Abstract
Decades of research identify the hippocampal formation as central to memory storage and recall. Events are stored via distributed population codes, the parameters of which (e.g., sparsity and overlap) determine both storage capacity and fidelity. However, it remains unclear whether the parameters governing information storage are similar between species. Because episodic memories are rooted in the space in which they are experienced, the hippocampal response to navigation is often used as a proxy to study memory. Critically, recent studies in rodents that mimic the conditions typical of navigation studies in humans and nonhuman primates (i.e., virtual reality) show that reduced sensory input alters hippocampal representations of space. The goal of this study was to quantify this effect and determine whether there are commonalities in information storage across species. Using functional molecular imaging, we observe that navigation in virtual environments elicits activity in fewer CA1 neurons relative to real-world conditions. Conversely, comparable neuronal activity is observed in hippocampus region CA3 and the dentate gyrus under both conditions. Surprisingly, we also find evidence that the absolute number of neurons used to represent an experience is relatively stable between nonhuman primates and rodents. We propose that this convergence reflects an optimal ensemble size for episodic memories.
SIGNIFICANCE STATEMENT One primary factor constraining memory capacity is the sparsity of the engram, the proportion of neurons that encode a single experience. Investigating sparsity in humans is hampered by the lack of single-cell resolution and differences in behavioral protocols. Sparsity can be quantified in freely moving rodents, but extrapolating these data to humans assumes that information storage is comparable across species and is robust to restraint-induced reduction in sensory input. Here, we test these assumptions and show that species differences in brain size build memory capacity without altering the structure of the data being stored. Furthermore, sparsity in most of the hippocampus is resilient to reduced sensory information. This information is vital to integrating animal data with human imaging navigation studies.
Keywords: neural coding, neuroethology, primate, rodent, spatial cognition, virtual reality
Introduction
The hippocampus, situated at the top of the hierarchy of the association cortices (Marr, 1971; McNaughton and Morris, 1987; Rolls and Treves, 1990; Buffalo, 2015), is central to both navigation and episodic memory, reflecting the deep connection between memory and place (O'Keefe and Nadel, 1978; Burgess et al., 2002; Buzsáki and Moser, 2013; Buffalo, 2015). Electrophysiological studies across species demonstrate that hippocampal activity integrates many features of the animal's external surroundings, as well as internal states and previous experience (Wilson and McNaughton, 1993; Ono et al., 1993; Rolls, 1999; Ekstrom et al., 2003; Wirth et al., 2003; Quiroga et al., 2008; MacDonald et al., 2011; Mankin et al., 2012; Thome et al., 2012). The mechanisms underlying hippocampal information storage have been the focus of extensive investigation (Treves and Rolls, 1994; Burgess et al., 2002; Waydo et al., 2006). Storage capacity depends on the number of coding units, the selectivity of individual neurons (population sparsity), and the pattern of connections between them (Marr, 1971; McNaughton and Morris, 1987; Rolls and Treves, 1990). The hippocampus is hypothesized to use sparse coding schemes to maximize memory storage capacity (Marr, 1971; McNaughton and Morris, 1987). Critically, although the number of place fields a neuron expresses depends in part on the size of the environment and the site of recording (Rich et al., 2014; Strange et al., 2014), functional imaging studies show that rodents reliably activate similar numbers of neurons across episodes. This stability in ensemble size suggests a mechanism that constrains the number of neurons involved in hippocampal memory encoding.
Several factors limit the degree to which these rodent data may generalize to primates. The vast majority of human and nonhuman primate (NHP) studies of navigation (and memory more generally) are performed while participants are physical restrained due to the requirements of the particular experimental technique. This is problematic because restricting self-motion information reduces the number and specificity of active neurons in rodent CA1 (Foster et al., 1989; Terrazas et al., 2005; Rich et al., 2014). Moreover, whereas electrophysiological studies in humans and NHPs can demonstrate precise spatial tuning of individual neurons (Hori et al., 2005; Waydo et al., 2006; Quiroga et al., 2008; Thome et al., 2012; Furuya et al., 2014), this method provides only limited sampling of the total cells required to represent an episode.
Many of these challenges may be overcome by monitoring the transcription of the immediate-early gene Arc (Guzowski et al., 1999). Arc expression reliably captures neuronal activity across the brain, providing a robust estimate of ensemble size. Importantly, Arc can also be examined in the same manner in restrained or freely moving animals. We trained six animals to forage for reward in real or virtual environments (Fig. 1, Movies 1 and 2). After completion of the task, we performed fluorescence in situ hybridization for Arc in the right posterior hippocampus (Fig. 2). This region shows reliable activation related to spatial/episodic memory in humans and NHPs (Colombo et al., 1998; Burgess et al., 2002; Engle et al., 2016) and is homologous to the rodent dorsal hippocampus (Amaral and Lavenex, 2007).
Figure 1.

Navigation in real and virtual environments. Monkeys foraging in real-world environments (A) moved freely around two visually distinct rooms while being followed by the experimenter. Random locations around the room were continuously baited with food reward by another experimenter. Monkeys foraging in a virtual environment (B) were seated in front of a wraparound display with access to a joystick (inset) and required to find and collide with large red boxes (visible on center screen) to receive a juice reward. See Movie 1 and Movie 2.
Figure 2.
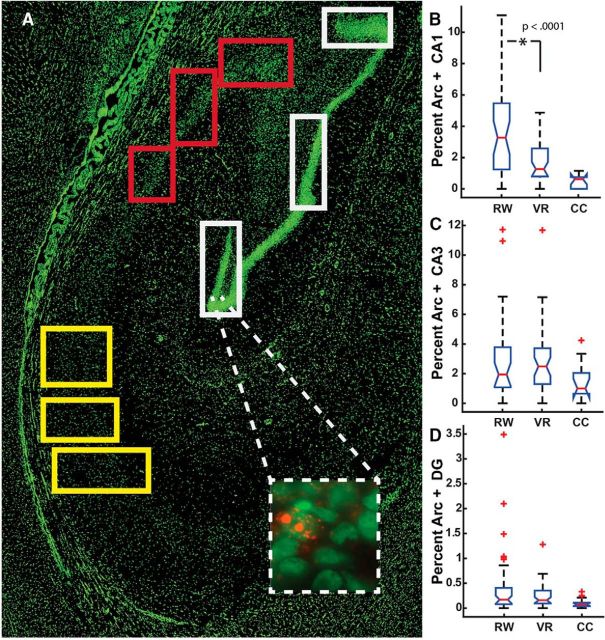
Functional molecular imaging of behaviorally driven hippocampal activity. A, Representative 10× image showing a section of right posterior hippocampus. Nuclei were labeled with Sytox green. Colored boxes (i.e., panels) demonstrate the sampling scheme (white, dentate gyrus; red, CA3; and yellow, CA1). Inset, 40× image showing a cluster of dentate granule cells. Red dots indicate sites of intranuclear transcription of Arc mRNA. B–D, Box-and-whisker plots of regional proportions of neurons between experimental conditions (RW, real-world; VR, virtual reality). Red line represents sample median, box edges represent 25th and 75th percentiles, and whiskers extend 2.7 SDs. Red + symbols represent outliers. Only the CA1 region (B) showed significant differences in proportion of cells activated by real and virtual navigation. All navigation conditions were significantly different from CC.
Real-world navigation in rhesus macaques. This movie depicts a rhesus macaque being guided through one of the two rooms that were used for navigation. The monkey can be seen traveling to foraging locations scattered throughout the room where researchers placed preferred foods.
Virtual navigation in rhesus macaques. This movie depicts a rhesus macaque seated in front of a wraparound display depicting a virtual environment. Within this environment, red cubes appear in pseudo-random locations, and the monkey can be seen using a joystick to navigate to several of these cubes in order to obtain fluid reward.
The aim of the current study was twofold. First, we tested the hypothesis that the degradation of self-motion signals during virtual navigation alters the composition of hippocampal ensembles. Second, we investigated whether, relative to smaller, well characterized mammals, the larger NHP hippocampus will recruit proportionately more neurons to represent a single event, thus keeping relative sparsity constant.
Materials and Methods
Subjects.
The present study used 8 adult male (weight, 9–15 pounds) rhesus macaques (Macacca mulatta). All procedures were approved by the Institutional Animal Care and Use Committee at the University of Arizona and followed the guidelines of the National Institutes of Health.
Behavioral training.
Two animals were trained exclusively on the real-world task, two exclusively on the virtual-foraging task, and an additional two received training on both the real-world and virtual-foraging tasks. Two further animals received no training and served as positive and negative control subjects. Animals in the behavioral groups were placed on fluid restriction and received extensive training for several months as described below.
Each session began with a 25 min rest period during which animals remained in a restraint chair in a light- and sound-isolating transport cage. Behavior during this time was monitored by a small infrared camera. This rest period ensured that all transcription associated with transport was translated and degraded by the time the monkey was killed. Animals trained on either real or virtual environments, returned to the transport chamber for 25 min, and then performed a second round of either real or virtual exploration.
Animals that were assigned to forage in real-world conditions only were randomly placed in either room A or room B, whereas those in the virtual-only condition foraged in the virtual environment. Animals in the mixed real-world and virtual condition were randomly assigned to room A or B, along with the virtual environment.
In the real-world condition, animals were trained to calmly forage while being followed by an experimenter holding a primate restraint poll (Movie 1) to facilitate the retrieval of the animal following training. During each training session, animals were exposed to one of two rooms with unique geometry and local cues. Experimenters continuously baited random locations in the room with small food items (e.g., watermelon, strawberries) to ensure that monkeys explored the entire room.
In the virtual navigation condition, animals were trained to navigate using a joystick to move around a virtual arena displayed across three 40 by 30 cm screens in search for large red squares placed randomly in the environment (Fig. 1B, Movie 2). Reward was delivered via a juice spigot near the animal's mouth as the subject collided with each square. Several cues were placed throughout the environment (i.e., trees, crates, and clouds) with a bounding border of a large, stone-textured wall. Clouds were simulated using a 512 × 512 sky-box (Terragen; Planetside Software). The virtual reality system was custom written (T.M.E. in OpenGL 1.2) and rendered using NVIDIA hardware (NVIDIA GeForce2 MX/MX400).
The duration of training trials was gradually reduced until all behavioral animals were placed on the following daily schedule: 25 min rest, 5 min navigation, 25 min rest, and 5 min navigation. The timing of each behavioral epoch was selected to correspond to the spatiotemporal dynamics of Arc expression.
Two monkeys served as negative and positive controls. The negative control received no behavioral exposure and was anesthetized in its home cage via intramuscular injection of ketamine (30 mg/kg). Data from the negative control animal are referred to as caged control (CC) in the text and figures. The positive control animal received electroconvulsive stimulation (ECS) using procedures typically used for human electroconvulsive therapy (Moscrip et al., 2004). The positive control animal was sedated with a combination of ketamine (15 mg/kg, IM) and atropine (0.5 mg/kg, IM), transported to the surgical suite, and then administered methohexital (1 mg/kg, IV) and succinylcholine (3.5 mg/kg, IV). Vital signs (blood pressure, endtidal CO2, pulse oximetry, and EKG) were monitored continuously. After full anesthesia, the monkey received 100% oxygen (positive pressure) and stimulating electrodes (Somatics) were placed in traditional bifrontotemporal configuration. Electrical stimulation was delivered for 1.9 s at 100 mC and the duration of the following seizure was monitored on a Spectrum 5000Q ECT device (MECTA). Manifestation of motor symptoms were monitored using the cuff technique (American Psychiatric Association, 2001). The monkey was then given an overdose of beuthanasia (sodium pentobarbital, 10 cc/10 lbs, IV).
Tissue processing.
After the behavioral exposure, animals were returned to the primate restraint chair, anesthetized rapidly via gas anesthesia, and then administered an overdose of beuthanasia. Their brains were then rapidly extracted, portioned into 1-cm-thick slices and flash frozen. Blocks from each animal containing right posterior hippocampus were serially sectioned at 25 μm and mounted onto 3-triethoxysilylpropylamine slides. Tissue sections were placed on slides so that each slide contained sections from two different animals, interleaving animals and conditions. Every fifth section was separately mounted for Nissl staining to aid in identifying anatomical landmarks. Slides were dried and stored at −80°C until use.
Fluorescence in situ hybridization was performed as described previously (Ramirez-Amaya et al., 2013). Deviations from this procedure are described below. The specificity of the Arc riboprobe (NCBI Gene ID: 102130416) was confirmed by tissue from the monkey that received ECS. Hybridized tissue was imaged using a 40× oil-immersion objective on a DeltaVision RT Deconvolution Microscope (Applied Precision). Using a rhesus macaque atlas (Paxinos et al., 1999), hippocampal subregions (CA1, CA3, and DG) were identified (Fig. 2A). Across all regions of the hippocampus, 265 image stacks were obtained at 40× (2–3 stacks/region in each of 5 sections/animal).
While the experimental design permits analysis of two behavioral epochs, the present study focuses on the final behavioral exposure, which is associated with intranuclear signals (Fig. 2A, inset) to maintain adequate statistical power. Arc+ neurons were counted using an unbiased brick (Howard et al., 1985) with 5 μm exclusion edges using FIJI (Schindelin et al., 2012) by two observers who were blind to the experimental conditions. To compare the values to similar studies in rodents, we performed a synthesis of the literature published in the last 16 years that provide Arc expression data for CA1 (Guzowski et al., 1999; Vazdarjanova et al., 2002; Vazdarjanova and Guzowski, 2004; Burke et al., 2005; Ramirez-Amaya et al., 2005; Rosi et al., 2005; Guzowski et al., 2006; Vazdarjanova et al., 2006; Marrone et al., 2008; Miyashita et al., 2009; Rosi et al., 2009; Penner et al., 2011; Gheidi et al., 2012; Marrone et al., 2012a,2012b; Hartzell et al., 2013; Marrone et al., 2014), CA3 (Vazdarjanova et al., 2002; Vazdarjanova and Guzowski, 2004; Ramirez-Amaya et al., 2005; Rosi et al., 2005; Vazdarjanova et al., 2006; Miyashita et al., 2009; Rosi et al., 2009; Gheidi et al., 2012; Marrone et al., 2012a; Marrone et al., 2014), or DG (Chawla et al., 2005; Ramirez-Amaya et al., 2005; Rosi et al., 2005; Ramirez-Amaya et al., 2006; Vazdarjanova et al., 2006; Alme et al., 2010; Penner et al., 2011,Marrone et al., 2012a,2012b; Ramirez-Amaya et al., 2013).
Results
Navigation in both real and virtual environments reliably induce Arc transcription
Relative to the CC condition, navigation (nav) in both real-world and virtual reality conditions reliably activated more neurons in CA1 (t(86) = 3.014, p = 0.001; nav = 2.8, CC = 0.499, Cohen's d = 1.22), CA3 (t(75) = 1.750, p < 0.05; nav = 2.86, CC = 1.48, d = 0.68), and DG (t(98) = 1.67, p < 0.05; nav = 0.324, CC = 0.103, d = 0.626). A two-way ANOVA revealed significant main effects of region (i.e., CA1, CA3, and DG; F(1,2) = 41.98, p < 0.001) and condition (real vs virtual; F(1,1) = 8.48, p < 0.005), and an interaction (F(1,2) = 6.91, p = 0.001). Tukey's HSD revealed that real navigation activated more neurons than did virtual navigation, but only in CA1 (p < 0.001, Fig. 2B), not in CA3 or DG (Fig. 2C,D). The current protocol successfully labeled 42–51% of principal cells in positive control conditions, suggesting that the differential Arc expression across hippocampal subregions result is not due to nonspecific binding of the riboprobe.
Comparisons with Arc expression in the rodent
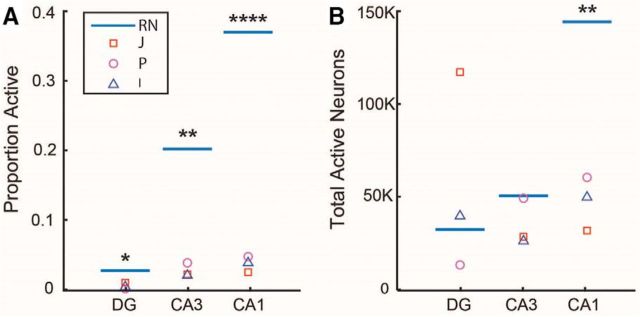
The observed sparseness of activity across the NHP hippocampus appears to deviate strongly from what has been reported in many rodent studies. To facilitate comparison of the current data collected from the NHPs with data collected in rodents, we conducted a meta-analysis of the existing rodent literature. Data were obtained from experiments that used comparable real-world conditions (i.e., open-field foraging) and used identical Arc imaging techniques (see Materials and Methods for references) to arrive at an average proportion of neurons activated per region in the rat (dorsal CA1: 37.0 ± 1.1%; dorsal CA3: 20.2 ± 1.0%; dorsal suprapyramidal DG: 2.7 ± 0.2%).
Across hippocampal regions, activity levels were approximately an order of magnitude smaller in NHPs than in rodents (DG: t(2) = −8.54, p = 0.013; CA3: t(2) = −29.91, p = 0.001; CA1: t(2) = −51.33, p < 0.0001; Fig. 3A). We next considered whether the total number of active neurons is conserved across species. Using published estimates of the number of principal cells in the rodent and NHP hippocampus (Amaral and Lavenex, 2007), we calculated the total number of activated neurons per subregion in both species (Fig. 3B). When quantified in this manner, the total neurons activated by navigation showed no significant species-related difference in the DG (t(2) = 0.776, p = 0.51) or CA3 (t(2) = −2.102, p = 0.17). The proportion of behaviorally driven neurons in CA1, however, was ∼3-fold lower in NHPs than in the rodent.
Figure 3.
Comparison of activity patterns in rodents and primates. A, Mean proportion of activated neurons reported in rodents (blue line, RN, Rattus norvegicus) and observed in individual primates (square, Monkey J; circle, Monkey P; triangle, Monkey I), highlighting the difference between species. B, Close concordance between the total numbers of neurons activated in the DG and CA3 regions. In contrast, the number of CA1 neurons activated in rats was significantly higher than in monkeys (*p < 0.05, **p < 0.01, ****p < 0.0001, monkeys vs rats in the same region).
Discussion
In rodents, only CA1 place cells have been examined under impoverished motor and vestibular input and, under these conditions (which mimic virtual reality), fewer cells respond and their place fields are correspondingly larger (Foster et al., 1989; Terrazas et al., 2005; Ravassard et al., 2013, Aghajan et al., 2015). Testing whether these same changes can be observed in the NHP is important given differences in the importance of different sensory inputs between species. The rat's sensory input is likely dominated by vibrissal input and olfactory input, which are dramatically deprived under virtual conditions. In contrast, in macaques, visual information is far more important. Because this input can be mimicked much more reliably during virtual navigation, it is possible that virtual foraging may produce a hippocampal response closer to real-world conditions in the NHP than in the rodent. The current data, however, suggest that this is not the case because the size of CA1 neuronal ensembles activated during navigation is reduced in both NHPs and rodents by the attenuated sensory input experienced in virtual environments. Moreover, these data are consistent with the idea that CA1 pyramidal cells in NHPs (and likely humans) exhibit a comparable expansion in place field size during navigation in virtual spaces relative to real-world conditions. A potentially important caveat, however, exists in extrapolating these data to the literature observing NHPs and humans navigating in virtual environments. Although the foraging conditions selected for behavior in the NHPs was deliberately made to match the typical conditions under which rodent place cells are recorded, they differ considerably from the typical conditions used to test navigation in virtual reality, particularly in humans. These conditions typically involve goal-directed navigation to one or more fixed locations and therefore require focused attention, goal-directed behavior, and often explicitly require episodic memory encoding and retrieval in a way that foraging does not. It should be noted, however, that although these differences in task demands are known to alter the dynamics of place cell maps (Muzzio et al., 2009), they are relatively subtle. Despite this caveat, it is clear that, when they are tested under the same conditions, cells in CA1 of both NHPs and rodents react in the same way as the reduced sensory input inherent in virtual navigation.
No experiments to date have investigated whether spatial representations in the DG or CA3 region are similarly degraded under reduced sensory input. The present data, however, suggest that (unlike in CA1) representations in these regions are robust under these conditions. That is, the size of neuronal ensembles activated during navigation is not changed in either the DG or area CA3 by the altered sensory conditions experienced in virtual environments (Fig. 2B–D). This resilience may have developed as a means to maximize the number of patterns that can be stored in the DG and CA3 networks.
By providing a robust estimate of population sparsity, these data can be combined with previous estimates of single neuron sparsity (Rolls and Treves, 1990; Strange et al., 2014) and synaptic connectivity (Amaral and Lavenex, 2007) to estimate storage capacity. Assuming that the activity in a single spatial context approximates the activity inherent in the encoding of a single event, the total number of events that can be stored within the neuronal network of each species can be determined using the estimate for binary Hebb–Marr memory (Marr, 1971; McNaughton and Morris, 1987; Rolls and Treves, 1990). These data show that the disparity in memory capacity between species increases dramatically at each stage of information processing along the trisynaptic loop (Table 1). That is, the DG networks have comparable capacity across species, but in CA1, the NHP network has nearly 100 times the capacity of the rodent network. Although these numbers are crude approximations, they demonstrate that the increase in both cell number and sparsity in the NHP hippocampus results in memory capacity that is at least an order of magnitude greater than in the rodent counterpart. This increased sparsity cannot be the result of repeated exposure to the contexts because hippocampal representations in NHPs do not undergo experience-dependent tuning or habituation (Thome et al., 2012) and thus are likely a stable species-related difference in hippocampal information processing.
Table 1.
Estimated storage capacity of the monkey and rat hippocampus
| CA1 |
CA3 |
DG |
|||||||
|---|---|---|---|---|---|---|---|---|---|
| Monkey | Rat | Ratioe | Monkey | Rat | Ratioe | Monkey | Rat | Ratioe | |
| Total cellsa | 1,300 | 390 | 3.3 | 1,270 | 250 | 5.1 | 12,000 | 1,200 | 10 |
| Active cellsb | 50.1 | 144.3 | 0.35 | 36.2 | 50.5 | 0.72 | 50.4 | 32.4 | 1.56 |
| Sparsityc | 0.04 | 0.37 | 9.25 | 0.03 | 0.20 | 6.67 | 0.04 | 0.03 | 1.33 |
| Storage capacityd | 467 | 5 | 93.4 | 854 | 17 | 50.24 | 39294 | 951 | 41.32 |
aEstimates (in thousands) obtained from Amaral and Lavenex (2007).
bEstimates (in thousands) are the product of the proportion of active cells obtained here and the estimate of total cell number.
cRatio of active cells to principal cells.
dStorage capacity estimate for Hebb–Marr memory,
eRatio is presented as monkey/rat except in the case of sparsity, which is presented as rat/monkey to demonstrate more clearly how much sparser the coding in the monkey is.
The most intriguing observation from this experiment is that the size of hippocampal neural ensembles engaged by navigation is not a fixed proportion of the population, but rather an absolute number (Fig. 3). The interpretation that rats and monkeys have different capacity for episodes of foraging experience, yet equivalent (or at least comparable) information content within each episode of experience is the most parsimonious one. It remains possible, however, that, although the hippocampus plays a similar role as an index to bind together disparate cortical representations in different species, the individual cortical targets may be quite different. If this is the case, then the cortical representations must negotiate the same trade-off between information content and storage capacity, but the large species difference in cortical size relative to hippocampal size does make it possible for this balance to be achieved differently in the neocortex than in the hippocampus. The absence of an increase in the hippocampal index, however, would place serious limitations on the size and distribution of a cortical ensemble that could be reliably recruited.
Despite these open questions, the current data support the novel hypothesis that, as mammals evolved and brains grew, evolutionary processes determined an optimal number of neurons necessary to establish stable episodic representations in the hippocampus and this region enlarged to build memory capacity.
Footnotes
This work was supported by the National Institutes of Health (Grant R01 AG003376, McKnight Brain Research Foundation, to C.A.B. and Grant MH060884 to S.H.L.), the Natural Sciences and Engineering Research Council of Canada (D.F.M. and B.L.M.), and Alberta Innovates Health Solution (Polaris Award to B.L.M.). We thank Tom Beach for surgical assistance, David Towers and Michael Montgomery for technical assistance, and Luann Snyder and Michelle Carroll for administrative assistance.
The authors declare no competing financial interests.
References
- Aghajan ZM, Acharya L, Moore JJ, Cushman JD, Vuong C, Mehta MR (2015) Impaired spatial selectivity and intact phase precession in two-dimensional virtual reality. Nat Neurosci 18:121–128. [DOI] [PubMed] [Google Scholar]
- Alme CB, Buzzetti RA, Marrone DF, Leutgeb JK, Chawla MK, Schaner MJ, Bohanick JD, Khoboko T, Leutgeb S, Moser EI, Moser MB, McNaughton BL, Barnes CA (2010) Hippocampal granule cells opt for early retirement. Hippocampus 20:1109–1123. 10.1002/hipo.20810 [DOI] [PubMed] [Google Scholar]
- Amaral DG, Lavenex P (2007) Hippocampal neuroanatomy. In: The hippocampus book (Andersen P, Morris RG, Amaral D, Bliss T, O'Keefe J, eds), pp. 37–114. New York: OUP. [Google Scholar]
- American Psychiatric Association (2001) Practice of electroconvulsive therapy: recommendations for treatment, training, and privileging. Arlington, VA: American Psychiatric Publishing. [Google Scholar]
- Buffalo EA. (2015) Bridging the gap between spatial and mnemonic views of the hippocampal formation. Hippocampus 25:713–718. 10.1002/hipo.22444 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burgess N, Maguire EA, O'Keefe J (2002) The human hippocampus and spatial and episodic memory. Neuron 35:625–641. 10.1016/S0896-6273(02)00830-9 [DOI] [PubMed] [Google Scholar]
- Burke SN, Chawla MK, Penner MR, Crowell BE, Worley PF, Barnes CA, McNaughton BL (2005) Differential encoding of behavior and spatial context in deep and superficial layers of the neocortex. Neuron 45:667–674. 10.1016/j.neuron.2005.01.042 [DOI] [PubMed] [Google Scholar]
- Buzsáki G, Moser EI (2013) Memory, navigation and theta rhythm in the hippocampal-entorhinal system. Nat Neurosci 16:130–138. 10.1038/nn.3304 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chawla MK, Guzowski JF, Ramirez-Amaya V, Lipa P, Hoffman KL, Marriott LK, Worley PF, McNaughton BL, Barnes CA (2005) Sparse, environmentally selective expression of Arc RNA in the upper blade of the rodent fascia dentata by brief spatial experience. Hippocampus 15:579–586. 10.1002/hipo.20091 [DOI] [PubMed] [Google Scholar]
- Colombo M, Fernandez T, Nakamura K, Gross CG (1998) Functional differentiation along the anterior-posterior axis of the hippocampus in monkeys. J Neurophysiol 80:1002–1005. [DOI] [PubMed] [Google Scholar]
- Ekstrom AD, Kahana MJ, Caplan JB, Fields TA, Isham EA, Newman EL, Fried I (2003) Cellular networks underlying human spatial navigation. Nature 425:184–188. 10.1038/nature01964 [DOI] [PubMed] [Google Scholar]
- Engle JR, Machado CJ, Permenter MR, Vogt JA, Maurer AP, Bulleri AM, Barnes CA (2016) Network patterns associated with navigation behavior are altered in aged nonhuman primates. J Neurosci 36:12217–12227. 10.1523/JNEUROSCI.4116-15.2016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foster TC, Castro CA, McNaughton BL (1989) Spatial selectivity of rat hippocampal neurons: dependence on preparedness for movement. Science 244:1580–1582. 10.1126/science.2740902 [DOI] [PubMed] [Google Scholar]
- Furuya Y, Matsumoto J, Hori E, Boas CV, Tran AH, Shimada Y, Ono T, Nishijo H (2014) Place-related neuronal activity in the monkey parahippocampal gyrus and hippocampal formation during virtual navigation. Hippocampus 24:113–130. 10.1002/hipo.22209 [DOI] [PubMed] [Google Scholar]
- Gheidi A, Satvat E, Marrone DF (2012) Experience-dependent recruitment of Arc expression in multiple systems during rest. J Neurosci Res 90:1820–1829. 10.1002/jnr.23057 [DOI] [PubMed] [Google Scholar]
- Guzowski JF, McNaughton BL, Barnes CA, Worley PF (1999) Environment-specific expression of the immediate-early gene Arc in hippocampal neuronal ensembles. Nat Neurosci 2:1120–1124. 10.1038/16046 [DOI] [PubMed] [Google Scholar]
- Guzowski JF, Miyashita T, Chawla MK, Sanderson J, Maes LI, Houston FP, Lipa P, McNaughton BL, Worley PF, Barnes CA (2006) Recent behavioral history modifies coupling between cell activity and Arc gene transcription in hippocampal CA1 neurons. Proc Natl Acad Sci U S A 103:1077–1082. 10.1073/pnas.0505519103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartzell AL, Burke SN, Hoang LT, Lister JP, Rodriguez CN, Barnes CA (2013) Transcription of the immediate-early gene Arc in CA1 of the hippocampus reveals activity differences along the proximodistal axis that are attenuated by advanced age. J Neurosci 33:3424–3433. 10.1523/JNEUROSCI.4727-12.2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hori E, Nishio Y, Kazui K, Umeno K, Tabuchi E, Sasaki K, Endo S, Ono T, Nishijo H (2005) Place-related neural responses in the monkey hippocampal formation in a virtual space. Hippocampus 15:991–996. 10.1002/hipo.20108 [DOI] [PubMed] [Google Scholar]
- Howard V, Reid S, Baddeley A, Boyde A (1985) Unbiased estimation of particle density in the tandem scanning reflected light microscope. J Microsc 138:203–212. 10.1111/j.1365-2818.1985.tb02613.x [DOI] [PubMed] [Google Scholar]
- MacDonald CJ, Lepage KQ, Eden UT, Eichenbaum H (2011) Hippocampal ‘time cells’ bridge the gap in memory for discontiguous events. Neuron 71:737–749. 10.1016/j.neuron.2011.07.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mankin EA, Sparks FT, Slayyeh B, Sutherland RJ, Leutgeb S, Leutgeb JK (2012) Neuronal code for extended time in the hippocampus. Proc Natl Acad Sci U S A 109:19462–19467. 10.1073/pnas.1214107109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marr D. (1971) Simple memory: a theory for archicortex. Philos Trans R Soc Lond B Biol Sci 262:23–81. 10.1098/rstb.1971.0078 [DOI] [PubMed] [Google Scholar]
- Marrone DF, Schaner MJ, McNaughton BL, Worley PF, Barnes CA (2008) Immediate-early gene expression at rest recapitulates recent experience. J Neurosci 28:1030–1033. 10.1523/JNEUROSCI.4235-07.2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marrone DF, Satvat E, Shaner MJ, Worley PF, Barnes CA (2012a) Attenuated long-term Arc expression in the aged fascia dentata. Neurobiol Aging 33:979–990. 10.1016/j.neurobiolaging.2010.07.022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marrone DF, Ramirez-Amaya V, Barnes CA (2012b) Neurons generated in senescence maintain capacity for functional integration. Hippocampus 22:1134–1142. 10.1002/hipo.20959 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marrone DF, Satvat E, Odintsova IV, Gheidi A (2014) Dissociation of spatial representations within hippocampal region CA3. Hippocampus 24:1417–1420. 10.1002/hipo.22367 [DOI] [PubMed] [Google Scholar]
- McNaughton BL, Morris RGM (1987) Hippocampal synaptic enhancement and information storage within a distributed memory system. Trends Neurosci 10:408–415. 10.1016/0166-2236(87)90011-7 [DOI] [Google Scholar]
- Miyashita T, Kubik S, Haghighi N, Steward O, Guzowski JF (2009) Rapid activation of plasticity-associated gene transcription in hippocampal neurons provides a mechanism for encoding of one-trial experience. J Neurosci 29:898–906. 10.1523/JNEUROSCI.4588-08.2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moscrip TD, Terrace HS, Sackeim HA, Lisanby SH (2004) A primate model of anterograde and retrograde amnesia produced by convulsive treatment. J ECT 20:26–36. 10.1097/00124509-200403000-00007 [DOI] [PubMed] [Google Scholar]
- Muzzio IA, Kentros C, Kandel E (2009) What is remembered? Role of attention on the encoding and retrieval of hippocampal representations. J Physiol 587:2837–2854. 10.1113/jphysiol.2009.172445 [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Keefe J, Nadel L (1978) The hippocampus as a cognitive map. New York: OUP. [Google Scholar]
- Ono T, Nakamura K, Nishijo H, Eifuku S (1993) Monkey hippocampal neurons related to spatial and nonspatial functions. J Neurophysiol 70:1516–1529. [DOI] [PubMed] [Google Scholar]
- Paxinos G, Huang XF, Toga AW (1999) The rhesus monkey brain in stereotaxic coordinates. San Diego: Academic. [Google Scholar]
- Penner MR, Roth TL, Chawla MK, Hoang LT, Roth ED, Lubin FD, Sweatt JD, Worley PF, Barnes CA (2011) Age-related changes in Arc transcription and DNA methylation within the hippocampus. Neurobiol Aging 32:2198–2210. 10.1016/j.neurobiolaging.2010.01.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quiroga RQ, Kreiman G, Koch C, Fried I (2008) Sparse but not ‘grandmother-cell’ coding in the medial temporal lobe. Trends Cogn Sci 12:87–91. 10.1016/j.tics.2007.12.003 [DOI] [PubMed] [Google Scholar]
- Ramírez-Amaya V, Vazdarjanova A, Mikhael D, Rosi S, Worley PF, Barnes CA (2005) Spatial exploration-induced Arc mRNA and protein expression: evidence for selective, network-specific reactivation. J Neurosci 25:1761–1768. 10.1523/JNEUROSCI.4342-04.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramirez-Amaya V, Marrone DF, Gage FH, Worley PF, Barnes CA (2006) Integration of new neurons into functional neural networks. J Neurosci 26:12237–12241. 10.1523/JNEUROSCI.2195-06.2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramirez-Amaya V, Angulo-Perkins A, Chawla MK, Barnes CA, Rosi S (2013) Sustained transcription of the immediate early gene Arc in the dentate gyrus after spatial exploration. J Neurosci 33:1631–1639. 10.1523/JNEUROSCI.2916-12.2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ravassard P, Kees A, Willers B, Ho D, Aharoni D, Cushman J, Aghajan ZM, Mehta MR (2013) Multisensory control of hippocampal spatiotemporal selectivity. Science 340:1342–1346. 10.1126/science.1232655 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rich PD, Liaw HP, Lee AK (2014) Large environments reveal the statistical structure governing hippocampal representations. Science 345:814–817. 10.1126/science.1255635 [DOI] [PubMed] [Google Scholar]
- Rolls ET. (1999) Spatial view cells and the representation of place in the primate hippocampus. Hippocampus 9:467–480. [DOI] [PubMed] [Google Scholar]
- Rolls ET, Treves A (1990) The relative advantages of sparse versus distributed encoding for associative neuronal networks in the brain. Network 1:407–421. 10.1088/0954-898X_1_4_002 [DOI] [Google Scholar]
- Rosi S, Ramirez-Amaya V, Vazdarjanova A, Worley PF, Barnes CA, Wenk GL (2005) Neuroinflammation alters the hippocampal pattern of behaviorally induced Arc expression. J Neurosci 25:723–731. 10.1523/JNEUROSCI.4469-04.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosi S, Ramirez-Amaya V, Vazdarjanova A, Esparza EE, Larkin PB, Fike JR, Wenk GL, Barnes CA (2009) Accuracy of hippocampal network activity is disrupted by neuroinflammation: rescue by memantine. Brain 132:2464–2477. 10.1093/brain/awp148 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schindelin J, Arganda-Carreras I, Frise E, Kaynig V, Longair M, Pietzsch T, Preibisch S, Rueden C, Saalfeld S, Schmid B, Tinevez JY, White DJ, Hartenstein V, Eliceiri K, Tomancak P, Cardona A (2012) FIJI: an open-source platform for biological-image analysis. Nat Methods 9:676–682. 10.1038/nmeth.2019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strange BA, Witter MP, Lein ES, Moser EI (2014) Functional organization of the hippocampal longitudinal axis. Nat Rev Neurosci 15:655–669. 10.1038/nrn3785 [DOI] [PubMed] [Google Scholar]
- Terrazas A, Krause M, Lipa P, Gothard KM, Barnes CA, McNaughton BL (2005) Self-motion and the hippocampal spatial metric. J Neurosci 25:8085–8096. 10.1523/JNEUROSCI.0693-05.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thome A, Erickson CA, Lipa P, Barnes CA (2012) Differential effects of experience on tuning properties of macaque MTL neurons in a passive viewing task. Hippocampus 22:2000–2011. 10.1002/hipo.22070 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Treves A, Rolls ET (1994) Computational analysis of the role of the hippocampus in memory. Hippocampus 4:374–391. 10.1002/hipo.450040319 [DOI] [PubMed] [Google Scholar]
- Vazdarjanova A, Guzowski JF (2004) Differences in hippocampal neuronal population responses to modifications of an environmental context: evidence for distinct, yet complementary, functions of CA3 and CA1 ensembles. J Neurosci 24:6489–6496. 10.1523/JNEUROSCI.0350-04.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vazdarjanova A, McNaughton BL, Barnes CA, Worley PF, Guzowski JF (2002) Experience-dependent coincident expression of the effector immediate-early genes arc and Homer 1a in hippocampal and neocortical neuronal networks. J Neurosci 22:10067–10071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vazdarjanova A, Ramirez-Amaya V, Insel N, Plummer TK, Rosi S, Chowdhury S, Mikhael D, Worley PF, Guzowski JF, Barnes CA (2006) Spatial exploration induces ARC, a plasticity-related immediate-early gene, only in calcium/calmodulin-dependent protein kinase II-positive principal excitatory and inhibitory neurons of the rat forebrain. J Comp Neurol 498:317–329. 10.1002/cne.21003 [DOI] [PubMed] [Google Scholar]
- Waydo S, Kraskov A, Quiroga Quiroga R, Fried I, Koch C (2006) Sparse representation in the human medial temporal lobe. J Neurosci 26:10232–10234. 10.1523/JNEUROSCI.2101-06.2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson MA, McNaughton BL (1993) Dynamics of the hippocampal ensemble code for space. Science 261:1055–1058. 10.1126/science.8351520 [DOI] [PubMed] [Google Scholar]
- Wirth S, Yanike M, Frank LM, Smith AC, Brown EN, Suzuki WA (2003) Single neurons in the monkey hippocampus and learning of new associations. Science 300:1578–1581. 10.1126/science.1084324 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Real-world navigation in rhesus macaques. This movie depicts a rhesus macaque being guided through one of the two rooms that were used for navigation. The monkey can be seen traveling to foraging locations scattered throughout the room where researchers placed preferred foods.
Virtual navigation in rhesus macaques. This movie depicts a rhesus macaque seated in front of a wraparound display depicting a virtual environment. Within this environment, red cubes appear in pseudo-random locations, and the monkey can be seen using a joystick to navigate to several of these cubes in order to obtain fluid reward.