Abstract
To study the regulation of colorectal adenocarcinoma progression by O-GlcNAc, we have focused on the O-GlcNAc-mediated epigenetic regulation of human colon cancer stem cells (CCSC). Xenograft tumors from colon tumor cells with O-linked N-acetylglucosamine transferase (OGT) knockdown grew significantly slower than those formed from control cells, indicating a reduced proliferation of tumor cells due to inhibition of OGT expression. Significant reduction of the CCSC population was observed in the tumor cells after OGT knockdown, whereas tumor cells treated with the O-GlcNAcase inhibitor showed an increased CCSC population, indicating that O-GlcNAc levels regulated the CCSC compartment. When grown in suspension, tumor cells with OGT knockdown showed a reduced ability to form tumorspheres, indicating a reduced self-renewal of CCSC due to reduced levels of O-GlcNAc. ChIP-sequencing experiments using an anti-O-GlcNAc antibody revealed significant chromatin enrichment of O-GlcNAc-modified proteins at the promoter of the transcription factor MYBL1, which was also characterized by the presence of H3K27me3. RNA-sequencing analysis showed an increased expression of MYBL1 in tumor cells with OGT knockdown. Forced overexpression of MYBL1 led to a reduced population of CCSC and tumor growth in vivo, similar to the effects of OGT silencing. Moreover, two CpG islands near the transcription start site of MYBL1 were identified, and O-GlcNAc levels regulated their methylation status. These results strongly argue that O-GlcNAc epigenetically regulates MYBL1, functioning similarly to H3K27me3. The aberrant CCSC compartment observed after modulating O-GlcNAc levels is therefore likely to result, at least in part, from the epigenetic regulation of MYBL1 expression by O-GlcNAc, thereby significantly affecting tumor progression.
Keywords: colon cancer, epigenetics, gene knockout, glycosylation, glycosyltransferase, O-linked N-acetylglucosamine (O-GlcNAc), stem cells
Introduction
The post-translational modification of O-linked N-acetylglucosamine (O-GlcNAc)3 is a unique type of glycosylation of serine and/or threonine residues of a wide variety of cytosolic, nuclear, and mitochondrial proteins. The product is a single β-linked N-acetylglycosamine residue that is not further elaborated (1–3). Unlike the glycosylation typical of membrane-bound or secreted proteins, such as N-glycans, mucin type O-glycans, or glycophosphatidyl anchors, these O-GlcNAc residues cycle on and off in a dynamic and inducible manner in response to cellular stimuli (4, 5). O-GlcNAcylation is catalyzed by a single enzyme, termed O-linked N-acetylglucosamine transferase (OGT), which transfers N-acetylglucosamine from UDP-GlcNAc to protein substrates (6), whereas the removal of O-GlcNAc modification is catalyzed by a single enzyme known as N-acetyl-β-glucosaminidase (OGA) (7). These enzymes dynamically regulate whether any protein will express O-GlcNAc and its function in response to various stimuli. O-GlcNAc-modified proteins have critical functions in the regulation of cellular processes such as nutrient sensing, insulin signaling, cell cycle control, transcriptional regulation of protein expression, protein trafficking, and protein-protein interactions (3, 8, 9).
The modulation of O-GlcNAc expression is important in oncogenic signaling and cancer phenotypes (10). For example, aberrant expression levels of O-GlcNAc have been found in human tumor tissues compared with matched adjacent normal tissue for cancers of the breast, lung, colon, and thyroid (9, 11–13). Metastatic breast cancer cell lines have high expression levels of both OGT and O-GlcNAc, and experimental silencing of OGT expression in these cell lines leads to inhibition of tumor growth both in vivo and in vitro through targeting of an oncogenic transcription factor FoxM1 (14). Knockdown of OGT in lung and colon cancer cells results in reduced tumor cell growth and invasion in vitro (13), whereas elevated O-GlcNAcylation was found to promote colonic inflammation and tumorigenesis by modulating NF-κB signaling (15). Another recent study showed that MAPK/ERK signaling positively regulates OGT expression and O-GlcNAcylation in various tumor cells, which, in turn, enhances the effects of ERK signaling on cancer cell proliferation and anchorage-independent growth (16). Moreover, O-GlcNAcylation regulates cancer cell metabolic reprogramming and survival stress signaling via regulation of HIF-1α (17), and increased glucose uptake promotes oncogenesis via O-GlcNAc pathways (18). All these results suggest that O-GlcNAcylation may represent a novel regulator of tumorigenesis and tumor progression.
O-GlcNAcylation also has important roles in the epigenetic regulation of embryonic development (19–21). A number of chromatin-associated proteins and transcription factors, such as RNA polymerase II, are O-GlcNAcylated, which play a pivotal role in regulating chromatin structure and the stability of particular transcription factors. O-GlcNAc dynamically regulates chromatin structure both in vivo and in vitro. O-GlcNAcylation of histone (H2A, H2B, and H4) changes during mitosis and with heat shock and is considered part of the histone code (22). Dynamic O-GlcNAc cycling in the promoter regions of genes linked to longevity, stress, and immunity has been revealed in Caenorhabditis elegans using ChIP-chip arrays (23). Interestingly, two reports (24, 25) independently demonstrated that Drosophila OGT is encoded by super sex combs (sxc), an essential component of the polycomb group (PcG) repressive complex, directly linking O-GlcNAcylation to PcG-mediated gene silencing by histone methylation, modifications that are necessary for early embryonic development, and stem cell maintenance (26, 27).
The role of O-GlcNAc in early development was investigated using mouse embryonic stem cells (ESC). Myers et al. (27) identified 142 modification sites of O-GlcNAc for over 60 proteins, many of which are involved in transcriptional regulation implicated in ESC pluripotency and self-renewal. In addition, they found that polycomb repressive complex 2 (PRC2) is necessary to maintain normal levels of OGT and for the correct cellular distribution of O-GlcNAc-modified proteins in ESC. Other studies showed that OGT was involved in the regulation of TET1, a family member of dioxygenases that catalyzes the hydroxylation of 5-methylcytosine and represses gene expression. OGT can positively regulate protein levels of TET1 (28), and it preferentially causes association of TET1 with gene promoters in close proximity of CpG-rich transcription start sites (29). In addition, GlcN-induced OGT activation mediated glucose production through cleaved Notch1 and FoxO1, which contributed to the regulation of maintenance of self-renewal in mouse ESCs (30). These results suggest that the O-GlcNAc pathway has a functional connection to polycomb repressive complex and is implicated in the regulation of stem cells.
It has been proposed that a variety of cancers, including colon cancer, are initiated and maintained by a small proportion of cells, called tumor-initiating cells or cancer stem cells (CSC), which display stem cell properties, including self-renewal and differentiation (31). There is a growing body of evidence that CSC play an important role in not only cancer development but also recurrence and metastasis of some tumors (32). Small numbers of CSC can rapidly produce tumors upon transplantation in animal models, whereas non-CSC produce tumors very slowly. CSC have been shown to have high resistance to chemotherapeutic treatment in some cancers, and the surviving CSC contribute to cancer recurrence (33). In addition, CSC acquire mesenchymal characteristics after undergoing epithelial-mesenchymal transition and migrate and invade target organs through blood and lymph vessels (32, 33). Therefore, the characterization of key signaling proteins and pathways that regulate the self-renewal and maintenance of colon CSCs (CCSC) is crucial for understanding how CSC regulate colon tumorigenesis and progression, as well as for the development of novel treatment strategies targeting the CSC. CCSC have been identified and isolated from both human colon cancer (34–37) and mouse colon adenoma (38). We recently reported that expression levels of β1,6-N-acetylglucosaminyltransferase V, which synthesizes N-glycans with β(1,6)-branched glycans on cell surface receptors, regulate the canonical Wnt/β-catenin signaling pathway by affecting the N-glycosylation of Wnt receptors, which further leads to altered colon adenoma progression in Apcmin/+ mice by modulating the self-renewal and tumorigenicity of CCSC (37).
Although studies have revealed an association between increased O-GlcNAc expression and colon tumor development, the mechanisms by which colon tumor progression was regulated by aberrant expression of O-GlcNAc have not been fully elucidated. In this study, the effects of O-GlcNAc expression levels on colorectal cancer stem cells and tumorigenesis were investigated. We provide evidence that the epigenetic modification can regulate the CSC population and colon tumor development. MYBL1, a transcription activator, was identified and implicated, at least in part, in the regulation of colon tumor development mediated by altered O-GlcNAcylation.
Results
Regulation of Colon Cancer Stem Cells and Colon Tumorigenesis by Expression Levels of O-GlcNAc
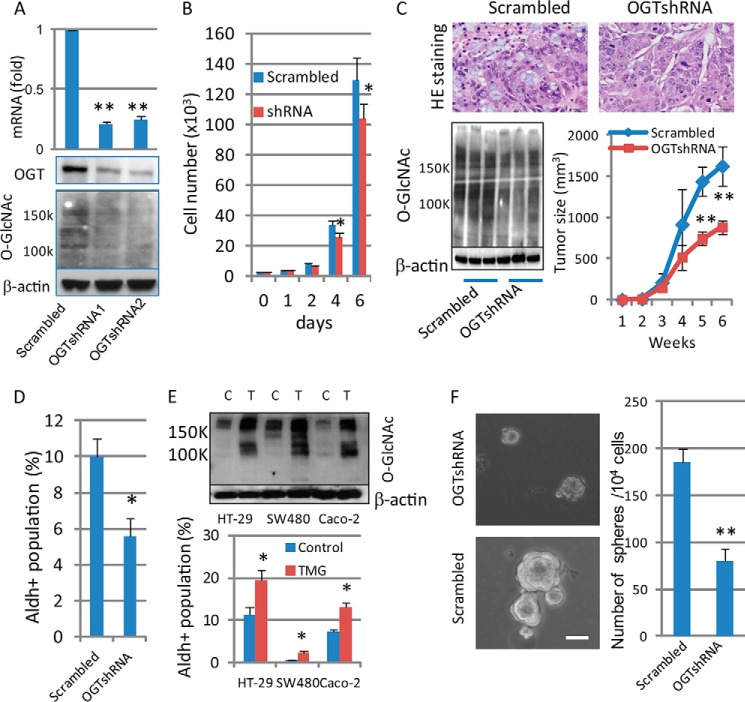
To determine the effects of altering OGT activity on colon cancer tumorigenesis and the cancer stem cell (CCSC) compartment, the expression of OGT was silenced in HT-29 cells by shRNA lentivirus infection and confirmed by RT-PCR and Western blotting (Fig. 1A). As shown in Fig. 1B, tumor cells with OGT-shRNA knockdown showed a 20% reduced growth in vitro after 6 days. Both OGT knockdown and control (scrambled) tumor cells formed tumors as xenografts when injected into NOD/SCID mice, confirmed by H&E staining (Fig. 1C, top). Tumors formed by injection of cells with OGT-shRNA expression showed decreased expression of O-GlcNAc detected by Western blotting (Fig. 1C, bottom left) and grew significantly slower than the tumors formed by injection of control cells (Fig. 1C, bottom right), indicating a reduced proliferation of tumor cells in vivo due to inhibition of OGT expression. Because increased activity of aldehyde dehydrogenase 1 (ALDH1), detected by the aldefluor assay, is a characteristic of CCSC, this assay has been used in the detection and enrichment of CCSC (36, 39). We utilized this methodology to demonstrate that CCSC were enriched in the aldefluor-positive cell population from two colon cancer cell lines (37). Significant reductions in the proportion of CCSC detected by the Aldefluor assay in the total tumor cell population were observed after OGT knockdown, compared with control cells (Fig. 1D). To test further the regulation of CCSC by O-GlcNAcylation levels, tumor cells were treated with Thiamet-G (TMG, 10 μm), a specific OGA inhibitor causing increased levels of O-GlcNAc (40), and the population of CCSC was measured. Tumor cells with TMG treatment showed an increased CCSC population in three different colon adenocarcinoma cell lines (Fig. 1E), indicating the regulation of the CCSC compartment by enhanced O-GlcNAc levels. These results demonstrated that manipulating O-GlcNAc levels regulated the relative size of the compartment of CCSC in either a positive (increased O-GlcNAc levels) or negative (decreased O-GlcNAc levels) direction. To identify a mechanism by which O-GlcNAc levels affect the population of CCSC, we next focused on the growth of tumorspheres in suspension culture. We reported that HT-29 cells, when grown in serum-free stem cell culture media for 7–10 days, formed typical tumorspheres (37), a measurement of the self-renewal of stem cells (41–43). We found that gene expression of aldehyde dehydrogenase-1 (ALDH1) was increased in tumorspheres compared with parental cells grown on plastic plates (37), supporting the enrichment of CCSC in tumorspheres. As shown in Fig. 1F, although both control and OGT knockdown cells showed the ability to form tumorspheres in suspension culture for 7–10 days, both the number and size of tumorspheres were significantly decreased in OGT knockdown cells compared with the control cells, consistent with the reduced population of CCSC after knockdown of OGT (Fig. 1D). These results demonstrate that O-GlcNAc levels regulate colon tumorigenesis by modulating the self-renewal of CCSC, which results in altered proportions of CCSC in the population of cultured colon cancer cells.
FIGURE 1.
Expression levels of O-GlcNAc regulate colon cancer stem cells and colon tumorigenesis. A, HT-29 cells were infected with scrambled or OGT-shRNAs lentivirus, and expressions of OGT and O-GlcNAc were detected by qRT-PCR (top) and Western blots (bottom). For each transcript, the values were normalized to control (GAPDH) and expressed as mean ± S.D. from three independent experiments. B, both control (scrambled) and OGT knockdown HT-29 cells (1 × 103) were split into 6-well plates and grown at 37 °C for the indicated days, and cell numbers were counted. Data are expressed as mean ± S.D. from three wells. C, tumor cells (2 × 106) with scrambled or OGT-shRNA expression were injected into SCID mice (n = 5), and secondary tumor growth was observed for up to 6 weeks. Tumors were dissected at week 6, and tumor tissues were collected for H&E staining (top) and Western blotting with anti-O-GlcNAc antibody using tumor tissues collected from three mice of each group (left bottom). Tumor size was measured every week and expressed as the mean with error bars indicating ±1 S.D. (n = 5; right bottom). D, Aldefluor assays were performed using HT-29 tumor cells with scrambled or OGT-shRNA expression, and the percentage of Aldefluor-positive cells was determined under similar gating criteria. Diethylaminobenzaldehyde, inhibitor of aldehyde dehydrogenase, was used to determine the background of the assay. Data are expressed as mean ± S.D. from three independent experiments. E, after treatment with OGA inhibitor (TMG, 10 μm) for 24 h, tumor cells were collected for immunoblotting using anti-O-GlcNAc (top) and Aldefluor assays (bottom). C, control; T, TMG treatment. F, control and OGT knockdown HT-29 cells (4000 cells/well) were grown in stem cell culture media in suspension for 7–10 days. Tumorspheres were formed (left panel), and the number of tumorspheres was counted and expressed as mean ± S.D. from 3 to 5 wells (right panel). Similar results were obtained from two independent experiments. Scale bars, 50 μm. *, p < 0.05; **, p < 0.01.
Identification of O-GlcNAc-bound Genes in HT-29 Cells
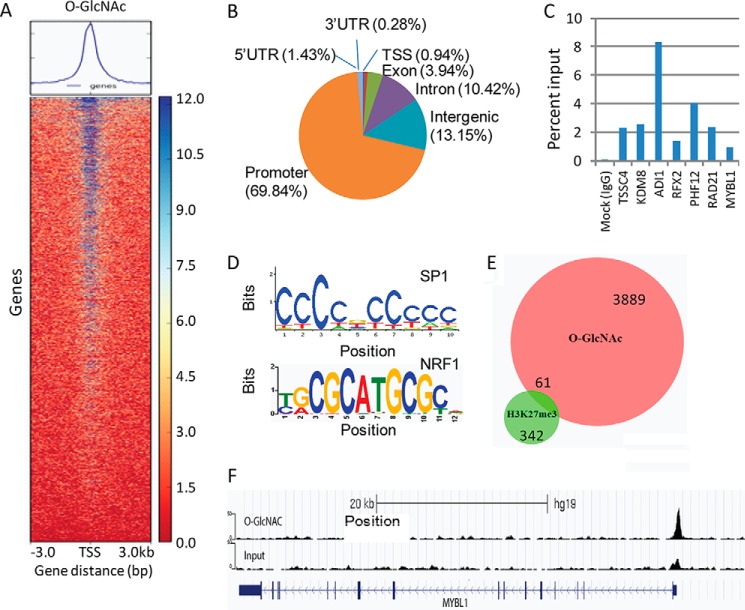
Because both O-GlcNAc and CSC have been implicated in tumorigenesis and tumor progression and the emerging roles of O-GlcNAc have been highlighted in development and epigenetic regulation, we asked the question whether O-GlcNAc expression levels are involved in the regulation of CCSC, thus regulating tumor development, and whether the regulation of CCSC by O-GlcNAc is through an epigenetic mechanism. To this end, we performed ChIP-seq using an anti-O-GlcNAc antibody along with an antibody against H3K27me3, a well known histone repression marker involved in PRC2-mediated repression (44). A total of 4184 significant O-GlcNAc peaks were identified after ChIP-seq using the anti-O-GlcNAc antibody (Fig. 2A). Among these sites, 2922 (69.8%) were located near transcription start sites (TSS) (Fig. 2B). Seven genes enriched in these peaks were randomly chosen for validation by ChIP-qPCR (Fig. 2C). Motif enrichment analysis of the O-GlcNAc-binding sites showed that the top two occurring motifs (CCCCGCCCH and CGCANGCGC) were most similar to known binding sites of SP1 and NRF1, respectively (Fig. 2D). Although only a total 459 H3K27me3 peaks were “called” by ChIP-seq using the anti-H3K27me3 antibody, we did observe several polycomb repressive complex target genes enriched in H3K27me3 peaks, such as CBX4, PAX7, FGF14, FOXB1, WNT11, as reported previously (45). Using a Venn diagram, 61 genes were identified (Fisher's exact test, p < 1.554e-04) from overlapped areas identified from H3K27me3 and O-GlcNAc ChIP-seq (Fig. 2E and Table 1), indicating an overlap of gene-binding sites utilized by O-GlcNAc and H3K27me3. These data strongly suggest that O-GlcNAc is likely be involved in the epigenetic regulation of gene expression, exerting a similar function to H3K27me3.
FIGURE 2.
Identification of O-GlcNAc-bound genes in HT-29 cells by ChIP-seq. A, BAM files from the sequencing reads aligned with Bowtie2.0 were uploaded to Deep Tools for promoter region score calculation, which was further used for the generation of a heat map from O-GlcNAc ChIP. B, distribution of O-GlcNAc peaks identified by ChIP-seq using anti-O-GlcNAc antibody. C, seven genes enriched in these peaks were randomly chosen for validation by ChIP-qPCR. Data were described as mean value of percentage of input from two independent ChIP experiments. D, de novo motif enrichment analysis of O-GlcNAc-binding sites identified top two motifs that showed similarities to known binding sites of transcription factors SP1 and NRF1, respectively. E, Venn diagram showing the overlapped genes enriched in antibody binding peaks that were occupied by both O-GlcNAc and H3K27me3 antibodies. F, both input and O-GlcNAc ChIP-Seq reads were aligned to human genome reference (Hg19) Bowtie2.0. Sequencing peaks at the promoter region of MYBL1 were viewed in the UCSC genome browser.
TABLE 1.
The list of genes identified by ChIP-seq using anti-O-GlcNAc that overlapped with genes from H3K27me3 ChIP-seq
FDR is false discovery rate.
| Gene | p value | FDR | Gene | p value | FDR |
|---|---|---|---|---|---|
| % | % | ||||
| RSG1 | 1.23492E-06 | 0.183298346 | SEC22B | 8.46227E-07 | 1.83856E-08 |
| NTPCR | 1.46463E-06 | 0.1263328 | TTC32 | 9.40929E-10 | 0.000343856 |
| EMX1 | 9.19534E-06 | 4.008194953 | MED12L | 8.47196E-05 | 3.601706652 |
| RAP2B | 1.29537E-06 | 0.570660568 | CCNI | 3.10862E-15 | 6.58364E-15 |
| ARHGAP24 | 8.47196E-05 | 1.165785414 | TRIO | 3.11086E-06 | 0.413906601 |
| LMNB1 | 1.28455E-07 | 0.000397906 | RPP40 | 1.11525E-11 | 1.18464E-21 |
| USP42 | 1.77353E-11 | 4.15416E-08 | FIGNL1 | 1.61369E-11 | 0.037522696 |
| GTPBP10 | 1.33227E-15 | 5.75E-12 | PNPLA8 | 3.06915E-07 | 0.009889463 |
| RBM28 | 9.67559E-13 | 1.51649E-10 | MNX1 | 3.37413E-07 | 4.608248287 |
| MYBL1 | 7.85261E-05 | 4.647272125 | MED30 | 3.13390E-08 | 0.016218826 |
| MRPL13 | 3.13390E-08 | 0.611435568 | SNTB1 | 3.33956E-05 | 2.044364544 |
| TRIB1 | 1.24906E-05 | 0.014482876 | ASAP1 | 2.96965E-05 | 1.080569959 |
| ST3GAL1 | 1.27569E-05 | 0.593707037 | ZFAT | 3.33998E-05 | 0.854230646 |
| COL22A1 | 6.72013E-05 | 0.655773401 | KCNK9 | 7.47744E-05 | 0.000329677 |
| EIF2C2 | 8.59442E-05 | 0.07040097 | PLEC | 3.30403E-10 | 0.000285762 |
| IDI1 | 1.26085E-07 | 0.00584926 | KLF6 | 4.44089E-16 | 1.37679E-10 |
| RRP8 | 3.61890E-10 | 0.510656388 | RBM4B | 5.11351E-10 | 4.70867E-08 |
| ARHGAP9 | 4.91897E-08 | 1.303E-05 | NDUFA12 | 0 | 9.74035E-08 |
| ZMYM2 | 0 | 1.66969E-07 | MTMR6 | 1.33368E-08 | 2.235485308 |
| RXFP2 | 9.63760E-05 | 0.326004955 | CCPG1 | 1.47331E-08 | 2.64487E-06 |
| ZNHIT3 | 8.28504E-13 | 0.00025914 | SRCIN1 | 0 | 1.16445E-20 |
| KPNA2 | 3.47843E-07 | 0.01720213 | ENPP7 | 7.64202E-05 | 1.480910916 |
| CBX4 | 2.39568E-05 | 4.130425379 | PTPN2 | 0 | 4.92844E-06 |
| C19orf70 | 3.90687E-13 | 2.22385E-14 | TDRD12 | 3.01183E-06 | 0.77294203 |
| SHKBP1 | 2.20817E-05 | 7.27095E-05 | FBXO46 | 0 | 1.11842E-16 |
| MYPOP | 0 | 8.75602E-93 | CCDC8 | 5.40018E-11 | 0.003746069 |
| STRN4 | 2.93344E-05 | 1.395307868 | PRR24 | 2.54327E-06 | 0.023498096 |
| TMC4 | 4.26387E-07 | 1.01104E-07 | ZNF865 | 2.74072E-11 | 4.1227E-06 |
| ZNF784 | 0 | 5.40558E-09 | UBE2 M | 9.83912E-09 | 0.004195453 |
| ZNF337 | 0 | 4.85132E-23 | CLDN5 | 6.47437E-05 | 2.430510618 |
| RANGAP1 | 8.20677E-12 | 4.56174E-09 |
Gene Expression Profiling Regulated by O-GlcNAc
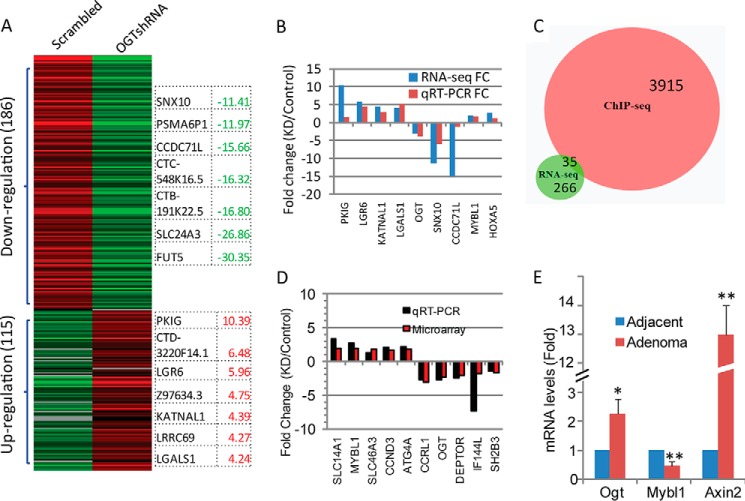
To study further whether altered OGT expression levels affect the expression of genes that were identified by ChIP-seq using the anti-O-GlcNAc antibody and were involved in the regulation of stem cells, gene expression profiling of tumor cells with OGT knockdown (shRNA) was determined in HT-29 cells by RNA-seq. Using a cutoff value of fold-change >1.5 and p < 0.05, a total 301 genes were identified to be differentially expressed in tumor cells with OGT knockdown, among which 115 genes were up-regulated and 186 genes were down-regulated (Fig. 3A). Twelve transcription factors were found among significantly affected genes by suppression of OGT (Table 2). The top differentially affected genes by OGT knockdown are listed in Fig. 3A. A validative qRT-PCR was performed for detection of some of these genes that showed changes by RNA-seq (Fig. 3B). By comparison of results from both RNA-seq and ChIP-seq, 35 out of 301 altered genes overlapped with genes identified by ChIP using anti-O-GlcNAc antibody (Fig. 3C and Table 3). The gene encoding transcription factor MYBL1 was then identified in a complex that was bound by the anti-O-GlcNAc antibody (Fig. 2F) and was up-regulated in cells with reduced OGT. Increased transcription of MYBL1 after knockdown of OGT was further confirmed by a gene expression microarray experiment (data not shown), and altered genes, including MYBL1, were chosen for validation by qRT-PCR (Fig. 3D). Interestingly, MYBL1 was also among the overlapped genes identified by ChIP-seq using both anti-O-GlcNAc and anti-H3K27me3 antibodies (Table 1). H3K27me3 is a well known repressive histone marker (44), indicating that MYBL1 was very likely involved in the epigenetic regulation of colon cancer stem cells and tumorigenesis mediated by O-GlcNAc. To further confirm the up-regulation of MYBL1 obtained from cell lines, we performed qRT-PCR experiments using pooled total RNA samples isolated from Apc mutation-induced mouse colon adenoma tissues (37). As shown in Fig. 3E, Ogt expression was increased in adenoma tissues, accompanied by increased expression of Axin2, a target gene of the Wnt/β-catenin signaling pathway that was activated in colon adenomas and carcinomas (37, 46–48), compared with adjacent normal tissues, whereas Mybl1 expression was significantly reduced in adenoma tissues (p < 0.01), which was consistent with the results from the cell lines that silencing of OGT increased MYBL1 gene expression. Also, these results were supported by a recent report showing decreased expression of both MYB and MYBL1 in human colorectal cancer tissues than adjacent normal tissues (49).
FIGURE 3.
Gene expression profiling regulated by O-GlcNAc. A, total RNA was isolated from scrambled and OGT-shRNA transfected HT-29 cells and used for RNA-seq analysis. A heat map of 301 transcripts differentially expressed (fold-change >1.5 and p < 0.05) between control and OGT knockdown cells was generated from the RNA-seq data. Genes showing the greatest fold-change were shown by the heat map. Red indicates a high expression level, and green indicates a low expression level compared with control cells. KD, OGT knockdown (shRNA). B, isolated total RNA was used for validation of transcript levels of altered genes using qRT-PCR. For each transcript, the values expressed as fold-change, calculated from three independent experiments, were compared with those from RNA-seq. C, Venn diagram showing the overlapped genes identified by ChIP-seq by O-GlcNAc and RNA-seq. D, total RNA was isolated from scrambled and OGT-shRNA transfected HT-29 cells and used for microarray analysis. The altered gene expression was validated using qRT-PCR and expressed by fold-change in comparison with those from microarray data. E, total RNA was pooled from four adjacent and adenoma tissues of Apcmin/+ mice, and expression of Ogt, Axin2, and Mybl1 were detected by qRT-PCR. For each transcript, the values were normalized to control (Gapdh) and expressed as mean ± S.D. from three independent experiments. *, p < 0.05; **, p < 0.01.
TABLE 2.
Regulated transcription factors by knockdown (shRNA) of OGT detected by RNA-seq
KD indicates knockdown (shRNA).
| Gene symbol | -Fold change (KD/control) | p value | Q value |
|---|---|---|---|
| CDX2 | −2.75 | 9.0207E-09 | 1.0971E-06 |
| EGR1 | −1.65 | 4.9268E-32 | 3.1278E-29 |
| FOXA2 | −1.51 | 6.4186E-12 | 1.2402E-09 |
| HOXA5 | 2.78 | 0 | 0 |
| HOXB9 | 1.86 | 0 | 0 |
| HR | −2.77 | 3.2045E-13 | 7.4648E-11 |
| KLF2 | −2.03 | 4.8405E-10 | 7.2445E-08 |
| MAFB | −1.79 | 1.2591E-07 | 1.2204E-05 |
| MYBL1 | 1.85 | 9.9399E-11 | 1.6454E-08 |
| TRIM29 | 1.51 | 0 | 0 |
| YBX2 | −2.26 | 1.1153E-26 | 6.0783E-24 |
| ZIC2 | −3.17 | 2.7692E-08 | 3.0706E-06 |
TABLE 3.
35 genes identified by RNA-seq are among genes identified by ChIP-seq using anti-O-GlcNAc
| Gene | -fold change (KD/control) | p value | Q value |
|---|---|---|---|
| AC002467.7 | 1.51 | 6.3024E-08 | 6.4785E-06 |
| AC011747.4 | −1.71 | 1.968E-07 | 1.8517E-05 |
| ANXA13 | −3.77 | 1.563E-201 | 1.254E-198 |
| ATP6V0E2-AS1 | 1.58 | 2.4349E-09 | 3.2487E-07 |
| BBS9 | 2.38 | 2.2204E-16 | 7.5904E-14 |
| DOCK4 | 2.13 | 2.2143E-10 | 3.524E-08 |
| EGR1 | −1.65 | 4.9268E-32 | 3.1278E-29 |
| FBXO48 | 2.04 | 7.4336E-12 | 1.4127E-09 |
| GMFG | 1.798E + 308 | 1.5299E-13 | 3.6223E-11 |
| HACE1 | 2.27 | 1.2464E-08 | 1.4695E-06 |
| IGSF8 | −1.95 | 9.9362E-15 | 2.7205E-12 |
| KATNAL1 | 4.40 | 1.8088E-12 | 3.7187E-10 |
| LYST | 1.62 | 4.428E-07 | 3.9295E-05 |
| MAFB | −1.79 | 1.2591E-07 | 1.2204E-05 |
| MGEA5 | −2.26 | 1.4649E-17 | 5.642E-15 |
| MRPL54 | −1.54 | 6.9279E-29 | 4.0427E-26 |
| MYBL1 | 1.85 | 9.9399E-11 | 1.6454E-08 |
| N6AMT2 | 1.56 | 0 | 0 |
| NDRG1 | −2.44 | 1.8993E-15 | 5.7749E-13 |
| NUDT7 | 1.85 | 9.6922E-08 | 9.5064E-06 |
| OGT | −3.20 | 3.8352E-13 | 8.8981E-11 |
| PCLO | 2.48 | 0 | 0 |
| PLEKHB1 | 2.15 | 4.7533E-08 | 5.0852E-06 |
| PRSS16 | −1.72 | 1.8037E-16 | 6.3927E-14 |
| RAB4B-EGLN2 | −1.72 | 3.9211E-28 | 2.1993E-25 |
| RP11–637A17.2 | 1.798E + 308 | 1.9756E-10 | 3.197E-08 |
| RP3–395M20.8 | −1.58 | 3.7513E-07 | 3.3704E-05 |
| RSU1 | 1.80 | 0 | 0 |
| SIGLEC12 | −3.96 | 8.6783E-29 | 5.0136E-26 |
| SLC23A2 | −1.53 | 5.0136E-26 | 4.7461E-22 |
| SMPD3 | −1.92 | 4.4591E-10 | 6.7324E-08 |
| TMED4 | 1.76 | 0 | 0 |
| TMEM180 | 1.74 | 1.2035E-13 | 2.8849E-11 |
| UBAC2-AS1 | 2.51 | 0 | 0 |
| ZNF613 | 2.20 | 4.9853E-09 | 6.3437E-07 |
Tumor-suppressive Functions of Transcription Factor MYBL1
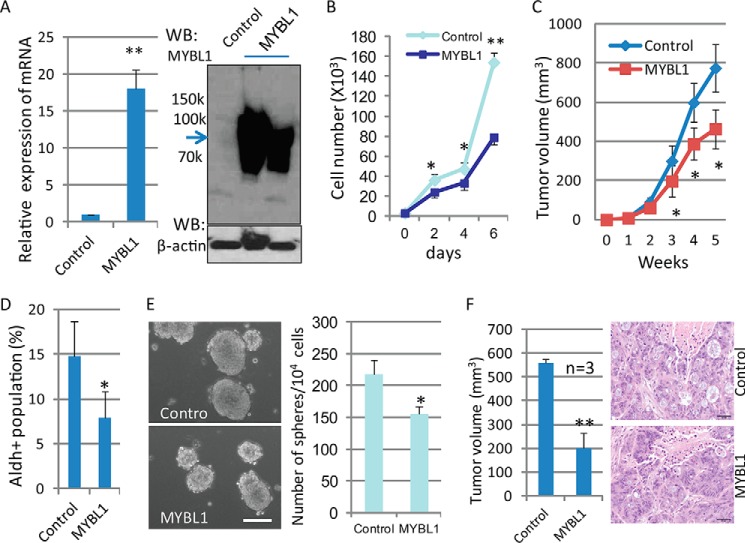
MYBL1, a Myb family member, is a strong transcriptional activator and has been implicated in the regulation of proliferation, differentiation, and apoptosis of hematopoietic cells (50, 51). To determine whether the differential expression of the MYBL1 following knockdown of OGT contributed to the reduction in the population of colon cancer stem cells and inhibited colon tumorigenesis, experiments for functional validation were performed in vitro and in vivo. As shown in Fig. 4, overexpression of MYBL1 in HT-29 cells, confirmed by qRT-PCR and Western blotting (Fig. 4A), significantly inhibited tumor cell growth compared with control cells (Fig. 4B), indicating an inhibition of cell proliferation in these cells due to the exogenous expression of the MYBL1 gene. To confirm further the ability of the MYBL1 gene to inhibit tumor progression in vivo, we next tested the ability of tumor cells overexpressing MYBL1 to form tumors in NOD/SCID mice. Slower tumor growth was observed in xenografts resulting from injection of tumor cells with MYBL1 overexpression, compared with control cells (Fig. 4C). Consistent with inhibited tumorigenesis, reduced populations of cancer stem cells as detected by the Aldefluor assay (Fig. 4D) and reduced stem cell self-renewal were also observed in MYBL1-expressing tumor cells (Fig. 4E). Rapid rates of tumor formation in NOD/SCID mice are another fundamental characteristic of CSC. To characterize the tumor-forming ability of CCSC purified from both control and MYBL1-overexpressing tumor cells, the same number of CCSC (103) was sorted from cell cultures using the Aldefluor assay and injected into NOD/SCID mice (Fig. 4F). Although CCSC isolated from both control and MYBL1-overexpressing cells formed epithelial tumors, as confirmed by H&E staining, tumor growth was significantly reduced in those mice that received CCSC with MYBL1 overexpression. Our results are supportive of the fact that MYBL1 functions to suppress tumor progression and suggest that the reduced cancer stem cell population and inhibited colon tumorigenesis observed after knockdown of OGT is likely due to enhanced expression of MYBL1.
FIGURE 4.
Tumor-suppressive functions of MYBL1. A, HT-29 cells were transfected with MYBL1 cDNA, and expression of MYBL1 was detected by qRT-PCR (left panel) and Western blot (WB) (right panel). For each transcript, the values were normalized to control (GAPDH) and expressed as mean ± S.D. from three independent experiments. B, both control and MYBL-expressing HT-29 cells (1 × 103) were split into 6-well plates and grown at 37 °C for the indicated days, and cell numbers were counted. Data were expressed as mean ± S.D. from three wells. C, tumor cells (2 × 106) with either control or MYBL1 expression were injected into SCID mice (n = 6), and secondary tumor growth was observed for up to 6 weeks. Tumor size was measured every week and expressed as mean ± S.D. (n = 6). D, Aldefluor assays were performed using control and MYBL1-transfected HT-29 tumor cells, and the percentage of Aldefluor-positive cells was determined using similar gating criteria. Data are expressed as mean ± S.D. from three independent experiments. E, control and MYBL1-expressing HT-29 cells (4000 cells/well) were grown in stem cell culture media in suspension for 7–10 days; tumorspheres were formed, and the number of spheres was counted and expressed as mean ± S.D. from 3 to 5 wells. Similar results were obtained from two independent experiments. Scale bars, 50 μm. F, Aldefluor-positive cells (3 × 103) isolated from control and MYBL1-expressing cells were injected subcutaneously into the backs of NOD/SCID mice (n = 3); secondary tumor growth was observed for up to 8 weeks (left), and tumor size was measured and expressed as the mean ± S.D. (n = 3). Tumors were dissected at week 8, and H&E staining was performed (right). *, p < 0.05; **, p < 0.01.
MYBL1 Was Epigenetically Regulated by O-GlcNAc
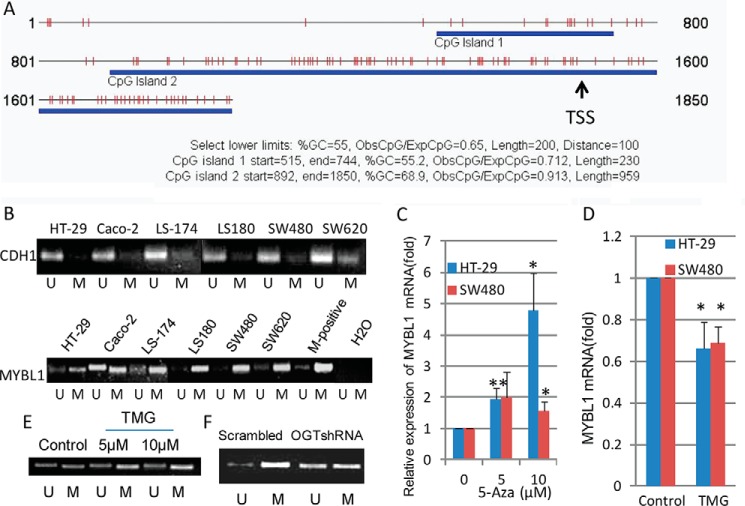
Epigenetic aberrations are frequent events in human colon cancer development (52, 53), and promoter methylation has been implicated in the epigenetic regulation of tumor-suppressive genes in colon cancer (54). To determine whether altered expression of MYBL1 in OGT-knockdown tumor cells was caused by promoter methylation differences, the promoter methylation status of the MYBL1 gene was analyzed. We first searched the human MYBL1 gene for CpG islands around the TSS (−1.5 to + 0.35 kb) using the program CpG Island Searcher. As shown in Fig. 5A, two CpG islands around the TSS area were found in MYBL1. To determine the levels of methylation, methylation-specific PCR targeting the area surrounding the TSS was performed on the MYBL1 gene with predicted CpG island(s) around its TSS using six colon tumor cell lines. Interestingly, CDH1, whose gene expression is silenced due to promoter hypermethylation in many human tumors, including primary colorectal carcinomas (55), showed very low promoter methylation in tumor cell lines (Fig. 5B, top), which was consistent with previous reports (55, 56). By contrast, very strong promoter methylation of MYBL1 was observed in all tumor cell lines, except the Caco-2 line in which a strong unmethylated band was also observed, likely suggesting a relatively low methylation of MYBL1 in Caco-2 cells (Fig. 5B, bottom). Treatment of tumor cells with the demethylation compound, 5-aza-2′-deoxycytidine, increased expression of MYBL1 (Fig. 5C), indicating that the transcription of the MYBL1 gene can be regulated by methylation status in two colon tumor cell lines. When treated with the OGA inhibitor, TMG, HT-29 cells showed increased levels of O-GlcNAc (Fig. 1E, top) and reduced expression of MYBL1 (Fig. 5D), which was accompanied by increased promoter methylation of MYBL1 (Fig. 5E). Concomitantly, significantly reduced promoter methylation of MYBL1 was observed in tumor cells with OGT knockdown, compared with control (scrambled) cells (Fig. 5F). All these results together strongly suggest that MYBL1 is regulated by O-GlcNAc levels, and methylation of the MYBL1 gene promoter is very likely involved in this epigenetic regulation.
FIGURE 5.
MYBL1 was epigenetically regulated by O-GlcNAc. A, presence of CpG islands was analyzed around TSS site (−1500 to +350) of MYBL1 using the CpG Island Searcher. B, after the genomic DNA was isolated and bisulfate-treated, methylation-specific PCR for CDH1 (top) and MYBL1 (bottom) genes was performed in different colon tumor cells. The presence of a PCR product band in lanes M or U indicates methylated or unmethylated genes, respectively. M-positive, methylated human DNA standard used as a methylation-positive control. Data are representative of two independent experiments. C, tumor cells were treated with the demethylating reagent 5-aza-2′-deoxycytidine (5-Aza) at two different concentrations for 3 days, and total RNA was isolated and used for detection of transcript levels of MYBL1. For each transcript, the values were normalized to control (GAPDH) and expressed as mean ± S.D. from three independent experiments. D, after treatment with OGA inhibitor (TMG, 10 μm) for 24 h, tumor cells were collected for detection of transcripts of MYBL1 by qRT-PCR. E, after genomic DNA was isolated from tumor cells treated with OGA inhibitor (TMG, 10 μm) for 24 h and bisulfate-treated, methylation-specific PCR for the MYBL1 gene was performed. F, genomic DNA was isolated from both scrambled and OGT-shRNA transfected HT-29 cells, and methylation-specific PCR for the MYBL1 gene was performed. Images were representative from two independent experiments. *, p < 0.05; **, p < 0.01.
Discussion
Studies have shown an association between altered O-GlcNAc expression and colon cancer progression; however, the underlying mechanisms remain unclear. In our study, we chose several human colon tumor lines to determine how colon tumorigenesis was regulated by aberrant expression of O-GlcNAc. We found that tumor cell growth in vitro was remarkably decreased after suppression of O-GlcNAc expression caused by OGT knockdown. In vivo tumor growth in NOD/SCID mice that resulted from injection of tumor cells with inhibited OGT expression was significantly inhibited, compared with that observed from injection of control cells. Based on these observations, it was concluded that O-GlcNAc expression levels were implicated in the regulation of tumor-related phenotypes.
Studies have highlighted the importance of cancer stem cells in regulating colon tumorigenesis and progression. CCSC have been isolated from both human and mouse colon tumors (34, 36–38, 57). Consistent with these reports, CCSC were detected in human colon cancer lines used in our study, demonstrated by using Aldefluor assay as the marker for stem cells. We found that the population of CCSC was significantly decreased in OGT-suppressed tumor cells using shRNA, whereas increased expression of O-GlcNAc, induced by treatment of tumor cells with an OGA inhibitor (TMG), up-regulated the proportion of CCSC, indicating the regulation of CCSC by O-GlcNAc levels. These results supported the hypothesis that the altered amounts of CCSC were involved in tumorigenesis and tumor progression regulated by OGT levels. CCSC can form tumorspheres in suspension culture, reflecting their ability for stem cell self-renewal. We found that self-renewal of CCSC was deregulated by OGT, as determined by stem cell suspension culture, consistent with a deregulated proportion of CCSC by OGT. Based on the fact that CSC play a pivotal role in tumor initiation and progression (31, 58), a decrease in the CCSC compartment could therefore contribute to inhibited colon tumor development observed in SCID mice after knockdown of OGT.
O-GlcNAc levels are responsible for the modification of various target proteins, including transcription factors, epigenetic modulators, and RNA polymerase II, demonstrating that O-GlcNAc functions as an epigenetic modifier of gene expression (59). Substantial evidence that O-GlcNAc is involved in epigenetic regulation is from two early reports that OGT is encoded by a PcG gene, Sxc, and is associated with polycomb repression in Drosophila (24, 25). PcG proteins form two groups of polycomb-repressive complexes (PRC1 and PRC2) and play a pivotal role in the regulation of stem cell differentiation and cancer development through DNA methylation and histone modifications (21, 60). To address whether the regulation of CCSC by O-GlcNAc occurs through an epigenetic mechanism, in this study we performed ChIP-seq in HT-29 cells using an anti-O-GlcNAc antibody along with an antibody against H3K27me3, a well known polycomb-repressive marker (44, 61). We focused mainly on identifying the genes that have an overlap of promoter occupancy with both O-GlcNAc and H3K27me3 because close proximity of O-GlcNAc and histone markers suggests recruitment to promoter areas as a complex, implicating the epigenetic role of O-GlcNAc. A total of 4184 significant O-GlcNAc peaks were identified after ChIP-seq using anti-O-GlcNAc antibody. Using motif enrichment analysis, the two most commonly occurring motifs were very similar to the known binding sites of SP1 and NRF1. Intriguingly, 69.8% of binding sites were located near TSS, and 61 genes were identified from overlapped peaks from H3K27me3 and O-GlcNAc ChIP-seq, suggesting an overlap of gene-binding sites by O-GlcNAc and H3K27me3. These results imply the involvement of O-GlcNAc, like H3K27me3, in epigenetic regulation of genes in colon tumor cells. To further determine which O-GlcNAc-bound genes were involved in the epigenetic regulation of colon tumors, our next effort was focused on identifying genes that were modified by O-GlcNAc as evident by ChIP-seq analysis and that also showed changes after O-GlcNAc levels were altered via OGT knockdown. By RNA-seq analysis, a total 301 genes, including 12 transcription factors, were identified to be differentially expressed in tumor cells with OGT knockdown, among which the transcription factor MYBL1 was identified as binding anti-O-GlcNAc antibody and was up-regulated in altered OGT-expressing cells. Most importantly, MYBL1 was also one of overlapped genes identified by ChIP-seq using both anti-O-GlcNAc and anti-H3K27me3 antibodies, strongly suggesting that MYBL1 was involved in the epigenetic regulation of colon cancer stem cells and tumorigenesis mediated by O-GlcNAc. Indeed, this postulation was further supported by both our qRT-PCR experiments showing increased OGT but significantly decreased MYBL1 expression in mouse colon adenoma tissues, compared with adjacent normal tissues and a recent report that MYBL1 was down-regulated in human colorectal cancer tissues (49).
MYBL1, also known as A-myb, is a member of the transcription factor myb gene family that includes the v-myb oncogene, c-myb proto-oncogene (MYB), and closely related genes A-myb and B-myb (62). Although these members share highly conserved DNA-binding domains and binding site specificities, they appear to have different biological roles (62, 63). Studies have shown that Myb proteins play important oncogenic roles in a variety of solid tumors (64), including colon tumor (65–68). MYBL1 is a strong transcriptional activator (69) and is often co-expressed with other members of the myb family (62), but its function is not as well studied as other members. Although some studies have reported the role of A-myb in regulation of both proliferation and differentiation of hematopoietic cells, A-myb appears to be non-oncogenic (62). However, recent studies have revealed that the chromosomal rearrangement of MYBL1 is likely involved in some tumors. For example, truncating rearrangements in MYBL1 were identified in diffuse pediatric low grade gliomas by using genomic analysis, although MYBL1 was not significantly amplified in these tumors (70). Different from full-length wild-type MYBL1, these truncated MYBL1 transcripts induced anchorage-independent growth in 3T3 cells and formed tumors in nude mice. MYBL1 gene fusions were also identified recently in salivary adenoid cystic carcinoma (71, 72). Interestingly, another study (73) identified MYBL1 as a transcriptional activator of E-cadherin, a suppressor of invasion and metastasis in many tumors (74), by using a large scale RNAi reporter screen for E-cadherin expression in pancreatic tumor cells. The proposed function of MYBL1 as an activator of E-cadherin was further confirmed by experimental validation showing that knockdown of MYBL1 by multiple distinct siRNAs significantly reduced CDH1 expression compared with control cells and is supported by another observation that the knockdown of MYB inhibited CDH1 expression (75). More recently, it was reported that MYBL1 is among the gene set that is used to predict the survival of patients with diffuse large B-cell lymphomas (76, 77), and its high expression levels have a favorable prognostic value for lymphoma patients (78). These results suggest strongly that MYBL1 can play a suppressive role in tumor development.
To understand further the role of altered gene expression of MYBL1 in the regulation of colon tumor development mediated by O-GlcNAc observed in our study, we stably overexpressed MYBL1 in HT-29 colon adenocarcinoma cells. Results obtained from these experiments strongly suggest that MYBL1 displays some features of genes that suppress or inhibit colon tumor development. First, in vitro cell growth was remarkably inhibited when MYBL1 was expressed in tumor cells. Second, in vivo xenograft tumor growth in NOD/SCID mice that resulted from injection of tumor cells expressing MYBL1 was significantly suppressed compared with that observed from injection of mock-transfected cells. Third, a reduced proportion of cancer stem cells was observed in tumor cell populations that showed MYBL1 overexpression, consistent with the inhibited tumor growth of these cells injected in NOD/SCID mice. Also, the cancer stem cells isolated from MYBL1-transfected tumor cells showed reduced tumorigenicity when injected into NOD/SCID mice compared with those from mock-transfected tumor cells. Based on these observations, it is reasonable to conclude that MYBL1 expression functions to inhibit some aspects of the transformed phenotype, which supports the suppressive role of MYBL1 observed in pancreatic tumors (73). Therefore, the decreased CCSC population and reduced colon tumorigenesis observed after knockdown of OGT can be attributed, at least in part, to the increased expression of transcription factor MYBL1.
Aberrant DNA methylation is an epigenetic change found as a ubiquitous event in many human cancers (79). Hypermethylation of promoter CpG islands, frequently observed in human colon tumors, is related to transcriptional silencing of some genes, including tumor suppressors (54). In our study, a promoter methylation mechanism appears to be involved in the regulation MYBL1 gene expression by O-GlcNAc, because we identified two CpG islands around the TSS area of the MYBL1 gene and observed high levels of promoter methylation of MYBL1 in most colon tumor cell lines using methylation-specific PCR. Also, tumor cells treated with a demethylation drug showed remarkable increases in expression of the MYBL1 gene, suggesting that MYBL1 can be regulated by methylation status. Most importantly, increased O-GlcNAc expression induced by the treatment of tumor cells with the OGA inhibitor resulted in enhanced promoter methylation of the MYBL1 gene, whereas reduced O-GlcNAc by OGT knockdown was shown to lower promoter methylation, strongly indicating an involvement of DNA methylation in the regulation of MYBL1 gene expression by O-GlcNAc levels. DNA methylation is catalyzed by a family of DNA methyltransferases (DNMT), including DNMT1, DNMT3a, and DNMT3b, that transfer a methyl group predominantly to the C5 position of cytosine of CpG dinucleotides located at the promoter regions of genes (80). Interestingly, no significant changes in expression of DNMT were observed in comparison with OGT knock-out and wild-type tumor cells by both RNA-seq and gene expression microarray analyses, indicating that altered DNA methylation at the MYBL1 promoter caused by reduced O-GlcNAc did not result from changed DNMT expression. O-GlcNAc modifications may affect the recruitment of DNMT to promoter regions and/or affect complex formation of DNMT with promoter regions of other proteins, like H3K9me2, that regulate DNA methylation (80, 81). Because we also noticed the overlap of binding peaks at promoter areas modified with O-GlcNAc and the repressive histone marker, H3K27me3, it is plausible to suggest that in colon adenocarcinoma O-GlcNAc associates with H3K27me3 (and/or H3K9me2) and epigenetically regulates gene expression through a methylation mechanism. Methylation sequencing is underway to further confirm the role of O-GlcNAc levels in regulating promoter methylation of the MYBL1 gene.
Analysis of methylation of MYBL1 and regions upstream and downstream of its gene body using the TCGA for 450 cases of colorectal adenocarcinoma revealed a very strong region of methylation in the 3′UTR of MYBL1, with lesser incidence of methylation in its promoter region and 5′UTR (data not shown). Because methylation in the gene body and 3′UTR can both alter gene expression (82–85), it is likely that the hypermethylation observed in the 3′UTR of MYBL1 is involved in the suppression of gene expression in colon carcinoma. Many studies have noted different patterns of gene methylation when primary tumors and in vitro cultured cell lines derived from similar tumors were compared (55, 56, 86–88).
It is unclear how increased expression of MYBL1 regulates CCSC and colon tumor progression. Because increasing degrees of differentiation and resulting inhibition of stemness of cancer stem cells has been shown to lead to a reduced population of CSC (89, 90), it is possible that MYBL1 may regulate cell differentiation-related signaling pathways inducing CSC differentiation. Indeed, previous studies have reported that the myb gene family plays an important role not only in cell proliferation but also in cell differentiation (91). For example, ectopic expression of c-Myb in murine myeloid progenitor cells (32Dcl3) results in inhibition of these cells to undergo G-CSF-induced terminal differentiation and increased proliferative potential, whereas overexpression of A-myb (MYBL1) results in growth arrest and concomitant terminal differentiation of these cells into granulocytes (92), suggesting a potential role of MYBL1 in regulating immature cell differentiation, rather than cell proliferation. This possibility is substantiated by a recent study in which the transcription factor CDX1-microRNA axis has been proposed to regulate colon cancer stem cell differentiation (89). In addition, MYBL1 is a strong transcriptional activator likely targeting directly or indirectly some tumor suppressors, like E-cadherin (73), which consequently leads to the inhibition of tumor development. Experiments are in progress to determine the mechanisms by which MYBL1 regulates colon cancer stem cells and colon tumor progression.
In addition, the post-translational modification by O-GlcNAc has been implicated in the regulation of up to 4000 target proteins with different cell functions (3, 93). In our study, more than 3000 genes were identified in colon tumor cells that were bound by O-GlcNAcylated proteins using O-GlcNAc ChIP-seq. However, what types of O-GlcNAc target proteins bound to identified genes in our study remains unknown. The global identification of these O-GlcNAcylated proteins may provide more information regarding the epigenetic regulation of colon tumor development by O-GlcNAc.
In summary, we demonstrate that an epigenetic mechanism is likely involved in the regulation of the CCSC population and colon tumor progression by the level of O-GlcNAcylation. We identified MYBL1, a transcription activator, as a downstream target that very likely regulates colon tumor progression mediated by altered O-GlcNAcylation. Our findings shed new light on the molecular mechanisms of O-GlcNAc modification on colon tumorigenesis and progression and support OGT as a promising target for an inhibitor with therapeutic utility for colon cancer.
Experimental Procedures
Cell Lines and Materials
Human colon cancer cell lines LS180, HT-29, Caco-2, LS-174, SW480, and SW620 were obtained from the American Type Culture Collection (Manassas, VA). TMG was from Cayman Chemical. 5-Aza-2′-deoxycytidine was from Sigma. The lentiviral pLKO.1 vectors containing scrambled and OGT shRNA sequences were from Addgene and Thermo Fisher Scientific, respectively. The sequence for scrambled shRNA was CTAAGGTTAAGTCGCCCTCGCTCGAGCGAGGGCGACTTAACCTTAGG and for OGT shRNA was CCGGGCCCTAAGTTTGAGTCCAAATCTCGAGAATTTGGACTCAAACTTAGGGCTTTTTG (TRCN0000035064) and CCGGGCCCTAAGTTTGAGTCCAAATCTCGAGATTTCTCGGAATACTGCTCAGCTTTTTG (TRCN0000035067). Pooled total RNA samples were isolated from adenoma and adjacent tissues of Apcmin/+ mice (37). pCMV6-XL4/MYBL1 plasmid was purchased from Origene Technologies, and the lentiviral pLX304/MYBL1 was from DNASU. Anti-OGT (H300) was obtained from Santa Cruz Biotechnology; anti-O-GlcNAc antibody (CTD110.6) was from Covance; rabbit anti-MYBL1 (HPA008791) and mouse anti-β-actin were from Sigma. ChIP-ITTM Express kit, ChIP-ITTM control kit, and anti-histone H3K27me3 antibody were purchased from Active Motif; anti-O-GlcNAc antibody (RL2) was from Abcam.
Cell Transfection and Lentiviral Infections
Cell transfections were performed with LipofectamineTM 2000 according to the manufacturer's instructions. Twenty four hours after transfection, cells were placed under G418 (800 μg/ml) selection for 3 weeks. Lentivirus was produced by transfection of HEK-293T cells using Lipofectamine 2000 as described (94). In brief, plasmids containing 0.75 μg of packaging plasmid (pCMV/8.91), 0.25 μg of envelope (pMD2.G), and 1 μg of lentiviral shRNA scrambled or lentiviral expression clone were mixed with Lipofectamine 2000 in Opti-MEM I medium and added to 293T cells in 6-well plates that were replenished with Opti-MEM I medium. The 293T cells were refed with fresh media the next day, and infectious lentivirus supernatant was then collected every 24 to 72 h. Polybrene was added to lentiviral supernatants at a final concentration of 8 μg/ml, and the virus was placed on cells to be infected. The virus entered cells during centrifugation at 2500 rpm for 30 min at 37 °C. Cells were fed with normal growth medium between infections. Two rounds of infection were performed prior to selection of infected cells using puromycin at 1 μg/ml.
Chromatin Immunoprecipitation Assay (ChIP)
ChIP was performed using the Active Motif ChIP kit (95) following the manufacturer's instructions. Briefly, both control and OGT knockdown tumor cells were cross-linked with 1% formaldehyde for 10 min at room temperature. Fixed cells were collected, and chromatin was isolated after sonication to shear the DNA to an average length of 300–500 bp. For immunoprecipitation, anti-O-GlcNAc, anti-H3K27me3 (repression marker), and negative control IgG (Active Motif) (5 μg/each) was added to the same amount of chromatin, respectively, and incubated at 4 °C overnight. Immunoprecipitates were washed, and the cross-linking was reversed. Eluted chromatin was purified and used for ChIP-sequencing and ChIP-PCR.
ChIP-seq
ChIP samples in duplicate were sent to the Genomic Services Lab at HudsonAlpha (Huntsville, AL) for construction of DNA libraries and sequencing. For data analysis of ChIP-seq, the sequence reads were aligned to the human Hg19 reference genome by Bowtie2.0 with one unique read allowed. The ChIP-seq enrichment peaks were called by MACVS1.4 (96) and then further annotated with genome distribution and ontology enrichment by Great 2.0.2. De novo motif discovery was performed with DREME and further aligned to known motif databases UniPROBE and JASPAR with TomTom. The heat map for scores associated with promoter regions was generated by Deep Tools (97). The results from the sequencing were further validated by ChIP-qPCR. Primers used in ChIP-qPCR were listed in Table 4. The ChIP-seq dataset has been deposited at the NCBI GEO Database with an accession number GSE93656.
TABLE 4.
Sequences of primers used in PCR
M is methylated, and U is unmethylated.
| Gene | Forward primer (5′ to 3′) | Reverse primer (5′ to 3′) | |
|---|---|---|---|
| qPCR (human) | |||
| PKIG | CTCGGACTTCATCTCCTGTGA | CTGAGTCTCCCTGGATGTCA | |
| LGR6 | TCAAGCCCTGTGAGTACCTC | CACGGAGAGCAACACGATG | |
| KATNAL1 | CCTCAGATCAGGCGTCCC | GCTCCTACTCCTGCCATTTC | |
| LGALS1 | CATCGTGTGCAACAGCAAG | AGGCTGGAAGGGAAAGACAG | |
| OGT | ACAAGGAAAGAGGGCAGTTG | AATCAGGTTTGAGACGCAATG | |
| SNX10 | GATAGCAGCCTTCACCTCTT | GAAACACACGCCTCAATGTC | |
| CCDC71L | GAGGAATGAACAAGTGTCAGTCG | ACAACCATCCGGCAACTTCTT | |
| HOXA5 | TGAGCGAGCAATTCAGGGAC | ATCCATGCCATTGTAGCCGT | |
| MYBL1 | TGGCAACAGTGAAGCTGTTC | ATGGCTGGTGGAGTGCTAAA | |
| SLC14A | GTGGTGCTCCAGTTCATTGA | GGGTTGTTGACGAACACCA | |
| SLC46A3 | TTGGATTCACCACTCTGCTG | GAGGCACTACCCAAAGCTGA | |
| CCND3 | CTGGTCCTAGGGAAGCTCAA | GAATGAAGGCCAGGAAATCA | |
| ATG4A | GGACCCTCATACAACCCAGA | AGGCAATGGAAAGTCTGGTC | |
| CCRL1 | ATCACCAGCTGCAACATGAG | GAGTGCGATGCTTTCTGTGA | |
| DEPTOR | CGAAGACTGATGGAGCTGCT | CTCAGGCAGAAGGGACTGTC | |
| IF144L | CAGCCGTCAGGGATGTACTAT | CCAAGCATAAAGGCTACAATCA | |
| SH2B3 | TCCCTTTCTCCCTTCCTCAC | GGAAGGCCCAACTCTGAAC | |
| GAPDH | TGGACCTGACCTGCCGTCTAG | CCTCCGACGCCTGCTTCAC | |
| qPCR (mouse) | |||
| Ogt | GAGGAGCATGTCAGGAGAGG | AACATCCATCCCTGTGGTGT | |
| Mybl1 | TCACCTGAACCCTGAAGTGA | GGCGCTTGTGTGCTTCATA | |
| Axin2 | GCAGCTCAGCAAAAAGGGAAAT | TACATGGGGAGCACTGTCTCGT | |
| Gapdh | ACGGGAAGCTCACTGGCATG | CCAGGCGGCACGTCAGATC | |
| ChIP-PCR | |||
| KDM8 | CGCTCTGAGAATGCAATTTAGC | AAAACCACTGCCAAGAGACG | |
| SIRT2 | CGCCCCAGTTAGGTAAGAAAG | CAATCAGAGTATTCGGGAGGAC | |
| PHF | CAGGAAGCTAGGGACTGTTCAT | TGATAGCCCATATCCCATTACC | |
| TSSC | GGTCTTGTTTCCCCAACAAAT | AGAACTACAGCTCCCACAAGGA | |
| RAD21 | GCGCAATGACTATTTCCTTTTC | CTAACACCGCAACTCTCAAAGA | |
| MYBL1 | AGGAAGGGGAAATTCCATTAAA | CCCAGAAATCAACCATCCTCTA | |
| RFX2 | CACTTATTTCACCTCCTGCGC | GAAGCAAGTGGGCCGGAG | |
| ADI1 | GCAACAGAAACGGACCGC | CAGCGAAGTTTACGGGGTTG | |
| MSP-PCR | |||
| MYBL1 | M | TTAGGGTAGGGAGGTTGATAAGC | AAAACAAAACGAAAAAACAACGA |
| U | TTAGGGTAGGGAGGTTGATAAGTG | CAAAACAAAACAAAAAAACAACAAA | |
| CDH1 | M | TTAGGTTAGAGGGTTATCGCGT | TAACTAAAAATTCACCTACCGAC |
| U | TAATTTTAGGTTAGAGGGTTATTGT | CACAACCAATCAACAACACA | |
RNA-seq
RNA-seq was performed as described (98). Briefly, poly(A) mRNA was isolated from HT-29 cells expressing control shRNA or shRNA directed at inhibiting OGT expression, purified, and fragmented. After first and second strand cDNA synthesis was performed, ends were repaired and adenylated, and adapters were then ligated. Fragments were enriched with PCR amplification. The library was validated with an Agilent Technologies 2100 Bioanalyzer (Agilent Technologies, Palo Alto, CA). The sequencing was performed using Illumina HiSEQ2K platform using PhiX as a reference control (Illumina, San Diego). The resultant FASTQ files were imported into CLC Genomics Workbench and aligned to the human hg19/B37 reference sequence. Total exon reads or transcript reads per kilobase pair per million were used as gene expression values. The R/Bioconductor package DESeq was used to test differential expression for sequence count data. The differential expression of each gene with respect to the sample type after accounting for treatment effect was tested, and the Benjamini and Hochberg method were used to correct the p value for false discovery rate. The fold-change cutoff of 1.5 and the p value of < 0.05 were used to identify differentially expressed genes. The RNA-seq data have been deposited at NCBI with an accession number GSE93656.
Microarray Analysis
Total RNA was isolated from tumor cells using TRIzol reagent and purified using RNeasy columns (Qiagen). Using a random hexamer incorporating a T7 promoter, double-stranded cDNA was synthesized from total RNA. cRNA was generated from the double-stranded cDNA template through an in vitro transcription reaction and purified using the Ambion WT Expression kit and sample cleanup module. cDNA was then regenerated through a random primed reverse transcription using a dNTP mixture containing dUTP. Fragmented and biotinylated cDNA was used for hybridization with an Affymetrix human GeneChip® gene 1.0 ST array according to the manufacturer's protocol. Gene expression alterations were determined using the PARTEK Genomics Suite. The CEL files were imported from the Affymetrix Expression Console, background-corrected, and quantile-normalized, and probe summarization was performed using Robust Multichip Analysis. A gene summarization was performed on the data that estimates the intensity of individual genes by averaging the intensities of all the probe sets comprising the gene followed by an n-way analysis of variance using a mixed model and methods of moment to equate analysis of variance mean sum of squares to their expected values. The data were then analyzed using a two-sample t test for significance at p ≤ 0.05 and a fold-change cutoff of 2.0.
Aldefluor Assay
The Aldefluor assay was performed using the ALDEFLUOR kit from StemCell Technologies (99). In brief, dissociated single cells (1 × 106 cells/ml) were incubated in Aldefluor assay buffer containing ALDH substrate, Bodipy-aminoacetaldehyde (1.5 μm) at 37 °C for 30 min. In each experiment, a fraction of cells was stained under identical conditions with a specific ALDH inhibitor, diethylaminobenzaldehyde (15 μm), as a negative control. After staining with propidium iodide, Alde-positive (CSC) and -negative cells (non-CSC) were analyzed using flow cytometry.
Self-renewal of Colon Cancer Stem Cells in Vitro
Single cell suspensions were plated in 6-well ultralow attachment plates (Costar) at a density of 3 × 103 to 2 × 104/ml. Colon spheroid cultures were grown in a serum-free epithelial growth medium containing B27 (Invitrogen), EGF (20 ng/ml), bFGF (20 ng/ml), and heparin (4 μg/ml) for 7–10 days (100, 101). The size and number of the spheres were quantified for each experiment.
Implantation of Tumor Cells in NOD/SCID Mice
All procedures were performed in accordance with the Guide for the Care and Use of Laboratory Animals (National Institutes of Health) and were approved by the Institutional Animal Care and Use Committee of the University of Georgia. NOD/SCID mice were purchased from The Jackson Laboratory. Subconfluent tumor cells were harvested and resuspended in serum-free Hanks' buffered saline solution in a 70-μl volume containing 1–3 × 106 cells. After mice were anesthetized with isofluorane, 70 μl of the single cell suspension mixed with 30 μl of Matrigel was injected subcutaneously into the back of NOD/SCID mice of 6–8 weeks age using a 27-gauge needle (37). Tumor growth and tumor size were measured using calipers once a week.
Quantitative PCR Analysis
Total RNA from tumor cells was isolated using TRIzol (Invitrogen). The reverse transcription reactions and qRT-PCR analyses were performed using iScriptTM cDNA synthesis kit or Superscript III as described previously (102) and iQTMSYBR®Green Supermix (Bio-Rad), respectively. Each PCR was performed in triplicate, and experiments were repeated at least three times. Primers used in the qRT-PCR analysis are listed in Table 4.
Methylation-specific PCR
Purification and bisulfite treatment of genomic DNA samples were performed using the DNeasy tissue kit and EZ DNA Methylation-GoldTM kit (Zymo Research), respectively, according to the manufacturer's instructions. Methylation-specific PCR was carried out using the following cycling conditions: for MYBL1, 95 °C for 5 min; 40 cycles at 95 °C for 30 s, 52 °C for 30 s, and 72 °C for 45 s; and a final cycle at 72 °C for 5 min; and for CDH1, 95 °C for 5 min; 35 cycles at 95 °C for 30 s, 57 °C for 30 s, and 72 °C for 30 s; a final cycle at 72 °C for 5 min for methylated primers and the annealing temperature was 53 °C for unmethylated primers (103). Universal Methylated Human DNA (enzymatically methylated by M.SssI Methyltransferase, Zymo Research) was used as a positive control for methylated alleles. The primer sequences used for methylation-specific PCR are listed in Table 4. The PCR products were isolated on a 1.5% agarose gel and visualized by ethidium bromide staining.
Statistical Analysis
Statistical analysis was performed using statistical software R. Significant differences between two groups were determined by the unpaired t test. p values <0.05 were considered statistically significant.
Author Contributions
H. G. and M. P. designed the research; H. G. performed most of the experiments; B. Z. and P. B. analyzed the ChIP-seq and RNA-seq data; A. V. N. and K. W. M. performed RT-PCR validation; T. N. performed pathological analysis of tumors; and H. G. and M. P. wrote and edited the manuscript.
Acknowledgments
We thank Drs. Liwei Xie and Hang Yin for help with figure preparation from ChIP-seq data, and Pam Kirby, Julie Nelson, Christopher Whitlock, Manasa Kadiyala, and Samuel Milam for technical assistance. We are grateful to Drs. Shaying Zhao, Liwei Xie, Jinkyu Lee, and Hong Qiu for the instructive discussion.
This work was supported, in whole or in part, by National Institutes of Health Grants U01CA128454 and P41-GM103490 (to M. P.). The authors declare that they have no conflicts of interest with the contents of this article. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
Accession number GSE93656 has been deposited in the NCBI GEO Database.
- O-GlcNAc
- O-linked N-acetylglucosamine
- seq
- sequencing
- TSS
- transcription start site
- OGT
- O-linked N-acetylglucosamine transferase
- CCSC
- colon cancer stem cell
- qRT
- quantitative RT
- ALDH
- aldehyde dehydrogenase
- CSC
- cancer stem cell
- OGA
- N-acetyl-β-glucosaminidase
- TMG
- Thiamet-G
- ESC
- embryonic stem cell
- DNMT
- DNA methyltransferase
- PcG
- polycomb group.
References
- 1. Torres C. R., and Hart G. W. (1984) Topography and polypeptide distribution of terminal N-acetylglucosamine residues on the surfaces of intact lymphocytes. Evidence for O-linked GlcNAc. J. Biol. Chem. 259, 3308–3317 [PubMed] [Google Scholar]
- 2. Holt G. D., and Hart G. W. (1986) The subcellular distribution of terminal N-acetylglucosamine moieties. Localization of a novel protein-saccharide linkage, O-linked GlcNAc. J. Biol. Chem. 261, 8049–8057 [PubMed] [Google Scholar]
- 3. Bond M. R., and Hanover J. A. (2015) A little sugar goes a long way: the cell biology of O-GlcNAc. J. Cell Biol. 208, 869–880 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Kearse K. P., and Hart G. W. (1991) Lymphocyte activation induces rapid changes in nuclear and cytoplasmic glycoproteins. Proc. Natl. Acad. Sci. U.S.A. 88, 1701–1705 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Hart G. W., Housley M. P., and Slawson C. (2007) Cycling of O-linked β-N-acetylglucosamine on nucleocytoplasmic proteins. Nature 446, 1017–1022 [DOI] [PubMed] [Google Scholar]
- 6. Haltiwanger R. S., Holt G. D., and Hart G. W. (1990) Enzymatic addition of O-GlcNAc to nuclear and cytoplasmic proteins. Identification of a uridine diphospho-N-acetylglucosamine:peptide β-N-acetylglucosaminyltransferase. J. Biol. Chem. 265, 2563–2568 [PubMed] [Google Scholar]
- 7. Dong D. L., and Hart G. W. (1994) Purification and characterization of an O-GlcNAc selective N-acetyl-β-d-glucosaminidase from rat spleen cytosol. J. Biol. Chem. 269, 19321–19330 [PubMed] [Google Scholar]
- 8. Hart G. W., Slawson C., Ramirez-Correa G., and Lagerlof O. (2011) Cross-talk between O-GlcNAcylation and phosphorylation: roles in signaling, transcription, and chronic disease. Annu. Rev. Biochem. 80, 825–858 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Slawson C., Copeland R. J., and Hart G. W. (2010) O-GlcNAc signaling: a metabolic link between diabetes and cancer? Trends Biochem. Sci. 35, 547–555 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Ma Z., and Vosseller K. (2014) Cancer metabolism and elevated O-GlcNAc in oncogenic signaling. J. Biol. Chem. 289, 34457–34465 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Krzeslak A., Pomorski L., and Lipinska A. (2010) Elevation of nucleocytoplasmic β-N-acetylglucosaminidase (O-GlcNAcase) activity in thyroid cancers. Int. J. Mol. Med. 25, 643–648 [DOI] [PubMed] [Google Scholar]
- 12. Gu Y., Mi W., Ge Y., Liu H., Fan Q., Han C., Yang J., Han F., Lu X., and Yu W. (2010) GlcNAcylation plays an essential role in breast cancer metastasis. Cancer Res. 70, 6344–6351 [DOI] [PubMed] [Google Scholar]
- 13. Mi W., Gu Y., Han C., Liu H., Fan Q., Zhang X., Cong Q., and Yu W. (2011) O-GlcNAcylation is a novel regulator of lung and colon cancer malignancy. Biochim. Biophys. Acta 1812, 514–519 [DOI] [PubMed] [Google Scholar]
- 14. Caldwell S. A., Jackson S. R., Shahriari K. S., Lynch T. P., Sethi G., Walker S., Vosseller K., and Reginato M. J. (2010) Nutrient sensor O-GlcNAc transferase regulates breast cancer tumorigenesis through targeting of the oncogenic transcription factor FoxM1. Oncogene 29, 2831–2842 [DOI] [PubMed] [Google Scholar]
- 15. Yang Y. R., Kim D. H., Seo Y. K., Park D., Jang H. J., Choi S. Y., Lee Y. H., Lee G. H., Nakajima K., Taniguchi N., Kim J. M., Choi E. J., Moon H. Y., Kim I. S., Choi J. H., et al. (2015) Elevated O-GlcNAcylation promotes colonic inflammation and tumorigenesis by modulating NF-κB signaling. Oncotarget 6, 12529–12542 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Zhang X., Ma L., Qi J., Shan H., Yu W., and Gu Y. (2015) MAPK/ERK signaling pathway-induced hyper-O-GlcNAcylation enhances cancer malignancy. Mol. Cell. Biochem. 410, 101–110 [DOI] [PubMed] [Google Scholar]
- 17. Ferrer C. M., Lynch T. P., Sodi V. L., Falcone J. N., Schwab L. P., Peacock D. L., Vocadlo D. J., Seagroves T. N., and Reginato M. J. (2014) O-GlcNAcylation regulates cancer metabolism and survival stress signaling via regulation of the HIF-1 pathway. Mol. Cell 54, 820–831 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Onodera Y., Nam J. M., and Bissell M. J. (2014) Increased sugar uptake promotes oncogenesis via EPAC/RAP1 and O-GlcNAc pathways. J. Clin. Invest. 124, 367–384 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Love D. C., Krause M. W., and Hanover J. A. (2010) O-GlcNAc cycling: emerging roles in development and epigenetics. Semin. Cell Dev. Biol. 21, 646–654 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Slawson C., and Hart G. W. (2011) O-GlcNAc signalling: implications for cancer cell biology. Nat. Rev. Cancer 11, 678–684 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Singh J. P., Zhang K., Wu J., and Yang X. (2015) O-GlcNAc signaling in cancer metabolism and epigenetics. Cancer Lett. 356, 244–250 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Sakabe K., Wang Z., and Hart G. W. (2010) β-N-acetylglucosamine (O-GlcNAc) is part of the histone code. Proc. Natl. Acad. Sci. U.S.A. 107, 19915–19920 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Love D. C., Ghosh S., Mondoux M. A., Fukushige T., Wang P., Wilson M. A., Iser W. B., Wolkow C. A., Krause M. W., and Hanover J. A. (2010) Dynamic O-GlcNAc cycling at promoters of Caenorhabditis elegans genes regulating longevity, stress, and immunity. Proc. Natl. Acad. Sci. U.S.A. 107, 7413–7418 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Sinclair D. A., Syrzycka M., Macauley M. S., Rastgardani T., Komljenovic I., Vocadlo D. J., Brock H. W., and Honda B. M. (2009) Drosophila O-GlcNAc-transferase (OGT) is encoded by the Polycomb group (PcG) gene, super sex combs (sxc). Proc. Natl. Acad. Sci. U.S.A. 106, 13427–13432 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Gambetta M. C., Oktaba K., and Müller J. (2009) Essential role of the glycosyltransferase sxc/Ogt in polycomb repression. Science 325, 93–96 [DOI] [PubMed] [Google Scholar]
- 26. Kerppola T. K. (2009) Polycomb group complexes–many combinations, many functions. Trends Cell Biol. 19, 692–704 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Myers S. A., Panning B., and Burlingame A. L. (2011) Polycomb repressive complex 2 is necessary for the normal site-specific O-GlcNAc distribution in mouse embryonic stem cells. Proc. Natl. Acad. Sci. U.S.A. 108, 9490–9495 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Shi F. T., Kim H., Lu W., He Q., Liu D., Goodell M. A., Wan M., and Songyang Z. (2013) Ten-eleven translocation 1 (Tet1) is regulated by O-linked N-acetylglucosamine transferase (Ogt) for target gene repression in mouse embryonic stem cells. J. Biol. Chem. 288, 20776–20784 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Vella P., Scelfo A., Jammula S., Chiacchiera F., Williams K., Cuomo A., Roberto A., Christensen J., Bonaldi T., Helin K., and Pasini D. (2013) Tet proteins connect the O-linked N-acetylglucosamine transferase Ogt to chromatin in embryonic stem cells. Mol. Cell 49, 645–656 [DOI] [PubMed] [Google Scholar]
- 30. Jeon J. H., Suh H. N., Kim M. O., Ryu J. M., and Han H. J. (2014) Glucosamine-induced OGT activation mediates glucose production through cleaved Notch1 and FoxO1, which coordinately contributed to the regulation of maintenance of self-renewal in mouse embryonic stem cells. Stem Cells Dev. 23, 2067–2079 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Wicha M. S., Liu S., and Dontu G. (2006) Cancer stem cells: an old idea–a paradigm shift. Cancer Res. 66, 1883–1890 [DOI] [PubMed] [Google Scholar]
- 32. Ishiwata T. (2016) Cancer stem cells and epithelial-mesenchymal transition: novel therapeutic targets for cancer. Pathol. Int. 66, 601–608 [DOI] [PubMed] [Google Scholar]
- 33. Sato R., Semba T., Saya H., and Arima Y. (2016) Concise review: stem cells and epithelial-mesenchymal transition in cancer: biological implications and therapeutic targets. Stem Cells 34, 1997–2007 [DOI] [PubMed] [Google Scholar]
- 34. Ricci-Vitiani L., Lombardi D. G., Pilozzi E., Biffoni M., Todaro M., Peschle C., and De Maria R. (2007) Identification and expansion of human colon-cancer-initiating cells. Nature 445, 111–115 [DOI] [PubMed] [Google Scholar]
- 35. Yeung T. M., Gandhi S. C., Wilding J. L., Muschel R., and Bodmer W. F. (2010) Cancer stem cells from colorectal cancer-derived cell lines. Proc. Natl. Acad. Sci. U.S.A. 107, 3722–3727 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Huang E. H., Hynes M. J., Zhang T., Ginestier C., Dontu G., Appelman H., Fields J. Z., Wicha M. S., and Boman B. M. (2009) Aldehyde dehydrogenase 1 is a marker for normal and malignant human colonic stem cells (SC) and tracks SC overpopulation during colon tumorigenesis. Cancer Res. 69, 3382–3389 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Guo H., Nagy T., and Pierce M. (2014) Post-translational glycoprotein modifications regulate colon cancer stem cells and colon adenoma progression in Apc(min/+) mice through altered Wnt receptor signaling. J. Biol. Chem. 289, 31534–31549 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Zeilstra J., Joosten S. P., Dokter M., Verwiel E., Spaargaren M., and Pals S. T. (2008) Deletion of the WNT target and cancer stem cell marker CD44 in Apc(Min/+) mice attenuates intestinal tumorigenesis. Cancer Res. 68, 3655–3661 [DOI] [PubMed] [Google Scholar]
- 39. Lin L., Liu A., Peng Z., Lin H. J., Li P. K., Li C., and Lin J. (2011) STAT3 is necessary for proliferation and survival in colon cancer-initiating cells. Cancer Res. 71, 7226–7237 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Yuzwa S. A., Macauley M. S., Heinonen J. E., Shan X., Dennis R. J., He Y., Whitworth G. E., Stubbs K. A., McEachern E. J., Davies G. J., and Vocadlo D. J. (2008) A potent mechanism-inspired O-GlcNAcase inhibitor that blocks phosphorylation of τ in vivo. Nat. Chem. Biol. 4, 483–490 [DOI] [PubMed] [Google Scholar]
- 41. Ponti D., Costa A., Zaffaroni N., Pratesi G., Petrangolini G., Coradini D., Pilotti S., Pierotti M. A., and Daidone M. G. (2005) Isolation and in vitro propagation of tumorigenic breast cancer cells with stem/progenitor cell properties. Cancer Res. 65, 5506–5511 [DOI] [PubMed] [Google Scholar]
- 42. Zucchi I., Sanzone S., Astigiano S., Pelucchi P., Scotti M., Valsecchi V., Barbieri O., Bertoli G., Albertini A., Reinbold R. A., and Dulbecco R. (2007) The properties of a mammary gland cancer stem cell. Proc. Natl. Acad. Sci. U.S.A. 104, 10476–10481 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Vermeulen L., Todaro M., de Sousa Mello F., Sprick M. R., Kemper K., Perez Alea M., Richel D. J., Stassi G., and Medema J. P. (2008) Single-cell cloning of colon cancer stem cells reveals a multi-lineage differentiation capacity. Proc. Natl. Acad. Sci. U.S.A. 105, 13427–13432 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Margueron R., and Reinberg D. (2011) The Polycomb complex PRC2 and its mark in life. Nature 469, 343–349 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Bracken A. P., Dietrich N., Pasini D., Hansen K. H., and Helin K. (2006) Genome-wide mapping of Polycomb target genes unravels their roles in cell fate transitions. Genes Dev. 20, 1123–1136 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Klaus A., and Birchmeier W. (2008) Wnt signalling and its impact on development and cancer. Nat. Rev. Cancer 8, 387–398 [DOI] [PubMed] [Google Scholar]
- 47. Wu Z. Q., Brabletz T., Fearon E., Willis A. L., Hu C. Y., Li X. Y., and Weiss S. J. (2012) Canonical Wnt suppressor, Axin2, promotes colon carcinoma oncogenic activity. Proc. Natl. Acad. Sci. U.S.A. 109, 11312–11317 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Yang Y. R., Jang H. J., Yoon S., Lee Y. H., Nam D., Kim I. S., Lee H., Kim H., Choi J. H., Kang B. H., Ryu S. H., and Suh P. G. (2014) OGA heterozygosity suppresses intestinal tumorigenesis in Apc(min/+) mice. Oncogenesis 3, e109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Wu Y., Wang X., Wu F., Huang R., Xue F., Liang G., Tao M., Cai P., and Huang Y. (2012) Transcriptome profiling of the cancer, adjacent non-tumor and distant normal tissues from a colorectal cancer patient by deep sequencing. PLoS ONE 7, e41001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Golay J., Broccoli V., Lamorte G., Bifulco C., Parravicini C., Pizzey A., Thomas N. S., Delia D., Ferrauti P., Vitolo D., and Introna M. (1998) The A-Myb transcription factor is a marker of centroblasts in vivo. J. Immunol. 160, 2786–2793 [PubMed] [Google Scholar]
- 51. Arsura M., Hofmann C. S., Golay J., Introna M., and Sonenshein G. E. (2000) A-myb rescues murine B-cell lymphomas from IgM-receptor-mediated apoptosis through c-myc transcriptional regulation. Blood 96, 1013–1020 [PubMed] [Google Scholar]
- 52. Okugawa Y., Grady W. M., and Goel A. (2015) Epigenetic alterations in colorectal cancer: emerging biomarkers. Gastroenterology 149, 1204–1225 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Hammoud S. S., Cairns B. R., and Jones D. A. (2013) Epigenetic regulation of colon cancer and intestinal stem cells. Curr. Opin. Cell Biol. 25, 177–183 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Ng J. M., and Yu J. (2015) Promoter hypermethylation of tumour suppressor genes as potential biomarkers in colorectal cancer. Int. J. Mol. Sci. 16, 2472–2496 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Lind G. E., Thorstensen L., Løvig T., Meling G. I., Hamelin R., Rognum T. O., Esteller M., and Lothe R. A. (2004) A CpG island hypermethylation profile of primary colorectal carcinomas and colon cancer cell lines. Mol. Cancer 3, 28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Paz M. F., Fraga M. F., Avila S., Guo M., Pollan M., Herman J. G., and Esteller M. (2003) A systematic profile of DNA methylation in human cancer cell lines. Cancer Res. 63, 1114–1121 [PubMed] [Google Scholar]
- 57. Merlos-Suárez A., Barriga F. M., Jung P., Iglesias M., Céspedes M. V., Rossell D., Sevillano M., Hernando-Momblona X., da Silva-Diz V., Muñoz P., Clevers H., Sancho E., Mangues R., and Batlle E. (2011) The intestinal stem cell signature identifies colorectal cancer stem cells and predicts disease relapse. Cell Stem Cell 8, 511–524 [DOI] [PubMed] [Google Scholar]
- 58. Ghiaur G., Gerber J., and Jones R. J. (2012) Concise review: cancer stem cells and minimal residual disease. Stem Cells 30, 89–93 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Lewis B. A., and Hanover J. A. (2014) O-GlcNAc and the epigenetic regulation of gene expression. J. Biol. Chem. 289, 34440–34448 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Aloia L., Di Stefano B., and Di Croce L. (2013) Polycomb complexes in stem cells and embryonic development. Development 140, 2525–2534 [DOI] [PubMed] [Google Scholar]
- 61. Kassis J. A., and Kennison J. A. (2010) Recruitment of polycomb complexes: a role for SCM. Mol. Cell. Biol. 30, 2581–2583 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Zhou Y., and Ness S. A. (2011) Myb proteins: angels and demons in normal and transformed cells. Front. Biosci. 16, 1109–1131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Rushton J. J., Davis L. M., Lei W., Mo X., Leutz A., and Ness S. A. (2003) Distinct changes in gene expression induced by A-Myb, B-Myb and c-Myb proteins. Oncogene 22, 308–313 [DOI] [PubMed] [Google Scholar]
- 64. George O. L., and Ness S. A. (2014) Situational awareness: regulation of the myb transcription factor in differentiation, the cell cycle and oncogenesis. Cancers 6, 2049–2071 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Ramsay R. G., Thompson M. A., Hayman J. A., Reid G., Gonda T. J., and Whitehead R. H. (1992) Myb expression is higher in malignant human colonic carcinoma and premalignant adenomatous polyps than in normal mucosa. Cell Growth Differ. 3, 723–730 [PubMed] [Google Scholar]
- 66. Ramsay R. G., and Gonda T. J. (2008) MYB function in normal and cancer cells. Nat. Rev. Cancer 8, 523–534 [DOI] [PubMed] [Google Scholar]
- 67. Kriegl L., Vieth M., Kirchner T., and Menssen A. (2012) Up-regulation of c-MYC and SIRT1 expression correlates with malignant transformation in the serrated route to colorectal cancer. Oncotarget 3, 1182–1193 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Malaterre J., Pereira L., Putoczki T., Millen R., Paquet-Fifield S., Germann M., Liu J., Cheasley D., Sampurno S., Stacker S. A., Achen M. G., Ward R. L., Waring P., Mantamadiotis T., Ernst M., and Ramsay R. G. (2016) Intestinal-specific activatable Myb initiates colon tumorigenesis in mice. Oncogene 35, 2475–2484 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Golay J., Loffarelli L., Luppi M., Castellano M., and Introna M. (1994) The human A-myb protein is a strong activator of transcription. Oncogene 9, 2469–2479 [PubMed] [Google Scholar]
- 70. Ramkissoon L. A., Horowitz P. M., Craig J. M., Ramkissoon S. H., Rich B. E., Schumacher S. E., McKenna A., Lawrence M. S., Bergthold G., Brastianos P. K., Tabak B., Ducar M. D., Van Hummelen P., MacConaill L. E., Pouissant-Young T., et al. (2013) Genomic analysis of diffuse pediatric low-grade gliomas identifies recurrent oncogenic truncating rearrangements in the transcription factor MYBL1. Proc. Natl. Acad. Sci. U.S.A. 110, 8188–8193 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Mitani Y., Liu B., Rao P. H., Borra V. J., Zafereo M., Weber R. S., Kies M., Lozano G., Futreal P. A., Caulin C., and El-Naggar A. K. (2016) Novel MYBL1 gene rearrangements with recurrent MYBL1-NFIB fusions in salivary adenoid cystic carcinomas lacking t(6;9) translocations. Clin. Cancer Res. 22, 725–733 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Brayer K. J., Frerich C. A., Kang H., and Ness S. A. (2016) Recurrent fusions in MYB and MYBL1 define a common, transcription factor-driven oncogenic pathway in salivary gland adenoid cystic carcinoma. Cancer Discov. 6, 176–187 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Adams R., Nicke B., Pohlenz H. D., and Sohler F. (2015) Deciphering seed sequence based off-target effects in a large-scale RNAi reporter screen for E-cadherin expression. PLoS ONE 10, e0137640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Jeanes A., Gottardi C. J., and Yap A. S. (2008) Cadherins and cancer: how does cadherin dysfunction promote tumor progression? Oncogene 27, 6920–6929 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Pieraccioli M., Imbastari F., Antonov A., Melino G., and Raschellà G. (2013) Activation of miR200 by c-Myb depends on ZEB1 expression and miR200 promoter methylation. Cell Cycle 12, 2309–2320 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Lossos I. S., Czerwinski D. K., Alizadeh A. A., Wechser M. A., Ishitani R., Botstein D., and Levy R. (2004) Prediction of survival in diffuse large-B-cell lymphoma based on the expression of six genes. N. Engl. J. Med. 350, 1828–1837 [DOI] [PubMed] [Google Scholar]
- 77. Alizadeh A. A., Eisen M. B., Davis R. E., Ma C., Lossos I. S., Rosenwald A., Boldrick J. C., Sabet H., Tran T., Yu X., Powell J. I., Yang L., Marti G. E., Moore T., Hudson J. Jr., et al. (2000) Distinct types of diffuse large B-cell lymphoma identified by gene expression profiling. Nature 403, 503–511 [DOI] [PubMed] [Google Scholar]
- 78. Zhao S., Dong X., Shen W., Ye Z., and Xiang R. (2016) Machine learning-based classification of diffuse large B-cell lymphoma patients by eight gene expression profiles. Cancer Med. 5, 837–852 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Jones P. A., and Baylin S. B. (2007) The epigenomics of cancer. Cell 128, 683–692 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Cedar H., and Bergman Y. (2009) Linking DNA methylation and histone modification: patterns and paradigms. Nat. Rev. Genet. 10, 295–304 [DOI] [PubMed] [Google Scholar]
- 81. Dong C., Wu Y., Yao J., Wang Y., Yu Y., Rychahou P. G., Evers B. M., and Zhou B. P. (2012) G9a interacts with Snail and is critical for Snail-mediated E-cadherin repression in human breast cancer. J. Clin. Invest. 122, 1469–1486 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Malumbres M., Pérez de Castro I., Santos J., Fernández Piqueras J., and Pellicer A. (1999) Hypermethylation of the cell cycle inhibitor p15INK4b 3′-untranslated region interferes with its transcriptional regulation in primary lymphomas. Oncogene 18, 385–396 [DOI] [PubMed] [Google Scholar]
- 83. Yang X., Han H., De Carvalho D. D., Lay F. D., Jones P. A., and Liang G. (2014) Gene body methylation can alter gene expression and is a therapeutic target in cancer. Cancer Cell 26, 577–590 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Maussion G., Yang J., Suderman M., Diallo A., Nagy C., Arnovitz M., Mechawar N., and Turecki G. (2014) Functional DNA methylation in a transcript specific 3′UTR region of TrkB associates with suicide. Epigenetics 9, 1061–1070 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. Fujii S., Shinjo K., Matsumoto S., Harada T., Nojima S., Sato S., Usami Y., Toyosawa S., Morii E., Kondo Y., and Kikuchi A. (2016) Epigenetic upregulation of ARL4C, due to DNA hypomethylation in the 3′-untranslated region, promotes tumorigenesis of lung squamous cell carcinoma. Oncotarget 7, 81571–81587 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86. Esteller M., Corn P. G., Baylin S. B., and Herman J. G. (2001) A gene hypermethylation profile of human cancer. Cancer Res. 61, 3225–3229 [PubMed] [Google Scholar]
- 87. Baysan M., Woolard K., Bozdag S., Riddick G., Kotliarova S., Cam M. C., Belova G. I., Ahn S., Zhang W., Song H., Walling J., Stevenson H., Meltzer P., and Fine H. A. (2014) Micro-environment causes reversible changes in DNA methylation and mRNA expression profiles in patient-derived glioma stem cells. PLoS ONE 9, e94045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88. Bhat K. P., Balasubramaniyan V., Vaillant B., Ezhilarasan R., Hummelink K., Hollingsworth F., Wani K., Heathcock L., James J. D., Goodman L. D., Conroy S., Long L., Lelic N., Wang S., Gumin J., et al. (2013) Mesenchymal differentiation mediated by NF-κB promotes radiation resistance in glioblastoma. Cancer Cell 24, 331–346 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89. Jones M. F., Hara T., Francis P., Li X. L., Bilke S., Zhu Y., Pineda M., Subramanian M., Bodmer W. F., and Lal A. (2015) The CDX1-microRNA-215 axis regulates colorectal cancer stem cell differentiation. Proc. Natl. Acad. Sci. U.S.A. 112, E1550–1558 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90. Hou Z. J., Luo X., Zhang W., Peng F., Cui B., Wu S. J., Zheng F. M., Xu J., Xu L. Z., Long Z. J., Wang X. T., Li G. H., Wan X. Y., Yang Y. L., and Liu Q. (2015) Flubendazole, FDA-approved anthelmintic, targets breast cancer stem-like cells. Oncotarget 6, 6326–6340 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91. Oh I. H., and Reddy E. P. (1999) The myb gene family in cell growth, differentiation and apoptosis. Oncogene 18, 3017–3033 [DOI] [PubMed] [Google Scholar]
- 92. Oh I. H., and Reddy E. P. (1997) Murine A-myb gene encodes a transcription factor, which cooperates with Ets-2 and exhibits distinctive biochemical and biological activities from c-myb. J. Biol. Chem. 272, 21432–21443 [DOI] [PubMed] [Google Scholar]
- 93. Ma J., and Hart G. W. (2014) O-GlcNAc profiling: from proteins to proteomes. Clin. Proteomics 11, 8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94. Zhao P., Nairn A. V., Hester S., Moremen K. W., O'Regan R. M., Oprea G., Wells L., Pierce M., and Abbott K. L. (2012) Proteomic identification of glycosylphosphatidylinositol anchor-dependent membrane proteins elevated in breast carcinoma. J. Biol. Chem. 287, 25230–25240 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95. Dey A., Seshasayee D., Noubade R., French D. M., Liu J., Chaurushiya M. S., Kirkpatrick D. S., Pham V. C., Lill J. R., Bakalarski C. E., Wu J., Phu L., Katavolos P., LaFave L. M., Abdel-Wahab O., et al. (2012) Loss of the tumor suppressor BAP1 causes myeloid transformation. Science 337, 1541–1546 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96. Zhang Y., Liu T., Meyer C. A., Eeckhoute J., Johnson D. S., Bernstein B. E., Nusbaum C., Myers R. M., Brown M., Li W., and Liu X. S. (2008) Model-based analysis of ChIP-Seq (MACS). Genome Biol. 9, R137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97. Ramírez F., Ryan D. P., Grüning B., Bhardwaj V., Kilpert F., Richter A. S., Heyne S., Dündar F., and Manke T. (2016) deepTools2: a next generation web server for deep-sequencing data analysis. Nucleic Acids Res. 44, W160–W165 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98. Weige C. C., Birtwistle M. R., Mallick H., Yi N., Berrong Z., Cloessner E., Duff K., Tidwell J., Clendenning M., Wilkerson B., Farrell C., Bunz F., Ji H., Shtutman M., Creek K. E., Banister C. E., and Buckhaults P. J. (2014) Transcriptomes and shRNA suppressors in a TP53 allele-specific model of early-onset colon cancer in African Americans. Mol. Cancer Res. 12, 1029–1041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99. Ginestier C., Hur M. H., Charafe-Jauffret E., Monville F., Dutcher J., Brown M., Jacquemier J., Viens P., Kleer C. G., Liu S., Schott A., Hayes D., Birnbaum D., Wicha M. S., and Dontu G. (2007) ALDH1 is a marker of normal and malignant human mammary stem cells and a predictor of poor clinical outcome. Cell Stem Cell 1, 555–567 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100. Dontu G., Abdallah W. M., Foley J. M., Jackson K. W., Clarke M. F., Kawamura M. J., and Wicha M. S. (2003) In vitro propagation and transcriptional profiling of human mammary stem/progenitor cells. Genes Dev. 17, 1253–1270 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101. Kanwar S. S., Yu Y., Nautiyal J., Patel B. B., and Majumdar A. P. (2010) The Wnt/β-catenin pathway regulates growth and maintenance of colonospheres. Mol. Cancer 9, 212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102. Nairn A. V., dela Rosa M., and Moremen K. W. (2010) Transcript analysis of stem cells. Methods Enzymol. 479, 73–91 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103. Chen C. L., Liu S. S., Ip S. M., Wong L. C., Ng T. Y., and Ngan H. Y. (2003) E-cadherin expression is silenced by DNA methylation in cervical cancer cell lines and tumours. Eur. J. Cancer 39, 517–523 [DOI] [PubMed] [Google Scholar]