Abstract
Prenyltransferases of the dimethylallyltryptophan synthase (DMATS) superfamily catalyze the attachment of prenyl or prenyl-like moieties to diverse acceptor compounds. These acceptor molecules are generally aromatic in nature and mostly indole or indole-like. Their catalytic transformation represents a major skeletal diversification step in the biosynthesis of secondary metabolites, including the indole alkaloids. DMATS enzymes thus contribute significantly to the biological and pharmacological diversity of small molecule metabolites. Understanding the substrate specificity of these enzymes could create opportunities for their biocatalytic use in preparing complex synthetic scaffolds. However, there has been no framework to achieve this in a rational way. Here, we report a chemoinformatic pipeline to enable prenyltransferase substrate prediction. We systematically catalogued 32 unique prenyltransferases and 167 unique substrates to create possible reaction matrices and compiled these data into a browsable database named PrenDB. We then used a newly developed algorithm based on molecular fragmentation to automatically extract reactive chemical epitopes. The analysis of the collected data sheds light on the thus far explored substrate space of DMATS enzymes. To assess the predictive performance of our virtual reaction extraction tool, 38 potential substrates were tested as prenyl acceptors in assays with three prenyltransferases, and we were able to detect turnover in >55% of the cases. The database, PrenDB (www.kolblab.org/prendb.php), enables the prediction of potential substrates for chemoenzymatic synthesis through substructure similarity and virtual chemical transformation techniques. It aims at making prenyltransferases and their highly regio- and stereoselective reactions accessible to the research community for integration in synthetic work flows.
Keywords: bioinformatics, computer modeling, enzyme catalysis, high-throughput screening (HTS), substrate specificity, chemoinformatics, prenyltransferases, reaction database, substrate prediction
Introduction
Prenylated primary and secondary metabolites, including indole alkaloids, flavonoids, coumarins, xanthones, quinones, and naphthalenes, are widely distributed in terrestrial and marine organisms. They exhibit a wide range of biological activities, including cytotoxic, antioxidant, and antimicrobial activities (1–3). Compared with their non-prenylated precursors, these compounds usually demonstrate distinct and often improved biological and pharmacological activities, which makes them promising candidates for drug discovery and development (1, 2, 4–6). These compounds could be considered hybrid molecules of prenyl moieties of different chain lengths (n·C5, where n is an integer number) and aromatic skeletons originating from various biosynthetic pathways (7, 8). Prenyl transfer reactions (i.e. the connections of prenyl moieties to the aromatic nucleus) are catalyzed by a diverse family of prenyltransferases. Interestingly, this step usually represents the key transformation in the biosynthesis of such compounds. A prenyl moiety can be attached by prenyltransferases via its C1 (regular prenylation) or C3 (reverse prenylation) to carbon, oxygen, or nitrogen atoms of an acceptor (Fig. 1, A and B) (7, 8). Together with the observed regiospecific prenylations at different positions of an acceptor molecule, prenyltransferases contribute significantly to the structural and biological diversity of natural products (7).
FIGURE 1.
A, exemplary transformation of brevianamide F (E1) to tryprostatin B (E3). B, regiochemistry of the nucleophilic attack on the prenyl moiety. For regular prenylation, bond formation occurs between C2 and the carbon adjacent to the pyrophosphate group. An attack of C2 on the tertiary carbon of DMAPP leads to the reversely prenylated product. C, illustration of the SMIRKS-like notation derived from A (generated by SMARTSviewer) (22). GA, general prenyl moiety acceptor; GD, general prenyl moiety donor; GB, general base; GP, general prenylation product; PP, pyrophosphate; PGB, protonated general base. D (left), a reactive epitope indicated around the reactive atom ([cH1:1]) with the atomic properties given in SMARTS nomenclature (brackets). D (right), reactive epitope as generated by SMARTSviewer.
Based on their amino acid sequences and biochemical and structural characteristics, prenyltransferases are categorized into different subgroups (7). In the last decade, significant progress has been achieved with the members of the dimethylallyltryptophan synthase (DMATS)5 superfamily, and >40 enzymes of this group were identified and characterized by mining of fungal and bacterial genomes (7). These enzymes catalyze transfer reactions of a prenyl moiety from prenyl diphosphate (e.g. dimethylallyl diphosphate (DMAPP)) to diverse acceptors, such as tryptophan, tyrosine, tryptophan-containing cyclic dipeptides, xanthones, tricyclic or tetracyclic aromatic moieties, or even non-aromatic compounds. Among the acceptors, indole derivatives, including tryptophan and tryptophan-containing cyclic dipeptides, are substrates of most of the DMATS enzymes investigated so far (7, 9).
The DMATS enzymes have already been demonstrated to display high substrate and catalytic promiscuity. They not only catalyze prenylation of their substrates and closely related compounds, but they also use structurally quite different compounds as prenyl acceptors (10). Therefore, these enzymes were successfully used for production of a large number of prenylated derivatives, including prenylated tryptophan and tyrosine analogs, tryptophan-containing peptides and derivatives thereof, hydroxyxanthones, hydroxynaphthalenes, flavonoids, indolocarbazoles, and acylphloroglucinols (10). For example, N1-, C4-, C5-, C6- and C7-prenylated tryptophan and N1-, C2-, C3-, C4- and C7-prenylated tryptophan-containing peptides and derivatives were obtained by using DMATS enzymes as biocatalysts (9, 10).
One of the challenges in discovery and use of DMATS enzymes as biocatalysts in a rational and targeted manner is the prediction of the acceptance of a putative substrate. On the one hand, the enzymes share similar structures, albeit often at low sequence identities, and catalyze, in many cases, similar reactions. On the other hand, different enzymes with similar natural substrates accept further non-native aromatic substances with clearly different activities (7). Therefore, bioinformatic and chemoinformatic approaches for the prediction of the catalytic activity of these enzymes are welcome and necessary to harness the full biosynthetic potential of this enzyme class.
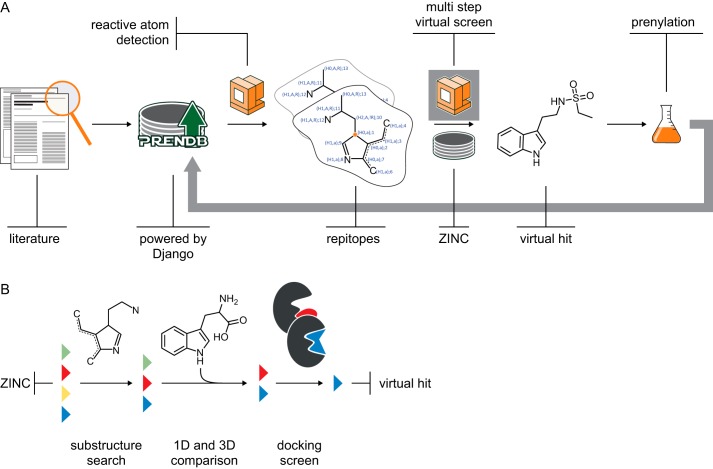
We describe in this work the creation and evaluation of a database that catalogs and stores prenyltransferase reaction information. Because storage of the reactions is automated, the database is not static but will grow with each new reaction described in the literature. Furthermore, we present an application of PrenDB, where we predict and validate putative substrates for prenyltransferases (Fig. 2).
FIGURE 2.
Schematic of the underlying work flow. A, reactions of prenyltransferases were digitalized and stored in PrenDB. The reactive atoms were detected algorithmically and reconstituted to reactive epitopes (repitopes). Prenylation candidates were selected by a multistep virtual screening approach, and their transformation potentials were evaluated experimentally on selected prenyltransferases. B, multistep virtual screening of database compounds (colored triangles): (i) repitope-based substructure search based on the substrate space covered in PrenDB; (ii) one-dimensional property and three-dimensional shape comparison with known substrates; and (iii) docking screen on three promiscuous prenyltransferases with available crystal structure.
Results
PrenDB Statistics
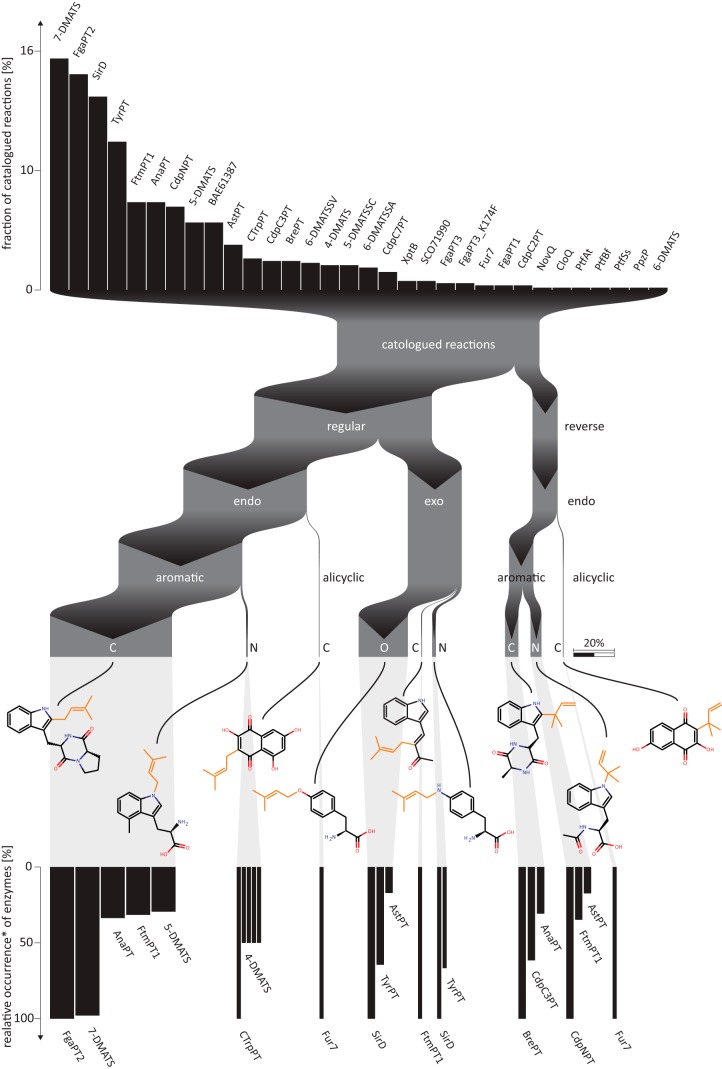
Digitalization and chemoinformatic encoding of enzymatic reactions of the DMATS superfamily allow for a deep analysis of their substrate space and reactivity toward distinct chemical epitopes (Fig. 3). In total, 32 unique enzymes were found throughout the literature examined. The three most prominent prenyltransferases in terms of the number of annotated transformations are 7-DMATS, FgaPT2, and SirD, accounting for 15, 14, and 13% of the reactions in the database, respectively. At the other end of the spectrum, there are seven enzymes for which only a single reaction has been published. With respect to promiscuity, the number of unique reactive epitopes–molecular substructures centered around the reactive atom and henceforth called repitopes in this work–was used as a descriptor (cf. Fig. 1D for an exemplary repitope). The enzymes 7-DMATS, FgaPT2, and AnaPT transfer prenyl moieties onto the broadest range of chemical epitopes. Together, these three enzymes contribute >65% of the repitope space. Knowledge of the reactive atom and its surroundings makes a further distinction of enzymatic transformations possible; the Sankey diagram in Fig. 3 shows how the cataloged reactions not only can be linked to their prenyltransferases, but also can be subdivided into types per the reactive atom. The clear majority of reactions (87%) corresponds to the regular type of prenyl moiety transfer (Fig. 1B, right), where the thermodynamically more stable regioisomer is formed. In 73% of all reactions and in all reverse attachments, the reactive atom is a member of a ring system (endo). Only a small part of regular prenylations occur at exocyclic atoms (exo, 26%). There, prenyl moieties are transferred onto oxygen and nitrogen atoms of tyrosine and aniline-like moieties by SirD, FtmPT2, or 4-DMATS. Reverse prenylation can be observed at carbon and nitrogen atoms only. They are incorporated in aromatic ring systems and less frequently also in alicyclic moieties, such as benzoquinones. More than 60% of all reactions occur at aromatic carbon atoms; derivatives of indole, including tryptophan, are the most frequent repitopes in this largest reaction subclass. Much rarer are prenylations at nitrogen (8%) or at exocyclic oxygen atoms (23%). By comparison, 5% (33 entries) of reactions of tryptophan-like moieties (e.g. at atom position C2 in brevianamide F (E1) (Fig. 1A) lead to formation of compounds with a fused ring system, where the atomic environment of the reactive atom becomes dearomatized during the reaction (FtmPT1, Fur7, and 4-DMATS).
FIGURE 3.
Illustrative interpretation of the analysis of PrenDB. Thirty-two enzymes contribute to the entirety of digitalized reactions (top histogram). The reactions can be further subdivided into categories based on the chemistry of the transformation (middle). Each subgroup corresponds to one or more enzymes (bottom histogram). *, the contribution of the enzyme with the highest contribution to a particular group was set to 100.
Substrate Space
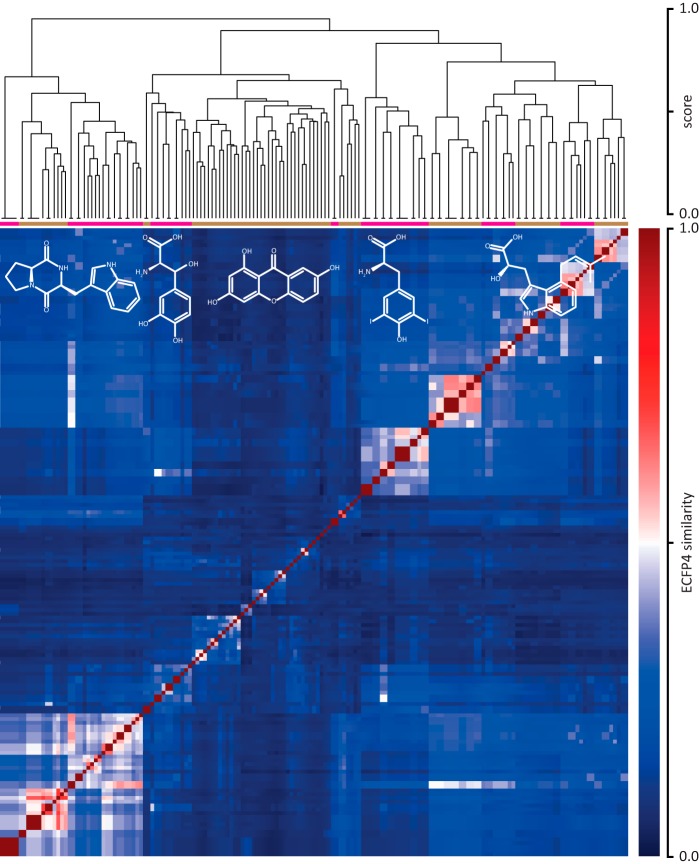
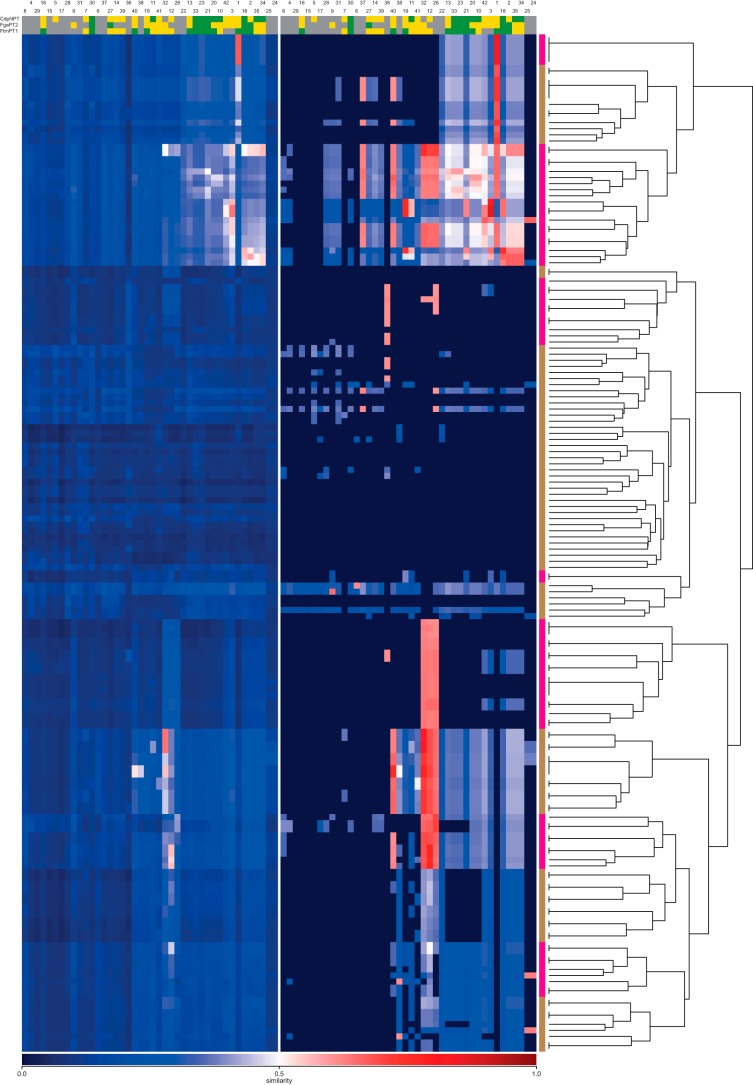
Throughout the analyzed literature (44 articles from 17 journals), 167 unique substrates were found. To analyze the substrate space diversity of prenyltransferases, a similarity matrix based on the pairwise ECFP4 (11) fingerprint molecular similarity was calculated (Fig. 4, Table 1, and supplemental Table S1), followed by a hierarchical clustering. This allows the grouping of substrates based on their chemical structure. From the corresponding dendrograms and supported by the reorganized distance matrix, five substrate classes distributed over 14 clusters can be deduced: (i) unsubstituted indoles, derivatives of tryptophan and proline-tryptophan diketopiperazines; (ii) derivatives of tyrosine with modifications on benzene and aliphatic atoms; (iii) naphthalene, quinone, and flavonoid derivatives; (iv) side chain-modified tyrosines; (v) side chain-modified and multiply prenylated tryptophans. More than 50% of the substrate space is covered by indole-containing compounds. Molecules with tyrosine and flavonoid or xanthone motifs contribute 18 and 26% to the substrate space, respectively. Furthermore, the space spanned by the fragments obtained via bond cleavage during the fragment-based subgraph isomorphism perception process (cf. “Materials and Methods”) is covered to an extent of 64% by tryptophan and diketopiperazine epitopes. This predominance of indoles can be explained by tryptophan and indole derivatives being the native substrates for 78% of the enzymes in PrenDB. This, combined with the DMATS bias in the literature, eventually leads to strongly indole-biased data.
FIGURE 4.
Hierarchical clustering of the substrates extracted from PrenDB. Individual clusters derived from the dendrograms are annotated with an image of the corresponding cluster representative. Magenta and brown bars indicate 14 detected clusters. Black horizontal lines on the leaves of the dendrogram indicate the number of molecules grouped together.
TABLE 1.
Cluster size and substrate space coverage derived from hierarchical clustering
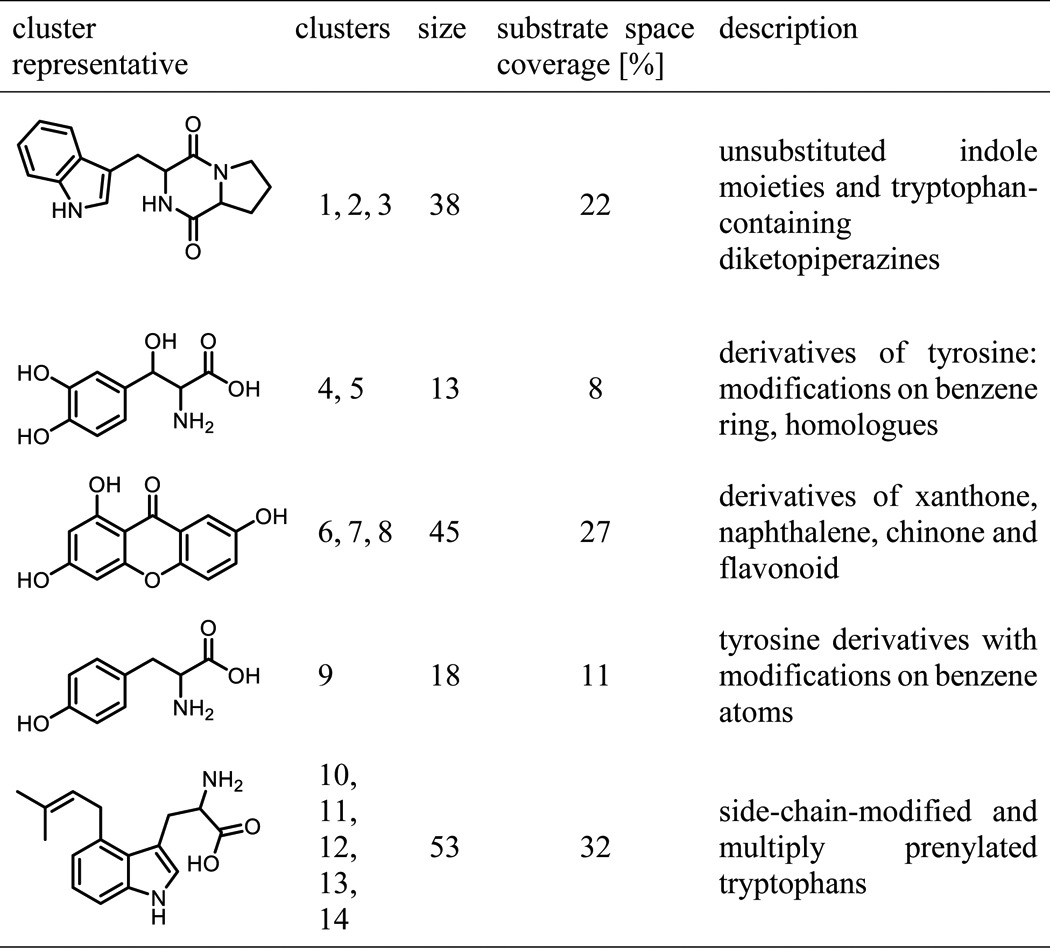
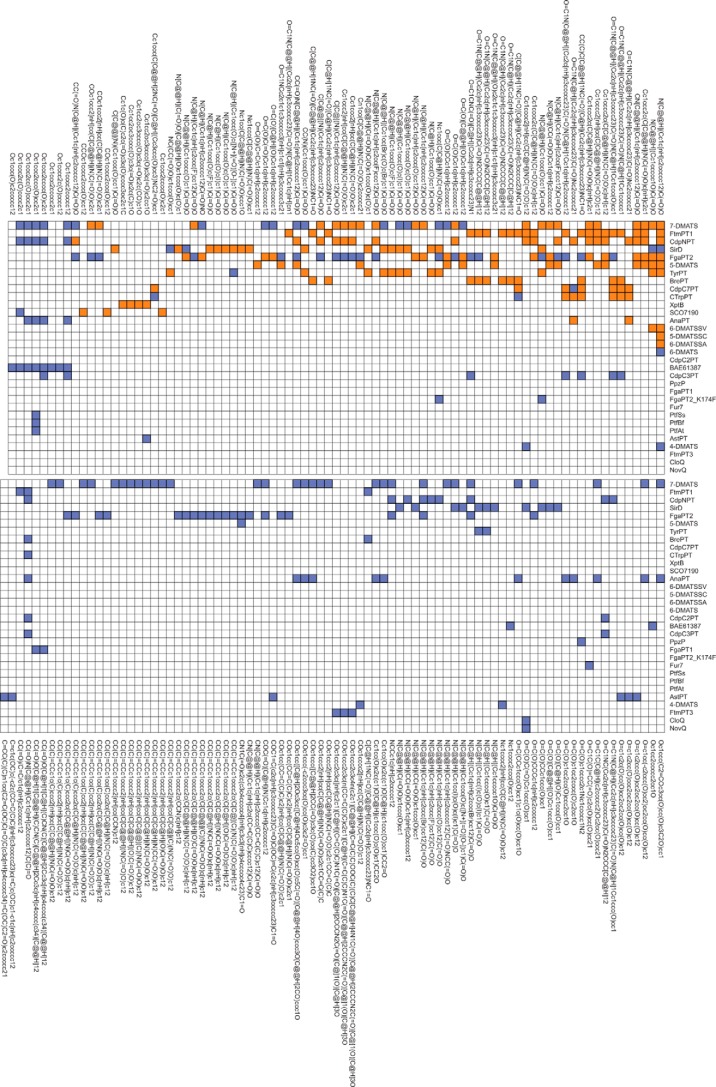
Fig. 5 shows the knowledge about prenyltransferase reactions, as digitalized and stored within PrenDB, in terms of catalyzed transformations (combinations of a particular substrate and an enzyme) and the corresponding yield achieved. In the top right corner of the matrix, transformations of the most abundant substrates (tryptophan, tyrosine, diketopiperazines, and their derivatives, respectively) together with the most promiscuous enzymes (7-DMATS, FtmPT1, CdpNPT, SirD, and FgaPT2) can be found. At the same time, the matrix is sparse (i.e. contains a lot of blanks). This sparsity is the result of the availability of data and thus represents the research focus of the prenyltransferase field in the past. It presents a challenging starting situation for model building.
FIGURE 5.
Coverage of enzyme-substrate space. Orange squares indicate transformation yields >10%, and cornflower squares show a yield <10% for the corresponding enzyme-substrate combination, respectively. White squares, absence of data.
Repitopes
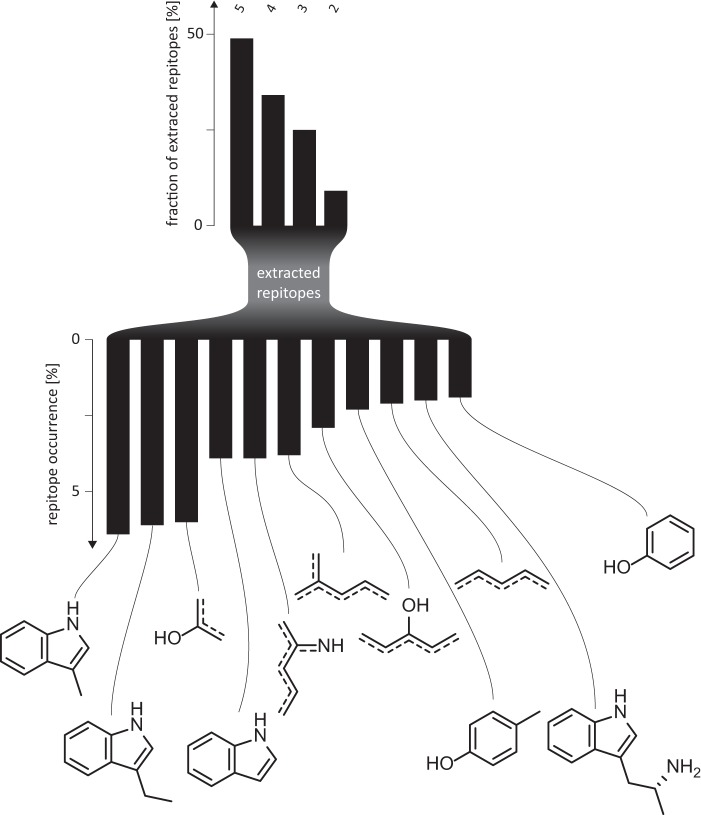
For each of the 665 cataloged reactions (each defined as a unique triplet of a substrate, product, and enzyme), a repitope (i.e. reactive epitope; see “Materials and Methods” for a complete definition) was extracted using the reactive atom detection and repitope reconstitution routines of the algorithm developed in this work (see “Materials and Methods”). Each repitope comprises four reconstitution depths (from 2 to 5 bond distances). Over all repitope depths, 276 unique repitopes, defined by their unique SMARTS string, were extracted. A SMARTS string is a one-dimensional encoding of chemical substructures and an efficient way to store a complete definition of each repitope (see “Materials and Methods”). A reconstitution depth of 5 delivered the largest contribution to the repitope space with 135 (49%) members. This is consistent with expectations, because larger depths will lead to more diverse descriptions. Depths 2, 3, and 4 account for 25 (9%), 69 (25%), and 94 (34%) repitopes, respectively. Interestingly, the sum over all amounts of each reconstitution level is greater than the total number of unique repitopes. This means that distinct combinations of the substrate molecule, its reactive atom, and the depth level are not mutually exclusive, thus resulting in duplicate entries. Fig. 6 shows the distribution of repitopes for the various depth levels of reconstitution, underlining the relationship between repitope size and diversity. Although the largest class of repitopes contributes the most to repitope space, also smaller, more general and ambiguous repitopes are among the top 10 most frequent repitopes; substructures of tyrosine, benzene, and indole are at ranks 3, 5, 6, 7, and 9, respectively, whereas complete and extended indole and tyrosine structures are at ranks 1, 2, and 4 and, at 8, 10, and 11.
FIGURE 6.
Distribution of repitopes of different depth in repitope space. Dashed lines, conjugated bonds.
Prediction of Novel Substrates via a Multistep Screening Procedure
In a sequential application of virtual screening tools (Fig. 2), beginning with prenylation prediction through repitopes stored in PrenDB and concluding with docking into three prenyltransferases with a known crystal structure (FgaPT2, FtmPT1, and CdpNPT), 38 virtual hits were selected through the following procedure. (i) A compound was considered as a virtual hit if any PrenDB repitope could be found within its molecular framework at least once. Supplemental Table S2 shows the number of repitopes matching a particular hit, with a high repitope hit rate indicating promiscuous compounds (i.e. molecules that are classified as substrates of multiple enzymes). Using repitopes based on a reconstitution depth of 3, 168,906 compounds were selected in this first step. (ii) Comparison of molecular properties with those of known substrates and removal of molecules outside the respective ranges (Table 2) reduced the number of virtual hits to 90,559. Going beyond one- and two-dimensional molecular descriptors and ensuring that the (iii) three-dimensional shape (judged by a high score in the OEChem shape congruency tool; see “Materials and Methods”) matched between putative and known substrates led to a selection of 451 compounds. This repitope-, property-, and shape-based determination of prenylation potential of the selected compounds was further refined by the (iv) docking results; for each compound, an optimal enzyme structure for docking was selected based on a compound's structural overlap with the co-crystallized substrate. The amount of this overlap was quantified by the same shape congruency methodology mentioned above but was automatically invoked from within the docking application HYBRID (see “Materials and Methods”). The generated poses, from which 38 molecules were selected for experimental validation, show a distinct geometrical consensus of the key interactions with the enzymes (Fig. 7): first, polar interactions with the general base Glu-89/102/116; second, occupation of the apolar indole-subpocket and hydrogen bond interactions in the vicinity of the opening of the active site, residues His-279 and Arg-244.
TABLE 2.
Range limits of physico-chemical properties derived from the substrate space stored in PrenDB
| Physico-chemical property | Minimum | Maximum |
|---|---|---|
| Molecular mass (Da) | 140 | 515 |
| No. of heavy atoms | 11 | 37 |
| No. of carbon atoms | 8 | 27 |
| No. of heteroatoms | 1 | 10 |
| No. of chiral centers | 0 | 5 |
| Hydrogen bond acceptors | 0 | 6 |
| Hydrogen bond donors | 1 | 6 |
| No. of atoms in a ring system | 6 | 25 |
| No. of rotatable bonds | 0 | 6 |
| No. of rigid bonds | 9 | 40 |
| XLogP | −3.94 | 3.76 |
| Minimal solubility attribute | Poorlya | |
| 2D polar surface area (Å2) | 20.0 | 1125 |
| Removal of known aggregators | Trueb |
a Solubility categories (insoluble, poorly, moderately, soluble, very, highly) are derived from reparametrized atom types from the XLogP algorithm within the OEChem toolkit.
b An aggregator is considered an exact match with one of approximately 400 published aggregators compiled in the OEChem toolkit.
FIGURE 7.
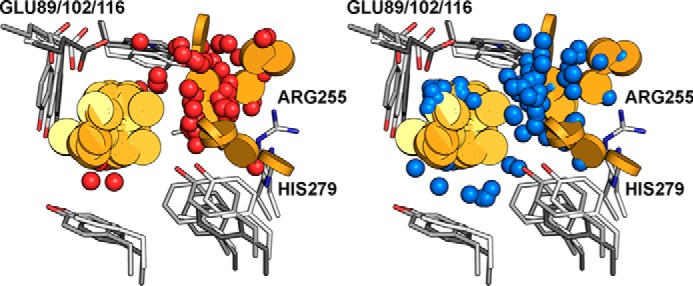
Molecular features extracted from poses of the selected virtual hits. Left, red spheres, hydrogen bond acceptors; yellow discs, aromatic moieties. The majority of acceptor functionalities can be found around the basic residues Arg-255 and His-279. Right, blue spheres indicate hydrogen bond donors. They are located around the highly conserved glutamate (Glu-89/102/116) and in the vicinity of backbone carbonyls (omitted for clarity).
Novel Substrates for Prenyltransferases FgaPT2, FtmPT1, and CdpNPT
To assess the predictive performance of our virtual screening, the 38 potential substrates were tested as prenyl acceptors in enzyme assays with the tryptophan prenyltransferase FgaPT2 and the two tryptophan-containing cyclic dipeptide prenyltransferases FtmPT1 and CdpNPT. The selected substances clearly differ structurally from the substrates for the DMATS prenyltransferases reported previously (7, 10). The reaction mixtures were analyzed with LC-MS to detect the formation of prenylated products. As shown in supplemental Table S2 and Fig. 8, 23 of these substances were accepted by FtmPT1, 22 by FgaPT2, and 25 by CdpNPT. In relation to the number of hits selected from our virtual screen, this corresponds to a hit rate of 60.5% for FtmPT1, 57.9% for FgaPT2, and 65.8% for CdpNPT. Product yields of >50% were observed for 12 substrates with FtmPT1, 7 with FgaPT2, and 10 with CdpNPT, respectively. The prenylated products can be detected in a straightforward manner as signals in their corresponding mass spectra; their [M + H]+ ions are shifted by 68 Da relative to their educts. Overall, we thus obtained high hit rates and yields >50% in the case of 29 reactions (25% of all attempted reactions).
FIGURE 8.
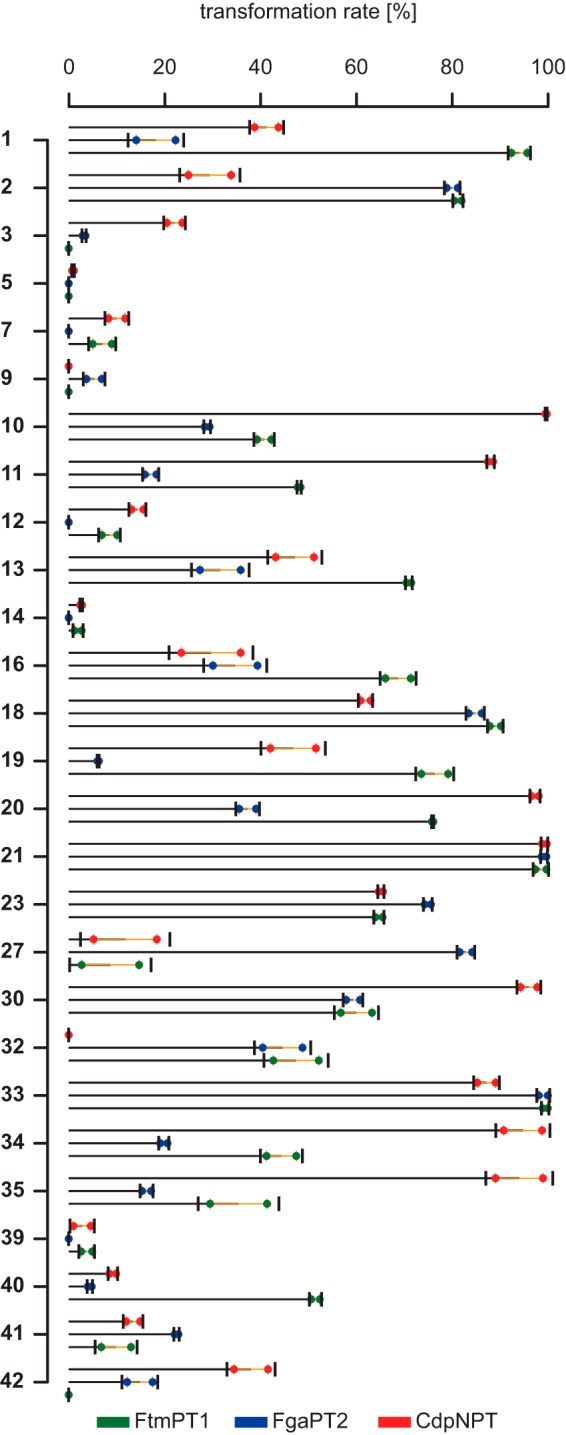
Transformation rates of virtual hits relative to l-tryptophan obtained for the three examined enzymes FtmPT1, CdpNPT, and FgaPT2. Horizontal bars, mean; vertical bars, S.D.; orange interval, S.E.; colored circles, data points.
Similarity Analysis of Known Substrates and Selected Compounds
To assess the novelty of the 38 selected compounds, the similarity with the substrate space cataloged within PrenDB (167 substrates) was calculated and visualized by generating a similarity matrix based on the ECFP4 fingerprint-based Tanimoto similarity (Fig. 9, left). The matrix shows an overall low similarity score between our selection and the known substrate space. This points toward the potential to access truly novel substrate space by employing repitopes. Of note, the similarity is higher in columns corresponding to compounds that were successfully prenylated in our assays by at least two of our test enzymes. The right panel of Fig. 9 shows similarity scores as calculated by our in-house fragment-based method RedFrag (12); in contrast to ECFP4, RedFrag compares the fragmental composition of molecules and the two-dimensional arrangement of fragments. RedFrag accentuates the commonalities and differences between known substrates and our selections. Compound 1, a tryptophan-homoproline-diketopiperazine (94.1% yield on FtmPT1), shows high similarity scores with tryptophan, its indole core derivatives, and, expectedly, tryptophan-tryptophan-, tryptophan-alanine-, tryptophan-glycine-, and tryptophan-proline-diketopiperazines from clusters 1, 2, and 3, respectively. Of note, RedFrag emphasizes the similarity of compound 1 and cluster 3 based on the presence of the indole scaffold. In contrast, ECFP4 emphasizes the dissimilarity of this compound originating from the absence of the diketopiperazine motif in the same cluster. Compounds 12, 26, and 32 show high similarity with substrates from cluster 3 (also 9, 10, and 11). Compounds 12 and 32 are regioisomers of a brominated tryptophan derivative. They show distinctly different yields: 14.4 and 8.5% for 12 in CdpNPT and FtmPT1; 47.5 and 44.7% for 32 in FtmPT1 and FgaPT2, respectively. It is evident that the position of the bromine atom has a major impact on the role of such compounds as substrates. The influence of regiochemistry of indole core substitutions or single-atom replacements at this core is further exemplified by compound 26. Its benzothiophene moiety (replacing the nitrogen atom in an indole by a sulfur atom) is not accepted as a substrate by any of the three test enzymes.
FIGURE 9.
Similarity matrix between the selected compounds and known substrates for prenyltransferases extracted from PrenDB. Left, ECFP4 fingerprint similarity. Middle, RedFrag scores calculated with ECFP4 fingerprints. Color coding (top), green, yield >50%; yellow, yield between 1 and 50%; gray, no transformation. Right, magenta and brown bars indicate 14 detected clusters. Black vertical lines on the leaves of the dendrogram indicate the number of molecules grouped together.
Selected compounds with low similarity but remarkable yields indicate novel substrate classes or motifs; compound 16 shows a good yield in FtmPT1 and moderate yields in FgaPT2 and CdpNPT (68.8, 34.8, and 29.7%, respectively). Its conjugated indole-4-imidazolin-2-one motif has no similar counterparts within the known substrate space. This is also true for compound 30 (yields of 60.1, 59.4, and 96.1%, respectively) and its benzylated hydroxyl-indole structure. Compound 27, a pyrimidine-indole, shows excellent yield in FgaPT2 (82.9%). Further examples with high yields but RedFrag similarity scores <0.6 are compounds 11, 13, 20, 23, and 33.
Structure Elucidation of the Products of Compounds 21 and 30
To investigate at which position within a given substrate the prenylation occurred, we carried out exemplary FtmPT1 incubations with two indole derivatives, indole 3-ethyl ethylsulfonamide (21) and 6-benzyloxyindole (30), which were very well (98.6% yield) and moderately (60.1% yield) accepted by this enzyme, respectively (Fig. 10 and supplemental Table S2). As shown in Fig. 11A, a single dominant peak was observed in the LC-MS chromatograms of the incubation mixtures, which were isolated on a Multospher 120 RP-18 column for structure elucidation. 1H NMR data revealed, surprisingly, the presence of two compounds in each reaction mixture. 21a and 21b originated from 21 in a ratio of 1:1 from the reaction mixture, and 30a and 30b originated in a ratio of 1:0.85 from 30. Further purification resulted in four pure products. Through NMR and MS analyses, the structure of 21a was subsequently elucidated as a regularly N1-prenylated derivative. The second product, 21b, was identified as a reversely C3-prenylated derivative with a simultaneous cyclization of C2 of the indole with the nitrogen atom of the side chain located at C3, resulting in the formation of a 6/5/5 fused ring system. Compounds 30a and 30b were proven to be regularly C2- and C5-prenylated derivatives, respectively. These results unequivocally proved specific prenylations at the indole ring without or with additional modifications, such as cyclization. Detailed studies of the relationships of enzymes, substrates listed in supplemental Table S2, and their products are under further investigation.
FIGURE 10.
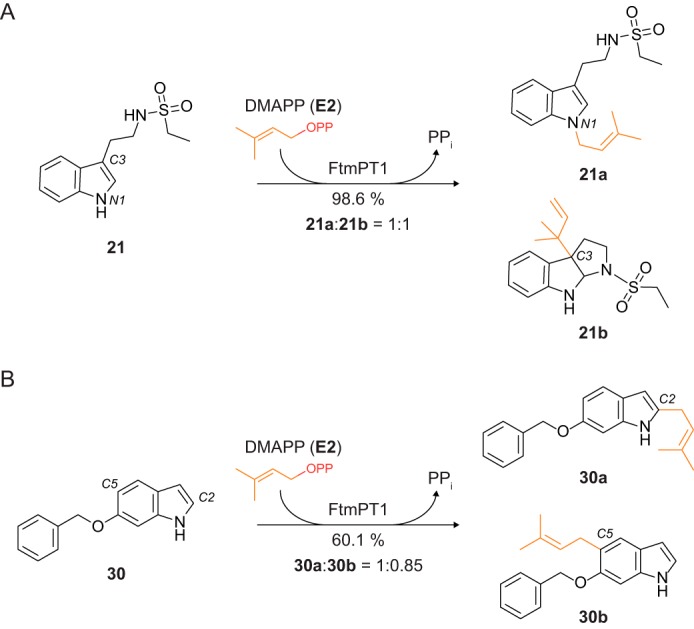
Transformations of virtual hits 21 and 30 by FtmPT1. A, indole 3-ethyl ethylsulfonamide (21) was regularly prenylated at position N1, leading to 21a, and reversely prenylated at position C3 with a simultaneous formation of a 6/5/5 fused ring system (21b). Typically for C3-prenylation at indole substructures, a dearomatization and intramolecular cyclization accompany the prenylation reaction. B, 6-benzyloxyindole (30) was regularly prenylated at positions C2 and C5.
FIGURE 11.
LC-MS analysis of the reaction mixtures and prenyl transfer reactions of FtmPT1 with 21 (A) and 30 (B). EIC, extracted ion chromatogram; mAU, milliabsorbance unit.
A comparison of the elucidated structures of the products 21a, 21b, 30a, and 30b with the PrenDB-predicted prenylation sites of their corresponding educts reveals that the prenylation site was correctly predicted in two of four cases. However, the responsible enzyme, FtmPT1, was only proposed for the prenylation of 21 to yield 21a. In the case of 30, the product 30b was predicted to originate from the enzyme CdpNPT or FgaPT2.
Discussion
This study demonstrates the power of systematically organizing and analyzing diverse and disparate experimental enzymatic data by means of chemoinformatic methods. Besides a comprehensive repository of the existing knowledge about prenyltransferase reactions, the determination of repitopes allowed us to predict novel substrates that are distinctly different from the ones that have been identified previously, both natural and synthetic. Moreover, we achieved an overall high hit rate of 71% in terms of molecules that were accepted by at least one prenyltransferase. However, it has to be noted that the repitopes stored in PrenDB are not yet accurate enough in all cases to precisely predict the correct enzyme and/or the correct reactive atom. This shortcoming is presumably correlated with the comparatively small number of instances in the database. Although the existing body of literature clearly represents a considerable experimental effort, chemistry and the biochemical reactivity of enzymes are so diverse that even higher numbers of substrate-enzyme-product triplets would be necessary to obtain more complete repitopes that also account for the different reactivity of certain substructures. The chemoinformatic strategy that we employed in this work is certainly flexible enough to accurately model more fine-grained patterns.
At the same time, a database such as PrenDB can provide excellent help in determining which reactions and substrates would be worthwhile to test next. On a basic level, one could simply be guided by the number of reactions already described for each enzyme and focus on the underrepresented ones. But also more sophisticated approaches can be envisioned; enzyme phylogenetic trees could be based not on amino acid sequence, but on substrate similarity. Further exploration would thus focus on filling in the missing links. Ultimately, such strategies might merge with machine learning approaches, where the algorithm itself would suggest which enzyme-substrate pairs to test next based on the maximum information gain of each investigation.
Nevertheless, despite some shortcomings, our database and resulting prediction algorithm are already useful for correctly predicting a large number of substrates and thereby aiding in the creation of novel chemical matter. The imprecisions could also be taken as a strength in this context, in that they allow for serendipitous discoveries (e.g. the reverse prenylation of 21 to 21b by FtmPT1).
Last, it has to be emphasized that the concept of repitopes and their fragment-based determination can easily be extended to other enzymatic reactions. The automatic processing of potentially large numbers of reactions and the concomitant conversion into the reaction principles (i.e. repitopes) will lead to facile systematizations and gain of knowledge from the analyses of the emerging data.
The high hit rates (58–66%) for each enzyme and the fact that one-fourth of the reactions had a yield of ≥50% demonstrate the excellent performance of our knowledge-based repitope approach. The combination of PrenDB and its ligand-based approach with protein structure-based tools, such as docking, therefore seems to constitute a powerful combination of strategies. Furthermore, these results prove the potential usefulness of the tested enzymes for the production of prenylated derivatives.
Materials and Methods
The prenylation reaction as conducted by the enzymes of the DMATS superfamily formally corresponds to a substitution reaction occurring on carbon, oxygen, and nitrogen atoms of small metabolites through the transfer of small apolar moieties (denoted as dma (short for dimethylallyl) in the following example). The leaving group is always a pyrophosphate (PPi) and a formal proton accepted by a general base. The reaction can be written in a symbolic way (SMIRKS notation, Daylight Chemical Information System, Inc. website, accessed March 16, 2016; see below); the atoms taking part in the chemical transformation are arranged in a one-line notation showing the bond cleavages and formations.
Square brackets enclose individual atoms, and adjacent atoms are taken to be linked by covalent bonds. Letters denote elements, gB is the general base, and PPi is pyrophosphate; commas represent a logical OR; and numbers are arbitrary labels to allow for unambiguous tracking of each atom. In the above example, it can be seen that the hydrogen atom with label 2 ([H:2]) is substituted by the dma group and moves from its adjacent carbon, oxygen, or nitrogen atom (labeled 1) to the general base (label 5). Fig. 1A illustrates this general transformation in a two-dimensional way exemplarily for the reaction between brevianamide F (E1) and DMAPP (E2) catalyzed by FtmPT1. With this symbolic notation and common chemoinformatic tools in hand, it is possible to virtually transform, for example, any carbon atom [C:1] bearing a hydrogen atom [H:2] (GA in Fig. 1C) into the prenylated product (GP), at the same time generating a protonated general base (PGB) and pyrophosphate (PP) as by-products. Although feasible in silico, a chemical transformation based solely on the reactive atom is unreasonable and ambiguous; in reality, only carbon, nitrogen, and oxygen atoms located within the correct atomic surroundings can undergo prenylation. Thus, the entire molecule, or at least a crucial motif within it, is necessary to completely characterize a reactive environment. We call such a set of atoms consisting of the reactive atom, to which the transferred moiety will be attached, and its neighboring atoms a “reactive epitope” (repitope for short). In the case of the transformation of E1, the corresponding repitope is shown in Fig. 1D. The specification of the carbon atom can now be extended to its full repitope notation,
where lowercase letters denote membership of an atom in an aromatic system, HX indicates the presence of X adjacent hydrogen atoms, and parentheses indicate branching of the molecular framework. From this notation, it can be concluded that the reactive atom is aromatic and is bound to one hydrogen atom; its direct neighbors are an aromatic nitrogen and another aromatic carbon without any attached hydrogen atoms. The second neighbor shell consists of two aromatic carbon atoms and one aliphatic carbon atom. This convenient one-line notation of chemical environments is called SMARTS (one-line molecular patterns; Daylight Chemical Information System, Inc. website; accessed March 16, 2016) and is widely used in the field of chemoinformatics, especially for substructure searches. Atoms, their properties, and binding characteristics are encoded with alphanumeric characters. Multiple molecule SMARTS together with the information about bond breakage and formation yield the SMIRKS of a reaction. With a repitope, such as the one described above, and the enzymatic transformation encoded in SMIRKS notation, it is possible to virtually transform any substrate molecule into its corresponding prenylation product or to easily search for putative substrates by invoking substructure-based virtual screens in publicly available vendor databases.
The corresponding SMARTS notation for each repitope could in principle be deduced by hand, given the chemical structures of the substrate and product molecules. To achieve an efficient handling of several hundred enzymatic transformations, with only the two-dimensional structures of substrate, product, and the transferred moiety as input, an automated procedure for the extraction of transformation SMARTS, and thus repitopes, appears to be as indispensable as it is difficult to accomplish. A fully specified repitope requires knowledge of the reactive atom as well as its surroundings. Repitope deduction can be accomplished by applying subgraph isomorphism-based algorithms followed by the reconstitution of the chemical environment. Both steps (reactive atom perception and repitope reconstitution) will be described in detail below. All coding was done in python. For chemoinformatic calculations, the python wrappers of the RDKit (open source toolkit for chemoinformatics; accessed March 16, 2016) library were utilized. Fingerprint-based similarity calculation was carried out with the OEChem toolkit (OpenEye Scientific Software, Inc., Santa Fe, NM).
Perception of the Reactive Atom
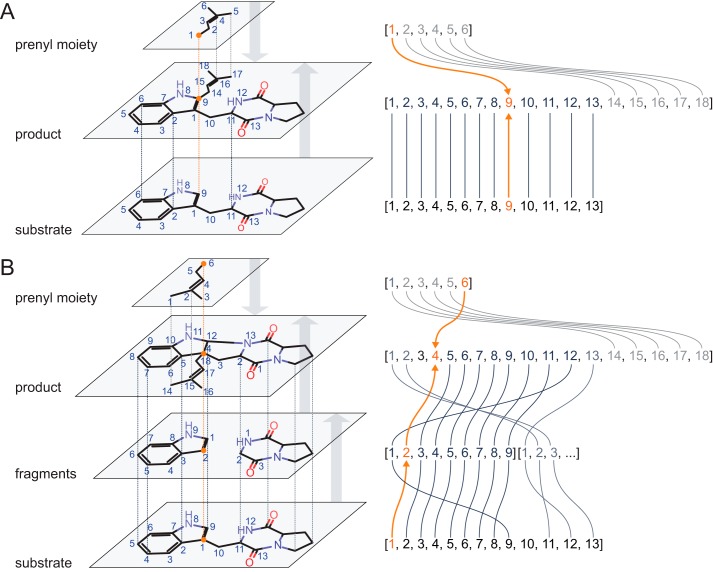
In the case of a simple linear substitution reaction, as depicted in Fig. 1A, the reactive atom can be found by mapping the molecular structure of the substrate molecule onto the molecular structure of the product. For non-symmetric molecules, this leads to a unique match with an atom-to-atom correspondence between substrate and product. Because the number of atoms in the product is always greater than the number in the substrate, the substrate is a substructure of the product (i.e. its complete molecular skeleton can be found within that of the product). With the same approach, the atom-to-atom correspondence between the transferred moiety (i.e. the prenyl group) and the product molecule can be obtained. The intersection of the atom-to-atom matched sets of the substrate and the transferred moiety consists of only one atom, the reactive atom (Fig. 12A). If, however, the enzymatic transfer of a moiety is accompanied by a subsequent (or concerted) rearrangement of the molecular skeleton of the product (e.g. a cyclization), the substrate cannot be considered to be a direct substructure of the product anymore. Thus, the reactive atom can no longer be determined through the atom-wise substrate-to-product mapping as described above. In such a case, a possible strategy for establishing a substructure correspondence (i.e. a subgraph isomorphism) would be to weaken atom or bond type matching criteria. The resulting atom-to-atom correspondences are allowed to be more general in that way but are often ambiguous at best. To circumvent this problem, we assumed that, although the entire substrate may undergo dramatic changes in its molecular skeleton, smaller structural motifs (molecular fragments) remain unaffected by such transformations and can therefore still be unambiguously mapped onto the substrate structure before and after the reaction. Fig. 12B illustrates the consecutive steps in this fragment-based substructure isomorphism approach. In contrast to the aforementioned subgraph isomorphism based on the entire substrate structure, an additional fragmentation step has to be performed. Breakable bonds (bonds connecting ring systems with other ring systems or with acyclic motifs (see Fig. 13 (A and B) for exemplary fragmentations and the breakable bond definition)) are cleaved, leading to a set of fragments. An intermediate filter step ensures that very small fragments (single atoms, linker moieties, terminal groups) are not considered further. The remaining fragments are mapped onto the product structure, and their atom-to-atom correspondence is investigated for an intersection with the atom mappings of the transferred moiety and the product molecule. By preserving the atom-to-atom correspondences between the substrate molecule and its fragments, the reactive atom can be identified by finding the intersecting atom of one of the matching fragments in the substrate.
FIGURE 12.
A, substrate-based subgraph isomorphism. The substrate structure matches the product as a whole. The intersection of atom overlaps between substrate and prenyl moiety delivers the reactive atom (orange arrows, index 9). B, fragment-based subgraph isomorphism. Substrate structure is fragmented into smaller epitopes preserving the substrate-fragment atom matchings. By matching fragments onto the product and analyzing the intersection with the prenyl moiety, the reactive atom can be found within the structure of the substrate (orange arrows, index 1).
FIGURE 13.
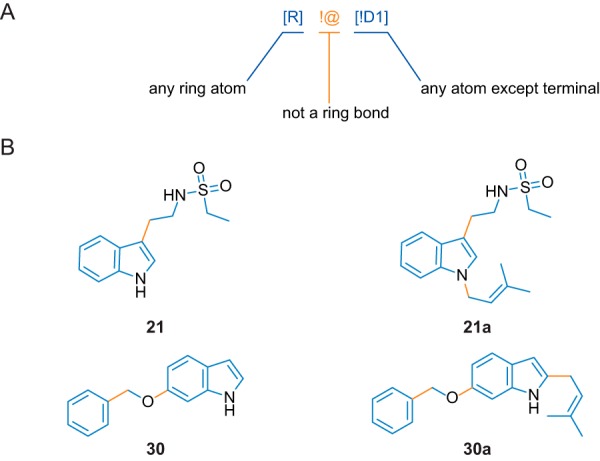
A, fragmentation rule expressed as SMARTS string. The rule consists of two atom definitions and a bond definition. Enclosed in square brackets (blue) are atoms connected by any bond type except for a ring bond (orange). The atom on the left can be of any type but must be a member of a ring system (hydrogen atoms are excluded indirectly because they are not allowed to form ring systems). On the right, the atom must not be a terminal atom (hydrogen atoms are excluded indirectly because they are always terminal). B, breakable bonds (orange) as defined by the fragmentation rule and the resulting fragments (blue) based on the substrates 21 and 30 and their corresponding prenylation products 21a and 30a.
Reconstitution of the Reactive Epitope
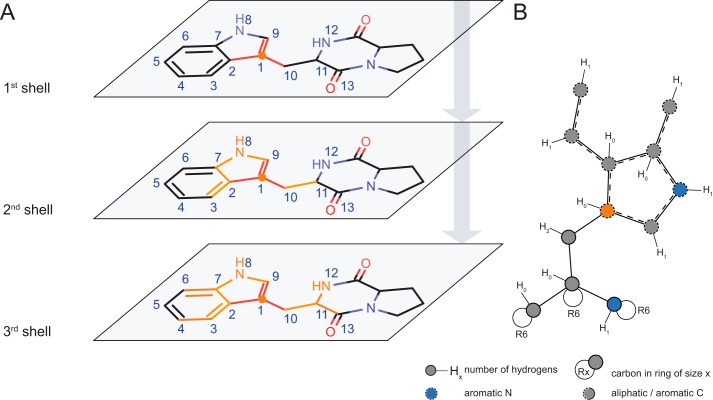
As already mentioned, knowledge of the reactive atom alone is only of limited use for substructure searches or virtual transformations, because both methods yield ambiguous results when only a single atom is given as input. It is therefore necessary to rebuild the chemical environment of the reactive atom to obtain a description of a particular transformation that has discriminative power. To obtain such a description, the reactive atom is augmented with additional atoms from its first, second, third, (etc.) neighbor shells (Fig. 14) (i.e. by traversing the atomic neighborhood of the reactive atom up to a fixed distance (i.e. number of bonds)). The traversed atoms are then extracted as a molecular subset and converted into a regular molecular object and, eventually, a SMARTS string. Different depths of reconstitution lead to either small and nonspecific repitopes (d = 1) or larger and more stringent ones for large depths (d > 3). In extremis, at the largest possible depth, the repitope becomes identical to the molecule itself. Thus, a balance has to be found, and the most useful repitopes are able to represent reasonable chemical environments for a particular reaction but still allow for certain flexibility and diversity in retrieving putative substrates.
FIGURE 14.
A, a repitope is generated by sequentially rebuilding the substrate molecule shell by shell with the reactive atom (index 1) as anchor point. Each iteration adds another neighbor shell to the repitope, resulting in a fully defined depth-3 repitope as depicted by SMARTSviewer (B).
Database of Prenylation Reactions
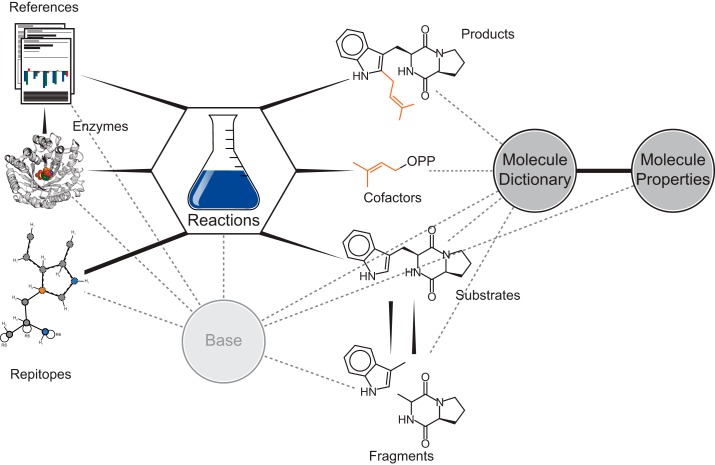
With the algorithmic tools to deconstruct a given transformation catalyzed by a prenyltransferase into a reaction SMARTS and the corresponding repitopes in hand, investigation of as many transformations as possible can readily be conducted. Hence, we decided to create a database (PrenDB) storing the known transformations in an efficiently browsable and queryable manner. For this purpose, a literature search was performed to extract substrates, products, enzymes, and available metadata (such as kinetics and yields) from 44 publications (full articles, reviews, and communications) across 17 journals. Each enzymatic reaction is represented by the SMILES strings (13) of the product and substrate molecules, combined with the preferred name of the involved enzyme. The advantage of using SMARTS is that each reaction can be visualized and processed with common chemoinformatic software. Furthermore, each reaction entry contains multiple repitopes, generated with the aforementioned algorithm and different environmental depths (2–5 bonds around the detected reactive atom). This reaction table (a table is a collection of database entries that are semantically equal) is supported by and connected with further tables holding metadata extracted from the literature and/or calculated with chemoinformatic tools (Fig. 15). The molecule dictionary table comprises all small molecules involved in the reaction: substrates, products, transferred moieties (such as DMAPP or benzylpyrophosphate), and fragments. Additionally, each entry comes with a molecular properties table, where basic physico-chemical properties can be looked up. The reference table contains the literature used for data extraction together with hyperlinks to articles and entries on PubMed and UniProtKB. PrenDB can be browsed and extended with python scripts bundled with the algorithms for repitope generation described in this work (or more conveniently via a web interface) in a straightforward manner. Because of access speed and portability considerations, we decided to use the sqlite3 backend as the underlying database architecture and the Django python package for middleware and frontend.
FIGURE 15.
Design of PrenDB. The database tables are related to each other in a one-to-one (reactions and repitopes), one-to-many (substrates and reactions), or many-to-many (substrates and fragments) relationship, reflecting their real world correspondence. The central reaction dictionary holds the necessary data to encode a reaction based on substrate, product, and cofactor molecules, the enzyme, and the resulting repitope. A reference table is added to supplement the database with metadata and enhance its usability. Dashed lines, abstract inheritance; solid wedged lines, one-to-many relationships (e.g. a molecule can act as substrate in as many reactions as an enzyme). A repitope belonging solely to one particular reaction is indicated by a straight solid line (one-to-one relationship).
Virtual Screen for Putative Substrates of Prenyltransferases
To predict novel substrates for transformation by a prenyltransferase, a multistep screening process was carried out with a subset of the ZINC database (14), which stores commercially available small molecules in ready-to-be-processed formats. First, the ZINC clean leads database, with a total of 5.1 million compounds, was filtered for the presence of any of the extracted repitopes from PrenDB. The repitope depth was 3. The screening was carried out as substructure searches using the python wrappers of the OEChem toolkit. Second, compounds were filtered utilizing the MolProp toolkit (OpenEye Scientific Software). Only compounds with physico-chemical properties within the range spanned by known substrates cataloged in PrenDB were allowed (Table 2). The remaining compounds were subsequently submitted to the shape congruency analysis based on the OEChem API. In short, for each compound, a low energy conformer was generated, and its 3-dimensional overlay with each known substrate was optimized. Compounds with an overlay score >0.9, reflecting excellent three-dimensional shape matching, were allowed for the next step. Fourth, the remaining compounds were docked into the three most promiscuous prenyltransferases for which a crystal structure had been determined (FgaPT2, FtmPT1, and CdpNPT; Protein Data Bank codes 3I4X, 3O2K, and 4E0U, respectively), employing the multi-target HYBRID (15) engine; for each compound, up to 200 conformers were generated with OMEGA (16). The ensemble of conformations of each molecule was then overlaid with the co-crystallized ligand in each of the three selected crystal structures to determine the best suited enzyme for the following exhaustive docking. The method for overlaying conformers is built directly into the HYBRID engine and is based on the same methodology as implemented in the OEChem API and the ROCS application (17). For the actual docking step (translational and rotational optimization of a compound conformer within the binding site of the protein), HYBRID scores for a given protein-ligand complex were calculated based on the shape and electrostatic complementarity of the ligand and protein's binding site (Fig. 16). Shape and electrostatic features are represented by Gaussian potentials. During optimization, the overlap between ligand and protein features is maximized. After docking, calculated poses were visually inspected to remove those that form improbable interactions that are not sufficiently penalized by present-day scoring functions, and the selected compounds were acquired from their respective vendors and experimentally tested.
FIGURE 16.
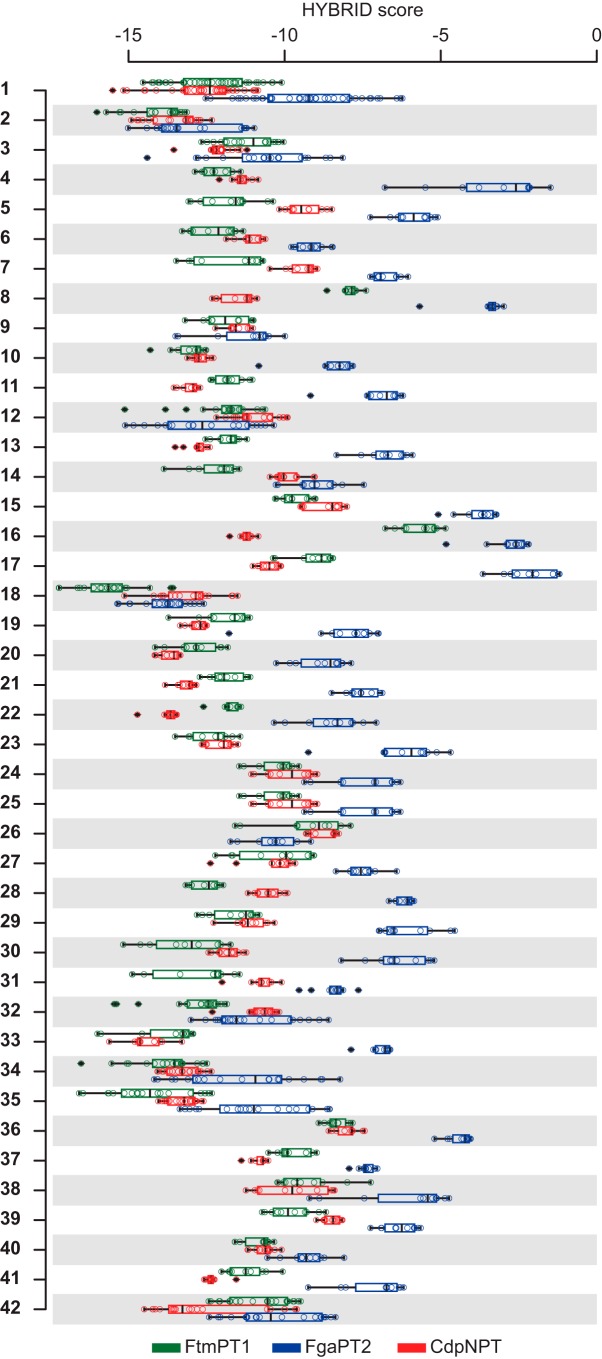
Distribution of docking scores for each of the 42 initially selected poses. Each distribution (represented as a box plot) shows 10 scores (circles)/compound. Boxes embrace 50% of the scores, and horizontal lines (whiskers) cover 99.3% of the scores. Circles with black diamonds, outliers.
Experimental Validation
Chemicals, Bacterial Strains, and Culture Conditions
DMAPP was synthesized according to the method described for geranyl diphosphate reported previously (18). The 38 tested substrates were purchased from Enamine Ltd. (Kiev, Ukraine), ChemBridge Corp. (San Diego, CA), MolPort (Riga, Latvia), Vitas-M Ltd. (Apeldoorn, Netherlands), and Mcule, Inc. (Budapest, Hungary).
Escherichia coli strains XL1 Blue MRF′ (Stratagene, Heidelberg, Germany) and E. coli BL21 (DE3) (Invitrogen, Karlsruhe, Germany) were used for protein overproduction. The strains with expression plasmids were cultivated in lysogeny broth or Terrific broth medium at 37 °C with 50 μg·ml−1 carbenicillin or 25 μg·ml−1 kanamycin as selection marker. Overproduction of FtmPT1 with pAG012, FgaPT2 with pIU18, and CdpNPT with pHL5 were carried out as reported previously (19–21).
Enzyme Assays with Recombinant Proteins
In the assays to determine the acceptance of the different substrates, the enzyme reaction mixtures contained 50 mm Tris-HCl, pH 7.5, 10 mm CaCl2, 2 mm DMAPP, 2–7.5% (v/v) glycerol, 1–2% (v/v) DMSO, 1 mm aromatic substrate, and 0.4 mg·ml−1 purified recombinant protein in a volume of 100 μl. The reaction mixtures were incubated at 37 °C for 16 h and terminated by the addition of an equal volume of methanol. The reaction mixtures were brought to dryness by vacuum evaporation and subsequently resuspended in 100 μl of methanol and centrifuged at 13,000 rpm for 15 min. Five μl of the supernatants were analyzed on LC-MS.
For isolation of the enzyme products, the reaction mixtures were scaled up to 10 ml, containing 50 mm Tris-HCl, pH 7.5, 10 mm CaCl2, 2 mm DMAPP, 2–7.5% (v/v) glycerol, 1–2% (v/v) DMSO, 1 mm aromatic substrate, and 0.4 mg·ml−1 purified recombinant protein, and incubated at 37 °C for 16 h. The reactions were terminated by the addition of 10 ml of methanol and brought to dryness by using a rotary evaporator at 37 °C. The residues were resuspended in 1 ml of methanol, centrifuged at 13,000 rpm for 15 min, and purified on an HPLC device.
LC-ESI-HRMS Analysis of the Reaction Mixtures
The treated enzyme reaction mixtures (5 μl) mentioned above were analyzed on an Agilent 1260 Infinity HPLC system (Böblingen, Germany) in combination with a photodiode array detector and a Bruker micrOTOF-Q III mass spectrometer. For separation, a Multospher 120 RP-18 column (250 × 2 mm, 5 μm, CS-Chromatographie Service, Langerwehe, Germany) with a flow rate of 0.25 ml·min−1 was used. Water (solvent A) and MeCN (solvent B), both containing 0.1% (v/v) formic acid, were used for a linear gradient of 5–100% (v/v) solvent B in A in 40 min. Subsequently, the column was washed with 100% solvent B for 5 min and equilibrated with 5% (v/v) solvent B for 10 min. The separations were monitored with the Bruker micrOTOF-Q III mass spectrometer using the positive ion ESI. HPLC and MS data were processed by using Bruker Compass DataAnalysis version 4.2 (build 383.1) software.
Isolation of Enzymatic Products
Isolation of the enzyme products was performed on an Agilent HPLC series 1200. The separation was carried out on a MultoHigh Chiral AM-RP column (250 × 10 mm, 5 μm, CS-Chromatographie Service) with a flow rate of 1 ml·min−1 and different linear gradients of methanol in water.
NMR Analysis
The isolated enzyme products were brought to dryness by using a rotary evaporator at 37 °C and dissolved in 0.7 ml of CD3OD. NMR spectra were recorded on a JEOL ECA 500-MHz spectrometer (JEOL Germany GmbH, Munich, Germany). The signal of CD3OD at 3.31 ppm was used as an internal reference for chemical shifts. Data processing was done by using MestReNova version 6.0.2-5475 software.
Compound 21a
1H NMR (methanol-d4, 500 MHz) δ = 7.55 (dt, J = 8.0, 0.9 Hz), 7.30 (dt, J = 8.2, 0.9 Hz), 7.12 (ddd, J = 8.2, 7.0, 0.9 Hz), 7.06 (s), 7.02 (ddd, J = 8.0, 7.0, 0.9 Hz), 5.35 (m), 4.70 (d, J = 6.8 Hz) Approx. 3.33 (t, J = 7.3 Hz, signal overlapping with those of solvent), 2.9 (t, J = 7.3 Hz), 2.89 (q, J = 7.4 Hz), 1.85 (s), 1.76 (s), 1.19 (t, J = 7.4 Hz); HR-ESI-MS: m/z = 321.1647, Calcd. for C17H25N2O2S, [M + H]+: 321.1631.
Compound 21b
1H NMR (methanol-d4, 500 MHz) δ = 7.14 (d, J = 7.6 Hz), 7.05 (td, J = 7.6, 1.1 Hz), 6.70 (td, J = 7.6, 0.9 Hz), 6.60 (d, J = 7.6 Hz), 6.07 (dd, J = 17.4, 10.9 Hz), 5.38 (s), 5.12 (dd, J = 10.9, 1.3 Hz), 5.08 (dd, J = 17.4, 1.3 Hz), 3.54 (dd, J = 10.0, 8.4 Hz), 3.10 (q, J = 7.4 Hz), 2.94 (ddd, J = 11.5, 9.7, 5.3 Hz), 2.40 (ddd, J = 12.1, 11.9, 7.9 Hz), 2.07 (dd, J = 12.3, 5.3 Hz), 1.29 (t, J = 7.4 Hz), 1.10 (s), 0.98 (s); HR-ESI-MS: m/z = 321.1642, Calcd. for C17H25N2O2S, [M + H]+: 321.1631.
Compound 30a
1H NMR (methanol-d4, 500 MHz) δ = 7.46 (br d, J = 7.5 Hz), 7.37 (d, J = 8.6 Hz), 7.35 (br t, J = 7.5 Hz), 7.30 (br t, J = 7.5 Hz), 6.92 (d, J = 2.2 Hz), 6.81 (s), 6.73 (dd, J = 8.6, 2.2 Hz), 5.40 (m), 5.08 (s), 3.38 (d, J = 7.0 Hz), 1.76 (s), 1.74 (s); HR-ESI-MS: m/z = 292.1703, Calcd. for C20H22NO, [M + H]+: 292.1696.
Compound 30b
1H NMR (methanol-d4, 500 MHz) δ = 7.48 (br d, J = 7.5 Hz), 7.37 (br t, J = 7.5 Hz), 7.29 (br t, J = 7.5 Hz), 7.24 (s), 7.04 (d, J = 3.2 Hz), 6.96 (s), 6.28 (dd, J = 3.2 Hz, 0.9), 5.35 (m), 5.09 (s), 3.39 (d, J = 7.5 Hz), 1.72 (s), 1.67 (s); HR-ESI-MS: m/z = 292.1704, Calcd. for C20H21NO, [M + H]+: 292.1696.
Author Contributions
J. G. and P. K. designed the study and wrote the paper. J. G. extracted the data of prenyltransferase reactions from the literature, designed and implemented PrenDB, analyzed cataloged reactions and the substrate space in an automatic fashion, programmed automatic repitope extraction, conducted the virtual screen for novel substrates, and conducted and analyzed the similarity study. F. K. designed, performed, and analyzed the enzyme assays with recombinant proteins; performed and analyzed LC-ESI-HRMS analysis of the reaction mixtures; performed the isolation and structure elucidation of compounds 21a, 21b, 30a, and 30b; and contributed to writing of the paper. S.-M. L. contributed to database design, designed enzyme assays, analyzed LC-ESI-HRMS spectra, and contributed to writing the paper. All authors reviewed the results and approved the final version of the manuscript.
Supplementary Material
Acknowledgments
We thank Frank Balzer for help with making PrenDB available online and Christian Raab for data processing during the implementation. S.-M. L. acknowledges the Deutsche Forschungsgemeinschaft for funding the Bruker micrOTOF-Q III mass spectrometer.
The authors declare that they have no conflicts of interest with the contents of this article.

This article contains supplemental Tables S1 and S2.
- DMATS
- dimethylallyltryptophan synthase
- DMAPP
- dimethylallyl diphosphate
- SMILES
- simplified molecular input line entry system
- SMARTS
- SMILES arbitrary target specification
- SMIRKS
- a hybrid language of SMILES and SMARTS
- ECFP
- extended connectivity fingerprint
- ESI
- electrospray ionization
- dma
- dimethylallyl
- HRMS
- high resolution MS.
References
- 1. Liu A. H., Liu D. Q., Liang T. J., Yu X. Q., Feng M. T., Yao L. G., Fang Y., Wang B., Feng L. H., Zhang M. X., and Mao S. C. (2013) Caulerprenylols A and B, two rare antifungal prenylated para-xylenes from the green alga Caulerpa racemosa. Bioorg. Med. Chem. Lett. 23, 2491–2494 [DOI] [PubMed] [Google Scholar]
- 2. Oya A., Tanaka N., Kusama T., Kim S. Y., Hayashi S., Kojoma M., Hishida A., Kawahara N., Sakai K., Gonoi T., and Kobayashi J. (2015) Prenylated benzophenones from Triadenum japonicum. J. Nat. Prod. 78, 258–264 [DOI] [PubMed] [Google Scholar]
- 3. Sunassee S. N., and Davies-Coleman M. T. (2012) Cytotoxic and antioxidant marine prenylated quinones and hydroquinones. Nat. Prod. Rep. 29, 513–535 [DOI] [PubMed] [Google Scholar]
- 4. Li S.-M. (2010) Prenylated indole derivatives from fungi: structure diversity, biological activities, biosynthesis and chemoenzymatic synthesis. Nat. Prod. Rep. 27, 57–78 [DOI] [PubMed] [Google Scholar]
- 5. Wollinsky B., Ludwig L., Hamacher A., Yu X., Kassack M. U., and Li S.-M. (2012) Prenylation at the indole ring leads to a significant increase of cytotoxicity of tryptophan-containing cyclic dipeptides. Bioorg. Med. Chem. Lett. 22, 3866–3869 [DOI] [PubMed] [Google Scholar]
- 6. Botta B., Vitali A., Menendez P., Misiti D., and Delle Monache G. (2005) Prenylated flavonoids: pharmacology and biotechnology. Curr. Med. Chem. 12, 717–739 [DOI] [PubMed] [Google Scholar]
- 7. Winkelblech J., Fan A., and Li S.-M. (2015) Prenyltransferases as key enzymes in primary and secondary metabolism. Appl. Microbiol. Biotechnol. 99, 7379–7397 [DOI] [PubMed] [Google Scholar]
- 8. Heide L. (2009) Prenyl transfer to aromatic substrates: genetics and enzymology. Curr. Opin. Chem. Biol. 13, 171–179 [DOI] [PubMed] [Google Scholar]
- 9. Mai P., Zocher G., Ludwig L., Stehle T., and Li S.-M. (2016) Actions of tryptophan prenyltransferases toward fumiquinazolines and their potential application for the generation of prenylated derivatives by combining chemical and chemoenzymatic syntheses. Adv. Synth. Catal. 358, 1639–1653 [Google Scholar]
- 10. Fan A., Winkelblech J., and Li S.-M. (2015) Impacts and perspectives of prenyltransferases of the DMATS superfamily for use in biotechnology. Appl. Microbiol. Biotechnol. 99, 7399–7415 [DOI] [PubMed] [Google Scholar]
- 11. Rogers D., and Hahn M. (2010) Extended-connectivity fingerprints. J. Chem. Inf. Model. 50, 742–754 [DOI] [PubMed] [Google Scholar]
- 12. Gunera J., and Kolb P. (2015) Fragment-based similarity searching with infinite color space. J. Comput. Chem. 36, 1597–1608 [DOI] [PubMed] [Google Scholar]
- 13. Weininger D. (1988) SMILES, a chemical language and information system 1: introduction to methodology and encoding rules. J. Chem. Inf. Comput. Sci. 28, 31–36 [Google Scholar]
- 14. Irwin J. J., and Shoichet B. K. (2005) ZINC: a free database of commercially available compounds for virtual screening. J. Chem. Inf. Model. 45, 177–182 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. McGann M. (2012) FRED and HYBRID docking performance on standardized datasets. J. Comput. Aided Mol. Des. 26, 897–906 [DOI] [PubMed] [Google Scholar]
- 16. Hawkins P. C. D., Skillman A. G., Warren G. L., Ellingson B. A., and Stahl M. T. (2010) Conformer generation with OMEGA: algorithm and validation using high quality structures from the Protein Databank and Cambridge Structural Database. J. Chem. Inf. Model. 50, 572–584 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Hawkins P. C. D., Skillman A. G., and Nicholls A. (2007) Comparison of shape-matching and docking as virtual screening tools. J. Med. Chem. 50, 74–82 [DOI] [PubMed] [Google Scholar]
- 18. Woodside A. B., Huang Z., and Poulter C. D. (1988) Trisammonium geranyl diphosphate. Org. Synth. 66, 211–215 [Google Scholar]
- 19. Grundmann A., and Li S.-M. (2005) Overproduction, purification and characterization of FtmPT1, a brevianamide F prenyltransferase from Aspergillus fumigatus. Microbiology 151, 2199–2207 [DOI] [PubMed] [Google Scholar]
- 20. Unsöld I. A., and Li S.-M. (2005) Overproduction, purification and characterization of FgaPT2, a dimethylallyltryptophan synthase from Aspergillus fumigatus. Microbiology 151, 1499–1505 [DOI] [PubMed] [Google Scholar]
- 21. Yin W.-B., Ruan H.-L., Westrich L., Grundmann A., and Li S.-M. (2007) CdpNPT, an N-prenyltransferase from Aspergillus fumigatus: overproduction, purification and biochemical characterisation. Chem. Bio Chem. 8, 1154–1161 [DOI] [PubMed] [Google Scholar]
- 22. Schomburg K., Ehrlich H.-C., Stierand K., and Rarey M. (2010) From structure diagrams to visual chemical patterns. J. Chem. Inf. Model. 50, 1529–1535 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.