Abstract
Cox23 is a known conserved assembly factor for cytochrome c oxidase, although its role in cytochrome c oxidase (CcO) biogenesis remains unresolved. To gain additional insights into its role, we isolated spontaneous suppressors of the respiratory growth defect in cox23∆ yeast cells. We recovered independent colonies that propagated on glycerol/lactate medium for cox23∆ cells at 37°C. We mapped these mutations to the mitochondrial genome and specifically to COX1 yielding an I101F substitution. The I101F Cox1 allele is a gain-of-function mutation enabling yeast to respire in the absence of Cox23. CcO subunit steady-state levels were restored with the I101F Cox1 suppressor mutation and oxygen consumption and CcO activity were likewise restored. Cells harboring the mitochondrial genome encoding I101F Cox1 were used to delete genes for other CcO assembly factors to test the specificity of the Cox1 mutation as a suppressor of cox23∆ cells. The Cox1 mutant allele fails to support respiratory growth in yeast lacking Cox17, Cox19, Coa1, Coa2, Cox14 or Shy1, demonstrating its specific suppressor activity for cox23∆ cells.
Keywords: cytochrome oxidase, mitochondria, COX1, COX23
INTRODUCTION
Cytochrome c oxidase (CcO) of the mitochondrial respiratory chain couples the reduction of molecular oxygen with proton translocation across the inner membrane (IM) to generate the membrane potential used to synthesize ATP. Mammalian CcO contains 14 subunits in which the 3 core subunits (Cox1-Cox3) are encoded by the mitochondrial genome 1,2. This catalytic core is surrounded by nuclear-encoded subunits, which confer enzyme stability and provide sites for the regulation of its activity 3,4. The fully assembled holoenzyme is further organized into supercomplexes with other respiratory complexes 5,6,7,8. Subunit 1 (Cox1) of CcO contains two heme a and one copper (Cu) ion as cofactors 9. One heme a functions in electron transfer, whereas the second heme a (heme a3) has an open coordinate site where O2 binds in a reaction center that also contains the CuB site. Cox2 contains a cysteine-bridged, binuclear Cu site (CuA) within a soluble globular domain that serves as the site of electron transfer from reduced cytochrome c.
The assembly of CcO requires a myriad of steps including the coordinated assembly of subunits translated on cytoplasmic and mitochondrial ribosomes and insertion of heme a and copper cofactors. Studies with yeast mutants impaired in heme a biosynthesis and CcO biogenesis have revealed that CcO assembly proceeds in a modular fashion with Cox1 maturation preceding independently of Cox2 or Cox3 maturation 10,11,12. Over 40 yeast accessory proteins have been found to be important for the assembly of CcO 13,14,15.
Hemylation and copper ion insertion are processes that occur within the intermembrane space (IMS) of mitochondria. The final step in heme a formation is catalyzed by Cox15, which has its catalytic domain projecting into the IMS. The mechanism of insertion of heme a into Cox1 is not resolved, but this process is assisted by the IM protein Shy1 16. Copper ion metallation of Cox1 and Cox2 initiates within the IMS by the Cu(I) donor protein Cox17 17. Cox17-mediated Cu(I) donation involves two accessory factors Cox11 and Sco1 that function in the metallation of the CuB site in Cox1 and CuA site in Cox2, respectively 18,19. Both Cox11 and Sco1 are inner membrane (IM)-associated proteins with Cu(I)-binding globular domains protruding into the IMS. Cox17-mediated Cu(I) transfer to Sco1 is followed by the subsequent transfer to Cox2 in a reaction dependent on a key redox role of Sco2 in metazoans 19,20. Likewise, Cox17-mediated Cu(I) transfer to Cox11 is believed to occur prior to transfer to Cox1 forming the CuB center 18,21,22.
Cox17 is part of a family of IMS proteins including CcO assembly factors Cox19, Cox23, Pet191 and Cmc1 that all possess a conserved twin Cx9C structural motif 17,23,24. Cox17 forms a helical hairpin conformation stabilized by two disulfide bonds of the twin Cx9C cysteines 25,26,27. Cox17 has 2 additional conserved Cys residues upstream of the first Cys of the twin Cx9C motif, and these vicinal thiolates bind Cu(I) in a bis-coordinate complex 27. A second of these twin Cx9C proteins Cox19 lacks the additional Cu(I) binding residues of Cox17 and was recently shown to interact with the inner membrane Cox11 protein and mediates the redox regulation of Cox11 28.
Cox23 also lacks the Cu-binding Cys residues and is not expected to bind Cu(I) in vivo. Yeast lacking Cox23 are CcO-deficient but residual levels of the enzyme persist 29. The respiratory defect in cox23∆ cells is partially suppressed by overexpression of Cox17 in cells only when cultured in 2 mM CuSO4 29. This observation led to speculation that Cox23 functions in Cu delivery to CcO during its biogenesis. This prediction is consistent with a recent study in human cells. The abundance of the human Cox23 ortholog is attenuated in fibroblasts or myoblasts isolated from patients with mutations in SCO1 or SCO2 30. Furthermore, the abundance of Cox23 was attenuated in control fibroblasts treated with a Cu chelator to deplete cellular copper 30.
In the absence of any clear functional data on Cox23, we screened for spontaneous suppressors of the respiratory defect of cox23∆ cells. In this report, we describe the isolation of a robust suppressor of the respiratory defect in cox23∆ cells that mapped to the mitochondrial-encoded Cox1 subunit.
RESULTS AND DISCUSSION
Yeast lacking Cox23 exhibit a partial growth defect on glycerol/lactate growth medium. The respiratory growth defect is more pronounced in BY4741 cells relative to the W303 genetic background 29. The growth impairment was sufficiently strong in BY4741 and BY4743 cells cultured at 37°C to permit a suppressor screen (Fig. 1C). We plated haploid and diploid cox23∆ cells (BY4741 and BY4743, respectively) at a density of ~107 cells per plate on glycerol/lactate medium at both 30 and 37°C. After 8 days a series of colonies appeared initially in the diploid null cells at 37°C relative to 30°C cultures (Fig. 1A). A series of respiratory competent colonies were collected and replated under respiratory conditions. Whereas the parent diploid cox23∆ cells failed to propagate at 37°C, the isolated colonies retained their ability to propagate at both 30 and 37°C temperatures (Fig. 1B). In serial dilution drop tests, the suppressors grew more robustly at 30°C, but growth at 37°C although apparent but reduced relative to WT cells (Fig. 1C). The addition of exogenous copper sulfate did not enhance respiratory growth (Fig. 1C), and the addition of the copper (I) chelator bathocuproine sulfonate (25 μM) impaired cox23∆ cells and the cox23∆ suppressor strain (data not shown).
Figure 1. FIGURE 1: Growth properties of cox23Δ suppressor.
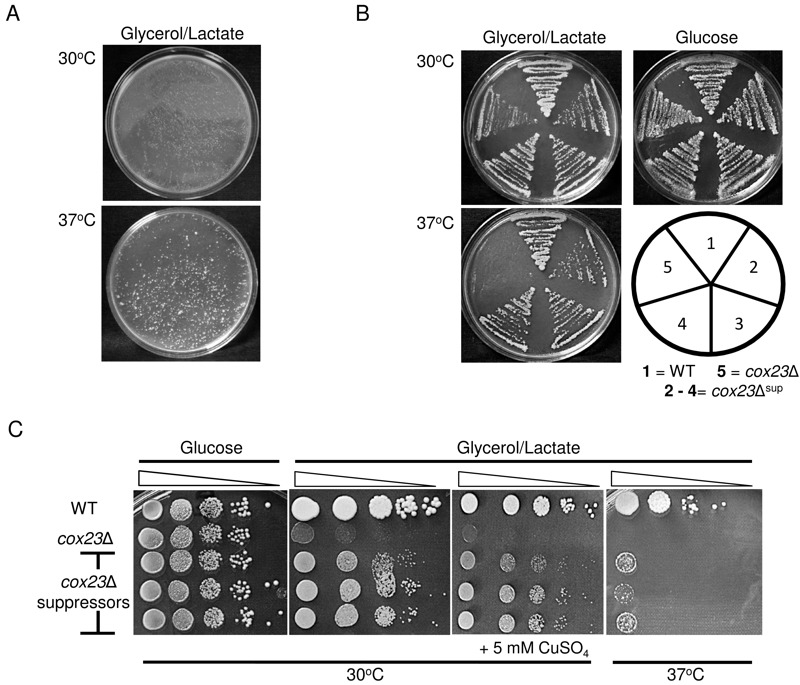
(A) Diploid cox23Δ yeast cells were plated at a density of ~107 cells/plate on YP glycerol/lactate (YPGL). The plates were photographed after 8 days of incubation at 30 and 37°C.
(B) BY4743, cox23Δ, and cox23Δ suppressor cells were streaked on YPGL or YP-glucose (YPD) and incubated at the indicated temperatures. (C) Serial dilutions of BY4743, cox23Δ, and cox23Δ suppressors spotted on YPD or YPGL plates and incubated at 30°C for 2 and 5 days and at 37°C for 7 days, respectively (left most lane is OD600 = 0.5).
Mitochondria isolated from the suppressor colonies grown in galactose medium at 30°C were used for evaluation of CcO function. As can be seen in Fig. 2A, mitochondria isolated from the suppressors showed a stabilization of Cox1, Cox2, Cox5a and Cox13 subunit levels relative to the diploid null cells. Consistent with the respiratory growth shown in Fig. 1, the restoration of CcO subunits was partial relative to WT cells. Consistent with the enhanced levels of CcO subunits, CcO enzymatic activity was elevated in the cox23∆ suppressors (Fig. 2B) and respiratory supercomplexes consisting of bc1 and CcO were increased in abundance relative to the parent cox23∆ null mutant (Fig. 2C). Cellular oxygen consumption was largely restored in the cox23∆ suppressors (Fig. 2D) and heme a/a3 levels were partially restored (Fig. 2E).
Figure 2. FIGURE 2: Functional characterization of cox23Δ suppressor.
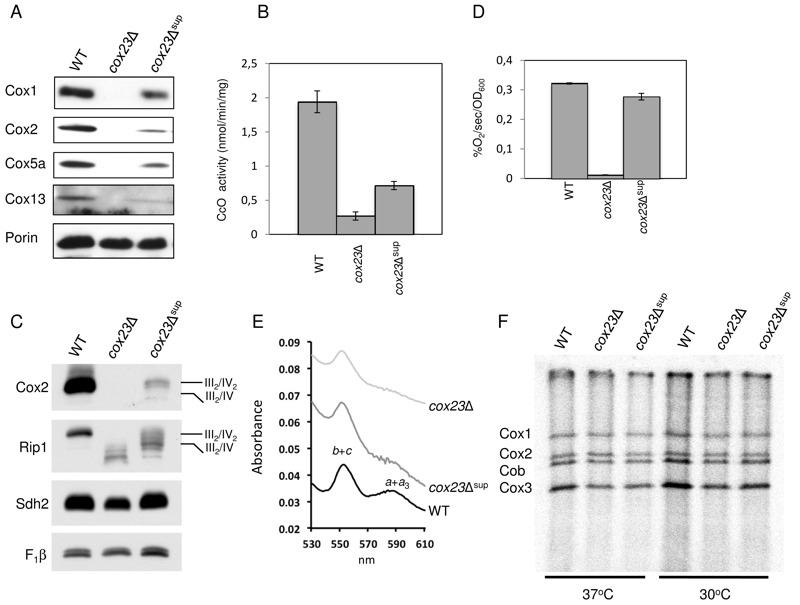
(A) Steady-state concentration of representative Complex IV (CIV) subunits. Total mitochondrial protein (20 μg) were separated on 12% SDS-PAGE, transferred to nitrocellulose and probed with CIV subunit specific antibodies and porin as loading control.
(B) Mitochondria from WT, cox23Δ, and cox23Δ suppressor cells grown in liquid YPGal were purified and assayed for CcO activity (nmol cytochrome c oxidized/min/mg protein). The data represents the average of four independent experiments (error bars indicate standard deviation).
(C) Total mitochondrial protein (30 μg) were solubilized in 1% digitonin, the mitochondrial protein complexes separated on a blue-native gel, then transferred to PVDF. The complexes were visualized using antibodies specific to subunits of the respiratory complexes.
(D) WT, cox23Δ, and cox23Δ suppressor cells were grown in liquid YP-Galactose (YPGal) at 30°C overnight and carbon-swapped to YPGL for 10 hours before oxygen consumption was measured. The data represents the average of three independent experiments (error bars indicate standard deviation). 1000 cells were plated in YPD-agar to confirm viability (data not shown).
(E) Pyridine hemochromes analysis of mitochondrial hemes. Optical absorbance spectra were recorded of reduced minus oxidized cytochromes in the shown strains.
(F) Mitochondrial proteins of wild-type (WT), cox23Δ, and cox23Δ suppressor cells were pulse-labeled with [35S]-methionine for 7.5 min at 30°C and 37°C. Total protein were extracted and separated on 15% polyacrylamide gel, then dried and exposed to x-ray film.
Mutant cells harboring the suppressor did not exhibit elevated levels of newly translated Cox1, Cox2 or Cox3 as seen in 35S-methionine labeling in a mitochondrial translation assay. Mitochondrial translation was equivalent between cox23∆ mutant cells and the suppressor strain at both 30°C and 37°C (Fig. 2F).
Since the suppressors arose from the parent diploid BY4743 cox23∆ strain, the suppressor mutation was likely either due to a dominant mutation or mitochondrial DNA mutation. To distinguish between these two scenarios, we conducted tetrad dissection of the diploid suppressors (Fig. 3A). Five tetrads shown exhibited the usual 2:2 segregation of LYS2 and MET15 markers in the diploid strain suggesting the tetrad dissection proceeded normally. However, all four spores in each suppressor were able to grow in glycerol/lactate medium. If a dominant nuclear mutation were responsible for the suppressor phenotype, then only two of the four spores would be expected to respire. Thus, we suspected the suppressor mutation was a mitochondrial DNA mutation. To test for this, we crossed a haploid cox23∆ suppressor clone to a haploid cox23∆ clone, which is derived from tetrad dissection of cox23∆ suppressor, to generate diploids. The diploids were plated on either glucose or glycerol/lactate medium. As can be seen in Fig. 3B, approximately 50% of the diploids were respiratory competent in being able to propagate on glycerol/lactate medium. This is expected if the suppressor mutation was mitochondrial, since the diploids would retain either mitochondrial genome of the starting haploids.
Figure 3. FIGURE 3:Confirmation of mtDNA origin of cox23Δ suppressor.
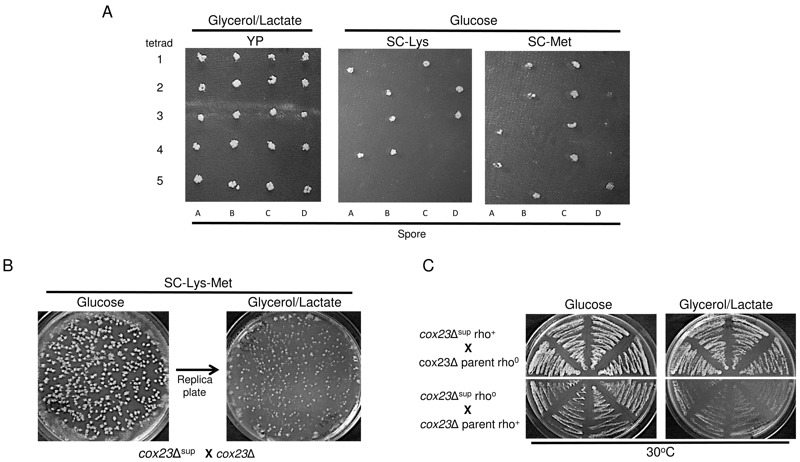
(A) BY4743 cox23Δ suppressors were grown in potassium acetate solution at room temperature to induce tetrad formation. After 4-5 days, tetrads were dissected and the spores were allowed to grow on YPD-Agar. The spores were replica plated into YPGL or SCD minus Lys or Met Agar plates to localize the suppressor DNA.
(B) cox23Δ suppressor spore was mated with cox23Δ haploid of the opposite mating type. Approximately 1000 cells were allowed to grow in SCD minus Lys and Met Agar plate. The diploid colonies were then replica plated into SCGL minus Lys and Met plate to confirm the mitochondrial location of the suppressor.
(C) cox23Δ suppressor spore and cox23Δ haploid cells were exposed to EtBr to induce rhoo status before mating with rho+ cox23Δ and cox23Δ suppressor cells of the opposite mating type, respectively. Four colonies from each mating plate were streaked on YPD and then replica plated in YPGL to confirm the cox23Δ mitochondrial suppressor.
The final proof of the mitochondrial origin of the cox23∆ suppressor was in generating diploids once again with a haploid cox23∆ suppressor clone and a haploid cox23∆ parent null clone, but this time starting with one of these two strains as a rhoo variant. The rhoo variants were generated by propagating the strain on ethidium bromide (EtBr) prior to conducting the cross. The resulting diploids were plated on glucose and glycerol/lactate. The only diploids capable of respiratory growth were those in which the mitochondrial DNA originated from the cox23∆ suppressor (Fig. 3C). The diploids obtaining mitochondrial DNA from the parental cox23∆ null failed to show growth on glycerol/lactate medium. This confirms that the mutation allowing respiratory growth of cox23∆ cells resides within the mitochondrial genome.
To identify the mitochondrial mutation, DNA sequencing was carried out on COX1, COX2 and COX3 as the most likely candidates. No mutations were identified in COX2 or COX3. In contrast, an A>T mutation was identified in codon 101 of COX1, which leads to an Ile to Phe substitution (Fig. 4A). This is a conserved residue position in Cox1 with either Ile or Met as the common residue. A Phe is found at this corresponding position in Schizosaccharomyces pombe and curiously this organism lacks Cox23 in its proteome. The position of this Met in the bovine Cox1 structure lies at the start of the third transmembrane helix and projects outward toward the interface of Cox1 and Cox3 near the matrix side of inner membrane (Fig. 4B).
Figure 4. FIGURE 4: Identification of Cox1 I101F as the cox23Δ suppressor.
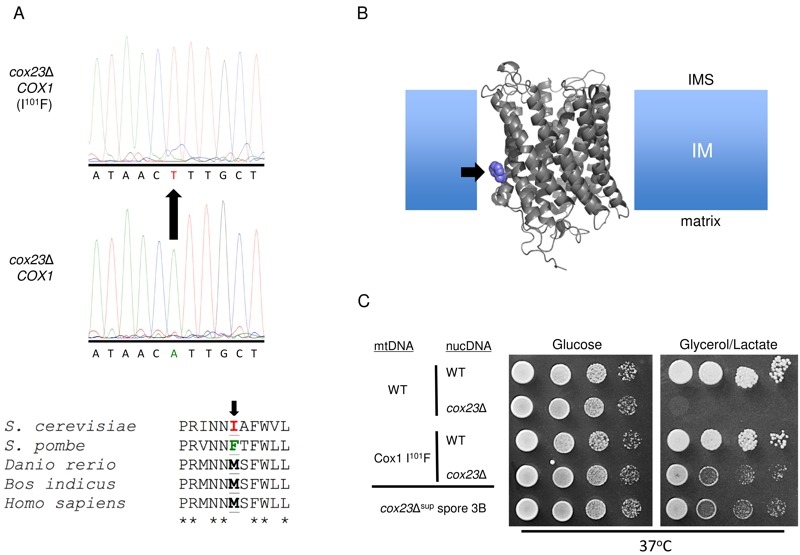
(A) Cox1 ORF cDNA was isolated by RT-PCR. Sanger sequencing led to the identification of an A → T mutation causing an Ile to Phe substitution in Cox1 at position 101 that curiously is wild-type in S. pombe (which lacks COX23 in its genome).
(B) Probable location of the Cox1 mutation in the inner mitochondrial membrane based on sequence alignment to the Bos taurus crystal structure (1OCC.pdb).
(C) Drop test on YPD and YPGL of Cox1 WT or I101F-bearing haploid cells with or without introduced cox23 deletion (rows 1-4) compared to the cox23Δ spore known to contain Cox1 I101F (bottom, row 5).
To confirm that the Ile101Phe substitution was responsible for conferring respiratory growth in cox23∆ cells, we isolated haploid cells containing the mutant mitochondrial genome encoding Cox1 I101F and subsequently deleted the COX23 locus (Fig. 4C). Cox1 I101F containing yeast were competent to propagate on glycerol/lactate medium at 37°C regardless of the presence of Cox23. The respiratory growth of the cox23∆ cells was similar to that of the starting cox23∆ suppressor cells.
We tested whether the Cox1 I101F substitution was a gain-of-function mutation specific for only cox23∆ cells. Cells with either a WT mitochondrial genome or the mutant COX1 genome were used to generate deletions in CcO biogenesis genes related to COX23. As mentioned, Cox23 is one of several soluble twin Cx9C proteins present within the IMS compartment, the other two being Cox17 and Cox19. Yeast harboring deletions in either COX17 or COX19 failed to propagate on glycerol/lactate medium regardless of whether Cox1 had the I101F substitution or not (Fig. 5A). We also tested whether Cox1 I101F will facilitate respiratory growth in other CcO assembly mutants including SHY1, COA1 and COA2. Yeast lacking Shy1, Coa1 or Coa2 are partially impaired in CcO biosynthesis and exhibit a respiratory defect (Fig. 5B). The presence of the mutant COX1 allele failed to restore respiratory growth in any of these mutants, although growth was restored by vector-encoded SHY1, COA1 or COA2 in their respective mutants (Fig. 5B, bottom panel).
Figure 5. FIGURE 5: Suppression by Cox1 I101F is restricted to cox23Δ cells.
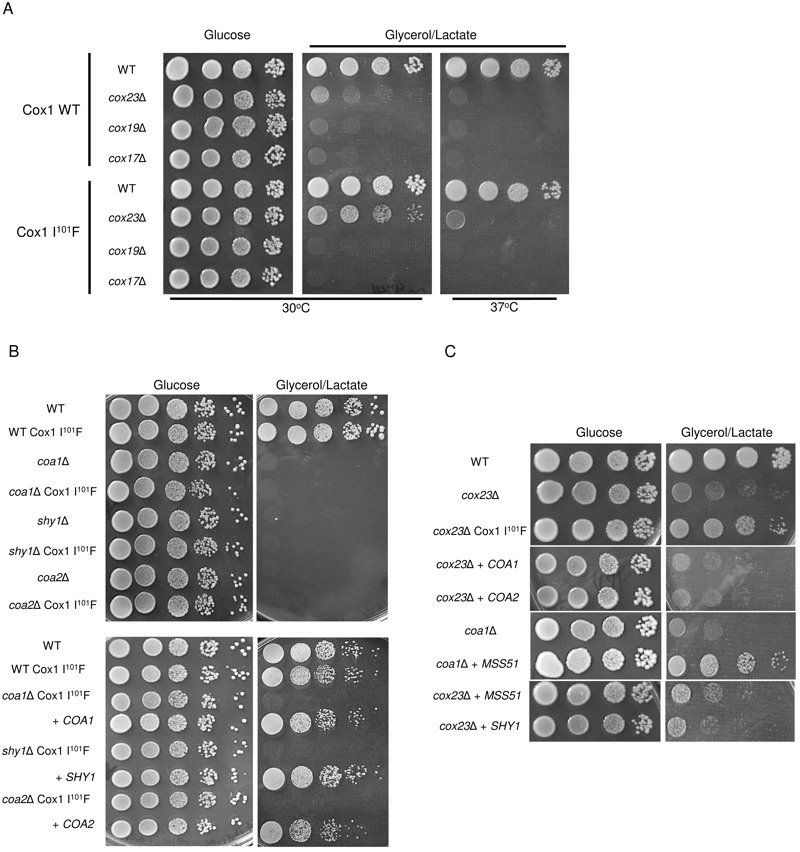
(A) cox17Δ or cox19Δ was introduced to BY haploid wild-type nuclear DNA with or without Cox1 I101F mtDNA mutation and plated on YPD and YPGL plates for 3 and 7 days at 30°C respectively.
(B) coa1Δ, coa2Δ or shy1Δ was introduced to BY haploid wild-type nuclear DNA with Cox1 I101F mtDNA mutation and plated on YPD and YPGL plates and incubated at 30°C for 3 and 7 days respectively.
(C) cox23Δ cells were transformed with MSS51, SHY1, COA1 or COA2 overexpression plasmids and plated on YPD and YPGL plates and incubated for 3 and 7 days at 30°C respectively.
Respiratory growth of certain CcO assembly mutants, e.g. shy1∆ and coa1∆ cells, can be partially restored by overexpression of the Cox1 translational activator Mss51 31. We tested whether cox23∆ cells are competent in respiratory growth upon overexpression of Mss51. Neither high levels of Mss51, Shy1, Coa1 or Coa2 were able to mediate enhanced growth, unlike the suppressor effect seen with elevated levels of Mss51 in coa1∆ cells (Fig. 5C). Thus, Cox1 I101F is a specific suppressor of cox23∆ cells.
Since S. pombe contains a Cox1 with a Phe at the corresponding sequence position as the Ile101 in yeast Cox1 and also lacks Cox14, we asked whether yeast containing the Cox1 I101F allele would be competent to respire in the absence of Cox14. To test this we introduced COX14 deletion in cox23∆ suppressor strain and put plasmids-borne COX23 back to the strain. Yeast lacking Cox14 but containing mitochondria with the Cox1 I101F allele were unable to propagate on glycerol/lactate rich medium (Fig. 6A). Overexpression of Cox23 from either a low or high copy vector failed to restore respiratory growth of cox14∆ cells containing the Cox1 Phe101 mitochondrial protein.
Figure 6. FIGURE 6: Cox1 I101F is unable to restore respiratory growth of cox14∆ cells.
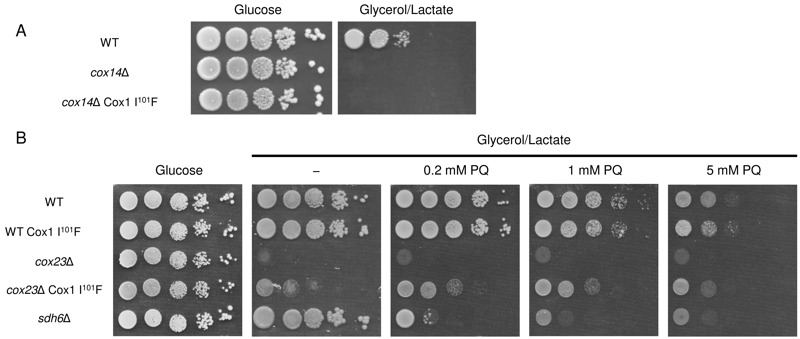
(A) COX14 was either deleted in a WT strain or in a cox23∆ suppressor strain. High-copy or low-copy plasmids containing COX23 were transformed to cox14∆ cox23∆ deletion strain to make the strain a cox14∆ containing COX1 suppressor mutation. The resulting cells were tested for respiratory growth.
(B) cox23∆ mutant cells with and without the Cox1 I101F allele were plated on glycerol/lactate medium containing increasing concentrations of paraquat (0 - 5 mM) at 30°C.
Since we recovered the cox23∆ suppressor under respiratory condition at 37°C in which more reactive oxygen species are generated, we tested how the suppressor behave under the reactive oxidative stress condition. Yeast harboring the Cox1 I101F allele did not exhibit any enhanced sensitivity or resistance to reactive oxygen stress induced by culturing the cells in increasing levels of paraquat (Fig. 6B), whereas the presence of Cox1 I101F in cox23∆ cells led to a slight resistance toward paraquat. This contrasts with the known paraquat sensitivity of sdh6∆ cells 32.
In summary, we recovered independent colonies that propagated on glycerol/lactate medium for cox23∆ cells. We mapped these mutations to the mitochondrial genome and specifically to COX1 yielding an I101F substitution. The I101F Cox1 allele is a gain-of-function mutation enabling yeast to respire in the absence of Cox23. CcO steady-state levels were restored with the I101F Cox1 suppressor mutation and oxygen consumption and CcO activity were likewise restored. The allele fails to support respiratory growth in yeast lacking Cox17, Cox19 Coa1, Coa2, Cox14 or Shy1, demonstrating its specific suppressor activity for cox23∆ cells.
Most species have an Ile, Leu or Met at the corresponding Ile101 sequence position. In contrast, S. pombe has a Phe at this position in Cox1. The gain-of-function I101F Cox1 allele in the S. cerevisiae cox23Δ deletion mutant may provide an explanation for the lack of Cox23 in S. pombe. Ile101 is situated at the start of TM3 in Cox1 near the matrix side of the IM. This residue in the bovine CcO structure projects outward packing against the first TM helix in Cox3 33 (Fig. 4B).
Despite the present studies, the function of Cox23 in CcO biogenesis remains unresolved. This work reveals a potential role in Cox1 maturation. Recently, the Cox23 homolog Cox19 was shown to shield the CuB metallochaperone Cox11 from oxidation on a membrane proximal cysteinyl residue 28. If Cox23 has a related function, the redox state of the remaining two Cu(I)-binding cysteinyl residues in Cox11 may be dependent on Cox23. Cox19 and Cox23 may have non-redundant roles with Cox11 in forming the Cox1 CuB site in many species, but the I101F Cox1 allele may permit Cox19 to perform both functions in S. pombe and the present S. cerevisiae mutant. Alternatively, Cox23 may have a novel role in the hemylation of Cox1 in an unresolved heme a transfer step. Further research is needed to resolve these scenarios.
MATERIALS AND METHODS
Yeast Strains and Vectors
The Saccharomyces cerevisiae yeast strains used in this study were from a yeast knockout collection (Invitrogen). The COA1, COA2, SHY1, and MSS51 ORFs were cloned into plasmid pRS413 and pRS416 under control of the MET25 promoter and CYC1 terminator. Yeast strains were transformed using lithium acetate. Yeast cells were cultured either in rich medium (YP) or synthetic complete (SC) medium lacking the appropriate nutrients for plasmid selection. Final concentration of carbon sources used (glucose, galactose, glycerol, lactate) in liquid media and agar plates is 2% except for raffinose (0.2%). Spores from diploid transformants were recovered after sporulation in 0.3% potassium acetate for 5 days at room temperature (RT). Rhoo cells were obtained after overnight incubation of yeast cells in YP-Glucose (YPD) with ethidium bromide (EtBr) at RT.
Table 1.
Genotype and sources of yeast strains.
| Strain | Genotype | Source |
| BY4743 | MATa/α his3Δ1/his3Δ1 leu2Δ0/leu2Δ0 LYS2/lys2Δ0 MET15/met15Δ0 ura3Δ0/ura3Δ0 | Invitrogen |
| BY4743 cox23Δ | MATa/α his3Δ1/his3Δ1 leu2Δ0/leu2Δ0 LYS2/lys2Δ0 MET15/met15Δ0 ura3Δ0/ura3Δ0 Δcox23::kanMX4 | Invitrogen |
| BY4741 | MATa his3Δ1 leu2Δ0 met15Δ0 ura3Δ0 | Invitrogen |
| BY4741 cox23Δ | MATa his3Δ1 leu2Δ0 met15Δ0 ura3Δ0 Δcox23::kanMX4 | Invitrogen |
| BY4741 cox19Δ | MATa his3Δ1 leu2Δ0 met15Δ0 ura3Δ0 Δcox19::kanMX4 | Invitrogen |
| BY4741 cox17Δ | MATa his3Δ1 leu2Δ0 met15Δ0 ura3Δ0 Δcox17::kanMX4 | Invitrogen |
| BY4741 coa1Δ | MATa his3Δ1 leu2Δ0 met15Δ0 ura3Δ0 Δcoa1::kanMX4 | Invitrogen |
| BY4741 coa2Δ | MATa his3Δ1 leu2Δ0 met15Δ0 ura3Δ0 Δcoa2::kanMX4 | Invitrogen |
| BY4741 shy1Δ | MATa his3Δ1 leu2Δ0 met15Δ0 ura3Δ0 Δshy1::kanMX4 | Invitrogen |
Mitochondrial Purification
Intact mitochondria were isolated from yeast as described previously 34. Total mitochondrial protein concentration was determined using Coomassie Plus Protein Assay Reagent (ThermoScientific).
Blue Native PAGE
Blue Native PAGE (BN-PAGE) was performed as previously described 35. Briefly, 20 to 30 μg isolated mitochondria was solubilized in sample buffer (1% digitonin, 0.5 M 6-aminocaproic acid, pH 7.0), incubated in ice for 20 min and then centrifuged (20,000 x g for 10 min at 4°C). Supernatants were mixed with 0.5 μl 5% Coomassie brilliant blue G250 and loaded on a NativePAGE NovexTM 3-12% gradient polyacrylamide gel (Invitrogen) alongside a high-mass protein marker (GE Healthcare).
Immunoblotting
BN-PAGE mass-resolved complexes were detected after transfer to a polyvinylidene difluoride (PVDF) membrane. Alternatively, mitochondrial proteins were detected after separation of 10 to 30 μg solubilized and reduced mitochondria on 12% SDS-PAGE gel and transfer to nitrocellulose. Proteins were visualized using Supersignal (ThermoScientific) to detect horseradish peroxidase-conjugated secondary antibodies. Primary antibodies used were either purchased or generous gifts: anti-Cox1 and anti-Cox2 (Mitoscience), anti-Porin (Molecular Probes), Anti-Sdh2 (21st Century Biochemicals), anti-F1 ATPase (Dr. A. Tzagoloff), anti-Cyt1 and anti-Cox5a (Dr. B. Meunier), and anti-Cox13 (Dr. P. Rehling).
Miscellaneous Assays
CcO activity in isolated mitochondria were determined spectrophotometrically by supplying reduced cytochrome c and following the initial rate of cytochrome c oxidation at 550 nm using an Agilent 8453 spectrophotometer. Reduced cytochrome c was prepared by adding equimolar amount of sodium hydrosulfite (Aldrich) to horse heart cytochrome c (Sigma) and desalting using a PD-10 gravity flow colum (GE Healthcare). The rate of oxygen consumption of cells grown in YP-Galactose (YPGal) media then carbon-swapped to YP-Glycerol/Lactate (YPGL) was determined from the linear response on a 5300A biological oxygen monitor (Yellow Springs Instruments Co.). Optical absorption spectroscopy was used to monitor mitochondrial heme pools. Two mg of purified mitochondria was suspended in 250 μl of distilled water. Same volume of a stock solution (200 mM NaOH, 40% pyridine) and 1.5 μl of 0.1 M K3Fe(CN)6 were added to the mitochondrial suspension. Each spectrum represents the calculated difference spectrum of the reduced (dithionite) minus oxidized (ferricyanide) cytochromes and was recorded by an Agilent 8453 spectrophotometer. Absorption maxima at 550 and 558 nm correspond to cytochromes b/c and a/a3, respectively.
For in vivo mitochondrial translation assay, cells were grown overnight in YPGal media to an OD600 of 1. [35S]-methionine labeling and sample preparation for 15% SDS-PAGE was performed as previously described 36. Gels were dried and radiolabelled mitochondrial proteins were visualized by overnight film autoradiography. Growth tests to determine the respiratory competency of yeast strains were performed on agar plates containing 2% glucose or 2% glycerol-2% lactate. Yeast cells were grown overnight in YPD medium and adjusted to an optical density at 600 nm of 0.5. Serial dilutions were spotted onto plates and incubated at 30°C for 2 days (glucose plates) or 4 to 8 days (glycerol-lactate plates). Bathocuproine sulfonate (BCS) and bathophenanthroline sulfonate (BPS) were purchased from Sigma.
Funding Statement
This work was supported by grant ES03817 from the National Institutes of Environmental Health Sciences NIH to D.R.W. R.D.C. was supported by training grant T32 DK007115 from the National Institutes of Health.
References
- 1.Yoshikawa S, Shinzawa-Itoh K, Tsukihara T. X-ray structure and the reaction mechanism of bovine heart cytochrome c oxidase. J Inorg Biochem. 2000;82(1-4):1–7. doi: 10.1016/s0162-0134(00)00137-9. [DOI] [PubMed] [Google Scholar]
- 2.Balsa E, Marco R, Perales-Clemente E, Szklarczyk R, Calvo E, Landazuri MO, Enriquez JA. NDUFA4 is a subunit of complex IV of the mammalian electron transport chain. Cell Metab. 2012;16(3):378–86. doi: 10.1016/j.cmet.2012.07.015. [DOI] [PubMed] [Google Scholar]
- 3.Fontanesi F, Soto IC, Barrientos A. Cytochrome c oxidase biogenesis: new levels of regulation. IUBMB Life. 2008;60(9):557–68. doi: 10.1002/iub.86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Fontanesi F, Soto IC, Horn D, Barrientos A. Assembly of mitochondrial cytochrome c-oxidase, a complicated and highly regulated cellular process. Am J Physiol Cell Physiol. 2006;291(6):C1129–C147. doi: 10.1152/ajpcell.00233.2006. [DOI] [PubMed] [Google Scholar]
- 5.Cruciat CM, Brunner S, Baumann F, Neupert W, Stuart RA. The cytochrome bc1 and cytochrome c oxidase complexes associate to form a single supracomplex in yeast mitochondria. J Biol Chem. 2000;275(24):18093–1808. doi: 10.1074/jbc.M001901200. [DOI] [PubMed] [Google Scholar]
- 6.Schagger H, Pfeiffer K. Supercomplexes in the respiratory chains of yeast and mammalian mitochondria. EMBO J. 2000;19(8):1777–183. doi: 10.1093/emboj/19.8.1777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Schagger H. Respiratory chain supercomplexes. IUBMB Life. 2001;52(3-5):119–28. doi: 10.1080/15216540152845911. [DOI] [PubMed] [Google Scholar]
- 8.Eubel H, Heinemeyer J, Sunderhaus S, Braun HP. Respiratory chain supercomplexes in plant mitochondria. Plant Physiol Biochem. 2004;42(12):937–42. doi: 10.1016/j.plaphy.2004.09.010. [DOI] [PubMed] [Google Scholar]
- 9.Tsukihara T, Aoyama H, Yamashita E, Tomizaki T, Yamaguchi H, Shinzawa-Itoh K, Hakashima R, Yaono R, Yoshikawa S. Structures of metal sites of oxidized bovine heart cytochrome c oxidase at 2. A. Science. 1995;269(5227):1069–174. doi: 10.1126/science.7652554. [DOI] [PubMed] [Google Scholar]
- 10.McStay GP, Su CH, Thomas SM, Xu JT, Tzagoloff A. Characterization of assembly intermediates containing subunit 1 of yeast cytochrome oxidase. J Biol Chem . 2013;288(37):26546–56. doi: 10.1074/jbc.M113.498592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.McStay GP, Su CH, Tzagoloff A. Modular assembly of yeast cytochrome oxidase. Mol Biol Cell . 2012;24(4):440–52. doi: 10.1091/mbc.E12-10-0749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Su CH, McStay GP, Tzagoloff A. The Cox3p assembly module of yeast cytochrome oxidase. Mol Biol Cell . 2014;25(7):965–76. doi: 10.1091/mbc.E13-10-0575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Tzagoloff A, Dieckmann CL. PET Genes of Saccharomyces cerevisiae. Microbiological Rev . 1990;54(3):211–25. doi: 10.1128/mr.54.3.211-225.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Barrientos A, Barros MH, Valnot I, Rotig A, Rustin P, Tzagoloff A. Cytochrome oxidase in health and disease. Gene . 2002;286(1):53–63. doi: 10.1016/S0378-1119(01)00803-4. [DOI] [PubMed] [Google Scholar]
- 15.Soto IC, Fontanesi F, Liu JI, Barrientos A. Biogenesis and assembly of eukaryotic cytochrome c oxidase catalytic core. Biochim Biophys Acta . 2012;1817(6):883–97. doi: 10.1016/j.bbabio.2011.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Khalimonchuk O, Bestwick M, Meunier B, Watts TC, Winge DR. Formation of the redox cofactor centers during Cox1 maturation in yeast cytochrome oxidase. Mol Cell Biol . 2010;30(4):1004–17. doi: 10.1128/MCB.00640-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Glerum DM, Shtanko A, Tzagoloff A. Characterization of COX17, a Yeast Gene Involved in Copper Metabolism and Assembly of Cytochrome Oxidase. Mol Cell Biol . 1996;271(24):14504–9. doi: 10.1074/jbc.271.24.14504. [DOI] [PubMed] [Google Scholar]
- 18.Horng YC, Cobine PA, Maxfield AB, Carr HS, Winge DR. Specific copper transfer from the Cox17 metallochaperone to both Sco1 and Cox11 in the assembly of yeast cytochrome c oxidase. J Biol Chem . 2004;279(34):35334–40. doi: 10.1074/jbc.M404747200. [DOI] [PubMed] [Google Scholar]
- 19.Banci L, Bertini I, Ciofi-Baffoni S, Hadjiloi T, Martinelli M, Palumaa P. Mitochondrial copper(I) transfer from Cox17 to Sco1 is coupled to electron transfer. Proc Natl Acad Sci U S A . 2008;105(19):6803–8. doi: 10.1073/pnas.0800019105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Leary SC, Sasarman F, Nishimura T, Shoubridge EA. Human SCO2 is required for the synthesis of CO II and as a thiol-disulphide oxidoreductase for SCO1. Hum Mol Genet . 2009;18(12):2230–40. doi: 10.1093/hmg/ddp158. [DOI] [PubMed] [Google Scholar]
- 21.Carr HS, George GN, Winge DR. Yeast Cox11, a protein essential for cytochrome c oxidase assembly, is a Cu(I) binding protein. J Biol Chem . 2002;277(34):31237–42. doi: 10.1074/jbc.M204854200. [DOI] [PubMed] [Google Scholar]
- 22.Banci L, Bertini I, Cantini F, Ciofi-Baffoni S, Gonnelli L, Mangani S. Solution structure of Cox11: A novel type of beta -immunoglobulin-like fold involved in CuB site formation of cytochrome c oxidase. J Biol Chem . 2004;279(33):34833–9. doi: 10.1074/jbc.M403655200. [DOI] [PubMed] [Google Scholar]
- 23.Khalimonchuk O, Winge DR. Function and redox state of mitochondrial localized cysteine-rich proteins important in the assembly of cytochrome c oxidase. Biochim Biophys Acta . 2007;1783(4):618–28. doi: 10.1016/j.bbamcr.2007.10.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Longen S, Bien M, Bihlmaier K, Kloeppel C, Kauff F, Hammermeister M, Westermann B, Hermmann JM, Riemer J. Systematic analysis of the twin cx(9)c protein family. J Mol Biol . 2009;393(2):356–68. doi: 10.1016/j.jmb.2009.08.041. [DOI] [PubMed] [Google Scholar]
- 25.Abajian C, Yatsunyk LA, Ramirez BE, Rosenzweig AC. Yeast Cox17 Solution Structure and Copper(I) Binding. J Biol Chem . 2004;279(51):53584–92. doi: 10.1074/jbc.M408099200. [DOI] [PubMed] [Google Scholar]
- 26.Arnesano F, Balatri E, Banci L, Bertini I, Winge DR. Folding studies of Cox17 reveal an important interplay of cysteine oxidase and copper binding. Structure . 2005;13(5):713–22. doi: 10.1016/j.str.2005.02.015. [DOI] [PubMed] [Google Scholar]
- 27.Banci L, Bertini I, Ciofi-Baffoni S, Janicka A, Martinelli M, Kozlowski H, Palumaa P. A structural-dynamical characterization of human Cox17. J Biol Chem . 2008;283(12):7912–20. doi: 10.1074/jbc.M708016200. [DOI] [PubMed] [Google Scholar]
- 28.Bode M, Woellhaf MW, Bohnert M, van der Laan M, Sommer F, Jung M, Zimmermann R, Schroda M, Hermann JM. Redox-regulated dynamic interplay between Cox19 and the copper-binding protein Cox11 in the intermembrane space of mitochondria facilitates biogenesis of cytochrome c oxidase. Mol Biol Cell . 2015;26(13):2385–401. doi: 10.1091/mbc.E14-11-1526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Barros MH, Johnson A, Tzagoloff A. Cox23, a homologue of COX17, is required for cytochrome oxidase assembly. J Biol Chem . 2004;279(30):31943–7. doi: 10.1074/jbc.M405014200. [DOI] [PubMed] [Google Scholar]
- 30.Leary SC, Cobine PA, Nishimura T, Verdijk RM, de Krijger R, de Coo R, Tarnopolsky MA, Winge DR, Shoubridge EA. COX19 mediates the transduction of a mitochondrial redox signal from SCO1 that regulates ATP7A-mediated cellular copper efflux. Mol Biol Cell . 2013;24(6):683–91. doi: 10.1091/mbc.E12-09-0705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Pierrel F, Bestwick M, Cobine P, Khalimonchuck O, Cricco J, Winge D. Coa1 links the Mss51 post-translational function to Cox1 cofactor insertion in cytochrome c oxidase assembly. EMBO J . 2007;26(20):4335–46. doi: 10.1038/sj.emboj.7601861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Na U, Yu W, Cox J, Bricker DK, Brockmann K, Rutter J, Thummel CS, Winge DR. The LYR factors SDHAF1 and SDHAF3 mediate maturation of the iron-sulfur subunit of succinate dehydrogenase. Cell Metab . 2014;20(2):253–66. doi: 10.1016/j.cmet.2014.05.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Tsukihara T, Aoyama H, Yamashita E, Tomizaki T, Yamaguichi H, Shinzawa-Itoh K, Nakashima R, Yaono R, Yoshikawa S. The whole structure of the 13-subunit oxidized cytochrome c oxidase at 2.8 A. Science . 1996;272(5265):1136–44. doi: 10.1126/science.272.5265.1136. [DOI] [PubMed] [Google Scholar]
- 34.Diekert K, De Kroon AI, Kispal G, Lill R. Isolation and subfractionation of mitochondria from the yeast Saccharomyces cerevisiae. Meth Cell Biol . 2001;65:37–51. doi: 10.1016/s0091-679x(01)65003-9. [DOI] [PubMed] [Google Scholar]
- 35.Wittig I, Braun HP, Schagger H. Blue native PAGE. Nat Protoc . 2006;1(1):418–28. doi: 10.1038/nprot.2006.62. [DOI] [PubMed] [Google Scholar]
- 36.Barrientos A, Korr D, Tzagoloff A. Shy1p is necessary for full expression of mitochondrial COX1 in the yeast model of Leigh's syndrome. EMBO J . 2002;21(1-2):43–52. doi: 10.1093/emboj/21.1.43. [DOI] [PMC free article] [PubMed] [Google Scholar]


