Abstract
We analyzed the effects of single-nucleotide polymorphisms (SNPs) on laryngeal carcinoma (LC) risk and overall survival (OS) in 170 Chinese male LC patients followed for 10 years. After assessment of clinical characteristics (age, laryngectomy, neck dissection, tumor differentiation, TNM status), the patients were genotyped for 24 SNPs associated with risk in multiple cancers. LC risk was assessed using log-rank test and Cox proportional hazard models. The median OS time was 48 months. By the follow-up deadline, OS was 41.2%. Kaplan-Meier analysis indicated 1-, 3-, and 5-year survival rates to be 84.7%, 57.2%, and 47.1%, respectively. Five LC clinicopathological characteristics, namely total laryngectomy (TL), low differentiation (LD), T3-T4, N1-N2, and clinical stage III-IV were associated with worse OS (HR: 2.35, p < 0.001; HR: 2.39, p = 0.02; HR: 2.17, p < 0.001; HR: 2.39, p < 0.001; and HR: 3.29, p < 0.001, respectively). Univariate cox regression analysis indicated that four SNPs were associated (p < 0.05) with LC OS in the codominant genetic model compared to patients with the homozygous wild-type genotype: rs10088262 G/A (HR = 1.57), rs1665650 A/G (HR = 0.65); rs3802842 C/C (HR = 2.18), and rs59336 T/A and T/T (HR = 0.61 and 2.61, respectively).
Keywords: laryngeal carcinoma, single-nucleotide polymorphism, overall survival, hazard ratio
INTRODUCTION
Laryngeal carcinoma (LC) is a common tumor of the head and neck. It accounts for 1% - 5% of all malignant tumors, comprises 3.3% - 8.1% of all the head and neck malignant tumors, and among these its incidence is only lower than that of nasopharyngeal carcinoma [1]. According to GLOBOCAN 2012 [2], 156,877 individuals were diagnosed with LC worldwide, while 20,014 cases were diagnosed in China. In this country, the age-adjusted incidence was 1.1 per 100,000, and the age-adjusted mortality rate was 0.7 per 100,000 for both sexes. LC generally affects people aged 50-70 years, especially males. In China, the ratio of men to women diagnosed with LC was 10.5:1 [2, 3]. In recent years, the incidence of LC has increased steadily because of multiple carcinogenic factors [4, 5], and in China is predicted to reach 55,900 new cases/year in the next 5 years. As with other cancers, the pathogenesis of laryngeal carcinoma involves the combined effects of environmental and genetic factors. To date, several genes have been implicated in the occurrence of LC and have been shown to affect its prognosis [6–8]. Recent studies have shown that single genes may be associated with many related cancers [9]. For example, the murine double minute 2 (MDM2) gene has been proposed to contribute to the emergence and development of many tumors, especially digestive carcinomas [10] and LC [1].
The association of single nucleotide polymorphisms (SNP), the most common form of genetic variation, with multiple cancers, has also been highlighted [11, 12]. While extensive studies have evaluated the relevance of clinicopathological parameters such as surgical treatment modalities, age, tumor stage, differentiation, and lymph node metastasis, as prognostic factors in head and neck cancers [13], only a few basic studies have revealed an essential role of specific genes in digestive tract cancers. Moreover, the association between SNPs within or near these genes and LC prognosis has not been fully investigated. In the present study, we analyzed the association of 24 SNPs related to digestive tract cancers with LC prognosis in 170 Han Chinese male patients, and identified four SNPs significantly associated with OS. These results shed light on the genetic component of LC and may prove useful to guide further studies addressing its pathogenesis.
RESULTS
Patient, tumor, and treatment characteristics
170 LC male patients with a mean age of 60.7 years (range, 32 to 82 years) were enrolled in this study; none of them presented distant metastases (i.e they were M0). According to primary tumor and lymph node stage, 40 cases (23.5%) were T1, 62 cases (36.5%) were T2, 50 cases (29.4%) were T3, and 18 cases (10.6%) were T4, while 116 cases (68.2%) were N0, 30 cases (17.6%) were N1, and 24 cases (16.1%) were N2. Follow-up time, survival status, tumor stage and age distribution are shown in Table 1. 37 patients underwent neck dissection and 133 patients had non-neck dissection. Surgical procedures are listed in Table 2.
Table 1. Clinicopathological characteristics of the patients included in this study.
| Patient Characteristics | No. | % |
|---|---|---|
| Total | 170(male, M0 c) | 100 |
| Min follow-up time (month) | 3 | - |
| Max follow-up time (month) | 122 | - |
| Median follow-up time (month) | 38 | - |
| Survival status | 70 survivors | 41.2 |
| 100 dead | 58.8 | |
| Mean Age | 60.75 | |
| Range | 32-82 | |
| <60 | 80 | 47.1 |
| ≥60 | 90 | 52.9 |
| Tumor Stage a | ||
| T1 | 40 | 23.5 |
| T2 | 62 | 36.5 |
| T3 | 50 | 29.4 |
| T4 | 18 | 10.6 |
| N0 | 116 | 68.2 |
| N1 | 30 | 17.6 |
| N2 | 24 | 14.1 |
| Clinical Stage b | ||
| I | 37 | 21.8 |
| II | 36 | 21.2 |
| III | 61 | 35.9 |
| IV | 36 | 21.2 |
| Differentiation degree | ||
| Low | 14 | 8.2 |
| Moderate | 125 | 73.5 |
| High | 31 | 18.2 |
| Merged Surgical Procedures | ||
| Neck Dissection | 37 | 21.8 |
| Non-Neck Dissection | 133 | 78.2 |
a: T-stage: tumor stage; N-stage: lymph node stage
b: Clinical stage reference: international unifying new TNM classification from Union for International Cancer Control
c: M0: No distant metastasis;
Table 2. Univariate Cox proportional hazards analysis of potential factors affecting survival.
| Variable | Wald | HR (95% CI) | p a | Details | Median | 1-, 3-, 5-year survival rate | Overall comparison | |||
|---|---|---|---|---|---|---|---|---|---|---|
| Estimate | 95%CI | Chi-Square | df | p b for log-rank | ||||||
| Total | 48.00 | 29.26-66.74 | 0.929, 0.788, 0.671 | |||||||
| Age | 0.55 | 1.16 (0.78-1.72) | 0.460 | <60 | 59.00 | 30.05-87.95 | 0.85, 0.584, 0.414 | |||
| ≥60 | 48.00 | 29.86-66.14 | 0.867, 0.633, 0.507 | 0.556 | 1 | 0.456 | ||||
| Laryngectomy | 17.64 | 2.35 (1.58-3.49) | <0.001* | PL | 73.00 | 50.84-95.16 | 0.885, 0.73, 0.632 | |||
| TL | 30.00 | 21.32-38.68 | 0.818, 0.468, 0.27 | 18.96 | 1 | <0.001* | ||||
| Neck dissection | 1.69 | 0.71 (0.43-1.19) | 0.194 | ND | 36.00 | - | 0.676, -, - | |||
| NND | 56.00 | 36.32-75.69 | 0.91, 0.752, 0.583 | 1.73 | 1 | 0.188 | ||||
| Differentiation | 8.99 | 0.011* | HD | 71.00 | 15.55-126.45 | 0.676, -, - | ||||
| 0.07 | 0.93 (0.57-1.57) | 0.794 | MD | 59.00 | 39.14-78.86 | 0.912, 0.712, 0.602 | ||||
| 5.44 | 2.39 (1.15-4.99) | 0.02* | LD | 15.00 | 0.00-33.33 | 0.214, -, - | 9.78 | 2 | 0.008* | |
| T | 14.07 | 2.17 (1.45-3.25) | <0.001* | T1-T2 | 77.00 | 56.18-97.82 | 0.882, 0.695, 0.616 | |||
| T3-T4 | 32.00 | 22.25-41.75 | 0.824, 0.483, 0.243 | 14.96 | 1 | <0.001* | ||||
| N | 17.04 | 2.39 (1.58-3.62) | <0.001* | N0 | 71.00 | 48.28-93.73 | 0.897, 0.706, 0.6 | |||
| N1-N2 | 26.00 | 14.08-37.92 | 0.796, 0.403, - | 18.33 | 1 | <0.001* | ||||
| Clinical stage | 26.86 | 3.29 (2.10-5.18) | <0.001* | I-II | 98.00 | 67.49-128.51 | 0.849, 0.732, 0.527 | |||
| III, IV | 32.00 | 24.2-39.8 | 0.897, 0.649, 0.418 | 30.15 | 1 | <0.001* | ||||
a. p for Wald test < 0.05 indicates statistical significance for individual coefficients
b. p for Log-rank Test < 0.05 indicates statistical significance for grouping variables
Abbreviations: PL: Partial laryngectomy; TL: Total laryngectomy; ND: Neck dissection; NND: Non-neck dissection; HD: High differentiation; MD: Moderate differentiation; LD: Low differentiation; df: degrees of freedom. *: indicates statistical significance
Overall survival analysis
At the median follow-up period of 38 months (range, 3 to 122 months), the mean and median survival times were 62.13 and 48 months, respectively. By the follow-up time deadline, OS was 41.2% (100 dead and 70 survivors). Kaplan-Meier statistical analysis indicated that 1-, 3-, and 5-year survival rates were 84.7%, 57.2% and 47.1%, respectively [14].
Analysis of clinical characteristics
We performed univariate Cox proportional hazards analysis to evaluate the association of age, laryngectomy, neck dissection, differentiation, T stage, N stage, and clinical stage with LC survival rates. Significant correlations were found for laryngectomy (HR: 2.35, 95% CI: 1.58-3.49, Wald-p < 0.001); tumor differentiation (HR: 2.39, 95%CI: 1.15-4.99, Wald-p = 0.02); T stage (HR: 2.17, 95% CI: 1.45-3.25, Wald-p < 0.001); N stage (HR: 2.39, 95% CI: 1.58-3.62, Wald p < 0.001); and clinical stage (HR: 3.29, 95% CI: 2.10-5.18, Wald-p < 0.001). The log-rank test further validated the significance of these five variables (Table 2). Total laryngectomy (TL) median survival time (30 months; 95% CI: 21.32-38.68) was significantly shorter than that of partial laryngectomy (PL; 73 months; 95% CI: 50.84-95.16; log-rank p <0.001) [14]. With respect to tumor differentiation status, a significant difference was detected between low differentiation (LD) and high differentiation (HD) groups (log-rank p = 0.008): median survival time was 71 months (95% CI: 15.55-126.45) for HD, 59 months (95% CI: 39.14-78.86) for moderate differentiation (MD), and 15 months (95% CI: 0.00-33.33) for LD [14]. Stratification based on primary tumor staging showed a significantly longer median survival time of 77 months (95% CI: 56.18-97.82) for T1-T2, compared to 32 months (95% CI: 22.25-41.75) for T3-T4 (log-rank p < 0.001) [14]. Lymph node staging analyses also showed a significant difference in the median survival time of N0 (71 months; 95% CI: 48.28-93.73) versus N1-N2 (26 months; 95% CI: 14.08-37.92; log-rank p < 0.001) [14]. Additionally, clinical stage (TNM status) subgroup analysis revealed a significant longer median survival time for stage I-II (98 months; 95% CI: 67.49-128.51) compared to III-IV (32 months; 95% CI: 24.2-39.8; log-rank p < 0.001) [14].
SNP analysis
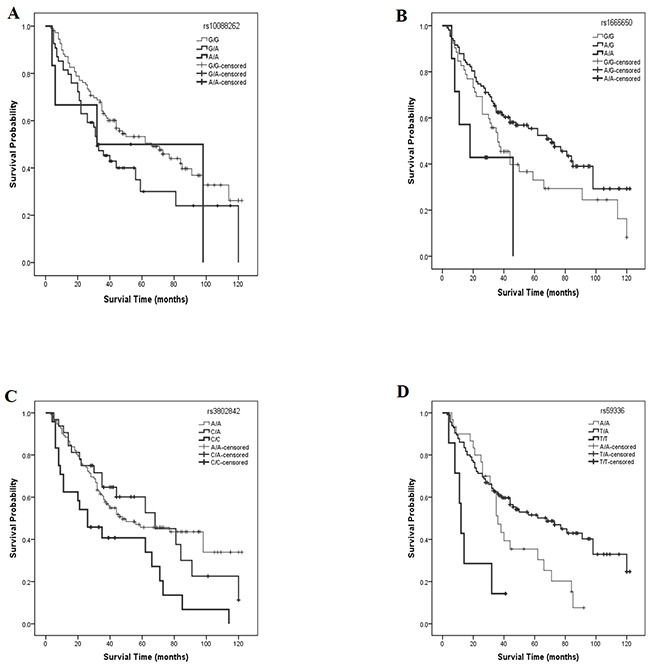
In SNP univariate analyses, the lower frequency allele was coded as the ‘risk’ allele. All SNP genotypes were coded as 0, 1, or 2, to represent the number of risk alleles they possessed for that SNP. The HR and 95% CI of levels 1 and 2 were compared with those for level 0 (reference genotype). Preliminary results showed significant differences for four SNPs, namely rs10088262, rs1665650, rs3802842 and rs59336 (Table 3 and Figure 1A). Among these, a significant overall effect on survival was detected for three SNPs. The median survival times for patients with rs1665650 genotypes 0, 1 or 2 were 36, 71, and 18 months (χ2 = 18.96, log-rank p < 0.001) respectively (Figure 1B); for rs3802842 genotypes 0, 1 or 2, median survival times were 48, 68, and 26 months (χ2 =10.06, log-rank p = 0.007) respectively (Figure 1C); for rs59336 genotypes 0, 1 or 2, median survival times were 36, 68, 12 months (χ2 = 15.21, log-rank p < 0.001) respectively (Figure 1D).
Table 3. Analysis of SNPs associated with OS in LC patients.
| SNP | Genotype | Total N | Variables in the Equation | Median | 95% CI | Chi-Square | p b for log-rank | |||
|---|---|---|---|---|---|---|---|---|---|---|
| Wald | HR | 95% CI | p a | |||||||
| rs10088262 | G/G (0) | 55 | 4.69 | 0.096 | 66.00 | 39.47-92.53 | ||||
| G/A (1) | 71 | 4.38 | 1.57 | 1.03-2.39 | 0.036* | 32.00 | 22.64-41.36 | |||
| A/A (2) | 34 | 0.76 | 1.57 | 0.57-4.34 | 0.382 | 32.00 | 0-87.21 | |||
| Overall | 160 | 48.00 | 30.56-65.44 | 4.84 | 0.809 | |||||
| rs1665650 | G/G (0) | 52 | 7.99 | 0.018* | 36.00 | 25.73-46.27 | ||||
| A/G (1) | 107 | 3.95 | 0.65 | 0.43-0.99 | 0.047* | 71.00 | 49.22-92.78 | |||
| A/A (2) | 7 | 2.00 | 1.98 | 0.77-5.09 | 0.157 | 18.00 | 0.04-35.94 | |||
| Overall | 166 | 48.00 | 30.54-65.46 | 8.56 | 0.014* | |||||
| rs3802842 | A/A (0) | 112 | 9.44 | 0.009* | 48.00 | 18.72-77.29 | ||||
| C/A (1) | 32 | 0.01 | 1.02 | 0.60-1.73 | 0.939 | 68.00 | 31.09-104.91 | |||
| C/C (2) | 24 | 9.03 | 2.18 | 1.31-3.61 | 0.003* | 26.00 | 9.00 -43.00 | |||
| Overall | 168 | 50.00 | 31.06-68.94 | 10.06 | 0.007* | |||||
| rs59336 | A/A (0) | 30 | 13.3 | 0.001* | 36.00 | 30.31-41.69 | ||||
| T/A (1) | 115 | 4.09 | 0.61 | 0.37-.985 | 0.043* | 68.00 | 38.56-97.44 | |||
| T/T (2) | 7 | 4.28 | 2.61 | 1.05-6.49 | 0.039* | 12.00 | 9.43-14.57 | |||
| Overall | 152 | 46.00 | 27.65-64.35 | 15.21 | <0.001* | |||||
p a for Wald test < 0.05 indicates statistical significance for individual coefficients
p b for Log-rank Test < 0.05 indicates statistical significance for grouping variables
*: indicates statistical significance
Figure 1. Survival rate curves for different SNP polymorphisms.
Multivariate Cox proportional hazards analysis
We performed multivariate Cox regression analysis by including the five clinical predictors that showed statistical significance in univariate analyses. Results indicated that two SNPs (rs10088262 and rs3802842) correlated significantly with OS in LC patients: compared with “GG” in rs10088262, the risk rate of genotype “G/A” was 1.969 (95% CI: 1.26-3.09, p = 0.003); on the other hand, the risk rate of rs3802842 (C/C vs. A/A) was 1.839 (95% CI: 1.10-3.08, p = 0.021) (Table 4).
Table 4. Multivariate Cox proportional hazards analysis of rs10088262 and rs3802842 adjusted for HD, MD, LD, T stage and clinical stage.
| SNP | Variable | Wald | df | HR | 95% CI | p a |
|---|---|---|---|---|---|---|
| rs10088262 | rs10088262(G/G) | 8.853 | 2 | 0.012* | ||
| rs10088262(G/A) | 8.719 | 1 | 1.969 | 1.26-3.09 | 0.003* | |
| rs10088262(A/A) | 0.661 | 1 | 1.534 | 0.55-4.29 | 0.416 | |
| rs3802842 | rs3802842(A/A) | 7.014 | 2 | 0.030* | ||
| rs3802842(C/A) | 0.510 | 1 | 0.820 | 0.48-1.41 | 0.475 | |
| rs3802842(C/C) | 5.353 | 1 | 1.839 | 1.10-3.08 | 0.021* |
Abbreviations: HD: High differentiation; MD: Moderate differentiation; LD: Low differentiation; df: degrees of freedom
*: indicates statistical significance.
DISCUSSION
Our study analyzed, during a ten-year follow-up, the prognostic association of clinical parameters and multiple cancer-related SNPs in 170 male LC patients from northwest China. We found that five clinical characteristics [laryngectomy, tumor differentiation, tumor status (T), regional lymph node status (N), and clinical (TNM) stage] were correlated with survival. Specifically, clinical outcome was affected by laryngectomy variants, as TL had poorer prognosis than PL, with a median survival of 30 months, compared with 73 months for PL. This conclusion, however, may be confounded by the fact that TL is more often performed to remove larger malignant tumors, which carry higher risk. The associations found for other clinical features were consistent with the findings of previous studies, as low tumor differentiation, and higher T, N, and clinical stages all correlated with worse prognoses [15, 16].
Of the 24 SNPs analyzed, four showed an association with LC prognosis in univariate analyses. After adjusting for significant clinical parameters, multivariate regression analysis revealed that two SNP genotypes, i.e. “G/A” of rs10088262 (HR: 1.969) and “C/C” of rs3802842 (HR: 1.839) were significantly associated with LC prognosis. The SNPs analyzed here have been shown to be associated with risk for diverse types of cancer. Rs10088262 is an intergenic variant located on chromosome 8q24.13; its minor allele “A” has been correlated with reduced risk of esophageal cancer [12]. However, in the present study the genotype “G/A” of this SNP indicated poor prognosis, with a median survival time of 32 months, compared with 66 months for the “G/G” genotype.
Carrying the “C” allele at rs3802842 has been associated with a lower risk for rectal tumors [17]. Another study, however, suggested that the “CC” genotype of rs3802842 may significantly increase colorectal cancer (CRC) risk in the recessive model [18]. Our analysis revealed that the median survival of genotype “C/C” of the rs3802842 polymorphism was only 26 months, significantly less than the homozygous wild genotype “A/A” (48 months). Rs3802842 maps to a region between “colorectal cancer associated 1” and “colorectal cancer associated 2” (C11orf92/COLCA1- C11orf93/COLCA2) genes, located on 11q23.1, which are arranged on opposite strands and share a regulatory region that contains genetic variants that are in high linkage disequilibrium with rs3802842. Expression levels of COLCA1 and COLCA2 transcripts correlate with rs3802842 genotypes. Genetic, expression and immunohistochemical data implicate COLCA1 and COLCA2 in the pathogenesis of colon cancer, whereas histologic analyses indicate the involvement of immune pathways [19].
MATERIALS AND METHODS
Patients and methods
Study population
170 male patients, aged 32-82 years (average age, 60.7 years), were enrolled at the First Affiliated Hospital of Xi’an Jiao Tong University from January 30, 2002 to April 7, 2003; patient follow-up ended on April 7, 2013. Patients received neither radiotherapy nor chemotherapy before enrollment, and in all cases LC diagnosis was confirmed by two pathologists. All participants were unrelated Han Chinese and had no other malignancy histories.
Patient demographics and blood collection
A standardized epidemiological questionnaire including residential region, age, smoking status, alcohol use, ethnicity, education status, and family history of cancer was used to collect personal information through in-person interviews. Related information was collected through a consultation with the treating physicians or from medical chart reviews. LC staging relies on the TNM system designed jointly by the Union International Cancer Control Version 7.0 (UICC 7.0). Venous blood samples (5 ml) and signed informed consent were obtained from each participant. All blood samples were quickly frozen in liquid nitrogen and stored at -80°C. This study was approved by the ethics committee of the First Affiliated Hospital of Xi’an Jiao Tong University.
SNP selection and genotyping
Using the HapMap database, 24 candidate SNPs with minor allele frequencies > 5% in the Asian population and previously published associations with other cancers were selected from chromosomes 8, 9, 10, 11, and 12. Basic information about the 24 SNPs is listed in Table 5. Genomic DNA was extracted from peripheral blood using phenol–chloroform, and its concentration was measured using a DU530 UV/VIS spectrophotometer (Beckman Instruments, Fullerton, CA, USA) according to the manufacturer’s protocol. MassARRAY Assay Design 3.0 Software (Sequenom, San Diego, California, USA) was used to design Multiplex SNP MassEXTEND assays[20]. Genotyping was performed using the Sequenom MassARRAY RS1000 following a standard protocol recommended by the manufacturer [20], and data were analyzed using Sequenom Typer 4.0 Software (Sequenom, San Diego, CA, USA) [20, 21].
Table 5. SNPs analyzed.
| SNP | Band | A/B | Gene | Cancer type | Ref |
|---|---|---|---|---|---|
| rs2439302 | 8p12 | C/G | NRG1 | Thyroid | [22] |
| rs7832232 | 8p11.22 | G/A | intergenic | Pancreatic | [23] |
| rs10088262 | 8q24.13 | A/G | intergenic | Esophageal | [12] |
| rs10505477 | 8q24.21 | T/C | intergenic | Gastric | [24] |
| rs6983267 | 8q24.21 | G/T | intergenic | Prostate | [25] |
| rs7014346 | 8q24.21 | A/G | POU5F1B | Colorectal | [26] |
| rs13294589 | 9p21.2 | G/A | intergenic | Esophageal | [12] |
| rs10114408 | 9q22.32 | T/A | intergenic | Colorectal | [27] |
| rs965513 | 9q22.33 | A/G | intergenic | Thyroid | [22] |
| rs10795668 | 10p14 | A/G | intergenic | Colorectal | [28] |
| rs2274223 | 10q23.33 | G/A | PLCE1 | Esophageal | [29] |
| rs1665650 | 10q25.3 | A/G | HSPA12A | Colorectal | [30] |
| rs12413624 | 10q26.11 | T/A | intergenic | Colorectal/Gastric | [31] |
| rs10500715 | 11p15.4 | G/T | SBF2 | Pancreatic | [32] |
| rs3824999 | 11q13.4 | C/A | POLD3 | Colorectal | [33] |
| rs3802842 | 11q23.1 | A/C | C11orf92-C11orf93 | Colorectal | [18] |
| rs10774214 | 12p13.32 | T/C | intergenic | Colorectal | [34] |
| rs3217901 | 12p13.32 | A/G | CCND2 | Colorectal | [34] |
| rs10879357 | 12q21.1 | G/A | TPH2 | Colorectal | [34] |
| rs671 | 12q24.12 | A/G | ALDH2 | Esophageal | [35] |
| rs4767364 | 12q24.13 | G/A | NAA25 | Aero-digestive tract | [36] |
| rs11066280 | 12q24.13 | A/T | C12orf51 | Gastric | [37] |
| rs59336 | 12q24.21 | T/A | TBX3 | Colorectal | [34] |
| rs7315438 | 12q24.21 | T/C | intergenic | Colorectal/Esophageal | [38] |
A/B stands for minor/major alleles on the sample frequencies.
Statistical analysis
Patients’ baseline characteristics, disease stage, and treatment modalities were summarized using descriptive statistics. The overall survival (OS) time was defined as the period from diagnosis until death of any cause or until the date of the last follow-up, at which data point was censored. All summary statistics on time-to-event variables were estimated according to the Kaplan-Meier method and compared using the log-rank test. Univariate and multivariate Cox proportional hazards regression models were used to calculate the hazard ratios (HR), and 95% confidence intervals (95% CI) of the effect of clinical variables and SNPs, respectively, on the overall survival (OS) of LC patients. SPSS software (version 21.0) was used for statistical analysis. A p value < 0.05 was considered significant.
CONCLUSION
We found that five clinicopathological charac-teristics, namely total laryngectomy, low differentiation, T3-T4, N1-N2, and clinical stage III-IV, were associated with survival in LC patients. Although four SNP were found to be significantly associated with OS in univariate cox regression analysis, multivariate analysis showed that two SNPs (rs10088262 and rs3802842) were associated with LC prognosis after adjustment for clinical factors. Combined with previous research, our study suggests an association for these SNPs with multiple cancers. Further larger studies are required to validate our findings and to assess the molecular mechanisms underlying the observed associations.
Acknowledgments
This study was funded by the China Postdoctoral Science Foundation (NO. 2015M572575) and Key Science and Technology Program of Shaanxi Province, China (NO. 2014K11-01-01-09). We are grateful to the patients for their participation in this study. We also thank the clinicians and hospital staff who contributed to the sample and data collection for this study.
Footnotes
CONFLICTS OF INTEREST
All authors declare that they have no conflicts of interest.
REFERENCES
- 1.Wang H, Ma K. Association between MDM2 rs769412 and rs937283 polymorphisms with alcohol drinking and laryngeal carcinoma risk. International journal of clinical and experimental pathology. 2015;8:7436–7440. [PMC free article] [PubMed] [Google Scholar]
- 2.Ferlay J, Soerjomataram I, Dikshit R, Eser S, Mathers C, Rebelo M, Parkin DM, Forman D, Bray F. Cancer incidence and mortality worldwide: sources, methods and major patterns in GLOBOCAN 2012. International journal of cancer. 2015;136:E359–386. doi: 10.1002/ijc.29210. [DOI] [PubMed] [Google Scholar]
- 3.Bray F, Ren JS, Masuyer E, Ferlay J. Global estimates of cancer prevalence for 27 sites in the adult population in 2008. International journal of cancer. 2013;132:1133–1145. doi: 10.1002/ijc.27711. [DOI] [PubMed] [Google Scholar]
- 4.Leon X, Quer M, Diez S, Orus C, Lopez-Pousa A, Burgues J. Second neoplasm in patients with head and neck cancer. Head & neck. 1999;21:204–210. doi: 10.1002/(sici)1097-0347(199905)21:3<204::aid-hed4>3.0.co;2-7. [DOI] [PubMed] [Google Scholar]
- 5.Mallis A, Jelastopulu E, Mastronikolis NS, Naxakis SS, Kourousis C, Papadas TA. Laryngeal cancer and passive smoking: the neglected factor? European archives of oto-rhino-laryngology. 2011;268:727–731. doi: 10.1007/s00405-010-1403-z. [DOI] [PubMed] [Google Scholar]
- 6.Simsek H, Han U, Onal B, Simisek G. The expression of EGFR, cerbB2, p16, and p53 and their relationship with conventional parameters in squamous cell carcinoma of the larynx. Turkish journal of medical sciences. 2014;44:411–416. [PubMed] [Google Scholar]
- 7.Shen Z, Zhan G, Deng H, Kang C, Guo J. Growth inhibitory effect of microRNA-519b-3p on larynx squamous Hep-2 cells. [Article in Chinese] Zhonghua er bi yan hou tou jing wai ke za zhi. 2014;49:151–156. [PubMed] [Google Scholar]
- 8.Chai D, Bao Z, Hu J, Ma L, Feng Z, Tao Y. Aberrant expression of CyclinE and p27 in laryngeal squamous cell carcinoma and the clinical significance. [Article in Chinese] Lin chuang er bi yan hou tou jing wai ke za zhi. 2014;28:165–169. 174. [PubMed] [Google Scholar]
- 9.Lu C, Xie M, Wendl MC, Wang J, McLellan MD, Leiserson MD, Huang KL, Wyczalkowski MA, Jayasinghe R, Banerjee T, Ning J, Tripathi P, Zhang Q, Niu B, Ye K, Schmidt HK, et al. Patterns and functional implications of rare germline variants across 12 cancer types. Nature communications. 2015;6:10086. doi: 10.1038/ncomms10086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Abderrahmane R, Louhibi L, Moghtit FZ, Boubekeur A, Benseddik K, Boudjema A, Benrrahal F, Aberkane M, Fodil M, Saidi-Mehtar N. TP53 Arg 72Pro and MDM2 SNP309 polymorphisms and colorectal cancer risk: a west Algerian population study. Pathology oncology research. 2015;21:629–635. doi: 10.1007/s12253-014-9867-6. [DOI] [PubMed] [Google Scholar]
- 11.Ek WE, Levine DM, D’Amato M, Pedersen NL, Magnusson PK, Bresso F, Onstad LE, Schmidt PT, Tornblom H, Nordenstedt H, Romero Y, Mayo Clinic Esophageal A. Barrett’s Esophagus Registry C. Chow WH, Murray LJ, Gammon MD, et al. Germline genetic contributions to risk for esophageal adenocarcinoma, Barrett’s esophagus, and gastroesophageal reflux. Journal of the National Cancer Institute. 2013;105:1711–1718. doi: 10.1093/jnci/djt303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wang J, Zhang B, Yang Z, Zhou L, Geng T, Li H, Fu X, Xue X, Liu M, Tong R, Jin T, Zhang Y. Association of gastrointestinal gland cancer susceptibility loci with esophageal carcinoma among the Chinese Han population: a case-control study. Tumour biology. 2015 doi: 10.1007/s13277-015-3945-6. [DOI] [PubMed] [Google Scholar]
- 13.Leoncini E, Vukovic V, Cadoni G, Pastorino R, Arzani D, Bosetti C, Canova C, Garavello W, La Vecchia C, Maule M, Petrelli L, Pira E, Polesel J, Richiardi L, Serraino D, Simonato L, et al. Clinical features and prognostic factors in patients with head and neck cancer: Results from a multicentric study. Cancer epidemiology. 2015;39:367–374. doi: 10.1016/j.canep.2015.02.004. [DOI] [PubMed] [Google Scholar]
- 14.Shen Z, Ren W, Bai Y, Chen Z, Li J, Li B, Jin T, Cao P, Shao Y. DIRC3 and near NABP1 genetic polymorphisms are associated laryngeal squamous cell carcinoma patient survival. Oncotarget. 2016;7:79596–04. doi: 10.18632/oncotarget.12865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bice TC, Tran V, Merkley MA, Newlands SD, van der Sloot PG, Wu S, Miller MC. Disease-Specific Survival with Spindle Cell Carcinoma of the Head and Neck. Otolaryngology--head and neck surgery. 2015;153:973–980. doi: 10.1177/0194599815594360. [DOI] [PubMed] [Google Scholar]
- 16.Dubal PM, Unsal AA, Echanique KA, Vazquez A, Reder LS, Baredes S, Eloy JA. Laryngeal adenosquamous carcinoma: A population-based perspective. The Laryngoscope. 2015 doi: 10.1002/lary.25704. [DOI] [PubMed] [Google Scholar]
- 17.Mates IN, Jinga V, Csiki IE, Mates D, Dinu D, Constantin A, Jinga M. Single nucleotide polymorphisms in colorectal cancer: associations with tumor site and TNM stage. Journal of gastrointestinal and liver diseases. 2012;21:45–52. [PubMed] [Google Scholar]
- 18.Duan X, Li X, Lou H, Geng T, Jin T, Liang P, Li S, Long Y, Chen C. Genetic association of PLCE1, C11orf92-C11orf93, and NOC3L with colorectal cancer risk in the Han population. Tumour biology. 2014;35:1813–1817. doi: 10.1007/s13277-013-1242-9. [DOI] [PubMed] [Google Scholar]
- 19.Peltekova VD, Lemire M, Qazi AM, Zaidi SH, Trinh QM, Bielecki R, Rogers M, Hodgson L, Wang M, D’Souza DJ, Zandi S, Chong T, Kwan JY, Kozak K, De Borja R, Timms L, et al. Identification of genes expressed by immune cells of the colon that are regulated by colorectal cancer-associated variants. International journal of cancer. 2014;134:2330–2341. doi: 10.1002/ijc.28557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gabriel S, Ziaugra L, Tabbaa D. SNP genotyping using the Sequenom MassARRAY iPLEX platform. Current protocols in human genetics. 2009 doi: 10.1002/0471142905.hg0212s60. Chapter 2: Unit 2 12. [DOI] [PubMed] [Google Scholar]
- 21.Thomas RK, Baker AC, Debiasi RM, Winckler W, Laframboise T, Lin WM, Wang M, Feng W, Zander T, MacConaill L, Lee JC, Nicoletti R, Hatton C, Goyette M, Girard L, Majmudar K, et al. High-throughput oncogene mutation profiling in human cancer. Nature genetics. 2007;39:347–351. doi: 10.1038/ng1975. [DOI] [PubMed] [Google Scholar]
- 22.Wang YL, Feng SH, Guo SC, Wei WJ, Li DS, Wang Y, Wang X, Wang ZY, Ma YY, Jin L, Ji QH, Wang JC. Confirmation of papillary thyroid cancer susceptibility loci identified by genome-wide association studies of chromosomes 14q13, 9q22, 2q35 and 8p12 in a Chinese population. Journal of medical genetics. 2013;50:689–695. doi: 10.1136/jmedgenet-2013-101687. [DOI] [PubMed] [Google Scholar]
- 23.Low SK, Kuchiba A, Zembutsu H, Saito A, Takahashi A, Kubo M, Daigo Y, Kamatani N, Chiku S, Totsuka H, Ohnami S, Hirose H, Shimada K, Okusaka T, Yoshida T, Nakamura Y, et al. Genome-wide association study of pancreatic cancer in Japanese population. PloS one. 2010;5:e11824. doi: 10.1371/journal.pone.0011824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Shen L, Du M, Wang C, Gu D, Wang M, Zhang Q, Zhao T, Zhang X, Tan Y, Huo X, Gong W, Xu Z, Chen J, Zhang Z. Clinical significance of POU5F1P1 rs10505477 polymorphism in Chinese gastric cancer patients receving cisplatin-based chemotherapy after surgical resection. International journal of molecular sciences. 2014;15:12764–12777. doi: 10.3390/ijms150712764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Fernandez P, Salie M, du Toit D, van der Merwe A. Analysis of Prostate Cancer Susceptibility Variants in South African Men: Replicating Associations on Chromosomes 8q24 and 10q11. Prostate cancer. 2015;2015:465184. doi: 10.1155/2015/465184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wei W, Jiang M, Luo L, Li Z, Wang P, Dong WQ. Colorectal cancer susceptibility variants alter risk of breast cancer in a Chinese Han population. Genetics and molecular research. 2013;12:6268–6274. doi: 10.4238/2013.December.4.14. [DOI] [PubMed] [Google Scholar]
- 27.Jiao S, Hsu L, Berndt S, Bezieau S, Brenner H, Buchanan D, Caan BJ, Campbell PT, Carlson CS, Casey G, Chan AT, Chang-Claude J, Chanock S, Conti DV, Curtis KR, Duggan D, et al. Genome-wide search for gene-gene interactions in colorectal cancer. PloS one. 2012;7:e52535. doi: 10.1371/journal.pone.0052535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Li FX, Yang XX, Hu NY, Du HY, Ma Q, Li M. Single-nucleotide polymorphism associations for colorectal cancer in southern chinese population. Chinese journal of cancer research. 2012;24:29–35. doi: 10.1007/s11670-012-0029-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Chen YZ, Cui XB, Pang XL, Li L, Hu JM, Liu CX, Cao YW, Yang L, Li F. Relationship between rs2274223 and rs3765524 polymorphisms of PLCE1 and risk of esophageal squamous cell carcinoma in a Kazakh Chinese population. [Article in Chinese] Zhonghua bing li xue za zhi. 2013;42:795–800. [PubMed] [Google Scholar]
- 30.Kantor ED, Hutter CM, Minnier J, Berndt SI, Brenner H, Caan BJ, Campbell PT, Carlson CS, Casey G, Chan AT, Chang-Claude J, Chanock SJ, Cotterchio M, Du M, Duggan D, Fuchs CS, et al. Gene-environment interaction involving recently identified colorectal cancer susceptibility Loci. Cancer epidemiology, biomarkers & prevention. 2014;23:1824–1833. doi: 10.1158/1055-9965.EPI-14-0062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Su Q, Wang Y, Zhao J, Ma C, Wu T, Jin T, Xu J. Polymorphisms of PRLHR and HSPA12A and risk of gastric and colorectal cancer in the Chinese Han population. BMC gastroenterology. 2015;15:107. doi: 10.1186/s12876-015-0336-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wu C, Kraft P, Stolzenberg-Solomon R, Steplowski E, Brotzman M, Xu M, Mudgal P, Amundadottir L, Arslan AA, Bueno-de-Mesquita HB, Gross M, Helzlsouer K, Jacobs EJ, Kooperberg C, Petersen GM, Zheng W, et al. Genome-wide association study of survival in patients with pancreatic adenocarcinoma. Gut. 2014;63:152–160. doi: 10.1136/gutjnl-2012-303477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Dunlop MG, Dobbins SE, Farrington SM, Jones AM, Palles C, Whiffin N, Tenesa A, Spain S, Broderick P, Ooi LY, Domingo E, Smillie C, Henrion M, Frampton M, Martin L, Grimes G, et al. Common variation near CDKN1A, POLD3 and SHROOM2 influences colorectal cancer risk. Nature genetics. 2012;44:770–776. doi: 10.1038/ng.2293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Jia WH, Zhang B, Matsuo K, Shin A, Xiang YB, Jee SH, Kim DH, Ren Z, Cai Q, Long J, Shi J, Wen W, Yang G, Delahanty RJ, Genetics, Epidemiology of Colorectal Cancer C et al. Genome-wide association analyses in East Asians identify new susceptibility loci for colorectal cancer. Nature genetics. 2013;45:191–196. doi: 10.1038/ng.2505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zhao T, Wang C, Shen L, Gu D, Xu Z, Zhang X, Xu Y, Chen J. Clinical significance of ALDH2 rs671 polymorphism in esophageal cancer: evidence from 31 case-control studies. OncoTargets and therapy. 2015;8:649–659. doi: 10.2147/OTT.S76526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.McKay JD, Truong T, Gaborieau V, Chabrier A, Chuang SC, Byrnes G, Zaridze D, Shangina O, Szeszenia-Dabrowska N, Lissowska J, Rudnai P, Fabianova E, Bucur A, Bencko V, Holcatova I, Janout V, et al. A genome-wide association study of upper aerodigestive tract cancers conducted within the INHANCE consortium. PLoS genetics. 2011;7:e1001333. doi: 10.1371/journal.pgen.1001333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zhang G, Gu D, Zhao Q, Chu H, Xu Z, Wang M, Tang C, Wu D, Tong N, Gong W, Zhou J, Xu Y, Zhang Z, Chen J. Genetic variation in C12orf51 is associated with prognosis of intestinal-type gastric cancer in a Chinese population. Biomedicine & pharmacotherapy. 2015;69:133–138. doi: 10.1016/j.biopha.2014.11.009. [DOI] [PubMed] [Google Scholar]
- 38.Geng TT, Xun XJ, Li S, Feng T, Wang LP, Jin TB, Hou P. Association of colorectal cancer susceptibility variants with esophageal cancer in a Chinese population. World journal of gastroenterology. 2015;21:6898–6904. doi: 10.3748/wjg.v21.i22.6898. [DOI] [PMC free article] [PubMed] [Google Scholar]


