Abstract
Radiation therapy is a mainstay of cancer treatment, yet the molecular determinants of clinical response are poorly understood. We identified exceptional responders to radiotherapy based on clinical response, and investigated the associated tumor sequencing data in order to identify additional patients with similar mutations. Among head and neck squamous cell cancer patients receiving palliative radiotherapy at our institution, we identified one patient with documented complete metabolic response. Targeted sequencing analysis of the tumor identified a somatic frame-shift mutation in ATM, a gene known to be associated with radio-sensitivity in the germline. To validate the association of somatic ATM mutation with radiotherapy response, we identified eight patients with ATM truncating mutations who received radiotherapy, all of whom demonstrated excellent responses with a median local control period of 4.62 years. Analysis of 22 DNA repair genes in The Cancer Genome Atlas (TCGA) data revealed mutations in 15.9% of 9064 tumors across 24 cancer types, with ATM mutations being the most prevalent. This is the first study to suggest that exceptional responses to radiotherapy may be determined by mutations in DNA repair genes. Sequencing of DNA repair genes merits attention in larger cohorts and may have significant implications for the personalization of radiotherapy.
Keywords: somatic ATM mutations, radiation therapy
INTRODUCTION
Patients with metastatic head and neck cancer or locally advanced disease and poor performance status often receive palliative radiotherapy (RT). Palliative RT often alleviates symptoms but rarely provides long-term loco-regional control or prolonged survival [1, 2]. Although full-dose RT may provide for more durable loco-regional control, the increased acute toxicity associated with standard treatment typically outweighs the benefits in patients with limited life expectancies. Occasionally, some patients respond better than anticipated to palliative RT and achieve long-term disease control. The mechanistic basis for these exceptional responses to palliative RT remains poorly understood.
Although there are several germline syndromes that predispose to marked radiation sensitivity, to our knowledge, alterations in genes associated with these syndromes have not been linked to tumor hypersensitivity to RT [3]. Here we examine patients at our institution who received palliative RT for head and neck squamous cell cancers (HNSCC) and identified a patient with long-term disease control (Figure 1a). Targeted sequencing analysis of the tumor revealed a frame-shift mutation in ATM, a gene centrally involved in the DNA damage response. To further investigate the role of ATM in tumor response, we then examined a separate institutional database of patients who underwent targeted sequencing analysis and identified eight patients with similar ATM mutations that received palliative RT, all of whom appeared to have excellent responses (Figure 1b).
Figure 1. Identification of long-term survivors with ATM frameshift or truncating mutations demonstrating good responses to radiotherapy.
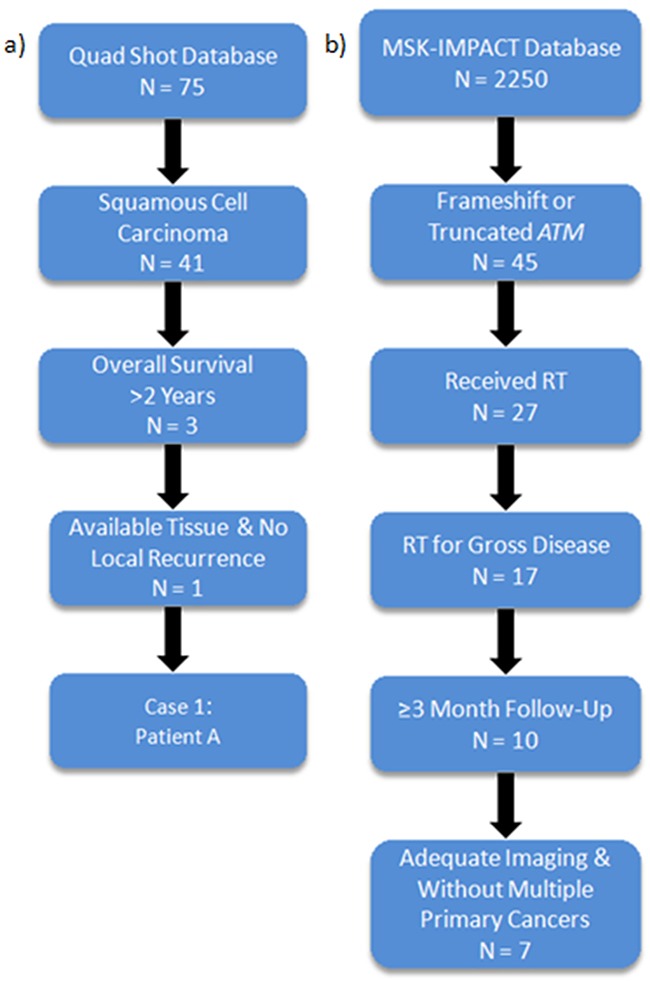
a. Institutional database of palliative Quad Shot RT revealed 1 SCC patient with long-term disease control. b. Institutional sequencing database (MSK-IMPACT) identifies 8 patients with good responses to RT.
RESULTS
Here we briefly describe the relevant clinical histories of three of the eight to demonstrate the exceptional responses to palliative radiotherapy observed. We also discuss genomic analysis of tumor sequencing data to identify patients with a DNA repair mutation that may be a determinant of clinical response.
Case 1: Patient A (HNSCC with long-term complete metabolic response)
Patient A is a 96 year old female with a p16 positive, moderately differentiated, invasive SCC of the right lateral oral tongue. Right-sided level I and II lymph nodes (SUV 2.3) were also present and were suspicious for metastatic disease (Figure 2a). Patient A was treated with cetuximab 500 mg/m2 for 7 months until disease progression. Patient A was then treated with 44.4 Gy of RT in 12 fractions with the “Quad Shot” regimen, with complete metabolic resolution of disease on a PET/CT performed 3 months post-RT (Figure 2b). A CT scan 8 months after RT and accompanying physical exam demonstrated no evidence of recurrence. She was last seen in clinic 34 months after RT with no evidence of recurrence and she has not needed additional therapy since that time.
Figure 2. Pre- and post-therapy imaging for patient A.
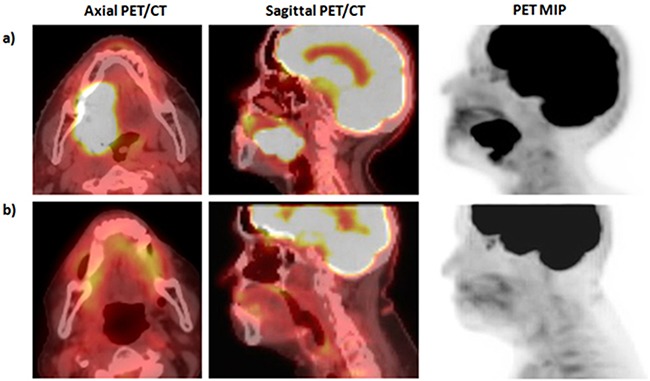
*MIP: Fludeoxyglucose (18F) positron emission tomography (FDG-PET) maximum intensity projection (MIP). a) Pre-radiotherapy PET/CT demonstrates primary oral tongue SCC with SUV of 35.4 with FDG avid level Ib and II lymph nodes. b) Three month post-RT PET/CT demonstrates complete resolution of FDG avid malignancy.
Targeted sequencing [4] using a next generation sequencing panel of cancer genes demonstrated that patient A had a frameshift mutation in the ATM gene at position 1455, along with 29 other somatic mutations. Of note, the tumor is also p53 wild-type with a mutation in RAD50 (R519H) and a frameshift mutation in MLH1 at position 64.
Institutional tumor sequencing database
Due to the strong evidence linking ATM to radiation sensitivity in the constitutional setting [5, 6], we searched our institutional MSK-IMPACT (Memorial Sloan Kettering-Integrated Mutation Profiling of Actionable Cancer Targets) database of 2,250 patients who underwent targeted panel sequencing to identify other patients with truncating mutations in ATM. Forty-five other patients, harboring a variety of primary malignancies, were found to have frame-shift or truncating ATM mutations. Of this cohort, seventeen patients underwent radiotherapy to grossly evident tumor. Ten of these patients had a minimum of 3 months of follow-up, of which 2 cases were excluded, one due to inadequate imaging and another due to multiple synchronous primary malignancies. The remaining eight patients demonstrated long-term disease control within the RT treatment field (Table 1). We review one of the above 8 cases with long-term loco-regional control from radiotherapy and briefly discuss another two of the remaining seven cases below.
Table 1. Characteristics of 8 Patients with ATM mutations identified from both the Quad Shot and MSK-IMPACT databases. LC: local control.
| Patient | Disease | ATM Mutation | RT Target | RT Dose / Fraction(Gy) | In-Field Recurrence | Duration of LC(Months) |
|---|---|---|---|---|---|---|
| A | Head and Neck Squamous Cell Cancer | I14531 | Oral Tongue | 44.4 / 12 | No | 34.5 |
| B | Endometrial Cancer | G1370¥ | L3-L4 | 36 / 12 | No | 52.5 |
| Right Presacral | 36 / 12 | No | 33.7 | |||
| C | Non-Small Cell Lung Cancer | Q711 | Whole Brain | 37.5 / 15 | Yes | 43.2 |
| D | Breast Cancer | D1548M1 | Whole Brain | 37.5 / 15 | Yes | 41.0 |
| E | Colon Cancer | R1875* | Abdominal Metastasis | 30 / 10 | No | 3.8 |
| F | Colon Cancer | R1898* | Pelvis | 37.5 / 15 | No | 3.9 |
| G | Non-Small Cell Lung Cancer | I14411Q1331H | Neck & Upper Mediastinum | 50.4 / 28 | No | 3.1 |
| H | Thyroid Cancer | L2738* | Thyroid & Upper Mediastinum | 70 / 33 | No | 5.16 |
°Note: Patient B had two separate sites treated at two different time points; *: nonsense mutation, 1: frameshift mutation, ¥: splice site mutation
Case 2: Patient B (Endometrial Cancer with long-term response)
Patient B is a 68 year old woman diagnosed with FIGO stage IB, Grade 2 endometrial cancer who underwent a total abdominal hysterectomy, bilateral salpingo-oophorectomy, and pelvic lymph node sampling. She recurred 2 years later and underwent gross total surgical resection.
Re-staging studies 3 years after initial diagnosis demonstrated a left para-spinal recurrence involving the L4 vertebral body (Figure 3a). She received palliative RT to 36 Gy in 12 fractions. Subsequent MR imaging demonstrated stable disease and a PET/CT scan demonstrated complete metabolic response to treatment. She was noted to have an out-of-field recurrence in a pre-sacral lymph node 1 year later (Figure 3b). She received 36Gy in 12 fractions to this second site of disease. Subsequent imaging demonstrated reduction in size of this lesion without evidence of recurrence (Figure 3c). She is currently without evidence of in-field recurrence of the L3-L4 lesion for over 4.3 years, and demonstrates stable disease of the right pre-sacral lesion for 2.8 years.
Figure 3. Pre- and post-therapy imaging for patient B.
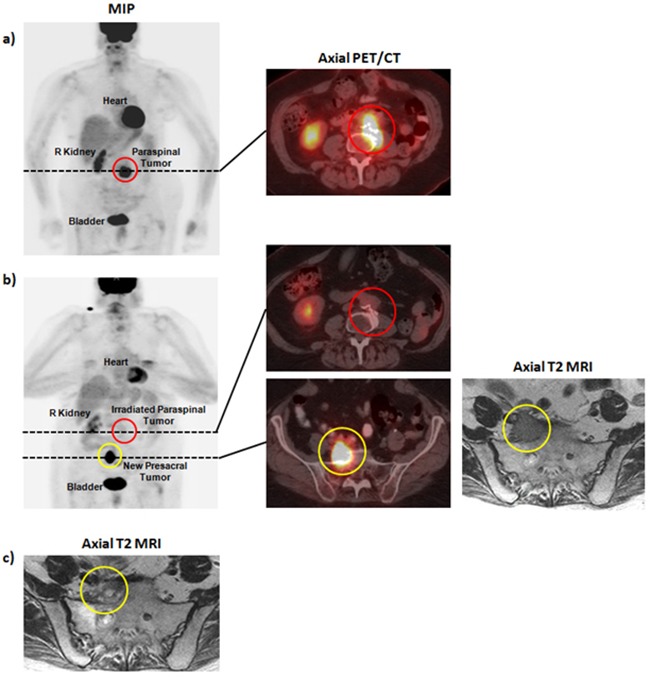
a. Pre-radiotherapy PET/CT demonstrating a para-spinal mass involving the L4 vertebral body. b. Post-radiotherapy PET/CT and MRI demonstrating treatment response at L3-L4 and an out-of-field recurrence in a right pre-sacral lymph node. d. Post-radiotherapy MRI demonstrating reduction in size of the right pre-sacral lymph node.
Case 3: Patient C (Lung Cancer with long-term response)
Patient C is a 64 year old woman diagnosed with EGFR mutated NSCLC. MR imaging of the brain demonstrated a 5 mm enhancing brain metastasis in the right temporal lobe. She received whole-brain radiotherapy (WBRT) to 37.5 Gy, and was subsequently enrolled on a clinical trial with a pan-HER inhibitor. She developed progression of disease in her pleura 3 years later. An MRI of the brain performed over 3.5 years after diagnosis unfortunately demonstrated a new left frontal lobe metastasis, documenting an in-field recurrence. She demonstrated local control for 43 months with an overall survival of 48 months. A graded prognostic assessment was utilized to predict expected survival outcomes for patients with brain metastasis based on age, KPS, presence of extracranial metastasis, and number of brain lesions. The assessment predicted an overall survival of 6.5 months for Patient C, as compared to the observed 48 months.
Genomic analysis of NHEJ pathway mutations in TCGA
We next examined The Cancer Genome Atlas (TCGA) data to identify the frequency of mutations in 22 DNA repair genes involved in the non-homologous end joining (NHEJ) pathway [8] across 24 cancer types (Table 2). Genes in NHEJ, similar to ATM, are strongly linked to in vitro sensitivity to IR across multiple cell types. Of 9,064 tumor samples, 15.9% exhibited at least one genetic alteration (germline loss of function mutation, somatic loss of function mutation, or somatic missense mutation) in one of the 22 NHEJ genes (Figure 4a) [9, 10]. Of these, ATM was the most highly mutated gene, with 19.6% of tumors with an NHEJ pathway alteration exhibiting some type of ATM mutation. Additionally, we examined the prevalence of NHEJ pathway alterations by cancer type and identified the most commonly mutated gene within the pathway, for each cancer type. Of 24 cancer types with NHEJ mutations, 15 (62.5%) cancer types revealed ATM to be the most commonly mutated gene (Figure 4b, Supplemental Figure 1).
Table 2. Twenty-two genes involved in the NHEJ pathway and their associated functions.
| Gene(HUGO) | Function |
|---|---|
| APLF | Chromatin-binding checkpoint protein [36] |
| ATM | DSB signaling [37] |
| DCLRE1C | End-processing [38] |
| LIG4 | Main DSB ligating enzyme [39] |
| MDC1 | DSB signaling; recruits DNA damage response elements [40] |
| MRE11A | DSB signaling of MRN complex [41] |
| NBN | DSB signaling of MRN complex [42] |
| NHEJ1 | Ligase accessory factor [43] |
| PARG | Catabolism of PAR [44] |
| POLM | Polymerase; gap-filling [45] |
| PRKDC | DSB signaling; DNA-dependent protein kinase catalytic subunit [46] |
| RAD50 | DSB signaling; MRN complex [47] |
| RNF168 | DSB signaling; 63-linked histone poly-ubiquitination of H2AX; (downstream RNF88, upstream BRCA1) [48] |
| RNF8 | Ubiquinates H2AX [49] |
| TP53BP1 | DSB signaling [50] |
| XRCC2 | DNA break and crosslink repair [51] |
| XRCC3 | DNA break and crosslink repair [51] |
| XRCC4 | Ligase accessory factor [52] |
| XRCC5 | Binds to DSB end/Ku heterodimer [53] |
| XRCC6 | Binds to DSB end/Ku heterodimer [54] |
| H2AFX | DSB signaling [55] |
Figure 4. Analysis of TCGA data to identify frequency of DNA repair mutations.
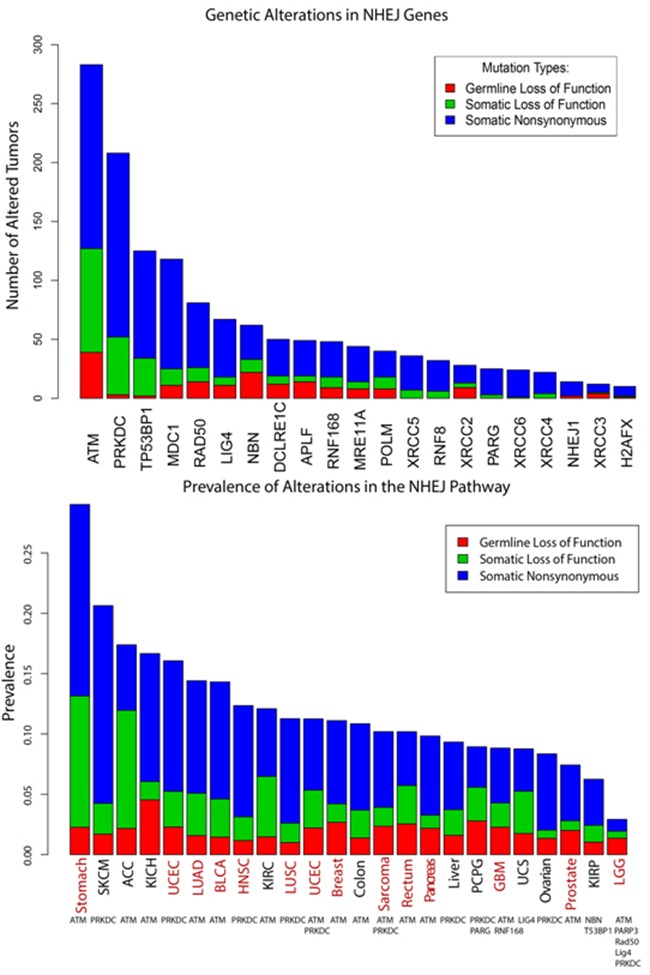
a. Analysis of 22 DNA repair related genes involved in NHEJ. a) 9064 tumor samples were examined across 24 cancer types, with 15.9% of tumors exhibiting at least one alteration among the 22 genes. ATM was the most highly mutated, with 19.6% of tumors exhibiting some type of ATM alteration. b. Prevalence of NHEJ pathway alterations are listed by cancer type. The most commonly mutated gene within the pathway, for each cancer type, is listed below the corresponding cancer type. ATM was the most commonly mutated gene in fifteen of the 24 cancer types (62.5%). Red text identifies cancers for which radiation is used as standard of care as either definitive or adjuvant treatment. Adrenocortical carcinoma [ACC], Bladder Urothelial Carcinoma [BLCA], Glioblastoma multiforme [GBM], Head and Neck squamous cell carcinoma [HNSC], Kidney Chromophobe [KICH], Kidney renal clear cell carcinoma [KIRC], Kidney renal papillary cell carcinoma [KIRP], Brain Lower Grade Glioma [LGG], Lung adenocarcinoma [LUAD], Lung squamous cell carcinoma [LUSC], Pheochromocytoma and Paraganglioma [PCPG], Skin Cutaneous Melanoma [SKCM], Uterine Corpus Endometrial Carcinoma [UCEC], Uterine Carcinosarcoma [UCS].
DISCUSSION
Ionizing radiation (IR) induces a number of DNA aberrations including base damage, single-strand breaks, and double-strand breaks (DSB) [11]. DSBs are thought to be the most lethal of these lesions, with each Gray of IR producing between 20-40 DSBs. ATM, a Ser/Thr kinase, serves as a key signaling hub in the DNA DSB damage response [12, 13]. ATM exists in the cell as an inactive homo-dimer which is activated by genotoxic stress, at which point the dimer dissociates and an ATM monomer is recruited to the site of the DSB by the Mre11-Rad50-NBS1 (MRN) complex [12, 14]. The activated ATM monomer phosphorylates H2AX and several other proteins in order to facilitate the recruitment of DSB repair machinery.
A wealth of evidence accumulated over the past 40 years indicates that ATM plays a critical role in the response to IR in experimental systems. First, cell lines derived from patients with ataxia-telangiectasia syndrome or carriers for the gene are known to exhibit marked sensitivity to IR [5, 6]. Knockdown of ATM is often used as a control in experiments to determine if other genes increase sensitivity to radiation [15]. Pharmacologic inhibition of ATM in vitro with several different selective inhibitors also leads to marked radio-sensitization and is ineffective in ATM deficient cell lines [16–18]. In an orthotopic xenograft model of glioblastoma multiforme, intra-tumoral injections of an ATM inhibitor led to significantly increased survival when combined with RT, although this effect appeared superior in p53-mutated cells [19].
A recent phase II trial of patients with metastatic prostate cancer demonstrated increased sensitivity of patients with ATM aberrations to a poly(adenosine diphosphate [ADP]-ribose) polymerase (PARP) inhibitor [20]. Although the germline ataxia-telangiectasia syndrome is due to compound heterozygosity, haplo-insufficiency in ATM is known to increase radio-sensitivity as well as increase risk of cancer development [6, 21, 22]. Interestingly, patients with mono-allelic ATM aberrations affecting the C-terminal phosphoinositide 3-kinase (PI3K) catalytic domain were identified in the aforementioned phase II study as sensitive to another class of DNA damaging agents, namely PARP inhibitors, suggesting a clinically relevant haplo-insufficient phenotype.
Rare germline genetic syndromes involving DSB repair genes have been associated with exquisite radio-sensitivity and severe adverse reactions to radiation therapy [3]. In addition to ATM, germline mutations in MRE11 has been shown to be predictive of radiotherapy response in bladder cancer [23, 24], while inherited defects in DSB repair genes NBS1 and Lig4 have been linked to fatal complications from low doses of radiotherapy [25–27]. High MRE11 expression levels on immunohistochemistry have been associated with improved cancer-specific survival in radiotherapy patients compared to high MRE11 expression in patients undergoing cystectomy alone, also suggesting a role for epigenetic alterations in MRE11 in radiotherapy response [23]. These cases suggest that common toxicities from radiotherapy may be heritable and has prompted a search for common and rare variants of inherited defects associated with toxicity [28]. However, tumor response to somatic genetic alterations in DNA repair genes have not been adequately explored. To our knowledge, this is the first report of multiple clinical cases with alterations in ATM demonstrating dramatic clinical responses to palliative radiotherapy.
ATM is one of only 22 genes recurrently mutated in 3 or more cancers (renal cell, lung adenocarcinoma, and prostate cancer) [8]. Furthermore, truncating mutations are present in at least 1% of tumors in over 12 different cancers. As more patients undergo sequencing as part of their workup, with some undergoing sequencing of multiple genes, identifying patients with alterations in DNA repair genes may offer novel opportunities for RT in previously palliative settings. These cases highlight the possibility that functional alterations in key DNA DSB repair genes may result in increased sensitivity to radiotherapy.
Here we have identified an unusual long-term response of an oral cavity SCC to palliative RT, with an associated frameshift ATM mutation. Patient A had a complete metabolic response to radiotherapy with biopsy confirmed absence of disease 1 year post-RT. Although p16 status is associated with an improved prognosis in oropharynx cancers, it is not associated with outcomes in squamous cell cancers of the oral cavity [29]. More importantly, p16 status in cancers outside the oropharynx does not appear to be linked to HPV infection [30]. We subsequently identified 8 patients with truncating mutations in ATM who received radiotherapy to gross disease and had tumor sequencing performed. The local control and overall survival rates in our cohort of patients with ATM mutations appear to be superior to historic controls, although these types of comparisons are limited. Median time to local recurrence for this population was 4.62 years, with two of the eight patients developing local recurrence within the radiation field (Figure 5). Patient C was treated with WBRT for a NSCLC brain metastasis and demonstrated disease control for more than 3.5 years. The other 6 patients included in this study exhibited similar exceptional responses to radiotherapy.
Figure 5. Kaplan-Meier analysis of local recurrence data for 8 patients with truncating and frameshift ATM mutations treated with radiotherapy to gross disease, in years.
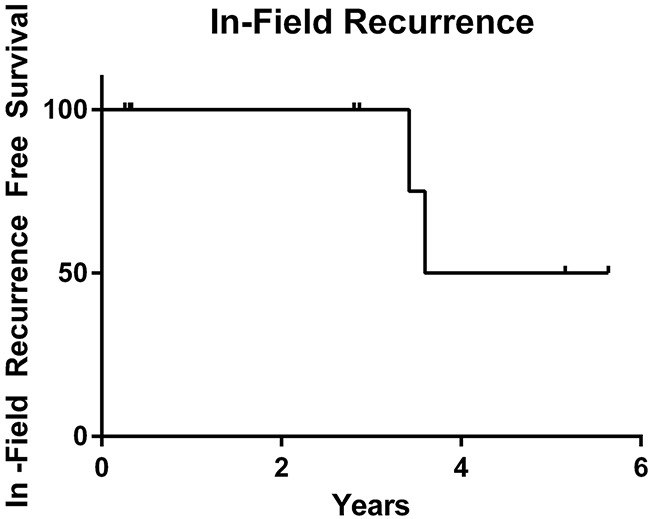
Median time to in-field local recurrence is 4.62 years (range: 0.26-5.64). Two of the eight patients developed an in-field recurrence.
These cases highlight the possibility of utilizing sequencing information to personalize radiotherapy treatment decisions in patients with genetic aberrations in DNA repair genes. The data presented here, along with evidence for mutations in DNA repair genes driving response to other DNA damaging agents, strongly supports further exploration of mutations in these genes in larger cohorts. Ultimately, data from this case series will need to be validated in a larger cohort of patients to definitively define the role of mutations in DNA repair genes in radiotherapy response. If more definitive evidence emerges suggesting radio-sensitivity in these patients, previously abandoned uses of large field, low-dose radiotherapy such as whole abdominal treatment or liver treatment could be reconsidered.
MATERIALS AND METHODS
The retrospective review of outcomes with corresponding genetic alterations was approved by the institutional review board (IRB).
Palliative radiotherapy for HNSCC
Our institutional preference for palliative radiotherapy to the head and neck is based on the RTOG 8502 regimen, which consists of two fractions of 3.7Gy per day for two days, and colloquially named the “Quad Shot.” The Quad Shot is typically repeated 3 to 4 times, with one cycle every 2-4 weeks [31, 32]. A database of patients who received palliative Quad Shot RT to the head and neck at our institution between 02/2005 and 06/2014 was used to identify patients who demonstrated long-term disease control [33].
Of the 75 patients who received palliative Quad Shot RT, 40 had squamous cell carcinoma (SCC) (Figure 1a). We focused on SCC and excluded other histologies such as thyroid and salivary carcinoma as they can have prolonged clinical courses. Of the 40 SCC patients, 6 patients had an overall survival of greater than 1 year, and 2 patients had an overall survival of greater than 2 years. Of the latter group, one patient had a cutaneous SCC and developed loco-regional recurrence shortly after RT. The remaining long-term survivor is discussed above.
Targeted sequencing
The MSK-IMPACT is an institutional effort to genotype patients for the determination of effective targeted therapies. It provides tumor genomic mutation profiling with a custom hybridization capture-based NGS assay [4]. The MSK-IMPACT targeted sequencing assay was performed on DNA from formalin-fixed, paraffin-embedded tumor samples with patient-matched normal blood samples. Bar-coded libraries from the tumor and normal blood samples were captured, sequenced, and subjected to a custom pipeline in order to identify somatic mutations. Deep sequencing of all exons and a custom 341 cancer-associated gene panel of selected introns was performed. An updated panel contained an expanded list of 410 genes. All exons had a minimum depth of coverage of 100X. Somatic mutations were identified via automated comparison of alterations from tumor and matched normal samples. Mutational load was calculated via determination of the number of somatic non-silent protein-coding mutations, excluding structural rearrangements and copy number gene alterations. The mutational burden was calculated based on the 341 gene panel for all patients, including those for whom the newer 410 gene panel was applied [34].
Analysis of TCGA data
Somatic mutation and copy number data for 24 cancer types analyzed by the TCGA was obtained from Broad Institute’s GDAC Firehose (http://gdac.broadinstitute.org). Two authors manually curated a list of DNA repair genes to identify genes that are associated with NHEJ (N.R., S.N.P) [8]. Germline events were obtained from a collaborator. Data analysis was performed in the R statistical environment version 3.2.4 using custom scripts.
Prevalence of altered tumors in the NHEJ pathway
We extracted from the TCGA Portal (https://gdc-portal.nci.nih.gov) all bam files that were used for WXS (Whole Exome Sequencing) in 24 cancer types, for a total of 9064 tumors. We then made a customized R script to calculate the number of tumors altered in the NHEJ pathway (using a list of 22 genes) per cancer type, and divided the alterations in three groups: germ line loss of function, somatic loss of function and somatic non-synonymous. The germ line alterations were extracted from a curated database (Ruomu et. Al., “Pan-cancer sequencing analysis reveals frequent germline mutations in cancer genes”, (2015), unpublished) [35] while the somatic alteration MAF files were extracted from the TCGA firehose (http://gdac.broadinstitute.org) for each cancer. For each cancer type, and for each alteration class, the number of corresponding altered tumors was divided by the total number of bam files assigned to that particular cancer type that were used for WXS. The resulting percentage, germ line loss of function, somatic loss of function, or somatic non-synonymous alterations is referred to as prevalence.
For each cancer type, we identified the most altered gene by counting the number of altered tumors per gene, across all three alteration types.
Statistics
Local recurrence was calculated from date of RT to date of death or last known follow-up. The Kaplan-Meier method was used to determine time to loco-regional recurrence within the radiation field.
SUPPLEMENTARY FIGURE
Footnotes
CONFLICTS OF INTEREST
The authors have no conflicts of interest to report.
GRANT SUPPORT
This research was funded in part through the NIH/NCI Cancer Center Support Grant P30 CA008748.
REFERENCES
- 1.Chen AM, Vaughan A, Narayan S, Vijayakumar S. Palliative radiation therapy for head and neck cancer: toward an optimal fractionation scheme. Head & neck. 2008;30:1586–1591. doi: 10.1002/hed.20894. [DOI] [PubMed] [Google Scholar]
- 2.Stevens CM, Huang SH, Fung S, Bayley AJ, Cho JB, Cummings BJ, Dawson LA, Hope AJ, Kim JJ, O’Sullivan B, Waldron JN, Ringash J. Retrospective study of palliative radiotherapy in newly diagnosed head and neck carcinoma. International journal of radiation oncology, biology, physics. 2011;81:958–963. doi: 10.1016/j.ijrobp.2010.06.055. [DOI] [PubMed] [Google Scholar]
- 3.Pollard JM, Gatti RA. Clinical radiation sensitivity with DNA repair disorders: an overview. International journal of radiation oncology, biology, physics. 2009;74:1323–1331. doi: 10.1016/j.ijrobp.2009.02.057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cheng DT, Mitchell TN, Zehir A, Shah RH, Benayed R, Syed A, Chandramohan R, Liu ZY, Won HH, Scott SN, Brannon AR, O’Reilly C, Sadowska J, et al. Memorial Sloan Kettering-Integrated Mutation Profiling of Actionable Cancer Targets (MSK-IMPACT): A Hybridization Capture-Based Next-Generation Sequencing Clinical Assay for Solid Tumor Molecular Oncology. J Mol Diagn. 2015;17:251–264. doi: 10.1016/j.jmoldx.2014.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Taylor AM, Harnden DG, Arlett CF, Harcourt SA, Lehmann AR, Stevens S, Bridges BA. Ataxia telangiectasia: a human mutation with abnormal radiation sensitivity. Nature. 1975;258:427–429. doi: 10.1038/258427a0. [DOI] [PubMed] [Google Scholar]
- 6.Chen PC, Lavin MF, Kidson C, Moss D. Identification of ataxia telangiectasia heterozygotes, a cancer prone population. Nature. 1978;274:484–486. doi: 10.1038/274484a0. [DOI] [PubMed] [Google Scholar]
- 7.Paul W, Sperduto NK, Roberge David, Xu Zhiyuan, Shanley Ryan, Luo Xianghua, Sneed Penny K, Chao Samuel T, Weil Robert J, Suh John, Bhatt Amit, Jensen Ashley W, Brown Paul D, Shih Helen A, Kirkpatrick John, Gaspar Laurie E, Fiveash John B, Chiang Veronica, Knisely Jonathan P.S, Sperduto Christina Maria, Lin Nancy, Mehta. Minesh. Summary Report on the Graded Prognostic Assessment: An Accurate and Facile Diagnosis-Specific Tool to Estimate Survival for Patients With Brain Metastases. J Clin Oncol. 2012;30:419–425. doi: 10.1200/JCO.2011.38.0527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Pearl LH SA, Ward SE, Al-Lazikani B, Pearl FM. Therapeutic opportunities within the DNA damage response. Nat Rev Cancer. 2015;15:166–180. doi: 10.1038/nrc3891. [DOI] [PubMed] [Google Scholar]
- 9.Cerami E GJ, Dogrusoz U, Gross BE, Sumer SO, Aksoy BA, Jacobsen A, Byrne CJ, Heuer ML, Larsson E, Antipin Y, Reva B, Goldberg AP, Sander C, Schultz N. The cBio cancer genomics portal: an open platform for exploring multidimensional cancer genomics data. Cancer Discov. 2012;2:401–404. doi: 10.1158/2159-8290.CD-12-0095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gao J AB, Dogrusoz U, Dresdner G, Gross B, Sumer SO, Sun Y, Jacobsen A, Sinha R, Larsson E, Cerami E, Sander C, Schultz N. Integrative analysis of complex cancer genomics and clinical profiles using the cBioPortal. Sci Signal. 2013:6. doi: 10.1126/scisignal.2004088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Liauw SL, Connell PP, Weichselbaum RR. New paradigms and future challenges in radiation oncology: an update of biological targets and technology. Science translational medicine. 2013;5:173sr172. doi: 10.1126/scitranslmed.3005148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Shiloh Y, Ziv Y. The ATM protein kinase: regulating the cellular response to genotoxic stress, and more. Nature reviews Molecular cell biology. 2013;14:197–210. [PubMed] [Google Scholar]
- 13.Cremona CA, Behrens A. ATM signalling and cancer. Oncogene. 2014;33:3351–3360. doi: 10.1038/onc.2013.275. [DOI] [PubMed] [Google Scholar]
- 14.Thompson LH. Recognition, signaling, and repair of DNA double-strand breaks produced by ionizing radiation in mammalian cells: the molecular choreography. Mutation research. 2012;751:158–246. doi: 10.1016/j.mrrev.2012.06.002. [DOI] [PubMed] [Google Scholar]
- 15.Mukhopadhyay UK SA, Ferbeyre G. RNA Silencing of Checkpoint Regulators Sensitizes p53-Defective Prostate Cancer Cells to Chemotherapy while Sparing Normal Cells. Cancer Res. 2005;65:2872–2881. doi: 10.1158/0008-5472.CAN-04-2502. [DOI] [PubMed] [Google Scholar]
- 16.Hickson I, Zhao Y, Richardson CJ, Green SJ, Martin NM, Orr AI, Reaper PM, Jackson SP, Curtin NJ, Smith GC. Identification and characterization of a novel and specific inhibitor of the ataxia-telangiectasia mutated kinase ATM. Cancer research. 2004;64:9152–9159. doi: 10.1158/0008-5472.CAN-04-2727. [DOI] [PubMed] [Google Scholar]
- 17.Rainey MD, Charlton ME, Stanton RV, Kastan MB. Transient inhibition of ATM kinase is sufficient to enhance cellular sensitivity to ionizing radiation. Cancer research. 2008;68:7466–7474. doi: 10.1158/0008-5472.CAN-08-0763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Golding SE, Rosenberg E, Valerie N, Hussaini I, Frigerio M, Cockcroft XF, Chong WY, Hummersone M, Rigoreau L, Menear KA, O’Connor MJ, Povirk LF, van Meter T, Valerie K. Improved ATM kinase inhibitor KU-60019 radiosensitizes glioma cells, compromises insulin, AKT and ERK prosurvival signaling, and inhibits migration and invasion. Molecular cancer therapeutics. 2009;8:2894–2902. doi: 10.1158/1535-7163.MCT-09-0519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Biddlestone-Thorpe L, Sajjad M, Rosenberg E, Beckta JM, Valerie NC, Tokarz M, Adams BR, Wagner AF, Khalil A, Gilfor D, Golding SE, Deb S, Temesi DG, et al. ATM kinase inhibition preferentially sensitizes p53-mutant glioma to ionizing radiation. Clin Cancer Res. 2013;19:3189–3200. doi: 10.1158/1078-0432.CCR-12-3408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mateo SC J, Sandhu S, Miranda S, Mossop H, Perez-Lopez R, Nava Rodrigues D, Robinson D, Omlin A, Tunariu N, Boysen G, Porta N, Flohr P, Gillman A, Figueiredo I, Paulding C, Seed G, Jain S, Ralph C, Protheroe A, Hussain S, Jones R, Elliott T, McGovern U, Bianchini D, Goodall J, Zafeiriou Z, Williamson C.T, Ferraldeschi R, Riisnaes R, Ebbs B, Fowler G, Roda D, Yuan W, Wu Y.-M, Cao X, Brough R, Pemberton H, A’Hern R, Swain A, Kunju L.P, Eeles R, Attard G, Lord C.J, Ashworth A, Rubin M.A, Knudsen K.E, Feng F.Y, Chinnaiyan A.M, Hall E, de Bono J.S. DNA-Repair Defects and Olaparib in Metastatic Prostate Cancer. N Engl J Med. 2015;373:1697–1708. doi: 10.1056/NEJMoa1506859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Thompson D, Duedal S, Kirner J, McGuffog L, Last J, Reiman A, Byrd P, Taylor M, Easton DF. Cancer risks and mortality in heterozygous ATM mutation carriers. Journal of the National Cancer Institute. 2005;97:813–822. doi: 10.1093/jnci/dji141. [DOI] [PubMed] [Google Scholar]
- 22.Umesako S, Fujisawa K, Iiga S, Mori N, Takahashi M, Hong DP, Song CW, Haga S, Imai S, Niwa O, Okumoto M. Atm heterozygous deficiency enhances development of mammary carcinomas in p53 heterozygous knockout mice. Breast cancer research. 2005;7:R164–170. doi: 10.1186/bcr968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Choudhury A NL, Teo MT, Chilka S, Bhattarai S, Johnston CF, Elliott F, Lowery J, Taylor CF, Churchman M, Bentley J, Knowles MA, Harnden P, Bristow RG, Bishop DT, Kiltie AE. MRE11 Expression Is Predictive of Cause-Specific Survival following Radical Radiotherapy for Muscle-Invasive Bladder Cancer. Cancer Res. 2010;70:7017–7026. doi: 10.1158/0008-5472.CAN-10-1202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Teo MT DL, Nsengimana J, Buchwald C, Snowden H, Morgan J, Jensen JB, Knowles MA, Taylor G, Barrett JH, Borre M, Orntoft TF, Bishop DT, Kiltie AE. Next-generation sequencing identifies germline MRE11A variants as markers of radiotherapy outcomes in muscle-invasive bladder cancer. Ann Oncol. 2014;25:877–883. doi: 10.1093/annonc/mdu014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gotoff SP, Amirmokri E, Liebner EJ. Ataxia telangiectasia. Neoplasia, untoward response to x-irradiation, and tuberous sclerosis. American journal of diseases of children. 1967;114:617–625. doi: 10.1001/archpedi.1967.02090270073006. [DOI] [PubMed] [Google Scholar]
- 26.Bakhshi S, Cerosaletti KM, Concannon P, Bawle EV, Fontanesi J, Gatti RA, Bhambhani K. Medulloblastoma with adverse reaction to radiation therapy in nijmegen breakage syndrome. Journal of pediatric hematology/oncology. 2003;25:248–251. doi: 10.1097/00043426-200303000-00013. [DOI] [PubMed] [Google Scholar]
- 27.Plowman PN, Bridges BA, Arlett CF, Hinney A, Kingston JE. An instance of clinical radiation morbidity and cellular radiosensitivity, not associated with ataxia-telangiectasia. The British journal of radiology. 1990;63:624–628. doi: 10.1259/0007-1285-63-752-624. [DOI] [PubMed] [Google Scholar]
- 28.Kerns SL, Ostrer H, Rosenstein BS. Radiogenomics: using genetics to identify cancer patients at risk for development of adverse effects following radiotherapy. Cancer discovery. 2014;4:155–165. doi: 10.1158/2159-8290.CD-13-0197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lassen P PH, Johansen J, Kristensen CA, Andersen E, Andersen LJ, Evensen JF, Eriksen JG, Overgaard J. Danish Head and Neck Cancer Group (DAHANCA). Impact of HPV-associated p16-expression on radiotherapy outcome in advanced oropharynx and non-oropharynx cancer. Radiother Oncol. :310–316. doi: 10.1016/j.radonc.2014.11.032. [DOI] [PubMed] [Google Scholar]
- 30.Zafereo ME XL, Dahlstrom KR, Viamonte CA, El-Naggar AK, Wei Q, Li G, Sturgis EM. Squamous cell carcinoma of the oral cavity often overexpresses p16 but is rarely driven by human papillomavirus. Oral Oncol. 2016:47–53. doi: 10.1016/j.oraloncology.2016.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Spanos W, Jr, Guse C, Perez C, Grigsby P, Doggett RL, Poulter C. Phase II study of multiple daily fractionations in the palliation of advanced pelvic malignancies: preliminary report of RTOG 8502. International journal of radiation oncology, biology, physics. 1989;17:659–661. doi: 10.1016/0360-3016(89)90120-x. [DOI] [PubMed] [Google Scholar]
- 32.Corry J, Peters LJ, Costa ID, Milner AD, Fawns H, Rischin D, Porceddu S. The ‘QUAD SHOT’--a phase II study of palliative radiotherapy for incurable head and neck cancer. Radiotherapy and oncology. 2005;77:137–142. doi: 10.1016/j.radonc.2005.10.008. [DOI] [PubMed] [Google Scholar]
- 33.Lok BH JG, Gutiontov S, Lanning RM, Sridhara S, Sherman EJ, Tsai CJ, McBride SM, Riaz N, Lee NY. Palliative head and neck radiotherapy with the RTOG 8502 regimen for incurable primary or metastatic cancers. Oral Oncol. 2015;51:957–962. doi: 10.1016/j.oraloncology.2015.07.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Stadler ZK BF, Middha S, Hechtman JF, Tran C, Cercek A, Yaeger R, Segal NH, Varghese AM, Reidy-Lagunes DL, Kemeny NE, Salo-Mullen EE, Ashraf A, Weiser MR, Garcia-Aguilar J, Robson ME, Offit K, Arcila ME, Berger MF, Shia J, Solit DB, Saltz LB. Reliable Detection of Mismatch Repair Deficiency in Colorectal Cancers Using Mutational Load in Next-Generation Sequencing Panels. J Clin Oncol. doi: 10.1200/JCO.2015.65.1067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Jiang R LW, Riaz N, Sander C, Mitchison TJ, Marks DS. In preparation. Pan-cancer sequencing analysis reveals frequent germline mutations in cancer genes. [Google Scholar]
- 36.Grundy GJ RS, Zeng Z, Arribas-Bosacoma R, Iles N, Manley K, Oliver A, Caldecott KW. APLF promotes the assembly and activity of non-homologous end joining protein complexes. EMBO J. 2013;32:112–125. doi: 10.1038/emboj.2012.304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Taylor AM HD, Arlett CF, Harcourt SA, Lehmann AR, Stevens S, Bridges BA. Ataxia telangiectasia: a human mutation with abnormal radiation sensitivity. Nature. 1975;258:427–429. doi: 10.1038/258427a0. [DOI] [PubMed] [Google Scholar]
- 38.Darroudi F WW, Meijers M, Friedl AA, van der Burg M, Fomina J, van Dongen JJ, van Gent DC, Zdzienicka MZ. Role of Artemis in DSB repair and guarding chromosomal stability following exposure to ionizing radiation at different stages of cell cycle. Mutat Res. 2007;615:111–124. doi: 10.1016/j.mrfmmm.2006.11.029. [DOI] [PubMed] [Google Scholar]
- 39.Adachi N IT, Ishii Y, Takeda S, Koyama H. DNA ligase IV-deficient cells are more resistant to ionizing radiation in the absence of Ku70: implications for DNA double-strand break repair. Proc Natl Acad Sci USA. 2001;98:12109–21213. doi: 10.1073/pnas.201271098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wang Z ZQ, Chen T, Liao K, Bu Y, Hong S, Hu G. Silencing NFBD1/MDC1 enhances the radiosensitivity of human nasopharyngeal cancer CNE1 cells and results in tumor growth inhibition. Cell Death Dis. 2015;6:1849. doi: 10.1038/cddis.2015.214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Teo MT DL, Nsengimana J, Buchwald C, Snowden H, Morgan J, Jensen JB, Knowles MA, Taylor G, Barrett JH, Borre M, Orntoft TF, Bishop DT, Kiltie AE. Next-generation sequencing identifies germline MRE11A variants as markers of radiotherapy outcomes in muscle-invasive bladder cancer. Ann Oncol. 2014;25:877–883. doi: 10.1093/annonc/mdu014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ogawa R IH, Kuwabara Y, Kimura M, Mitsui A, Mori Y, Mori R, Tomoda K, Katada T, Harada K, Fujii Y. Identification of candidate genes involved in the radiosensitivity of esophageal cancer cells by microarray analysis. Dis Esophagus. 2008;21:288–297. doi: 10.1111/j.1442-2050.2007.00759.x. [DOI] [PubMed] [Google Scholar]
- 43.Fattah FJ KJ, Wang Y, Lee EH, Kan Y, Lichter N, Weisensel N, Hendrickson EA. A role for XLF in DNA repair and recombination in human somatic cells. DNA Repair (Amst) 2014;15:39–53. doi: 10.1016/j.dnarep.2013.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Shirai H FH, Gunji A, Maeda D, Hirai T, Poetsch AR, Harada H, Yoshida T, Sasai K, Okayasu R, Masutani M. Parg deficiency confers radio-sensitization through enhanced cell death in mouse ES cells exposed to various forms of ionizing radiation. Biochem Biophys Res Commun. 2013;435:100–106. doi: 10.1016/j.bbrc.2013.04.048. [DOI] [PubMed] [Google Scholar]
- 45.Chayot R DA, Montagne B, Ricchetti M. Lack of DNA polymerase mu affects the kinetics of DNA double-strand break repair and impacts on cellular senescence. DNA Repair. 2010;9:1187–1199. doi: 10.1016/j.dnarep.2010.09.001. [DOI] [PubMed] [Google Scholar]
- 46.Ruis BL FK, Hendrickson EA. The catalytic subunit of DNA-dependent protein kinase regulates proliferation, telomere length, and genomic stability in human somatic cells. Mol Cell Biol. 2008;28:6182–6195. doi: 10.1128/MCB.00355-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Gatei M JB, Chen P, Kijas AW, Becherel OJ, Gueven N, Birrell G, Lee JH, Paull TT, Lerenthal Y, Fazry S, Taucher-Scholz G, Kalb R, Schindler D, Waltes R, Dork T, Lavin MF. ATM protein-dependent phosphorylation of Rad50 protein regulates DNA repair and cell cycle control. J Biol Chem. 2011;286:31542–31556. doi: 10.1074/jbc.M111.258152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Bohgaki T BM, Cardoso R, Panier S, Zeegers D, Li L, Stewart GS, Sanchez O, Hande MP, Durocher D, Hakem A, Hakem R. Genomic instability, defective spermatogenesis, immunodeficiency, and cancer in a mouse model of the RIDDLE syndrome. PLoS Genet. 2011;7:e1001381. doi: 10.1371/journal.pgen.1001381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kolas NK CJ, Nakada S, Ylanko J, Chahwan R, Sweeney FD, Panier S, Mendez M, Wildenhain J, Thomson TM, Pelletier L, Jackson SP, Durocher D. Orchestration of the DNA-damage response by the RNF8 ubiquitin ligase. Science. 2007;318:1637–1640. doi: 10.1126/science.1150034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Riballo E KM, Rief N, Doherty A, Smith GC, Recio MJ, Reis C, Dahm K, Fricke A, Krempler A, Parker AR, Jackson SP, Gennery A, Jeggo PA, Lobrich M. A pathway of double-strand break rejoining dependent upon ATM, Artemis, and proteins locating to gamma-H2AX foci. Mol Cell. 2004:16. doi: 10.1016/j.molcel.2004.10.029. [DOI] [PubMed] [Google Scholar]
- 51.Takata M SM, Tachiiri S, Fukushima T, Sonoda E, Schild D, Thompson LH, Takeda S. Chromosome instability and defective recombinational repair in knockout mutants of the five Rad51 paralogs. Mol Cell Biol. 2001;21:2858–2866. doi: 10.1128/MCB.21.8.2858-2866.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Schulte-Uentrop L E-AR, Schliecker L, Willers H, Dahm-Daphi J. Distinct roles of XRCC4 and Ku80 in non-homologous end-joining of endonuclease-and ionizing radiation-induced DNA double-strand breaks. Nucleic Acids Res. 2008;36:2561–2569. doi: 10.1093/nar/gkn094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Mansour WY RT, Dahm-Daphi J. The alternative end-joining pathway for repair of DNA double-strand breaks requires PARP1 but is not dependent upon microhomologies. Nucleic Acids Res. 2010;38:6065–6077. doi: 10.1093/nar/gkq387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Mumbrekar KD GH, Vadhiraja BM, Bola Sadashiva SR. Polymorphisms in double strand break repair related genes influence radiosensitivity phenotype in lymphocytes from healthy individuals . DNA Repair (Amst) 2016;40:27–34. doi: 10.1016/j.dnarep.2016.02.006. [DOI] [PubMed] [Google Scholar]
- 55.Bassing CH CK, Sekiguchi J, Suh H, Whitlow SR, Fleming JC, Monroe C, Ciccone DN, Yan C, Vlasakova K, Livingston DM, Ferguson DO, Scully R, Alt FW. Increased ionizing radiation sensitivity and genomic instability in the absence of histone H2AX 8178. Proc Natl Acad Sci USA. 2002;99:8173. doi: 10.1073/pnas.122228699. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


