Abstract
The vast majority of well studied transmembrane channels, secondary carriers, primary active transporters and group translocators are believed to have arisen vis intragenic duplication events from simple channel-forming peptides with just 1–3 transmembrane α-helical segments, found ubiquitously in nature. Only a few established channel-forming proteins appear to have evolved via other pathways. The proposed pathway for the evolutionary appearance of the five types of transport proteins involved intragenic duplication of transmembrane pore-forming peptide-encoding genes, giving rise to channel proteins. These gave rise to single protein secondary carriers which upon superimposition of addition protein domains and proteins, including energy-coupling proteins and extracytoplasmic receptors, gave rise to multidomain, multicomponent carriers, primary active transporters and group translocators. Some of the largest and best characterized superfamilies of these transmembrane transport proteins are discussed from topological and evolutionary standpoints.
Keywords: Integral membrane transport proteins, Evolution, Sequence, Topology, Internal repeats, Duplication, Fusion
Introduction
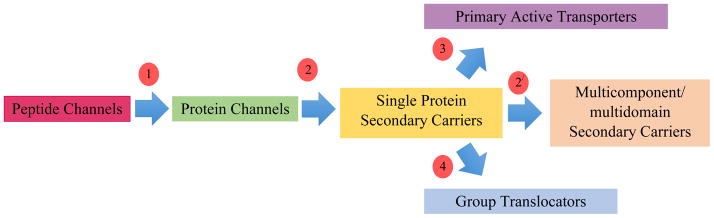
Transmembrane transport systems are vital for all aspects of cellular physiology including uptake and export of nutrients, end products of metabolism, drugs, toxins and macromolecules [1–4]. Our laboratory has studied these proteins for several decades, and have developed and maintain the IUBMB-approved Transporter Classification Database (TCDB; www.tcdb.org) [5–8]. TCDB provides a functional/phylogenetic system of classification with four well-defined classes, (1) Channels, (2) Secondary Carriers, (3) Primary Active Transporters and (4) Group Translocators. The first class includes both small oligomeric peptide channels and larger protein channels. The second class includes both single protein and multidomain, multicomponent, secondary carriers with the precursor channel protein providing the transport pathway. Primary active transporters and group translocators are usually multi-component systems in which the energy-coupling proteins are superimposed on the transporters. Only group translocators modify their substrates during transport, and these also usually require the participation of enzymes superimposed upon and mechanistically coupled to the transporters. However in a few recognized cases, enzymes have become transmembrane, gaining transport functions, and in other cases, transporters have evolved enzyme catalytic activities [9]. Thus, we believe that most transporters evolved in a sequential fashion from class 1 through class 2 to classes 3 and 4 via the pathway shown in Figure 1 [42]. This pathway and the functional diversity exhibited by members of a few large transport protein superfamilies will be the focus of this article [10,11].
Figure 1.
Proposed pathway for the evolution of most transport proteins found in living organisms. Genes encoding simple oligomeric pore-forming peptides (red) underwent intragenic duplication, triplication or quadruplication as well as gene fusional events to add auxiliary domains, generating protein channels (green) with fewer subunits (step 1). These then mutated to achieve stereospecific recognition of solutes as well as an “alternating access” transport mechanism by which the substrate binding site can shuffle between an outwardly open conformation and an inwardly open conformation with one or more intermediate occluded states, yielding a single protein secondary carrier (yellow) (step 2). In some cases, extra domains (for regulation and protein-protein interactions) and extra subunits (for substrate recognition, biogenesis and facilitation of catalysis) were added (step 2′) (tan). Finally, by the superimposition of energy coupling proteins (often enzymes) onto the carrier, primary active transporters (purple) and group translocating porters (blue) evolved (steps 3 and 4, respectively). In some cases, alternative pathways may have been followed, as for light-driven microbial rhodopsins and certain ion-translocating electron transfer proteins, where the primary active transport mechanism did not result from the superimposition of enzymes upon the carrier, but instead involved incorporation of the energy-coupling mechanism into the transport protein or vice versa. Such alternative events provided a much less common mechanism for generating primary active transporters and group translocators. Circles enclosing numbers indicate evolutionary steps leading to transporter types of the TC class, also indicated by the number.
In addition to α-type integral membrane transport proteins, transmembrane β-barrel proteins form a distinct class of channel proteins, called porins or outer membrane pore-forming proteins (OMPPs). They form channels in the outer membranes of many Gram-negative and Gram-positive bacteria, mitochondria, chloroplasts and certain other eukaryotic organelles such as peroxisomes [12,13]. The family and superfamily relationships of these OMPPs and the evolutionary pathways that gave rise to them have been considered in detail [14,15] and will not be discussed further here.
The precursors of most α-helical type transporters: channel-forming peptides
Virtually all types of organisms synthesize peptides designed to insert into membranes of either self or non-self to create oligomeric ion-conducting pores. None of these small peptides form carriers or more complex transporters. Many families and superfamilies of peptide pore-formers are known and recorded in TCDB. They include bacteriocins and archaeocins, made by bacteria and archaea, respectively. Many bacteriocins function to attack other organisms (>12 families in TC subclass 1.C) [16–18]. Eukaryotic “defensins” and “cecropins” (>12 families, also in subclass 1.C) serve as the first line of bodily defense, destroying infectious agents of disease such as envelope viruses, bacteria, fungi and parasites [19]. Ubiquitous organismal holins, “hole-formers”, (TC subclass 1.E) insert into the cytoplasmic membranes of the producing organisms, where they allow phage-induced cell lysis, programmed cell death, toxin release or development, depending on the system (>60 families) [20]. Two holin families (TC#’s 1.E.18 and 1.E.36) have been found to contain both 2 and 4 transmembrane segment (TMS) homologues, and the latter have arisen from the former by intragenic duplication [21]. Viroporins and viral fusion pore-forming proteins (in TC subclasses 1.A and 1.G, respectively) (>40 families) act at various stages during the virus infection cycle. It is clear that such transmembrane peptides are found ubiquitously throughout living organisms and could have provided the starting materials for the generation of more complex transport systems.
Intragenic multiplication to generate large protein channels and pores
Many α-type channel proteins (TC subclass 1.A) and β-barrel porins (TC subclass 1.B) have apparently arisen via intragenic duplication events as well as gene fusion events [11,12]. The best characterized of the former include members of the recently expanded Voltage-gated Ion Channel (VIC) Superfamily.
The Voltage-gated Ion Channel (VIC) Superfamily
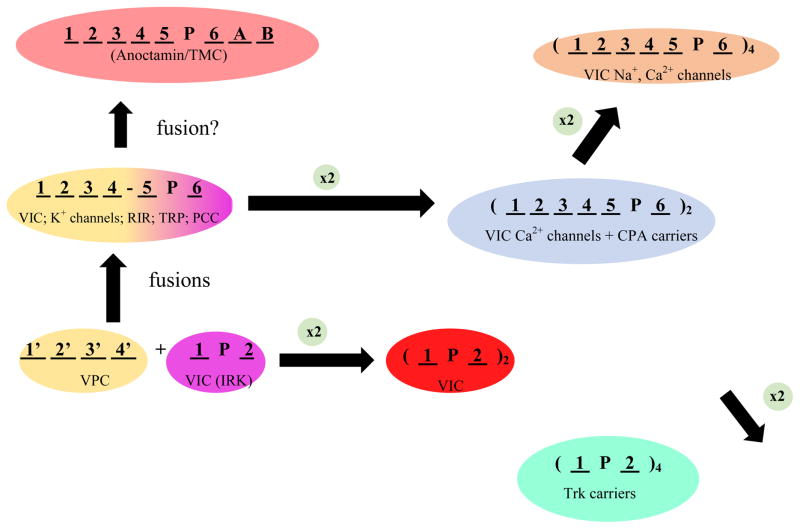
Figure 2 shows our current conception of how the VIC Superfamily evolved to its present level of complexity. Some members (TC# 1.A.1 and 1.A.2) are small 2 TMS proteins, each with a central reentrant “Pore” or “P”-loop, and these small proteins form homo- or hetero-tetrameric channels with a total of 8 TMSs. Another family, the Voltage-gated Proton Channel (VPC) Family (TC# 1.A.51), consists of proteins with 4 TMSs that serve both as voltage sensors and H+ channels [22]. Such 4 TMS domains evidently fused N-terminal to the 2 TMS VIC channels to yield 6 TMS proteins. The N-terminal 4 TMS domain confers voltage sensitivity to the C-terminal 2 TMS domain with central P-loop which retains its ion conductivity function. Although the sensor domains in the 6 TMS VIC Family proteins have lost their H+ channel properties, a few amino acid substitutions in this 4 TMS domain restore primordial proton channel function [23].
Figure 2.
Proposed pathway for the diversification of the Volatge-gated Ion Channel (VIC) Superfamily. The two precursors of 6 TMS VIC family members were (1) a 4 TMS voltage-gated H+ channel (VPC, TC# 1.A.51; yellow, lower left) and (2) a 2 TMS plus pore loop (P) ion (K+) channel (VIC, TC# 1.A.1 and IRK 1.A.2; purple, lower left), which fused with the former N-terminal and the latter C-terminal, to give the voltage-sensor and the channel, respectively, of typical 6 TMS VIC channels (central left; yellow and purple, respectively). Some of these channels gained extra domains by domain fusion events yielding large proteins with the usual 6 TMS topology (RIR, TC# 1.A.3; TRP, TC# 1.A.4 and PCC, TC# 1.A.5). Both the resultant 6 TMS channel and the original 2 TMS channel sometimes duplicated to give 12 TMS proteins (blue; VIC Ca2+ channels; TC# 1.A.1.11) as well as monovalent cation:proton antiporters (blue; CPA; TC# 2.A.36 and TC# 2.A.37), and 4 TMS proteins (red; some VIC K+ channels, TC# 1.A.1.9), respectively (central and lower right, respectively). While the 6 and 2 TMS proteins must tetramerize to form an active channel, the duplicated 12 and 4 TMS VIC channels form active dimers. Most VIC family calcium channels (TC# 1.A.1.11) and possibly all such sodium channels (TC# 1.A.1.10) in animals underwent a second duplication event to form monomeric 24 TMS channel proteins with four 6 TMS internal repeats (brown; top right). In prokaryotes, characterized Na+ and Ca2+ channel proteins of the VIC family usually have 6 TMSs with the P-loop between TMSs 5 and 6, and they must tetramerize to form active channels (not shown). The anoctamin family (TC# 1.A.17; orange) may have arisen from the 6 TMS VIC-like proteins as shown (upper left), although the positions of the P-loops in these proteins are still controversial. Trk K+ carriers (TC #2.A.8) arose following quadruplication of the simple 2 TMS + P-loop unit (green; bottom left). Ion specificity in higher eukaryotes may have evolved via the pathway: K+→ Ca2+→ Na+.
Most VIC K+ channels consist of tetramers of the basic 2 or 6 TMS subunit, but some of these channels have duplicated the 2 TMS element to give 4 TMS proteins that must dimerize to form a functional channel [24] (Figure 2). Animal Ca2+ channels are sometimes duplicated to give 12 TMS proteins, but most Ca2+ channels and all Na+ channels of animals (but not prokaryotes) have duplicated the 12 TMS dimers to give full length 24 TMS monomeric channels [25]. We suggest that in animals, K+ channels gave rise to Ca2+ channels which were the direct precursors of Na+ channels. Further, several different fusional events have given rise to the large 6 or more TMS proteins of the Ryanodine/Inositol triphosphate Receptor (RIR) Channels (TC# 1.A.3), the Transient Receptor Potential (TRP) Channels (1.A.4), and the Polycystin Cation (PCC) Channels (1.A.5) [26]. More recently, the Glutamate-gated Ion Channels (GIC) with a glutamate binding protein (originally from prokaryotes) fused to the channel domain (1.A.10) to give ligand-gated channels [27], and the Anoctamin/TMC/CSC superfamily (TC# 1.A.17) with a variety of fused domains of unknown function, linked to the probable 6 TMS VIC-like domain, have been shown to belong to the VIC superfamily (Z. Ye et al., manuscript in preparation).
Interestingly, two families of carriers, the Trk family of K+ transporters (TC# 2.A.38) and the CPA superfamily of cation:proton antiporters (TC# 2.A.36 and 2.A.37), appear to belong to the VIC superfamily. Trk proteins have 8 TMSs with 4 repeats derived from the 2 TMS channel-forming domain of VIC proteins with a usual P-loop between each pair of TMSs. The CPA family may have originated from a 6 TMS VIC protein with a single duplication giving rise to 12 TMS proteins that subsequently gained or lost one or more TMSs.
Secondary carriers - The Major Facilitator Superfamily (MFS)
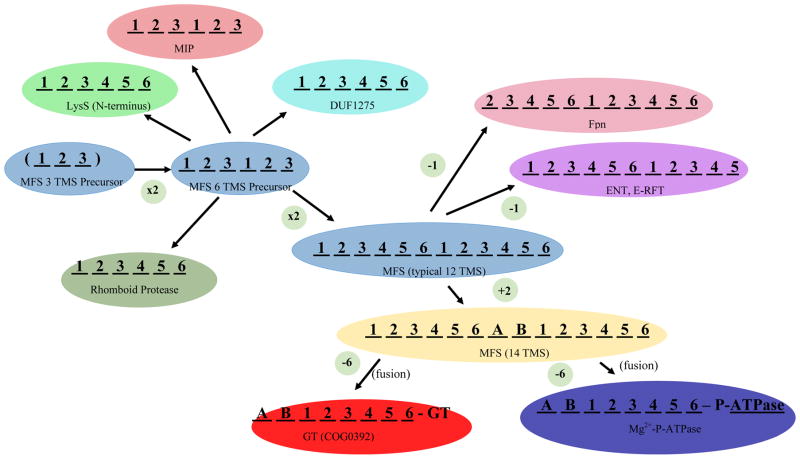
Just as the VIC superfamily is the largest group of homologous channel proteins found in nature, the Major Facilitator Superfamily (MFS) is the largest superfamily of secondary carriers [28,29]. MFS proteins arose via two sequential duplications: 3 --> 6 --> 12 TMSs, with some members gaining two more TMSs centrally located between the two 6 TMS repeat units, possibly by duplication of an adjacent 2 TMS hairpin structure [30] (Figure 3). But what was the origin of the primordial 3 TMS precursor? We have provided preliminary evidence that the simple 2 TMS + central P-loop VIC family channel was the precursor of this 3TMS repeat unit [31]. In this process, the last TMS may have retained its original orientation in the membrane, from out-to-in, the moderately hydrophobic reentrant P-loop became transmembrane, and the first TMS in the 2 TMS VIC precursor inverted, going from the original in-to-out orientation to the opposite out-to-in orientation [31]. Regardless of the veracity of this proposal, high resolution 3-D x-ray crystallographic structures have provided firm evidence for the 3 TMS precursor model for the evolution of MFS porters [32] (Figure 3).
Figure 3.
The proposed precursor for all members of the MFS was a 3 TMS peptide (blue; center left), possibly derived from a 2 TMS plus P-loop voltage-gated ion channel [31] (see text). This 3 TMS unit duplicated twice, first to a 6 TMS protein, and then again to give a 12 TMS protein, characteristic of most MFS carriers (TC# 2.A.1) (light blue; central left and middle). In a few cases, 12 TMS MFS carriers have duplicated or fused to give 24 TMS proteins catalyzing related but different transport processes (not shown). Six TMS products of the proposed 3 TMS duplication also gave rise to members of the MIP family of aquaporins and glycerol facilitators (TC# 1.A.8) (orange; upper left), the N-terminal 6 TMS domains of LysS family members (Lysyl-tRNA syntheases; TC# 9.B.111) (light green; upper left) proteins of the DUF1275 family (TC# 9.B.143) of unknown function (dark green; upper center), and the 6 TMS Rhomboid proteases (pea green; lower left). The basic 12 TMS MFS backbone lost an N-terminal TMS (Ferroportin (Fpn) Family; TC# 2.A.100; pink, upper right), or a C-terminal TMS (the ENT and E-RFT families, TC# 2.A.57 and 2.A.125, respectively) (purple; upper right). Two TMSs (A and B), found in several MFS families, were inserted between the two 6 TMS repeat units. TMSs A and B are believed to have arisen independently in several different MFS families, possibly by intragenic duplication of the adjacent α-helical hairpin-encoding genetic element (yellow; lower right) [30]. From the last 8 TMSs of a 14 TMS protein, possibly of the MFS family, DHA2 (TC# 2.A.1.3), transmembrane regions of glycosyl transferases (GT) (red; bottom center) and the N-terminal domains of some P-type Mg2+-transporting ATPases (TC# 3.A.3.4) (dark blue; bottom left) arose.
Recent studies suggest that the MFS includes more families of secondary carriers than previously believed (nearly 100), but most surprisingly, it appears to include catalytic enzymes such as rhomboid proteases that cleave their substrate proteins within a membrane [33], glycosyl transferases that translocate the growing carbohydrate chain across the membrane as it is being synthesized [34], and N-terminal domains of unknown function within Mg2+-transporting P-type ATPases and lysyl-tRNA synthetases (S. Wang et al., manuscript in preparation). Also, it seems that the 6 TMS Major Intrinsic Protein (MIP) Family (1.A.8) of aquaporins and glycerol facilitators [35], known to have a 3 TMS duplicated sequence, may be distantly related to MFS porters. Phylogenetic trees suggest which families are most closely related, and they also provide evidence that the duplication of the 6 TMS unit to give 12 TMS MFS carriers occurred multiple times during evolution of the superfamily (S. Wang et. al., manuscript in preparation).
Secondary carriers- the Transporter-Opsin-G-protein Receptor (TOG) Superfamily
A second superfamily with unexpected functional diversity is the Transporter - Opsin -G-protein Receptor (TOG) Superfamily [36]. All members of this superfamily apparently arose from a 4 TMS precursor protein that in turn arose from a 2 TMS peptide that duplicated due to an early intragenic duplication event. Then, to generate a full length transporter, a second duplication event gave rise to 8 TMS proteins, many of which lost a single TMS at their N-termini, but a few which lost the C-terminal TMS, to yield the most common 7 TMS topology for the superfamily. The proposed pathway is thus: 2 → 4 → 8 → 7 TMS proteins.
A majority of familial members of the TOG superfamily are secondary carriers, but there are interesting exceptions. One family, the ubiquitous microbial rhodopsin family (TC# 4.E.1), includes light-driven ion (proton or chloride) pumps and light-activated ion channels. Another TOG family, found only in eukaryotes, includes 7 TMS animal rhodopsins and G-protein-coupled receptors, members of the GPCR Family (TC# 9.A.14). Still others include Mg2+ channel protein families with minimally 4 TMSs, the single repeat unit that is duplicated in most other TOG superfamily members (F. Ghazi et al., manuscript in preparation). Finally, one TOG family, the DsbD family (TC#5.1.1), is a transmembrane electron carrier, transferring an electron pair from a cytoplasmic electron donor to a periplasmic or extracellular acceptor [37]. This DsbD family is best known for its function in disulfide bond formation in extracytoplasmic proteins, but other members of this family catalyze cytochrome c biogenesis, thiosulfate oxidation, methylamine utilization, heavy metal (mercury) resistance and peroxide reduction. While most of the earlier work on superfamily expansion indicated that transporters comprise a protein class that evolved independently of most other classes of proteins, such as enzymes, structural proteins and regulatory proteins, it now seems clear that transport protein families have diversified in function to a much greater degree than previously recognized. They have evolved to serve functions, in addition to or instead of transport.
Primary active transporters: The ATP-binding Cassette (ABC) Superfamily
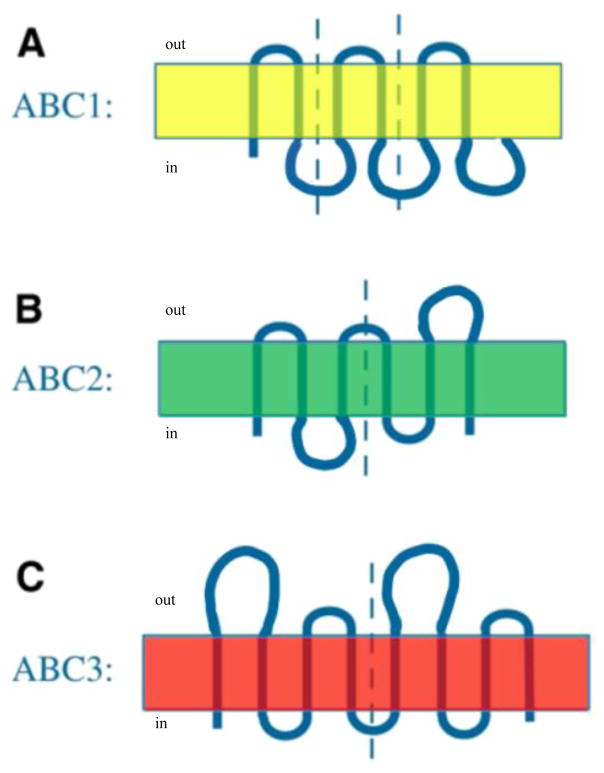
The largest and most diverse of all the “superfamilies” of primary active transport systems is the ATP-binding Cassette (ABC) superfamily (TC# 3.A.1) [38]. Normally, a family or superfamily in TCDB is defined on the basis of the integral membrane protein(s) that provide(s) the transmembrane transport pathway. However, in the case of the ABC superfamily, it was originally defined on the basis of the ATP-hydrolyzing energy-coupling proteins, and once established, such a convention is difficult to change. Subsequent bioinformatic analyses of the integral membrane proteins of ABC export systems revealed that they fell into at least three distinct families, ABC1, ABC2 and ABC3, which apparently evolved independently of each other. Primary sequence analyses revealed that ABC1 porters evolved from a 2 TMS precursor peptide by triplication to yield 6 TMS proteins as the dominant species; ABC2 proteins evolved by duplication of a 3 TMS precursor to yield a very dissimilar 6 TMS porter, and ABC3 proteins evolved by duplication of a 4 TMS element to yield either 8 or 10 putative TMS products (see Figure 4) [39,40]. The 10 TMS homologues of ABC3 systems appear to have the extra two TMSs in the middle of these proteins, between the two 4 TMS repeat units. Thus the “ABC Superfamily” can be considered to be misnomer since it consists of three distinct families of export-type transporters.
Figure 4.
Three topological types of ABC exporters, illustrating the type of internal repeats present in each one [40]. In all cases, vertical dashed lines separate the repeat units. A. ABC1: a six-TMS topology resulting from intragenic triplication of a primordial 2-TMS-encoding genetic element (yellow). The three hairpin repeats have the same orientation in the membrane. B. ABC2: a six-TMS topology resulting from intragenic duplication of a primordial 3 TMS-encoding genetic element (green). The two 3 TMS repeats have opposite orientation in the membrane. C. ABC3: an eight-TMS topology resulting from intragenic duplication of a primordial 4 TMS-encoding genetic element (red). The two 4 TMS repeats have the same orientation in the membrane. ABC3 porters apparently occur in two topological types, 8 TMS proteins with two 4 TMS repeats as shown, and 10 TMS proteins where the two extra TMSs are located in the middle of the polypeptide chain, between the two 4 TMS repeat units [39] (not shown). For assignment of TC families in the ABC functional superfamily to one of these three topological types, see reference 40 and TCDB.
A recently published comparison of 3-D structures of the membrane constituents of ABC uptake systems suggested to the authors that they too are polyphyletic. These uptake systems include integral membrane porters of extremely varied topologies, with numbers of TMSs ranging between 5 and 20. Examining fourteen high resolution structures of ABC importers revealed that the integral membrane uptake proteins exhibit at least three different structural folds. In fact, it was suggested that they are evolutionarily unrelated, although it should be kept in mind that the demonstration of different folds does not establish independent origin [41]. It is nevertheless possible that the membrane porters of both ABC superfamily uptake and efflux systems are polyphyletic.
ter Beek et al [41] referred to the three structural types of uptake systems as Type I, Type II and ECF-type. For Types I and II, the two membrane subunits are either identical (homodimers) or homologous (heterodimers). However, for ECF-type systems, different folds are observed for the two subunits which also may have different functions, the substrate binding (S) subunit and the energy transducing (T) subunit. Interestingly, evidence suggests that some ECF systems can function either as secondary carriers, dependent only on the S subunit, or as ATP-energized systems, dependent on both the S and T subunits as well as the A (ATPase) subunit [42]. Thus, at least 3 evolutionarily distinct types of ABC efflux systems and 3 types of import systems may be present in Nature. Nevertheless, a single mechanism (alternating access) appears to be operative for all of them [41]. This last conclusion as well as the ability of S subunits to function alone suggests that secondary carriers were the evolutionary precursors of ABC primary active transporters.
Group translocators: the prokaryotic sugar-transporting phosphotransferase system (PTS)
The phosphoenolpyruvate (PEP)-dependent sugar transporting phosphotransferase system of group translocators (PTS) phosphorylates its sugar substrates concomitantly with transmembrane transport [43]. Thus, while a sugar is taken up from the medium, a sugar-phosphate is released into the cytoplasm. [44]. The PTS also plays regulatory roles, influencing many aspects of cell physiology [44–46].
Bioinformatic analyses first suggested that the PTS, like the ABC functional superfamily, is a chimeric system with several dissimilar origins for the different sugar-specific integral membrane transport proteins. By contrast, the general energy-coupling phosphoryl transferase proteins of the PTS, Enzyme I and HPr, each form a single superfamily required for the group translocation of all sugar substrates of the system.
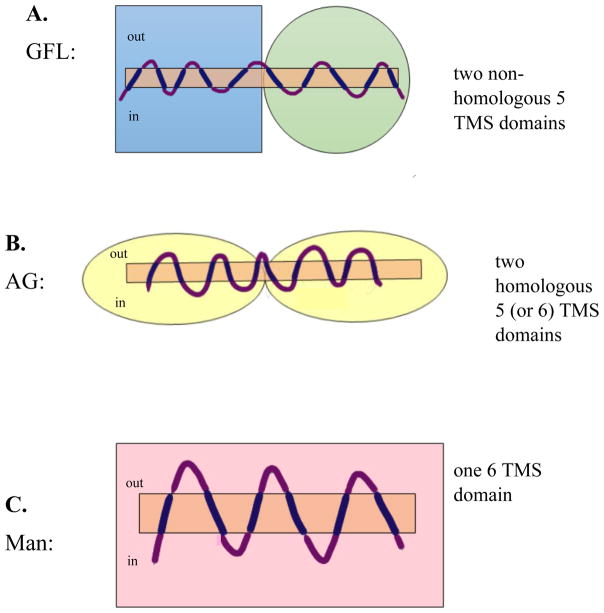
The sugar-specific PTS constituents, the Enzyme IIA, IIB and IIC proteins or protein domains, are not all homologous, and based on high resolution 3-D folds, each probably arose at least three times independently. The largest superfamily of IIC transporters is the Glucose-Fructose-Lactose-Glucitol (PTS-GFL) Superfamily which includes four phylogenetically distant families (TC#s 4.A.1–4). These proteins probably have 10 TMSs in a 5 + 5 TMS double domain arrangement, revealed by x-ray crystallographic analyses of one such system [43]. Interestingly, these two domains serve different functions, and based on 3-D structural characteristics, they are apparently not homologous to each other [47] (figure 5). In this respect, they may resemble ABC ECF-type systems. It is, of course, possible that the members of the four different families within the GFL superfamily, although homologous, have somewhat varied topologies [48]. Further experimentation will be required to examine this possibility.
Figure 5.
Three topological types of nonhomologous PTS group translocators (IIC domains). A. the Glucose/Fructose/Lactose (GFL) Superfamily. B. the Ascorbate/Galactitol (AG) Superfamily, and C. the Mannose (Man) Superfamily. All PTS Enzyme II complexes have IIA, IIB and IIC proteins or protein domains, but Man family complexes also have a IID constituent. PTS IIC domains have apparently evolved 3 times independently of each other, giving rise to the three predicted topological types shown. Boxes, circles, ovals and rectangles indicate the four different structural folds believed to represent the three independently evolving superfamilies of PTS porters (IIC proteins and protein domains).
The second largest family of PTS group translocators is the L-Ascorbate-Galactitol (PTS-AG) superfamily (TC# 4.A.5 and 4.A.7). These proteins appear to contain an internal duplication of a 5 or 6 TMS repeat unit that shows little or no structural similarity to the Enzymes II of the PTS-GFL superfamily [9,49,50]. Interestingly, the transporters of both the GFL and AG superfamilies use a variety of structurally dissimilar but functionally equivalent IIA and IIB proteins that do not correlate with the phylogenies of the corresponding IIC constituents. Evidently, the 3 constituents of the Enzyme II complexes have undergone shuffling relative to each other during their evolution [9].
Finally, porters of the Mannose (PTS-Man) Family (TC# 4.A.6) catalyze uptake of various hexo-ketoses and aldoses. The constituents of the Man Enzyme II complexes are not demonstrably homologous to any of the sugar-specific PTS constituents in the GFL and AG Superfamilies, and in addition to the usual IIA, IIB and IIC proteins or protein domains, there is an additional protein, the IID constituent, not present in other PTS systems. In this case, the IIC transport proteins are believed to have 6 TMSs, possibly arising from a 3 TMS precursor by intragenic duplication. However, no high resolution x-ray structure is available to confirm these suggestions. Thus, like the ABC superfamily, the PTS is a “functional” superfamily consisting of at least three types of integral membrane transport proteins, each of which appears to have evolved independently of the others.
Conclusions and Perspectives
We have been able to put most integral membrane transport proteins into a unified evolutionary framework. The precursors of these proteins are believed to have been pore-forming peptides which gained repeat sequences as a result of intragenic duplication events, one of the most common mechanisms for generating complexity in proteins during evolution [11]. Fusional events and the superimposition of catalytic proteins have added to the level of system complexity. Gene duplications, leading to large sets of paralogues, also occurred commonly during protein evolution, especially in multicellular eukaryotes that have proliferated paralogues into the hundreds to allow developmentally controlled gene expression in a cell type-specific fashion [51,52]. In the case of transporters, however, intragenic duplications and gene fusions promoted mechanistic complexity. This allowed oligomeric peptide channels to evolve into single peptide protein channels and then into stereospecific electrochemical energy-coupled carriers. Finally, with the superimposition of energy-coupling proteins, they assumed the properties of primary active and group translocating transporters (see figure 1). Relatively few transport systems evolved via other pathways. The usual pathway, from simple to complex, is exemplified by the evolutionary proposals presented here.
Highlights.
Transport proteins evolved from peptides via intragenic duplication and gene fusion.
Secondary carriers arose from large channel proteins by introducing point mutations.
More complex transporters arose by superimposition of energy coupling proteins.
The pathways taken for the appearance of large transport protein families are described.
Acknowledgments
The research reported in this communication was supported by NIH grants GM077402 and GM094610. I thank Faezeh Ghazi, Ida Javadi-Razaz, Steven Wang and Zach Ye for conducting the unpublished analyses described in this report, Professor Tsai-Tien Tseng for suggestions for manuscript improvement, and Joshua Asiaban and Sabrina Phan for assistance with manuscript preparation.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
• of special interest
•• of outstanding interest
- 1.Kinne RK. Endothelial and epithelial cells: general principles of selective vectorial transport. Int J Microcirc Clin Exp. 1997;17:223–230. doi: 10.1159/000179234. [DOI] [PubMed] [Google Scholar]
- 2.Busch W, Saier MH., Jr The transporter classification (TC) system, 2002. Crit Rev Biochem Mol Biol. 2002;37:287–337. doi: 10.1080/10409230290771528. [DOI] [PubMed] [Google Scholar]
- 3.Saier MH., Jr Computer-aided analyses of transport protein sequences: gleaning evidence concerning function, structure, biogenesis, and evolution. Microbiol Rev. 1994;58:71–93. doi: 10.1128/mr.58.1.71-93.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- •4.Nikaido H, Saier MH., Jr Transport proteins in bacteria: common themes in their design. Science. 1992;258:936–942. doi: 10.1126/science.1279804. Presents an overall (somewhat outated) view of transport protein design. [DOI] [PubMed] [Google Scholar]
- 5.Saier MH, Jr, Tran CV, Barabote RD. TCDB: the Transporter Classification Database for membrane transport protein analyses and information. Nucleic Acids Res. 2006;34:D181–186. doi: 10.1093/nar/gkj001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Saier MH, Jr, Yen MR, Noto K, Tamang DG, Elkan C. The Transporter Classification Database: recent advances. Nucleic Acids Res. 2009;37:D274–278. doi: 10.1093/nar/gkn862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Saier MH, Jr, Reddy VS, Tamang DG, Vastermark A. The transporter classification database. Nucleic Acids Res. 2014;42:D251–258. doi: 10.1093/nar/gkt1097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Saier MH, Jr, Reddy VS, Tsu BV, Ahmed MS, Li C, Moreno-Hagelsieb G. The Transporter Classification Database (TCDB): recent advances. Nucleic Acids Res. 2016 doi: 10.1093/nar/gkv1103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ••9.Saier MH, Hvorup RN, Barabote RD. Evolution of the bacterial phosphotransferase system: from carriers and enzymes to group translocators. Biochem Soc Trans. 2005;33:220–224. doi: 10.1042/BST0330220. Presents the early evidence for the evolution of the PTS. [DOI] [PubMed] [Google Scholar]
- •10.Chang AB, Lin R, Keith Studley W, Tran CV, Saier MH., Jr Phylogeny as a guide to structure and function of membrane transport proteins. Mol Membr Biol. 2004;21:171–181. doi: 10.1080/09687680410001720830. Describes an early view of integral membrane transport protein evolution. [DOI] [PubMed] [Google Scholar]
- •11.Lam VH, Lee JH, Silverio A, Chan H, Gomolplitinant KM, Povolotsky TL, Orlova E, Sun EI, Welliver CH, Saier MH., Jr Pathways of transport protein evolution: recent advances. Biol Chem. 2011;392:5–12. doi: 10.1515/BC.2011.018. Updates the Chang et al. (2004) paper on transporter evolution. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Nikaido H. Molecular basis of bacterial outer membrane permeability revisited. Microbiol Mol Biol Rev. 2003;67:593–656. doi: 10.1128/MMBR.67.4.593-656.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Brennan PJ, Nikaido H. The envelope of mycobacteria. Annu Rev Biochem. 1995;64:29–63. doi: 10.1146/annurev.bi.64.070195.000333. [DOI] [PubMed] [Google Scholar]
- 14.Remmert M, Linke D, Lupas AN, Soding J. HHomp--prediction and classification of outer membrane proteins. Nucleic Acids Res. 2009;37:W446–451. doi: 10.1093/nar/gkp325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ••15.Reddy BL, Saier MH., Jr Properties and Phylogeny of 76 Families of Bacterial and Eukaryotic Organellar Outer Membrane Pore-Forming Proteins. PLoS One. 2016;11:e0152733. doi: 10.1371/journal.pone.0152733. In this paper, families and superfamilies of outer membrane pore-forming proteins (OMPPs) are described from structural and functional standpoints, and their evolutionary origins are considered. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cavera VL, Arthur TD, Kashtanov D, Chikindas ML. Bacteriocins and their position in the next wave of conventional antibiotics. Int J Antimicrob Agents. 2015;46:494–501. doi: 10.1016/j.ijantimicag.2015.07.011. [DOI] [PubMed] [Google Scholar]
- 17.Etayash H, Azmi S, Dangeti R, Kaur K. Peptide Bacteriocins - Structure Activity Relationships. Curr Top Med Chem. 2015;16:220–241. doi: 10.2174/1568026615666150812121103. [DOI] [PubMed] [Google Scholar]
- 18.Besse A, Peduzzi J, Rebuffat S, Carre-Mlouka A. Antimicrobial peptides and proteins in the face of extremes: Lessons from archaeocins. Biochimie. 2015;118:344–355. doi: 10.1016/j.biochi.2015.06.004. [DOI] [PubMed] [Google Scholar]
- 19.Yi HY, Chowdhury M, Huang YD, Yu XQ. Insect antimicrobial peptides and their applications. Appl Microbiol Biotechnol. 2014;98:5807–5822. doi: 10.1007/s00253-014-5792-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- •20.Saier MH, Jr, Reddy BL. Holins in bacteria, eukaryotes, and archaea: multifunctional xenologues with potential biotechnological and biomedical applications. J Bacteriol. 2015;197:7–17. doi: 10.1128/JB.02046-14. The first published review summarizing the multiple cellular functions of holins considered across phylogenetic lines. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Reddy BL, Saier MH., Jr Topological and phylogenetic analyses of bacterial holin families and superfamilies. Biochim Biophys Acta. 2013;1828:2654–2671. doi: 10.1016/j.bbamem.2013.07.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Okamura Y, Fujiwara Y, Sakata S. Gating mechanisms of voltage-gated proton channels. Annu Rev Biochem. 2015;84:685–709. doi: 10.1146/annurev-biochem-060614-034307. [DOI] [PubMed] [Google Scholar]
- 23.Chanda B, Bezanilla F. A common pathway for charge transport through voltage-sensing domains. Neuron. 2008;57:345–351. doi: 10.1016/j.neuron.2008.01.015. [DOI] [PubMed] [Google Scholar]
- 24.Gonzalez W, Valdebenito B, Caballero J, Riadi G, Riedelsberger J, Martinez G, Ramirez D, Zuniga L, Sepulveda FV, Dreyer I, et al. K(2)p channels in plants and animals. Pflugers Arch. 2015;467:1091–1104. doi: 10.1007/s00424-014-1638-4. [DOI] [PubMed] [Google Scholar]
- 25.Nelson RD, Kuan G, Saier MH, Jr, Montal M. Modular assembly of voltage-gated channel proteins: a sequence analysis and phylogenetic study. J Mol Microbiol Biotechnol. 1999;1:281–287. [PubMed] [Google Scholar]
- 26.Takeshima H, Venturi E, Sitsapesan R. New and notable ion-channels in the sarcoplasmic/endoplasmic reticulum: do they support the process of intracellular Ca(2+) release? J Physiol. 2015;593:3241–3251. doi: 10.1113/jphysiol.2014.281881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Regan MC, Romero-Hernandez A, Furukawa H. A structural biology perspective on NMDA receptor pharmacology and function. Curr Opin Struct Biol. 2015;33:68–75. doi: 10.1016/j.sbi.2015.07.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- •28.Marger MD, Saier MH., Jr A major superfamily of transmembrane facilitators that catalyse uniport, symport and antiport. Trends Biochem Sci. 1993;18:13–20. doi: 10.1016/0968-0004(93)90081-w. The first description of the Major Facilitator Superfamily (MFS) [DOI] [PubMed] [Google Scholar]
- 29.Pao SS, Paulsen IT, Saier MH., Jr Major facilitator superfamily. Microbiol Mol Biol Rev. 1998;62:1–34. doi: 10.1128/mmbr.62.1.1-34.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Reddy VS, Shlykov MA, Castillo R, Sun EI, Saier MH., Jr The major facilitator superfamily (MFS) revisited. FEBS J. 2012;279:2022–2035. doi: 10.1111/j.1742-4658.2012.08588.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hvorup RN, Saier MH., Jr Sequence similarity between the channel-forming domains of voltage-gated ion channel proteins and the C-terminal domains of secondary carriers of the major facilitator superfamily. Microbiology. 2002;148:3760–3762. doi: 10.1099/00221287-148-12-3760. [DOI] [PubMed] [Google Scholar]
- 32.Keller R, Ziegler C, Schneider D. When two turn into one: evolution of membrane transporters from half modules. Biol Chem. 2014;395:1379–1388. doi: 10.1515/hsz-2014-0224. [DOI] [PubMed] [Google Scholar]
- 33.Freeman M. The rhomboid-like superfamily: molecular mechanisms and biological roles. Annu Rev Cell Dev Biol. 2014;30:235–254. doi: 10.1146/annurev-cellbio-100913-012944. [DOI] [PubMed] [Google Scholar]
- ••34.Weigel PH. Hyaluronan Synthase: The Mechanism of Initiation at the Reducing End and a Pendulum Model for Polysaccharide Translocation to the Cell Exterior. Int J Cell Biol. 2015;2015:367579. doi: 10.1155/2015/367579. Provides evidence that some glycosyl transferases function as group translocators by coupling glycosyl transfer to transmembrane polysaccharide export. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Park JH, Saier MH., Jr Phylogenetic characterization of the MIP family of transmembrane channel proteins. J Membr Biol. 1996;153:171–180. doi: 10.1007/s002329900120. [DOI] [PubMed] [Google Scholar]
- ••36.Yee DC, Shlykov MA, Vastermark A, Reddy VS, Arora S, Sun EI, Saier MH., Jr The transporter-opsin-G protein-coupled receptor (TOG) superfamily. FEBS J. 2013;280:5780–5800. doi: 10.1111/febs.12499. Provides experimental and bioinformatic evidence for the functionally diverse TOG superfamily. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Denoncin K, Collet JF. Disulfide bond formation in the bacterial periplasm: major achievements and challenges ahead. Antioxid Redox Signal. 2013;19:63–71. doi: 10.1089/ars.2012.4864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Higgins CF. ABC transporters: from microorganisms to man. Annu Rev Cell Biol. 1992;8:67–113. doi: 10.1146/annurev.cb.08.110192.000435. [DOI] [PubMed] [Google Scholar]
- 39.Khwaja M, Ma Q, Saier MH., Jr Topological analysis of integral membrane constituents of prokaryotic ABC efflux systems. Res Microbiol. 2005;156:270–277. doi: 10.1016/j.resmic.2004.07.010. [DOI] [PubMed] [Google Scholar]
- ••40.Wang B, Dukarevich M, Sun EI, Yen MR, Saier MH., Jr Membrane porters of ATP-binding cassette transport systems are polyphyletic. J Membr Biol. 2009;231:1–10. doi: 10.1007/s00232-009-9200-6. Provides bioinformatic evidence for the independent evolution of three families of integral membrane ABC transport proteins catalyzing exports of small molecules as well as macromolecules. [DOI] [PubMed] [Google Scholar]
- ••41.ter Beek J, Guskov A, Slotboom DJ. Structural diversity of ABC transporters. J Gen Physiol. 2014;143:419–435. doi: 10.1085/jgp.201411164. Presents structural evidence that ABC uptake systems are polyphyletic. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ••42.Hebbeln P, Rodionov DA, Alfandega A, Eitinger T. Biotin uptake in prokaryotes by solute transporters with an optional ATP-binding cassette-containing module. Proc Natl Acad Sci U S A. 2007;104:2909–2914. doi: 10.1073/pnas.0609905104. Provides evidence that the S-subunit of an ABC-ECF transport system can function either alone as a secondary transporter, or with other subunits as a primary active transporter. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Postma PW, Lengeler JW, Jacobson GR. Phosphoenolpyruvate:carbohydrate phosphotransferase systems of bacteria. Microbiol Rev. 1993;57:543–594. doi: 10.1128/mr.57.3.543-594.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Deutscher J, Francke C, Postma PW. How phosphotransferase system-related protein phosphorylation regulates carbohydrate metabolism in bacteria. Microbiol Mol Biol Rev. 2006;70:939–1031. doi: 10.1128/MMBR.00024-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Barabote RD, Saier MH., Jr Comparative genomic analyses of the bacterial phosphotransferase system. Microbiol Mol Biol Rev. 2005;69:608–634. doi: 10.1128/MMBR.69.4.608-634.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Vastermark A, Saier MH., Jr The involvement of transport proteins in transcriptional and metabolic regulation. Curr Opin Microbiol. 2014;18:8–15. doi: 10.1016/j.mib.2014.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Cao Y, Jin X, Levin EJ, Huang H, Zong Y, Quick M, Weng J, Pan Y, Love J, Punta M, et al. Crystal structure of a phosphorylation-coupled saccharide transporter. Nature. 2011;473:50–54. doi: 10.1038/nature09939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.McCoy JG, Levin EJ, Zhou M. Structural insight into the PTS sugar transporter EIIC. Biochim Biophys Acta. 2015;1850:577–585. doi: 10.1016/j.bbagen.2014.03.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Hvorup R, Chang AB, Saier MH., Jr Bioinformatic analyses of the bacterial L-ascorbate phosphotransferase system permease family. J Mol Microbiol Biotechnol. 2003;6:191–205. doi: 10.1159/000077250. [DOI] [PubMed] [Google Scholar]
- 50.Luo P, Yu X, Wang W, Fan S, Li X, Wang J. Crystal structure of a phosphorylation-coupled vitamin C transporter. Nat Struct Mol Biol. 2015;22:238–241. doi: 10.1038/nsmb.2975. [DOI] [PubMed] [Google Scholar]
- 51.Borday C, Chatonnet F, Thoby-Brisson M, Champagnat J, Fortin G. Neural tube patterning by Krox20 and emergence of a respiratory control. Respir Physiol Neurobiol. 2005;149:63–72. doi: 10.1016/j.resp.2005.02.014. [DOI] [PubMed] [Google Scholar]
- 52.LoVerso PR, Cui F. Cell type-specific transcriptome profiling in mammalian brains. Front Biosci (Landmark Ed) 2016;21:973–985. doi: 10.2741/4434. [DOI] [PMC free article] [PubMed] [Google Scholar]