Abstract
Strain QLW-P1FAT50C-4T, isolated from a shallow acidic freshwater pond located in the Austrian Alps at an altitude of 1300 m, was characterized by investigation of phenotypic, chemotaxonomic and genomic traits. As shown previously, phylogenetic analyses based on 16S rRNA gene sequences place the strain in the cryptic species complex PnecC within the genus Polynucleobacter. The major fatty acids of the strain were C16:1 ω7c and C18:1 ω7c. The strain has a genome of 2.23 Mbp with a GC content of 44.9 mol%. The strain encodes a seemingly complete gene cluster for anoxygenic photosynthesis but lacks typical genes for CO2 assimilation. In order to resolve the phylogenetic position of the strain within the species complex PnecC, concatenated partial sequences of eight housekeeping genes were analysed. The obtained phylogenetic reconstruction did not place strain QLW-P1FAT50C-4T close to any of the five previously described species within subcluster PnecC. Pairwise average nucleotide identity (ANI) comparisons of whole genome sequences suggested that strain QLW-P1FAT50C-4T (DSM 24008T =CIP 111100T) represents a novel species for which we propose the name Polynucleobacter wuianus sp. nov.
The genus Polynucleobacter and the species Polynucleobacter necessarius were proposed by K. Heckmann and H. J. Schmidt (1987) for bacterial endosymbionts of benthic freshwater ciliates affiliated with the genus Euplotes. Later, it was discovered that free-living strains, closely related to those endosymbiotic strains, represent important planktonic freshwater bacteria (Bahr et al., 1996; Hahn, 2003; Percent et al., 2008; Zwart et al., 2002). The genus Polynucleobacter can be subdivided in at least four subclusters labelled PnecA, PnecB, PnecC, and PnecD (Hahn, 2003). Subcluster PnecC, which includes the endosymbiotic P. necessarius and free-living species, was recognized as being a cryptic species complex, which means that this seemingly very species-rich subcluster cannot be resolved by 16S rRNA gene phylogenies (Hahn et al., 2016a; Hahn et al., 2016b). Taxonomic research on strains affiliated with subcluster PnecC is hampered by the lack of a type strain of the genus (Hahn et al., 2016a). Reference material representing the species P. necessarius was contained in the Euplotes aediculatus ‘stock 15’ culture (=E24 =ATCC 30859) (Heckmann and Schmidt, 1987), which is unfortunately not any more available. However, recently, it was shown (Hahn et al., 2016a) that the lost endosymbionts used for the species description are well represented by another endosymbiotic P. necessarius strain, i.e. STIR1, for which a complete genome sequence is available (Boscaro et al., 2013).
Here we describe strain QLW-P1FAT50C-4T affiliated with subcluster PnecC, which was isolated from an acidic freshwater pond, and propose to establish for this strain the species name P. wuianus sp. nov.
Home habitat and isolation
Strain QLW-P1FAT50C-4T shares with the type strain of P. asymbioticus (Hahn et al., 2016a) the same home habitat. Both strains were isolated from water from Pond-1 (geographic coordinates 47°44'23.37"N, 13°18'6.03"E) by Q. L. Wu in October 2003 (Hahn et al., 2005). This pond is located in the Austrian Alps at an altitude of 1300 m (Hahn et al., 2012b). It is a small, shallow (max depth < 1 m) natural freshwater pond, which was usually (years 2003-2013) characterized by a slightly acidic pH in the range of 4.5-5.5 and a quite low conductivity of usually 5-15 µS cm-1.
Isolation of the strain was described in detail elsewhere (Hahn et al., 2005). Briefly, the strain was isolated by using the filtration-acclimatization method and NSY medium (Hahn et al., 2005; Hahn et al., 2004).
Phenotypic and chemotaxonomic characterization
Cells of strain QLW-P1FAT50C-4T are short sometimes slightly curved rods of small to medium size. Cell size depends on growth stage and medium used for cultivation. While size in NSY medium is typically 1 µm cell length (50 cells measured; SD 0.2; range 0.6-1.7 µm) and 0.5 µm cell widths (n, 50; SD, 0.1; range 0.3-0.6 µm), cell size dropped to 0.5 x 0.3 µm when cells were grown in sterile water from their home habitat (Table 1). This cell size equals to a cell volume of 0.03 µm3 placing the strain among the smaller cells in freshwater bacterioplankton (Hahn et al., 2003). Strain QLW-P1FAT50C-4T forms small circular, convex colonies with shiny surface on NSY agar plates and lacks pigmentation. Growth at different temperatures and growth under anoxic conditions in an anaerobic chamber were examined by using NSY agar plates as described previously (Hahn et al., 2009). Furthermore, growth rates of the strain in liquid NSY medium were determined after appropriate thermal acclimatization at four different temperatures. Salinity (NaCl) tolerance was determined using NSY agar supplemented with various NaCl concentrations as described previously (Hahn et al., 2009). The strain showed no anaerobic growth, grew at temperatures up to 34°C and tolerated salt concentrations up to 0.5% (Table 1). Rates of growth in NSY medium determined at 8, 12, 22, and 28°C increased with temperatures and were 0.063 h-1, 0.128 h-1, 0.198 h-1, and 0.273 h-1, respectively.
Table 1.
Traits characterizing the five investigated Polynucleobacter taxa affiliated with subcluster PnecC. All type strains have the following characteristics in common. In motility tests negative; catalase oxidase positive; assimilation of acetate and pyruvate; weak assimilation of D-galacturonic acid; no assimilation of glycolate, oxalate, citrate and L-serine. n.d., not determined; w, weak reaction.
| P. wuianus sp. nov. | P. asymbioticus | P. duraquae | P. sinensis | P. yangtzensis | |
|---|---|---|---|---|---|
| QLW-P1FAT50C-4T | QLW-P1DMWA-1T | MWH-MoK4T | MWH-HuW1T | MWH-JaK3T | |
| (DSM 24008T) | (DSM 18221T) | (DSM 21495T) | (DSM 21492T) | (DSM 21493T) | |
| Cell morphology | short rods | short rods | curved rods | short curved rods | short rods |
| Cell length (µm) | 0.6-1.7 | 0.7-1.2 | 0.9-2.9 | 0.6-1.4 | 0.5-1.5 |
| Cell width (µm) | 0.3-0.6 | 0.4-0.5 | 0.4-0.5 | 0.4-0.5 | 0.3-0.5 |
| Temperature range of growth (°C) | 5 - 34 | 5 - 34(w) | 5 - 30 | 5 - 35 | 5 - 35 |
| NaCl tolerance (%NaCl, w/v) | 0 - 0.5 | 0 - 0.5(w) | 0 - 0.3 | 0 - 0.5 | 0 - 0.3(w) |
| Anaerobic growth | - | + | - | - | + |
| Assimilation of: | |||||
| Glyoxylic acid | w | w | - | - | - |
| Propionic acid | ➕ | + | - | + | + |
| Malonic acid | w | w | - | + | w |
| Oxaloacetic acid | ➕ | - | + | + | + |
| Malic acid | ➕ | + | w | + | + |
| Fumaric acid | ➕ | + | w | + | + |
| Succinic acid | ➕ | + | + | + | + |
| Levulinic acid | w | w | - | - | - |
| D-Mannose | ➖ | w | - | - | - |
| D-Glucose | ➖ | w | w | - | - |
| D-Galactose | ➖ | w | - | - | - |
| D-Lyxose | ➖ | w | w | - | w |
| D-Fructose | w | w | w | - | w |
| L-Fucose | ➖ | w | - | - | w |
| D-Sorbitole | ➖ | w | - | - | - |
| L-Glutamate | ➕ | + | - | + | - |
| L-Histidine | ➕ | n.d. | n.d. | n.d. | n.d. |
| L-Aspartate | ➕ | + | - | - | - |
| L-Cysteine | w | + | + | w | + |
| L-Alanine | ➕ | w | - | - | - |
| L-Asparagine | w | w | - | - | - |
| L-Leucine | ➖ | n.d. | n.d. | n.d. | n.d. |
| Betaine | - | w | - | - | - |
Utilization of various substrates was investigated in the same way as for previously described Polynucleobacter species (Hahn et al., 2011a; Hahn et al., 2011b; Hahn et al., 2009; Hahn et al., 2012a; Hahn et al., 2010). Briefly, growth enabled by utilization of a specific substrate was determined by comparison of OD at 575 nm established in liquid one tenth-strength NSY medium (0.3 g l-1) with and without 0.5 g l-1 test substrate. Differences of < 10 %, 10–50% and >50% of the OD obtained in the test treatments compared to the OD obtained without test substrate (i.e. in 0.3 g l-1 NSY medium) were scored after 10 days of growth as no utilization (-), weak utilization (w) and good utilization (+), respectively (Table 1).
The analysis of the whole-cell fatty acid composition was carried out as described previously (Hahn et al., 2012a). Biomass was harvested from R2A agar plates. Plates were inoculated with 1 ml cell suspension, incubated while keeping the agar surface moist and inspected for growth daily, starting the third day after inoculation. Once a biomass film was well visible, the cell mass was harvested. The results (Table 2) underpin the membership of strain QLW-P1FAT50C-4T in subcluster PnecC, for example by the presence of n-dodecanoic acid, which is lacking in members of subcluster PnecD (P. cosmopolitanus).
Table 2.
Major fatty acid compositions of the Polynucleobacter strains representing subcluster PnecC. Strains were grown on R2A agar. Data for strains QLW-P1DMWA-1T, MWH-HuW1T, MWH-JaK3T were taken from (Hahnet al., 2009) and data for MWH-Mok4T from Hahn et al. (2016b).
| Fatty acid | QLW-P1FAT50C-4T | QLW-P1DMWA-1T | MWH-MoK4T | MWH-HuW1T | MWH-JaK3T |
|---|---|---|---|---|---|
| (DSM 24008T) | (DSM 18221T) | (DSM 21495T) | (DSM 21492T) | (DSM 21493T) | |
| C12:0 | 3.3 | 3.4 | 3.8 | 5.5 | 3.7 |
| C14:0 | 0.3 | 0.9 | 0.3 | 0.3 | 1.2 |
| C15:0 | 0.2 | 0.3 | - | 0.3 | - |
| C16:0 | 18.6 | 22.2 | 15.9 | 29.6 | 15.5 |
| C17:0 | - | - | - | 0.5 | - |
| C18:0 | 0.5 | 1.2 | 0.5 | 2.4 | 0.5 |
| C20:0 | - | 1.1 | - | - | - |
| C14:1 ω5c | - | - | - | 0.2 | 0.6 |
| C15:1 ω6c | - | - | - | 0.6 | - |
| C16:1 ω5c | 0.4 | 0.9 | 0.4 | - | 0.4 |
| C16:1 ω7c | 40.8 | 41.3 | 38.6 | 45.0 | 35.6 |
| C18:1 ω 9c | - | - | 0.3 | 0.4 | - |
| C18:1 ω 7c | 20.4 | 12.9 | 19.8 | 1.1 | 20.4 |
| 11-methyl C18:1 ω7c | 4.6 | 3.1 | 4.2 | 1.1 | 8.1 |
| C12:0 2-OH | 0.3 | 2.5 | 1.3 | 1.3 | 2.2 |
| C16:0 2-OH | 1.5 | - | 1.8 | - | - |
| Feature 1 | - | 0.4 | - | 1.0 | 0.5 |
| Feature 2 | 7.1 | 9.6 | 11.9 | 9.9 | 9.2 |
| Feature 7 | 1.2 | 0.4 | - | 0.3 | 2.0 |
Summed features represent groups of two fatty acids which could not be separated by GLC and the MIDI system, such as summed feature 2 containing C16:1 isoI and C14:0-3OH and summed feature 7 containing C19:1 ω6c and an unknown compound with an ECL of 18.846, or represent compounds with uncertain identity such as summed feature 1 probably containing C12:0 ALDE?.
Genomic characterization
DNA used for genome sequencing was extracted from biomass grown in liquid NSY medium as described previously (Meincke et al., 2012). Two libraries were sequenced by an Illumina and a Roche system, respectively, and the obtained reads were de novo assembled. A shotgun library was paired–end sequenced on GS FLX by using Titanium chemistry (Beckman Coulter Genomics SA, Grenoble, France). Sequencing resulted in 212,176 filtered reads with a mean length of 361 nt. A 8 kb library was mate pair sequenced to receive long jumping distance pair sequences by using a Illumina MiSeq instrument (Eurofins Genomics, Munich, Germany). This sequencing resulted in 570,724 filtered reads with a mean length of 97 nt. A de novo hybrid assembly was conducted using an in-house pipeline that incorporates the software tool newbler 2.9 (Eurofins Genomics). The assembly resulted in 16 contigs contained in nine scaffolds. Some gaps could be closed by in silico analyses and all remaining gaps but one were closed by PCR amplification of gap regions and subsequent Sanger sequencing of amplicons. These efforts resulted in a closed genome sequence with a single gap. Primers constructed in order to bridge and sequence this last gap, resulted in PCR products but repeated efforts to obtain sequences from this amplicon did not result in sequences suitable for gap closure. The obtained PCR products, however, suggested a gap size of < 1000 bp. The obtained genome sequence has a coverage of about 62x and is characterized by a length of 2.23 Mbp and a G+C content of 44.9 mol% (Table 3). The genome sequence was annotated using the IMG/ER annotation pipeline (Markowitz et al., 2012) and deposited in DDBJ/EMBL/GenBank (Accession Number CP015922). Gene finding resulted in 2274 protein coding and 51 RNA genes. The genome of strain QLW-P1FAT50C-4T and the genomes of type strains of all four other previously described free-living Polynucleobacter species affiliated with subcluster PnecC (Hahn et al., 2016a) differ in coding density from the only yet sequenced endosymbiotic P. necessarius strain (Boscaro et al., 2013). While the five free-living strains possess coding densities in the range of 93.1-93.5%, the endosymbiotic strain is characterized by a coding density of less than 78.5%. Furthermore, the endosymbiotic and the free-living strains differ in genome size, consisting of 1.56 Mbp and 2.03-2.32 Mbp, respectively (Table 3).
Table 3.
Genome characteristics of the investigated Polynucleobacter strains.
| Strain | Life style | Genome size (Mbp) | Scaffolds | G+C content (mol%) | DDBJ/EMBL/GenBank accession number | IMG Genome ID | Reference |
|---|---|---|---|---|---|---|---|
| QLW-P1FAT50C-4T (=DSM 24008T) | free-living | 2.23 | 1 | 44.9 | CP015922 | 2596583503 | This study |
| QLW-P1DMWA-1T (=DSM 18221T) | free-living | 2.16 | 1 | 44.8 | CP000655 | 640427129 | Meincke et al., 2012 |
| MWH-MoK4T (=DSM 21495T) | free-living | 2.03 | 1 | 45.2 | CP007501 | 2505313000 | Hahn et al., 2016b |
| MWH-HuW1T (=DSM 21492T) | free-living | 2.32 | 19 | 45.5 | LOJJ00000000 | 2630969031 | Hahn et al., 2016a |
| MWH-JaK3T (=DSM 21493T) | free-living | 2.05 | 42 | 45.4 | LOJI00000000 | 2608642177 | Hahn et al., 2016as |
| STIR1 (Euplotes aediculatus) | endosymbiont | 1.56 | 1 | 45.6 | CP001010 | 2503982034 | Boscaro et al., 2013 |
While all five compared free-living strains possess similar genome sizes and G+C contents, they differ from each other pronouncedly in gene content (Table 4). Strain QLW-P1FAT50C-4T shares, for instance, the presence of various genes putatively involved in acquisition of inorganic nitrogen with the type strains of P. asymbioticus and P. yangtzensis but shares the presence of a large gene cluster putatively encoding an anoxygenic photosynthesis system only with the type strain of P. duraquae. The gene content listed in Table 4 represents only an incomplete list of content differences, however, this selection shows that all five type strains possess unique combinations of genes not shared by any other so far sequenced Polynucleobacter strain. Currently, it is unknown if listed genes present in a type strain are also present in all strains affiliated with the species represented by the type strain.
Table 4. Comparison of the presence and absence of selected genes.
| Genes putatively encoding | P. wuianus sp. nov. | P. asymbioticus | P. duraquae | P sinensis | P. yangtzensis |
|---|---|---|---|---|---|
| QLW-P1FAT50C-4T | QLW-P1DMWA-1T | MWH-MoK4T | MWH-HuW1T | MWH-JaK3T | |
| Inorganic nutrients | |||||
| ABC-type Fe3+ transport system | ➖ | ➖ | ➕ | ➕ | ➕ |
| feoAB genes (uptake of Fe2+) | ➕ | ➕ | ➖ | ➕ | ➕ |
| ABC-type Nitrate/Nitrite/Cyanate transporter | ➕ | ➕ | ➖ | ➖ | ➕ |
| Nitrate reductase (assimilatory) | ➕ | ➕ | ➖ | ➖ | ➕ |
| Nitrite reductase (assimilatory) | ➕ | ➕ | ➖ | ➖ | ➕ |
| Cyanate lyase (releases NH3 and CO2 from cyanate) | ➕ | ➕ | ➖ | ➖ | ➕ |
| Urease and ABC-type urease transporter | ➕ | ➕ | ➖ | ➖ | ➖ |
| Oxidative phosphorylation/Energy metabolism | |||||
| Cytochrome bd-I terminal oxidase (CydAB) | ➕ | ➕ | ➖ | ➕ | ➖ |
| Fumarate reductase | ➕ | ➖ | ➕ | ➖ | ➕ |
| Carbon monoxide dehydrogenase | ➕ | ➖ | 2 clusters | ➖ | ➕ |
| Acetate permease actP | ➖ | ➕ | ➖ | ➖ | ➖ |
| Anoxygenic photosynthesis | |||||
| Photosynthesis gene cluster | ➕ | ➖ | ➕ | ➖ | ➖ |
| Motility | |||||
| Flagella genes | ➖ | ➖ | ➕ | ➖ | ➖ |
| Oxidative stress | |||||
| Catalase | ➖ | 2 genes | ➖ | ➖ | 1 gene |
| Other | |||||
| Cellulose synthase operon protein C | ➖ | ➕ | ➖ | ➖ | ➖ |
| Cellulose synthase catalytic subunit [UDP-forming] | ➖ | ➕ | ➖ | ➖ | ➖ |
Phylogeny
The 16S rRNA gene sequence and the 16S-23S internal transcribed spacer (ITS) sequences of strain QLW-P1FAT50C-4T determined by previous PCR amplicon sequencing and by genome sequencing were identical, respectively. The 16S rRNA gene sequences of the strain and closely related strains (Hahn et al., 2005) is characterized by the lack of a base in E. coli numbering position 1007. All other investigated Polynucleobacter strains (109 strains) possess a guanine base in this position, which is in secondary structure models of the molecule base-pairing with an adenine base in E. coli numbering position 1022. Interestingly, QLW-P1FAT50C-4T and closely related strains share with all other investigated Polynucleobacter strains the presence of the adenine base in position 1022.
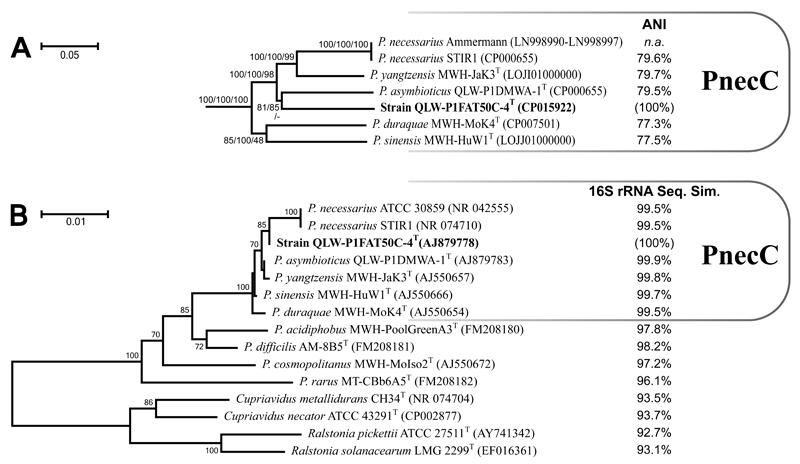
Phylogenetic reconstructions based on 16S rRNA gene sequences (Fig. 1B) place strain QLW-P1FAT50C-4T in subcluster PnecC (Hahn, 2003) of the genus Polynucleobacter (Hahn et al., 2016a). As reported previously, strains affiliated with this species complex share 16S rRNA genes with sequence similarities ≥ 99% (Fig. 1B). In order to better resolve the phylogenetic position of strain QLW-P1FAT50C-4T a tree based on concatenated multilocus sequences was calculated (Fig. 1A). Partial sequences of eight loci (rpoB, trpE, icdA, glnA, mdh, fbp, msbA, and gyrA) defined previously (Hahn et al., 2016a) were extracted from genome sequences (Table 3), concatenated and aligned by using the software MEGA7 (Kumar et al., 2016). This resulted in a total alignment length of 6359 bp. The same software was used for calculation of neighbour-joining (NJ), maximum likelihood (ML) and maximum parsimony (MP) trees (Fig. 1A). In previous pairwise comparisons, sequence similarity values of these concatenated sequences correlated well with average nucleotide identify (ANI) values of whole genome sequences (Hahn et al., 2016a). This suggests that this locus selection is somehow representative for the entire genomes. The phylogenetic reconstruction based on this multilocus sequence set also placed strain QLW-P1FAT50C-4T in subcluster PnecC but did not suggest that the strain is affiliated with any previously described species.
Fig. 1. Phylogenetic position of strain QLW-P1FAT50C-4T.
A, neighbour-joining (NJ) tree calculated with concatenated multilocus sequences of eight protein-encoding genes. The tree was rooted with sequences of two Cupriavidus strains (not shown). Bootstrap values are shown from left to right for NJ, maximum likelihood, and maximum parsimony trees calculated with the same sequence set. Percent values behind the strain names indicate ANI values obtained in comparison with strain QLW-P1FAT50C-4T. B, NJ tree calculated with 16S rRNA gene sequences. Percent values behind the strain names indicate 16S rRNA sequence similarity values obtained in comparison with strain QLW-P1FAT50C-4T.
Ecology
Until recently, all free-living Polynucleobacter strains affiliated with subcluster PnecC including strain QLW-P1FAT50C-4T were considered to belong to P. necessarius subspecies asymbioticus (Hahn et al., 2009). All these free-living strains have a planktonic lifestyle in common, that is, they dwell freely suspended in the water column of freshwater habitats (Wu & Hahn, 2006). Jezbera and colleagues, however, recognized that subcluster PnecC represents an ecologically heterogeneous taxon not resolvable by methods targeting 16S rRNA sequences (Jezbera et al., 2011). They could characterize a couple of subgroups within subcluster PnecC by using signature sequences contained in the 16S-23S ITS sequences of the strains and demonstrated differences in ecological preferences among the subgroups by using a cultivation-independent detection method. A major ecological difference between some groups was the preferences for either acidic or alkaline habitats. Strain QLW-P1FAT50C-4T belongs to subgroup F4 characterized by the signature sequence 5’-ACCATCAGCAGCAGTGATA-3’ (Jezbera et al., 2011). In a cultivation-independent survey covering a large area located in Central Europe (Austria and Czech Republic), this subgroup was only detected in acidic (pH 4 – 6.5) ponds located at altitudes > 1000 m in the Austrian Alps (Jezbera et al., 2011), however, strains sharing the diagnostic sequence of group F4 were also isolated from habitats in Denmark and Finland (Hahn et al., 2015; Jezbera et al., 2011). The distribution of subgroup F4 was found to be negatively correlated with pH and conductivity of freshwater habitats, and positively with a proxy for the concentration of dissolved humic matter (Jezbera et al., 2011). Currently, it is unknown if subgroup F4 consists only of a single or multiple species. A preference of strain QLW-P1FAT50C-4T for acidic habitats is also suggested by the presence of genes putatively encoding a Fe(II) transporter (Table 4) and the absence of genes encoding Fe(III) transporters (Hahn et al., 2016b).
Proposal of the new species Polynucleobacter wuianus sp. nov.
We tested if strain QLW-P1FAT50C-4T has to be considered to be affiliated with one of the four previously described free-living species affiliated with subcluster PnecC by performing average nucleotide identity (ANI) analyses with whole genome sequences by using the IMG/ER system (Markowitz et al., 2012). This resulted in all pairwise comparisons in ANI values of 77.3-79.7% (Fig. 1A), which suggests that the strain is not affiliated with any of these four species (Kim et al., 2014; Konstantinidis & Tiedje, 2005; Konstantinidis & Tiedje, 2007; Konstantinidis et al., 2006; Rosselló-Móra & Amann, 2015). A comparison of strain QLW-P1FAT50C-4T with the type material of the species P. necessarius cannot be performed due to the lack of a genome sequence or pure genomic DNA of the endosymbionts, however, previous investigations suggested that the genome sequence of the endosymbiont P. necessarius STIR1 has to be considered to share a highly similar genome sequence with the endosymbiont described as P. necessarius (Hahn et al., 2016a). An ANI comparison of the genomes of QLW-P1FAT50C-4T and STIR1 resulted in an ANI value of 79.6%, which is very similar to those values obtained for the four free-living type strains (Fig. 1A). Thus, strain QLW-P1FAT50C-4T has to be considered to represent a new species affiliated with subcluster PnecC of the genus Polynucleobacter.
Strain QLW-P1FAT50C-4T can be discriminated from the type strains of Polynucleobacter species not affiliated with subcluster PnecC by chemotaxonomic traits. As for other species affiliated with subcluster PnecC, strain QLW-P1FAT50C-4T can be discriminated from the type strains of P. rarus (Hahn et al., 2011a), P. acidiphobus (Hahn et al., 2011b) and P. difficilis (Hahn et al., 2012a) based on the G+C content of their DNA (Hahn et al., 2016a). The discrimination of strain QLW-P1FAT50C-4T from P. cosmopolitanus is possible by the absence of the fatty acid C12:03-OH (Hahn et al., 2010), which was so far only found in Polynucleobacter strains affiliated with the species P. cosmopolitanus. Furthermore, strain QLW-P1FAT50C-4T, like all other PnecC strains, contains the signature sequence 5’-GAGCCGGTGTTTCTTCCC-3’ at Escherichia coli position 445–463 of the 16S rRNA, which is absent in Polynucleobacter strains not affiliated with this subcluster (Hahn et al., 2016a). A feature distinguishing strain QLW-P1FAT50C-4T from previously described type strains of species affiliated with subcluster PnecC is the assimilation of L-alanine (Table 1). Only this strain showed a substantial assimilation of this amino acid while the other type strains showed no or only a weak assimilation.
Description of Polynucleobacter wuianus sp. nov.
Polynucleobacter wuianus (wu.i.a′nus. N.L. masc. adj. wuianus, of Wu, named after Qinglong L. Wu, Chinese microbiologist and microbial ecologist, who isolated the type strain and strongly contributed to research on diversity of Polynucleobacter and other freshwater bacteria).
Contains free-living Polynucleobacter strains dwelling in the water column of acidic or circum-neutral freshwater systems. Never found in alkaline waters (Jezbera et al., 2011). Cells are short sometimes slightly curved rods, 0.5-1.7 µm in length and 0.3–0.6 µm in width, depending on cultivation conditions. Chemo-organotrophic, aerobic, anaerobic growth was not observed. The type strain encodes a gene cluster for anoxygenic photosynthesis but pigmentation of cells or colonies indicating expression of a photosynthesis system was not observed so far. Does not encode genes for synthesis of flagella. Colonies grown on NSY agar are non-pigmented, circular and convex with smooth surface. Growth occurs up to 34 °C. Growth occurs in 0–0.5% (w/v) NaCl. Assimilates acetate, propionate, pyruvate, oxaloacetate, malate, fumarate, succinate, L-glutamate, L-aspartate, L-histidine, and L-alanine. Does not assimilate glycolate, oxalate, citrate, D-mannose, D-glucose, D-galactose, D- lyxose, L-fucose, D-sorbitol, L-leucine, L-serine, or betaine. Major cellular fatty acids are C16:1 ω7c and C18:1 ω7c. The type strain is QLW-P1FAT50C-4T (=DSM 24008T = CIP 111100T), which was isolated from acidic Pond-1 located in the Austrian Alps. The genome of the type strain is characterized by a size of 2.23 Mbp and a G+C content of 44.9 mol%. Genome and 16S rRNA gene sequences characterizing the type strain are available under the accession numbers CP015922 and AJ879778, respectively.
Acknowledgements
We thank Gabriele Pötter for carrying out the fatty acid measurements. This study was supported by the Austrian Science Fund (FWF) project I482-B09 (Ecological diversification in Polynucleobacter), and the European Science Foundation (ESF) project FREDI.
Footnotes
References
- Bahr M, Hobbie JE, Sogin ML. Bacterial diversity in an arctic lake: A freshwater SAR11 cluster. Aquat Microb Ecol. 1996;11:271–277. [Google Scholar]
- Boscaro V, Felletti M, Vannini C, Ackerman MS, Chain PSG, Malfatti S, Vergez LM, Shin M, Doak TG, et al. Polynucleobacter necessarius, a model for genome reduction in both free-living and symbiotic bacteria. Proc Natl Acad Sci USA. 2013;110:18590–18595. doi: 10.1073/pnas.1316687110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hahn MW. Isolation of strains belonging to the cosmopolitan Polynucleobacter necessarius cluster from freshwater habitats located in three climatic zones. Appl Environ Microbiol. 2003;69:5248–5254. doi: 10.1128/AEM.69.9.5248-5254.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hahn MW, Pockl M, Wu QL. Low intraspecific diversity in a Polynucleobacter subcluster population numerically dominating bacterioplankton of a freshwater pond. Appl Environ Microbiol. 2005;71:4539–4547. doi: 10.1128/AEM.71.8.4539-4547.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hahn MW, Stadler P, Wu QL, Pockl M. The filtration-acclimatization method for isolation of an important fraction of the not readily cultivable bacteria. J Microbiol Methods. 2004;57:379–390. doi: 10.1016/j.mimet.2004.02.004. [DOI] [PubMed] [Google Scholar]
- Hahn MW, Lang E, Tarao M, Brandt U. Polynucleobacter rarus sp. nov., a free-living planktonic bacterium isolated from an acidic lake. Int J Syst Evol Microbiol. 2011a;61:781–787. doi: 10.1099/ijs.0.017350-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hahn MW, Lang E, Brandt U, Sproeer C. Polynucleobacter acidiphobus sp. nov., a representative of an abundant group of planktonic freshwater bacteria. Int J Syst Evol Microbiol. 2011b;61:788–794. doi: 10.1099/ijs.0.023929-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hahn MW, Koll U, Jezberova J, Camacho A. Global phylogeography of pelagic Polynucleobacter bacteria: Restricted geographic distribution of subgroups, isolation by distance, and influence of climate. Environ Microbiol. 2015;17:829–840. doi: 10.1111/1462-2920.12532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hahn MW, Lang E, Brandt U, Wu QL, Scheuerl T. Emended description of the genus Polynucleobacter and the species Polynucleobacter necessarius and proposal of two subspecies, P. necessarius subsp. necessarius subsp. nov. and P. necessarius subsp. asymbioticus subsp. nov. Int J Syst Evol Microbiol. 2009;59:2002–2009. doi: 10.1099/ijs.0.005801-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hahn MW, Minasyan A, Lang E, Koll U, Sproeer C. Polynucleobacter difficilis sp. nov., a planktonic freshwater bacterium affiliated with subcluster B1 of the genus Polynucleobacter. Int J Syst Evol Microbiol. 2012a;62:376–383. doi: 10.1099/ijs.0.031393-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hahn MW, Schmidt J, Pitt A, Taipale SJ, Lang E. Reclassification of four Polynucleobacter necessarius strains as Polynucleobacter asymbioticus comb. nov., Polynucleobacter duraquae sp. nov., Polynucleobacter yangtzensis sp. nov., and Polynucleobacter sinensis sp. nov., and emended description of the species Polynucleobacter necessarius. Int J Syst Evol Microbiol. 2016a;66:2883–2892. doi: 10.1099/ijsem.0.001073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hahn MW, Jezberová J, Koll U, Saueressig-Beck T, Schmidt J. Complete ecological isolation and cryptic diversity in Polynucleobacter bacteria not resolved by 16S rRNA gene sequences. ISME J. 2016b;10:1642–1655. doi: 10.1038/ismej.2015.237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hahn MW, Lang E, Brandt U, Luensdorf H, Wu QL, Stackebrandt E. Polynucleobacter cosmopolitanus sp. nov., free-living planktonic bacteria inhabiting freshwater lakes and rivers. Int J Syst Evol Microbiol. 2010;60:166–173. doi: 10.1099/ijs.0.010595-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hahn MW, Lunsdorf H, Wu QL, Schauer M, Hofle MG, Boenigk J, Stadler P. Isolation of novel ultramicrobacteria classified as Actinobacteria from five freshwater habitats in Europe and Asia. Appl Environ Microbiol. 2003;69:1442–1451. doi: 10.1128/AEM.69.3.1442-1451.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hahn MW, Scheuerl T, Jezberova J, Koll U, Jezbera J, Simek K, Vannini C, Petroni G, Wu QLL. The passive yet successful way of planktonic life: Genomic and experimental analysis of the ecology of a free-living Polynucleobacter population. Plos One. 2012b;7 doi: 10.1371/journal.pone.0032772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heckmann K, Schmidt HJ. Polynucleobacter necessarius gen. nov., sp. nov., an obligately endosymbiotic bacterium living in the cytoplasm of Euplotes aediculatus. Int J Syst Bacteriol. 1987;37:456–457. [Google Scholar]
- Jezbera J, Jezberova J, Brandt U, Hahn MW. Ubiquity of Polynucleobacter necessarius subspecies asymbioticus results from ecological diversification. Environ Microbiol. 2011;13:922–931. doi: 10.1111/j.1462-2920.2010.02396.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim M, Oh H-S, Park S-C, Chun J. Towards a taxonomic coherence between average nucleotide identity and 16S rRNA gene sequence similarity for species demarcation of prokaryotes. Int J Syst Evol Microbiol. 2014;64:346–351. doi: 10.1099/ijs.0.059774-0. [DOI] [PubMed] [Google Scholar]
- Konstantinidis K, Tiedje J. Genomic insights that advance the species definition for prokaryotes. Proc Natl Acad Sci USA. 2005;102:2567–2572. doi: 10.1073/pnas.0409727102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Konstantinidis KT, Tiedje JM. Prokaryotic taxonomy and phylogeny in the genomic era: advancements and challenges ahead. Curr Opin Microbiol. 2007;10:504–509. doi: 10.1016/j.mib.2007.08.006. [DOI] [PubMed] [Google Scholar]
- Konstantinidis KT, Ramette A, Tiedje JM. The bacterial species definition in the genomic era. Philosophical Transactions of the Royal Society B: Biological Sciences. 2006;361:1929–1940. doi: 10.1098/rstb.2006.1920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar S, Stecher G, Tamura K. MEGA7: Molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol Biol Evol. 2016 doi: 10.1093/molbev/msw054. (in press) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Markowitz VM, Chen IMA, Palaniappan K, Chu K, Szeto E, Grechkin Y, Ratner A, Jacob B, Huang JH, et al. IMG: the integrated microbial genomes database and comparative analysis system. Nucleic Acids Res. 2012;40:D115–D122. doi: 10.1093/nar/gkr1044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meincke L, Copeland A, Lapidus A, Lucas S, Berry KW, Del Rio TG, Hammon N, Dalin E, Tice H, et al. Complete genome sequence of Polynucleobacter necessarius subsp. asymbioticus type strain (QLW-P1DMWA-1T) Stand Genomic Sci. 2012;6:74–83. doi: 10.4056/sigs.2395367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Percent SF, Frischer ME, Vescio PA, Duffy EB, Milano V, McLellan M, Stevens BM, Boylen CW, Nierzwicki-Bauer SA. Bacterial community structure of acid-impacted lakes: What controls diversity? Appl Environ Microbiol. 2008;74:1856–1868. doi: 10.1128/AEM.01719-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosselló-Móra R, Amann R. Past and future species definitions for Bacteria and Archaea. Syst Appl Microbiol. 2015;38:209–216. doi: 10.1016/j.syapm.2015.02.001. [DOI] [PubMed] [Google Scholar]
- Wu QL, Hahn MW. Differences in structure and dynamics of Polynucleobacter communities in a temperate and a subtropical lake, revealed at three phylogenetic levels. FEMS Microbiol Ecol. 2006;57:67–79. doi: 10.1111/j.1574-6941.2006.00105.x. [DOI] [PubMed] [Google Scholar]
- Zwart G, Crump BC, Agterveld M, Hagen F, Han SK. Typical freshwater bacteria: an analysis of available 16S rRNA gene sequences from plankton of lakes and rivers. Aquat Microb Ecol. 2002;28:141–155. [Google Scholar]