocal microscope using a ×63 1.2W C-Apochromat water objective (Leica). Images were taken with a pin hole of 0.6 to minimize the z-section. Pixel dimension was 0.07 μm × 0.07 μm. The morphometric analysis was conducted using ImageJ. Focal adhesions of individual cells were identified by edge finding and then converted into a binary image. Particle analysis was then applied to determine morphometric values including numbers, area, perimeter and shape factor. Focal adhesion greater than 0.5 μm2 was included in the final analysis.
Flow cytometry
Cell surface expression of β1-integrins was analysed by flow cytometry 72 h post-transfection. Cell surface levels of active b1-integrin were determined using the HUTS-21 antibody (1:100), which specifically binds the active form of β1-integrins. Total levels of β1-integrins were determined by co-incubating the HUTS-21 antibody with 0.5 mM MnCl2, a stimuli that forces integrins into an active conformation. Cells were stained with secondary Alexa488-labelled antibody (1:500) and analysed by flow cytometry (FACSCalibur; BD Biosciences). All steps were performed on ice to limit the analysis to cell surface expression. The ratio of active versus total β1-integrin levels was calculated.
Experimental metastasis assays
Fluorescently tagged MDA-MB-231 cells were injected into the tail vein of ∼6-week-old female nude mice. No statistical method was used to predetermine the sample size. No method of randomization was used and no blind experiments were conducted. All animal experiments were conducted in accordance with the Home Office Animals (Scientific Procedures) Act of 1986. Two days before injection either the mCherry- or GFP-positive cells were transfected with siRNA targeting the gene of interest, and the other coloured cells were transfected with a control siRNA. Mice were always injected in a 1:1 mixture of GFP- and mCherry-expressing MDA-MB-231 (1 million cells were injected). Additional controls for the ratio of mCherry and GFP cells were conducted by seeding 10 μl of the 1:1 cell suspension into a glass-bottom dish coated with poly-lysine (Sigma); after 2 h, cells were fixed in 4% PFA and imaged with a Zeiss LSM 780 confocal microscope using a Plan-Neofluar 10 × 0.3 objective. The mice were euthanized after either 2 or 48 h, and evaluated for fluorescent tumour cells in the lungs using confocal imaging and the same settings and laser power as used for the control plates. The ratio between red and green fluorescent cell area of each lung set as well as the control plate were then determined by automated quantification using Volocity software. The metastatic index was then calculated taking into the account the initial ratio at the time of injection (control plates on poly-lysine or lungs after 2 h) and the ratio observed within the lungs at 48 h.
Quantification of spontaneous metastasis
Fluorescently tagged MDA-MB-231 cells were injected into the fat pad of ∼6-week-old female nude mice. No method of randomization was used and no blind experiments were conducted. All animal experiments were conducted in accordance with the Home Office Animals (Scientific Procedures) Act of 1986. Mice were always injected in a 1:1 mixture of GFP- and mCherry-expressing MDA-MB-231. When tumours were 7–10 mm in diameter the mice were euthanized and imaged using intravital imaging. Spontaneous metastases were quantified by determining the ratio of GFP- versus mCherry-positive cell area in the inguinal lymph nodes and normalizing it to the ratio within the primary tumour. The ratio between red and green fluorescent cell area was determined by automated quantification using Volocity software.
Analysis of clinical data sets
Kaplan Meier plots for distant metastasis-free survival (Gene set for Breast cancer) were generated using the online resource http://kmplot.com/analysis/ with auto cutoff selected60. Expression data for different breast cancer sub-types (gene set analysis tumours) were obtained using the Lund University online resource http://co.bmc.lu.se/gobo/gsa.pl (ref. 61). Frequencies of mutations, deletions, amplifications, and multiple alterations in FAM40A/STRIP1 and FAM40B/STRIP2 in all cancer types were generated using the online resource http://www.cbioportal.org/public-portal/index.do (ref. 62, 63).
Statistical analyses
All statistical tests were performed using one-way ANOVA, Sidak’s multiple comparison test or unpaired Student’s t-test (two-tailed), *P < 0.05, **P < 0.01, ***P < 0.001. No statistical method was used to predetermine sample size. The experiments were not randomized. The investigators were not blinded to allocation during experiments and outcome assessment apart from the different siRNA screens that were blinded.
Supplementary Material
Acknowledgement
CDM, SH, MT, AB, GF, PAB, BT and ES are financially supported by Cancer Research UK. CDM was further supported by a FEBS long-term fellowship. We thank laboratory members for help and advice throughout this work. We thank Nicola O’Reilly and Sven Kjaer for help with peptide synthesis and protein purification, and members of the BRU for help with metastasis assays.
Footnotes
Author contribution
CDM and ES carried out all the experiments except those noted otherwise. SH performed all the kinase assays and immuno-precipitations assays. GF and BT performed the fly screen. MT, and PAB made the mathematical model. AB helped with Structured Illumination Microscopy. CDM and E.S. conceived the study and wrote the manuscript.
Competing financial interests
The authors declare no competing financial interests.
References
- 1.Friedl P, Gilmour D. Collective cell migration in morphogenesis, regeneration and cancer. Nature reviews Molecular cell biology. 2009;10:445–457. doi: 10.1038/nrm2720. [DOI] [PubMed] [Google Scholar]
- 2.Kalluri R, Weinberg RA. The basics of epithelial-mesenchymal transition. The Journal of clinical investigation. 2009;119:1420–1428. doi: 10.1172/JCI39104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Keren K, Yam PT, Kinkhabwala A, Mogilner A, Theriot JA. Intracellular fluid flow in rapidly moving cells. Nature cell biology. 2009;11:1219–1224. doi: 10.1038/ncb1965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Charras G, Paluch E. Blebs lead the way: how to migrate without lamellipodia. Nature reviews Molecular cell biology. 2008;9:730–736. doi: 10.1038/nrm2453. [DOI] [PubMed] [Google Scholar]
- 5.Lammermann T, Sixt M. Mechanical modes of 'amoeboid' cell migration. Current opinion in cell biology. 2009;21:636–644. doi: 10.1016/j.ceb.2009.05.003. [DOI] [PubMed] [Google Scholar]
- 6.Vicente-Manzanares M, Ma X, Adelstein RS, Horwitz AR. Non-muscle myosin II takes centre stage in cell adhesion and migration. Nature reviews Molecular cell biology. 2009;10:778–790. doi: 10.1038/nrm2786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wyckoff JB, Pinner SE, Gschmeissner S, Condeelis JS, Sahai E. ROCK- and myosin-dependent matrix deformation enables protease-independent tumor-cell invasion in vivo. Current biology : CB. 2006;16:1515–1523. doi: 10.1016/j.cub.2006.05.065. [DOI] [PubMed] [Google Scholar]
- 8.Sahai E, Marshall CJ. Differing modes of tumour cell invasion have distinct requirements for Rho/ROCK signalling and extracellular proteolysis. Nature cell biology. 2003;5:711–719. doi: 10.1038/ncb1019. [DOI] [PubMed] [Google Scholar]
- 9.Sanz-Moreno V, Marshall CJ. The plasticity of cytoskeletal dynamics underlying neoplastic cell migration. Current opinion in cell biology. 2010;22:690–696. doi: 10.1016/j.ceb.2010.08.020. [DOI] [PubMed] [Google Scholar]
- 10.Madsen CD, Sahai E. Cancer dissemination--lessons from leukocytes. Developmental cell. 2010;19:13–26. doi: 10.1016/j.devcel.2010.06.013. [DOI] [PubMed] [Google Scholar]
- 11.Friedl P, Wolf K. Plasticity of cell migration: a multiscale tuning model. The Journal of cell biology. 2010;188:11–19. doi: 10.1083/jcb.200909003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sanz-Moreno V, et al. Rac activation and inactivation control plasticity of tumor cell movement. Cell. 2008;135:510–523. doi: 10.1016/j.cell.2008.09.043. [DOI] [PubMed] [Google Scholar]
- 13.Matsumura F, Hartshorne DJ. Myosin phosphatase target subunit: Many roles in cell function. Biochemical and biophysical research communications. 2008;369:149–156. doi: 10.1016/j.bbrc.2007.12.090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hartshorne DJ, Ito M, Erdodi F. Role of protein phosphatase type 1 in contractile functions: myosin phosphatase. The Journal of biological chemistry. 2004;279:37211–37214. doi: 10.1074/jbc.R400018200. [DOI] [PubMed] [Google Scholar]
- 15.Grassie ME, Moffat LD, Walsh MP, MacDonald JA. The myosin phosphatase targeting protein (MYPT) family: a regulated mechanism for achieving substrate specificity of the catalytic subunit of protein phosphatase type 1delta. Archives of biochemistry and biophysics. 2011;510:147–159. doi: 10.1016/j.abb.2011.01.018. [DOI] [PubMed] [Google Scholar]
- 16.Parsons JT, Horwitz AR, Schwartz MA. Cell adhesion: integrating cytoskeletal dynamics and cellular tension. Nature reviews Molecular cell biology. 2010;11:633–643. doi: 10.1038/nrm2957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Fehon RG, McClatchey AI, Bretscher A. Organizing the cell cortex: the role of ERM proteins. Nature reviews Molecular cell biology. 2010;11:276–287. doi: 10.1038/nrm2866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Fukata Y, et al. Association of the myosin-binding subunit of myosin phosphatase and moesin: dual regulation of moesin phosphorylation by Rho-associated kinase and myosin phosphatase. The Journal of cell biology. 1998;141:409–418. doi: 10.1083/jcb.141.2.409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Janssens V, Goris J. Protein phosphatase 2A: a highly regulated family of serine/threonine phosphatases implicated in cell growth and signalling. The Biochemical journal. 2001;353:417–439. doi: 10.1042/0264-6021:3530417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Basu S. PP2A in the regulation of cell motility and invasion. Current protein & peptide science. 2011;12:3–11. doi: 10.2174/138920311795659443. [DOI] [PubMed] [Google Scholar]
- 21.Butler T, Paul J, Europe-Finner N, Smith R, Chan EC. Role of serine-threonine phosphoprotein phosphatases in smooth muscle contractility. American journal of physiology Cell physiology. 2013;304:C485–504. doi: 10.1152/ajpcell.00161.2012. [DOI] [PubMed] [Google Scholar]
- 22.Bai SW, et al. Identification and characterization of a set of conserved and new regulators of cytoskeletal organization, cell morphology and migration. BMC biology. 2011;9:54. doi: 10.1186/1741-7007-9-54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chen HW, et al. CKA, a novel multidomain protein, regulates the JUN N-terminal kinase signal transduction pathway in Drosophila. Molecular and cellular biology. 2002;22:1792–1803. doi: 10.1128/MCB.22.6.1792-1803.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Goudreault M, et al. A PP2A phosphatase high density interaction network identifies a novel striatin-interacting phosphatase and kinase complex linked to the cerebral cavernous malformation 3 (CCM3) protein. Molecular & cellular proteomics : MCP. 2009;8:157–171. doi: 10.1074/mcp.M800266-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Glatter T, Wepf A, Aebersold R, Gstaiger M. An integrated workflow for charting the human interaction proteome: insights into the PP2A system. Molecular systems biology. 2009;5:237. doi: 10.1038/msb.2008.75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ribeiro PS, et al. Combined functional genomic and proteomic approaches identify a PP2A complex as a negative regulator of Hippo signaling. Molecular cell. 2010;39:521–534. doi: 10.1016/j.molcel.2010.08.002. [DOI] [PubMed] [Google Scholar]
- 27.Herzog F, et al. Structural probing of a protein phosphatase 2A network by chemical cross-linking and mass spectrometry. Science. 2012;337:1348–1352. doi: 10.1126/science.1221483. [DOI] [PubMed] [Google Scholar]
- 28.Hwang J, Pallas DC. STRIPAK complexes: structure, biological function, and involvement in human diseases. The international journal of biochemistry & cell biology. 2014;47:118–148. doi: 10.1016/j.biocel.2013.11.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Preisinger C, et al. YSK1 is activated by the Golgi matrix protein GM130 and plays a role in cell migration through its substrate 14-3-3zeta. The Journal of cell biology. 2004;164:1009–1020. doi: 10.1083/jcb.200310061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lu TJ, et al. Inhibition of cell migration by autophosphorylated mammalian sterile 20-like kinase 3 (MST3) involves paxillin and protein-tyrosine phosphatase-PEST. The Journal of biological chemistry. 2006;281:38405–38417. doi: 10.1074/jbc.M605035200. [DOI] [PubMed] [Google Scholar]
- 31.Matsuki T, et al. Reelin and stk25 have opposing roles in neuronal polarization and dendritic Golgi deployment. Cell. 2010;143:826–836. doi: 10.1016/j.cell.2010.10.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.ten Klooster JP, et al. Mst4 and Ezrin induce brush borders downstream of the Lkb1/Strad/Mo25 polarization complex. Developmental cell. 2009;16:551–562. doi: 10.1016/j.devcel.2009.01.016. [DOI] [PubMed] [Google Scholar]
- 33.Gloerich M, et al. Rap2A links intestinal cell polarity to brush border formation. Nature cell biology. 2012;14:793–801. doi: 10.1038/ncb2537. [DOI] [PubMed] [Google Scholar]
- 34.Sugden PH, McGuffin LJ, Clerk A. SOcK, MiSTs, MASK and STicKs: the GCKIII (germinal centre kinase III) kinases and their heterologous protein-protein interactions. The Biochemical journal. 2013;454:13–30. doi: 10.1042/BJ20130219. [DOI] [PubMed] [Google Scholar]
- 35.Montell DJ, Rorth P, Spradling AC. slow border cells, a locus required for a developmentally regulated cell migration during oogenesis, encodes Drosophila C/EBP. Cell. 1992;71:51–62. doi: 10.1016/0092-8674(92)90265-e. [DOI] [PubMed] [Google Scholar]
- 36.Montell DJ. Border-cell migration: the race is on. Nature reviews Molecular cell biology. 2003;4:13–24. doi: 10.1038/nrm1006. [DOI] [PubMed] [Google Scholar]
- 37.Montell DJ, Yoon WH, Starz-Gaiano M. Group choreography: mechanisms orchestrating the collective movement of border cells. Nature reviews Molecular cell biology. 2012;13:631–645. doi: 10.1038/nrm3433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Silver DL, Montell DJ. Paracrine signaling through the JAK/STAT pathway activates invasive behavior of ovarian epithelial cells in Drosophila. Cell. 2001;107:831–841. doi: 10.1016/s0092-8674(01)00607-9. [DOI] [PubMed] [Google Scholar]
- 39.Duchek P, Rorth P. Guidance of cell migration by EGF receptor signaling during Drosophila oogenesis. Science. 2001;291:131–133. doi: 10.1126/science.291.5501.131. [DOI] [PubMed] [Google Scholar]
- 40.Duchek P, Somogyi K, Jekely G, Beccari S, Rorth P. Guidance of cell migration by the Drosophila PDGF/VEGF receptor. Cell. 2001;107:17–26. doi: 10.1016/s0092-8674(01)00502-5. [DOI] [PubMed] [Google Scholar]
- 41.Somogyi K, Rorth P. Evidence for tension-based regulation of Drosophila MAL and SRF during invasive cell migration. Developmental cell. 2004;7:85–93. doi: 10.1016/j.devcel.2004.05.020. [DOI] [PubMed] [Google Scholar]
- 42.Kean MJ, et al. Structure-function analysis of core STRIPAK Proteins: a signaling complex implicated in Golgi polarization. The Journal of biological chemistry. 2011;286:25065–25075. doi: 10.1074/jbc.M110.214486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Amano M, Nakayama M, Kaibuchi K. Rho-kinase/ROCK: A key regulator of the cytoskeleton and cell polarity. Cytoskeleton. 2010;67:545–554. doi: 10.1002/cm.20472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Croft DR, et al. Conditional ROCK activation in vivo induces tumor cell dissemination and angiogenesis. Cancer research. 2004;64:8994–9001. doi: 10.1158/0008-5472.CAN-04-2052. [DOI] [PubMed] [Google Scholar]
- 45.Tozluoglu M, et al. Matrix geometry determines optimal cancer cell migration strategy and modulates response to interventions. Nature cell biology. 2013;15:751–762. doi: 10.1038/ncb2775. [DOI] [PubMed] [Google Scholar]
- 46.Friedl P, Wolf K. Plasticity of cell migration: a multiscale tuning model. The Journal of cell biology. 2010;188:11–19. doi: 10.1083/jcb.200909003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Gyorffy B, Lanczky A, Szallasi Z. Implementing an online tool for genome-wide validation of survival-associated biomarkers in ovarian-cancer using microarray data from 1287 patients. Endocrine-related cancer. 2012;19:197–208. doi: 10.1530/ERC-11-0329. [DOI] [PubMed] [Google Scholar]
- 48.Fidalgo M, et al. Adaptor protein cerebral cavernous malformation 3 (CCM3) mediates phosphorylation of the cytoskeletal proteins ezrin/radixin/moesin by mammalian Ste20-4 to protect cells from oxidative stress. The Journal of biological chemistry. 2012;287:11556–11565. doi: 10.1074/jbc.M111.320259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Eto M. Regulation of cellular protein phosphatase-1 (PP1) by phosphorylation of the CPI-17 family, C-kinase-activated PP1 inhibitors. The Journal of biological chemistry. 2009;284:35273–35277. doi: 10.1074/jbc.R109.059972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wolf K, et al. Compensation mechanism in tumor cell migration: mesenchymal-amoeboid transition after blocking of pericellular proteolysis. The Journal of cell biology. 2003;160:267–277. doi: 10.1083/jcb.200209006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Carragher NO, et al. Calpain 2 and Src dependence distinguishes mesenchymal and amoeboid modes of tumour cell invasion: a link to integrin function. Oncogene. 2006;25:5726–5740. doi: 10.1038/sj.onc.1209582. [DOI] [PubMed] [Google Scholar]
- 52.Voss K, et al. Functional analyses of human and zebrafish 18-amino acid in-frame deletion pave the way for domain mapping of the cerebral cavernous malformation 3 protein. Human mutation. 2009;30:1003–1011. doi: 10.1002/humu.20996. [DOI] [PubMed] [Google Scholar]
- 53.Zheng X, et al. CCM3 signaling through sterile 20-like kinases plays an essential role during zebrafish cardiovascular development and cerebral cavernous malformations. The Journal of clinical investigation. 2010;120:2795–2804. doi: 10.1172/JCI39679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Faurobert E, et al. CCM1-ICAP-1 complex controls beta1 integrin-dependent endothelial contractility and fibronectin remodeling. The Journal of cell biology. 2013;202:545–561. doi: 10.1083/jcb.201303044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Davoli T, et al. Cumulative haploinsufficiency and triplosensitivity drive aneuploidy patterns and shape the cancer genome. Cell. 2013;155:948–962. doi: 10.1016/j.cell.2013.10.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Lorentzen A, Bamber J, Sadok A, Elson-Schwab I, Marshall CJ. An ezrin-rich, rigid uropod-like structure directs movement of amoeboid blebbing cells. Journal of cell science. 2011;124:1256–1267. doi: 10.1242/jcs.074849. [DOI] [PubMed] [Google Scholar]
- 57.Petrie RJ, Koo H, Yamada KM. Generation of compartmentalized pressure by a nuclear piston governs cell motility in a 3D matrix. Science. 2014;345:1062–1065. doi: 10.1126/science.1256965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Dietzl G, et al. A genome-wide transgenic RNAi library for conditional gene inactivation in Drosophila. Nature. 2007;448:151–156. doi: 10.1038/nature05954. [DOI] [PubMed] [Google Scholar]
- 58.Hirata E, et al. In vivo fluorescence resonance energy transfer imaging reveals differential activation of Rho-family GTPases in glioblastoma cell invasion. J Cell Sci. 2012;125:858–868. doi: 10.1242/jcs.089995. [DOI] [PubMed] [Google Scholar]
- 59.Yeung T, et al. Effects of substrate stiffness on cell morphology, cytoskeletal structure, and adhesion. Cell Motil Cytoskeleton. 2005;60:24–34. doi: 10.1002/cm.20041. [DOI] [PubMed] [Google Scholar]
- 60.Gyorffy B, et al. An online survival analysis tool to rapidly assess the effect of 22,277 genes on breast cancer prognosis using microarray data of 1,809 patients. Breast Cancer Res Treat. 2010;123:725–731. doi: 10.1007/s10549-009-0674-9. [DOI] [PubMed] [Google Scholar]
- 61.Ringner M, Fredlund E, Hakkinen J, Borg A, Staaf J. GOBO: gene expression-based outcome for breast cancer online. PLoS ONE. 2011;6:e17911. doi: 10.1371/journal.pone.0017911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Cerami E, et al. The cBio cancer genomics portal: an open platform for exploring multidimensional cancer genomics data. Cancer Discov. 2012;2:401–404. doi: 10.1158/2159-8290.CD-12-0095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Gao J, et al. Integrative analysis of complex cancer genomics and clinical profiles using the cBioPortal. Sci Signal. 2013;6:pl1. doi: 10.1126/scisignal.2004088. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.