Abstract
RecQ helicases are essential in the maintenance of genome stability. Five paralogues (RecQ1, Bloom, Werner, RecQ4, and RecQ5) are found in human cells, with distinct but overlapping roles. Mutations in human RecQ4 give rise to three distinct genetic disorders (Rothmund-Thomson, RAPADILINO, and Baller-Gerold syndromes), characterized by genetic instability, growth deficiency, and predisposition to cancer. Previous studies suggested that RecQ4 was unique because it did not seem to contain a RecQ C-terminal region (RQC) found in the other RecQ paralogues; such a region consists of a zinc domain and a winged helix domain and plays an important role in enzyme activity. However, our recent bioinformatic analysis identified in RecQ4 a putative RQC. To experimentally confirm this hypothesis, we report the purification and characterization of the catalytic core of human RecQ4. Inductively coupled plasma-atomic emission spectrometry detected the unusual presence of two zinc clusters within the zinc domain, consistent with the bioinformatic prediction. Analysis of site-directed mutants, targeting key RQC residues (putative zinc ligands and the aromatic residue predicted to be at the tip of the winged helix β-hairpin), showed a decrease in DNA binding, unwinding, and annealing, as expected for a functional RQC domain. Low resolution structural information obtained by small angle X-ray scattering data suggests that RecQ4 interacts with DNA in a manner similar to RecQ1, whereas the winged helix domain may assume alternative conformations, as seen in the bacterial enzymes. These combined results experimentally confirm the presence of a functional RQC domain in human RecQ4.
Keywords: DNA helicase, DNA repair, mutagenesis in vitro, protein purification, protein-DNA interaction, recombinant protein expression, small angle X-ray scattering (SAXS), structural model
Introduction
RecQ helicases belong to a ubiquitous family of DNA unwinding enzymes that are essential to maintain the genome stability by acting at the interface of DNA replication, recombination, and repair. RecQ helicases have been proposed to be involved in various cellular activities including stabilization and repair of damaged DNA replication fork, telomere maintenance, base excision repair, homologous recombination, and DNA damage checkpoint signaling (1–3). Whereas all cellular organisms possess at least one RecQ helicase, humans have five different paralogues called RecQ1, BLM, WRN, RecQ4, and RecQ5. They share a similarity in their helicase domain but also have some additional domains and unique features. Genetic mutations in three paralogues (BLM, WRN, and RecQ4) result in rare genetic disorders, namely Bloom, Werner, and Rothmund-Thomson, RAPADILINO, and Baller-Gerold syndromes. In addition to the common characteristics of genetic instability, short stature, and predisposition to cancer, the disorders have specific clinical symptoms unique to each mutation, suggesting that the five RecQ helicases have distinct, as well as partially overlapping roles within the cell (4–6). Although RecQ4-deficient Rothmund-Thomson syndrome patients have an elevated risk for osteosarcoma, an increased level of expression of RecQ4 has been reported in human sporadic osteosarcoma, prostate tumor, and breast tumor samples (7–10).
The human RecQ4 helicase encodes 1208 amino acid residues. Despite the importance of human RecQ4 in DNA replication and repair and its association to three autosomal recessive disorders and cancer, some of the structural and biochemical aspects of the protein are still hazy. In addition to the catalytic core, RecQ4 comprises an N-terminal domain that has some homology with the yeast DNA replication factor Sld2 and includes a zinc knuckle (11). A puzzling feature of RecQ4 was the lack of a RQC2 domain, which is an essential part of the catalytic core of all the RecQ helicases that have been characterized. This appeared consistent with initial reports (12, 13), suggesting that RecQ4 had no helicase activity. However, several laboratories (14–16) have clearly demonstrated that the recombinant protein is an active DNA helicase, and the presence of a RQC domain has been suggested by a more detailed bioinformatic analysis (17). The putative RQC domain comprises a zinc-binding domain and a winged helix (WH) domain (see Fig. 1, A and B). In most of the RecQ helicases, an aromatic residue present at the tip of a β-hairpin in WH domain has been reported to be involved in DNA binding and unwinding (18–22).
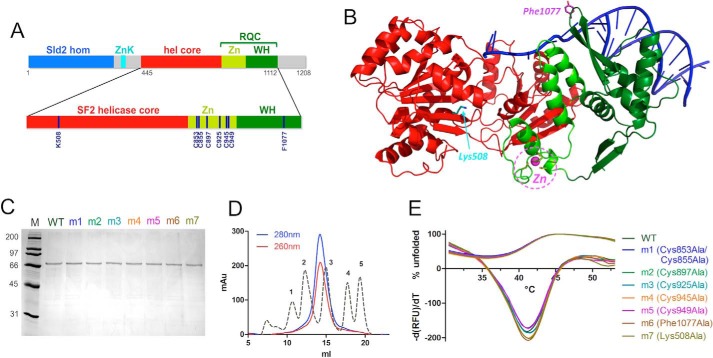
FIGURE 1.
Expression and purification of human RecQ4. A, domain organization of human RecQ4 showing the Sld2 homologous region at the N terminus (blue) followed by a zinc knuckle (cyan), a helicase core (red), and RQC domain (consisting of a zinc-binding domain (bright green) and a winged helix domain (dark green)). Enlarged at the bottom of the panel is the catalytic core of RecQ4 (helicase and RQC domain) with the positions of the residues mutated in this work. B, the crystal structure of RecQ1 (PDB code 2WWY) with the same color code; critical residues are highlighted. C, SDS-PAGE gel of purified proteins. M, molecular markers; m1, C853A/C855A; m2, C897A; m3, C925A; m4, Cs945A; m5, C949A; m6, F1077A; m7, K508A. D, size exclusion chromatography indicates that the recombinant protein is a monomer. The calibration peaks correspond to protein markers: peak 1, ferritin, 440 kDa; peak 2, aldolase, 158 kDa; peak 3, ovalbumin, 44 kDa; peak 4, ribonuclease A, 13.7 kDa; peak 5, aprotinin, 6.5 kDa. E, heat denaturation profiles of RecQ4 variants. The curves depicting the percentage of unfolded proteins (positive y quadrant, 0–100) and the −(dRFU)/dT curves are plotted together (mostly in the negative y quadrant). The experiment suggests that all the mutants are folded and stable.
Here we report the biochemical and structural characterization of the catalytic core of human RecQ4 helicase. A number of site-directed mutants, mostly targeting conserved residues within the putative RQC domain, were purified and characterized. Their effect on the catalytic activities of the protein confirms the bioinformatic analysis and the key role of the RQC domain in DNA binding, annealing, and unwinding. Numerous missense/nonsense mutations of Rothmund-Thomson, Baller-Gerold, and RAPADILINO syndromes map onto this region. Our biochemical and structural analysis of the RQC domain provides an essential framework to rationalize the patient mutations. Low resolution structural information suggests that RecQ4 interacts with DNA in a manner similar to RecQ1, whereas the WH domain may assume alternative conformations.
Results and Discussion
Expression and Purification of Recombinant hRecQ4-HelRQC
We produced in bacterial cells a deletion mutant of hRecQ4 corresponding to the helicase core plus putative RQC domain (hRecQ4-HelRQC; Fig. 1A) as a fusion protein with a N-terminal His-SUMO tag. The tag was cleaved with the SUMO protease, and the protein was purified to homogeneity (Fig. 1C). Size exclusion chromatography indicates that the protein is a monomer in solution (Fig. 1D).
The canonical RQC domain found in all or most of the RecQ proteins contains a zinc domain; a bioinformatic analysis carried out on RecQ4 (17) suggested the presence of up to eight putative zinc ligands (seven Cys and one His) and therefore the possibility of two zinc binding sites (Fig. 1A). We have therefore used inductively coupled plasma-atomic emission spectrometry (ICP-AES) to measure the amount of zinc in the purified protein; because the cysteine residue can also bind iron and a subset of helicases contain iron-sulfur clusters, an analysis of the iron content was also carried out. The analysis showed the presence of two zinc atoms for each molecule of protein, whereas no iron was detected, confirming that RecQ4 proteins have an anomalous RQC zinc domain with two rather than one Zn2+ ion (Table 1).
TABLE 1.
The amounts of zinc present in the RecQ4 variants
| Protein | Protein concentration | Zinc concentration | Protein:zinc (molar ratio) |
|---|---|---|---|
| μm | μm | ||
| WT | 2.5 | 4.9 | 1:2 |
| m1 (C853A/C855A) | 0.3 | 0.34 | 1:1 |
| m2 (C897A) | 2.0 | 2.4 | 1:1 |
| m3 (C925A) | 1.5 | 1.6 | 1:1 |
| m4 (C945A) | 1.5 | 1.7 | 1:1 |
| m5 (C949A) | 2.0 | 2.1 | 1:1 |
| m6 (F1077A) | 1.2 | 2.5 | 1:2 |
| m7 (K508A) | 1.5 | 2.9 | 1:2 |
Biochemical Characterization of the hRecQ4-HelRQC
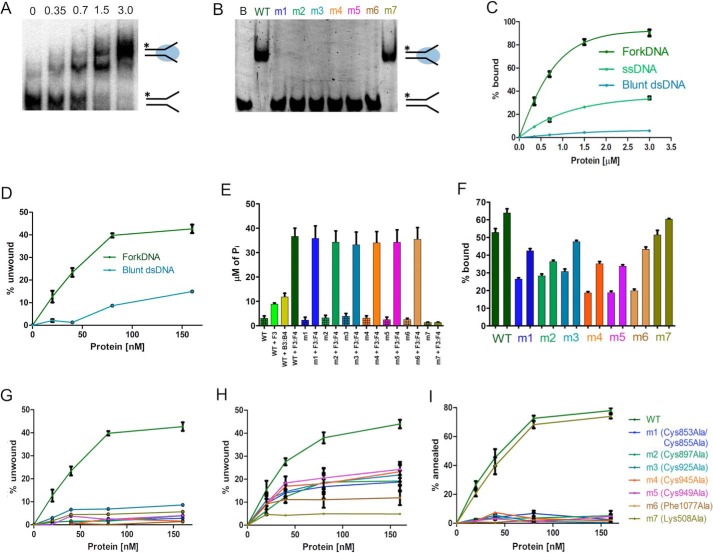
The recombinant protein was characterized in terms of DNA binding, ATP hydrolysis, strand annealing, and helicase activity. To test for DNA binding, we used EMSAs; a fluorescent DNA probe (ssDNA, blunt dsDNA, and fork DNA) was incubated with increasing protein concentrations (0–3 μm), and the mixture was analyzed by polyacrylamide gel electrophoresis to monitor the formation of the protein-DNA complex (Fig. 2A). The protein shows the highest affinity toward a fork DNA probe, compared with ssDNA and blunt dsDNA (Fig. 2C).
FIGURE 2.
Biochemical activities of the purified recombinant WT and mutant proteins. A, EMSA gel showing the binding activity of increasing amounts (0–3 μm) of WT RecQ4 with 10 nm forked DNA (F1:F4). B, EMSA gel showing the DNA binding activity of the mutants at the highest concentration (3 μm), with 10 nm DNA substrate. Lane B, fork DNA (F1:F4); m1, C853A/C855A; m2, C897A; m3, C925A; m4, C945A; m5, C949A; m6, F1077A; m7, K508A. All the RQC mutants have no affinity in limiting substrate concentration. C, comparison of DNA binding activity of the WT protein on three substrates: blunt dsDNA (B1:B4), ssDNA (F1), and fork DNA (F1:F4) (10 nm). The protein has a preference for forked substrates. The one-site total function of GraphPad Prism was used to fit the data points. D, comparison of helicase activity of the WT protein on two substrates (10 nm): blunt dsDNA (B1:B2) and fork DNA (F1:F2). Forked substrates are preferred, although there is some activity on blunt substrates at higher protein concentrations. E, ATPase assay of WT and mutant proteins in the presence of 500 nm nucleic acid. ATP hydrolysis by WT protein is highly stimulated by fork DNA (F3:F4) compared with other substrates (ssDNA F3 and blunt dsDNA B3:B4). Only the Walker A mutant affects the hydrolysis. F, DNA binding activity of 0.5 and 1 μm of proteins (WT and mutants) at high fork DNA (F3:F4) concentration (100 nm). The bar graph shows the percentage of DNA bound by the proteins at respective concentrations. At this DNA concentration, the RQC mutants show a partial binding activity. G, comparison of helicase activity of mutants and WT at 10 nm DNA substrate (F1:F2). All the mutations abolish the DNA helicase activity. H, comparison of helicase activity of mutants and WT at 100 nm DNA substrate (F1:F2). Partial activity can be observed for the RQC mutants, consistent with the impairment in DNA binding observed in F, suggesting that the effect of the mutants is due to impairment in DNA binding. I, comparison of annealing activity of WT and mutants at 10 nm DNA. All the RQC mutants show a loss of DNA annealing activity. Each experiment was repeated at least three times.
At higher protein concentration, two bands appear in the EMSA (Fig. 2A), suggesting the presence of complexes with different stoichiometries. These results could be explained either by a second RecQ4 helicase binding to the DNA substrate or by the formation of a 2:2 protein-DNA complex, as seen in the crystal structure of RecQ1 (20). Although we cannot rule out that under certain conditions RecQ4 may dimerize, in our hand the truncated protein always behaved as a monomer in size exclusion chromatography; moreover, SAXS data on the protein alone and the protein-DNA complex are consistent with a monomeric form (see below). A previously published biochemical study on the Drosophila RecQ4 also suggests the predominant presence of monomeric form in solution (14). Because the EMSA forked substrate includes 22 base pairs plus two 15-nucleotide-long tails and the protein at high concentrations does bind to ssDNA (Fig. 2C) and shows helicase activity toward blunt substrates, we suggest that the higher molecular weight band on Fig. 2A may be explained as a second monomer assembling on the substrate. We need to stress that the DNA binding experiment was carried out in the presence of a large excess of protein. We have performed further DNA binding assays at higher DNA concentrations (100 nm) by diluting 10 nm of fluorescent DNA with unlabeled substrate (supplemental Fig. S1). A significant decrease in the higher band was observed, validating the explanation discussed above. It has to be stressed that in the SAXS experiment, the protein:DNA molar ratio was 1:1. No evidence of cooperativity was observed (supplemental Fig. S2).
The ATPase activity was evaluated by measuring the inorganic phosphate released during the hydrolysis, in the presence of ssDNA (F3), blunt dsDNA (B3:B4), and fork DNA (F3:F4) substrates to stimulate the ATP hydrolysis. Fork DNA stimulated the activity significantly better than the other substrates (Fig. 2E).
The helicase activity of the protein was assessed by a FRET-based method, using both fork DNA and blunt dsDNA as substrates. The result shows that fork DNA is significantly preferred (Fig. 2D), consistent with the binding affinities and the stimulatory effects on ATP hydrolysis. However, a lower level of activity was observed also with the blunt dsDNA substrate. Although in general blunt duplexes are poor substrates for RecQ helicases, unwinding of blunt dsDNA has been occasionally reported for bacterial and yeast enzymes (23, 24). Indeed the crystal structure of the Werner WH domain with DNA shows the β-hairpin aromatic residue wedging into a blunt substrate (18), partially opening the double helix, providing a structural insight into the unwinding of a blunt substrate.
The strand annealing activity of the protein was analyzed using a FRET-based assay, by monitoring the decrease in fluorescence when two complementary strands carrying a fluorophore and a quencher come together. The protein showed a robust annealing activity in a range of concentrations comparable with the helicase activity (Fig. 2I). The result shows that the two strands were annealed to 80% by 160 nm protein in 30 min.
To confirm that the activities observed are indeed due to the hRecQ4-HelRQC recombinant protein and not to a contaminant, we generated a site-directed mutant where the key lysine residue within the Walker A motif was mutated to alanine (K508A, mutant m7), thus affecting the ATP binding site. The protein was expressed and purified in a similar way to the wild type (Fig. 1C) and exhibited a similar behavior when subjected to heat denaturation experiments (Fig. 1E), confirming that the overall fold has not been affected. As expected, the K508A mutant behaved similarly to the wild type in DNA binding experiments (Fig. 2B), but both the ATPase and the helicase activity were negligible (Fig. 2, E and G). The annealing activity was unaffected (Fig. 2I), consistent with the notion that annealing does not requires ATP binding or hydrolysis.
Characterization of RQC Mutants Confirms the Results of the Bioinformatic Analysis and the Role of the RQC Domain in the Helicase Activity
To validate the presence of the RQC domain predicted by in silico analysis (17), several putative functional residues were mutated to alanine. Within the zinc domain, the presence of two insertions makes it difficult to uniquely identify in RecQ4 the residues corresponding to the four cysteines that have been shown to act as zinc ligands in other RecQ proteins. Two cysteines are absolutely conserved in RecQ4 (Cys925 and Cys945) with the other residue (Cys949) only partially conserved. However, other three potentially conserved zinc ligands are detected in an insertion (Cys853, Cys855, and Cys897). We have therefore generated a number of mutants (C853A/C855A, C897A, C925A, C945A, and C949A) to verify their role in zinc binding and enzymatic activity. Previous biochemical and structural studies carried out on eukaryotic RecQ proteins have highlighted the key role of an aromatic residue located at the tip of a β-hairpin of the winged helix domain, stacking on top of the nucleic acid bases at the ss/dsDNA junction (22); based on the proposed sequence alignment, in RecQ4 this position is occupied by Phe1077; an additional mutant was therefore produced, with this residue mutated to alanine.
All the corresponding proteins were expressed and purified similarly to the wild-type protein (Fig. 1C). To evaluate the folding and stability of the purified proteins, heat denaturation experiments were performed using SYPRO Orange fluorescent dye, whose fluorescence emission intensity increases when it binds to the hydrophobic patches of the thermally unfolded protein (ThermoFluor assay). This property was used to determine the melting temperatures of the purified proteins by monitoring their heat denaturation curves. The wild type and mutant proteins show the same heat denaturation profiles (Fig. 1E), and their melting temperatures were nearly identical (Table 2), suggesting a comparable stability of the wild type and mutants and indicating that the mutations did not affect the 3D structure of the proteins.
TABLE 2.
The melting temperature of RecQ4 variants
The values were obtained from heat denaturation curves of protein. The experiment was carried out in triplicate, and the errors are ±0.3 °C. ΔTm = Tm (mutant) − Tm (wild type).
| Protein | Tm | ΔTm |
|---|---|---|
| °C | °C | |
| WT | 41.7 | |
| m1 (C853A/C855A) | 41.4 | −0.3 |
| m2 (C897A) | 41.6 | −0.1 |
| m3 (C925A) | 41.4 | −0.3 |
| m4 (C945A) | 41.4 | −0.3 |
| m5 (C949A) | 41.8 | + 0.1 |
| m6 (F1077A) | 41.9 | + 0.2 |
| m7 (K508A) | 41.8 | + 0.1 |
When metal binding analysis using ICP-AES was carried out on the mutant proteins, whereas the wild-type protein, the WH β-hairpin mutant (F1077A) and the Walker A mutant (K508A) showed the presence of Zn2+ with a 2:1 molar ratio (2 zinc ions per protein molecule), all the cysteine mutants bound only 1 Zn2+ per protein molecule (Table 1). These results suggests that the zinc-binding domain within the RQC includes two independent Zn2+-binding regions: most likely one region is located in the insertion between the second and third helix within the zinc domain, it includes Cys853, Cys855, and Cys897 and is unique to RecQ4 proteins, whereas the second is located between the third helix and the WH domain and is equivalent to what is seen in RecQ1 and other RecQ helicases (including Cys925, Cys945, and Cys949). As expected, mutation of the Walker A or β-hairpin residues does not affect the amount of Zn2+ bound.
When the ability to bind fork DNA was assessed, both the cysteine mutants and the β-hairpin mutant abolished the affinity toward DNA at low nucleic acid concentrations (Fig. 2B), confirming the essential role of the RQC domain in substrate recognition. On the contrary, the Walker A mutant (mut7) behaved similarly to the wild type protein, as expected (Fig. 2B). When DNA binding was carried out at higher DNA concentration (100 nm; Fig. 2F), the binding was impaired but still measurable.
To see the effect of the mutations on ATP hydrolysis, ATPase assays were performed on the mutant proteins using fork DNA substrate as a stimulant. All the RQC mutants behaved similarly to the wild-type protein, with variation well within the statistical error, when in the presence of excess DNA (Fig. 2E). This is a further confirmation that the RQC mutants are properly folded and able to display a catalytic activity. There is an apparent discrepancy between the fact that mutation in the RQC impairs DNA affinity (Fig. 2B) and the fact that in the ATPase assay fork DNA stimulates the ATP hydrolysis activity in the mutants. However, the DNA binding experiments in Fig. 2B are carried out at low concentration of substrate (10 nm), to better appreciate the differences among the mutants, whereas an excess of DNA substrate (500 nm) was used in the ATPase assay. When the ATPase assay was carried out at 100 nm DNA (rather than 500 nm; supplemental Fig. S3), a partial ATPase activity can be observed, consistent with the partial DNA binding abilities of the mutants (Fig. 2F), thus explaining the stimulatory effect of high concentration of DNA on the ATPase activity. Nucleic acid unwinding was negligible for all the RQC mutants at 10 nm substrate (Fig. 2G). When helicase activity is measured at higher substrate concentration (100 nm; Fig. 2H), partial activity can be observed for the RQC mutants. This confirms the importance of the zinc domain and the key role of Phe1077 in the WH β-hairpin for the enzyme activity; furthermore it suggests that the main role of the RQC domain is to interact with nucleic acids. Annealing was also abolished for all the RQC mutants (Fig. 2H).
These results experimentally confirm the presence of a fully functional RQC domain and its essential role in the DNA binding and thus enzyme activity of RecQ4 proteins. The results obtained for the mutant involving the aromatic residue at the end of the WH β-hairpin (F1077A) not only validate the accuracy of the bioinformatic analysis (17) but also confirm its essential role in opening up the dsDNA, as seen in other RecQ helicases. There is some discrepancy as to the role of the β-hairpin in the annealing activity: whereas mutation of the aromatic residue of RecQ4 does abolish annealing (Fig. 2H), the same mutation in RecQ1 does not (20). However, the annealing activity in RecQ1 appears to be subjected to a complex regulation, requiring the full-length protein and the formation of a tetrameric assembly, whereas the monomeric truncated form of RecQ4 here described exhibits annealing activity.
Even more interesting are the results on the cysteine mutants. The role of the zinc domain and the conserved cysteines has not been clearly defined. When two of the Escherichia coli RecQ zinc ligands are mutated to Asn, the DNA binding and helicase activity is abolished, whereas ATPase activity is retained (25); similarly, mutations of the zinc ligands in BLM result in either insoluble or biochemically compromised protein, where DNA binding, ATPase, and helicase activities are compromised (26). A more recent paper suggests that mutations in three of the cysteine residues that coordinate zinc affect ATP binding, DNA binding, ATPase, and helicase activity but not strand annealing (27); furthermore the same three cysteine mutants exhibit enhanced thermal denaturation. Our results overlap only in part with previous work, showing that the cysteine mutants in RecQ4 have the same thermal stability as the wild type, do not impair the ATPase activity (in excess of stimulatory DNA), but affect nucleic acid binding, annealing, and helicase activity (Fig. 2), consistent with the role of the cysteines in the E. coli protein (25). Moreover, the RecQ4 zinc domain is unusual in having two independent zinc clusters, and we find that they are both critical for the helicase activity.
Mapping the missense/nonsense mutations of Rothmund-Thomson, Baller-Gerold, and RAPADILINO syndromes onto the protein sequence (28) reveals a number of single amino acid mutants (including L926R, L927R, R1021TW, I1051V, and S1079I), as well as truncations (Arg826* and Arg1072*) located in the putative RQC domain, pointing to an important role of the RQC in the regulation of the enzyme activity and in the development of these rare diseases.
SAXS Data Reveal the Architecture of the RecQ4-DNA Complex
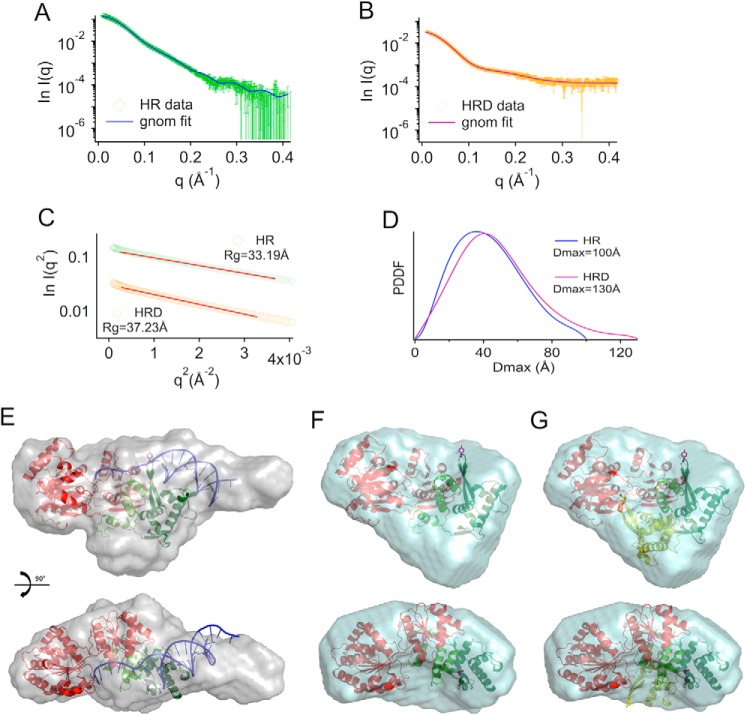
To carry out a low resolution structural characterization of the protein, SAXS data were collected in the presence and absence of a fork DNA substrate. Experimental scattering curves for both samples (Fig. 3, A and B) show good signal to noise ratios, with resolutions of ∼25–30 Å. Guinier's approximation showed no particle aggregation or radiation damage (Fig. 3C). The main structural parameters calculated from the experimental scattering curves show a bigger volume of the envelope of protein-DNA complex compared with that of protein alone (Table 3). The plot of the Pr function extrapolated using GNOM shows a more elongated shape in the presence of nucleic acid, with a Dmax = 130 Å with respect to Dmax = 100 Å for the protein alone (Fig. 3D).
FIGURE 3.
SAXS analysis of human RecQ4. A, experimental SAXS profile (green) compared with the theoretical scattering curve calculated from the ab initio model (blue line) for the catalytic core of human RecQ4 (helicase-RQC (HR)). B, experimental SAXS profile (orange) compared with the theoretical scattering curve calculated from the ab initio model (magenta line) for the catalytic core of human RecQ4-DNA complex (HRD). C, Guinier's fits with experimental Rg values for HR and HRD in the allowed region (q < 1.3 Rg). D, pair distance distribution function (PDDF) for HR and HRD calculated in the allowed region (q > Π/Dmax). E, final model reconstructed from the scattering curve for the protein-DNA complex (HRD). The crystal structure of the RecQ1-DNA complex (PDB code 2WWY) was fitted onto the SAXS envelope. A long protrusion roughly corresponds to the location of DNA in the model. Some unaccounted density could correspond to two long insertions (90 residues in total) within the zinc domain. F, final model reconstructed from the scattering curve for the protein alone. In the absence of DNA, the envelope is more globular and lacks details, so that it is less easy to fit the RecQ1 model. G, to check for the possibility that the WH domain assumes an alternative conformation, the orientation of the WH seen in RecQ1 (dark green) has been overlapped with an alternative conformation (light green) based on the E. coli structure (PDB code 1OYY), suggesting that the protein in solution is a mixture of multiple states.
TABLE 3.
SAXS parameters for HR (RecQ4) and HRD (RecQ4-DNA complex)
| Sample | q range | Rg | Dmax | Volume |
|---|---|---|---|---|
| Å−1 | Å | Å | Å3 | |
| HR | 0.012–0.412 | 33.190 ± 0.008 | 100 | 119,800 |
| HRD | 0.012–0.412 | 37.231 ± 0.024 | 130 | 133,800 |
The molecular envelope obtained in the presence of the nucleic acid substrate (Fig. 3E) shows an elongated extension that can be fitted using the crystal structure of RecQ1-DNA complex. The “extended” DNA configuration does indeed match that seen in the RecQ1 complex, whereas the crystal structures of the DNA complexes of Bloom or a bacterial RecQ helicase show a less protruding nucleic acid substrate, binding at a different orientation. Some “empty” space in the envelope most likely corresponds to the presence of two insertions (with a total of 90 amino acid residues) within the zinc domain, including the second zinc cluster.
In the absence of nucleic acid substrate, the envelope obtained (Fig. 3F) is more globular and lacks details, so that it is less easy to fit a molecular model. A possible explanation is that, in the absence of DNA, the WH domain (that has been shown to be flexible in other RecQ helicases, assuming two configurations that are almost 90° apart) does not assume a well defined position, and the protein in solution is a mixture of multiple states (Fig. 3G).
In both cases (protein alone and protein-DNA complex) the radius of gyration and SAXS envelope are consistent with a monomeric protein; no evidence of the presence of a dimer was detected. This confirms the size exclusion chromatography data.
In conclusion, we report a comprehensive biochemical and structural analysis of the catalytic core of the human RecQ4 helicase, and in particular we focus on the architecture and function of the RQC domain. We experimentally validate a previous bioinformatic analysis (17) and confirm the prediction of two, rather than one, zinc clusters within the zinc domain. Mutagenesis of key residues within the RQC domain demonstrates that this region is essential for the enzymatic activities and suggests that its main function is in nucleic acid interaction. Low resolution SAXS data show that RecQ4 binds DNA similarly to RecQ1, whereas the WH domain may be flexible in the absence of substrates. These results provide a useful framework to understand the physiological role of RecQ4 and the disease-causing mutations.
Experimental Procedures
Cloning and Site-directed Mutagenesis
The nucleotide sequence encoding the helicase and RQC domain (encompassing amino acids 445–1112) was PCR-amplified from a plasmid containing the full-length protein (16), using the Platinum Pfx DNA polymerase (Invitrogen) and following primers: 5′-ATTGAGGCTCACAGAGAACAGATTGGTGGTAGCCTGGACCCCACCGTG-3′ and 5′-TTTGCGCCGAATAAATACCTAAGCTTGTCTTCATTCCTCTTCCTCAAAGTAGCG-3′. The amplified fragment was subcloned into pET-SUMO/CAT vector (Invitrogen) through a restriction free cloning method (29) for protein expression as a fusion with a N-terminal six-histidine-SUMO tag cleavable by SUMO protease.
Site-directed mutagenesis was carried out to mutate the conserved cysteines in the putative zinc-binding domain (mutant m1, C853A/C855A; mutant m2, C897A; mutant m3, C925a; mutant m4, C945A; and mutant m5, C949A), the conserved aromatic residue in the β–hairpin of the putative winged-helix domain (mutant m6, F1077A), and the Walker A catalytic lysine (mutant m7, K508A).
Protein Expression and Purification
The wild-type and mutant proteins were expressed in Rosetta 2 (DE3) cells, using the autoinduction method (30), grown at 17 °C for 48 h, and harvested by centrifugation. The cells were lysed by sonication in 50 mm Hepes, pH 8.0, 0.5 m NaCl, 1 mm tris-(2-carboxyethyl) phosphine hydrochloride, 5% glycerol (v/v), 10 mm imidazole, 1 tablet of Complete EDTA-free protease inhibitor mixture (Roche), and DNase I (Sigma). Proteins were purified by nickel affinity chromatography, with a 2 m NaCl washing step to eliminate DNA contamination, followed by tag cleavage with SUMO protease, and an additional step of metal affinity to eliminate the His-SUMO tag and the His-tagged protease. The protein purification was further optimize by heparine chromatography and size exclusion chromatography in 20 mm Tris, pH 8.0, 250 mm NaCl, 5% glycerol (v/v), and 5 mm β-mercaptoethanol on a Superdex-200 column. Protein concentration was determined by measuring the absorbance at 280 nm, and protein purity was analyzed on SDS-PAGE. Metal binding analysis was carried out by ICP-AES. Purifications of all the mutant proteins were performed following the same protocol.
ThermoFluor Assay
To determine the protein stability and to compare the various mutants, a heat denaturation analysis was carried out using a CFX96 Touch real time quantitative PCR system (Bio-Rad). Protein stability measurements were performed in a buffer containing 20 mm Tris, pH 8.0, 250 mm NaCl, 5% glycerol (v/v), and 5 mm β-mercaptoethanol. The 25 μl of reaction mix contained 0.5 μm protein and SYPRO Orange (Invitrogen) at 20× concentration. Heat denaturing curves were observed within the temperature range of 20–80 °C, at a ramp rate of 1.8 °C/min, collecting data every 10 s. The data obtained was normalized using GraphPad Prism software.
Oligonucleotide Preparation
All the oligonucleotides (unlabeled; 5′ 6FAM-labeled; 3′ BHQ1-labeled) were chemically synthesized and purified by reverse phase high pressure liquid chromatography (Sigma-Aldrich). Each oligonucleotide was then resuspended in Tris-EDTA buffer (10 mm Tris-HCl, pH 7.5, 1 mm EDTA). The oligonucleotide sequences used in this work are reported in Table 4. For the helicase assay the dsDNA substrates (F1:F2 and B1:B2) were prepared with the fluorescent strand and the complementary strand at a 1:2 m ratio in 10 mm Tris, pH 7.5, 50 mm NaCl, 1 mm EDTA by briefly heating at 95 °C, followed by slow cooling to room temperature. The substrates for the ATPase assay and EMSA were prepared as above.
TABLE 4.
Oligonucleotides used in this study (6FAM label at 5′ end and BHQ1 label at 3′end)
| Name | Sequence (5′ → 3′) |
|---|---|
| F1 | [6FAM] CTACTACCCCCACCCTCACAACCTTTTTTTTTTTTTT |
| F2 | TTTTTTTTTTTTTTGGTTGTGAGGGTGGGGGTAGTAG [BHQ1] |
| B1 | [6FAM] CTACTACCCCCACCCTCACAACC |
| B2 | GGTTGTGAGGGTGGGGGTAGTAG [BHQ1] |
| Capture | CTACTACCCCCACCCTCACAACC |
| F3 | CTACTACCCCCACCCTCACAACCTTTTTTTTTTTTTT |
| F4 | TTTTTTTTTTTTTTGGTTGTGAGGGTGGGGGTAGTAG |
| B3 | CTACTACCCCCACCCTCACAACC |
| B4 | GGTTGTGAGGGTGGGGGTAGTAG |
Electrophoretic Mobility Shift Assay
The fluorescent DNA binding assay was performed in 20 mm Tris-HCl, pH 7.5, 5 mm MgCl2, 50 mm KCl, 8 mm DTT, 0.1 mg/ml BSA, and 5% glycerol v/v with 10 nm substrates (F1, F1:F4 and B1:B4; Table 4) and increasing concentrations (0–3 μm) of the purified proteins in 20 μl of reaction volume, incubated at room temperature for 30 min. The reaction mixture was then loaded on a 6% non-denaturing polyacrylamide gel (v/v) and run at 4 °C in Tris/borate/EDTA buffer. Fluorescent labeled nucleic acid fragments were detected by fluorescent scanner (ImageQuant; GE Healthcare), and quantification of protein-nucleic acid complexes was performed with ImageQuant image analysis software (GE Healthcare). To measure DNA binding in the presence of 100 nm fork DNA substrate, 10 nm labeled DNA (F1:F4) was mixed with 90 nm unlabeled substrate (F3:F4).
ATPase Assays
The Malachite Green assay kit (Cayman) was used to determine the ATPase activity by measuring the inorganic phosphate released during ATP hydrolysis (31, 32). The method is based on the change in absorbance at A620 of the complex formed between malachite green (MG) molybdate and free orthophosphate under acidic conditions (molybdophosphoric acid complex). The assay was carried out in 50-μl reaction volumes, containing 20 mm Tris-HCl, pH 7.5, 5 mm MgCl2, 50 mm KCl, 8 mm DTT, 0.1 mg/ml BSA, and 5% glycerol (v/v) with 500 nm purified protein, 3 mm ATP, and 100 or 500 nm of nucleic acid (F3, F3:F4, and B3:B4; Table 4). The ATPase reactions were incubated at 37 °C for 30 min. After incubation, 5 μl of MG acidic solution was added and incubated at room temperature for 10 min, followed by addition of 15 μl of MG molybdenum reagent and incubation at room temperature for further 5 min. The absorption at 620 nm was then measured using a Lambda25 UV/VIS Spectrometer (PerkinElmer Life Sciences). The concentrations of inorganic phosphate were determined by comparison with a standard curve obtained in the presence of known phosphate concentrations.
Helicase Assays
The helicase activity was based on FRET, by using a double-stranded substrate (Table 4) with a fluorophore (6FAM) on one strand and a quencher (BHQ1) on the other, and by monitoring the fluorence emitted when the two strands are separated by the helicase action (33). The assay was performed in 20 mm Tris-HCl, pH 7.5, 5 mm MgCl2, 50 mm KCl, 8 mm DTT, 0.1 mg/ml BSA, and 5% glycerol (v/v) with 10 or 100 nm dsDNA substrate (F1:F2 and B1:B2), 3 mm ATP, and 125 nm capture strand (to prevent reannealing) in 30 μl of reaction volume. The unwinding reaction was started by incubating increasing concentrations (0–160 nm) of the purified proteins in the reaction mixture at 37 °C for 30 min. The fluorescence intensity was recorded using the Infinite F200 PRO microplate reader (TECAN). To have a measurement corresponding to 100% unwinding, the reaction was incubated at 95 °C. Each assay was done in triplicate. The percentage of unwinding was calculated and plotted using GraphPad Prism software.
Strand Annealing Assays
A FRET-based strand annealing assay was carried out by monitoring the decrease in fluorescence when the two complementary oligonucleotides (one labeled with a fluorophore and the other with a quencher) anneals. The assay was performed in 20 mm Tris-HCl, pH 7.5, 5 mm MgCl2, 50 mm KCl, 8 mm DTT, 0.1 mg/ml BSA, and 5% glycerol with 10 nm 6FAM-labeled oligonucleotide (F1) and 25 nm BHQ1-labeled oligonucleotide (F2) in 30 μl of reaction volume. The reaction was started by incubating increasing concentrations (0–160 nm) of the purified proteins in the reaction mixture at 37 °C for 30 min. The fluorescence intensity was recorded using the Infinite F200 PRO microplate reader (TECAN). To have a measurement corresponding to 100% annealing, the fluorescence of the annealed substrate (F1:F2) was measured. Each assay was done in triplicate. The percentage of annealing was calculated and plotted using GraphPad Prism software.
Small Angle X-ray Scattering
The Synchrotron scattering data were collected at the SWING Beamline (34) at Soleil both from the protein alone and in complex with DNA. For the latter an equimolar amount of fork DNA (F3:F4) was added to the protein and incubated at room temperature for 60 min. The protein and protein-DNA complex were then purified using size exclusion chromatography. Samples corresponding to 0.5, 1.5, and 3 mg/ml were automatically loaded using a sample changer thermostatted at 4 °C; data were collected at a wavelength of 1.54 Å, and the distance between the sample and the PCCD170170 (AVIEX) detector was set to 4.78 m, q was in the range from 0.012 to 0.412 Å−1. After calibration, all data were averaged, normalized, and background-subtracted using the Soleil software Foxtrot. Absolute scale was determined using the reference curve collected from BSA at 5 mg/ml. Guinier's approximation was used to check for particle aggregation and radiation damage in the sample. Structural parameters were calculated from the experimental curves using Primus (35). The maximum length of the model (Dmax) was estimated using GNOM (36) looking at the best P(r) function shape. Final SAXS envelopes were obtained by averaging 20 ab initio DAMMIF (37) models with 20 ab initio DAMMIN (38) models using SUPCOMB (39) and DAMAVER (40).
Author Contributions
A. M. and S. O. designed the experiments; A. M. and F. M. performed cloning, protein expression, and purification; A. M. carried out all the biochemical experiments and analyzed the data; M. D. M. and A. M. performed the SAXS data collection; M. D. M. analyzed the SAXS data; and A. M. and S. O. wrote the manuscript with input from all the authors.
Supplementary Material
Acknowledgments
We thank Aurelien Thureau (Soleil, SWING Beamline) for help in SAXS data collection and Alessandro Vindigni for useful discussion.
This work was supported by Associazione Italiana per la Ricerca sul Cancro Grants IG10646 and IG14718, the ICTP TRIL fellowship programme, the Cross-Border Cooperation Programme Italy-Slovenia 2007–2013 by the European Regional Development Fund and national funds (PROTEO), and the European Community's Seventh Framework Programme (2007–2013) under BioStruct-X Grant Agreement 283570. The authors declare that they have no conflicts of interest with the contents of this article.

This article contains supplemental Figs. S1–S3.
- RQC
- RecQ C-terminal domain
- WH
- winged helix
- SAXS
- small angle X-ray scattering
- ICP-AES
- inductively coupled plasma-atomic emission spectrometry
- ssDNA
- single-stranded DNA
- MG
- malachite green
- 6-FAM
- 6-carboxyfluorescein
- PDB
- Protein Data Bank
- HR
- helicase-RQC
- HRD
- human RecQ4-DNA complex.
References
- 1. Croteau D. L., Popuri V., Opresko P. L., and Bohr V. A. (2014) Human RecQ helicases in DNA repair, recombination, and replication. Annu. Rev. Biochem. 83, 519–552 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Keijzers G., Maynard S., Shamanna R. A., Rasmussen L. J., Croteau D. L., and Bohr V. A. (2014) The role of RecQ helicases in non-homologous end-joining. Crit. Rev. Biochem. Mol. Biol. 49, 463–472 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Larsen N. B., and Hickson I. D. (2013) RecQ helicases: conserved guardians of genomic integrity. Adv. Exp. Med. Biol. 767, 161–184 [DOI] [PubMed] [Google Scholar]
- 4. Oshima J., Sidorova J. M., and Monnat R. J. Jr. (2016) Werner syndrome: clinical features, pathogenesis and potential therapeutic interventions. Ageing Res. Rev. pii, S1568–S1637 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. de Renty C., and Ellis N. A. (2017) Bloom's syndrome: why not premature aging?: a comparison of the BLM and WRN helicases. Ageing Res. Rev. 33, 36–51 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Suhasini A. N., and Brosh R. M. (2013) DNA helicases associated with genetic instability, cancer, and aging. Adv. Exp. Med. Biol. 767, 123–144 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Su Y., Meador J. A., Calaf G. M., Proietti De-Santis L., Zhao Y., Bohr V. A., and Balajee A. S. (2010) Human RecQL4 helicase plays critical roles in prostate carcinogenesis. Cancer Res. 70, 9207–9217 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Fang H., Nie L., Chi Z., Liu J., Guo D., Lu X., Hei T. K., Balajee A. S., and Zhao Y. (2013) RecQL4 helicase amplification is involved in human breast tumorigenesis. PLoS One 8, e69600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Arora A., Agarwal D., Abdel-Fatah T. M., Lu H., Croteau D. L., Moseley P., Aleskandarany M. A., Green A. R., Ball G., Rakha E. A., Chan S. Y., Ellis I. O., Wang L. L., Zhao Y., Balajee A. S., et al. (2016) RECQL4 helicase has oncogenic potential in sporadic breast cancers. J. Pathol. 238, 495–501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Mo D., Fang H., Niu K., Liu J., Wu M., Li S., Zhu T., Aleskandarany M. A., Arora A., Lobo D. N., Madhusudan S., Balajee A. S., Chi Z., and Zhao Y. (2016) Human helicase RECQL4 drives cisplatin resistance in gastric cancer by activating an AKT-YB1-MDR1 signaling pathway. Cancer Res. 76, 3057–3066 [DOI] [PubMed] [Google Scholar]
- 11. Marino F., Mojumdar A., Zucchelli C., Bhardwaj A., Buratti E., Vindigni A., Musco G., and Onesti S. (2016) Structural and biochemical characterization of an RNA/DNA binding motif in the N-terminal domain of RecQ4 helicases. Sci. Rep. 6, 21501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Yin J., Kwon Y. T., Varshavsky A., and Wang W. (2004) RECQL4, mutated in the Rothmund-Thomson and RAPADILINO syndromes, interacts with ubiquitin ligases UBR1 and UBR2 of the N-end rule pathway. Hum. Mol. Genet. 13, 2421–2430 [DOI] [PubMed] [Google Scholar]
- 13. Macris M. A., Krejci L., Bussen W., Shimamoto A., and Sung P. (2006) Biochemical characterization of the RECQ4 protein, mutated in Rothmund-Thomson syndrome. DNA Repair 5, 172–180 [DOI] [PubMed] [Google Scholar]
- 14. Capp C., Wu J., and Hsieh T. S. (2009) Drosophila RecQ4 has a 3′-5′ DNA helicase activity that is essential for viability. J. Biol. Chem. 284, 30845–30852 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Suzuki T., Kohno T., and Ishimi Y. (2009) DNA helicase activity in purified human RECQL4 protein. J. Biochem. 146, 327–335 [DOI] [PubMed] [Google Scholar]
- 16. Rossi M. L., Ghosh A. K., Kulikowicz T., Croteau D. L., and Bohr V. A. (2010) Conserved helicase domain of human RecQ4 is required for strand annealing-independent DNA unwinding. DNA Repair (Amst.) 9, 796–804 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Marino F., Vindigni A., and Onesti S. (2013) Bioinformatic analysis of RecQ4 helicases reveals the presence of a RQC domain and a Zn knuckle. Biophys. Chem. 177–178, 34–39 [DOI] [PubMed] [Google Scholar]
- 18. Kitano K., Kim S. Y., and Hakoshima T. (2010) Structural basis for DNA strand separation by the unconventional winged-helix domain of RecQ helicase WRN. Structure 18, 177–187 [DOI] [PubMed] [Google Scholar]
- 19. Lucic B., Zhang Y., King O., Mendoza-Maldonado R., Berti M., Niesen F. H., Burgess-Brown N. A., Pike A. C., Cooper C. D., Gileadi O., and Vindigni A. (2011) A prominent β-hairpin structure in the winged-helix domain of RECQ1 is required for DNA unwinding and oligomer formation. Nucleic Acids Res. 39, 1703–1717 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Pike A. C., Gomathinayagam S., Swuec P., Berti M., Zhang Y., Schnecke C., Marino F., von Delft F., Renault L., Costa A., Gileadi O., and Vindigni A. (2015) Human RECQ1 helicase-driven DNA unwinding, annealing, and branch migration: insights from DNA complex structures. Proc. Natl. Acad. Sci. U.S.A. 112, 4286–4291 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Kitano K. (2014) Structural mechanisms of human RecQ helicases WRN and BLM. Front. Genet. 5, 366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Bhattacharyya B., and Keck J. L. (2014) Grip it and rip it: structural mechanisms of DNA helicase substrate binding and unwinding. Protein Sci. 23, 1498–1507 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Cejka P., and Kowalczykowski S. C. (2010) The full-length Saccharomyces cerevisiae Sgs1 protein is a vigorous DNA helicase that preferentially unwinds Holliday junctions. J. Biol. Chem. 285, 8290–8301 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Umezu K., Nakayama K., and Nakayama H. (1990) Escherichia coli RecQ protein is a DNA helicase. Proc. Natl. Acad. Sci. U.S.A. 87, 5363–5367 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Liu J. L., Rigolet P., Dou S. X., Wang P. Y., and Xi X. G. (2004) The zinc finger motif of Escherichia coli RecQ is implicated in both DNA binding and protein folding. J. Biol. Chem. 279, 42794–42802 [DOI] [PubMed] [Google Scholar]
- 26. Guo R. B., Rigolet P., Zargarian L., Fermandjian S., and Xi X. G. (2005) Structural and functional characterizations reveal the importance of a zinc binding domain in Bloom's syndrome helicase. Nucleic Acids Res. 33, 3109–3124 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Sami F., Gary R. K., Fang Y., and Sharma S. (2016) Site-directed mutants of human RECQ1 reveal functional importance of the zinc binding domain. Mutat. Res. 790, 8–18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Larizza L., Roversi G., and Volpi L. (2010) Rothmund-Thomson syndrome. Orphanet J. Rare Dis. 5, 2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Unger T., Jacobovitch Y., Dantes A., Bernheim R., and Peleg Y. (2010) Applications of the restriction free (RF) cloning procedure for molecular manipulations and protein expression. J. Struct. Biol. 172, 34–44 [DOI] [PubMed] [Google Scholar]
- 30. Studier F. W. (2005) Protein production by auto-induction in high density shaking cultures. Protein Expr. Purif. 41, 207–234 [DOI] [PubMed] [Google Scholar]
- 31. Chan K. M., Delfert D., and Junger K. D. (1986) A direct colorimetric assay for Ca2+-stimulated ATPase activity. Anal. Biochem. 157, 375–380 [DOI] [PubMed] [Google Scholar]
- 32. Huang Y., and Liu Z. R. (2002) The ATPase, RNA unwinding, and RNA binding activities of recombinant p68 RNA helicase. J. Biol. Chem. 277, 12810–12815 [DOI] [PubMed] [Google Scholar]
- 33. Tani H., Fujita O., Furuta A., Matsuda Y., Miyata R., Akimitsu N., Tanaka J., Tsuneda S., Sekiguchi Y., and Noda N. (2010) Real-time monitoring of RNA helicase activity using fluorescence resonance energy transfer in vitro. Biochem. Biophys. Res. Commun. 393, 131–136 [DOI] [PubMed] [Google Scholar]
- 34. Fourme R., Thompson A., Girard E., Legrand P., Perez J., Dumas P., and Refregiers M. (2004) The Biology Programme at SOLEIL, 8th International Conference on Biology and Synchrotron Radiation (BSR8), September 7–11, 2004, Himeji, Japan [Google Scholar]
- 35. Konarev P. V., Volkov V. V., Sokolova A. V., Koch M. H., and Svergun D. I. (2003) PRIMUS: a Windows-PC based system for small-angle scattering data analysis. J. Appl. Crystallogr. 36, 1277–1282 [Google Scholar]
- 36. Svergun D. I. (1992) Determination of the regularization parameter in indirect-transform methods using perceptual criteria. J. Appl. Crystallogr. 25, 495–503 [Google Scholar]
- 37. Franke D., and Svergun D. I. (2009) DAMMIF, a program for rapid ab-initio shape determination in small-angle scattering. J. Appl. Crystallogr. 42, 342–346 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Svergun D. I. (1999) Restoring low resolution structure of biological macromolecules from solution scattering using simulated annealing. Biophys. J. 76, 2879–2886 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Kozin M., and Svergun D. I. (2001) Automated matching of high- and low-resolution structural models. J. Appl. Crystallogr. 34, 33–41 [Google Scholar]
- 40. Volkov V. V., and Svergun D. I. (2003) Uniqueness of ab-initio shape determination in small-angle scattering. J. Appl. Crystallogr. 36, 860–864 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.