Abstract
Transforming growth factor-β (TGF-β) signals through both SMAD and non-SMAD pathways to elicit a wide array of biological effects. Existing data have shown the association and coordination between STATs and SMADs in mediating TGF-β functions in hepatic cells, but it is not clear how STATs are activated under these circumstances. Here, we report that JAK1 is a constitutive TGFβRI binding protein and is absolutely required for phosphorylation of STATs in a SMAD-independent manner within minutes of TGF-β stimulation. Following the activation of SMADs, TGF-β also induces a second phase of STAT phosphorylation that requires SMADs, de novo protein synthesis, and contribution from JAK1. Our global gene expression profiling indicates that the non-SMAD JAK1/STAT pathway is essential for the expression of a subset of TGF-β target genes in hepatic stellate cells, and the cooperation between the JAK1-STAT3 and SMAD pathways is critical to the roles of TGF-β in liver fibrosis.
Keywords: hepatic stellate cell (HSC), Janus kinase (JAK), SMAD transcription factor, STAT3, transforming growth factor beta (TGF-B), Liver fibrosis, Smad3
Introduction
Transforming growth factor-β (TGF-β) and its related factors have a broad array of regulatory functions ranging from specifying tissue pattern formation during embryonic development to maintaining physiological homeostasis in the adults (1). The function of TGF-β varies and even can be opposing in different cells and at different developmental and disease stages. One example of this contextuality is the dichotomy of the roles of TGF-β in liver diseases. TGF-β contributes to all stages of chronic liver disease development, from liver injuries caused by inflammation and fibrosis to cirrhosis and hepatocellular carcinoma (2). TGF-β has cytostatic and apoptotic effects on hepatocytes and consequently is critical for controlling liver mass during development and liver regeneration. Loss of TGF-β signaling in the liver thus is frequently associated with hyperproliferative disorders and cancer. However, high levels of TGF-β, as a consequence of liver injury and damage, activate hepatic stellate cells (HSCs),3 promote myofibroblast transdifferentiation, and stimulate production of extracellular matrix. All of the above lead to fibrosis and cirrhosis, upon which hepatocellular carcinoma and hepatic failure develop.
TGF-β signals by binding to a core complex of heteromeric type I (TGFβRI) and type II (TGFβRII) transmembrane receptors, each equipped with an intracellular Ser/Thr kinase domain (3). The ligand binding to the receptor complex leads to phosphorylation and thereby activation of TGFβRI by the cytoplasmic kinase domains of TGFβRII. The activated TGFβRI further phosphorylates and activates SMAD2 and SMAD3, which then form a heteromeric complex with SMAD4 and accumulate in the nucleus to regulate target gene expression. Activated TGF-β receptors also signal through a multitude of other signal transducers that are collectively known as non-SMAD pathways (4). The receptor-activated, non-SMAD transducers can mediate signaling responses as stand-alone pathways, in conjunction with SMADs, or converge onto SMADs to control SMAD activities. For instance, simultaneous inputs from SMAD and p38 MAPK pathways are required to generate a full-fledged apoptosis in hepatocytes; both of these conduits are important for the tumor suppresser role of TGF-β (5).
To broaden our understanding of how TGF-β signal is conveyed in liver cells, particularly through the non-SMAD pathways, we sought novel TGFβRI binding partners using stable isotope labeling by amino acids in cell culture (SILAC) (6). Here, we describe the identification of JAK1 as a TGFβRI-interacting protein and show that JAK1 activates STAT3 in both SMAD-independent and -dependent manners in response to TGF-β in hepatic cells. We further demonstrate that STAT3 is required to cooperate with SMAD3 for mediating TGF-β-induced fibrotic response in hepatic stellate cells.
Results
Identification of JAK1 as a TGFβRI-interacting Protein
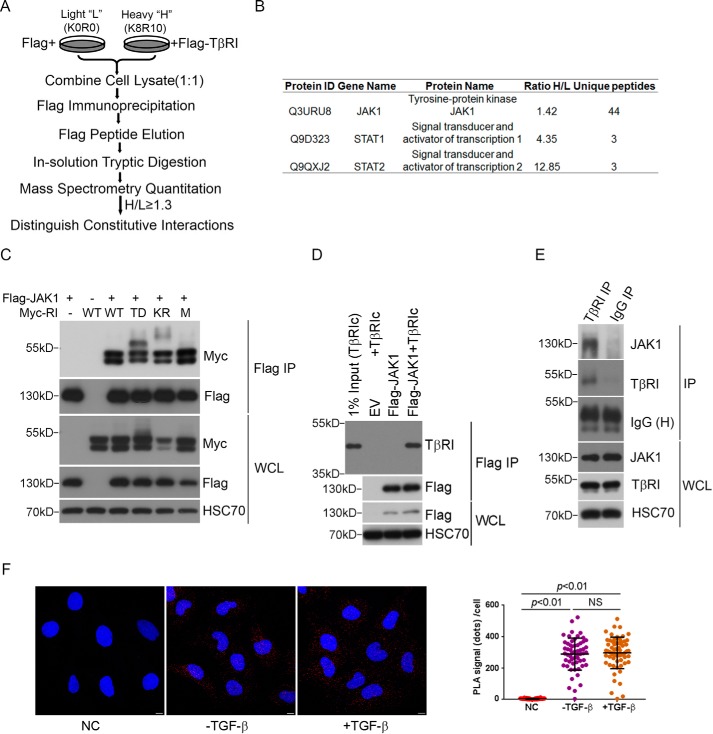
To search for additional protein effectors that mediate the SMAD-independent, noncanonical TGF-β signaling, we sought new binding partners of TGFβRI using SILAC (Fig. 1A). The AML12 mouse hepatocytes were chosen in this experiment for their robust responses to TGF-β through both the SMAD and non-canonical JNK and p38 MAPK pathways (7). Of 325 proteins that were identified with this approach (supplemental Tables S1 and S2), 15, including FKBP5, STRAP, SPTBN1, and TRAP1, were previously known TGFβRI-interacting proteins (Table 1). Also among these 15 known proteins are TAK1 and TAB1, which have been specifically implicated in TGF-β-induced JNK and p38 MAPK activation (7–9). However, we were intrigued by the presence of JAK1, STAT1, and STAT2 in the larger list of potential TGFβRI-interacting proteins (supplemental Table S2 and Fig. 1B), especially JAK1, because it implies that the JAK-STAT pathway may play a direct role in mediating TGF-β signaling.
FIGURE 1.
Identification of JAK1 as a TGFβRI-interacting protein by quantitative proteomic analysis. A, SILAC experiment to discover TβRI-interacting proteins. AML12 cells were labeled with l-lysine and l-arginine (K0R0, light) or (K8R10, heavy), respectively. The cells cultured in K8R10 medium (heavy) were transfected with FLAG-TGFβRI, whereas the cells cultured in K0R0 medium (light) were transfected with an empty vector. The cell lysates were combined at equal protein amounts and subjected to MS quantitation. A cut-off of 1.3 heavy/light (H/L) protein ratio was set to weed out background contaminants. B, JAK1, STAT1, and STAT2 were identified as potential TGFβRI-interacting proteins from the screen. C, JAK1 interacts with Myc-tagged wild type TGFβRI as well as TGFβRI mutants in HEK293 cells. TD, constitutively active; KR, kinase-dead; M (mL45), SMAD binding-defective. D, JAK1 interacts with purified cytoplasmic domain of TGFβRI in vitro. E, JAK1 interacts with TGFβRI endogenously. AML12 cells were subjected to immunoprecipitation by TGFβRI antibody or control IgG. The presence of JAK1 in the immunoprecipitated protein complex was detected by Western blotting. F, PLA was used to detect the in vivo interaction of JAK1 and TGFβRI in LX-2 cells. LX-2 cells were treated with ±4 ng/ml TGF-β for 5 min, and the cells were incubated with both JAK1 monoclonal antibody and TGFβRI polyclonal antibody. Negative control (NC) cells were incubated with TGFβRI polyclonal antibody and normal mouse IgG. Scale bar, 10 μm.
TABLE 1.
List of known TGFβRI-interacting proteins identified in the SILAC study
AML12 cells cultured in K8R10 medium (heavy) and K0R0 medium (light) were transfected with FLAG-TGFβRI or control FLAG-vector, respectively. Cell lysates from the above were combined and subjected to FLAG immunoprecipitation, trypsin digestion, and MS analysis. Proteins that were identified by at least two unique peptides were quantified, and those whose heavy/light ratio was ≥1.3 were considered as potential TGFβRI-interacting proteins.
| Protein ID | Gene name | Protein name | Heavy/light ratio | Unique peptides |
|---|---|---|---|---|
| Q8CF89 | Tab1 | TGF-β-activated kinase 1 and MAP3K7-binding protein 1 | 1.41 | 9 |
| Q3UZT7 | Ctnnb1 | Catenin β-1 | 1.50 | 7 |
| Q8CG19 | Ltbp1 | Latent-transforming growth factor β-binding protein 1 | 1.54 | 35 |
| B2RUC7 | Strap | Serine-threonine kinase receptor-associated protein | 1.57 | 3 |
| Q9JKF1 | Iqgap1 | Ras GTPase-activating-like protein IQGAP1 | 1.61 | 19 |
| Q9QZ06 | Tollip | Toll-interacting protein | 1.62 | 3 |
| A2AP93 | Map3k7 | Mitogen-activated protein kinase kinase kinase 7 | 1.68 | 4 |
| Q7TQI3 | Otub1 | Ubiquitin thioesterase OTUB1 | 1.80 | 3 |
| Q8R5H1 | Usp15 | Ubiquitin C-terminal hydrolase 15 | 2.08 | 9 |
| Q8BTJ0 | Prpf4 | U4/U6 small nuclear ribonucleoprotein Prp4 | 2.16 | 4 |
| Q549A5 | Clu | Clusterin | 5.07 | 4 |
| Q62261 | Sptbn1 | Spectrin β chain, non-erythrocytic 1 | 6.61 | 48 |
| Q922Z3 | Trap1 | Heat shock protein 75 kDa, mitochondrial | 1.31 | 3 |
| A8CVP4 | Lims1 | LIM and senescent cell antigen-like-containing domain protein 1 | 2.41 | 3 |
| Q4FJN2 | Fkbp5 | Peptidyl-prolyl cis/trans-isomerase FKBP5 | 1.50 | 4 |
To validate the authenticity of the interaction between JAK1 and TGFβRI, we co-expressed FLAG-tagged JAK1 with Myc-tagged TGFβRI and its constitutively active (TD), kinase-deficient (KR), and SMAD binding-defective (mL45) variants in HEK293 cells (10). Co-immunoprecipitation and Western blotting analyses indicated that all forms of Myc-TGFβRI interacted specifically with FLAG-JAK1 (Fig. 1C), suggesting that this interaction occurs constitutively. To determine whether JAK1 directly interacts with TGFβRI, we conducted an affinity pull-down assay using the cytoplasmic fragment of TGFβRI (TβRIc) produced and purified from the baculovirus-insect cell expression system and the full-length FLAG-JAK1 produced and purified from transiently transfected HEK293 cells, and the result confirmed the binding (Fig. 1D). Interaction between endogenous TGFβRI and JAK1 was also confirmed by immunoprecipitation in AML12 cells, in which JAK1 was specifically co-precipitated by anti-TGFβRI antibody (Fig. 1E) and visualized using a proximity ligation assay (PLA) in human hepatic stellate LX-2 cells (Fig. 1F). The binding between JAK1 and TGFβRI appeared to be constitutive in nature because FLAG-tagged JAK1 interacted indiscriminately with Myc-tagged WT, TD, KR, or mL45 mutant forms of TGFβRI (Fig. 1C), and TGF-β treatment did not alter the outcome of JAK1 and TGFβRI interaction in PLA experiments (Fig. 1F).
JAK1 Mediates the TGF-β-induced Early Phase Phosphorylation of STAT3
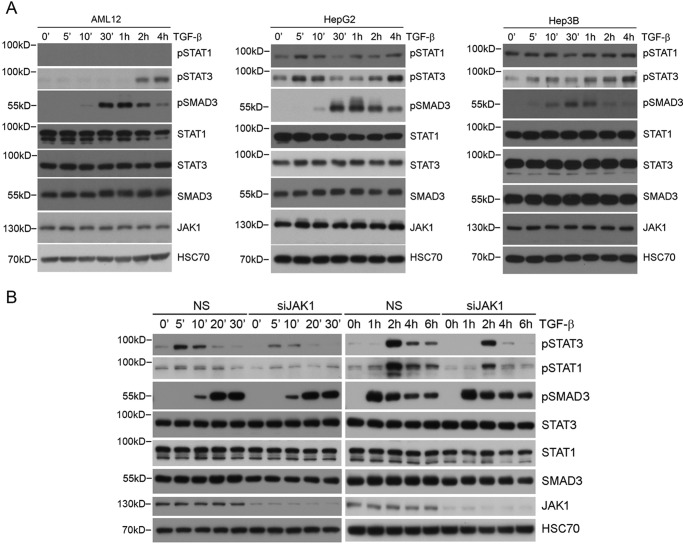
In classical cytokine signaling pathways, binding of various ligands, such as interferons, interleukins, and growth factors, to their cognate receptors activates the corresponding JAKs, which in turn recruits STAT proteins to the receptor and activates them through tyrosine phosphorylation (11). To determine whether any of the STAT proteins are phosphorylated in response to TGF-β, we treated AML12 as well as human hepatocellular carcinoma HepG2 and Hep3B cells with TGF-β and examined the levels of tyrosine-phosphorylated STAT1 and STAT3 over the time course as indicated. The results showed two phases of phosphorylation of both STAT1 and STAT3 in HepG2 cells with an initial rapid activation peaking at 5 min after the ligand stimulation, followed by a late phase that began 1 h after and progressively reached a plateau 4 h after the stimulation (Fig. 2A, middle). Interestingly, the temporal order of this bi-phase activation straddled over the time course of SMAD3 activation, which peaked about 0.5–1 h after the ligand stimulation (Fig. 2A). In AML12 cells, the level of phosphorylated STAT1 was too low to be detected, but a robust late phase activation of STAT3 was readily observed (Fig. 2A, left). Once again, according to the paradigm of cytokine signaling, the attenuation of initial STAT1 and STAT3 phosphorylation is causally associated with expression of SOCS (suppressor of cytokine signaling), which physically binds JAKs and inhibits their kinase activities and STAT phosphorylation (12). Indeed, in Hep3B cells, which are deficient for SOCS1 (13), we observed STAT3 phosphorylation being sustained at a high level 5 min after the ligand stimulation, whereas the level of phosphorylated STAT1 was persistently high even before the stimulation (Fig. 2A, right). This biphasic pattern of STAT1 and STAT3 phosphorylation induced by TGF-β was also observed in human hepatic stellate LX-2 cells, although the initial phase of STAT1 phosphorylation was much weaker compared with that of STAT3 (Fig. 2B). To determine whether JAK1 is required for STAT3 phosphorylation induced by TGF-β, we depleted JAK1 in LX-2 cells using siRNA and found that the initial rapid phase of STAT3 phosphorylation was diminished, whereas the late phase still persisted, albeit to a lesser extent (Fig. 2B). This result indicated that JAK1 is at least required for the early phase phosphorylation of STAT3, whereas other kinases might contribute to the late phase phosphorylation of STAT3.
FIGURE 2.
TGF-β-induced STAT3 early phosphorylation requires JAK1. A, TGF-β induced STAT3 phosphorylation in different hepatic cells. B, TGF-β induced biphasic phosphorylation of STAT3 in LX-2 cells, and the early phase of STAT3 phosphorylation is dependent on JAK1. LX-2 cells transfected with non-silencing (NS) control or JAK1 siRNA were treated with 4 ng/ml TGF-β for the indicated times.
TGF-β Induces the Early Phase of STAT3 Phosphorylation Independent of SMADs
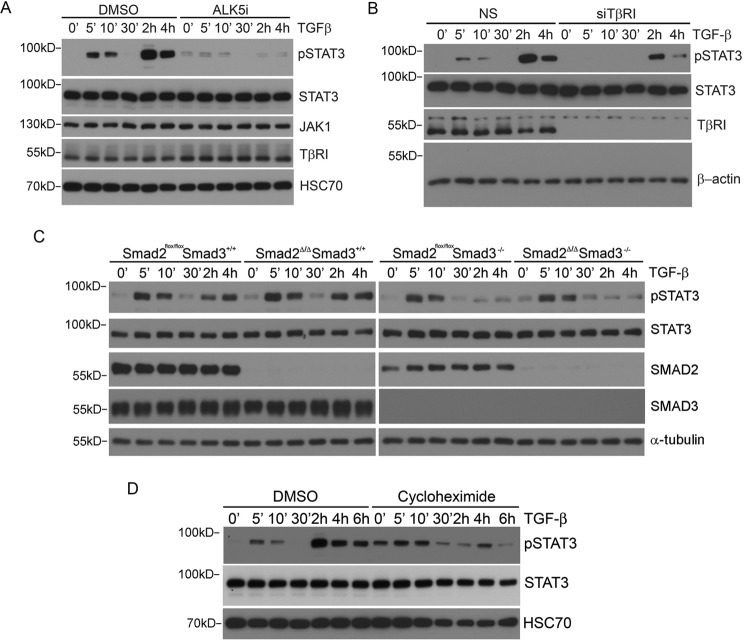
To formally demonstrate that the TGF-β-induced phosphorylation of STATs is part of the TGFβRI-mediated signaling response, we incubated LX-2 cells with SB431542 (ALK5i), a chemical inhibitor of TGFβRI, before the addition of the ligand. This treatment abolished both the early and late phase phosphorylation of STAT3 (Fig. 3A). We further knocked down the expression of TGFβRI in LX-2 cells and found that this experimental manipulation also led to a drastic reduction of STAT3 phosphorylation in both phases following TGF-β stimulation (Fig. 3B).
FIGURE 3.
TGF-β-induced STAT3 early phosphorylation requires TGFβRI but not SMADs. A, kinase activity of TGFβRI is required for TGF-β-induced STAT3 phosphorylation. LX-2 cells were preincubated with 10 μm SB431542 (ALK5i) for 4 h and then treated with 4 ng/ml TGF-β for the indicated times. B, knockdown of TGFβRI in LX-2 cells decreased TGF-β-induced STAT3 phosphorylation. LX-2 cells transfected with the indicated siRNA were treated with 4 ng/ml TGF-β, and cell lysates were subjected to Western blotting. C, the early phase of TGF-β-induced STAT3 phosphorylation is independent of SMAD2/3, but the second phase of activation requires SMAD3. Smad2flox/floxSmad3+/+ or Smad2flox/floxSmad3−/− MEFs were infected with either adeno-GFP or adeno-CRE to generate control or cells devoid of SMAD2 (Smad2Δ/Δ). Two days after adenovirus infection, cells were treated with 4 ng/ml TGF-β for the indicated times, and phospho-STAT3 was detected by Western blotting. D, de novo protein synthesis is required for the TGF-β-induced second phase of STAT3 phosphorylation. LX-2 cells were preincubated with 10 ng/ml cycloheximide (CHX) for 2 h, and the cells were treated with TGF-β for the indicated times.
The facts that SMAD-binding defective TGFβRI (10) is capable of binding JAK1 and that TGF-β-induced SMAD phosphorylation follows the early but precedes the late phase of STAT phosphorylation raise an interesting possibility of SMAD-independent STAT3 phosphorylation in the early phase. To test whether this was the case, we took advantage of Smad2flox/floxSmad3+/+ and Smad2flox/floxSmad3−/− MEFs to create cells that lack SMAD2 only (Smad2Δ/ΔSmad3+/+) or lack both SMAD2 and SMAD3 (Smad2Δ/ΔSmad3−/−) after infecting them with adenoviruses expressing the CRE recombinase. The results indicated that regardless of the absence of SMAD2 alone or SMAD2 and SMAD3 in combination, the early phase phosphorylation of STAT3 was unaffected, but whenever SMAD3 was removed, the late phase phosphorylation of STAT3 was blocked (Fig. 3C). Further experiments in LX-2 cells using cycloheximide (CHX) showed that blocking protein synthesis also inhibited the late phase phosphorylation of STAT3 (Fig. 3D). Thus, in keeping with the paradigm set by interferon and interleukin signaling responses, TGF-β-induced early phase phosphorylation of STATs is a SMAD-independent, non-canonical signaling response, whereas the late phase phosphorylation requires SMAD3-mediated transcription and de novo protein synthesis.
Significance of TGFβRI-activated STAT3 in TGF-β Signaling
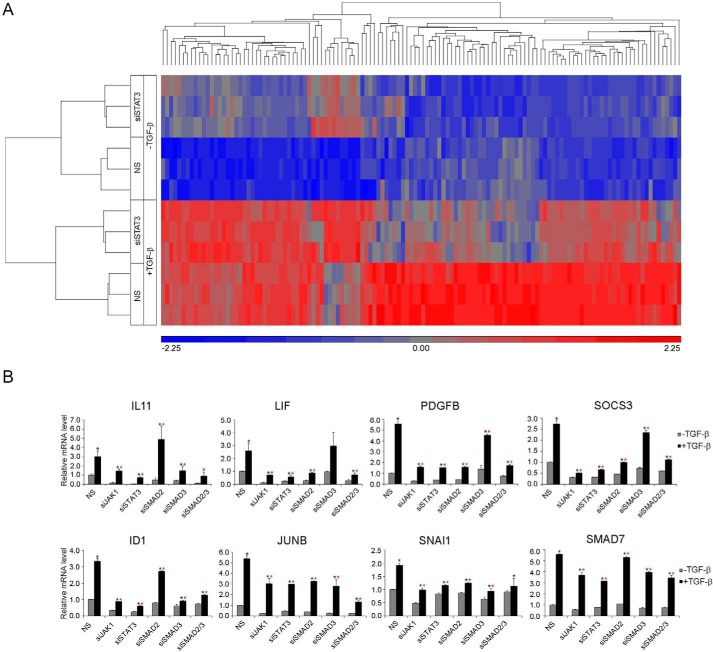
To address the significance of TGFβRI-activated STAT3 in TGF-β signaling, we compared the gene expression profiles in LX-2 cells associated with silencing STAT3 expression using siSTAT3 following 2 h of TGF-β treatment (Fig. 4A). We found 202 genes in the non-silenced control cells whose activity registered at least a 1.5-fold increase in response to TGF-β stimulation (supplemental Table S3). Of these 202 genes, 126 in the siSTAT3-silenced cells showed at least a 40% reduction in their response to TGF-β (Fig. 4A and supplemental Table S4). We also noticed that the basal expression of two genes, IL11 and SLC19A2, was reduced significantly in the silenced cells, although the -fold induction by TGF-β did not change much (supplemental Table S4). These 128 genes constitute potential targets of STAT3-dependent TGF-β signaling. Many well known targets of TGF-β and SMAD-mediated transcription targets, including JUNB, SMAD7, SNAI1, and ID1, were identified in this cohort of STAT3-dependent genes. This list also includes known ligand IL11, LIF, and PDGFB, whose expression could be essential for promoting STAT3 phosphorylation in the late phase. In addition, SOCS3, the classical negative feedback regulator of the JAK-STAT pathway, which plays an important role in the rapid attenuation of STAT3 phosphorylation at the end of the first phase, was also present in the list. The requirement of JAK1 and STAT3 for the induction of these genes by TGF-β was confirmed by quantitative RT-PCR (qRT-PCR), as was that of SMAD2 and/or SMAD3, although each gene showed a varying degree of reliance on SMAD2 or SMAD3 (Fig. 4B). For instance, knockdown of SMAD3, which is required for late phase STAT3 phosphorylation, had no effect on LIF expression and only slightly affected PDGFB and SOCS3 expression (Fig. 4B), suggesting that the early phase of STAT3 activation plays an important role in some TGF-β target genes. Overall, these results also indicated that the JAK1-STAT3 pathway is essential for eliciting TGF-β signaling responses in a subset of target genes.
FIGURE 4.
STAT3 is required for TGF-β-induced transcription of a subset of genes in LX-2 cells. A, hierarchical clustering of TGF-β-responsive and STAT3-dependent genes. Triplicates for each group are shown in the graph. Cells were treated with or without TGF-β for 2 h. B, qRT-PCR results of representative STAT3-dependent TGF-β target genes. Results are shown as relative expression ± S.D. (error bars) (n = 3). Statistically significant difference (p < 0.05) after TGF-β treatment is indicated by a black asterisk; statistically significant differences (p < 0.05) between control and knockdown cells after TGF-β treatment are indicated by a red asterisk.
The Interplay between STAT3 and SMAD3 in TGF-β-induced Transcriptional Responses
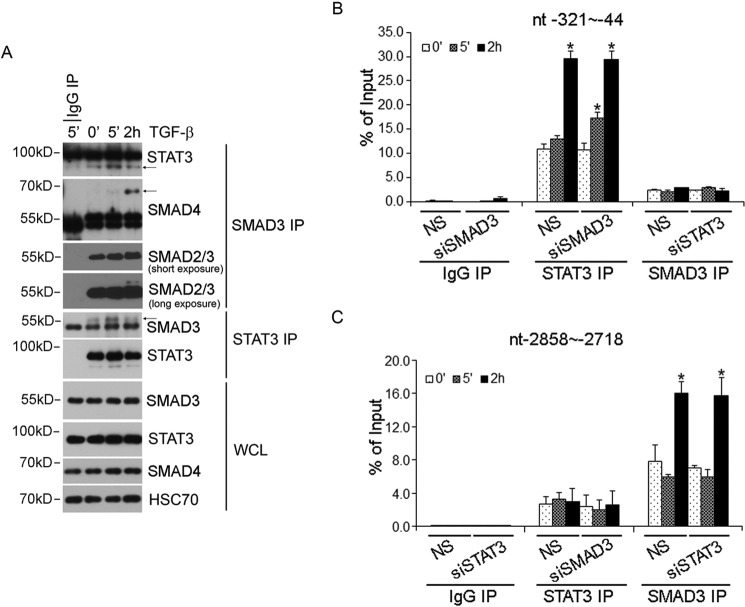
Functional interaction between SMADs and STAT3 has been reported to either enhance or antagonize STAT3-mediated transcription in hepatoma cells (14, 15). Physical interaction between SMAD3 and STAT3 was also reported (16). Moreover, unphosphorylated SMAD3 was also shown to be capable of recruiting PIAS3 (protein inhibitor of activated STAT3) to inhibit STAT3-dependent transcription by blocking DNA binding (17). To reconcile this mixed bag of information in the setting of biphasic activation of the JAK1-STAT3 axis that we observed here, we first examined the physical interaction between STAT3 and SMAD3 in LX-2 cells. We observed an enhancement of affinity between endogenous STAT3 and SMAD3 following 5 min of TGF-β stimulation; however, this interaction was weakened after 2 h of TGF-β stimulation, when SMAD3 became associated with SMAD4 (Fig. 5A). In lieu of the time course of SMAD3 phosphorylation in LX-2 cells (Fig. 2B), this result implied that the association of SMAD3 with SMAD4 dislodged the C-terminal phosphorylated SMAD3 from STAT3, which prefers the unphosphorylated SMAD3 as a binding partner. To assess the functional cooperation of STAT3 and SMAD3 in transcription, we used chromatin immunoprecipitation (ChIP) to examine their ability to bind the JUNB promoter. The JUNB promoter contains a well characterized proximal STAT3 binding site (18, 19) and a well characterized distal SMAD3 binding site (20). Our results showed that although robust and specific binding of STAT3 or SMAD3 to their respective sites was observed after 2 h of TGF-β stimulation, little STAT3 was found associated with the proximal site in the JUNB promoter in the first 5 min of activation, unless SMAD3 expression was knocked down with siSMAD3 (Fig. 5, B and C). These results suggested that unphosphorylated SMAD3 indeed possesses the ability to inhibit STAT3 binding to DNA, but it only does so during the early phase of activation of the JAK1-STAT3 axis. Our results also indicated that knockdown of STAT3 or SMAD3 did not affect the binding of the other party at its cognate site in the JUNB promoter in the late phase of activation (Fig. 5, B and C), suggesting that STAT3 or SMAD3 acts independently to coordinate JUNB transcription.
FIGURE 5.
Interplay between STAT3 and SMAD3 in LX-2 cells. A, physical interaction between STAT3 with SMAD3 in LX-2 cells. LX-2 cells were treated with 4 ng/ml TGF-β for 5 min or 2 h, and SMAD3 or STAT3 protein complex was immunoprecipitated by SMAD3 or STAT3 antibody. B, ChIP analysis of SMAD3 and STAT3 binding to the STAT3 DNA binding sites (bp −321 to −44) in the JUNB promoter after TGF-β stimulation. C, ChIP analysis of SMAD3 and STAT3 binding to the SMAD3 DNA binding sites (bp −2858 to −2718) in the JUNB promoter after TGF-β stimulation. Rabbit IgG was used as a negative control. *, statistically significant differences (p < 0.05) upon TGF-β stimulation. NS, not significant. Error bars, S.D.
Both STAT3 and SMAD3 Are Required for TGF-β-induced HSC Proliferation and Myofibroblast Differentiation
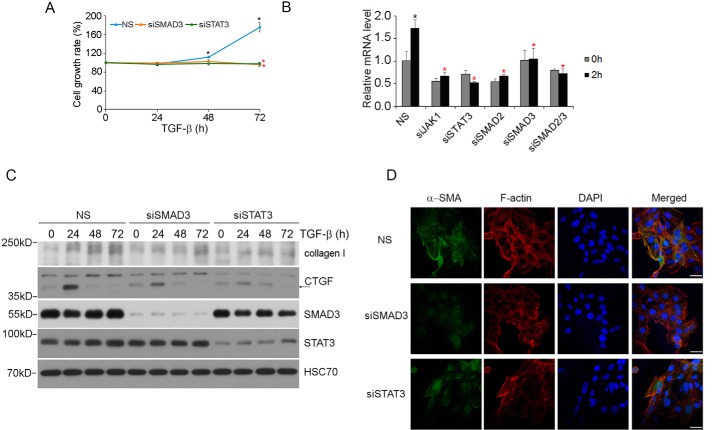
TGF-β is a strong driving force that compels HSCs to differentiate toward myofibroblast during liver damage and fibrosis (2, 21). This process is characterized by increased proliferation, synthesis of extracellular matrix components, and formation of α-smooth muscle actin (α-SMA) stress fibers. To assess the role of STAT3 in the TGF-β-induced fibrosis, we first examined the effect of TGF-β on the proliferation of STAT3-depleted or SMAD3-depleted LX-2 cells. The results indicated that whereas non-silenced control cells mounted a normal proliferative response to TGF-β, depleting either STAT3 or SMAD3 rendered cells unresponsive (Fig. 6A). The requirement of STAT3 and SMAD3 in TGF-β-induced proliferative response correlates with their role in induction of MYC expression (Fig. 6B). We next examined the TGF-β-induced extracellular matrix production in LX-2 cells using CTGF and collagen I expression as markers and found that both of them were significantly decreased after depleting either SMAD3 or STAT3 (Fig. 6C). Finally, we conducted immunofluorescence staining of α-SMA stress fibers, which marks the differentiation of hepatic stellate cells and the onset of liver fibrosis (22). In this experiment, LX-2 cells were transfected with siSTAT3 or siSMAD3 and treated with TGF-β before immunostaining with α-SMA antibody. The results showed a drastic decrease in α-SMA expression and formation of α-SMA fibers in either STAT3- or SMAD3-depleted cells (Fig. 6D), indicating that both STAT3 and SMAD3 are positively required for mediating the fibrogenic response of TGF-β.
FIGURE 6.
STAT3 is required for TGF-β-induced proliferation and fibrosis in LX-2 cells. A, TGF-β-induced proliferation in LX-2 cells is dependent on both SMAD3 and STAT3. LX-2 cells were transfected with the indicated siRNA and then treated with 2 ng/ml TGF-β for 24, 48, or 72 h, and viable cells at each time points were measured. Statistically significant difference (p < 0.05) after TGF-β treatment in siNS control cells is indicated by a black asterisk; statistically significant differences (p < 0.05) between control siNS cells and siSMAD3 or siSTAT3 cells is indicated by a red asterisk. B, qRT-PCR results of TGF-β-induced MYC gene expression in LX-2 cells. Cells were treated with or without TGF-β for 2 h. Data are shown as in Fig. 4B. C, STAT3 and SMAD3 are required for TGF-β-induced collagen I and CTGF expression. LX-2 cells transfected with the indicated siRNA were treated with 4 ng/ml TGF-β, and cell lysates were analyzed by Western blotting. D, STAT3 and SMAD3 are required for TGF-β-induced α-SMA stress fiber formation. LX-2 cells transfected with the indicated siRNA were treated with 4 ng/ml TGF-β for 4 days. Scale bar, 50 μm. NS, not significant. Error bars, S.D.
Discussion
Over the past decade, several reports have described the cross-talk between JAK-STAT and TGF-β pathways in hepatic cells (14, 23). TGF-β has been shown to potentiate IL-6-induced STAT3 activation in hepatoma cells (14) and activate STAT3 in hepatocytes and HSC cells (24, 25). Moreover, elevated STAT3 phosphorylation was reported in fibrosis and cirrhosis patient samples in conjunction with TGF-β and SMAD3 activation (24, 26). However, these cases of STAT3 activation were observed hours after TGF-β stimulation and therefore were attributed to secondary effects of cytokine or growth factor release triggered by TGF-β. Here, we report a physical interaction between TGFβRI and JAK1 and show that TGF-β activates STAT3 directly through JAK1 within minutes of stimulation. Our data also suggest that this early phase of TGF-β-induced, SMAD-independent activation of STAT3 primes the cells for further and stronger activation of STAT3 in the late phase, possibly by triggering the expression of various cytokines and growth factors.
The transcriptional cooperation between SMAD3 and STAT3 through p300 in hepatocytes was reported previously (14). This observation is consistent with our finding that activated SMAD3 and STAT3 bind to their respective DNA binding sites in the JUNB promoter to enhance TGF-β-mediated transcription in HSCs. However, unlike their functional cooperation in transcription, the direct physical interaction between SMAD3 and STAT3 usually resulted in antagonizing each other. For instance, SMAD3 could bring PIAS3 to STAT3, which inhibits STAT3 DNA binding (17, 27), whereas overexpression of hyperactive STAT3 traps SMAD3 from SMAD3-SMAD4 complex formation (16). It seems that the phosphorylation status of SMAD3 determines whether it inhibits or cooperates with STAT3 in transcription.
Like other SMAD-independent non-canonical pathways, TGF-β-induced activation of JAK-STAT is also cell context-dependent. The rapid early activation of STAT3 was observed in HSCs and fibroblasts but not in normal hepatocytes. This cell contextual difference in STAT3 activation could have implications in liver fibrosis, in which TGF-β1 is considered a pivotal profibrogenic cytokine. Like TGF-β, STAT3 activation was detected in all rodent models for liver injury and fibrosis. STAT3 in hepatocytes was shown to play a protective role in preventing liver fibrosis (28–30); however, it facilitates fibrogenesis in HSCs because mice devoid of STAT3 in HSCs (GFAPStat3−/− mice) are less susceptible to BDL- or CCL4-induced liver fibrosis (31). Similarly, in LX-2 cells, we observed that STAT3 is required for TGF-β-induced HSC proliferation, collagen I expression, and α-SMA stress fiber organization. These findings suggest that in addition to SMAD3, STAT3 directly participates in HSC activation and transdifferentiation in response to TGF-β and subsequent hepatic fibrosis.
Experimental Procedures
Antibodies and Reagents
Anti-FLAG-peroxidase (A8592, Sigma), anti-SMAD3 (ab40854, Abcam), anti-Ser(P)-423/Ser(P)-425 of SMAD3 (600-401-919, Rockland), anti-SMAD2 (ab33875, Abcam), anti-Myc (2278, Cell Signaling Technology), anti-α-tubulin (T9026, Sigma), anti-β-actin (ab8226, Abcam), anti-HSC70 (sc-7298, Santa Cruz Biotechnology, Inc.), anti-STAT3 (9139, Cell Signaling Technology), anti-STAT3 (Tyr(P)-705) (9131, Cell Signaling Technology), anti-JAK1 (3344, Cell Signaling Technology), anti-TGFβRI (sc-398, Santa Cruz Biotechnology), anti-CTGF (ab6992, Abcam), anti-collagen I (MA1-141, Thermo Fisher Scientific), rhodamine phalloidin (R415, Thermo Fisher Scientific), and anti-α-SMA (A2547, Sigma) were used for Western blotting, immunoprecipitation, or immunofluorescence. Cycloheximide (catalog no. 239763) was purchased from Millipore. Knockdown experiments were performed using the following predesigned siRNAs: non-silencing control siNS (1027310, Qiagen), siTGFβRI (SI02223627, Qiagen), siJAK1 (SI00605514, Qiagen), and siSTAT3 (6582S, Cell Signaling Technology). Validated siSMAD2 and siSMAD3 were described previously (5).
Cell Culture, TGF-β Stimulation, and Immunoprecipitation
AML12 cells were cultured in DMEM/F-12 supplemented with 10% fetal bovine serum (FBS), 0.005 mg/ml insulin, 0.005 mg/ml transferrin, 5 ng/ml selenium, and 40 ng/ml dexamethasone. Smad2flox/floxSmad3+/+ and Smad2flox/floxSmad3−/− MEFs were isolated from embryonic day 14.5 embryos of breeding Smad2flox/floxSmad3+/− mice (32, 33). All mice were maintained and handled according to protocols approved by the Animal Care and Use Committee of NCI, National Institutes of Health. LX-2 and MEFs were cultured in DMEM supplemented with 10% FBS. Hep3B and HepG2 cells were cultured in MEM supplemented with 1% non-essential amino acids and 10% FBS. Before 4 ng/ml TGF-β1 (100-21B, Peprotech) treatment, cells were starved overnight in cell medium containing 0.2% FBS. Cell proliferation was measured by Cell Counting Kit-8 (Dojindo Molecular Technologies).
For the anti-TGFβRI immunoprecipitation (IP) experiment, cells were harvested in FLAG lysis buffer containing 25 mm Tris-HCl, pH 7.5, 300 mm NaCl, and 1% Triton with protease and phosphatase inhibitors. The TGFβRI protein complex was immunoprecipitated by anti-TGFβRI antibody, and the presence of JAK1 in the protein complex was detected by Western blotting.
In Vitro Binding Assay
HEK293 cells transfected with FLAG-JAK1 (DU4539, University of Dundee) or control FLAG vector were precipitated by anti-FLAG-agarose (A2220, Sigma-Aldrich). After thorough washing, the agarose beads were incubated with purified TβRIc (34) for 2 h at 4 ºC. After a brief spin, supernatants were removed, and agarose beads were thoroughly washed before FLAG-JAK1 was eluted with FLAG peptide (F3290, Sigma-Aldrich). If there was an interaction between TβRIc and JAK1, TβRIc would be also eluted. The elution fraction was subjected to Western blotting analysis.
SILAC and Mass Spectrometric Analysis
For the SILAC experiment, AML12 cells were labeled with normal l-lysine and l-arginine (K0R0, light) or l-[13C6-15N2]lysine and l-[U-13C6-15N4]arginine (K8R10, heavy) (6). The cells cultured in K8R10 medium (heavy) were transfected with FLAG-TGFβRI, whereas the cells cultured in K0R0 medium (light) were transfected with a control FLAG vector. The cell lysates were combined at equal protein amounts. The combined cell lysates were subjected to FLAG immunoprecipitation and eluted with FLAG peptides. The eluted proteins were reduced, alkylated, and trypsin-digested overnight following a filter-aided digestion procedure using a FASP digestion kit (Protein Discovery, San Diego, CA) according to the vendor's protocol. Tryptic peptides were desalted, lyophilized, and reconstituted in 25% acetonitrile with 0.1% formic acid and further fractionated using strong cation exchange chromatography. The strong cation exchange fractions of the samples were pooled into nine fractions each, lyophilized, and reconstituted in 0.1% trifluoroacetic acid to be analyzed by LC-MS.
The LC-MS/MS data were processed using MaxQuant software (version 1.5.2.8). MS/MS data were searched by the Andromeda search engine against a Uniprot Mouse database containing both forward and reverse sequences as well as a common contaminant database in MaxQuant. The false discovery rate was set to 0.01 for both peptide and protein identifications. Protein identifications and quantitation results were further filtered and processed using the Perseus program (version 1.5.1.6) to generate normalized protein ratios. Only proteins that were identified by two unique peptides were used in further analysis. If a protein had a heavy/light ratio of <1.3, it was categorized as a background contaminant. The list of potential TGFβRI-interacting proteins whose heavy/light ratios were ≥1.3 was further filtered to remove “common contaminants” that are often found in FLAG IP affinity purification as defined by the CRAPome (35).
RNA Extraction, Microarray Analysis, and qRT-PCR
Total RNA from LX-2 cells was extracted by the RNeasy minikit (74104, Qiagen) according to the manufacturer's instructions. Microarray experiments were performed on Human Gene ST2.0 arrays according to the standard Affymetrix GeneChip protocol at the Affymetrix service core in the Frederick National Laboratory for Cancer Research. The array data were deposited in the Gene Expression Omnibus with the accession number GSE92638. Partek Genomics Suite version 6.6 was used for the microarray data analysis and hierarchical clustering. For qRT-PCR, RNA was converted to cDNA with a reverse transcription kit (4374966, Invitrogen), and the relative gene expression was measured by qRT-PCR. All samples were run in three biological replicates, each with 2–3 technical replicates. -Fold changes were calculated using the 2−ΔΔCt method, the calculated threshold values were determined by the maximum curvature, and ΔCt was calculated as Ctcontrol − Ctsample. The primers used for the RT-PCR experiment were as follows: ID1, CTGCTCTACGACATGAACGG (forward) and GAAGGTCCCTGATGTAGTCGAT (reverse); IL11, CGAGCGGACCTACTGTCCTA (forward) and GCCCAGTCAAGTGTCAGGTG (reverse); JUNB, ACGACTCATACACAGCTACGG (forward) and GCTCGGTTTCAGGAGTTTGTAGT (reverse); SNAI1, TCGGAAGCCTAACTACAGCGA (forward) and AGATGAGCATTGGCAGCGAG (reverse); LIF, CCAACGTGACGGACTTCCC (forward) and TACACGACTATGCGGTACAGC (reverse); PDGFB, CTCGATCCGCTCCTTTGATGA (forward) and CGTTGGTGCGGTCTATGAG (reverse); SMAD7, CAAGAGGCTGTGTTGCTGTG (forward) and GGCCTTCCATCCAACTCTCT (reverse); SOCS3, CCTGCGCCTCAAGACCTTC (forward) and GTCACTGCGCTCCAGTAGAA (reverse); MYC, GGCTCCTGGCAAAAGGTCA (forward) and CTGCGTAGTTGTGCTGATGT (reverse); β-actin, AACTCCATCATGAAGTGTGACG (forward) and GATCCACATCTGCTGGAAGG (reverse); 18S rRNA, GATATGCTCATGTGGTGTTG (forward) and AATCTTCTTCAGTCGCTCCA (reverse).
PLA
The PLA was performed using the Duolink® system (DUO92101, Sigma-Aldrich) according to the manufacturer's instructions. Briefly, LX-2 cells were grown on BD Falcon 4-chamber slides. After fixation and permeabilization, cells were incubated with anti-JAK1 (monoclonal) and anti-TGFβRI (polyclonal) primary antibodies together, or anti-TGFβRI antibody and normal mouse IgG as a negative control. After washing, cells were sequentially incubated with secondary antibodies with PLA probes, ligation solution, and detection solution with thorough washes between each step. PLA signals were visualized using a Leica TCS SP8 confocal system. Image analyses of PLA data were performed using Blobfinder (36).
ChIP
ChIP was performed using STAT3 antibody (sc-482 X, Santa Cruz Biotechnology), SMAD3 antibody (ab28379, Abcam), or control rabbit IgG plus Protein G beads using the HighCell# ChIP kit (C01010063, Diagenode). Briefly, cells were fixed with formaldehyde, lysed, and sonicated to shear the chromatin. The immunoprecipitated chromatin fragments were eluted and treated with proteinase K. The eluted DNA fragments were quantified by qPCR using primers specific to the JUNB promoter region, bp −321 to −44 (forward, CCAGTGGACTCCAGGGAAATC; reverse, GCGCTAGTCAGCCACGGAAG) or bp −2858 to −2718 (forward, CTGAATTACTGTGGCCTCCT; reverse, GCTAATAACTGCAGCTGACATC). Values are expressed as the -fold enrichment of the percentage of input.
Statistical Analysis
Statistical differences were calculated by using Student's t test.
Author Contributions
L.-Y. T. and Y. E. Z. conceived the project; L.-Y. T. performed most of experiments and analyzed data; Z. M., M. Z., and L.-R. Y. performed MS analyses; Y. T. isolated MEFs; M. H. helped with IP experiments; and L.-Y. T. and Y. E. Z. wrote the manuscript.
Supplementary Material
Acknowledgments
We thank Drs. C. Stuelten and M. Weinstein for Smad2flox and C-X. Deng for Smad3+/− mice. We also thank J. Wang and N. Sova for assistance.
This work was supported by the intramural research program of the National Institutes of Health, NCI, Center for Cancer Research (to Y. E. Z.). The authors declare that they have no conflicts of interest with the contents of this article. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.

This article contains supplemental Tables S1–S4.
- HSC
- hepatic stellate cell
- TGFβRI
- type I TGF-β receptor
- TGFβRII
- type II TGF-β receptor
- SILAC
- stable isotope labeling by amino acids in cell culture
- K0R0
- normal lysine and arginine
- K8R10
- l-[13C6-15N2]lysine and l-[U-13C6-15N4]arginine
- PLA
- proximity ligation assay
- α-SMA
- α-smooth muscle actin
- qRT-PCR
- quantitative RT-PCR
- IP
- immunoprecipitation.
References
- 1. Morikawa M., Derynck R., and Miyazono K. (2016) TGF-β and the TGF-β family: context-dependent roles in cell and tissue physiology. Cold Spring Harb. Perspect. Biol. 8, a021873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Fabregat I., Moreno-Càceres J., Sánchez A., Dooley S., Dewidar B., Giannelli G., Ten Dijke P., and IT-LIVER Consortium (2016) TGF-β signalling and liver disease. FEBS J. 283, 2219–2232 [DOI] [PubMed] [Google Scholar]
- 3. Hata A., and Chen Y. G. (2016) TGF-β signaling from receptors to Smads. Cold Spring Harb. Perspect. Biol. 8, a022061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Zhang Y. E. (2016) Non-Smad signaling pathways of the TGF-β family. Cold Spring Harb. Perspect. Biol. 9, a022129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Yang Y. A., Zhang G. M., Feigenbaum L., and Zhang Y. E. (2006) Smad3 reduces susceptibility to hepatocarcinoma by sensitizing hepatocytes to apoptosis through downregulation of Bcl-2. Cancer Cell 9, 445–457 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Ong S. E., Blagoev B., Kratchmarova I., Kristensen D. B., Steen H., Pandey A., and Mann M. (2002) Stable isotope labeling by amino acids in cell culture, SILAC, as a simple and accurate approach to expression proteomics. Mol. Cell. Proteomics 1, 376–386 [DOI] [PubMed] [Google Scholar]
- 7. Yamashita M., Fatyol K., Jin C., Wang X., Liu Z., and Zhang Y. E. (2008) TRAF6 mediates Smad-independent activation of JNK and p38 by TGF-β. Mol. Cell 31, 918–924 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Shibuya H., Iwata H., Masuyama N., Gotoh Y., Yamaguchi K., Irie K., Matsumoto K., Nishida E., and Ueno N. (1998) Role of TAK1 and TAB1 in BMP signaling in early Xenopus development. EMBO J. 17, 1019–1028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Sorrentino A., Thakur N., Grimsby S., Marcusson A., von Bulow V., Schuster N., Zhang S., Heldin C. H., and Landström M. (2008) The type I TGF-β receptor engages TRAF6 to activate TAK1 in a receptor kinase-independent manner. Nat. Cell Biol. 10, 1199–1207 [DOI] [PubMed] [Google Scholar]
- 10. Yu L., Hébert M. C., and Zhang Y. E. (2002) TGF-β receptor-activated p38 MAP kinase mediates Smad-independent TGF-β responses. EMBO J. 21, 3749–3759 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Harrison D. A. (2012) The Jak/STAT pathway. Cold Spring Harb. Perspect. Biol. 4, a011205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Inagaki-Ohara K., Kondo T., Ito M., and Yoshimura A. (2013) SOCS, inflammation, and cancer. JAKSTAT 2, e24053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Kusaba M., Nakao K., Goto T., Nishimura D., Kawashimo H., Shibata H., Motoyoshi Y., Taura N., Ichikawa T., Hamasaki K., and Eguchi K. (2007) Abrogation of constitutive STAT3 activity sensitizes human hepatoma cells to TRAIL-mediated apoptosis. J. Hepatol. 47, 546–555 [DOI] [PubMed] [Google Scholar]
- 14. Yamamoto T., Matsuda T., Muraguchi A., Miyazono K., and Kawabata M. (2001) Cross-talk between IL-6 and TGF-β signaling in hepatoma cells. FEBS Lett. 492, 247–253 [DOI] [PubMed] [Google Scholar]
- 15. Zauberman A., Lapter S., and Zipori D. (2001) Smad proteins suppress CCAAT/enhancer-binding protein (C/EBP) β- and STAT3-mediated transcriptional activation of the haptoglobin promoter. J. Biol. Chem. 276, 24719–24725 [DOI] [PubMed] [Google Scholar]
- 16. Wang G., Yu Y., Sun C., Liu T., Liang T., Zhan L., Lin X., and Feng X. H. (2016) STAT3 selectively interacts with Smad3 to antagonize TGF-β. Oncogene 35, 4388–4398 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Yoon J. H., Sudo K., Kuroda M., Kato M., Lee I. K., Han J. S., Nakae S., Imamura T., Kim J., Ju J. H., Kim D. K., Matsuzaki K., Weinstein M., Matsumoto I., Sumida T., and Mamura M. (2015) Phosphorylation status determines the opposing functions of Smad2/Smad3 as STAT3 cofactors in TH17 differentiation. Nat. Commun. 6, 7600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Coffer P., Lutticken C., van Puijenbroek A., Klop-de Jonge M., Horn F., and Kruijer W. (1995) Transcriptional regulation of the junB promoter: analysis of STAT-mediated signal transduction. Oncogene 10, 985–994 [PubMed] [Google Scholar]
- 19. Kojima H., Nakajima K., and Hirano T. (1996) IL-6-inducible complexes on an IL-6 response element of the junB promoter contain Stat3 and 36 kDa CRE-like site binding protein. Oncogene 12, 547–554 [PubMed] [Google Scholar]
- 20. Jonk L. J., Itoh S., Heldin C. H., ten Dijke P., and Kruijer W. (1998) Identification and functional characterization of a Smad binding element (SBE) in the JunB promoter that acts as a transforming growth factor-β, activin, and bone morphogenetic protein-inducible enhancer. J. Biol. Chem. 273, 21145–21152 [DOI] [PubMed] [Google Scholar]
- 21. Cong M., Iwaisako K., Jiang C., and Kisseleva T. (2012) Cell signals influencing hepatic fibrosis. Int. J. Hepatol. 2012, 158547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Carpino G., Morini S., Ginanni Corradini S., Franchitto A., Merli M., Siciliano M., Gentili F., Onetti Muda A., Berloco P., Rossi M., Attili A. F., and Gaudio E. (2005) α-SMA expression in hepatic stellate cells and quantitative analysis of hepatic fibrosis in cirrhosis and in recurrent chronic hepatitis after liver transplantation. Dig. Liver Dis. 37, 349–356 [DOI] [PubMed] [Google Scholar]
- 23. Tang Y., Kitisin K., Jogunoori W., Li C., Deng C. X., Mueller S. C., Ressom H. W., Rashid A., He A. R., Mendelson J. S., Jessup J. M., Shetty K., Zasloff M., Mishra B., Reddy E. P., et al. (2008) Progenitor/stem cells give rise to liver cancer due to aberrant TGF-β and IL-6 signaling. Proc. Natl. Acad. Sci. U.S.A. 105, 2445–2450 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Liu Y., Liu H., Meyer C., Li J., Nadalin S., Königsrainer A., Weng H., Dooley S., and ten Dijke P. (2013) Transforming growth factor-β (TGF-β)-mediated connective tissue growth factor (CTGF) expression in hepatic stellate cells requires Stat3 signaling activation. J. Biol. Chem. 288, 30708–30719 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Yang Y., Pan X., Lei W., Wang J., Shi J., Li F., and Song J. (2006) Regulation of transforming growth factor-β 1-induced apoptosis and epithelial-to-mesenchymal transition by protein kinase A and signal transducers and activators of transcription 3. Cancer Res. 66, 8617–8624 [DOI] [PubMed] [Google Scholar]
- 26. Xu M. Y., Hu J. J., Shen J., Wang M. L., Zhang Q. Q., Qu Y., and Lu L. G. (2014) Stat3 signaling activation crosslinking of TGF-β1 in hepatic stellate cell exacerbates liver injury and fibrosis. Biochim. Biophys. Acta 1842, 2237–2245 [DOI] [PubMed] [Google Scholar]
- 27. Chung C. D., Liao J., Liu B., Rao X., Jay P., Berta P., and Shuai K. (1997) Specific inhibition of Stat3 signal transduction by PIAS3. Science 278, 1803–1805 [DOI] [PubMed] [Google Scholar]
- 28. Kroy D. C., Beraza N., Tschaharganeh D. F., Sander L. E., Erschfeld S., Giebeler A., Liedtke C., Wasmuth H. E., Trautwein C., and Streetz K. L. (2010) Lack of interleukin-6/glycoprotein 130/signal transducers and activators of transcription-3 signaling in hepatocytes predisposes to liver steatosis and injury in mice. Hepatology 51, 463–473 [DOI] [PubMed] [Google Scholar]
- 29. Mair M., Zollner G., Schneller D., Musteanu M., Fickert P., Gumhold J., Schuster C., Fuchsbichler A., Bilban M., Tauber S., Esterbauer H., Kenner L., Poli V., Blaas L., Kornfeld J. W., et al. (2010) Signal transducer and activator of transcription 3 protects from liver injury and fibrosis in a mouse model of sclerosing cholangitis. Gastroenterology 138, 2499–2508 [DOI] [PubMed] [Google Scholar]
- 30. Wang H., Lafdil F., Kong X., and Gao B. (2011) Signal transducer and activator of transcription 3 in liver diseases: a novel therapeutic target. Int. J. Biol. Sci. 7, 536–550 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Meng F., Wang K., Aoyama T., Grivennikov S. I., Paik Y., Scholten D., Cong M., Iwaisako K., Liu X., Zhang M., Osterreicher C. H., Stickel F., Ley K., Brenner D. A., and Kisseleva T. (2012) Interleukin-17 signaling in inflammatory, Kupffer cells, and hepatic stellate cells exacerbates liver fibrosis in mice. Gastroenterology 143, 765–776.e1–3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Liu Y., Festing M. H., Hester M., Thompson J. C., and Weinstein M. (2004) Generation of novel conditional and hypomorphic alleles of the Smad2 gene. Genesis 40, 118–123 [DOI] [PubMed] [Google Scholar]
- 33. Yang X., Letterio J. J., Lechleider R. J., Chen L., Hayman R., Gu H., Roberts A. B., and Deng C. (1999) Targeted disruption of SMAD3 results in impaired mucosal immunity and diminished T cell responsiveness to TGF-β. EMBO J. 18, 1280–1291 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Millet C., Yamashita M., Heller M., Yu L. R., Veenstra T. D., and Zhang Y. E. (2009) A negative feedback control of transforming growth factor-β signaling by glycogen synthase kinase 3-mediated Smad3 linker phosphorylation at Ser-204. J. Biol. Chem. 284, 19808–19816 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Mellacheruvu D., Wright Z., Couzens A. L., Lambert J. P., St-Denis N. A., Li T., Miteva Y. V., Hauri S., Sardiu M. E., Low T. Y., Halim V. A., Bagshaw R. D., Hubner N. C., Al-Hakim A., Bouchard A., et al. (2013) The CRAPome: a contaminant repository for affinity purification-mass spectrometry data. Nat. Methods 10, 730–736 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Allalou A., and Wählby C. (2009) BlobFinder, a tool for fluorescence microscopy image cytometry. Comput. Methods Programs Biomed. 94, 58–65 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.