Abstract
Human RecQ-like helicase 4 (RECQL4) plays crucial roles in replication initiation and DNA repair; however, the contextual regulation of its unwinding activity is not fully described. Mutations in RECQL4 have been linked to three diseases including Rothmund-Thomson syndrome, which is characterized by osteoskeletal deformities, photosensitivity, and increased osteosarcoma susceptibility. Understanding regulation of RECQL4 helicase activity by interaction partners will allow deciphering its role as an enzyme and a signaling cofactor in different cellular contexts. We became interested in studying the interaction of RECQL4 with ribosomal protein S3 (RPS3) because previous studies have shown that RPS3 activity is sometimes associated with phenotypes mimicking those of mutated RECQL4. RPS3 is a small ribosomal protein that also has extraribosomal functions, including apurnic-apyrimidinic endonuclease-like activity suggested to be important during DNA repair. Here, we report a functional and physical interaction between RPS3 and RECQL4 and show that this interaction may be enhanced during cellular stress. We show that RPS3 inhibits ATPase, DNA binding, and helicase activities of RECQL4 through their direct interaction. Further domain analysis shows that N-terminal 1–320 amino acids of RECQL4 directly interact with the C-terminal 94–244 amino acids of RPS3 (C-RPS3). Biochemical analysis of C-RPS3 revealed that it comprises a standalone apurnic-apyrimidinic endonuclease-like domain. We used U2OS cells to show that oxidative stress and UV exposure could enhance the interaction between nuclear RPS3 and RECQL4. Regulation of RECQL4 biochemical activities by RPS3 along with nuclear interaction during UV and oxidative stress may serve to modulate active DNA repair.
Keywords: base excision repair (BER), DNA helicase, DNA repair, genomic instability, oxidative stress, protein-protein interaction
Introduction
The RecQ class of helicases function at various stages of DNA metabolism. The presence of at least one homolog in each species evidences their crucial role in maintaining genomic stability. In humans, three of five RecQ helicases have been associated to different genetic disorders. WRN and BLM have been linked with Werner's syndrome and Bloom's syndrome, respectively, and RECQL4 has been linked with Rothmund-Thomson syndrome (RTS),2 Baller-Gerald syndrome, and RAPADILINO syndrome. Of these, RECQL4 is unique on the basis of its domain structure and function. The N terminus of this protein represents the only human homolog of Saccharomyces cerevisiae essential replication initiation protein Sld2 (1, 2). The helicase domain is conserved across the RecQ class of helicases. Initial reports suggested RECQL4 lacked of helicase activity (3), but annealing, strand exchange and helicase activities of RECQL4 were later demonstrated (4–7). Human RECQL4 was shown to unwind up to 22-bp double-stranded regions through its helicase activity (6, 7). ATPase and annealing activities of human RECQL4 have also been extensively studied (7, 8). Robust helicase and annealing activities on substrates of various lengths have been reported for Drosophila melanogaster RecQ4 (DmRecQ4) (4). There is a significant difference in the N-terminal regions of human RECQL4 and DmRecQ4 that might contribute to the stronger helicase activity of DmRecQ4 and its functional necessity (9). Recent studies analyzing domain specific substrate binding preferences (8) and new structural features within known domains (10) of RECQL4 also emphasize its wide substrate range.
Studying the primary functions of a pleiotropic enzyme like RECQL4 requires systematic analysis of its enzymatic properties, assessment of its functional binding partners, and analyzing how these binding partners influence activity in relevant biological contexts. The role of RECQL4 in replication initiation process has been well characterized by assessing some of its important interaction partners such as MCM10 and TOPBP1 (11, 12). However, its role in repair processes like double-stranded break repair (13), base excision repair (14), and nucleotide excision repair (15) warrants more analysis. Characterization of its functional interactions with repair proteins will likely reveal its particular roles in these processes and delineate the contributions of RECQL4 as a signaling cofactor and as an active enzyme.
Extraribosomal functions of ribosomal proteins have been a topic of intense interest in recent years. Ribosomal protein S3 (RPS3) is a component of the 40 S ribosomal subunit (16). However, nuclear localization of RPS3, with or without different stimulations (including hydrogen peroxide, methyl methane sulfonate, and TNF) has been reported (17–19). A mass spectrometry study revealed RPS3 as an interaction partner of WRN (20). Various moonlighting functions have been reported for RPS3. Its activity as an AP endonuclease (21, 22) was shown to play an important role in xeroderma pigmentosum group D individuals. These patients also show increased sensitivity to UV light, which is also characteristics of RTS patients (22, 23). Drosophila RPS3 exhibits 8-oxoguanine glycosylase activity along with potent AP endonuclease activity (24). Other accessory roles of RPS3 include acting as a NF-κB complex subunit (25) and serving as an assistant for microtubules (26).
RTS patient cell lines and fibroblasts depleted of RECQL4 exhibit defects in the oxidative DNA damage repair (14). In contrast, knockdown of RPS3 enhances survival of cells treated with hydrogen peroxide (27). The common features of RTS, Baller-Gerald syndrome, and RAPADILLINO syndrome include multiple skin abnormalities, osteoskeletal deformities, and osteosarcoma (9). Increased RPS3 expression has been linked with enhanced invasion and migration of GLI2-mediated osteosarcoma cells (28). RPS3 is also overexpressed in melanoma, and its knockdown suppresses cell growth and induces apoptosis in melanoma cells (29).
In this study, we characterize the interaction between RPS3 and RECQL4. We show functional interaction of an active AP endonuclease domain of human RPS3 (C-RPS3, 94–244 aa) with N-terminal (N-RECQL4, 1–320 aa) portion of human RECQL4 and reveal how this interaction affects ATPase, helicase, and DNA binding activities of RECQL4. Furthermore, we evaluate this novel association in the context of UV and hydrogen peroxide exposure.
Results
Purification of Human RECQL4 and Helicase-dead K508N-RECQL4 Using a Baculovirus System
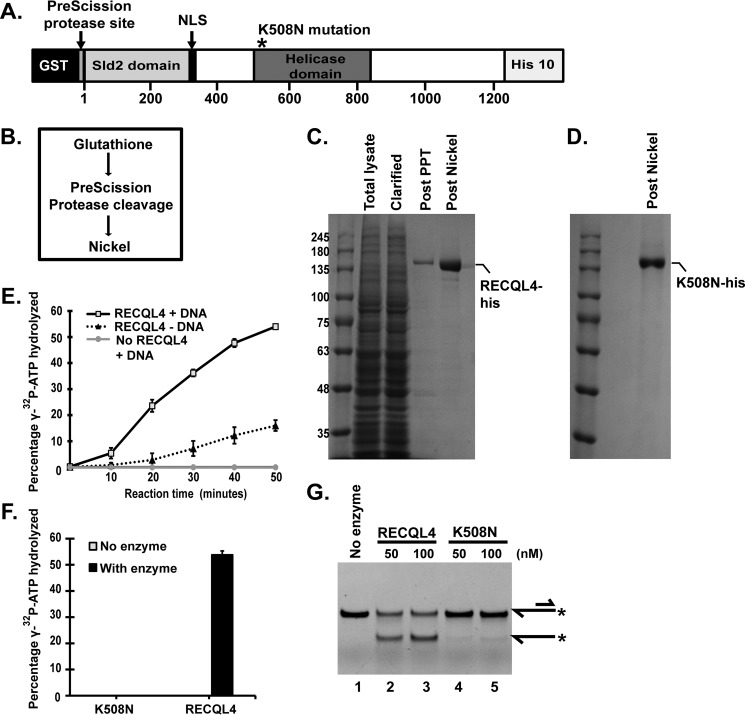
Purification of human RECQL4 using bacterial and baculovirus expression systems has been reported previously (3, 5–7). We used a baculovirus expression system for purification of RECQL4 and helicase-dead K508N-RECQL4 mutant. We expressed full-length human RECQL4 or the K508N-RECQL4 mutant with an N-terminal GST tag and a C-terminal His10 tag (Fig. 1A) in SF9 cells. We purified full-length human RECQL4 and its helicase-dead mutant K508N-RECQL4 using a baculovirus expression system and a simple three-step strategy modified from (4) (Fig. 1B). Cells expressing RECQL4 or K508N-RECQL4 mutant constructs after respective recombinant baculovirus infections were lysed and used for purification of RECQL4 helicase (Fig. 1C) or K508N-RECQL4 (Fig. 1D). The GST tag was removed by PreScission protease cleavage. This step effectively removed most of the lysate impurities, whereas the subsequent nickel column step allowed concentration of the protein (Fig. 1C).
FIGURE 1.
Purification and biochemical analysis of human RECQL4 helicase and K508N-RECQL4 mutant. A, schematic of the recombinant human RECQL4 construct expressed in SF9 cells with a C-terminal His tag and an N-terminal GST tag attached to RECQL4 by the PreScission protease cleavage sequence. * represents position of the K508N mutation for K508N-RECQL4 purification. B, purification strategy for RECQL4 and K508N-RECQL4. C, purification stage fractions of RECQL4 protein. Total lysate, total lysate of protein expressing SF9 cells; Clarified lysate, total lysate supernatant after centrifugation; Post PPT, clarified lysate applied to glutathione column followed by PreScission protease treatment (PPT); Post Nickel, nickel column concentration of the protein in the supernatant of the post PPT fraction. D, final post nickel purification fraction of K508N-RECQL4 (purification columns and process, the same as RECQL4). E, ATPase assay. The plot depicts percentage of [γ-32P]ATP hydrolyzed in the presence of 40 nm RECQL4 and DNA, as well as with no-enzyme and no-DNA controls at indicated time points assayed by TLC. The data points are the mean values of triplicate experiments with S.D. shown as error bars. F, bar graph showing percentages of [γ-32P]ATP hydrolyzed in the presence of 40 nm RECQL4 or the helicase-dead mutant K508N. G, polyacrylamide gel showing DNA helicase activity of RECQL4 and K508N on 5′-FAM-labeled 3′ overhang substrate (20 nm). Substrate and product structures are illustrated on the right side of the gel. * represents the position of FAM label on the substrate.
Human RECQL4 Exhibits DNA Stimulated ATPase Activity and Robust Helicase Activity
Different substrates with varying ratios of single-stranded and double-stranded regions including fork, bubble, circular substrates, and duplexes were used to analyze the helicase activity of human RECQL4. However, robust ATP-dependent helicase activity for human RECQL4 has not been demonstrated for substrates with double-stranded regions longer than 22 bp (3, 5–7, 30). Typically demonstration of strand exchange activity has been reported for substrates with longer double-stranded regions in the presence of excess single-stranded traps. Here we assayed the ATPase activity of purified RECQL4 and the K508N-RECQL4 mutant and observed hydrolysis of [γ-32P]ATP by incubating it with RECQL4. The presence of DNA stimulated the ATPase activity of human RECQL4 (Fig. 1E). No ATP hydrolysis activity was observed from the K508N mutant (Fig. 1F), confirming that the ATP hydrolysis activity is mainly driven by Walker A motif in the helicase domain.
We demonstrated robust helicase activity of human RECQL4 enzyme on the 3′ overhang substrate with 30 bp of double-stranded region (Fig. 1G, lanes 2 and 3). We confirmed that the observed helicase activity was attributable to the RECQL4 helicase and not to other minor contaminant proteins by assaying the known helicase-ATPase-dead mutant, K508N-RECQL4 (7), which did not exhibit helicase activity (Fig. 1G, lanes 4 and 5). Thus we demonstrated DNA-stimulated ATPase activity and robust helicase activity of human RECQL4, which can unwind 3′ overhang substrate with a 30-bp double-stranded region and a 30-nucleotide-long single-stranded tail.
Human RECQL4 Helicase Physically Interacts with Human RPS3
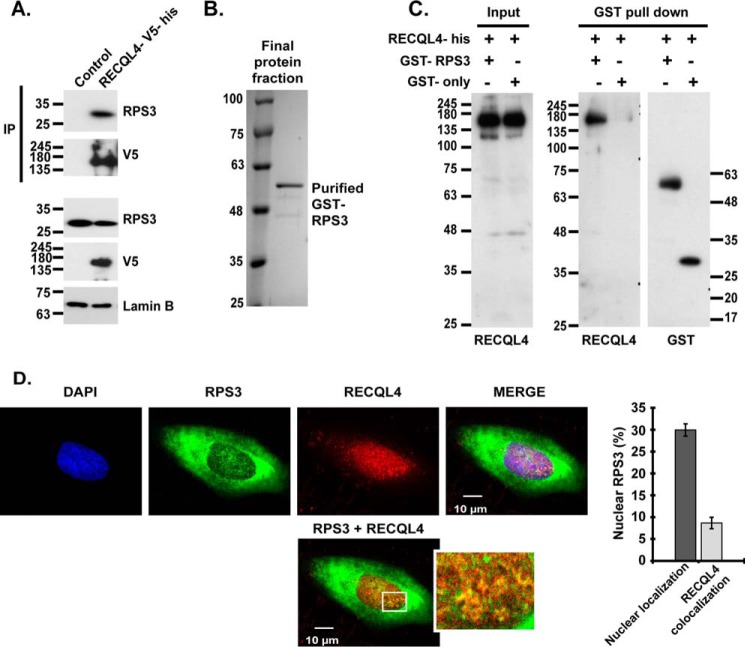
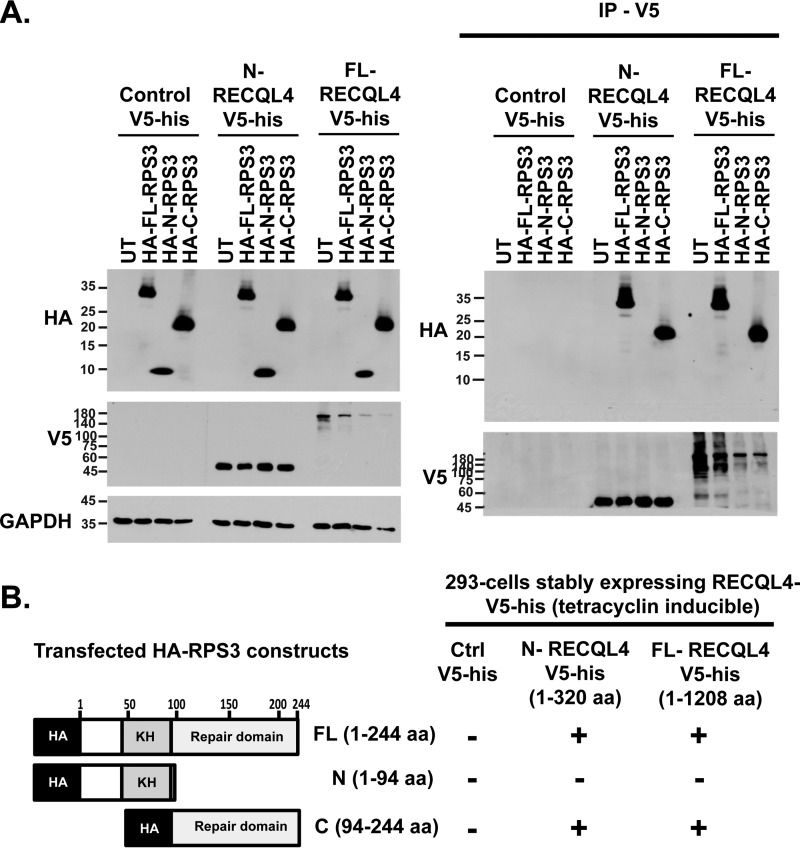
To investigate the functional interaction of RECQL4 and RPS3, we used nuclear lysates of 293 cell lines stably expressing control vector or full-length RECQL4 with a V5 and a histidine tag at its C terminus. Immunoprecipitation (IP) of nuclear lysates using V5 agarose beads followed by Western blotting analysis demonstrated interaction between RPS3 and RECQL4 (Fig. 2A). We then purified full-length RPS3 with an N-terminal GST tag (Fig. 2B). To evaluate whether RPS3 physically interacts with RECQL4 helicase, we carried out a GST pulldown assay. Incubation of GST-RPS3 protein or GST-only protein with RECQL4-his protein and subsequent Western blotting analysis revealed a direct physical interaction between RPS3 and RECQL4 helicase (Fig. 2C). To investigate probable context of this interaction, we immunostained U2OS cells with anti-RPS3 and anti-RECQL4 antibodies followed by confocal imaging of immunostained cells. We observed limited level of colocalization in these two proteins mostly in the nuclear compartment (Fig. 2D). Nuclear localization of RPS3 has been reported previously (18, 19, 31).
FIGURE 2.
RECQL4 physically interacts with RPS3. A, nuclear lysates of 293 cell lines stably expressing RECQL4-V5-his or control plasmid are used for V5 pulldown followed by anti-RPS3, anti-V5 and anti-Lamin B immunoblotting. B, Coomassie Blue-stained SDS-PAGE depicting final purified protein fraction of GST-FL-RPS3 after BiorexTM70 and glutathione-Sepharose purification steps. C, GST pulldown assay. Anti-RECQL4 and anti-GST blots show input and pulldown fractions of GST pulldown assay carried out by incubating RECQL4-his with GST-RPS3 or GST only proteins. D, confocal images of representative U2OS cells immunostained by anti-RECQL4, anti-RPS3 antibodies, and DAPI. The plot shows nuclear RPS3 (%) and nuclear colocalization of RPS3-RECQL4 (%) quantified from three-dimensional reconstructions of Z section confocal images, using Imaris software. The data are shown as the means ± S.D. of triplicate experiments.
RPS3 Inhibits Helicase and ATPase Activity of RECQL4
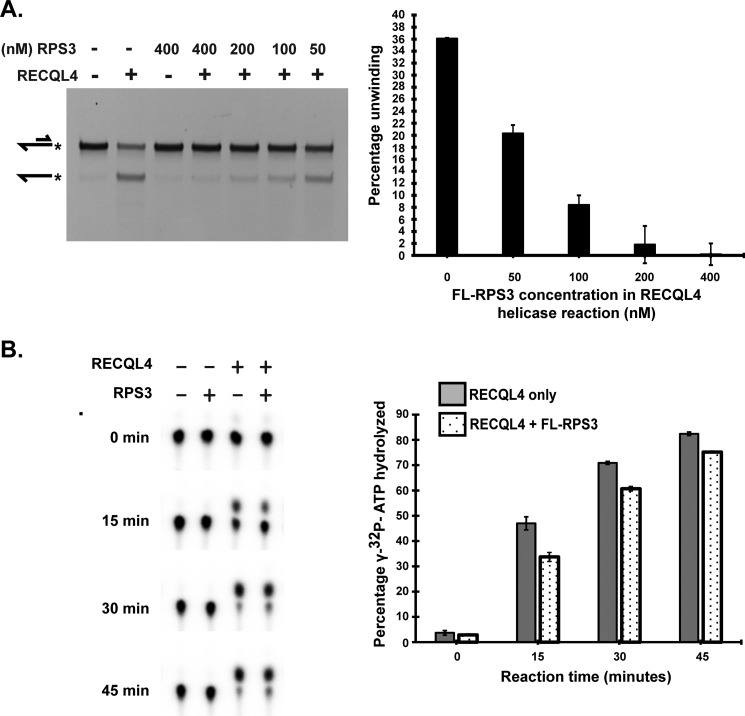
To establish the functionality of this interaction, we investigated whether it affects the biochemical activities of RECQL4 helicase. We conducted a RECQL4 helicase activity assay in the presence of differing concentrations of RPS3 protein. RPS3 inhibited helicase activity of RECQL4 protein in a dose-dependent manner (Fig. 3A). Helicase activity of RECQL4 is mediated through ATP hydrolysis. Hence we inspected whether RPS3 directly inhibits the ATPase activity of RECQL4. We performed an ATPase time course activity assay for RECQL4 helicase in the presence or absence of RPS3. RECQL4 ATPase activity was decreased consistently in the presence of RPS3 along the reaction time course (Fig. 3B). These results suggest that physical interaction with RPS3 negatively regulates the ATPase and helicase activities of RECQL4. However, RPS3-mediated inhibition of RECQL4 helicase activity is much more pronounced compared with inhibition of RECQL4 ATPase activity. Thus we tested whether RPS3 also affects RECQL4 binding with the substrate by depleting RECQL4 from the reaction through their direct interaction. An electrophoretic mobility shift assay showed that increasing concentration of RPS3 inhibits binding of RECQL4 with single-stranded substrate in a dose-dependent manner (supplemental Fig. S3). Together these results suggest that inhibition of RECQL4 unwinding activity results from RPS3 inhibition of both RECQL4-DNA binding and ATPase activity.
FIGURE 3.
RPS3 inhibits helicase and ATPase activity of RECQL4. A, representative PAGE showing the RECQL4 helicase assay performed in the presence or absence of indicated concentrations of FL-RPS3 (1–244 aa). All helicase reactions contain 50 nm RECQL4 protein and 20 nm 5′FAM-labeled 3′ overhang substrate. Substrate and product structures are illustrated on the left side of the gel. * represents position of the FAM label on the substrate. Bar graph depicting percentage unwinding against the concentration of RPS3 protein in the helicase reaction. The data points shown are the means of triplicate experiments with S.D. indicated as error bars. B, ATPase activity of RECQL4 assayed using TLC. Of a total 20-μl reaction, 1 μl (with 400 nm RPS3, 80 nm of RECQL4, and 3.4 nm [γ-32P]ATP substrate) was taken for TLC analysis at indicated time points. Bar graph showing percentage of [γ-32P]ATP hydrolyzed in the presence (dotted bars) or absence (shaded bars) of FL-RPS3 plotted against the reaction time course. The data points shown are the means of triplicate experiments with S.D. indicated as error bars.
The N-terminal 1–320 aa of RECQL4 (N-RECQL4) Interacts with the C-terminal 94–244 aa of RPS3 (C-RPS3)
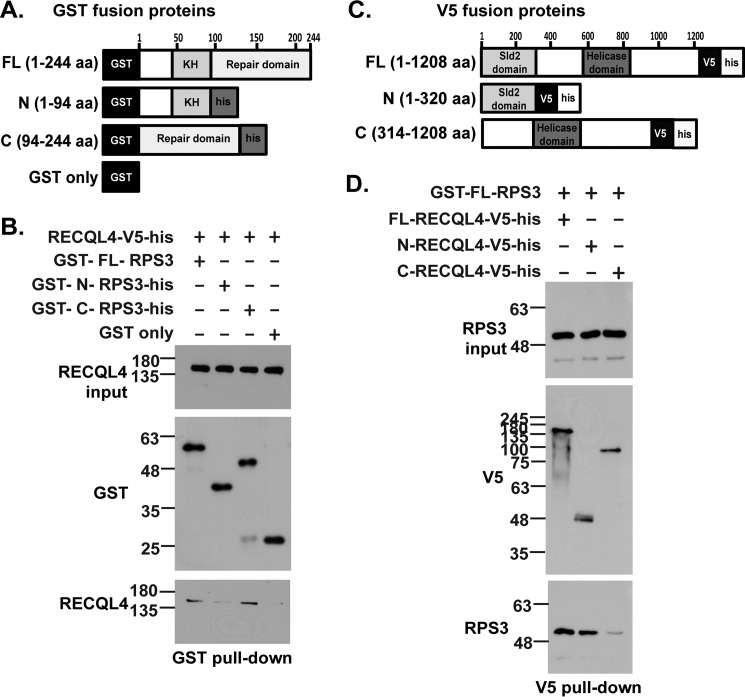
We investigated domain interactions between these two proteins to further characterize enzymatic interaction between RPS3 and RECQL4. N-terminal GST-tagged FL-RPS3 (1–244 aa), N-RPS3 (1–94 aa), and C-RPS3 (94–244 aa) purified proteins (Fig. 4A and supplemental Fig. S1A) were each incubated with FL-RECQL4-V5-his protein (Fig. 4A and supplemental Fig. S1B), and GST pulldown was performed. Pulldown using glutathione beads revealed that FL-RPS3 and C-RPS3 interact with FL-RECQL4 protein with almost similar intensities (Fig. 4B).
FIGURE 4.
The C-terminal (94–244 aa) of RPS3 and N-terminal (1–320 aa) of RECQL4 are the main interaction domains of the two proteins. A, schematic showing GST-tagged proteins used for the pulldown assay: GST-FL-RPS3 (1–244 aa), GST-N-RPS3-his (1–94 aa), GST-C-RPS3-his (94–244 aa), and GST only. B, GST pulldown assay. Anti-RECQL4 and anti-GST immunoblots show input and pulldown fractions when FL-RECQL4-V5-his was incubated with GST tagged FL-RPS3, N-RPS3, C-RPS3, or GST only proteins. C, schematic showing V5 tagged proteins used for pulldown assay: FL-RECQL4-V5-his (1–1208 aa), N-RECQL4-V5-his (1–320 aa), and C-RECQL4-V5-his (314–1208 aa). D, V5 pulldown assay. Anti-V5 and anti-RPS3 immunoblots show input and pulldown fractions when GST-FL-RPS3 was incubated with FL-RECQL4-V5-his, N-RECQL4-V5-his, or C-RECQL4-V5-his proteins.
We also sought to determine which RECQL4 domain interacts with FL-RPS3 protein using a V5 pulldown assay. We incubated FL-RECQL4-V5-his (1–1208 aa), N-RECQL4-V5-his (1–320 aa), and C-RECQL4-V5-his (314–1208 aa) (Fig. 4C and supplemental Fig. S1B) with GST-FL-RPS3 (1–244 aa) (Fig. 4C and supplemental Fig. S1A). Subsequent V5 pulldown followed by immunoblotting with anti-RPS3 and anti-V5 antibodies revealed that N-RECQL4 (1–320 aa) and FL-RECQL4 (1–1208 aa) interact with FL-RPS3 (1–244 aa) (Fig. 4D). Of note, the truncated RPS3 proteins used here and in the following activity assays were purified using a bacterial expression system (supplemental Fig. S1A). Truncated and full-length RECQL4 proteins with C-terminal V5 and histidine tags—FL-RECQL4, N-RECQL4, and C-RECQL4—were purified from lysates of 293 cell lines stably expressing respective proteins (supplemental Fig. S1B).
We confirmed the in vivo interaction of these domains by using an IP approach. We transfected N-terminal HA-tagged RPS3 protein constructs HA-N-RPS3 (1–94 aa), HA-C-RPS3 (94–244 aa) and HA-FL-RPS3 (1–244 aa) into 293 cell lines stably expressing FL-RECQL4-V5-his, N-RECQL4-V5-his and control vector (V5-his only). With V5, IP of whole cell lysates showed that HA-C-RPS3 (94–244 aa) and HA-FL-RPS3 (1–244 aa) interact with N-RECQL4-V5 his and FL-RECQL4-V5-his (Fig. 5, A and B). Together, these data confirmed that the N-terminal 1–320 aa of RECQL4 (N-RECQL4) and the C-terminal 94–244 aa of RPS3 (C-RPS3) are the main interaction domains. Human N-RECQL4 (1–320 aa) is known to have a Sld2 homologous region (9), whereas the C-RPS3 (94–244 aa) domain has a putative hairpin-helix-hairpin (HhH) motif known for AP endonuclease activity (24).
FIGURE 5.
C-RPS3 interacts with N-RECQL4 in vivo. A, 293 cells stably expressing FL-RECQL4-V5-his or N-RECQL4-V5-his transfected with HA-FL-RPS3 (1–244 aa), HA-N-RPS3 (1–94 aa), and HA-C-RPS3 (94–244 aa) plasmid constructs. Whole cell lysates were subjected to V5-IP followed by anti-HA, anti-V5, and anti-GAPDH Western blotting analysis of lysate and IP fractions. B, schematic showing results of V5 coimmunoprecipitation experiments.
The RECQL4 Interaction Domain of RPS3 (94–244 aa) Is Sufficient to Inhibit RECQL4 Helicase Activity
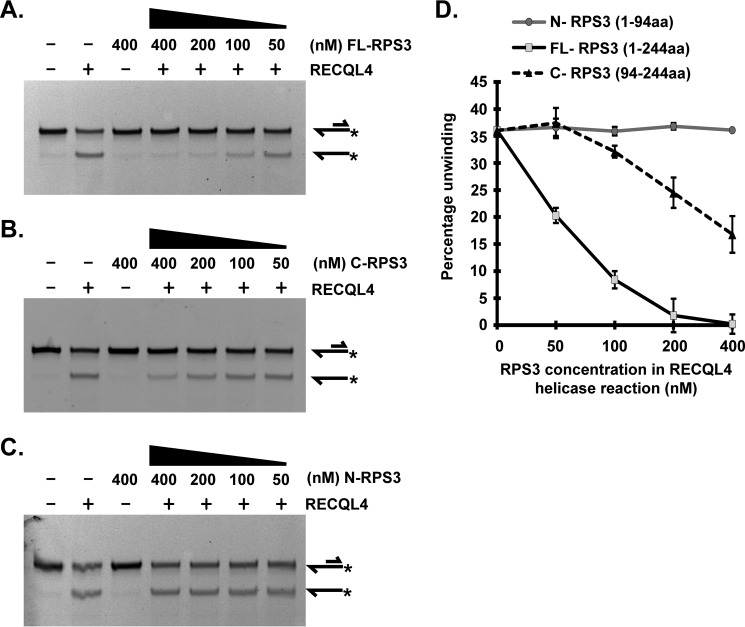
We investigated whether C-RPS3 alone is sufficient to inhibit RECQL4 helicase activity. We titrated various concentrations of FL-RPS3 (Fig. 6A), C-RPS3 (Fig. 6B), and N-RPS3 (Fig. 6C) in a RECQL4 helicase reaction. C-RPS3 did indeed inhibit RECQL4 helicase activity (Fig. 6B). However, the level of inhibition was not highly comparable with that of full-length protein (FL-RPS3) (Fig. 6D). Helicase activity of RECQL4 against 20 nm of the 3′ overhang substrate was almost completely inhibited in presence of 400 nm FL-RPS3 (Fig. 6A), whereas 400 nm C-RPS3 partially inhibited RECQL4 helicase activity (Fig. 6B). There was no significant effect of N-RPS3 on the helicase activity of RECQL4 (Fig. 6C). Quantitative illustration of these helicase assays using various concentrations of FL-RPS3, C-RPS3, and N-RPS3 is shown in Fig. 6D. Additionally, C-RPS3 demonstrated RECQL4 ATPase activity inhibition, but N-RPS3 did not (supplemental Fig. S2). Again, inhibition of RECQL4 ATPase activity by C-RPS3 was less than that of FL-RPS3. Although the effect of RPS3 on RECQL4 ATP hydrolysis activity was not as pronounced as that for its helicase inhibition, the former was still significant according to a Student's t test of triplicate experiments (supplemental Fig. S2). Direct inhibition of RECQL4 ATPase activity by RPS3 suggests enzymatic inhibition of RECQL4 rather than mere competitive substrate inhibition.
FIGURE 6.
C-RPS3 (94–244 aa), but not N-RPS3 (1–94 aa), contributes to RECQL4 helicase activity inhibition. All helicase reactions contain 50 nm RECQL4 protein and 20 nm 5′FAM-labeled 3′ overhang substrate. Substrate and product structures are illustrated on the right side of the gels. * represents position of FAM label on the substrate. A, representative PAGE showing the RECQL4 helicase assay performed in the presence or absence of indicated concentrations of full-length FL-RPS3 (1–244 aa). B, representative PAGE showing the RECQL4 helicase assay performed in the presence or absence of indicated concentrations of C-RPS3 (94–244 aa). C, representative PAGE showing RECQL4 helicase assay performed in the presence or absence of indicated concentrations of N-RPS3 (1–94 aa). D, graph depicting percentage unwinding by RECQL4 plotted against final concentration of FL-RPS3, N-RPS3, or C-RPS3 in RECQL4 helicase reaction. The data points shown are the means of triplicate experiments with S.D. indicated as error bars.
The C-terminal of RPS3 Shows AP Endonuclease Activity on Circular Double-stranded Substrates
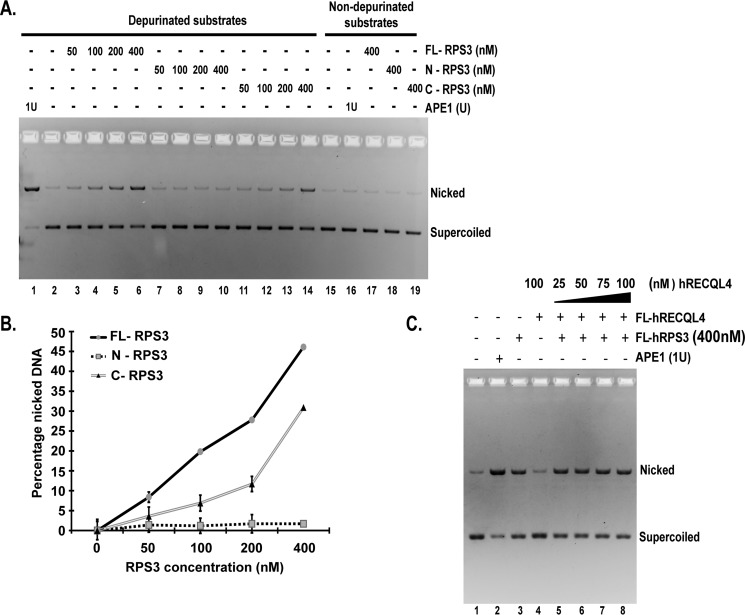
C-RPS3 (94–244 aa) is soluble and can be stably purified (supplemental Fig. S1A). After revealing this domain as the major RECQL4 interaction domain responsible for helicase activity inhibition, we studied whether the C-RPS3 is responsible for AP endonuclease activity of human RPS3. This domain was previously reported to have homology with HhH motif of the Escherichia coli AP endonuclease, endonuclease III (24). We assessed the AP endonuclease activity of purified FL-RPS3, N-RPS3, and C-RPS3 on circular double-stranded depurinated substrates using a plasmid nicking assay. Depurination of plasmid substrates under acidic conditions (pH 4.5) results in generation of AP sites, and this approach has been used to assay the AP endonuclease activity of various enzymes including yeast and human RPS3 (31, 32). In this assay AP sites on the substrate molecules are converted into single-stranded breaks or nicks by active AP endonucleases. Non-depurinated substrates used as negative controls (Fig. 7A, lanes 15–19) confirmed selective AP site generation on depurinated substrates (Fig. 7A, lanes 1–14). Depurinated supercoiled plasmid substrates showed a dose-dependent increase in nicked form DNA after incubation with increasing concentrations of FL-RPS3 (Fig. 7A, lanes 3–6). Human APE1, an AP-endonuclease, was used as the positive control to confirm generation of AP sites on depurinated substrates (Fig. 7A, lanes 1 and 16). We analyzed the AP endonuclease activity of N-RPS3 and C-RPS3 in a similar way. C-RPS3 exhibited a level of AP endonuclease activity comparable with that of FL-RPS3 (Fig. 7A, lanes 11–14 and 19). N-RPS3 did not show any significant AP endonuclease activity (Fig. 7A, lanes 7–10 and 18). Fig. 7B illustrates quantification of triplicate plasmid nicking assays with FL-RPS3, N-RPS3, and C-RPS3.
FIGURE 7.
FL-RPS3 and C-RPS3 exhibit comparable AP endonuclease activities. A, representative gel showing plasmid nicking assay. 4 nm depurinated pHC624 substrate was incubated with indicated concentrations of FL-RPS3 (lanes 3–6), N-RPS3 (lanes 7–10), C-RPS3 (lanes 11–14), and APE1 (lanes 1 and 16). Non-depurinated plasmid substrates (4 nm) were used as negative controls (lanes 15–19). Nicked indicates product amount, and Supercoiled indicates substrate amount in the reaction. B, plot showing percentage product formed against concentration of RPS3 proteins in the reaction (representative experiment in A). The data points shown are the means of triplicate experiments with S.D. indicated as error bars. C, gel showing plasmid nicking assay. The gel reveals the effect of indicated concentrations of RECQL4 (lanes 5–8) on AP endonuclease activity of RPS3 along with control reactions. APE1 acted as a positive control (lane 2). 1 unit (1U) is equal to 2 nm final concentration.
To reveal whether RECQL4 has any effect on this enzymatic property of RPS3, we titrated various concentrations of FL-RECQL4 along with FL-RPS3 in a plasmid nicking assay (Fig. 7C). There was no significant difference in the AP endonuclease activity of RPS3 in the presence of RECQL4 (Fig. 7C, lanes 5–8).
Oxidative Stress and UV Exposure Increases Nuclear RPS3-RECQL4 Interaction in a Dose-dependent Manner
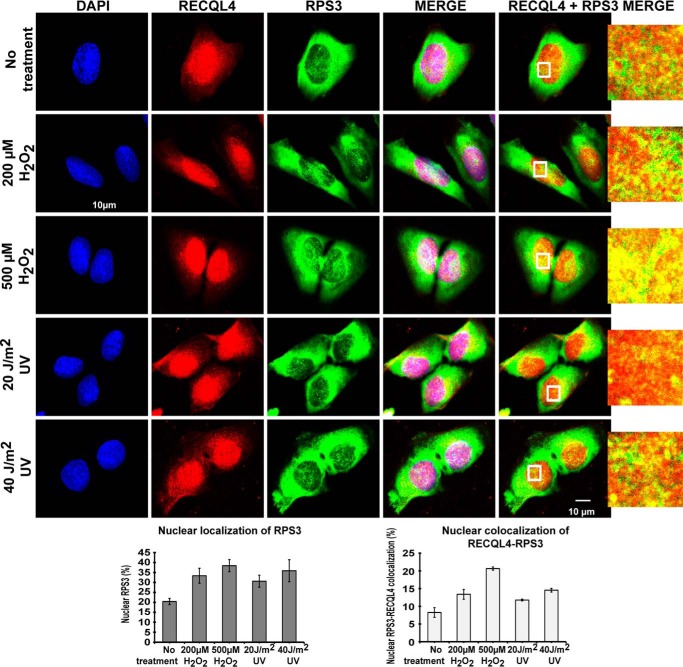
The helicase domain of RECQL4 has been shown to be important for various DNA repair functions (13–15), and this domain is frequently mutated in RTS, Baller-Gerald Syndrome, and RAPADILLINO syndrome patients (23). RTS patients are more prone to osteoskeletal deformities and osteosarcoma (23), and some studies have shown them to have increased sensitivity to UV (15) and oxidative stress (14, 33). RPS3 is also important for the progression of osteosarcoma (28). We used U2OS cells to evaluate the effect of hydrogen peroxide- and UV-induced DNA damage on the RECQL4-RPS3 interaction. Both treatments induced a dose-dependent increase in nuclear RPS3 as well as an increase in overlapping immunostaining signals of RPS3-RECQL4 inside the nuclear compartment (quantified using imaris software) (Fig. 8). The effect of hydrogen peroxide-induced DNA damage on nuclear localization of RPS3 was more pronounced than that of UV-induced DNA damage (Fig. 8). Together, our results hint at a possible role of the RPS3-RECQL4 interaction in regulating the DNA repair induced by similar genotoxic stresses (28, 34).
FIGURE 8.
Nuclear RECQL4-RPS3 interaction is enhanced under hydrogen peroxide- and UV-induced genotoxic stress in U2OS cells. U2OS cells subjected to mock, hydrogen peroxide (200 or 500 μm), or UV (20 or 40 J/m2) treatment were immunostained with anti-RECQL4 and anti-RPS3 antibodies. Representative confocal images of cells after treatments are shown in the top panels. Plots at the bottom show quantification of nuclear RPS3 (%) and nuclear colocalization of RECQL4-RPS3 (%) carried out using Imaris 8.3.1 software on z sections of confocal images taken from immunostained U2OS cells. The data points shown are the means of triplicate experiments with S.D. indicated as error bars.
Discussion
In this study we identify RPS3 as a novel physical and functional interaction partner of RECQL4 helicase and suggest that this interaction may have relevance for RECQL4-mediated DNA repair. RECQL4 is associated with DNA replication and repair based on its unique domain structure. The conserved RecQ type helicase domain of RECQL4 is important for DNA repair function and aging (35, 36). Mutations in RTS patients (23) are particularly concentrated in this domain (34). Because the N-terminal Sld2-like domain is sufficient to partially rescue lethal replication defects (1, 2), RECQL4 helicase activity is likely to be more important for its DNA repair function.
We purified human RECQL4 protein and showed its robust helicase activity on a 3′ overhang substrate with a 30-bp-long double-stranded region, by far the longest double-stranded region yet shown to be unwound by human RECQL4 by virtue of its helicase activity. Loss of helicase and ATPase activities by the K508N-RECQL4 mutant protein expressed and purified in identical way proves that those activities are mostly attributed to the intact Walker A motif of the helicase domain. These results also show that human RECQL4 can serve as an independent helicase potentially acting on substrates like intermediates formed during DNA repair and other processes. We also take into consideration the probable role of post-translational modification on the helicase activity of RECQL4. Unlike our study, most previous reports used bacterial expression system for purification (6, 7). Furthermore, another study using baculovirus system purified human RECQL4 also showed relatively weaker helicase activity (5). We attribute these differences in the activity of RECQL4 to the variations in substrate type, buffer conditions, purification strategies, and overall quality of final protein fractions.
Various extraribosomal functions of nuclear RPS3 have been reported such as its role as an NF-κB subunit in apoptosis and as an AP endonuclease in base excision repair (19, 24). RPS3 localizes to nucleus, both with or without various stimulations (25, 27, 31). Our finding of a dose-dependent increase in nuclear localization of RPS3 under conditions of oxidative and UV stress (Fig. 8) is consistent with previous report (17). We did not observe discrete foci formation between RPS3 and RECQL4 possibly because of homogenous RECQL4 expression. Therefore, we used Imaris software to quantify the signal overlap between these two proteins inside the nuclear compartment using three-dimensional reconstruction of cells obtained from Z sections of confocal images (Imaris 8.3.1). Such quantification showed dose-dependent accumulation of RPS3 in the nucleus, providing opportunities to enhance interaction but also revealed portion of nuclear RPS3 that does not interact with RECQL4 (Fig. 8). Thus our results together with previous reports suggest that RPS3 could play multiple roles inside the nucleus under stress conditions. We show here that the C-RPS3 (94–244 aa, containing a region homologous to E. coli endonuclease III) directly interacts with the N-RECQL4 domain. This C-RPS3 domain also hosts other key interactions related to DNA repair, for example mediating interactions with MDM2 and p53 (37). Similarly, the N-RECQL4-Sld2-like domain is well known for its interaction with proteins such as TOPBP1 and MCM10 (11, 12). It would be intriguing to analyze how RPS3 inhibits the helicase activity of RECQL4 when its major interaction is via N-RECQL4 (1–320 aa) and not via the helicase domain. Interestingly another important interaction partner of RECQL4, MCM10 is reported to inhibit RECQL4 helicase activity through its interaction with RECQL4's N-terminal 1–240 aa (11). Hence this evidence suggests that the N-terminal domain of RECQL4 may act as a scaffold for various interactions that allows helicase domain to perform its unwinding function. This gives a glimpse of how unique structural features of RECQL4 allow it to perform diverse duties in various biological processes. Further structural studies will be helpful to determine how these interaction partners interfere with the ATPase and helicase activity of RECQL4.
Mouse models devoid of active RECQL4 helicase domain have similar symptomatic profiles to RTS patients and yet they manage to survive (38). This suggests that helicase activity of RECQL4 has major role in DNA repair and maintenance of genome stability, but it may not be essential for replication role of RECQL4. We have shown that RPS3 inhibits DNA binding and ATPase and helicase activities of RECQL4, and furthermore, C-RPS3 itself has AP endonuclease activity. Hence, RPS3-RECQL4 association is more likely to be important to its role in DNA repair. We suggest two different mechanisms by which RPS3 can influence biological role of RECQL4 unwinding activity. First, RPS3 constrains unwinding of AP sites containing double-stranded substrates by inhibiting the helicase activity of RECQL4, thereby preventing further processing of unrepaired AP site containing double strand substrates. Thus RPS3 may repair the AP site lesions by its standalone activity. The human AP endonuclease APE1 reportedly inhibits helicase activity of WRN in similar way (39). Second, RECQL4 has been reported to stimulate enzymatic activity of base excision repair proteins like APE1, FEN1, and DNA polymerase β (14). RPS3 can compete with APE1 to occupy AP site substrates. Because RPS3 interacts with RECQL4 via its active AP endonuclease domain, it is possible that the presence of RECQL4 directly stimulates APE1 catalytic activity and simultaneously reduces competitive inhibition of APE1 by directly engaging free RPS3 through physical interaction and thus keeping it away from AP sites.
The AP endonuclease activity of RPS3 is not affected by RECQL4 (Fig. 7C). We cannot rule out inefficient RECQL4 helicase activity on such circular double-stranded substrate, because there is no continuous stretch of single-stranded DNA available for its activity. The C-RPS3 domain primarily contributes to the AP endonuclease activity of RPS3, in agreement with reports showing the presence of putative HhH motif homologous to E. coli endonuclease III (24). Our findings together with previous studies also suggest that the AP endonuclease activity of RPS3 could be different from that of APE1 in terms of substrate binding, processing, and enzyme potency (Fig. 7A) (40, 41). Because AP sites can be generated after the removal of a variety of modified bases by various DNA glycosylases, it is also possible that RPS3 may work as a standalone AP endonuclease in some cases. Alternatively, because AP sites are structurally unstable, binding of human RPS3 with weak endonuclease activity may serve to protect against further mutagenic effects. Reports of a high affinity of RPS3 for AP sites support this hypothesis (42). Thus we also cannot rule out a role for RPS3 as a negative regulator at the early stage of the base excision repair. DNA repair may be directly inhibited by occupying substrates like AP sites or indirectly by occupying RECQL4, which acts as positive regulator for early stage base excision repair proteins (14) or nucleotide excision repair proteins like XPA (15). Our results showing increased RECQL4-RPS3 association under conditions of hydrogen peroxide and UV-induced nuclear stress together with reported observations of improved protection against genotoxic stress in RPS3 knockdown cells (27, 29) support this model. Our results in U2OS cells indicate that this catalytic collaboration could particularly affect skeletal tissues. Increased sensitivity of RTS patients with skeletal deformities to UV and oxidative exposure is well described (23). Furthermore the AP endonuclease activity of RPS3 on UV-irradiated DNA was one of the initial lines of evidence for its role in DNA repair (22). These reports, along with our results, hint toward a probable involvement of the RPS3-RECQL4 interaction in maintaining the genome stability in skeletal tissue and osteosarcoma progression. In conclusion, we illustrate how physical interaction of catalytically active RPS3 with RECQL4 helicase alters biochemical properties of the helicase without interfering with the independent repair functions of the ribosomal protein (RPS3).
Experimental Procedures
Cells and DNA
Stable cell lines expressing either control vector or N-RECQL4-V5-his, C-RECQL4-V5-his, and FL-RECQL4-V5-his were prepared using the Flp-InTM-293 cell line from Invitrogen as per the manufacturer's instructions. Cell lines 293 and U2OS cells were maintained in DMEM with 10% fetal bovine serum and 1% penicillin streptomycin. SF9 insect cells were maintained in SF9-ESF serum-free medium.
For the purification of human RECQL4, using a Bac-to-Bac baculovirus expression system (Invitrogen), human RECQL4 cDNA was cloned into pFastBac1 vector with an N-terminal GST tag (separated from the RECQL4 sequence by the PreScission protease cleavage site sequence) and a C-terminal 10 histidine tag (His10) (Fig. 1A). RECQL4-K508N mutant was prepared by site-directed mutagenesis of pFastBac1-RECQL4 plasmid. For the expression of full-length RPS3 (1–244 aa), the full-length cDNA sequence of human RPS3 was cloned into modified pET41a vector (Novagen) with an N-terminal GST tag. For the expression of truncated N-RPS3 and C-RPS3, cDNA sequences of 1–94 and 94–244 aa of human RPS3 were cloned into pET41a vector (Novagen) with an N-terminal GST tag and a C-terminal six-histidine tag and modified pGW1 vector with an N-terminal HA tag.
Purification of Human RECQL4 Using a Baculovirus Expression System
Preparation of recombinant RECQL4 or RECQL4-K508N baculovirus stocks and their amplification was carried out using SF9 cells as per the manufacturer's instructions (Invitrogen). SF9 suspension cell culture (1 × 106 cells/ml) was infected with multiplicity of infection of 5 by virus stocks of RECQL4 or RECQL4-K508N. The culture was incubated at 27 °C for 72 h. The cells were pelleted, snap frozen, and stored at −80 °C. The cell pellet was lysed in 10 mm Tris-HCl, pH 7.8, 20 mm KCl, 2 mm EDTA, and 1× protease inhibitor cocktail (Roche). After lysis, NaCl and glycerol were slowly added to a final concentrations of 350 mm and 25%, respectively, and constantly stirred at 4 °C. Thereafter, the steps were performed at 4 °C unless otherwise specified. The lysate was clarified at 13,000 × g for 30 min. The supernatant was applied to glutathione-Sepharose (GE Healthcare) for 6 h followed by three wash steps with washing buffer (50 mm Tris-HCl, pH 7.8, 10% glycerol, 0.02% Triton X-100, 1 m NaCl, 2 mm EDTA, and protease inhibitor cocktail). The Sepharose was resuspended in 150 mm NaCl washing buffer (50 mm Tris-HCl, pH 7.8, 10% glycerol, 0.02% Triton X-100 150 mm NaCl, 2 mm EDTA, and protease inhibitor cocktail) and incubated with 40 units of PreScission protease (GE Healthcare)/1 liter of culture for 14 h. The flowthrough of the glutathione column was used as input for a 1-ml equilibrated nickel column (GE Healthcare). After one washing step with buffer (50 mm Tris-HCl, pH 7.8, 350 mm NaCl, 10% glycerol, 0.02% Triton X-100, and protease inhibitor cocktail), RECQL4 was eluted in a single step elution using nickel elution buffer (50 mm Tris-HCl, pH 7.8, 150 mm NaCl, 10% glycerol, 400 mm imidazole, 0.02% Triton X-100, and protease inhibitor cocktail). Elution fractions were snap frozen and stored at −80 °C.
Purification of Full-length and Truncated Human RPS3 Proteins
E. coli (Rosetta 2, DE3, pLysS) cultures expressing pET41a-GST-FL-RPS3 (1–244 aa) or pET41a-GST-N-RPS3-his (1–94 aa) or pET41a-GST-C-RPS3-his (94–244 aa) were grown to an optical density of 0.6 at 37 °C and induced using 1 mm isopropyl β-d-thiogalactopyranoside at 18 °C for 14 h. Induced cultures were harvested and snap frozen. Clarified lysates were fractionated as described below.
Purification of FL-RPS3 was carried out in two steps using BiorexTM70 beads (Bio-Rad) and glutathione-Sepharose (GE Healthcare). The harvested cell pellet of the FL-RPS3 culture was resuspended in lysis buffer (50 mm sodium phosphate, pH 7.2, 150 mm NaCl, 10% glycerol, 0.2% Triton X-100, 2.5 mm β-mercaptoethanol (2ME), 1 mm PMSF, and protease inhibitor cocktail) and lysed by sonication. The lysate was clarified by centrifugation (20,000 × g, 30 min), and the supernatant of the cell lysate was applied to equilibrated BiorexTM70 column. Elution of protein fractions was carried out using a 130–800 mm linear NaCl gradient. Peak fractions of elution were pooled and applied to equilibrated glutathione column. Elution was carried out using reduced glutathione elution buffer (50 mm Tris-HCl, pH 8.0, 200 mm NaCl, 20 mm reduced glutathione, and protease inhibitor cocktail). The elution fraction was dialyzed in 50% (v/v) glycerol, 50 mm Tris-HCl, pH 8.0, 200 mm NaCl, 1 mm PMSF, and 2.5 mm 2ME before storing at −20 °C.
Purification of truncated RPS3 proteins N-RPS3 (1–94 aa) and C-RPS3 (94–244 aa) was done in two steps using nickel and glutathione-Sepharose. Induced cell pellets were lysed in 50 mm Tris-HCl, pH 8.0, 200 mm NaCl, 10% glycerol, 0.2% Triton X-100, 2.5 mm 2ME, 1 mm PMSF, and protease inhibitor cocktail). Clarified lysates were applied to equilibrated nickel-Sepharose. Washing was conducted using 50 mm imidazole buffer, and elution was performed using 400 mm imidazole buffer. Peak elution fractions were dialyzed in 50% (v/v) glycerol, 50 mm Tris-HCl, pH 8.0, 200 mm NaCl, 1 mm PMSF, and 2.5 mm 2ME and stored at −20 °C.
Plasmid Nicking Assay
Depurinated circular DNA substrates were prepared by incubating pHC624 plasmid in depurination buffer (10 mm sodium citrate, pH 4.5, and 100 mm NaCl) at 70 °C for 5 min. The pH of reactions was neutralized to 8.0 by adding Tris-HCl, pH 8.0. Non-depurinated substrates were processed similarly to depurinated substrates using non-depurination buffer (10 mm Tris-HCl, pH 8.0, 100 mm NaCl, and 50 ng/μl pHC624 plasmid DNA) instead of depurination buffer. The same volume of Tris-HCl, pH 8.0, as that for the depurination reaction was added to non-depurination reactions.
For the assay, 4 nm of depurinated or non-depurinated substrates were incubated with indicated concentrations of RPS3 or APE1 in 40 mm Tris-HCl, pH 8.0, 35 mm KCl, 40 mm MgCl2 and in the absence or presence (for RPS3 + RECQL4) of 2 mm ATP at 37 °C for 30 min. The reactions were stopped by adding 0.2% SDS, 5% sucrose, 10 mm EDTA, 0.1 mg/ml bromphenol blue, and 1 mg/ml proteinase K before incubating at 45 °C for an additional 30 min. The reactions were resolved on 1.5% agarose gels with 10 μg/ml ethidium bromide at 100 V for 2 h. Imaging was carried out using ImageQuant LAS4000 (GE Healthcare). APE1 enzyme was purchased from NEB (M0282).
Helicase and ATPase Assay
For the helicase assay, 20 nm 5′ end-labeled 3′ overhang substrate (A, FAM-CGAAG GCCAT GATTG CGCAC TGAAT ACATC CTGCC CTGTT ATTAA TTACG TTATC TTACG; B, GATGT ATTCA GTGCG CAATC ATGGC CTTCG) was incubated with indicated concentrations of RECQL4 and/or RPS3 in 20 μl reaction volume (2.5 mm Tris-HCl, pH 7.5, 2.5 mm KCl, 4 mm MgCl2, 2 mm ATP, and 100 μg/ml BSA) for 30 min at 37 °C. The reaction was stopped by adding 0.2% SDS, 5% sucrose, 10 mm EDTA, and 0.1 mg/ml bromphenol blue at final concentration. The reactions were analyzed on 16% polyacrylamide gel in 1× Tris borate buffer. Imaging was carried out using Sybr-Green filter on ImageQuant LAS4000 (GE Healthcare).
For the ATPase assay, the indicated concentrations of RECQL4 in the absence or presence of RPS3 proteins were incubated with 1 mm ATP (3.4 nm [γ-32P]ATP) for the indicated time points at 37 °C in 2.5 mm Tris-HCl, pH 7.5, 2.5 mm KCl, 4 mm MgCl2, 50 μg/ml single-stranded circular DNA, and 100 μg/ml BSA. At the indicated time points, 1 μl of the reaction was spotted on Polygram CEL 300 PEI thin layer chromatography plates (Macherey-Nagel) that were prerun in distilled water. The plates were developed in 0.5 m LiCl and 1 m formic acid to separate inorganic phosphate from ATP. Developed plates were dried, and phosphorimaging was performed using TyphoonTMFLA7000 system (GE Healthcare).
Immunostaining and Cell Imaging
U2OS cells were seeded (10,000 cells/well) and grown for 6–8 h in 8-well chamber slides. The cells were fixed in 4% paraformaldehyde. Fixed cells were immunostained by mouse anti-RPS3 antibody (sc-376098) and rabbit anti-RECQL4 antibody. Fluorophore-conjugated secondary antibodies used are Alexa 488 anti-mouse (Invitrogen) and Alexa 647 anti-rabbit (Invitrogen). Coverslips were mounted on stained slides using ProLongGold antifade reagent with DAPI (Life Technologies). Confocal images were collected on a TCS-SP5-AOBS-MP microscope (Leica, Wetzlar, Germany) using a 63× oil lens. Quantification of nuclear RPS3 (%) and nuclear colocalization of RPS3-RECQL4 (%) was carried out on three-dimensional images from reconstituted Z sections using Imaris 8.3.1 (BItplane) software.
Immunoprecipitation
For whole cell lysates, the cells were lysed in 20 mm HEPES-KOH, pH 7.4, 300 mm NaCl, 10% glycerol, 1 mm EDTA, 0.2% Nonidet P-40, 1× protease inhibitor mix (Complete-Roche), and 1× phosphatase inhibitor mix (PhosStop-Roche) by sonication using a Bioruptor® Picosonication device (Diagenode). Cell lysates were centrifuged at 13,000 × g for 30 min, and from the clarified lysate, 1 mg of total protein was used for V5-IP by using equilibrated Anti-V5 conjugated beads (Sigma). IP and input fractions were analyzed by Western blotting using anti-RPS3 (sc-376098), anti-RECQL4, and anti-GAPDH (GTX28245) antibodies.
For the preparation of nuclear cell lysates, cell pellets were first lysed by cytoplasmic lysis buffer (20 mm HEPES-KOH, pH 7.4, 150 mm NaCl, 2 mm MgCl2, and 1× protease-phosphatase inhibitor mix) and centrifuged at 1000 × g for 5 min to pellet down the nuclear fraction. The nuclear pellet was then extracted in 20 mm HEPES-KOH, pH 7.4, 400 mm NaCl, 10% glycerol, 0.5% Nonidet P-40, 1× phosphatase, and protease inhibitor mix) after washing the pellet once with cytoplasmic buffer. Extracted nuclear lysate was centrifuged at 16,000 × g for 30 min, and supernatant nuclear fractions were diluted to 200 mm NaCl final concentrations for IP using V5 antibody-conjugated beads (Sigma). The beads were incubated with lysates for 12 h in the presence of 100 μg/ml ethidium bromide followed by three washes with nuclear extraction buffer before being eluted in 1× sample loading buffer. All the IP and input fractions were resolved on 10% SDS-PAGE and analyzed by Western blot using anti-RPS3 (sc-376098), anti-RECQL4, and anti-lamin B (sc-6216) antibodies.
Pulldown Assays
GST Pulldown Assay
RECQL4-His was incubated with GST-FL-RPS3 or GST-N-RPS3 or GST-C-RPS3 or GST only proteins in binding buffer (10 mm disodium phosphate, 1.8 mm monopotassium phosphate, 300 mm NaCl, 2.7 mm potassium phosphate, 0.3% Nonidet P-40, and 1× protease inhibitor mix) in a final reaction volume of 500 μl. The protein complexes were immobilized on equilibrated glutathione beads (GE Healthcare) for 6 h. The beads were washed three times with 1 ml of washing buffer (10 mm disodium phosphate, 1.8 mm monopotassium phosphate, 450 mm NaCl, 2.7 mm potassium phosphate, 0.5% Nonidet P-40, and 1× protease inhibitor mix). Input and pulldown fractions were analyzed by anti-RECQL4 and anti-GST Western blots.
V5 Pulldown Assay
GST-FL-RPS3 was incubated with FL-RECQL4-V5-his or N-RECQL4-V5-his or C-RECQL4-V5-his proteins in binding buffer (10 mm disodium phosphate, 1.8 mm monopotassium phosphate, 300 mm NaCl, 2.7 mm potassium phosphate, 0.3% Nonidet P-40, and protease inhibitors) in a final volume of 500 μl. The complexes of proteins were immobilized on V5 beads (Sigma) for 6 h. The beads were washed three times with 1 ml of washing buffer (10 mm disodium phosphate, 1.8 mm monopotassium phosphate, 450 mm NaCl, 2.7 mm potassium phosphate, 0.5% Nonidet P-40, and 1× protease inhibitor mix). Input and pulldown fractions were analyzed by anti-V5 and anti-GST Western blots.
Electrophoretic Mobility Shift Assay
For the EMSA, 50 nm 5′ end-labeled single-stranded DNA substrate (A, FAM-CGAAG GCCAT GATTG CGCAC TGAAT ACATC) was incubated with indicated concentrations of RPS3 and/or RECQL4 in 10 μl reaction volume (2.5 mm Tris-HCl, pH 7.5, 2.5 mm KCl, 4 mm MgCl2, 2 mm ATP, and 100 μg/ml BSA) for 5 min at 37°C. The reactions were stopped by adding 5% sucrose and 0.1 mg/ml bromphenol blue. The reactions were analyzed on 6% polyacrylamide gel in 1× Tris borate buffer. Imaging was carried out using a Sybr-Green filter on ImageQuant LAS4000 (GE Healthcare).
Author Contributions
A. V. P. and T.-S. H. designed the research; A. V. P. performed all the experiments; A. V. P. and T.-S. H. analyzed the data; and A. V. P. and T.-S. H. wrote the paper.
Supplementary Material
Acknowledgments
We are grateful to Dr. Ting-Fang Wang and Dr. Meng-Chao Yao (Institute of Molecular Biology, Academia Sinica, Taipei, Taiwan) for suggestions and support. We thank Wen-Hsiang Tsai for technical assistance. We thank John O'Brien and Marcus J. Calkins (Academia Sinica) for suggestions on the manuscript as native English speakers.
This work was supported by Ministry of Science and Technology Taiwan Grant MOST-105-2321-B-001-006 and Academia Sinica intramural funding Grant AS-105-TP-B04 (to T.-S. H.). The authors declare that they have no conflicts of interest with the contents of this article.

This article contains supplemental Figs. S1–S3.
- RTS
- Rothmund-Thomson syndrome
- RECQL4
- human RecQ-like helicase 4
- RPS3
- ribosomal protein S3
- AP
- apurnic-apyrimidinic
- aa
- amino acid(s)
- IP
- immunoprecipitation
- HhH
- helix-hairpin-helix
- 2ME
- β-mercaptoethanol
- FAM
- 5' 6-FAM (fluorescein).
References
- 1. Sangrithi M. N., Bernal J. A., Madine M., Philpott A., Lee J., Dunphy W. G., and Venkitaraman A. R. (2005) Initiation of DNA replication requires the RECQL4 protein mutated in Rothmund-Thomson syndrome. Cell 121, 887–898 [DOI] [PubMed] [Google Scholar]
- 2. Matsuno K., Kumano M., Kubota Y., Hashimoto Y., and Takisawa H. (2006) The N-terminal noncatalytic region of Xenopus RecQ4 is required for chromatin binding of DNA polymerase α in the initiation of DNA replication. Mol. Cell. Biol. 26, 4843–4852 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Macris M. A., Krejci L., Bussen W., Shimamoto A., and Sung P. (2006) Biochemical characterization of the RECQ4 protein, mutated in Rothmund-Thomson syndrome. DNA Repair 5, 172–180 [DOI] [PubMed] [Google Scholar]
- 4. Capp C., Wu J., and Hsieh T. S. (2009) Drosophila RecQ4 has a 3′-5′ DNA helicase activity that is essential for viability. J. Biol. Chem. 284, 30845–30852 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Suzuki T., Kohno T., and Ishimi Y. (2009) DNA helicase activity in purified human RECQL4 protein. J. Biochem. 146, 327–335 [DOI] [PubMed] [Google Scholar]
- 6. Xu X., and Liu Y. (2009) Dual DNA unwinding activities of the Rothmund-Thomson syndrome protein, RECQ4. EMBO J. 28, 568–577 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Rossi M. L., Ghosh A. K., Kulikowicz T., Croteau D. L., and Bohr V. A. (2010) Conserved helicase domain of human RecQ4 is required for strand annealing-independent DNA unwinding. DNA Repair 9, 796–804 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Keller H., Kiosze K., Sachsenweger J., Haumann S., Ohlenschläger O., Nuutinen T., Syväoja J. E., Görlach M., Grosse F., and Pospiech H. (2014) The intrinsically disordered amino-terminal region of human RecQL4: multiple DNA-binding domains confer annealing, strand exchange and G4 DNA binding. Nucleic Acids Res. 42, 12614–12627 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Capp C., Wu J., and Hsieh T. S. (2010) RecQ4: the second replicative helicase? Crit. Rev. Biochem. Mol. Biol. 45, 233–242 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Marino F., Mojumdar A., Zucchelli C., Bhardwaj A., Buratti E., Vindigni A., Musco G., and Onesti S. (2016) Structural and biochemical characterization of an RNA/DNA binding motif in the N-terminal domain of RecQ4 helicases. Sci. Rep. 6, 21501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Xu X., Rochette P. J., Feyissa E. A., Su T. V., and Liu Y. (2009) MCM10 mediates RECQ4 association with MCM2–7 helicase complex during DNA replication. EMBO J. 28, 3005–3014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Ohlenschläger O., Kuhnert A., Schneider A., Haumann S., Bellstedt P., Keller H., Saluz H. P., Hortschansky P., Hänel F., Grosse F., Görlach M., and Pospiech H. (2012) The N-terminus of the human RecQL4 helicase is a homeodomain-like DNA interaction motif. Nucleic Acids Res. 40, 8309–8324 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Singh D. K., Karmakar P., Aamann M., Schurman S. H., May A., Croteau D. L., Burks L., Plon S. E., and Bohr V. A. (2010) The involvement of human RECQL4 in DNA double-strand break repair. Aging Cell 9, 358–371 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Schurman S. H., Hedayati M., Wang Z., Singh D. K., Speina E., Zhang Y., Becker K., Macris M., Sung P., Wilson D. M. 3rd, Croteau D. L., and Bohr V. A. (2009) Direct and indirect roles of RECQL4 in modulating base excision repair capacity. Hum. Mol. Genet. 18, 3470–3483 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Fan W., and Luo J. (2008) RecQ4 facilitates UV light-induced DNA damage repair through interaction with nucleotide excision repair factor xeroderma pigmentosum group A (XPA). J. Biol. Chem. 283, 29037–29044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Warner J. R., and McIntosh K. B. (2009) How common are extraribosomal functions of ribosomal proteins? Mol. cell 34, 3–11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Yadavilli S., Hegde V., and Deutsch W. A. (2007) Translocation of human ribosomal protein S3 to sites of DNA damage is depandant on ERK-mediated phosphorylation following genotoxic stress. DNA Repair 6, 1453–1462 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Kim T. S., Kim H. D., and Kim J. (2009) PKCdelta-dependent functional switch of rpS3 between translation and DNA repair. Biochim. Biophys. Acta 1793, 395–405 [DOI] [PubMed] [Google Scholar]
- 19. Wan F., Weaver A., Gao X., Bern M., Hardwidge P. R., and Lenardo M. J. (2011) IKKβ phosphorylation regulates RPS3 nuclear translocation and NF-κB function during infection with Escherichia coli strain O157:H7. Nat. Immunol. 12, 335–343 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Lachapelle S., Gagné J. P., Garand C., Desbiens M., Coulombe Y., Bohr V. A., Hendzel M. J., Masson J. Y., Poirier G. G., and Lebel M. (2011) Proteome-wide identification of WRN-interacting proteins in untreated and nuclease-treated samples. J. Proteome Res. 10, 1216–1227 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Kim S. H., Lee J. Y., and Kim J. (2005) Characterization of a wide range base-damage-endonuclease activity of mammalian rpS3. Biochem. Biophys. Res. Commun. 328, 962–967 [DOI] [PubMed] [Google Scholar]
- 22. Kim J., Chubatsu L. S., Admon A., Stahl J., Fellous R., and Linn S. (1995) Implication of mammalian ribosomal protein S3 in the processing of DNA damage. J. Biol. Chem. 270, 13620–13629 [DOI] [PubMed] [Google Scholar]
- 23. Larizza L., Roversi G., and Volpi L. (2010) Rothmund-Thomson syndrome. Orphanet J. Rare Dis. 5, 2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Hegde V., Kelley M. R., Xu Y., Mian I. S., and Deutsch W. A. (2001) Conversion of the bifunctional 8-oxoguanine/β-δ apurinic/apyrimidinic DNA repair activities of Drosophila ribosomal protein S3 into the human S3 monofunctional β-elimination catalyst through a single amino acid change. J. Biol. Chem. 276, 27591–27596 [DOI] [PubMed] [Google Scholar]
- 25. Wan F., Anderson D. E., Barnitz R. A., Snow A., Bidere N., Zheng L., Hegde V., Lam L. T., Staudt L. M., Levens D., Deutsch W. A., and Lenardo M. J. (2007) Ribosomal protein S3: a KH domain subunit in NF-κB complexes that mediates selective gene regulation. Cell 131, 927–939 [DOI] [PubMed] [Google Scholar]
- 26. Jang C. Y., Kim H. D., Zhang X., Chang J. S., and Kim J. (2012) Ribosomal protein S3 localizes on the mitotic spindle and functions as a microtubule associated protein in mitosis. Biochem. Biophys. Res. Commun. 429, 57–62 [DOI] [PubMed] [Google Scholar]
- 27. Hegde V., Yadavilli S., and Deutsch W. A. (2007) Knockdown of ribosomal protein S3 protects human cells from genotoxic stress. DNA Repair 6, 94–99 [DOI] [PubMed] [Google Scholar]
- 28. Nagao-Kitamoto H., Setoguchi T., Kitamoto S., Nakamura S., Tsuru A., Nagata M., Nagano S., Ishidou Y., Yokouchi M., Kitajima S., Yoshioka T., Maeda S., Yonezawa S., and Komiya S. (2015) Ribosomal protein S3 regulates GLI2-mediated osteosarcoma invasion. Cancer Lett. 356, 855–861 [DOI] [PubMed] [Google Scholar]
- 29. Tian Y., Qin L., Qiu H., Shi D., Sun R., Li W., Liu T., Wang J., Xu T., Guo W., Kang T., Huang W., Wang G., and Deng W. (2015) RPS3 regulates melanoma cell growth and apoptosis by targeting Cyto C/Ca2+/MICU1 dependent mitochondrial signaling. Oncotarget 6, 29614–29625 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Ghosh A. K., Rossi M. L., Singh D. K., Dunn C., Ramamoorthy M., Croteau D. L., Liu Y., and Bohr V. A. (2012) RECQL4, the protein mutated in Rothmund-Thomson syndrome, functions in telomere maintenance. J. Biol. Chem. 287, 196–209 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Lee S. B., Kwon I. S., Park J., Lee K. H., Ahn Y., Lee C., Kim J., Choi S. Y., Cho S. W., and Ahn J. Y. (2010) Ribosomal protein S3, a new substrate of Akt, serves as a signal mediator between neuronal apoptosis and DNA repair. J. Biol. Chem. 285, 29457–29468 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Seong K. M., Jung S. O., Kim H. D., Kim H. J., Jung Y. J., Choi S. Y., and Kim J. (2012) Yeast ribosomal protein S3 possesses a β-lyase activity on damaged DNA. FEBS Lett. 586, 356–361 [DOI] [PubMed] [Google Scholar]
- 33. Werner S. R., Prahalad A. K., Yang J., and Hock J. M. (2006) RECQL4-deficient cells are hypersensitive to oxidative stress/damage: insights for osteosarcoma prevalence and heterogeneity in Rothmund-Thomson syndrome. Biochem. Biophys. Res. Commun. 345, 403–409 [DOI] [PubMed] [Google Scholar]
- 34. Ng A. J., Walia M. K., Smeets M. F., Mutsaers A. J., Sims N. A., Purton L. E., Walsh N. C., Martin T. J., and Walkley C. R. (2015) The DNA helicase recql4 is required for normal osteoblast expansion and osteosarcoma formation. PLoS Genet. 11, e1005160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Croteau D. L., Singh D. K., Hoh Ferrarelli L., Lu H., and Bohr V. A. (2012) RECQL4 in genomic instability and aging. Trends Genet. 28, 624–631 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Lu H., Fang E. F., Sykora P., Kulikowicz T., Zhang Y., Becker K. G., Croteau D. L., and Bohr V. A. (2014) Senescence induced by RECQL4 dysfunction contributes to Rothmund-Thomson syndrome features in mice. Cell Death Dis. 5, e1226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Yadavilli S., Mayo L. D., Higgins M., Lain S., Hegde V., and Deutsch W. A. (2009) Ribosomal protein S3: a multi-functional protein that interacts with both p53 and MDM2 through its KH domain. DNA Repair 8, 1215–1224 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Hoki Y., Araki R., Fujimori A., Ohhata T., Koseki H., Fukumura R., Nakamura M., Takahashi H., Noda Y., Kito S., and Abe M. (2003) Growth retardation and skin abnormalities of the Recql4-deficient mouse. Hum. Mol. Genet. 12, 2293–2299 [DOI] [PubMed] [Google Scholar]
- 39. Ahn B., Harrigan J. A., Indig F. E., Wilson D. M. 3rd, Bohr V. A. (2004) Regulation of WRN helicase activity in human base excision repair. J. Biol. Chem. 279, 53465–53474 [DOI] [PubMed] [Google Scholar]
- 40. Yacoub A., Augeri L., Kelley M. R., Doetsch P. W., and Deutsch W. A. (1996) A Drosophila ribosomal protein contains 8-oxoguanine and abasic site DNA repair activities. EMBO J. 15, 2306–2312 [PMC free article] [PubMed] [Google Scholar]
- 41. Wilson D. M. 3rd, Takeshita M., Grollman A. P., and Demple B. (1995) Incision activity of human apurinic endonuclease (Ape) at abasic site analogs in DNA. J. Biol. Chem. 270, 16002–16007 [DOI] [PubMed] [Google Scholar]
- 42. Hegde V., Wang M., Mian I. S., Spyres L., and Deutsch W. A. (2006) The high binding affinity of human ribosomal protein S3 to 7,8-dihydro-8-oxoguanine is abrogated by a single amino acid change. DNA Repair 5, 810–815 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.