Abstract
Polycystin complexes, or TRPP-PKD complexes, made of transient receptor potential channel polycystin (TRPP) and polycystic kidney disease (PKD) proteins, play key roles in coupling extracellular stimuli with intracellular Ca2+ signals. For example, the TRPP2-PKD1 complex has a crucial function in renal physiology, with mutations in either protein causing autosomal dominant polycystic kidney disease. In contrast, the TRPP3-PKD1L3 complex responds to low pH and was proposed to be a sour taste receptor candidate. It has been shown previously that the protein partners interact via association of the C-terminal or transmembrane segments, with consequences for the assembly, surface expression, and function of the polycystin complexes. However, the roles of extracellular components, especially the loops that connect the transmembrane segments, in the assembly and function of the polycystin complex are largely unknown. Here, with an immunoprecipitation method, we found that extracellular loops between the first and second transmembrane segments of TRPP2 and TRPP3 associate with the extracellular loops between the sixth and seventh transmembrane segments of PKD1 and PKD1L3, respectively. Immunofluorescence and electrophysiology data further confirm that the associations between these loops are essential for the trafficking and function of the complexes. Interestingly, most of the extracellular loops are also found to be involved in homomeric assembly. Furthermore, autosomal dominant polycystic kidney disease-associated TRPP2 mutant T448K significantly weakened TRPP2 homomeric assembly but had no obvious effect on TRPP2-PKD1 heteromeric assembly. Our results demonstrate a crucial role of these functionally underexplored extracellular loops in the assembly and function of the polycystin complexes.
Keywords: cell surface receptor, ion channel, protein assembly, protein-protein interaction, transient receptor potential channels (TRP channels), PKD, TRPP, autosomal dominant polycystic kidney disease, extracellular loop, polycystin
Introduction
Autosomal dominant polycystic kidney disease (ADPKD)4 is one of the most common inherited diseases, affecting 1 in every 400–1000 individuals (1). Mutations in integral membrane proteins TRPP2 and PKD1 are the cause of almost all clinically identified ADPKD cases (2, 3). For this reason, proteins in both families of TRPP2 and PKD1 are named “polycystins.” TRPP2 belongs to the transient receptor potential polycystin (TRPP) subfamily of the TRP superfamily of cation channels (4, 5). All TRPP proteins, including TRPP2 (polycystin-2), TRPP3 (polycystin-2L1), and TRPP5 (polycystin-2L2), have six transmembrane domains and intracellular N and C termini (2). PKD1 is a member of the polycystic kidney disease (PKD) protein family. Proteins in this family, including PKD1 (polycystin-1), PKD1L1 (polycystin-1L1), PKD1L2 (polycystin-1L2), PKD1L3 (polycystin-1L3), and PKDREJ (polycystin-REJ), all have 11 putative transmembrane domains, a large extracellular N terminus, and a relatively short intracellular C terminus (6–8). The last six transmembrane domains of PKD proteins share significant sequence similarity with TRPP proteins.
TRPP proteins can assemble as homotetrameric ion channels by themselves (9–14) and as heteromeric receptor-ion channel complexes with PKD proteins (15). Although it is expected that more polycystin complexes will be identified in the future, four of them have been functionally studied so far. The TRPP2-PKD1 complex was proposed to function as a sensor that couples extracellular stimuli with intracellular Ca2+ signals on the primary cilia of renal epithelial cells (6, 15, 16). The TRPP2-PKD1L1 complex is expressed in the primary cilia of embryonic nodal cells, where it senses nodal flow and determines left-right asymmetry in the early development of vertebrate animals (17, 18). The TRPP3-PKD1L3 complex is expressed in a subgroup of taste cells and has been proposed to function as a sour taste candidate (19–22), although a contradictory result was reported with the PKD1L3 knock-out mouse (23). The last one, the TRPP3-PKD1L1 complex, is a Ca2+-permeable channel in primary cilia, contributing to the ciliary Ca2+ signaling (24). It is generally believed that, in the polycystin complexes, TRPP subunits form an ion channel, whereas the PKD subunit receives the extracellular signal as a sensor or receptor (2, 6). However, our recent study showed that PKD1L3 is also a channel pore-forming subunit (25), suggesting that other PKD proteins may also function as an ion channel subunit in their complexes with TRPPs.
Previous studies on the assembly of the TRPP2-PKD1 and TRPP3-PKD1L3 complexes have extended our understanding of these essential molecules. It has been found that both TRPP2 and PKD1 have intracellular C-terminal coiled-coil domains, which are involved in the association between TRPP2 and PKD1 (26, 27). Further evidence showed that the coiled-coil domains from three TRPP2 subunits form a tightly bundled trimer in both solution and protein crystal (28, 29). The downstream region of this trimer forms the PKD1-binding site and binds to one copy of the C-terminal coiled-coil domain of PKD1, determining a 3:1 (TRPP2/PKD1) subunit stoichiometry of the full-length complex (29, 30). Our recent study concluded that this 3:1 stoichiometry is shared by the TRPP3-PKD1L3 complex (25), suggesting a common assembly mechanism among all polycystin complexes. Although several other domains and amino acids were found to be involved in TRPP2 homomeric assembly (31, 32), so far, the C-terminal coiled-coil domain is the only site shown to be directly involved in its assembly with PKD1. The association between TRPP3 and PKD1L3 differs from that of TRPP2 and PKD1. Since PKD1L3 lacks the C-terminal coiled-coil domain (33), the C-terminal interaction may not exist in the TRPP3-PKD1L3 complex. Instead, the transmembrane domains of TRPP3 and PKD1L3 were shown to mediate their association (34).
PKD and TRPP proteins have unique extracellular components in their structures. TRPP proteins have a large extracellular loop located between the first and the second transmembrane domains (S1-S2 loop), which has 224 amino acids in human TRPP2 (5). As a significant portion of TRPP proteins, S1-S2 loops span 23% (in TRPP2), 23% (in TRPP3), or 37% (in TRPP5) of the total protein length (5, 35, 36). As for the PKD proteins, besides the long extracellular N terminus (>3000 amino acids in human PKD1), they all have a relatively large extracellular loop between the sixth and seventh transmembrane domains (S6-S7 loop), which is composed of 211 amino acids in human PKD1 (7, 8). Considering the fact that the last six transmembrane domains of PKD proteins have sequence similarity with the six transmembrane domains of TRPP proteins, the S1-S2 loop of TRPP and the S6-S7 loop of PKD are located at homologous positions (Fig. 1, A and B). Many ADPKD pathogenic mutations are located in these loops (ADPKD Mutation Database), indicating a notable role of this loop in TRPP2 structure and function. Despite their large size and potential functional importance, very little was known about these extracellular loops until recently. During the preparation of this paper, three TRPP2 cryo-electron microscopy (cryo-EM) structures were reported (9, 10, 14). From these structures, it can be seen that the TRPP2 extracellular loops are involved in the homomeric assembly of the TRPP2 channel and may also be important for ion permeability and channel function regulation (9, 10). In this study, we hypothesized that TRPP and PKD proteins also associate through extracellular loops and tested this hypothesis with biochemical and biophysical methods, using the TRPP2-PKD1 and TRPP3-PKD1L3 complexes as model molecules. Our results indicate that the S1-S2 loop of the TRPP proteins and the S6-S7 loop of the PKD proteins bind to each other and play an essential role in the assembly and function of the polycystin complexes.
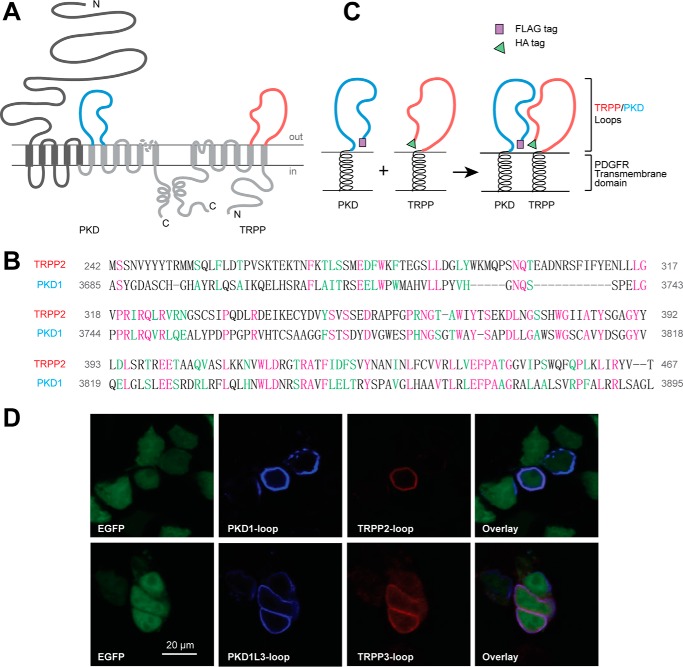
FIGURE 1.
PKD and TRPP loop constructs, which were cloned into pDisplay vector and transfected into HEK 293T cells, expressed on the extracellular surface of the plasma membrane. A, the putative transmembrane topology of PKD (left) and TRPP (right) proteins, showing the position of extracellular loops between the sixth and seventh transmembrane domains of PKD proteins (S6-S7 loop, colored in blue) and between the first and second transmembrane domains of TRPP proteins (S1-S2 loop, colored in red). B, protein sequence alignment between the S1-S2 loop of human TRPP2 and the S6-S7 loop of human PKD1, two examples in the families. Purple, identical; green, conserved. Identity was 57 of 230 (25.0%), and similarity was 97 of 230 (42%). Amino acids positions in the full-length protein are labeled on both sides. C, schematic diagram of the structures of the expressed loop protein constructs whose association was confirmed in this study. cDNA of the loops were inserted into modified pDisplay vector (Life Technologies), which generated extracellular loop fragments with N-terminal FLAG or HA tag and C-terminal PDGFR transmembrane domain. D, surface co-localization of the FLAG-tagged PKD1 loop and HA-tagged TRPP2 loop and of the FLAG-tagged PKD1L3 loop and HA-tagged TRPP3 loop in HEK 293T cells. EGFP was co-transfected to label the cytosol. PKD loops and TRPP loops were stained first with anti-FLAG or anti-HA antibodies followed by Cy5 or Texas Red-labeled secondary antibodies on non-permeabilized cells.
Results
DNA Constructs and Cell Surface Expression of the TRPP and PKD Extracellular Loops
To eliminate the interference of other regions and to mimic their native localization, we expressed the isolated loop fragments on the extracellular side of the plasma membrane and kept it tethered to the membrane. The DNA constructs were made by cloning the cDNA of the loop fragments into the pDisplay vector (Invitrogen). Proteins expressed in this vector have an Ig κ-chain leader sequence fused to the N terminus, which directs the protein to the secretory pathway, and a platelet-derived growth factor receptor (PDGFR) transmembrane domain fused to the C terminus, which anchors the protein to the plasma membrane (Fig. 1C). An HA tag is inserted between the Ig κ-chain leader sequence and the TRPP2 (or TRPP3) loop fragment. Similarly, a FLAG tag is placed between the Ig κ-chain leader sequence and the PKD1 (or PKD1L3) loop fragment (Fig. 1C). As a signal peptide, the Ig κ-chain leader sequence will be cleaved from the N terminus after expression, and the HA or FLAG tag will be exposed for detection.
The expression of the recombinant proteins on the extracellular surface of the plasma membrane was then confirmed by the immunofluorescence experiment (37). When either TRPP2 S1-S2 and PKD1 S6-S7 loops or TRPP3 S1-S2 and PKD1L3 S6-S7 loops were coexpressed in HEK 293T cells, we detected cell surface expression of both pairs of proteins (Fig. 1D). These results set the basis of our further tests.
The TRPP S1-S2 Loops Specifically Associate with the PKD S6-S7 Loops
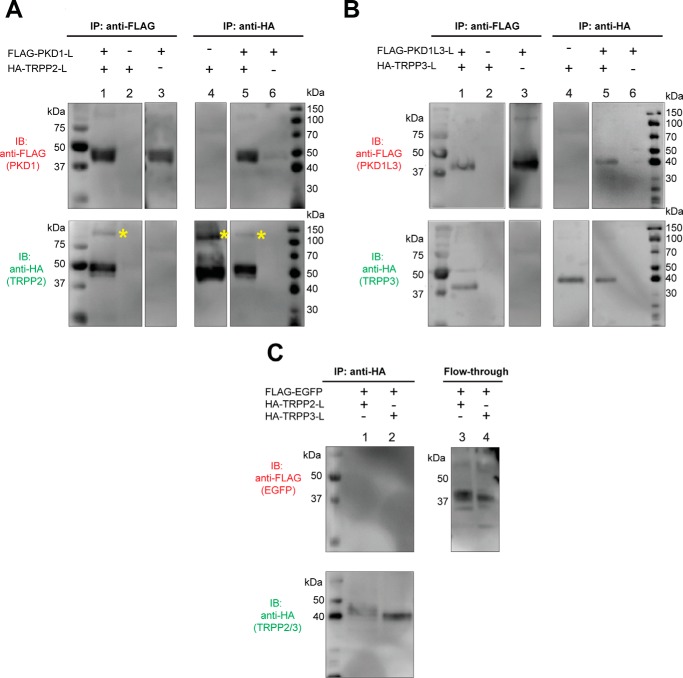
We then tested the association between TRPP S1-S2 loops and PKD S6-S7 loops with the co-immunoprecipitation (co-IP) method. All loop proteins migrate to the 40–50 kDa range on the SDS-polyacrylamide gel (Fig. 2). When expressed in HEK 293T cells, the HA-TRPP2 loop was immunoprecipitated by anti-FLAG antibody only when the FLAG-PKD1 loop was present (Fig. 2A, lanes 1–3 in both the top and bottom gels), indicating the association between the two loops. This result was further verified by using anti-HA antibody in co-IP. The FLAG-PKD1 loop was immunoprecipitated together with the HA-TRPP2 loop, which only happens when both proteins are coexpressed (Fig. 2A, lanes 4–6). It is worth noting that the TRPP2 loop gave another weak band with the size of about 100–120 kDa (Fig. 2A, labeled with asterisks). We also observed similar bands for other loops in some of the following experiments. Judging from the molecular weight, these bands most likely represent dimers of the loop proteins that were not completely separated by SDS treatment. However, we cannot rule out the possibility that the loops form complexes with endogenously expressed proteins. The homomeric assembly of the loops was tested later in Fig. 3.
FIGURE 2.
Specific association between the extracellular loops of PKD1 and TRPP2, and of PKD1L3 and TRPP3. A, co-IP shows the association between the FLAG-tagged PKD1 loop (FLAG-PKD1-L) and the HA-tagged TRPP2-loop (HA-TRPP2-L). Co-IP was done with either anti-FLAG (left) or anti-HA (right) antibody. With the N-terminal FLAG tag and the C-terminal PDGFR transmembrane domain, the PKD1 loop had an apparent molecular mass of about 45 kDa on SDS-PAGE. The HA-tagged TRPP2 loop with the PDGFR transmembrane domain has an apparent molecular mass of about 50 kDa. Besides the monomers, a small amount of the TRPP2 loop protein also migrated at a higher molecular mass position (labeled with yellow asterisks). B, co-IP shows the association between the FLAG-tagged PKD1L3 loop (FLAG-PKD1L3-L) and the HA-tagged TRPP3 loop (HA-TRPP3-L). Co-IP was done with either anti-FLAG (left) or anti-HA (right) antibody. Both proteins are about 40 kDa on SDS-PAGE. C, in the control experiment, when EGFP replaced the PKD loops, no association was found between the FLAG-tagged EGFP and the HA-tagged TRPP2 or TRPP3 loops. Co-IP was done with the anti-HA antibody (left). The flow-through samples (right) confirm overall expression of the FLAG-EGFP.
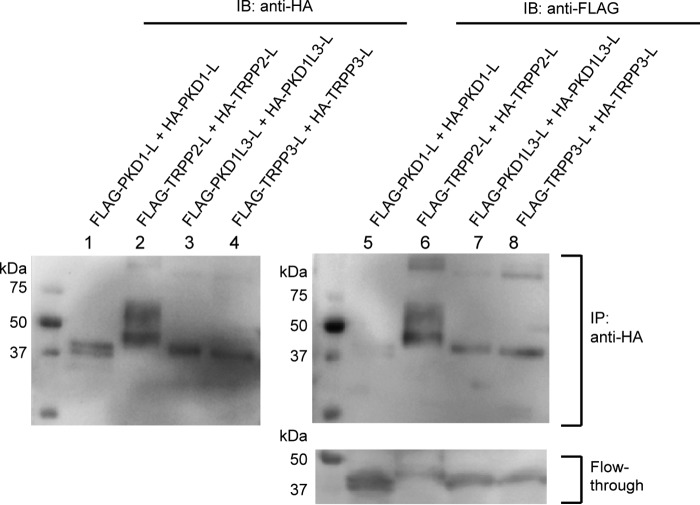
FIGURE 3.
Homomeric association between the extracellular loops. Co-IP followed by Western blotting (IB) shows the homomeric interaction between the FLAG- and HA-tagged TRPP2 loops (lanes 2 and 6 of the top two gels), the FLAG- and HA-tagged PKD1L3 loops (lanes 3 and 7 of the top two gels), and the FLAG- and HA-tagged TRPP3 loops (lanes 4 and 8 of the top two gels). However, the FLAG-PKD1 loop was not immunoprecipitated with the HA-PKD1 loop (lanes 1 and 5 of the top two gels), although it expressed well in cells, as can be seen in the flow-through sample (lane 5 in the bottom gel). Co-IP was done with the anti-HA antibody-coated beads.
Similar results were obtained when co-IP was done between the S1-S2 loop of TRPP3 and the S6-S7 loop of PKD1L3 (Fig. 2B). With either anti-FLAG (Fig. 2B, lanes 1–3) or anti-HA antibody (Fig. 2B, lanes 4–6), the HA-TRPP3 loop and the FLAG-PKD1L3 loop were co-immunoprecipitated only when both proteins were coexpressed. The data in Fig. 2 suggest that the association between the extracellular S1-S2 loop of TRPP proteins and the S6-S7 loop of PKD proteins may be common within the complexes formed between these two families. To test the specificity of the loop association, we used the enhanced green fluorescence protein (EGFP) as the negative control to replace the PKD loops in the co-IP experiments. When FLAG-EGFP was coexpressed with the HA-TRPP2 loop or the HA-TRPP3 loop, it was not co-immunoprecipitated with either HA-tagged loop protein (Fig. 2C, lanes 1 and 2), although it was expressed well in the cells, as seen in flow-through samples (Fig. 2C, lanes 3 and 4). These results indicate that the S1-S2 loops of TRPP proteins associate with the S6-S7 loops of their PKD partners.
Homomeric Assembly of the Extracellular Loops
Both the TRPP2-PKD1 and TRPP3-PKD1L3 complexes consist of three TRPP subunits and one PKD subunit (25, 29). The homomeric trimers of TRPP2 and TRPP3 are assembled through their C termini (28–30, 38–41). Since TRPP2 and TRPP3 are also able to form homotetrameric channels (9–13), there should be other domains responsible for the tetrameric assembly. We speculated on whether the extracellular S1-S2 loops also contributed to the homomeric assembly of TRPP and whether the extracellular S6-S7 loops contributed to any possible homomeric assembly of PKD proteins. To test this, we generated FLAG-tagged and HA-tagged versions of each loop protein and cotransfected two versions into HEK 293T cells. The co-IP experiments were then conducted with an anti-HA antibody, and immunoprecipitated proteins were detected with both anti-HA and anti-FLAG antibodies. Thus, the presence of the FLAG-tagged protein in the immunoprecipitated sample means that it has a homomeric association with its HA-tagged version (Fig. 3, top right gel). Unique among the four pairs, the PKD1 loop has the weakest, if any, homomeric interaction (Fig. 3, lanes 1 and 5), whereas the other three pairs of loop proteins demonstrate clear homomeric interaction (Fig. 3, lanes 2–4 and 6–8). The co-IP results of the TRPP loops indicate that, besides the interaction with PKD partners, the S1-S2 loops of these proteins are also involved in homomeric assembly, consistent with the homotetramer formed by this loop in the TRPP2 cryo-EM structures (9, 10, 14). The homomeric assembly of PKD1L3 is not expected because there is only one copy of PKD1L3 in its complex with TRPP3 (25). However, it is still possible that PKD1L3 can form a homomeric complex when it is not associated with TRPP3. This feature is largely missing in PKD1, consistent with the previous report showing that purified PKD1 is a monomer (42).
Extracellular Loops Assemble with Full-length Partners
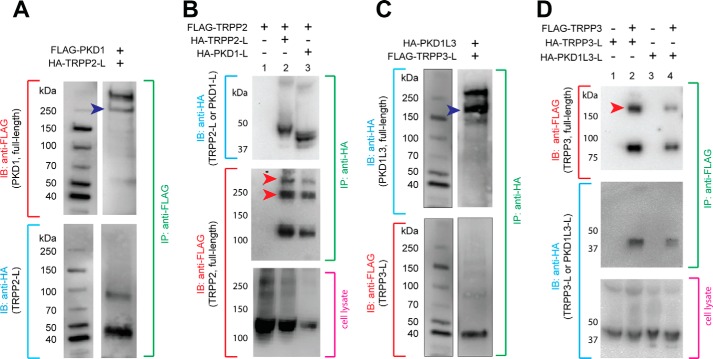
Thus far, we have demonstrated that the isolated S1-S2 loops of TRPP2 and TRPP3 interact with the isolated S6-S7 loops of their PKD partners. To further prove that the interaction happens naturally and has physiological relevance, we checked whether these loops assemble with the full-length binding partners and obtained positive results. First, when the HA-tagged TRPP2 S1-S2 loop was coexpressed with the FLAG-tagged full-length PKD1, it was co-immunoprecipitated with the latter by the anti-FLAG antibody (Fig. 4A). Second, the FLAG-tagged full-length TRPP2 was co-immunoprecipitated with the HA-tagged TRPP2 S1-S2 loop or PKD1 S6-S7 loop by the anti-HA antibody when they were coexpressed (Fig. 4B, lanes 2 and 3). Similarly, when the FLAG-tagged TRPP3 S1-S2 loop was coexpressed with HA-tagged full-length PKD1L3, it was co-immunoprecipitated with the latter by the anti-HA antibody (Fig. 4C). At the same time, the FLAG-tagged full-length TRPP3 was co-immunoprecipitated with the HA-tagged TRPP3 S1-S2 loop or PKD1L3 S6-S7 loop by the anti-FLAG antibody (Fig. 4D, lanes 2 and 4). These results confirmed that the S1-S2 loop of TRPP proteins and the S6-S7 loop of the PKD proteins are involved in the assembly of the full-length polycystin complexes and that the TRPP loops are also involved in the assembly of full-length TRPP homomeric complexes.
FIGURE 4.
Extracellular loops of TRPP and PKD associate with full-length proteins. A, co-IP of the HA-tagged TRPP2 S1-S2 loop (HA-TRPP2-L) and the full-length FLAG-tagged PKD1 protein in HEK 293T cells stably expressing FLAG-PKD1. Blue arrowhead, PKD1 N-terminal fragment, which is cleaved from the full-length protein at the G-protein-coupled receptor proteolytic site (GPS) (47). B, co-IP of the HA-tagged TRPP2 S1-S2 loop or the HA-tagged PKD1 S6-S7 loop with FLAG-tagged full-length TRPP2. Red arrowheads, oligomers of TRPP2 that occasionally show up on the SDS-polyacrylamide gel. FLAG-TRPP2 was not immunoprecipitated when loops were absent in control experiments (lane 1). C, co-IP of the FLAG-tagged TRPP3 S1-S2 loop (FLAG-TRPP3-L) and the full-length HA-tagged PKD1L3 in HEK 293T cells stably expressing HA-PKD1L3. Blue arrowhead, PKD1L3 N-terminal fragment, which is cleaved from the full-length protein (25). D, co-IP of the HA-tagged TRPP3 S1-S2 loop or the HA-tagged PKD1L3 S6-S7 loop with FLAG-tagged full-length TRPP3. Red arrowheads, the oligomers of TRPP3 that always show up on the SDS-polyacrylamide gel (25). Loops were not immunoprecipitated when FLAG-TRPP3 was absent in control experiments (lanes 1 and 3). IB, immunoblotting.
Effect of the ADPKD Pathogenic Mutations on the Assembly of the Extracellular Loops
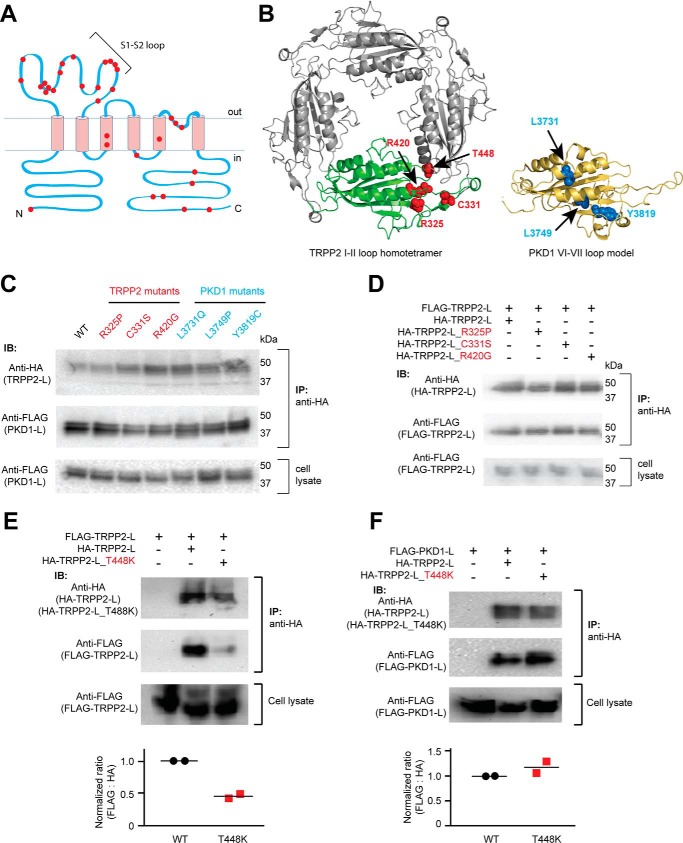
Both the S1-S2 loop of TRPP2 and the S6-S7 loop of PKD1 are mutation hot spots for ADPKD. For example, most clinically identified single-point pathogenic mutations of TRPP2 happen within the S1-S2 loop (Fig. 5A). The recently reported TRPP2 homotetrameric structures also indicated that many of these mutated amino acids are potentially important for stabilizing the structure of the S1-S2 loop or the intersubunit interaction between TRPP2 subunits (9, 10, 14). Therefore, we tested the effects of some mutations on the heteromeric assembly between the TRPP2 and PKD1 loops and the homomeric assembly between TRPP2 loops (Fig. 5B). In the first round of experimentation, three TRPP2 mutants (R325P, C331S, and R420G) and three PKD1 mutants (L3731Q, L3749P, and Y3819C) were tested. The three TRPP2 amino acids have been shown to be important for stabilizing the TRPP2 S1-S2 loop structure (9, 10, 14). The three PKD1 amino acids are also predicted to be crucial for protein structure, as seen in a structural model generated based on the TRPP2 cryo-EM structure (Fig. 5B). However, none of these TRPP2 or PKD1 mutations had a clear effect on the association between TRPP2 and PKD1 loops in the co-IP experiments (Fig. 5C). A further test revealed that the three TRPP2 mutations did not affect the homomeric TRPP2 loop assembly either (Fig. 5D). These results demonstrate that the structure of the loops and their association are quite stable, and these single-point mutations do not cause disease by directly disrupting the loop interaction. However, these mutations may alter local conformation, which leads to the pathological defect in the TRPP2-PKD1 complex. In contrast, T448K of TRPP2, a mutation occurring in the intersubunit interface (Fig. 5B), significantly weakened the homomeric TRPP2 loop assembly, as shown by the co-IP results (Fig. 5E), although it had no effect on the assembly between TRPP2 and PKD1 loops (Fig. 5F). These results indicate that the heteromeric assembly between the TRPP2 and PKD1 loops may have a different interface or interaction mechanism from that of the homomeric TRPP2 loop assembly.
FIGURE 5.
Effects of the ADPKD pathogenic mutations on the assembly of the extracellular loops. A, transmembrane topology of TRPP2 showing that most of the clinically identified single-point mutations (including substitution and deletion, indicated with red dots) are located in the S1-S2 loop. B, localization of the tested mutations in the loop structures. Left, amino acids shown as red van der Waals spheres indicate the four TRPP2 mutations mapped on the TRPP2 S1-S2 loop homotetramer viewing from the top (adapted from the cryo-EM structure of TRPP2, Protein Data Bank code 5T4D (9)). Right, amino acids in blue spheres show the three PKD1 mutations mapped on a structure model of the PKD1 S6-S7 loop made based on the TRPP2 cryo-EM structure. The model was generated on the SWISS-MODEL server (48). Structural graphics were prepared with the program PyMOL (49). C, three TRPP2 mutations in the S1-S2 loop and three PKD1 mutations in the S6-S7 loop had no effect on the assembly between TRPP2 and PKD1 loops. Co-IP was done between WT or mutant loop fragments proteins with anti-HA antibody-coated beads (same for all results in this figure). D, co-IP results indicate that the three mutations have no effect on homomeric assembly of the TRPP2 S1-S2 loop. E, pathogenic mutation T448K significantly weakened the homomeric assembly between TRPP loops. The scatter plot on the bottom shows the normalized ratios of the relative band intensity of the co-immunoprecipitated FLAG-TRPP2 loop to the indicated HA-TRPP2 loops. Data from two independent experiments and the mean (black bars) are shown. F, T448K has no effect on the assembly between TRPP2 and PKD1 loops. The scatter plot shows the normalized ratios of the relative band intensity of the co-immunoprecipitated FLAG-PKD1 loop to the indicated HA-TRPP2 loops. Data from two independent experiments and the mean (black bars) are shown. IB, immunoblotting.
Assembly of the Extracellular Loops Is Essential for the Trafficking of the Polycystin Complexes
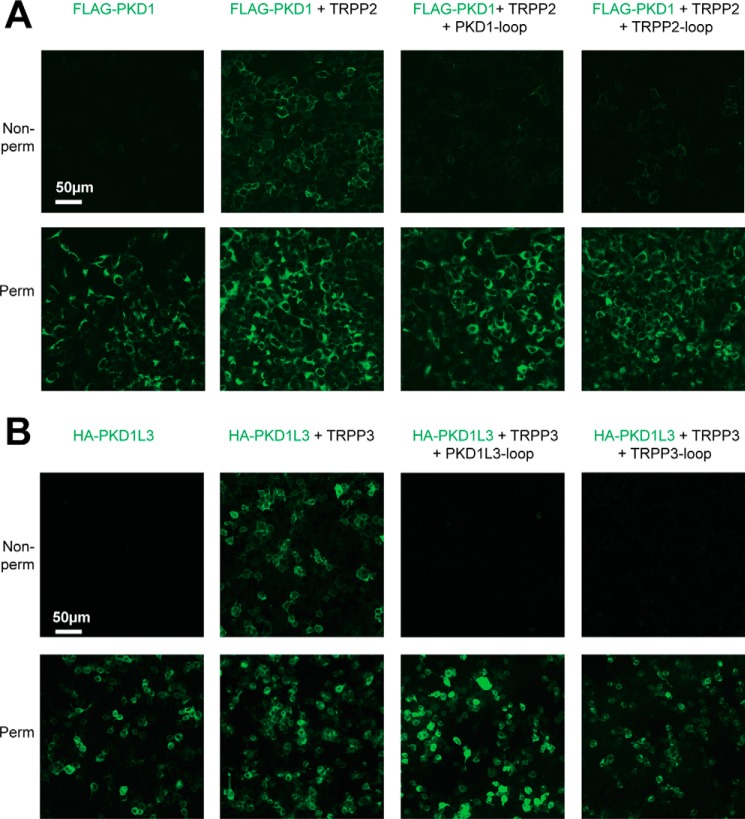
Up to now, results showed that the S1-S2 loop of TRPP proteins and the S6-S7 loop of PKD proteins associate with each other and are involved in the assembly of the polycystin complexes. If this association is critical, then when the loop fragments are coexpressed with full-length proteins, a dominant negative effect on the assembly and trafficking of the complexes is expected. Because it has been reported that the plasma membrane expression of PKD1 and PKD1L3 relies on forming complexes with TRPP2 and TRPP3, respectively (15, 21, 25, 29, 43), detecting the cell surface expression of PKD1 and PKD1L3 will allow us to monitor the assembly and trafficking of the corresponding polycystin complexes. In our experiments, FLAG-tagged full-length PKD1 was expressed on the plasma membrane only when coexpressed with full-length TRPP2 protein in HEK 293T cells (Fig. 6A, two columns on the left), consistent with previous reports (29, 43). However, coexpression of the PKD1 S6-S7 loop or TRPP2 S1-S2 loop greatly reduced the plasma membrane expression of full-length PKD1 (Fig. 6A, two columns on the right). Similar results were obtained with the TRPP3-PKD1L3 complex. HA-tagged full-length PKD1L3 expressed on the plasma membrane only when full-length TRRP3 was coexpressed in HEK 293T cells (Fig. 6B, two columns on the left), consistent with previous reports (21, 25, 44). Coexpression of the PKD1L3 S6-S7 loop or TRPP3 S1-S2 loop fragments caused the dominant negative effect on the plasma membrane trafficking of PKD1L3 (Fig. 6B, two columns on the right).
FIGURE 6.
Dominant negative effect of the external loops on the cell surface expression and TRPP2-PKD1 and TRPP3-PKD1L3 complexes. A, representative images showing that the surface trafficking of the TRPP2-PKD1 complex was blocked by coexpression of the loop protein fragments. DNA combinations (indicated above the pictures) were transfected into HEK 293T cell stably expressing FLAG-PKD1. Cells were stained with the anti-FLAG monoclonal antibody (to show PKD1) at non-permeabilized (top row, showing surface proteins) or permeabilized (bottom row, showing overall expression) conditions. B, representative images showing that the surface trafficking of the TRPP3-PKD1L3 complex was blocked by coexpression of the loop protein fragments. HEK 293T cells were transfected with the indicated DNA combinations and stained with anti-HA monoclonal antibody to show PKD1L3.
Coexpression of the Extracellular Loops Exerted a Dominant Negative Effect on the Channel Current of the Gain-of-function TRPP2 Channel and the TRPP3-PKD1L3 Complex
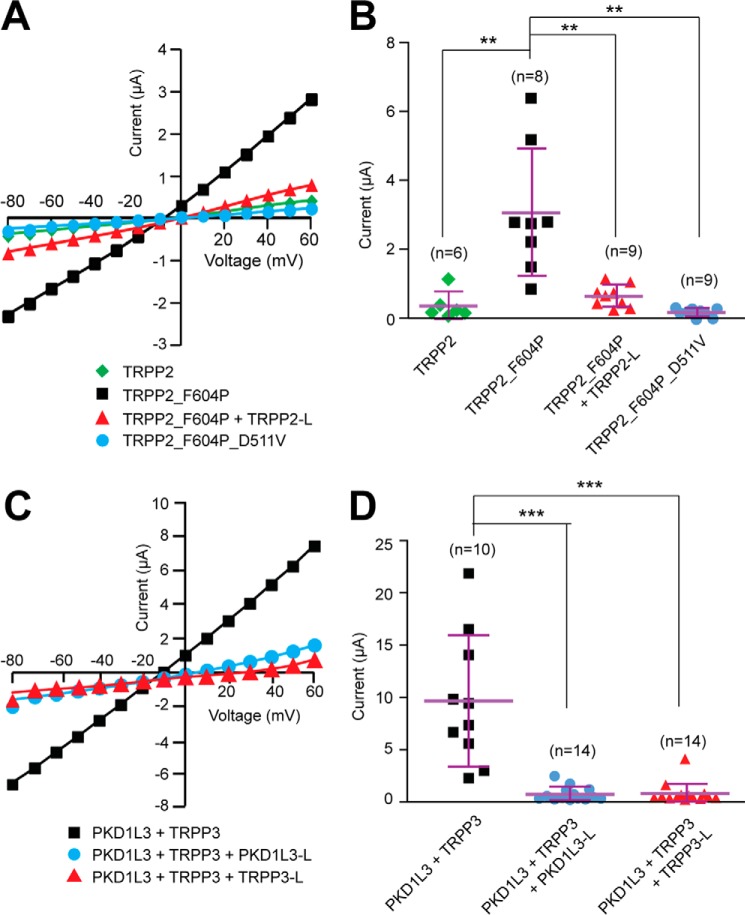
Next, we tested whether coexpression of the extracellular loop fragments causes dominant negative effects on the function of the polycystin complexes. Because the activation mechanism of the TRPP2-PKD1 complex is not yet known, we examined the effect of TRPP2 S1-S2 loop expression on the function of TRPP2_F604P, a gain-of-function mutant of TRPP2 generated in our previous study (45). Our results clearly showed that whereas TRPP2_F604P, compared with TRPP2 WT channel, gave robust current when expressed in Xenopus oocytes, coexpression with the TRPP2 S1-S2 loop abolished its current (Fig. 7, A and B). ADPKD-causing mutation D511V was introduced into TRPP2_F604P to serve as a negative control because it is known to abolish the TRPP2 current (Fig. 7, A and B) (11, 45).
FIGURE 7.
Dominant negative effect of the external loops on the channel activity of gain-of-function TRPP2_F604P channel and TRPP3-PKD1L3 complex. A, representative current-voltage curves of the currents of the indicated proteins expressed in Xenopus oocytes, indicating that the coexpression of TRPP2 S1-S2 loop inhibits the current of TRPP2_F604P. B, scatter plot showing that the TRPP2 loop exerts a dominant negative effect on the current of TRPP2_F604P. **, p < 0.01. Currents at +60 mV are shown. Mean and S.D. are shown with purple bars. C, representative current-voltage curves of acid-induced off-response currents in Xenopus oocytes expressing indicated proteins, revealing that the coexpression of either PKD1L3 S6-S7 loop or TRPP3 S1-S2 loop inhibits the current of the TRPP3-PKD1L3 complex. D, scatter plot showing that both PKD1L3 and TRPP3 loops exert a dominant negative effect on the acid-induced current of the TRPP3-PKD1L3 complex. Currents at +60 mV are shown. ***, p < 0.001. Error bars, S.D.
Previous reports have demonstrated that when the TRPP3-PKD1L3 complex is expressed in HEK 293T cells or Xenopus oocytes, it can be activated by acid through an off-response mechanism (currents appear when acid is washed out) (21, 25, 44). In the current study, when TRPP3 and PKD1L3 were coexpressed in Xenopus oocytes, robust channel currents were recorded by applying a pH 2.8 solution, followed by neutralization with a pH 7.5 solution (Fig. 7, C and D). However, when the TRPP3 S1-S2 loop or PKD1L3 S6-S7 loop was coexpressed at the same time, the acid-induced current of the TRPP3-PKD1L3 complex was greatly inhibited (Fig. 7, C and D). In both experiments of TRPP2_F604P and TRPP3-PKD1L3, the reduction of channel activity was most likely due to less surface expression of these proteins caused by coexpression of the loops, as we showed in Fig. 6. Thus, the results of the cell surface expression (Fig. 6) and channel activity (Fig. 7) clearly prove that the extracellular S1-S2 loop of TRPP and the S6-S7 loop of PKD are essential for polycystin complex assembly and trafficking.
Discussion
Previous studies have indicated the involvement of several different regions in the homomeric assembly of TRPP and heteromeric assembly between TRPP and PKD proteins. The C-terminal coiled-coil domain (26–29, 41), the N terminus (32), and a cysteine residue in the outer pore region (Cys632) (31) were all found to play roles in TRPP2 homomeric assembly. In TRPP3, a homologous C-terminal coiled-coil domain and a C1 domain, which is located upstream of the coiled-coil domain, have been shown to be essential for its homomeric assembly (25, 38). The TRPP2 coiled-coil domain is also directly involved in binding to the C-terminal coiled-coil domain of PKD1 and plays a key role in the TRPP2-PKD1 complex assembly (26, 27, 29, 30, 41). In contrast, transmembrane segments were found to play a crucial role in the assembly of the TRPP3-PKD1L3 complex (34). In this study, we discovered a new association site between the extracellular S1-S2 loops of TRPP proteins and the extracellular S6-S7 loops of PKD proteins. Although the association was only tested here in the TRPP2-PKD1 and TRPP3-PKD1L3 complexes, it may also exist in other polycystin complexes, such as TRPP2-PKD1L1 and TRPP3-PKD1L1. Thus, the assembly of the TRPP proteins and their PKD partners occurs at multiple sites, including at least the extracellular loops, the transmembrane domains, and, in some cases, the intracellular C termini. The intensive interaction between these two groups of proteins indicates a close functional coupling between them. This result is consistent with the finding that PKD1L3 is part of the pore-forming subunit of the TRPP3-PKD1L3 complex (25). It is worth noting that when the C-terminal coiled-coil interaction is abolished, the assembly between the full-length TRPP2 and PKD1 is greatly reduced, although the extracellular loops and the transmembrane domains are intact (29, 30, 41). These results indicate that the C-terminal interaction may serve as the initiating site for the assembly of the TRPP2-PKD1 complex, although our current data clearly showed the essential roles of these extracellular loops in complex assembly.
The extracellular loops studied here are unique in TRPP and PKD proteins and critical for their functions. TRP proteins in the seven subfamilies (TRPC, TRPV, TRPM, TRPN, TRPA, TRPP, and TRPML) all share sequence and structural similarities, especially in their transmembrane domains (4). However, within the whole family, only members in the TRPP and TRPML subfamilies have the large S1-S2 loop. The S1-S2 loops of TRPP proteins are homologous to the extracellular S6-S7 loops of PKD proteins. Several lines of evidence have suggested the importance of their roles in the function of TRPP and PKD proteins. First, these loops contain a highly conserved polycystin motif, which is unique to both PKD and TRPP proteins (33). Second, many ADPKD pathogenic mutations fall within these loops. In the case of TRPP2, besides the truncation mutations, there are 32 known single-point pathogenic mutations, including substitutions and deletions (ADPKD Mutation Database). Of these 32 mutations, 18 (56%) are located in the S1-S2 loop (Fig. 5A). Considering that the S1-S2 loop is only 23% of the total length of TRPP2, it is clearly a hot spot for pathogenic mutations, suggesting the essential roles that the S1-S2 loop plays in TRPP2 function. Third, in the cryo-EM structure of the TRPP2 homotetramer, the S1-S2 loops from four individual subunits form a tightly bound donut-like structure (9, 10, 14). The extensive interaction formed between neighboring loops indicates a significant role of this loop in channel assembly, consistent with our results of the homomeric interaction between TRPP2 S1-S2 loops (Fig. 3). In the structures, it can also be seen that the S1-S2 loops form a wide opening right on the extracellular side of the channel pore, suggesting possible involvement in ion permeability and channel regulation by extracellular stimuli (9, 10, 14).
The effects of the pathogenic mutations indicate a versatile function of these loops. In a study conducted with the gain-of-function TRPP2, the mutation R325P was found to have no effect on the channel current, whereas two other mutations in the S1-S2 loop, W414G and R420G, abolished channel activity (45). In another report, W414G was shown to affect the trafficking of TRPP2 to cilia when expressed in LLC-PK1 cells (46). Here we showed that R325P, C331S, and R420G did not affect homomeric TRPP2 loop assembly and the heteromeric TRPP2-PKD1 loop assembly (Fig. 5, C and D). However, T448K, another mutation that was predicted to affect TRPP2 loop assembly based on the structural data (9), greatly weakened TRPP2 homomeric loop assembly but did not affect the association between TRPP2 and PKD1 loops (Fig. 5, E and F). Thus, the TRPP2 S1-S2 loop is involved in the assembly, trafficking, and functional regulation of the TRPP2 channel and the TRPP2-PKD1 complex. Also, the result of T448K suggests that the interaction interface between the TRPP2 and PKD1 loops is different from that of the homomerically assembled TRPP2 loops.
The homomeric TRPP2 channel is a tetramer, and the S1-S2 loops from four subunits tightly assemble with each other (9, 10). The TRPP2-PKD1 and TRPP3-PKD1L3 complexes are also tetramers that contain three TRPP subunits and one PKD subunit (25, 29). Moreover, the coiled-coil domains of TRPP2 or TRPP3 form trimers and determine the 3:1 subunit stoichiometry of the TRPP2-PKD1 and TRPP3-PKD1L3 complexes (25, 29). Now we show that the extracellular loops assemble in polycystin complexes. It will be interesting to investigate whether the extracellular loops of TRPP and PKD proteins also assemble in a 3:1 stoichiometry as the C termini and full-length proteins do in the polycystin complexes.
Experimental Procedures
DNA Constructs
cDNAs for human TRPP2, TRPP3, PKD1, and mouse PKD1L3 were used in this study. For expressing isolated extracellular loops, the cDNAs that code the protein fragment Asn245–Thr468 of TRPP2, Ser123–Asp349 of TRPP3, Ala3683–Leu3896 of PKD1, and Ala1678–Ala1895 of PKD1L3 were cloned into pDisplay vector (Life Technologies, Inc.). The final constructs expressed the loop fragments with a fused murine Ig κ-chain leader sequence followed by an HA tag at the N terminus and a fused PDGFR transmembrane domain at the C terminus. The same loop fragments and EGFP were also cloned into a modified pDisplay vector with HA tag replaced by FLAG tag to generate N-terminal FLAG-tagged proteins. Full-length HA-tagged TRPP2 and FLAG-tagged TRPP3 were cloned into pCDNA3.1(−) vector. FLAG-tagged PKD1 and HA-tagged PKD1L3 were cloned into a pIRESpuro2 vector for generating stable cell lines. Their final constructs have the Ig κ-chain leader sequences to replace the original signal peptide. All constructs were made with PCR and verified by DNA sequencing. For RNA synthesis via in vitro transcription, cDNA was cloned into a modified pGEMHE2 vector.
Co-IP
DNAs were transfected into HEK 293T cells with LipoD293 transfection reagent (SignaGen Laboratories). 36-40 h after transfection, cells were harvested and lysed at 4 °C for 1 h with a lysis buffer containing 1% n-dodecyl-β-maltoside, 1 mm EDTA, and protease inhibitor (Sigma) in PBS. The cell lysate was centrifuged at 13,000 rpm for 30 min, and the supernatant was then incubated with either EZview anti-FLAG or anti-HA affinity beads (Sigma) overnight at 4 °C. The beads were collected by centrifuge at 6,000 rpm for 30 s and washed three times by gently shaking in a wash solution containing 0.1% n-dodecyl-β-maltoside, 1 m NaCl, 0.5% Trion X-100 in PBS for 5 min each time. The bound proteins were eluted with 0.1 m glycine-HCl, pH 2.6, and neutralized with 50 mm Tris-HCl, pH 7.5. Samples were analyzed by SDS-PAGE followed by Western blotting. Stable cell lines expressing FLAG-PKD1 or HA-PKD1L3 were generated with a protocol similar to one described previously (29). All co-IP experiments have been confirmed by at least two independent experiments. When cell lysates were included in Western blotting as a control, the volume of cell lysate loaded on the gel was ∼⅜ of the volume that was used in obtaining the corresponding co-IP bands.
SDS-PAGE and Western Blotting
Co-IP samples were run on 4–12% SDS-polyacrylamide gels (Life Technologies) and blotted with mouse monoclonal anti-FLAG (Sigma) or anti-HA (Covance) primary antibodies and HRP-conjugated goat anti-mouse secondary antibody. Western blotting images were visualized with a Molecular Imager Chemi Doc XRS+ imaging system (Bio-Rad). The relative intensity of Western blotting bands was measured with ImageJ (National Institutes of Health) and analyzed with Student's t test.
Immunofluorescence
36 h after the HEK 293T cells were transfected with LipoD293 transfection reagent (SignaGen Laboratories), cells were washed twice with PBS solution and fixed with 4% fresh paraformaldehyde in PBS for 15 min. After three 5-min washes with PBS solution, cells were permeabilized with 0.25% Triton X-100 in PBS for 20 min followed by another three 5-min washes (Triton X-100 was omitted when non-permeabilized cells were needed). Cells were then blocked with 2% goat serum in PBS for 1 h and incubated with the primary antibody in the same blocking solution at room temperature for 1 h. After three 10-min washes with PBST (0.1% Tween 20 in PBS), the cells were incubated with fluorescence-conjugated secondary antibody and 2% goat serum in PBS for 1 h at room temperature. After another three 10-min washes with PBS, cells were mounted on slides and imaged with a Zeiss LSM 700 confocal microscope. Where only one antigen was detected, mouse monoclonal anti-FLAG (Sigma) or monoclonal anti-HA (Covance) primary antibody and Alexa Fluor 488 goat anti-mouse secondary antibody were used. For co-localization analysis, mouse anti-FLAG (Sigma) and rabbit anti-HA (Santa Cruz Biotechnology, Inc.) primary antibodies and Cy5-conjugated goat anti-mouse (Life Technologies), and Alexa Fluor 594-conjugated goat anti-rabbit secondary antibodies were used.
Electrophysiology
DNA constructs in the pGEMHE2 vector were linearized, and RNA was synthesized with T7 RNA polymerase. 50 ng of total RNA was injected into every Xenopus oocytes, and the oocytes were then incubated at 18 °C for 3–5 days before recording. Channel currents were recorded with the two-electrode voltage clamp method. For recording TRPP2-F604P currents, bath solution containing 100 mm NaCl, 2 mm HEPES, pH 7.5, was used. Oocytes were clamped at −60 mV, and 50-ms voltage steps from −80 to +60 mV were applied in 10-mV increments to measure current-voltage relationships. Recording of the acid-induced off-response of the TRPP3-PKD1L3 complex was described in detail previously (25). Standard bath solution used contained 100 mm NaCl, 0.5 mm MgCl2, 2 mm HEPES, pH 7.5. The acid solution was generated by adding citric acid into the standard bath solution until pH reached 2.8. During the recording, oocytes were clamped at −60 mV in the standard bath solution, and voltage ramps from −80 to +60 mV were applied to monitor the current. The acid solution was applied to oocytes for 12 s, followed by washing with the standard bath solution until the current reached its peak amplitude. Subsequently, the 50-ms voltage steps from −80 to +60 mV were applied in 10-mV increments to measure current-voltage relationships. All recording results were been repeated with at least two batches of oocytes.
Author Contributions
Yong Yu conceived and coordinated the study and wrote the paper. Z. S. N. and B. L. conducted the cloning, co-IP, immunofluorescence, and recording experiments and analyzed the results. V. V., H. M., Yang Yu, C. N., Z. W., and P. K. helped with experiments. All authors analyzed the results and approved the final version of the manuscript.
Acknowledgments
We thank members of the Yu laboratory for commenting on the manuscript.
This work was supported by National Institutes of Health Grant DK102092 (to Yong Yu). The authors declare that they have no conflicts of interest with the contents of this article. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
- ADPKD
- autosomal dominant polycystic kidney disease
- TRPP
- transient receptor potential polycystin
- PKD
- polycystic kidney disease
- IP
- immunoprecipitation
- EGFP
- enhanced GFP.
References
- 1. Harris P. C., and Torres V. E. (2009) Polycystic kidney disease. Annu. Rev. Med. 60, 321–337 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Semmo M., Köttgen M., and Hofherr A. (2014) The TRPP subfamily and polycystin-1 proteins. Handb. Exp. Pharmacol. 222, 675–711 [DOI] [PubMed] [Google Scholar]
- 3. Wu G., and Somlo S. (2000) Molecular genetics and mechanism of autosomal dominant polycystic kidney disease. Mol. Genet. Metabol. 69, 1–15 [DOI] [PubMed] [Google Scholar]
- 4. Ramsey I. S., Delling M., and Clapham D. E. (2006) An introduction to TRP channels. Annu. Rev. Physiol. 68, 619–647 [DOI] [PubMed] [Google Scholar]
- 5. Mochizuki T., Wu G., Hayashi T., Xenophontos S. L., Veldhuisen B., Saris J. J., Reynolds D. M., Cai Y., Gabow P. A., Pierides A., Kimberling W. J., Breuning M. H., Deltas C. C., Peters D. J., and Somlo S. (1996) PKD2, a gene for polycystic kidney disease that encodes an integral membrane protein. Science 272, 1339–1342 [DOI] [PubMed] [Google Scholar]
- 6. Zhou J. (2009) Polycystins and primary cilia: primers for cell cycle progression. Annu. Rev. Physiol. 71, 83–113 [DOI] [PubMed] [Google Scholar]
- 7. Hughes J., Ward C. J., Peral B., Aspinwall R., Clark K., San Millán J. L., Gamble V., and Harris P. C. (1995) The polycystic kidney disease 1 (PKD1) gene encodes a novel protein with multiple cell recognition domains. Nat. Genet. 10, 151–160 [DOI] [PubMed] [Google Scholar]
- 8. International Polycystic Kidney Disease Consortium (1995) Polycystic kidney disease: the complete structure of the PKD1 gene and its protein. Cell 81, 289–298 [DOI] [PubMed] [Google Scholar]
- 9. Shen P. S., Yang X., DeCaen P. G., Liu X., Bulkley D., Clapham D. E., and Cao E. (2016) The Structure of the polycystic kidney disease channel PKD2 in lipid nanodiscs. Cell 167, 763–773.e11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Grieben M., Pike A. C., Shintre C. A., Venturi E., El-Ajouz S., Tessitore A., Shrestha L., Mukhopadhyay S., Mahajan P., Chalk R., Burgess-Brown N. A., Sitsapesan R., Huiskonen J. T., and Carpenter E. P. (2017) Structure of the polycystic kidney disease TRP channel Polycystin-2 (PC2). Nat. Struct. Mol. Biol. 24, 114–122 [DOI] [PubMed] [Google Scholar]
- 11. Koulen P., Cai Y., Geng L., Maeda Y., Nishimura S., Witzgall R., Ehrlich B. E., and Somlo S. (2002) Polycystin-2 is an intracellular calcium release channel. Nat. Cell Biol. 4, 191–197 [DOI] [PubMed] [Google Scholar]
- 12. Vassilev P. M., Guo L., Chen X. Z., Segal Y., Peng J. B., Basora N., Babakhanlou H., Cruger G., Kanazirska M., Ye Cp., Brown E. M., Hediger M. A., and Zhou J. (2001) Polycystin-2 is a novel cation channel implicated in defective intracellular Ca2+ homeostasis in polycystic kidney disease. Biochem. Biophys. Res. Commun. 282, 341–350 [DOI] [PubMed] [Google Scholar]
- 13. Chen X. Z., Vassilev P. M., Basora N., Peng J. B., Nomura H., Segal Y., Brown E. M., Reeders S. T., Hediger M. A., and Zhou J. (1999) Polycystin-L is a calcium-regulated cation channel permeable to calcium ions. Nature 401, 383–386 [DOI] [PubMed] [Google Scholar]
- 14. Wilkes M., Madej M. G., Kreuter L., Rhinow D., Heinz V., De Sanctis S., Ruppel S., Richter R. M., Joos F., Grieben M., Pike A. C., Huiskonen J. T., Carpenter E. P., Kühlbrandt W., Witzgall R., and Ziegler C. (2017) Molecular insights into lipid-assisted Ca2+ regulation of the TRP channel polycystin-2. Nat. Struct. Mol. Biol. 24, 123–130 [DOI] [PubMed] [Google Scholar]
- 15. Hanaoka K., Qian F., Boletta A., Bhunia A. K., Piontek K., Tsiokas L., Sukhatme V. P., Guggino W. B., and Germino G. G. (2000) Co-assembly of polycystin-1 and -2 produces unique cation-permeable currents. Nature 408, 990–994 [DOI] [PubMed] [Google Scholar]
- 16. Nauli S. M., Alenghat F. J., Luo Y., Williams E., Vassilev P., Li X., Elia A. E., Lu W., Brown E. M., Quinn S. J., Ingber D. E., and Zhou J. (2003) Polycystins 1 and 2 mediate mechanosensation in the primary cilium of kidney cells. Nat. Genet. 33, 129–137 [DOI] [PubMed] [Google Scholar]
- 17. Kamura K., Kobayashi D., Uehara Y., Koshida S., Iijima N., Kudo A., Yokoyama T., and Takeda H. (2011) Pkd1l1 complexes with Pkd2 on motile cilia and functions to establish the left-right axis. Development 138, 1121–1129 [DOI] [PubMed] [Google Scholar]
- 18. Field S., Riley K. L., Grimes D. T., Hilton H., Simon M., Powles-Glover N., Siggers P., Bogani D., Greenfield A., and Norris D. P. (2011) Pkd1l1 establishes left-right asymmetry and physically interacts with Pkd2. Development 138, 1131–1142 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Horio N., Yoshida R., Yasumatsu K., Yanagawa Y., Ishimaru Y., Matsunami H., and Ninomiya Y. (2011) Sour taste responses in mice lacking PKD channels. PLoS One 6, e20007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. LopezJimenez N. D., Cavenagh M. M., Sainz E., Cruz-Ithier M. A., Battey J. F., and Sullivan S. L. (2006) Two members of the TRPP family of ion channels, Pkd1l3 and Pkd2l1, are co-expressed in a subset of taste receptor cells. J. Neurochem. 98, 68–77 [DOI] [PubMed] [Google Scholar]
- 21. Ishimaru Y., Inada H., Kubota M., Zhuang H., Tominaga M., and Matsunami H. (2006) Transient receptor potential family members PKD1L3 and PKD2L1 form a candidate sour taste receptor. Proc. Natl. Acad. Sci. U.S.A. 103, 12569–12574 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Huang A. L., Chen X., Hoon M. A., Chandrashekar J., Guo W., Tränkner D., Ryba N. J., and Zuker C. S. (2006) The cells and logic for mammalian sour taste detection. Nature 442, 934–938 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Nelson T. M., Lopezjimenez N. D., Tessarollo L., Inoue M., Bachmanov A. A., and Sullivan S. L. (2010) Taste function in mice with a targeted mutation of the pkd1l3 gene. Chem. Senses 35, 565–577 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. DeCaen P. G., Delling M., Vien T. N., and Clapham D. E. (2013) Direct recording and molecular identification of the calcium channel of primary cilia. Nature 504, 315–318 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Yu Y., Ulbrich M. H., Li M. H., Dobbins S., Zhang W. K., Tong L., Isacoff E. Y., and Yang J. (2012) Molecular mechanism of the assembly of an acid-sensing receptor ion channel complex. Nat. Commun. 3, 1252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Tsiokas L., Kim E., Arnould T., Sukhatme V. P., and Walz G. (1997) Homo- and heterodimeric interactions between the gene products of PKD1 and PKD2. Proc. Natl. Acad. Sci. U.S.A. 94, 6965–6970 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Qian F., Germino F. J., Cai Y., Zhang X., Somlo S., and Germino G. G. (1997) PKD1 interacts with PKD2 through a probable coiled-coil domain. Nat. Genet. 16, 179–183 [DOI] [PubMed] [Google Scholar]
- 28. Molland K. L., Narayanan A., Burgner J. W., and Yernool D. A. (2010) Identification of the structural motif responsible for trimeric assembly of the C-terminal regulatory domains of polycystin channels PKD2L1 and PKD2. Biochem. J. 429, 171–183 [DOI] [PubMed] [Google Scholar]
- 29. Yu Y., Ulbrich M. H., Li M. H., Buraei Z., Chen X. Z., Ong A. C., Tong L., Isacoff E. Y., and Yang J. (2009) Structural and molecular basis of the assembly of the TRPP2/PKD1 complex. Proc. Natl. Acad. Sci. U.S.A. 106, 11558–11563 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Zhu J., Yu Y., Ulbrich M. H., Li M. H., Isacoff E. Y., Honig B., and Yang J. (2011) Structural model of the TRPP2/PKD1 C-terminal coiled-coil complex produced by a combined computational and experimental approach. Proc. Natl. Acad. Sci. U.S.A. 108, 10133–10138 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Feng S., Rodat-Despoix L., Delmas P., and Ong A. C. (2011) A single amino acid residue constitutes the third dimerization domain essential for the assembly and function of the tetrameric polycystin-2 (TRPP2) channel. J. Biol. Chem. 286, 18994–19000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Feng S., Okenka G. M., Bai C. X., Streets A. J., Newby L. J., DeChant B. T., Tsiokas L., Obara T., and Ong A. C. (2008) Identification and functional characterization of an N-terminal oligomerization domain for polycystin-2. J. Biol. Chem. 283, 28471–28479 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Li A., Tian X., Sung S. W., and Somlo S. (2003) Identification of two novel polycystic kidney disease-1-like genes in human and mouse genomes. Genomics 81, 596–608 [DOI] [PubMed] [Google Scholar]
- 34. Ishimaru Y., Katano Y., Yamamoto K., Akiba M., Misaka T., Roberts R. W., Asakura T., Matsunami H., and Abe K. (2010) Interaction between PKD1L3 and PKD2L1 through their transmembrane domains is required for localization of PKD2L1 at taste pores in taste cells of circumvallate and foliate papillae. FASEB J. 24, 4058–4067 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Veldhuisen B., Spruit L., Dauwerse H. G., Breuning M. H., and Peters D. J. (1999) Genes homologous to the autosomal dominant polycystic kidney disease genes (PKD1 and PKD2). Eur. J. Hum. Genet. 7, 860–872 [DOI] [PubMed] [Google Scholar]
- 36. Nomura H., Turco A. E., Pei Y., Kalaydjieva L., Schiavello T., Weremowicz S., Ji W., Morton C. C., Meisler M., Reeders S. T., and Zhou J. (1998) Identification of PKDL, a novel polycystic kidney disease 2-like gene whose murine homologue is deleted in mice with kidney and retinal defects. J. Biol. Chem. 273, 25967–25973 [DOI] [PubMed] [Google Scholar]
- 37. Lam C., Pavel M. A., Kashyap P., Salehi-Najafabadi Z., Valentino V., and Yu Y. (2014) Detection of CXCR2 cytokine receptor surface expression using immunofluorescence. Methods Mol. Biol. 1172, 193–200 [DOI] [PubMed] [Google Scholar]
- 38. Zheng W., Hussein S., Yang J., Huang J., Zhang F., Hernandez-Anzaldo S., Fernandez-Patron C., Cao Y., Zeng H., Tang J., and Chen X. Z. (2015) A novel PKD2L1 C-terminal domain critical for trimerization and channel function. Sci. Rep. 5, 9460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Yang Y., Keeler C., Kuo I. Y., Lolis E. J., Ehrlich B. E., and Hodsdon M. E. (2015) Oligomerization of the polycystin-2 C-terminal tail and effects on its Ca2+-binding properties. J. Biol. Chem. 290, 10544–10554 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Molland K. L., Paul L. N., and Yernool D. A. (2012) Crystal structure and characterization of coiled-coil domain of the transient receptor potential channel PKD2L1. Biochim. Biophys. Acta 1824, 413–421 [DOI] [PubMed] [Google Scholar]
- 41. Giamarchi A., Feng S., Rodat-Despoix L., Xu Y., Bubenshchikova E., Newby L. J., Hao J., Gaudioso C., Crest M., Lupas A. N., Honoré E., Williamson M. P., Obara T., Ong A. C., and Delmas P. (2010) A polycystin-2 (TRPP2) dimerization domain essential for the function of heteromeric polycystin complexes. EMBO J. 29, 1176–1191 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Oatley P., Stewart A. P., Sandford R., and Edwardson J. M. (2012) Atomic force microscopy imaging reveals the domain structure of polycystin-1. Biochemistry 51, 2879–2888 [DOI] [PubMed] [Google Scholar]
- 43. Chapin H. C., Rajendran V., and Caplan M. J. (2010) Polycystin-1 surface localization is stimulated by polycystin-2 and cleavage at the G protein-coupled receptor proteolytic site. Mol. Biol. Cell 21, 4338–4348 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Inada H., Kawabata F., Ishimaru Y., Fushiki T., Matsunami H., and Tominaga M. (2008) Off-response property of an acid-activated cation channel complex PKD1L3-PKD2L1. EMBO Rep. 9, 690–697 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Arif Pavel M., Lv C., Ng C., Yang L., Kashyap P., Lam C., Valentino V., Fung H. Y., Campbell T., Møller S. G., Zenisek D., Holtzman N. G., and Yu Y. (2016) Function and regulation of TRPP2 ion channel revealed by a gain-of-function mutant. Proc. Natl. Acad. Sci. U.S.A. 113, E2363–2372 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Cai Y., Fedeles S. V., Dong K., Anyatonwu G., Onoe T., Mitobe M., Gao J. D., Okuhara D., Tian X., Gallagher A. R., Tang Z., Xie X., Lalioti M. D., Lee A. H., Ehrlich B. E., and Somlo S. (2014) Altered trafficking and stability of polycystins underlie polycystic kidney disease. J. Clin. Invest. 124, 5129–5144 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Qian F., Boletta A., Bhunia A. K., Xu H., Liu L., Ahrabi A. K., Watnick T. J., Zhou F., and Germino G. G. (2002) Cleavage of polycystin-1 requires the receptor for egg jelly domain and is disrupted by human autosomal-dominant polycystic kidney disease 1-associated mutations. Proc. Natl. Acad. Sci. U.S.A. 99, 16981–16986 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Arnold K., Bordoli L., Kopp J., and Schwede T. (2006) The SWISS-MODEL workspace: a web-based environment for protein structure homology modelling. Bioinformatics 22, 195–201 [DOI] [PubMed] [Google Scholar]
- 49. DeLano W. L. (2010) The PyMOL Molecular Graphics System, version 1.3, Schrödinger, LLC, New York [Google Scholar]