Abstract
RNA editing is a cellular process that precisely alters nucleotide sequences, thus regulating gene expression and generating protein diversity. Over 60% of human transcripts undergo adenosine to inosine RNA editing, and editing is required for normal development and proper neuronal function of animals. Editing of one adenosine in the transcript encoding the glutamate receptor subunit B, glutamate receptor ionotropic AMPA 2 (GRIA2), modifies a codon, replacing the genomically encoded glutamine (Q) with arginine (R); thus this editing site is referred to as the Q/R site. Editing at the Q/R site of GRIA2 is essential, and reduced editing of GRIA2 transcripts has been observed in patients suffering from glioblastoma. In glioblastoma, incorporation of unedited GRIA2 subunits leads to a calcium-permeable glutamate receptor, which can promote cell migration and tumor invasion. In this study, we identify adenosine deaminase that acts on RNA 3 (ADAR3) as an important regulator of Q/R site editing, investigate its mode of action, and detect elevated ADAR3 expression in glioblastoma tumors compared with adjacent brain tissue. Overexpression of ADAR3 in astrocyte and astrocytoma cell lines inhibits RNA editing at the Q/R site of GRIA2. Furthermore, the double-stranded RNA binding domains of ADAR3 are required for repression of RNA editing. As the Q/R site of GRIA2 is specifically edited by ADAR2, we suggest that ADAR3 directly competes with ADAR2 for binding to GRIA2 transcript, inhibiting RNA editing, as evidenced by the direct binding of ADAR3 to the GRIA2 pre-mRNA. Finally, we provide evidence that both ADAR2 and ADAR3 expression contributes to the relative level of GRIA2 editing in tumors from patients suffering from glioblastoma.
Keywords: cancer, double-stranded RNA (dsRNA), glioblastoma, glutamate receptor, RNA editing, ADAR, ADAR3, GRIA2, adenosine deaminase, inosine
Introduction
Adenosine deaminases that act on RNA (ADARs)2 catalyze the hydrolytic deamination of adenosine residues within double-stranded RNA (dsRNA) structures that form by base pairing of nearby complementary sequences (1–3). This RNA editing event creates a non-canonical nucleoside, inosine, which base pairs similarly to guanosine (4). RNA editing affects gene expression through modification of codons to create novel protein isoforms (5). Furthermore, editing can alter microRNA and siRNA binding sites, splice acceptor or splice donor sites, or RNA structure and stability (6–9). A majority of the targets of RNA editing are found in the nervous system, and RNA editing is critical for maintaining proper neuronal function (10). Furthermore, transcripts encoding proteins involved in neurotransmission are often targets of RNA editing, which alters the protein amino acid sequence and physiological function of these ion channels and receptors (11, 12).
In mammals, three ADAR proteins, ADAR1, ADAR2, and ADAR3, and two ADAR-like proteins, ADAD1 and ADAD2, have been identified (13–17). The ADAR and ADAR-like protein family members contain several common modular domains that are important for function (18, 19). Most strikingly, the human ADAR and ADAR-like proteins contain at least one dsRNA binding domain (dsRBD), which are evolutionarily conserved among dsRNA-binding proteins (dsRBPs) and confer the ability to interact with dsRNA substrates (20). In addition, all ADAR proteins contain a conserved C-terminal deaminase domain; however, ADAR1 and ADAR2 (ADARB1) are the major deaminases and have millions of known editing targets (21–27). ADAR3 (ADARB2), although having high sequence similarity to ADAR1 and ADAR2, has not been shown to have deaminase activity in vitro and has no known in vivo targets (28). However, ADAR3 is known to bind to dsRNA via its dsRBDs, and ADAR3 contains a unique R-domain consisting of a series of arginine residues that are required for binding to single-stranded RNA in vitro (28). ADAD1 and ADAD2 contain only one dsRNA binding domain in addition to the deaminase domain, and little is known about their functional significance (9).
Both ADAR1 and ADAR2 are expressed ubiquitously (29–31), whereas ADAR3 is expressed primarily in the brain (16). Mutations in ADARs have been shown to contribute to disease including Aicardi-Goutières syndrome and dyschromatosis symmetrica hereditaria, and aberrant RNA editing has been implicated in prostate cancer, hepatocellular carcinoma, and chronic myeloid leukemia (19, 32, 33). Furthermore, changes in RNA editing are often associated with diseases of the central nervous system including neurodegenerative diseases and astrocytomas (11, 34); in fact, one commonality between the neurological disease amyotrophic lateral sclerosis (ALS) and glioblastoma (also referred to as grade IV astrocytoma) is reduced editing of one particular transcript, GRIA2 (9, 35).
The glutamate receptor subunit B (GRIA2) transcript was one of the first identified ADAR targets and encodes one subunit in the AMPA-type glutamate receptor (36, 37). Editing of GRIA2 at one specific adenosine (the Q/R site) modifies the genomically encoded glutamine (Q) codon to arginine (R) (38). The edited, arginine-containing GRIA2 can be incorporated into the AMPA receptor, rendering it impermeable to calcium; whereas the unedited glutamine-containing GRIA2 subunits form AMPA receptors that are permeable to calcium (35). ADAR2 is the primary editor at the Q/R site of GRIA2 (39). In fact, ADAR2 knock-out mice die shortly after birth from epileptic seizures caused by increased neuronal calcium influx (40), but these mice can be rescued from lethality through complementary DNA (cDNA) expression of the edited, R-form of GRIA2 (41, 42). Strikingly, altered editing at the Q/R site of GRIA2 has been associated with epilepsy, ALS, schizophrenia, and ischemia (34, 43). For these neuropathological diseases, it is thought that reduced GRIA2 editing results in an increase in glutamate receptors that are permeable to calcium, leading to excitotoxicity and cell death of neurons (44). However, reduced editing of GRIA2 has also been observed in malignant gliomas (45). In these cell lines, expression of protein translated from unedited GRIA2 transcripts promotes cell migration and invasion (46). It is unclear how editing of GRIA2 is reduced in glioblastoma as ADAR2 mRNA levels do not directly correlate with the editing level at the Q/R site of GRIA2 (45); however, it has been shown that transgenic overexpression of ADAR2 in glioblastoma cell lines can increase editing of GRIA2 and slow glioblastoma tumor growth in mice orthotopically injected with these cell lines (47).
Properly regulated RNA editing provides flexibility to the genetic code during development, in response to environmental conditions, or between cell or tissue types; however, this regulation often goes awry in disease (48–50). ADAR2 is able to self-regulate through an alternative splicing negative feedback loop in which ADAR2 edits an adenosine within its own transcript, leading to production of a shortened and editing-deficient protein isoform (51, 52). Furthermore, Pin1 and WWP2 can negatively regulate editing via cytoplasmic sequestration or ubiquitination and degradation of ADAR2, respectively (53). These global regulators affect RNA editing of ADAR2-edited sites over the entire transcriptome. However, editing by ADARs is also regulated on the transcript level by the landscape of RNA-binding proteins present on a given mRNA. For example, the RNA-binding proteins SRFS9, RPS14, and DDX15 were shown to inhibit RNA editing of specific target mRNAs (54). Interestingly, these proteins decrease in expression during brain development in mice, and the expression levels of SFRS9 and DDX15 are altered upon neuronal stimulation. Moreover, not all RNA-binding proteins inhibit RNA editing as evidenced by a recent study showing that heterogeneous nuclear ribonucleoprotein A2/B1 has the ability to promote editing of specific transcripts (55). Despite many advances in identifying regulators of RNA editing and the importance of GRIA2 editing in several disease pathologies, a specific regulator of editing in GRIA2 has not been identified.
In this study, we sought to identify the cellular regulator of GRIA2 editing that is altered in glioblastoma. Here, we provide evidence that the deaminase-deficient ADAR family member ADAR3 regulates editing of GRIA2. Obtaining data from a panel of astrocyte- and grade IV astrocytoma-derived cell lines, we suggest that ADAR2 expression alone cannot account for differences in GRIA2 editing between cells lines and that ADAR3 expression is consistent with what would be expected for an inhibitor of GRIA2 editing. Furthermore, expressing ADAR3 in one of these cell lines, we demonstrated that ADAR3 represses editing of GRIA2. To interrogate the biological mechanism of ADAR3 inhibition of RNA editing, we took a two-pronged approach, expressing ADAR3 with mutations in annotated domains to determine its mode of action and immunoprecipitating ADAR3 to elucidate protein and RNA interactions. Our findings indicate that the dsRNA binding domains of ADAR3 are required for inhibition of RNA editing and that ADAR3 binds directly to GRIA2 precursor mRNA (pre-mRNA) transcripts. Finally, our data implicate ADAR3 in the context of glioblastoma; measuring both ADAR2 and ADAR3 expression and GRIA2 editing in six glioblastoma tumors and matched adjacent brain tissue, we found increased expression of ADAR3 in nearly all the tumors compared with adjacent tissue and observed decreased GRIA2 editing in all the tumors.
Results
The Expression of ADAR2 Relative to ADAR3 Correlates with the Extent of GRIA2 Editing
To identify cellular factors that affect the ability of ADAR2 to edit GRIA2, we identified a panel of cell lines that had variable levels of editing of the Q/R site of GRIA2. As reduced editing of the Q/R editing site has been reported in glioblastoma patients (45), we analyzed endogenous GRIA2 editing in immortalized normal human astrocyte (NHA) cells and grade IV astrocytoma cell lines U87-MG and U118. The NHA cell line is immortalized through the expression of the catalytic component of human telomerase, E6 (an inhibitor of p53), and E7 (an inhibitor of Rb); therefore, although these NHA cells are not derived from glioblastoma and do not form tumors in vivo, the line is not perfectly representative of normal astrocytes (56). RNA was isolated from two independent biological replicates of each cell line. Following reverse transcription, PCR amplification, and Sanger sequencing, editing was quantitatively measured. At the Q/R site of GRIA2, U87 and NHA cells exhibited ∼65% editing, and U118 had 0% editing (Fig. 1, A and B). This is consistent with previous studies that showed 0% Q/R editing in U118 cells (57) and 50% Q/R editing in U87 cells (39). From these same cell lines, we detected ADAR2 expression by Western blotting. NHA cells had greater ADAR2 protein expression than U87 and U118, which had a similar, lower level of ADAR2 (Fig. 1C). Comparing ADAR2 protein expression and GRIA2 editing among the cell lines, we found that ADAR2 protein expression does not directly correlate with GRIA2 editing. Specifically our data indicate that U118 cells exhibit 0% editing at the Q/R site, and U87 cells have 65% editing at the Q/R site of GRIA2 mRNA while displaying a similar level of ADAR2 protein expression. Furthermore, NHA cells have elevated ADAR2 protein levels compared with U87 cells, but GRIA2 Q/R editing is not significantly different between these cell lines. These data suggest that both NHA cells and U118 cells express a negative regulator of GRIA2 editing. To test this possibility, we overexpressed ADAR2 in the NHA cell line (Fig. 1D) and measured editing of GRIA2. ADAR2 overexpression increased editing at the Q/R site to 100% (Fig. 1E). Consistent with our results, a previous study demonstrated that overexpression of ADAR2 in U118 cells results in increased editing at the Q/R site (57). Together, these results suggest that a negative regulator of RNA editing exists in these cells lines and can be outcompeted by overexpressing ADAR2.
FIGURE 1.
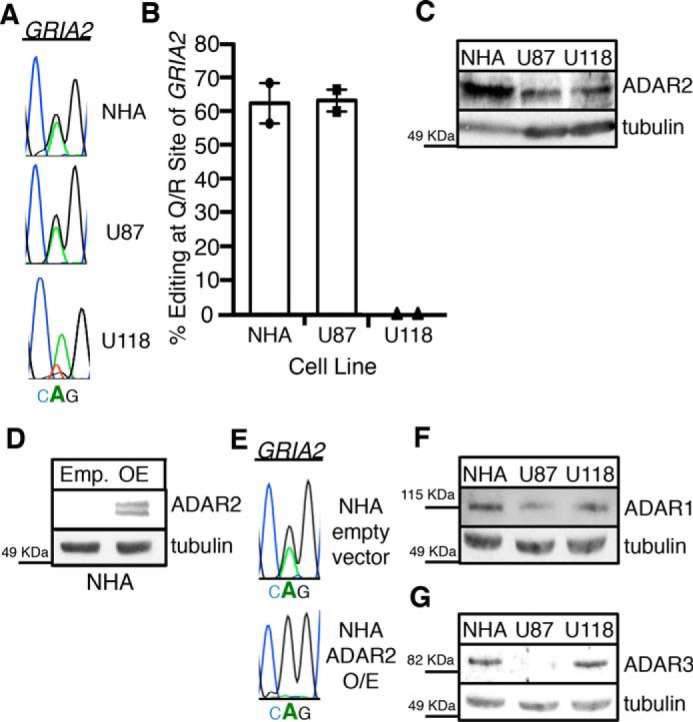
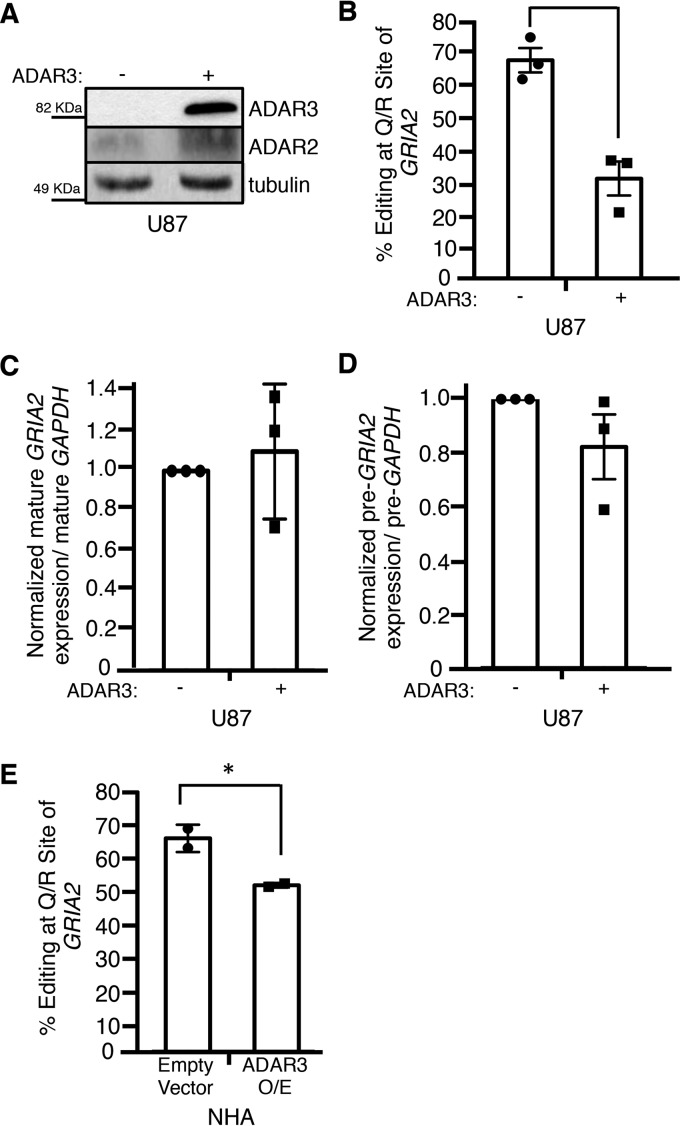
ADAR3 expression is consistent with an inhibitor of GRIA2 editing in astrocyte- and grade IV astrocytoma-derived cell lines. A, chromatograms of the Q/R site of GRIA2 in NHA, U87, and U118 cells. Black indicates transcripts with guanosine (edited), and green indicates transcripts with adenosine (unedited). B, editing levels at the Q/R site of GRIA2 were measured in the indicated cell lines (n = 2). Error bars represent S.E. C, equivalent amounts of cell lysates were determined by Bradford assay and subjected to SDS-PAGE and immunoblotting for ADAR2. Tubulin is the loading control. D, lysates from NHA cells transduced with retrovirus containing a neomycin resistance vector with no protein (Emp.) or human ADAR2 expressed from the CMV promoter (OE) were subjected to SDS-PAGE and immunoblotted for ADAR2. Tubulin is the loading control. E, chromatograms of the Q/R site of GRIA2 in the cell lines show in D. Black indicates transcripts with guanosine (edited), and green indicates transcripts with adenosine (unedited). Bold A indicates the genomically encoded adenosine at the Q/R site of GRIA2. F and G, equivalent amounts of cell lysates were determined by Bradford assay and subjected to SDS-PAGE and immunoblotting for ADAR1 (F) or ADAR3 (G). Tubulin is the loading control.
A previous study has shown that simultaneous overexpression of ADAR2 and ADAR1 reduces RNA editing of GRIA2 through the formation of ADAR1/ADAR2 heterodimers (57). To this end, we measured the expression of ADAR1 protein in the three cell lines and found that the expression level among the cell lines is relatively equal with slightly higher expression in NHA (Fig. 1F). However, this small expression difference seemed unlikely to fully account for the inhibition of editing, especially in regards to the striking differences in GRIA2 editing between the U87 and U118 cell lines. Interestingly, the third ADAR family member, ADAR3, which is catalytically inactive, has also been shown to inhibit editing of GRIA2 in vitro (28). We measured ADAR3 protein expression by Western blotting and found that ADAR3 expression is dramatically elevated in NHA and U118 compared with U87 cells (Fig. 1G). These data are consistent with ADAR3 functioning as a negative regulator of editing in NHA and U118 cells and suggest that both ADAR2 and ADAR3 expression contributes to the overall editing level at the Q/R site of GRIA2 in human cell lines.
ADAR3 Negatively Regulates Editing by ADAR2 at the Q/R Site of GRIA2
To directly test the hypothesis that ADAR3 negatively regulates editing by ADAR2, we expressed ADAR3 in the U87 cell line and measured GRIA2 editing. The U87 cell line was chosen as it lacked detectable ADAR3 expression (Fig. 1G). Using retroviral transduction, we generated U87 cells containing a neomycin resistance vector with no protein or human ADAR3 expressed from the CMV promoter and confirmed expression of ADAR3 by Western blotting (Fig. 2A). RNA editing at the Q/R site of GRIA2 was measured in three independent biological replicates of the control cells and cells stably expressing ADAR3. Expression of ADAR3 results in an ∼40% decrease in transcripts edited at the Q/R site of GRIA2 compared with control cells (Fig. 2B). Importantly, this was not due to a reduction in GRIA2 mRNA levels (Fig. 2, C and D) or a reduction in ADAR2 expression (Fig. 2A). We also tested the ability of ADAR3 to negatively regulate editing by ADAR2 in NHA cells, which have higher endogenous ADAR3 expression (Fig. 1G). Overexpression of ADAR3 in this cell line also resulted in an ∼15% decrease in editing of GRIA2 (Fig. 2E). As U87 cells have lower endogenous ADAR3 expression compared with NHA cells (Fig. 1G), the smaller decrease in editing observed upon ADAR3 overexpression in NHA cells suggests that endogenous ADAR3 expression is responsible for inhibition of GRIA2 editing. Together, these results indicate that ADAR3 is a negative regulator of GRIA2 editing.
FIGURE 2.
ADAR3 inhibits RNA editing at the Q/R site of GRIA2. A, equivalent amounts of lysates from U87 cells transduced with a retrovirus containing a neomycin resistance vector with no protein (−) or human ADAR3 expressed from the CMV promoter (+) were determined by Bradford assay and subjected to SDS-PAGE and immunoblotting for ADAR2 and ADAR3. Tubulin antibody was used as a loading control. B, editing levels at the Q/R site of GRIA2 were measured in U87 cells described in A for three independent biological replicates. Error bars represent S.E., and a significant (p value <0.005) change in editing level is denoted by the asterisk. C and D, the levels of mature (C) and precursor (D) GRIA2 mRNA and control GAPDH mRNA were determined by qRT-PCR for three independent biological replicates. The bar height represents the relative ratio of GRIA2 transcript to GAPDH transcripts normalized to the value obtained for the U87 cells transduced with a retrovirus containing a neomycin resistance vector with no protein (−). Error bars represent S.E. E, editing levels at the Q/R site of GRIA2 were measured in NHA cells transduced with retrovirus containing a neomycin resistance vector with no protein (Empty Vector) or human ADAR3 expressed from the CMV promoter (ADAR3 O/E) for two independent biological replicates. Error bars represent S.E., and a significant (p value <0.05) change in editing level is denoted by the asterisk.
The Ability to Bind dsRNA Is Required for ADAR3 to Regulate GRIA2 Editing
To determine the mechanism of action utilized by ADAR3 to inhibit RNA editing, mutations in the three known domains of ADAR3 were constructed (Fig. 3A). Although ADAR3 has not been observed to edit dsRNA in vitro (28), the region of ADAR3 with homology to the ADAR deaminase domain does contain the histidine-alanine-glutamate (HAE) motif that is known to be required for deaminase activity. As mutation of the glutamate in the HAE motif to alanine abolishes editing by other ADAR family members (58), the analogous mutation was made in ADAR3. A second domain of ADAR3 referred to as the R-domain (28) consists of a series of six arginine (R) residues. This domain is also present in an alternatively spliced exon of ADAR2 (59) and has been shown to be required for the ability of ADAR3 to bind single-stranded RNA in vitro (28). In the ADAR3 R-domain mutant construct, this series of arginines was mutated to six consecutive alanines. In the last construct, the KKXX(K/R) motif (where X is any amino acid) within both dsRNA binding domains of ADAR3 was mutated to EAXXA. Previous studies of ADAR3 demonstrated that mutation of these lysine residues to glutamate (E) and alanine (A) disrupts dsRNA binding in vitro (60). Western blotting of U87 cells transduced with retroviruses expressing the ADAR3 mutants driven by the CMV promoter resulted in similar expression levels as the U87 cells stably expressing wild-type ADAR3 driven by the CMV promoter (Fig. 3B). RNA editing at the Q/R site of GRIA2 in control and mutant ADAR3-expressing cells was measured for three independent biological replicates. Our results indicate that expressing ADAR3 with mutations in the deaminase domain and the R-domain had no effect on the ability of ADAR3 to inhibit GRIA2 editing (Fig. 3C). However, the ADAR3 construct with mutations in the dsRBDs was unable to inhibit editing of GRIA2 at the Q/R site (Fig. 3C). These findings indicate that the dsRBDs are required for repression of GRIA2 editing and that the deaminase and R-domains are not essential for ADAR3-mediated regulation of GRIA2 editing.
FIGURE 3.
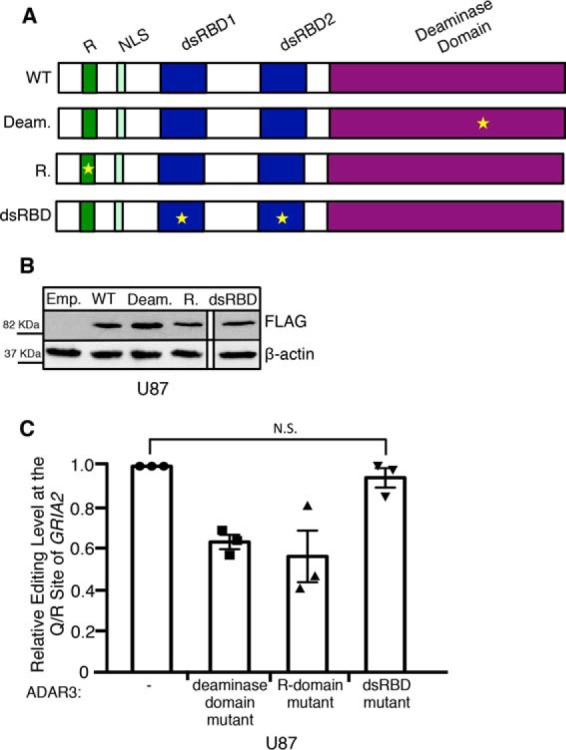
The dsRBDs of ADAR3 are required for inhibition of editing at the Q/R site of GRIA2. A, schematic of wild-type ADAR3 and ADAR3 with mutations in annotated domains. Regions annotated are the R-domain (R), nuclear localization signal (NLS), dsRBD1 and dsRBD2, and deaminase domain. Stars indicate the location of point mutation(s) in each mutant construct. B, Western blot of lysates from U87 cells expressing an integrated neomycin resistance vector with no protein (emp.) or expressing CMV-driven human wild-type ADAR3 (WT), ADAR3 with a point mutation to the deaminase domain (Deam.), ADAR3 with mutations to the R-domain (R.), or ADAR3 with mutations to the dsRBDs (dsRBD mutant lysate was run together on the same gel, but one lane is cropped out) subjected to SDS-PAGE and immunoblotting. All ADAR3 expression constructs contain an N-terminal FLAG tag and were detected with FLAG antibody. Actin is the loading control. C, editing levels at the Q/R site of GRIA2 were measured in the indicated cell lines for three independent biological replicates. The bar height represents the percentage of GRIA2 transcripts edited in each cell line normalized to U87 cells expressing a neomycin resistance vector with no protein (−). Error bars represent S.E. N.S. is not significant (p > 0.3), calculated using a two-tailed t test.
ADAR3 Binds to GRIA2 Pre-mRNA to Regulate Editing Levels
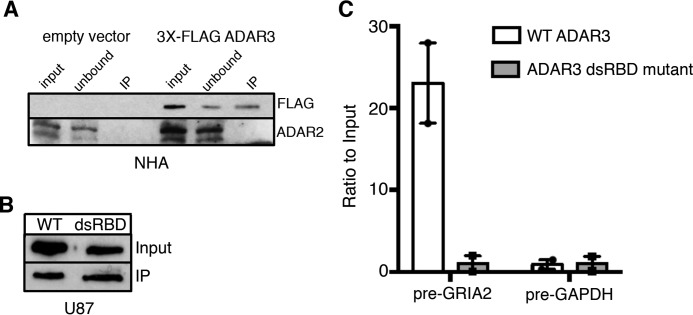
As mutations to the dsRBDs have been shown to disrupt ADAR3 binding to dsRNA in vitro (28), we hypothesized that the inability of the ADAR3 dsRBD mutant to repress GRIA2 editing was due to lack of binding to the dsRNA surrounding the Q/R site in vivo. However, studies of other dsRBPs have indicated roles for dsRBDs in mediating protein-protein interactions (61–63). Therefore, to discriminate between these possibilities, we first tested whether ADAR3 directly interacts with ADAR2 using an immunoprecipitation assay from NHA cells as these cells had the highest endogenous ADAR2 expression of the cell lines we examined (Fig. 1C) and ADAR3 inhibited GRIA2 editing in these cells (Fig. 2E). To facilitate immunoprecipitation, we expressed ADAR3 in NHA cells with an N-terminal 3X-FLAG epitope tag. After incubation of cell lysates with FLAG-conjugated magnetic resin, ADAR3 was detected in the immunoprecipitates from cells expressing the FLAG-ADAR3 protein but not the control cells (Fig. 4A). In contrast, although ADAR2 was expressed in both control and FLAG-ADAR3-expressing cells, ADAR2 was not present in immunoprecipitates from either cell line (Fig. 4A). This result is further supported by mass spectrometric analysis of the immunoprecipitates in which no peptides corresponding to ADAR2 were detected; however, peptides from multiple other proteins were detected in both biological replicates (supplemental Table S1). Therefore, our results suggest that ADAR3 does not directly interact with ADAR2.
FIGURE 4.
ADAR3 binds directly to GRIA2 transcripts. A, lysates from NHA cells transduced with a retrovirus containing a neomycin resistance vector with no protein (empty vector) or 3X-FLAG-tagged human ADAR3 expressed from the CMV promoter were subjected to incubation with α-FLAG magnetic beads. Input represents lysates before incubation with beads (0.5%, 3%:FLAG, ADAR2 blots), unbound represents lysates after immunoprecipitation (0.5%, 3%:FLAG, ADAR2 blots), and IP represents protein bound to FLAG beads (10%, 50%:FLAG, ADAR2 blots). Immunoblotting for the FLAG epitope (ADAR3) and ADAR2 was performed. B, Western blot of input lysates and immunoprecipitation lysates from 3X-FLAG-ADAR3 (WT) and 3X-FLAG-ADAR3 with mutations to the dsRBD. C, after treating immunoprecipitates from B with proteinase K, RNA was extracted and reverse transcribed with gene-specific primers. Bar heights indicate the ratio of cDNA in the immunoprecipitates over the inputs relative to input cDNA in U87 cells expressing wild-type ADAR3 and ADAR3 with mutations to the dsRNA binding domains normalized to the average of the two experiments (n = 2; error bars represent S.E.).
To test whether ADAR3 inhibits editing through direct binding to the GRIA2 transcript, an RNA immunoprecipitation (RIP) assay was performed. Briefly, U87 cells expressing 3X-FLAG-tagged ADAR3 or 3X-FLAG-tagged ADAR3 with mutations to the dsRBDs were UV-irradiated to cross-link protein with nucleic acids, and the resulting cell lysates were incubated with FLAG-conjugated magnetic resin. Western blotting confirmed equal immunoprecipitation of wild-type and dsRBD mutant ADAR3 (Fig. 4B). After immunoprecipitation, bound FLAG-ADAR3 proteins were degraded with proteinase K to release bound transcripts, and qRT-PCR was performed to determine relative enrichment of transcripts. As editing of the Q/R site occurs on pre-mRNA (64), qRT-PCR was performed for GRIA2 pre-mRNA and, as a negative control, pre-mRNA for the housekeeping gene GAPDH. Our findings indicate that GRIA2 pre-mRNA transcripts are significantly enriched in immunoprecipitates containing wild-type ADAR3 but not those containing the ADAR3 dsRBD mutant (Fig. 4C; p = 0.047), whereas the single-stranded control pre-mRNA encoding GAPDH is not enriched. These data suggest that ADAR3 inhibits editing of the Q/R site by binding to the GRIA2 pre-mRNA.
ADAR3 Is Overexpressed in GBM Patient Tumor Samples and Correlates with Decreased GRIA2 Editing
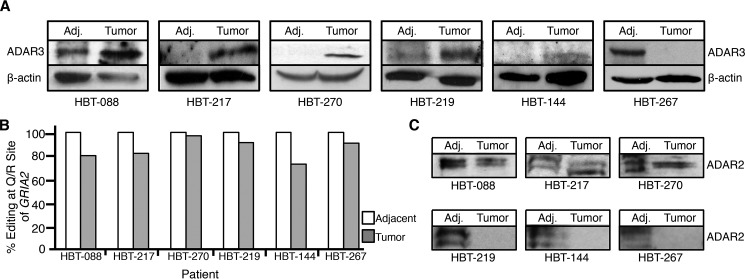
To investigate the relationship between GRIA2 editing and ADAR3 expression in the context of cancer, we obtained fresh frozen tumors and adjacent brain tissue from six patients suffering from grade IV, primary glioblastoma. In each of these tissue samples, ADAR3 expression was examined by Western blotting, and editing at the Q/R site of GRIA2 was measured as described above. Increased ADAR3 protein expression in the tumor tissue compared with adjacent tissue was observed in five of six patient samples (Fig. 5A and supplemental Fig. S1). Consistent with previous studies, all six sets of samples demonstrated 100% editing of the Q/R site in the adjacent normal tissue and reduced editing, ranging from 73 to 97%, in the glioblastoma tumor samples (Fig. 5B). These data suggest that overexpression of ADAR3 in glioblastoma tumors may contribute to decreased editing at the Q/R site of GRIA2.
FIGURE 5.
ADAR3 expression is elevated in a majority of glioblastoma patient tumors. A, lysates from glioblastoma tissue (Tumor) and matched adjacent brain tissue (Adj.) were subjected to SDS-PAGE and immunoblotting for ADAR3. Actin is the loading control. B, percentage of mRNAs edited at the Q/R site of GRIA2 in glioblastoma tissue and matched adjacent brain tissue from six glioblastoma patients. C, lysates from glioblastoma tissue (Tumor) and matched adjacent brain tissue (Adj.) were subjected to SDS-PAGE and immunoblotting for ADAR2.
Our data from the cell lines suggest that the level of editing at the Q/R site of GRIA2 is impacted by both the ADAR2 and ADAR3 protein expression levels (Fig. 1); therefore, the impact of ADAR3 expression on GRIA2 editing in tumor tissue needs to be assessed in tumors with little change in ADAR2 expression. To specifically address this concern, the protein expression of ADAR2 was also measured in each set of matched tumor and adjacent tissue (Fig. 5C and supplemental Fig. S1). ADAR2 expression remained constant between the adjacent and tumor tissue for half of the samples (Fig. 5C, top three blots), and decreased ADAR2 expression was observed in half of the samples (Fig. 5C, bottom three blots). The tumors that have decreased ADAR2 expression still exhibit relatively high editing levels (average 85%) at the Q/R site of GRIA2. As ADAR2 is required for high levels of editing of this site in vivo and even mice heterozygous for ADAR2 exhibit Q/R editing levels of ∼90% (41), we suggest that ADAR2 is expressed in these tumors but decreased relative to the adjacent tissue.
Focusing on the three tumors that exhibit increased ADAR3 expression and relatively constant ADAR2 expression, the average editing of the Q/R site of GRIA2 is 86% compared with 100% in the adjacent normal tissue. These results suggest that increased ADAR3 expression contributes to reduced GRIA2 editing in tumors of patients suffering from glioblastoma.
Discussion
RNA editing profoundly affects gene expression and cellular function, and alterations in editing have been implicated in cancer and neurological disease (11, 34, 65, 66). However, expression levels of the ADAR enzymes do not always correlate with the extent of editing (67, 68). In addition, the speed of RNA synthesis by RNA polymerase, the efficiency of pre-mRNA splicing, and the landscape of RNA-binding proteins can affect ADAR accessibility to target adenosines (54, 69–71). In this study, we explore the function of ADAR3, a dsRNA-binding but deaminase-deficient member of the ADAR protein family, and provide evidence for a regulatory role in inhibiting editing of GRIA2 transcripts. We dissect the mechanism of action for ADAR3 and determine that ADAR3 is able to inhibit RNA editing through direct binding to GRIA2 transcripts, which requires functional dsRNA binding domains in ADAR3. Furthermore, dysregulation of ADAR3 expression is observed in a majority of human glioblastoma patients, correlating with reduced GRIA2 editing in these tumors.
Our data suggest a competition model between ADAR2 and ADAR3 in which ADAR3 binding to GRIA2 prevents pre-mRNA binding by ADAR2 and subsequent editing activity. This RNA-mediated inhibition mechanism is in contrast to the previously reported role for ADAR1 in inhibition of editing of the Q/R site of GRIA2 via a protein-protein interaction of ADAR1 and ADAR2, which sequesters ADAR2 from the GRIA2 pre-mRNA (57). Our findings suggest that ADAR3 has the unique ability to regulate specific sites within the transcriptome rather than sequestering and inhibiting ADAR2 globally. Although ADAR1 and ADAR2 have also been shown to reciprocally inhibit each other's editing activity (28), this was tested on a transcript, the serotonin receptor mRNA, which has high affinity sites for both ADAR1 and ADAR2. The Q/R site of GRIA2 is unique in that ADAR1 is unable to bind or edit (14, 72), implicating the need for another form of regulation, which ADAR3 fulfills. Our preliminary data further support this specificity model in that after measuring RNA editing at a handful of other sites we were unable to identify any other transcripts where editing was affected upon ADAR3 expression (supplemental Fig. S2). However, future transcriptome-wide studies, in particular RIP-sequencing analysis of ADAR3, will be important for determining whether regulation by ADAR3 is specific to the GRIA2 transcript.
An alternative possibility is that ADAR3 binds to the GRIA2 transcript and facilitates splicing of the unedited pre-mRNA, which in turn prevents ADAR2 from editing. Early studies of ADAR2 editing of GRIA2 demonstrated that lack of Q/R editing resulted in accumulation of incompletely spliced transcripts (41). In support of a connection between splicing and editing, it has recently been shown for a number of mammalian mRNAs that splicing rates affect editing levels (50). However, in our hands, although we did detect a significant (40%) decrease in Q/R editing in the presence of ADAR3, this did not lead to an overall change in GRIA2 pre-mRNA or mature mRNA levels (Fig. 2, C and D).
It is notable that as most of the key residues in the deaminase domain are conserved among the human ADARs, ADAR3 was originally proposed to possess deaminase activity but have an unknown substrate specificity (73). Our data indicate that the deaminase domain does not function in ADAR3-mediated regulation of GRIA2 editing, raising questions about the functional significance of the domain. Interestingly, a recent structural study identified a key arginine residue in the RNA binding loop of the deaminase domain that is conserved in ADAR2 and ADAR1 but is a glutamine in ADAR3 (74). As mutation of this arginine to glutamine in human ADAR2 reduced editing activity 10-fold, these data provide an explanation for the lack of deamination activity observed for ADAR3. Despite lacking RNA editing activity, the regulatory role of deaminase-deficient ADARs is conserved in other organisms. The deaminase-deficient ADAR family member in Caenorhabditis elegans, ADR-1, also alters editing levels and has most recently been shown to inhibit RNA editing in the neurons of worms (75, 76).
In addition to the mechanistic implications of ADAR3 as a dsRBP that regulates editing, our data raise interesting questions about the physiological function of ADAR3 as both edited and unedited GRIA2 subunits are observed in the developing brain (77). Interestingly, ADAR3 is specifically expressed in the brain and central nervous system (16, 28) and may act as one of many brain-specific regulators of editing. Recently, several RBPs including SRFS9 and RPS14 were shown to alter editing of specific target mRNAs, and expression of these RBPs changed during embryonic development as well as upon neuronal stimulation (54). An interesting possibility is that ADAR3 expression varies during development or in specific brain regions and results in differential GRIA2 editing in the brain. In fact, recent studies have demonstrated that a rapid increase in Q/R editing is associated with human neural progenitor cells differentiating into neurons (78, 79), and another study demonstrated differential editing of GRIA2 at the Q/R site in different regions of the human brain (80), suggesting a need for both temporal and local regulation of GRIA2 editing in the nervous system. A preliminary study in our laboratory suggests that ADAR3 expression varies during normal development in the mouse brain cortex.3 Perhaps ADAR3 expression during development may provide an important form of substrate-specific regulation of RNA editing in the brain. Interestingly, ADAR3 has recently been implicated in the neurodegenerative disorder ALS, a disease that has long been known to be associated with reduced GRIA2 editing (81). Donnelly et al. (82) determined that ADAR3 was sequestered in long dsRNA repeats in the nuclei of ALS patients. Furthermore, upon knockdown of ADAR3, neuronally induced pluripotent stem cells were unable to survive in the presence of glutamate. These data, in combination with our finding that ADAR3 inhibits editing of the glutamate receptor in astrocytes, suggest that ADAR3 may play a critical role in regulating neuronal RNA editing and may be a major contributor to ALS physiology.
To our knowledge, this study is the first to analyze ADAR3 and ADAR2 protein expression levels in conjunction with Q/R editing in primary grade IV astrocytoma tumors and matched adjacent normal brain tissue from the same patient. Our study found that Q/R editing was reduced in all tumors compared with adjacent brain tissue. As recent studies have reported that editing at the Q/R site of GRIA2 varies depending on the region of the brain (44), we wanted to compare the editing levels in the tumors in conjunction with normal adjacent brain tissue. In all of the adjacent tissues sampled, editing at the Q/R site was 100%; therefore, we can report with high confidence that decreased editing in a majority of our tumor samples is not due to differences in editing in different brain regions.
Additionally, as we detected increased expression of ADAR3 in glioblastoma tumors compared with adjacent tissue, our findings raise a critical question regarding whether ADAR3 might causally contribute to oncogenesis. Throughout the course of our studies, we have consistently found that ADAR3 mRNA expression does not correlate with protein expression except when ADAR3 is expressed from a transgenic cDNA construct. Interestingly, the human genome expresses an antisense RNA complementary to an intronic region of ADAR3, suggesting the possibility that ADAR3 protein expression may be controlled post-transcriptionally. This confounding factor may be why ADAR3 has not been identified as an important oncogene in transcriptome-wide studies in cancers, such as The Cancer Genome Atlas, as these data are often based on differential mRNA expression. Future studies investigating the potential role of ADAR3 in oncogenesis might uncover an interesting and currently understudied player in glioblastoma progression.
Experimental Procedures
Cells and Viral Infection
U118, U87-MG, NHA, and HEK293T Epstein-Barr nuclear antigen cells were maintained in Dulbecco's modified Eagle's medium (DMEM) (Mediatech) supplemented with 10% fetal bovine serum (FBS) (Sigma), 100 μg/ml penicillin, and 100 units/ml streptomycin (Mediatech). Overexpression of ADAR3 or ADAR2 was performed using a retroviral infection with retroviruses produced in the laboratory. Briefly, for retroviral production, the overexpression plasmid of interest was co-transfected with plasmids expressing vesicular stomatitis virus-G glycoprotein and gag/pol packaging plasmids into HEK293T Epstein-Barr nuclear antigen cells using Trans-IT LT1 (Mirus) to generate retroviral particles. The retroviral particles were collected in DMEM, 10% FBS, 100 μg/ml penicillin, 100 units/ml streptomycin, and 10 mm HEPES. For viral transduction, the retroviral particles were added to cells of interest along with 8 μg/ml Polybrene (Sigma). After 24 h of incubation with the viral particles, fresh medium with the selection drug (either puromycin or neomycin/G418) was added. Cell selection was continued over 1 week and monitored by cell death of untransduced cells that had the same amount of selection drug in the medium. After selection, cells were maintained in the presence of the same amount of selection drug. Overexpression was confirmed by Western blotting using antibodies to ADAR2 (Sigma) or FLAG antibody (Sigma).
Tumor and Control Tissues
We included nine tissue samples from adult patients (ages 39–70) with primary grade IV glioblastoma. Malignancy grade (according to the World Health Organization guidelines) of all tumor tissue samples was confirmed by pathology. Control samples of normal tissue were dissected adjacent to the tumor during surgery. All tissue samples were flash frozen and stored by the Indiana University Simon Cancer Center Tissue Bank. Approval for work with the samples was obtained through the Indiana University Institutional Review Board (IRB number 1204008446).
Construction of ADAR3 and ADAR2 Expression Plasmids
ADAR3 with a 1X- or 3X-FLAG tag added in-frame at the N terminus were PCR-amplified from a plasmid obtained from Dr. Kazuko Nishikura (Wistar Institute) and cloned in to the pLNCX2 retroviral vector. Mutations to the R-domain, double-stranded RNA binding domain, and deaminase domain were created using site-directed mutagenesis. In the R-domain, six consecutive arginines were mutated to alanine. In the dsRBDs, the KKXXK motifs were mutated to EAXXA. In the deaminase domain, the HAE motif was mutated to HAA. The mutations at each site were confirmed by direct sequencing. ADAR2 with a His tag at the N terminus was cloned into the pLNCX2 retroviral vector.
Western Analysis
To determine protein expression of ADAR1, ADAR2, and ADAR3 in cultured cell lines, cells were suspended in 4% sodium dodecyl sulfate (SDS) and sonicated. Protein concentrations were determined by Bradford assay (Sigma), and equivalent amounts of lysate were subjected to SDS-PAGE and Western blotting with antibodies to ADAR3 (Santa Cruz Biotechnology), ADAR1 (a kind gift from Brenda Bass), ADAR2 (Sigma), tubulin (Sigma), and β-actin (Cell Signaling Technology). Specificity of ADAR2 and ADAR3 antibodies was confirmed by detection of bands with a similar mobility in cells overexpressing cDNA for each protein.
RNA Isolation, Editing Assays, and qRT-PCR
Total RNA was isolated from cell pellets using TRIzol (Invitrogen). RNA was purified by treatment with TURBO DNase (Ambion) and isolated using the RNeasy Extraction kit (Qiagen). cDNA was synthesized from 2 μg of DNase-treated total RNA with SuperscriptIII reverse transcriptase (Invitrogen). For GRIA2 editing assays, a gene-specific primer (5′-CAAGGATGTAGAATACTCCAGCAACG-3′) was used for reverse transcription of RNA. Resulting cDNA was amplified using Pfx Platinum DNA polymerase (Invitrogen) using nested PCR. Oligonucleotides were as follows: outer primers, 5′-CCTTTAGCCTATGAGATCTGGATGTGC-3′ and 5′-CAAGGATGTAGAATACTCCAGCAACG-3′; inner primers, 5′-GGCACACTGAGGAGTTTGAAGATGGAAGAG-3′ and 5′-CAGAATTCGTGTAGGAGGAGATTATGATCAGG-3′. PCR primers were designed to specifically amplify mRNA but not genomic DNA or pre-mRNA. PCR products were gel-purified and subjected to Sanger sequencing. For all editing assays, negative controls were conducted without SuperscriptIII to ensure that all DNA sequenced resulted from cDNA amplification. For qRT-PCR analysis, either gene-specific primers (GRIA2 mature mRNA, 5′-CAAGGATGTAGAATACTCCAGCAACG-3′; GRIA2 pre-mRNA, 5′-CTCTCCCATACCATTTCCATGAACTAATGA-3′) or a mixture of random hexamer primers and oligo(dT) were used to synthesize cDNA. cDNA levels were measured in an Eppendorf Realplex instrument using KAPA SYBR Fast Universal Master Mix. qRT-PCR primers used for analysis of GRIA2 mature mRNA were 5′-GGCTGCAGAAATCGCCAAACATTGTGGGTT-3′ and 5′-CATGATAGATATCCCGAGGCTCATGAAGGG-3′. qRT-PCR primers for analysis of GRIA2 pre-mRNA were 5′-CACCATGACTCCAGGTACTATTACTTTCCT-3′ and 5′-CATGATAGATATCCCGAGGCTCATGAAGGG-3′. Significance was calculated using an unpaired t test.
RIP Assay
After washing with PBS (137 mm NaCl, 2.7 mm KCl, 10 mm Na2HPO4, and 0.44 g of KH2PO4), cells adherent to a plate were covered with a thin layer of PBS and subjected to 150 mJ/cm2 UV radiation using a Spectrolinker (Spectronic Corp). Cells were then resuspended in hypotonic solution (20 mm Tris-HCl, pH 7.5, 15 mm NaCl, 10 mm EDTA, 0.5% Nonidet P-40, and 0.1% Triton X-100), incubated for 10 min on ice, and then lightly sonicated to lyse cells. The crude lysate was subjected to centrifugation to remove insoluble material, and the cleared lysates were added to FLAG magnetic beads (Sigma). After incubation for 1 h at 4 °C, the beads were washed with ice-cold hypotonic solution and resuspended in TBS (16 mm Tris-HCl, pH 7.5, and 110 mm NaCl), 1 μl of RNasin (Promega), and 0.5 μl of 20 mg/ml proteinase K (Sigma) and incubated at 42 °C for 15 min to degrade protein and release RNA. Protein samples were subjected to Western blotting (as described above), and RNA samples were isolated (as described above). Following DNase treatment, qRT-PCR for GAPDH and GRIA2 pre-mRNA was preformed as described above. Significance was calculated using an unpaired t test.
Tissue Homogenization
Flash frozen patient tumor samples and adjacent normal tissue were placed in TRIzol (Sigma) and ground in a mortar and pestle on dry ice. Following the manufacturer's protocol, RNA, genomic DNA, and protein were isolated from each sample. RNA editing assays for GRIA2 and Western blotting for the ADARs were performed as described above.
Author Contributions
E. O. and H. A. H. designed the experiments and wrote the manuscript. E. O. performed the majority of the experimental work. A. A. performed qRT-PCR experiments and experiments in the NHA cells. H. A. H. performed glioblastoma patient tissue homogenization and Western blotting. A. C.-G. provided glioblastoma patient tissues. All authors approved the manuscript.
Supplementary Material
Acknowledgments
We thank Angela Gallo (Bambino Gesù Children's Hospital, Italy) and Russell Pieper (University of California, San Francisco) for the U118 and NHA cell lines, respectively. We thank Peter Hollenhorst (Indiana University School of Medicine) for the retroviral packaging plasmids, pLNCX2 expression plasmid, and U87-MG cell line. We thank Kazuko Nishikura (Wistar Institute) for the ADAR3 cDNA plasmid that was used to clone the ADAR3 expression plasmids in this study. We thank Hundley laboratory members Nick McCrory for cloning the ADAR2 retroviral expression construct and Aidan Manning for technical assistance. We thank the Indiana University School of Medicine Proteomics Core Facility for performing the mass spectrometric analysis of ADAR3 and control immunoprecipitates.
This work was supported by American Cancer Society Institutional Research Grant IRG-84-002-28 (to H. A. H.) and American Cancer Society Research Scholar Award RSG-15-051-RMC (to H. A. H.). The authors declare that they have no conflicts of interest with the contents of this article.

This article contains supplemental Figs. S1 and S2 and Table S1.
E. Oakes and H. A. Hundley, unpublished results.
- ADAR
- adenosine deaminase acting on RNA
- dsRNA
- double-stranded RNA
- dsRBD
- dsRNA binding domain
- dsRBP
- dsRNA-binding protein
- GRIA2
- glutamate receptor ionotropic AMPA 2
- qRT-PCR
- quantitative real time PCR
- ALS
- amyotrophic lateral sclerosis
- NHA
- normal human astrocyte
- RIP
- RNA immunoprecipitation.
References
- 1. Nishikura K. (2010) Functions and regulation of RNA editing by ADAR deaminases. Annu. Rev. Biochem. 79, 321–349 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Yang Y., Zhou X., and Jin Y. (2013) ADAR-mediated RNA editing in non-coding RNA sequences. Sci. China Life Sci. 56, 944–952 [DOI] [PubMed] [Google Scholar]
- 3. Savva Y. A., Rieder L. E., and Reenan R. A. (2012) The ADAR protein family. Genome Biol. 13, 252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Bass B. L. (2002) RNA editing by adenosine deaminases that act on RNA. Annu. Rev. Biochem. 71, 817–846 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Pullirsch D., and Jantsch M. F. (2010) Proteome diversification by adenosine to inosine RNA editing. RNA Biol. 7, 205–212 [DOI] [PubMed] [Google Scholar]
- 6. Samuel C. E. (2011) Adenosine deaminases acting on RNA (ADARs) are both antiviral and proviral. Virology 411, 180–193 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Hundley H. A., and Bass B. L. (2010) ADAR editing in double-stranded UTRs and other noncoding RNA sequences. Trends Biochem. Sci. 35, 377–383 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Rosenthal J. J. (2015) The emerging role of RNA editing in plasticity. J. Exp. Biol. 218, 1812–1821 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Nishikura K. (2016) A-to-I editing of coding and non-coding RNAs by ADARs. Nat. Rev. Mol. Cell Biol. 17, 83–96 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Behm M., and Öhman M. (2016) RNA editing: a contributor to neuronal dynamics in the mammalian brain. Trends Genet. 32, 165–175 [DOI] [PubMed] [Google Scholar]
- 11. Tariq A., and Jantsch M. F. (2012) Transcript diversification in the nervous system: a to I RNA editing in CNS function and disease development. Front. Neurosci. 6, 99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Rosenthal J. J., and Seeburg P. H. (2012) A-to-I RNA editing: effects on proteins key to neural excitability. Neuron 74, 432–439 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Kim U., Wang Y., Sanford T., Zeng Y., and Nishikura K. (1994) Molecular cloning of cDNA for double-stranded RNA adenosine deaminase, a candidate enzyme for nuclear RNA editing. Proc. Natl. Acad. Sci. U.S.A. 91, 11457–11461 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Melcher T., Maas S., Herb A., Sprengel R., Seeburg P. H., and Higuchi M. (1996) A mammalian RNA editing enzyme. Nature 379, 460–464 [DOI] [PubMed] [Google Scholar]
- 15. O'Connell M. A., Krause S., Higuchi M., Hsuan J. J., Totty N. F., Jenny A., and Keller W. (1995) Cloning of cDNAs encoding mammalian double-stranded RNA-specific adenosine deaminase. Mol. Cell. Biol. 15, 1389–1397 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Melcher T., Maas S., Herb A., Sprengel R., Higuchi M., and Seeburg P. H. (1996) RED2, a brain-specific member of the RNA-specific adenosine deaminase family. J. Biol. Chem. 271, 31795–31798 [DOI] [PubMed] [Google Scholar]
- 17. Schumacher J. M., Lee K., Edelhoff S., and Braun R. E. (1995) Distribution of Tenr, an RNA-binding protein, in a lattice-like network within the spermatid nucleus in the mouse. Biol. Reprod. 52, 1274–1283 [DOI] [PubMed] [Google Scholar]
- 18. Goodman R. A., Macbeth M. R., and Beal P. A. (2012) ADAR proteins: structure and catalytic mechanism. Curr. Top. Microbiol. Immunol. 353, 1–33 [DOI] [PubMed] [Google Scholar]
- 19. Mannion N., Arieti F., Gallo A., Keegan L. P., and O'Connell M. A. (2015) New insights into the biological role of mammalian ADARs; the RNA editing proteins. Biomolecules 5, 2338–2362 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Barraud P., and Allain F. H. (2012) ADAR proteins: double-stranded RNA and Z-DNA binding domains. Curr. Top. Microbiol. Immunol. 353, 35–60 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Bazak L., Haviv A., Barak M., Jacob-Hirsch J., Deng P., Zhang R., Isaacs F. J., Rechavi G., Li J. B., Eisenberg E., and Levanon E. Y. (2014) A-to-I RNA editing occurs at over a hundred million genomic sites, located in a majority of human genes. Genome Res. 24, 365–376 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Vesely C., Tauber S., Sedlazeck F. J., Tajaddod M., von Haeseler A., and Jantsch M. F. (2014) ADAR2 induces reproducible changes in sequence and abundance of mature microRNAs in the mouse brain. Nucleic Acids Res. 42, 12155–12168 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Ekdahl Y., Farahani H. S., Behm M., Lagergren J., and Öhman M. (2012) A-to-I editing of microRNAs in the mammalian brain increases during development. Genome Res. 22, 1477–1487 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Alon S., Mor E., Vigneault F., Church G. M., Locatelli F., Galeano F., Gallo A., Shomron N., and Eisenberg E. (2012) Systematic identification of edited microRNAs in the human brain. Genome Res. 22, 1533–1540 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Wang I. X., So E., Devlin J. L., Zhao Y., Wu M., and Cheung V. G. (2013) ADAR regulates RNA editing, transcript stability, and gene expression. Cell Rep. 5, 849–860 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Bahn J. H., Ahn J., Lin X., Zhang Q., Lee J. H., Civelek M., and Xiao X. (2015) Genomic analysis of ADAR1 binding and its involvement in multiple RNA processing pathways. Nat. Commun. 6, 6355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Ohlson J., Ensterö M., Sjöberg B. M., and Ohman M. (2005) A method to find tissue-specific novel sites of selective adenosine deamination. Nucleic Acids Res. 33, e167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Chen C. X., Cho D. S., Wang Q., Lai F., Carter K. C., and Nishikura K. (2000) A third member of the RNA-specific adenosine deaminase gene family, ADAR3, contains both single- and double-stranded RNA binding domains. RNA 6, 755–767 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Patterson J. B., and Samuel C. E. (1995) Expression and regulation by interferon of a double-stranded-RNA-specific adenosine deaminase from human cells: evidence for two forms of the deaminase. Mol. Cell. Biol. 15, 5376–5388 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Sansam C. L., Wells K. S., and Emeson R. B. (2003) Modulation of RNA editing by functional nucleolar sequestration of ADAR2. Proc. Natl. Acad. Sci. U.S.A. 100, 14018–14023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Desterro J. M., Keegan L. P., Lafarga M., Berciano M. T., O'Connell M., and Carmo-Fonseca M. (2003) Dynamic association of RNA-editing enzymes with the nucleolus. J. Cell Sci. 116, 1805–1818 [DOI] [PubMed] [Google Scholar]
- 32. Zipeto M. A., Jiang Q., Melese E., and Jamieson C. H. (2015) RNA rewriting, recoding, and rewiring in human disease. Trends Mol. Med. 21, 549–559 [DOI] [PubMed] [Google Scholar]
- 33. Slotkin W., and Nishikura K. (2013) Adenosine-to-inosine RNA editing and human disease. Genome Med. 5, 105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Li J. B., and Church G. M. (2013) Deciphering the functions and regulation of brain-enriched A-to-I RNA editing. Nat. Neurosci. 16, 1518–1522 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Barbon A., and Barlati S. (2011) Glutamate receptor RNA editing in health and disease. Biochemistry 76, 882–889 [DOI] [PubMed] [Google Scholar]
- 36. Yang J. H., Sklar P., Axel R., and Maniatis T. (1997) Purification and characterization of a human RNA adenosine deaminase for glutamate receptor B pre-mRNA editing. Proc. Natl. Acad. Sci. U.S.A. 94, 4354–4359 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Maas S., Melcher T., Herb A., Seeburg P. H., Keller W., Krause S., Higuchi M., and O'Connell M. A. (1996) Structural requirements for RNA editing in glutamate receptor pre-mRNAs by recombinant double-stranded RNA adenosine deaminase. J. Biol. Chem. 271, 12221–12226 [DOI] [PubMed] [Google Scholar]
- 38. Wright A., and Vissel B. (2012) The essential role of AMPA receptor GluR2 subunit RNA editing in the normal and diseased brain. Front. Mol. Neurosci. 5, 34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Yamashita T., Tadami C., Nishimoto Y., Hideyama T., Kimura D., Suzuki T., and Kwak S. (2012) RNA editing of the Q/R site of GluA2 in different cultured cell lines that constitutively express different levels of RNA editing enzyme ADAR2. Neurosci. Res. 73, 42–48 [DOI] [PubMed] [Google Scholar]
- 40. Brusa R., Zimmermann F., Koh D. S., Feldmeyer D., Gass P., Seeburg P. H., and Sprengel R. (1995) Early-onset epilepsy and postnatal lethality associated with an editing-deficient GluR-B allele in mice. Science 270, 1677–1680 [DOI] [PubMed] [Google Scholar]
- 41. Higuchi M., Maas S., Single F. N., Hartner J., Rozov A., Burnashev N., Feldmeyer D., Sprengel R., and Seeburg P. H. (2000) Point mutation in an AMPA receptor gene rescues lethality in mice deficient in the RNA-editing enzyme ADAR2. Nature 406, 78–81 [DOI] [PubMed] [Google Scholar]
- 42. Feldmeyer D., Kask K., Brusa R., Kornau H. C., Kolhekar R., Rozov A., Burnashev N., Jensen V., Hvalby O., Sprengel R., and Seeburg P. H. (1999) Neurological dysfunctions in mice expressing different levels of the Q/R site-unedited AMPAR subunit GluR-B. Nat. Neurosci. 2, 57–64 [DOI] [PubMed] [Google Scholar]
- 43. Maas S., Kawahara Y., Tamburro K. M., and Nishikura K. (2006) A-to-I RNA editing and human disease. RNA Biol. 3, 1–9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Kwak S., and Kawahara Y. (2005) Deficient RNA editing of GluR2 and neuronal death in amyotropic lateral sclerosis. J. Mol. Med. 83, 110–120 [DOI] [PubMed] [Google Scholar]
- 45. Maas S., Patt S., Schrey M., and Rich A. (2001) Underediting of glutamate receptor GluR-B mRNA in malignant gliomas. Proc. Natl. Acad. Sci. U.S.A. 98, 14687–14692 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Ishiuchi S., Tsuzuki K., Yoshida Y., Yamada N., Hagimura N., Okado H., Miwa A., Kurihara H., Nakazato Y., Tamura M., Sasaki T., and Ozawa S. (2002) Blockage of Ca2+-permeable AMPA receptors suppresses migration and induces apoptosis in human glioblastoma cells. Nat. Med. 8, 971–978 [DOI] [PubMed] [Google Scholar]
- 47. Galeano F., Rossetti C., Tomaselli S., Cifaldi L., Lezzerini M., Pezzullo M., Boldrini R., Massimi L., Di Rocco C. M., Locatelli F., and Gallo A. (2013) ADAR2-editing activity inhibits glioblastoma growth through the modulation of the CDC14B/Skp2/p21/p27 axis. Oncogene 32, 998–1009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Deffit S. N., and Hundley H. A. (2016) To edit or not to edit: regulation of ADAR editing specificity and efficiency. Wiley Interdiscip. Rev. RNA 7, 113–127 [DOI] [PubMed] [Google Scholar]
- 49. Hong H., Lin J. S., and Chen L. (2015) Regulatory factors governing adenosine-to-inosine (A-to-I) RNA editing. Biosci. Rep. 35, e00182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Licht K., and Jantsch M. F. (2016) Rapid and dynamic transcriptome regulation by RNA editing and RNA modifications. J. Cell Biol. 213, 15–22 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Lai F., Chen C. X., Carter K. C., and Nishikura K. (1997) Editing of glutamate receptor B subunit ion channel RNAs by four alternatively spliced DRADA2 double-stranded RNA adenosine deaminases. Mol. Cell. Biol. 17, 2413–2424 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Rueter S. M., Dawson T. R., and Emeson R. B. (1999) Regulation of alternative splicing by RNA editing. Nature 399, 75–80 [DOI] [PubMed] [Google Scholar]
- 53. Marcucci R., Brindle J., Paro S., Casadio A., Hempel S., Morrice N., Bisso A., Keegan L. P., Del Sal G., and O'Connell M. A. (2011) Pin1 and WWP2 regulate GluR2 Q/R site RNA editing by ADAR2 with opposing effects. EMBO J. 30, 4211–4222 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Tariq A., Garncarz W., Handl C., Balik A., Pusch O., and Jantsch M. F. (2013) RNA-interacting proteins act as site-specific repressors of ADAR2-mediated RNA editing and fluctuate upon neuronal stimulation. Nucleic Acids Res. 41, 2581–2593 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Garncarz W., Tariq A., Handl C., Pusch O., and Jantsch M. F. (2013) A high-throughput screen to identify enhancers of ADAR-mediated RNA-editing. RNA Biol. 10, 192–204 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Sonoda Y., Ozawa T., Hirose Y., Aldape K. D., McMahon M., Berger M. S., and Pieper R. O. (2001) Formation of intracranial tumors by genetically modified human astrocytes defines four pathways critical in the development of human anaplastic astrocytoma. Cancer Res. 61, 4956–4960 [PubMed] [Google Scholar]
- 57. Cenci C., Barzotti R., Galeano F., Corbelli S., Rota R., Massimi L., Di Rocco C., O'Connell M. A., and Gallo A. (2008) Down-regulation of RNA editing in pediatric astrocytomas: ADAR2 editing activity inhibits cell migration and proliferation. J. Biol. Chem. 283, 7251–7260 [DOI] [PubMed] [Google Scholar]
- 58. Liu Y., and Samuel C. E. (1996) Mechanism of interferon action: functionally distinct RNA-binding and catalytic domains in the interferon-inducible, double-stranded RNA-specific adenosine deaminase. J. Virol. 70, 1961–1968 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Maas S., and Gommans W. M. (2009) Novel exon of mammalian ADAR2 extends open reading frame. PLoS One 4, e4225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Valente L., and Nishikura K. (2007) RNA binding-independent dimerization of adenosine deaminases acting on RNA and dominant negative effects of nonfunctional subunits on dimer functions. J. Biol. Chem. 282, 16054–16061 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Hitti E. G., Sallacz N. B., Schoft V. K., and Jantsch M. F. (2004) Oligomerization activity of a double-stranded RNA-binding domain. FEBS Lett. 574, 25–30 [DOI] [PubMed] [Google Scholar]
- 62. Ota H., Sakurai M., Gupta R., Valente L., Wulff B. E., Ariyoshi K., Iizasa H., Davuluri R. V., and Nishikura K. (2013) ADAR1 forms a complex with Dicer to promote microRNA processing and RNA-induced gene silencing. Cell 153, 575–589 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Gleghorn M. L., Gong C., Kielkopf C. L., and Maquat L. E. (2013) Staufen1 dimerizes through a conserved motif and a degenerate dsRNA-binding domain to promote mRNA decay. Nat. Struct. Mol. Biol. 20, 515–524 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Melcher T., Maas S., Higuchi M., Keller W., and Seeburg P. H. (1995) Editing of α-amino-3-hydroxy-5-methylisoxazole-4-propionic acid receptor GluR-B pre-mRNA in vitro reveals site-selective adenosine to inosine conversion. J. Biol. Chem. 270, 8566–8570 [DOI] [PubMed] [Google Scholar]
- 65. Galeano F., Tomaselli S., Locatelli F., and Gallo A. (2012) A-to-I RNA editing: the “ADAR” side of human cancer. Semin. Cell Dev. Biol. 23, 244–250 [DOI] [PubMed] [Google Scholar]
- 66. Dominissini D., Moshitch-Moshkovitz S., Amariglio N., and Rechavi G. (2011) Adenosine-to-inosine RNA editing meets cancer. Carcinogenesis 32, 1569–1577 [DOI] [PubMed] [Google Scholar]
- 67. Shtrichman R., Germanguz I., Mandel R., Ziskind A., Nahor I., Safran M., Osenberg S., Sherf O., Rechavi G., and Itskovitz-Eldor J. (2012) Altered A-to-I RNA editing in human embryogenesis. PLoS One 7, e41576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Wahlstedt H., Daniel C., Ensterö M., and Ohman M. (2009) Large-scale mRNA sequencing determines global regulation of RNA editing during brain development. Genome Res. 19, 978–986 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Rieder L. E., Staber C. J., Hoopengardner B., and Reenan R. A. (2013) Tertiary structural elements determine the extent and specificity of messenger RNA editing. Nat. Commun. 4, 2232. [DOI] [PubMed] [Google Scholar]
- 70. Licht K., Kapoor U., Mayrhofer E., and Jantsch M. F. (2016) Adenosine to inosine editing frequency controlled by splicing efficiency. Nucleic Acids Res. 44, 6398–6408 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Ryman K., Fong N., Bratt E., Bentley D. L., and Ohman M. (2007) The C-terminal domain of RNA Pol II helps ensure that editing precedes splicing of the GluR-B transcript. RNA 13, 1071–1078 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Wang Q., Miyakoda M., Yang W., Khillan J., Stachura D. L., Weiss M. J., and Nishikura K. (2004) Stress-induced apoptosis associated with null mutation of ADAR1 RNA editing deaminase gene. J. Biol. Chem. 279, 4952–4961 [DOI] [PubMed] [Google Scholar]
- 73. Keegan L. P., Leroy A., Sproul D., and O'Connell M. A. (2004) Adenosine deaminases acting on RNA (ADARs): RNA-editing enzymes. Genome Biol. 5, 209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Matthews M. M., Thomas J. M., Zheng Y., Tran K., Phelps K. J., Scott A. I., Havel J., Fisher A. J., and Beal P. A. (2016) Structures of human ADAR2 bound to dsRNA reveal base-flipping mechanism and basis for site selectivity. Nat. Struct. Mol. Biol. 23, 426–433 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Washburn M. C., and Hundley H. A. (2016) Trans and cis factors affecting A-to-I RNA editing efficiency of a noncoding editing target in C. elegans. RNA 22, 722–728 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Washburn M. C., Kakaradov B., Sundararaman B., Wheeler E., Hoon S., Yeo G. W., and Hundley H. A. (2014) The dsRBP and inactive editor ADR-1 utilizes dsRNA binding to regulate A-to-I RNA editing across the C. elegans transcriptome. Cell Rep. 6, 599–607 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Burnashev N., Monyer H., Seeburg P. H., and Sakmann B. (1992) Divalent ion permeability of AMPA receptor channels is dominated by the edited form of a single subunit. Neuron 8, 189–198 [DOI] [PubMed] [Google Scholar]
- 78. Whitney N. P., Peng H., Erdmann N. B., Tian C., Monaghan D. T., and Zheng J. C. (2008) Calcium-permeable AMPA receptors containing Q/R-unedited GluR2 direct human neural progenitor cell differentiation to neurons. FASEB J. 22, 2888–2900 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Pachernegg S., Münster Y., Muth-Köhne E., Fuhrmann G., and Hollmann M. (2015) GluA2 is rapidly edited at the Q/R site during neural differentiation in vitro. Front. Cell. Neurosci. 9, 69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Kawahara Y., Ito K., Sun H., Kanazawa I., and Kwak S. (2003) Low editing efficiency of GluR2 mRNA is associated with a low relative abundance of ADAR2 mRNA in white matter of normal human brain. Eur. J. Neurosci. 18, 23–33 [DOI] [PubMed] [Google Scholar]
- 81. Kwak S., Hideyama T., Yamashita T., and Aizawa H. (2010) AMPA receptor-mediated neuronal death in sporadic ALS. Neuropathology 30, 182–188 [DOI] [PubMed] [Google Scholar]
- 82. Donnelly C. J., Zhang P. W., Pham J. T., Haeusler A. R., Mistry N. A., Vidensky S., Daley E. L., Poth E. M., Hoover B., Fines D. M., Maragakis N., Tienari P. J., Petrucelli L., Traynor B. J., Wang J., et al. (2013) RNA toxicity from the ALS/FTD C9ORF72 expansion is mitigated by antisense intervention. Neuron 80, 415–428 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.